Abstract
Background:
The use of a polygenic risk score (PRS) to improve risk prediction of coronary heart disease (CHD) events has been demonstrated to have clinical utility in the general adult population. However, the prognostic value of a PRS for CHD has not been examined specifically in older populations of individuals aged ≥70 years, who comprise a distinct high-risk subgroup. The objective of this study was to evaluate the predictive value of a PRS for incident CHD events in a prospective cohort of older individuals without a history of cardiovascular events.
Methods:
We used data from 12,792 genotyped, healthy older individuals enrolled into the ASPREE trial, a randomized double-blind placebo-controlled clinical trial investigating the effect of daily 100mg aspirin on disability-free survival. Participants had no previous history of diagnosed atherothrombotic cardiovascular events, dementia, or persistent physical disability at enrolment. We calculated a PRS (metaGRS) consisting of 1.7 million genetic variants. The primary outcome was a composite of incident myocardial infarction or CHD death over 5 years.
Results:
At baseline, the median population age was 73.9 years and 54.9% were female. In total, 254 incident CHD events occurred. When the PRS was added to conventional risk factors, it was independently associated with CHD (hazard ratio 1.24 [95% confidence interval [CI] 1.08–1.42], p=0.002). The AUC of the conventional model was 70.53 (95%CI 67.00–74.06), and after inclusion of the PRS increased to 71.78 (95%CI 68.32–75.24, p=0.019), demonstrating improved prediction. Reclassification was also improved, as the continuous net reclassification index after adding PRS to the conventional model was 0.25 (95%CI 0.15–0.28).
Conclusion:
A PRS for CHD performs well in older people and improves prediction over conventional cardiovascular risk factors. Our study provides evidence that genomic risk prediction for CHD has clinical utility in individuals aged 70 years and older.
Keywords: Polygenic risk score, PRS, CHD, coronary heart disease, risk prediction
Introduction
An increasing number of recent studies have suggested the potential clinical utility of using a polygenic risk score (PRS) to improve the prediction of coronary heart disease (CHD) events in the general popualtion1–8. It is now well established that adults with a high genetic risk score will have higher risk for CHD events, compared to those with a low score3. Furthermore, the addition of a PRS has been shown to significantly improve CHD risk prediction when added to a risk model comprised of conventional risk factors.4 Notably, a PRS comprising 1.7 million variants (metaGRS) has been shown to increase CHD risk with a hazard ratio of 1.7 per standard deviation among >450,000 adults from the general UK population (mean age 56.6 years).4 Additional independent validation studies of the metaGRS have also shown consistent performance for CHD risk prediction5–11, including in ethnically diverse populations5, 6
The use of genomics in CHD risk prediction has important clinical implications, given the burden of CHD remains high in most countries, despite significant improvements in prevention and treatment. Improved approaches to risk prediction and early intervention may help to address the burden of CHD, and genomics presents a new opportunity. However, prior studies investigating a genomic risk score for CHD risk prediction have mostly included individuals with a mean age ranging from 50 to 60 years or younger. The use of PRS as a risk factor for CHD has not previously been investigated specifically in older individuals ≥70 years, who are themselves a distinct high-risk population.
In addition to the potential differences in PRS performance, older individuals may also require customized CHD risk prediction models with regards to conventional clinical risk factors12. Prediction models used to estimate the risk of future CHD events are usually derived from younger populations and based on conventional risk factors such as blood pressure, diabetes, smoking or blood lipids.13, 14 These risk models do not fully explain individual risk in older people and are not recommended for clinical usage in patients aged ≥70 years. They require calibration based on data from studies of older individuals. Here, we sought to investigate the prognostic value of a recently derived PRS for CHD in a population of older individuals without a history of CHD events, when added to a conventional risk factor model which we constructed, and also in comparison with the SCORE2-OP risk score, specifically optimised for prediction of cardiovascular events in older people.15 The objective of our study was to determine whether the potential clinical utility of a PRS for CHD would extend to older individuals aged 70 years and older.
Methods
The full methods are available in the supplemental material. Requests for data access will be via the ASPREE Principal Investigators with details for applications provided through the web site, www.ASPREE.org. The study was approved by local Ethics Committees and registered on Clinicaltrials.gov (NCT01038583). All participants provided informed written consent.
Results
Baseline characteristics
The median age of the 12,792 genotyped participants was 73.9 years (interquartile range 71.7, 77.3, Table 1); 7,027 (54.9%) were female, 391 (3.1%) were current smokers and 1,186 (9.3%) had diabetes. Comparing the 12,792 genotyped participants with non-genotyped participants of the ASPREE trial found only minor differences in baseline characteristics (Table I). The PRS showed a normal distribution (Figure III), and the mean value was −1.16 (SD 0.45). There was no relevant correlation of the PRS with other continuous variables within the data set (Figure IV). In a multivariable linear regression model, the PRS was significantly associated with age, gender, systolic blood pressure, non-HDL-cholesterol, HDL-cholesterol, diabetes, and family history of MI (Table II). During follow-up, 254 (2.0%) of genotyped participants had incident CHD events (169 in males, 85 in females). This included 226 incident cases of myocardial infarction and 50 cases of CHD death. The incidence rate was 3.11 CHD events per 1,000 person-years in PRS tertile 1, 4.29 CHD events per 1,000 person-years in PRS tertile 2 and 4.97 CHD events per 1,000 person-years in PRS tertile 3 (Table 2).
Table 1:
Baseline characteristics
| Overall Population | |
|---|---|
| Number of participants | 12,792 |
| Age (median (IQR)) | 73.9 (71.7, 77.3) |
| Age categories (%) | |
| 70–74 | 7,698/12,792 (60.2) |
| 75–79 | 3,271/12,792 (25.6) |
| 80–84 | 1,414/12,792 (11.1) |
| >85 | 409/12,792 (3.2) |
| Female (%) | 7,027/12,792 (54.9) |
| Current Smoker (%) | 391/12,792 (3.1) |
| Systolic Blood Pressure, mmHg (mean (SD)) | 139.46 (16.27) |
| Diastolic Blood Pressure, mmHg (mean (SD)) | 77.17 (9.97) |
| Diabetes (%) | 1,186/12,792 (9.3) |
| Body Mass Index, kg/m2 (mean (SD)) | 27.97 (4.55) |
| HDL-c, mmol/L (mean (SD)) | 1.59 (0.46) |
| Non-HDL-c, mmol/L (mean (SD)) | 3.69 (0.93) |
| Fasting Glucose, mg/dL (mean (SD)) | 98.29 (17.12) |
| Creatinine, mg/dL (mean (SD)) | 0.90 (0.22) |
| Family history of MI (%) | 340/12,792 (2.7) |
| Polygenic Risk Score (mean (SD)) | −1.16 (0.45) |
Missing values for continuous variables were: 341 for creatinine, 331 for non-HDL-c, 330 for HDL-c, 250 for fasting glucose and 56 for body mass index. Abbreviations: IQR = inter quartile range, SD = standard deviation, HDL-c = high density lipoprotein cholesterol, MI = myocardial infarction.
Table 2:
Incidence rate of CHD events per PRS tertiles
| PRS tertile | N | CHD events | Incidence rate per 1,000 person-years |
|---|---|---|---|
| 1 | 4,263 | 56 | 3.11 |
| 2 | 4,264 | 77 | 4.29 |
| 3 | 4,264 | 88 | 4.97 |
PRS for risk prediction
In the conventional model, all variables except systolic blood pressure and diabetes were found to be independent predictors of CHD events (Table 3). When the PRS was added as a continuous variable to the conventional model, it was found to be an independent predictor of outcome (Hazard ratio [HR] 1.24 [95% Confidence Interval [CI] 1.08–1.42], p=0.002). The HR of the PRS per SD was comparable to that reported by five other published studies of younger adults where the same PRS was used (Table III).
Table 3:
Hazard ratios for the conventional model, conventional model + continuous PRS and conventional model + categorical PRS
| Conventional model | Conventional model + continuous PRS | Conventional model + categorical PRS | SCORE2-OP + continuous PRS | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HR | 95%CI | p-value | HR | 95%CI | p-value | HR | 95%CI | p-value | HR | 95%CI | p-value | |
| Age | 1.09 | (1.06–1.12) | <0.001 | 1.09 | (1.06–1.12) | <0.001 | 1.09 | (1.06–1.12) | <0.001 | |||
| Female Sex | 0.48 | (0.34–0.67) | <0.001 | 0.46 | (0.33–0.65) | <0.001 | 0.47 | (0.34–0.66) | <0.001 | |||
| Current Smoking | 2.00 | (1.09–3.68) | 0.025 | 2.02 | (1.10–3.71) | 0.024 | 2.01 | (1.09–3.69) | 0.025 | |||
| SBP per 10 mmHg increase | 1.04 | (0.96–1.13) | 0.34 | 1.04 | (0.96–1.13) | 0.37 | 1.04 | (0.96–1.13) | 0.33 | |||
| Non-HDL-c | 1.35 | (1.17–1.56) | <0.001 | 1.35 | (1.17–1.55) | <0.001 | 1.34 | (1.17–1.55) | <0.001 | |||
| HDL-c | 0.65 | (0.44–0.95) | 0.026 | 0.65 | (0.44–0.95) | 0.027 | 0.64 | (0.44–0.94) | 0.024 | |||
| Diabetes | 0.82 | (0.49–1.38) | 0.45 | 0.81 | (0.48–1.36) | 0.42 | 0.81 | (0.48–1.36) | 0.42 | |||
| Creatinine | 1.83 | (1.03–3.26) | 0.040 | 1.81 | (1.01–3.23) | 0.045 | 1.82 | (1.02–3.24) | 0.043 | |||
| SCORE2-OP per 1% risk increase | 1.13 | (1.10; 1.16) | <0.001 | |||||||||
| PRS (continuous per SD) | 1.24 | (1.08–1.42) | 0.002 | 1.24 | (1.09; 1.42) | 0.001 | ||||||
| PRS 1st Tertile | 1.00 | (Reference) | ||||||||||
| PRS 2nd Tertile | 1.48 | (1.04–2.09) | 0.029 | |||||||||
| PRS 3rd Tertile | 1.64 | (1.16–2.33) | 0.005 | |||||||||
Abbreviations: SBP = systolic blood pressure, HDL-c = high density lipoprotein cholesterol, PRS = polygenic risk score, HR = hazard ratio, CI = confidence interval.
Using PRS tertiles as a predictor, CHD risk increased as the PRS category increased from the first to third tertile. When compared to the first PRS tertile (low risk group) the second tertile had a HR for CHD risk of 1.48 (95%CI 1.04–2.09, p=0.029) and the third PRS tertile had a HR of 1.64 (95%CI 1.16–2.33, p=0.005). Kaplan-Meier curves illustrated that individuals in the higher and middle PRS tertiles had a higher incidence of CHD events compared with lower PRS tertile (p=0.02, Figure 1). Furthermore, the continuous PRS was a significant predictor of outcome, when added to the SCORE2-OP risk model (HR 1.24, 95%CI 1.09–1.42, p=0.001).
Figure 1:
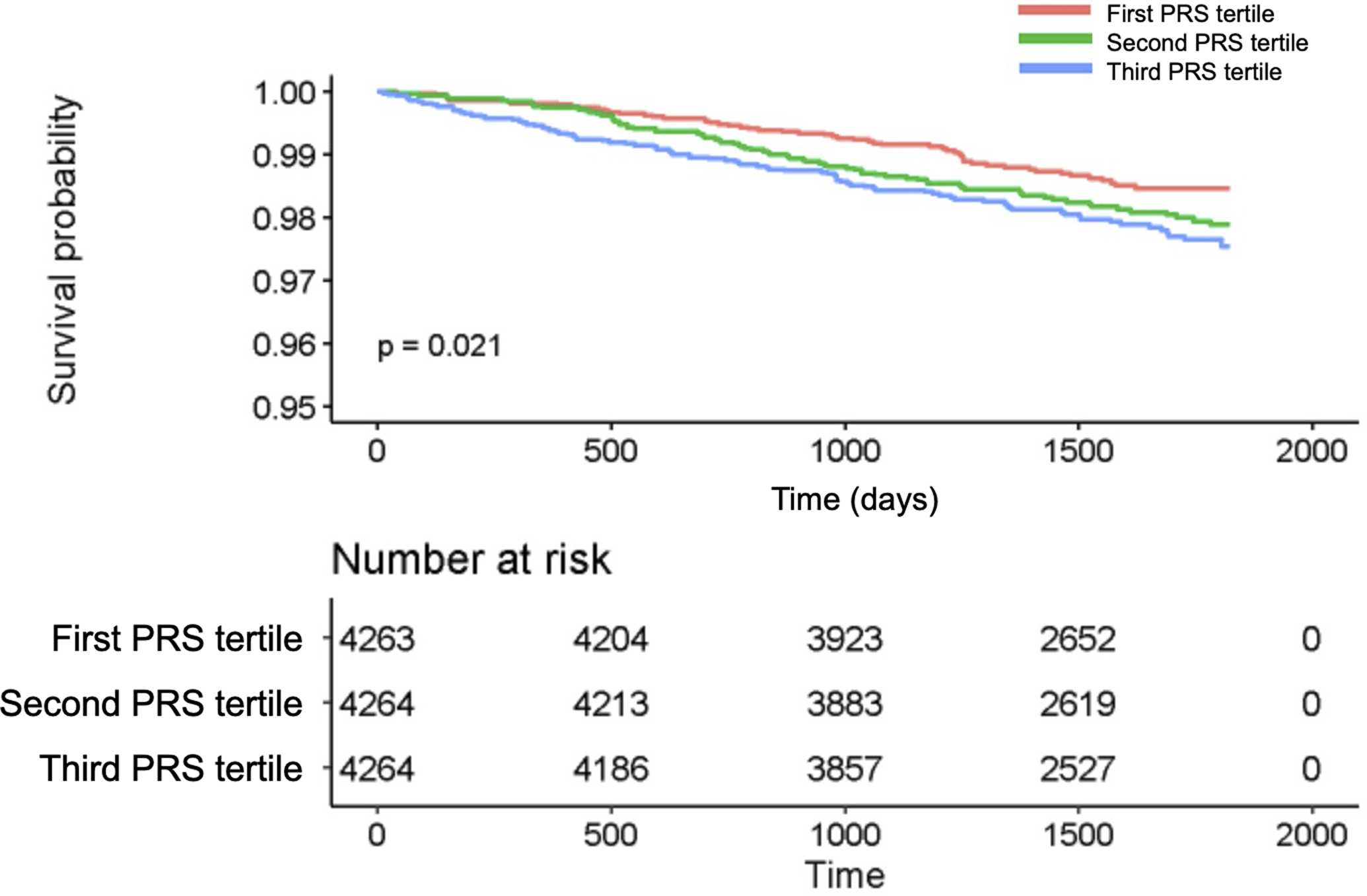
Kaplan-Meier curve for CHD events according to PRS tertiles
The figure provides the probability of a CHD event according to tertiles of the PRS, based on Kaplan-Meier estimates, and the individuals at risk.
Evaluation of each single predictor using receiver-operating-characteristics showed that sex (AUC 62.88%, 95%CI 59.58–66.17), HDL-cholesterol (AUC 61.56%, 95%CI 57.51–65.61), serum creatinine (AUC 61.39%, 95%CI 57.53–65.24) and age (AUC 57.50%, 95%CI 52.98–62.05) were the strongest predictors of incident CHD events (Figure 2). The PRS alone resulted in an AUC of 55.72% (95%CI 51.74–59.72). The AUC for the conventional model was 70.53% (95%CI 67.00–74.06) and significantly improved to 71.78% (95%CI 68.32–75.24) after adding the PRS as a continuous variable (p=0.019, Table 4, Figure V). The calibration plot showed a good agreement between predicted and observed CHD events (Figure VI). The SCORE2-OP risk model alone resulted in an AUC of 68.32% (95%CI 64.70–71.94) and increased to 69.73% (95%CI 66.17–73.30, p=0.081) after adding the continuous PRS.
Figure 2:
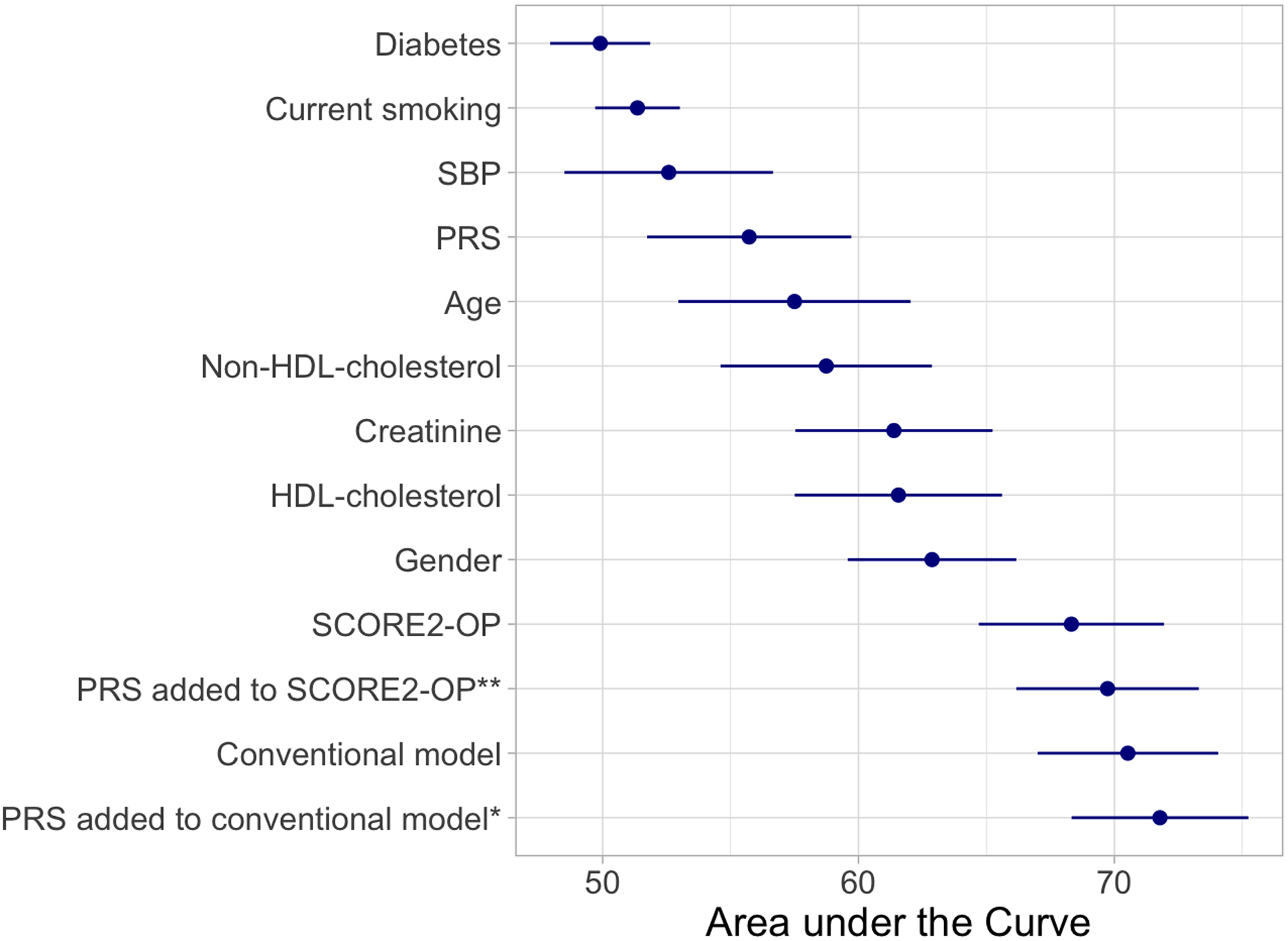
AUC for each predictor, the conventional model, SCORE2-OP and the PRS added to the models.
*p-value compared the Conventional Model = 0.019, **p-value compared the SCORE2-OP = 0.081. Abbreviations: SBP = systolic blood pressure, HDL = high density lipoprotein, PRS = polygenic risk score, AUC = area under the curve, CI = confidence interval.
Table 4:
Categorical net reclassification improvement table after adding PRS to the conventional model to predict the risk of a 5-year CHD event.
| Standard Model + Polygenic Risk Score | |||||
|---|---|---|---|---|---|
| Standard Model | < 1.5% | 1.5 to 2.49% | ≥ 2.5% | Total No. (%) of participants | |
| CHD events | < 1.5% | 37 | 9 | 0 | 46 (22) |
| 1.5 to 2.49% | 6 | 35 | 12 | 53 (25) | |
| ≥ 2.5% | 0 | 8 | 103 | 111 (53) | |
|
Total No. (%)
of participants |
43 (20) | 52 (25) | 115 (55) | 210 (100) | |
| CHD non-events | < 1.5% | 2248 | 157 | 2 | 2407 (49) |
| 1.5 to 2.49% | 204 | 854 | 149 | 1207 (25) | |
| ≥ 2.5% | 1 | 187 | 1114 | 1302 (26) | |
|
Total No. (%)
of participants |
2453 (50) | 1198 (24) | 1265 (26) | 4916 (100) | |
In this table the reclassification of participants from one risk category (<1.5% risk, 1-5-2.49% risk, ≥2.5% risk) to another category is described, when adding the Polygenic risk score to the standard model. The table is further by CHD events and CHD non-events. Light gray shaded boxes indicate the correct reclassification into a different risk category, while dark gray shaded boxes indicate the incorrect reclassification into a different risk category.
Reclassification analyses
In reclassification analyses, the continuous NRI was 0.25 (95%CI 0.15–0.28), when the PRS was added to the conventional model (Table IV). More individuals were to a higher risk category (NRI+ 0.16, 95%CI 0.08–9.20), than downwards (NRI- 0.09, 95%CI 0.04–0.10). For measurement of the categorical NRI, CHD risk categories of <1.5%, <2.5% and ≥2.5% were chosen based on the observed risk within ASPREE (Table IV, Table 3). Here, addition of the PRS to the conventional model resulted in a categorical reclassification of 0.063 (95%CI 0.001–0.129), with an upwards classification of 0.044 (95%CI of −0.007–0.105) and a downwards classification of 0.019 (95%CI 0.003–0.032).
Subgroup analyses
When comparing males and females, we only observed minor differences in baseline characteristics (Table V). Adding the continuous PRS to the conventional model, it was an independent predictor in males, but not in females (males HR 1.27 [95%CI 1.08–1.50], p=0.005 versus females HR 1.18 [95%CI 0.92–1.49], p=0.19, Table VI+VII). The same finding was observed when assessing the categorical PRS. The conventional model resulted in a lower AUC in males compared to females (males AUC 66.58%, females AUC 70.07%), but the incremental value of adding the PRS to the conventional model was greater in males compared with females (males AUC 68.18%, females AUC 71.00%, Table VIII).
In subgroup analyses by PRS tertile, baseline characteristics were similar for participants within the highest compared to the lowest PRS tertile (Table IX). The conventional model resulted in a lower AUC in individuals from the highest, compared to individuals from the lowest PRS tertile (highest tertile AUC 73.21%, lowest tertile AUC 76.62%), but the incremental value of addition of the PRS to the conventional model was similar in both groups (Table X).
Results of sensitivity analyses after adding use of antihypertensive drugs, statins, and genetic ethnicity PCAs to the model are reported in the supplemental results (Tables XI + XII). Interaction effects between sex and model covariables were examined, but no interaction between sex and PRS was found (HR 0.93, 95%CI 0.69–1.24, p=0.60; Table XIII).
Finally, we investigated the interaction of aspirin treatment (as per ASPREE randomization) with the PRS but did not find a significant interaction (p = 0.58, Table XIV).
Discussion
In this study, we evaluated the prognostic value of a previously derived polygenic risk score (metaGRS) to predict future CHD events in a population of healthy older individuals from the ASPREE trial. We demonstrated robust PRS performance in this older population and can confirm that addition of the PRS to a conventional cardiovascular risk model improved risk prediction (Figure 3). Our study suggests that the potential clinical utility of a PRS for CHD risk prediction extends to older individuals aged 70 years and older, who comprise an important high-risk group. Our study also represents an independent validation of a recently derived PRS, in a well-characterized older population. Our findings add further support to the growing body of evidence that supports the use of genomic risk information to improve CHD risk prediction, and our results indicate that the prognostic value of a genomic risk score for CHD extends to older individuals, who comprise an important high-risk group.
Figure 3:
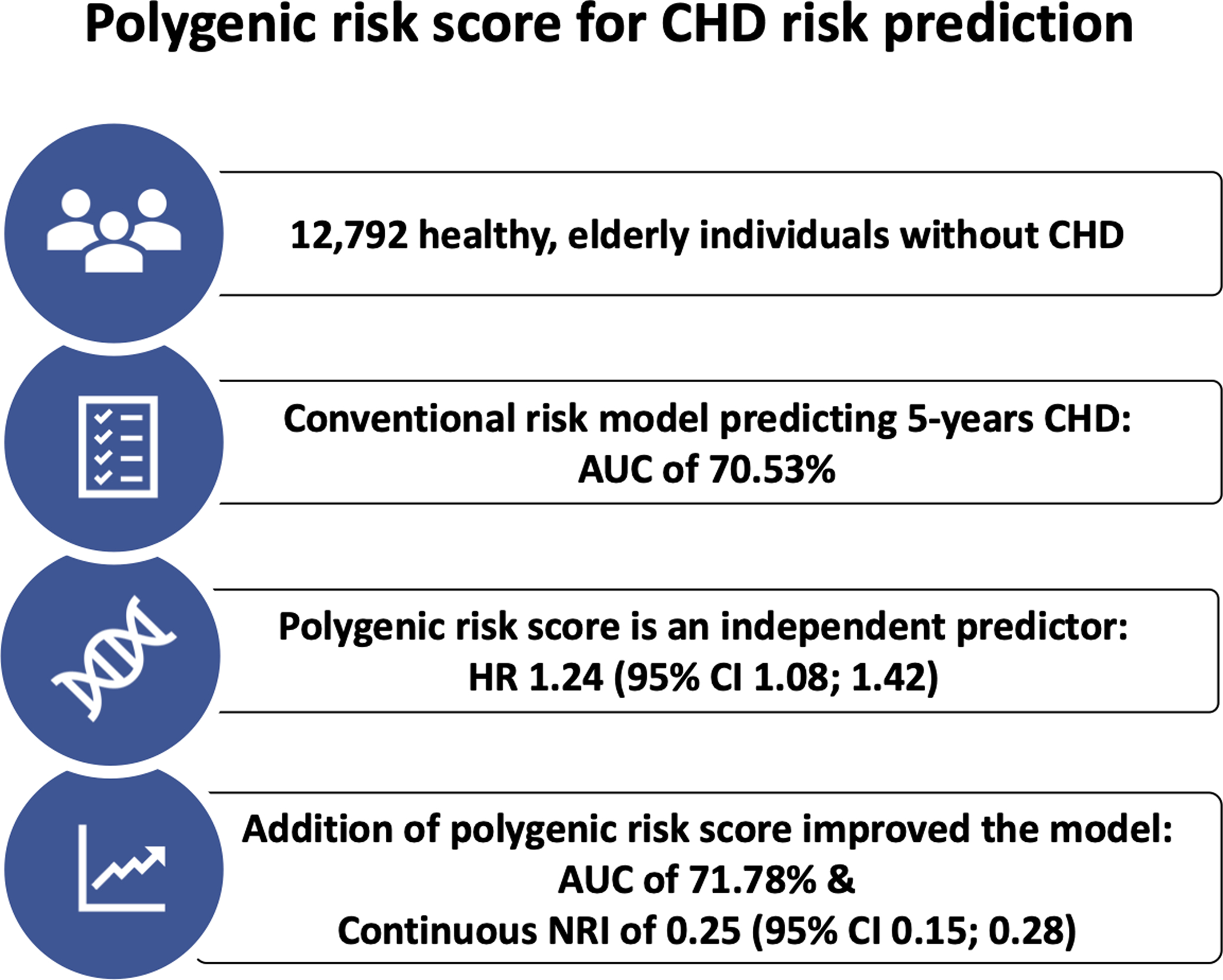
Central figure summarizing the main study findings
We evaluated the prognostic accuracy of a previously derived polygenic risk score (metaGRS) to predict 5 years CHD events in a population of healthy elderly individuals.
Abbreviations: CHD = coronary heart disease, AUC = area under the curve, HR = hazard ratio, CI = confidence interval.
The metaGRS used in our study was derived using data from a range of different CHD studies from younger adult populations, then validated in the UK Biobank population of around 500,000 British individuals, with mean age 56.5 years.4 The score has since been validated in a range of other external validation studies of younger adult populations (Table III).5, 6, 10, 11 The ASPREE population differs in several aspects. Firstly, and most notably, the median age of ASPREE participants at enrolment was far older at 73.9 years, nearly 20 years older than the UK Biobank. Yet, the HR of the PRS remained similar. Second, ASPREE is a highly ascertained clinical trial population, in which participants met strict inclusion criteria, with no history of CHD events at enrolment. Third, major CHD events in ASPREE were adjudicated as part of a randomized trial but did not include coronary revascularization. Given these important differences, it is noteworthy that the metaGRS still performed in a robust manner in the older ASPREE population, with similar HR and AUC compared with studies of younger populations. Similar to previous studies5, 6, our findings demonstrate a polygenic model derived from the UK Biobank generalizes well to other cohorts of European ancestry.
CHD accounts for a large proportion of deaths in older people. Accurate identification of older individuals at increased risk for CHD is therefore clinically important, particularly those not identified as high-risk by conventional risk factors. Due to a lack of evidence in individuals aged 70 years and older, the value of adding genetic information for CHD risk prediction in older people has not previously been tested robustly. Our study therefore provides the first evidence of its kind to suggest the predictive value and potential clinical utility of a PRS for CHD extends to individuals aged 70 years and older. We observed comparable predictive performance of the PRS versus younger population-based cohorts4–6, 10, 11 and demonstrated that addition of the PRS to a conventional risk factor model we constructed, and to the recently derived SCORE2-OP clinical risk model, improved prediction.
Notably, we found that the PRS alone (considered independently as a CHD risk factor) had similar discriminative power compared to conventional CHD risk factors used in routine practice. However, in our analyses the AUC of sex, HDL-cholesterol, creatinine, non-HDL-cholesterol, and age were stronger discriminators than the PRS alone. This emphasizes the importance of these risk factors as predictors in an older population, alongside a genetic risk score. Nevertheless, it is noteworthy that the PRS was found to predict CHD events independently of conventional risk factors, not showing correlation with the nine conventional risk factors examined (Figure IV). These unique properties of the genetic risk score (i.e., relatively strong predictive performance and independent effect) help demonstrate its future clinical potential for CHD risk prediction in populations of older adults.
Currently, the availability of PRS as a clinical tool for CHD prediction at large remains limited, with unresolved questions related to cost-effectiveness and implementation. Furthermore, some recent studies have provided conflicting results regarding the incremental value of adding genetic information to conventional CHD risk factors in younger populations.7, 8 Whilst the magnitude of improved CHD risk prediction achieved by the PRS may be small or incremental in an individual study, when the effects are extrapolated to a far larger population (e.g. an entire country, comprising millions of older adults) effects are substantial. In the future, individual genotyping will become more widely available and at lower cost, potentially facilitating improved CHD event prediction and risk stratification at the population level. Here we show that genetic risk is still highly relevant at older ages, and that a PRS for CHD still performs well, and may have potential clinical utility for preventive strategies in older people. However, further studies of more phenotypically and ethnically diverse elderly populations are required to generalize these results.
Specific findings of our study warrant further discussion. First, we did not find diabetes to be an independent predictor for CHD events, despite 9.3% of ASPREE participants having diabetes at baseline. Other studies have reported the relevance of diabetes regarding CHD risk in the elderly.12 This observation could be explained by the pre-selection of a healthy ASPREE population, in whom the duration of diabetes might be shorter, compared to the general population. A second notable finding of our study was that results were not confirmed in subgroup analyses for females. This finding was likely due to limited power because the majority of CHD events in ASPREE occurred in males. Further, we found no interaction effect between sex and PRS, and other studies have reported similar performance for CHD polygenic scores in both sexes.16 Third, we investigated the interaction of aspirin treatment with the metaGRS in exploratory analyses but found no significant interaction (p=0.58). This suggested that in the ASPREE trial, participants with a high CHD PRS did not benefit more from low-dose aspirin use, versus participants with a low PRS, for primary prevention of CHD events. Further studies are required to determine whether other genotypic sub-sets of the population may benefit from aspirin use.
Strengths of our study include a well-characterized, unique study population with incident cardiovascular events clinically adjudicated as part of a randomized trial. No other large clinical trial has recruited this number of healthy older individuals without a prior history of CHD events, with genotyping. All ASPREE participants received medical assessments by general practitioners at enrolment, to confirm eligibility for the trial, and to rule out previous diagnoses of CHD events. This provided confidence that participants were CHD event-free at enrolment, to examine the value of PRS in the context of primary prevention in the elderly. A range of conventional risk factor variables were also available in ASPREE, to examine alongside polygenic risk.
Limitations of our study include a rather short follow-up period (average 4.6 years per participant) contributing to the relatively small number of CHD events. Continued follow-up will provide more power for future analyses. We also acknowledge the potential healthy-volunteer effect (ascertained bias) and survivorship bias of the ASPREE trial population. ASPREE did not collect information related to revascularization, which is an important CHD endpoint used in metaGRS derivation dataset. Our findings may not be generalizable to other ancestries or more diverse populations.
In conclusion, we report a potential clinical benefit of using a PRS for improved risk prediction of CHD events in older people. Our study provides some of the first evidence that use of PRS for CHD prediction is robust across a diverse range of populations and age groups, including individuals aged 70 years and older which are a distinct high-risk group.
Supplementary Material
Acknowledgements:
We thank the ASPREE trial staff, participants, and general practitioners, and the Ramaciotti Centre for Genomics.
Funding:
The trial was supported by the National Institute on Aging and the National Cancer Institute at the National Institutes of Health (grant numbers U01AG029824 and U19AG062682); the National Health and Medical Research Council of Australia (grant numbers 334047, 1127060); Monash University and the Victorian Cancer Agency. Genotyping supported by Bioplatforms Australia, National Framework Initiative (2018–2020). J.N. is recipient of a fellowship by the Deutsche Forschungsgemeinschaft (NE 2165/1-1). C.M.R. is supported through a NHMRC Principal Research Fellowship (APP 1136372). P.L is supported by a National Heart Foundation Future Leader Fellowship (102604).
Non-Standard abbreviations and Acronyms
- PRS
Polygenic risk score
- ASPREE
ASPirin in Reducing Events in the Elderly
- Non-HDL
non-high-density-lipoprotein
- PCA
principal component analysis
- NRI
net reclassification improvement
Footnotes
Clinical Trial Registration: www.clinicaltrials.gov (NCT01038583)
Disclosures: No conflicts were reported.
Supplemental Material
References
- 1.Marenberg ME, Risch N, Berkman LF, Floderus B and de Faire U. Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med. 1994;330:1041–6. [DOI] [PubMed] [Google Scholar]
- 2.Lloyd-Jones DM, Nam BH, D’Agostino RB, Sr., Levy D, Murabito JM, Wang TJ, Wilson PW and O’Donnell CJ. Parental cardiovascular disease as a risk factor for cardiovascular disease in middle-aged adults: a prospective study of parents and offspring. JAMA. 2004;291:2204–11. [DOI] [PubMed] [Google Scholar]
- 3.Khera AV, Emdin CA, Drake I, Natarajan P, Bick AG, Cook NR, Chasman DI, Baber U, Mehran R, Rader DJ, et al. Genetic Risk, Adherence to a Healthy Lifestyle, and Coronary Disease. N Engl J Med. 2016;375:2349–2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Inouye M, Abraham G, Nelson CP, Wood AM, Sweeting MJ, Dudbridge F, Lai FY, Kaptoge S, Brozynska M, Wang T, et al. Genomic Risk Prediction of Coronary Artery Disease in 480,000 Adults: Implications for Primary Prevention. J Am Coll Cardiol. 2018;72:1883–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wunnemann F, Sin Lo K, Langford-Avelar A, Busseuil D, Dube MP, Tardif JC and Lettre G. Validation of Genome-Wide Polygenic Risk Scores for Coronary Artery Disease in French Canadians. Circ Genom Precis Med. 2019;12:e002481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dikilitas O, Schaid DJ, Kosel ML, Carroll RJ, Chute CG, Denny JA, Fedotov A, Feng Q, Hakonarson H, Jarvik GP, et al. Predictive Utility of Polygenic Risk Scores for Coronary Heart Disease in Three Major Racial and Ethnic Groups. Am J Hum Genet. 2020;106:707–716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Elliott J, Bodinier B, Bond TA, Chadeau-Hyam M, Evangelou E, Moons KGM, Dehghan A, Muller DC, Elliott P and Tzoulaki I. Predictive Accuracy of a Polygenic Risk Score-Enhanced Prediction Model vs a Clinical Risk Score for Coronary Artery Disease. JAMA. 2020;323:636–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mosley JD, Gupta DK, Tan J, Yao J, Wells QS, Shaffer CM, Kundu S, Robinson-Cohen C, Psaty BM, Rich SS, et al. Predictive Accuracy of a Polygenic Risk Score Compared With a Clinical Risk Score for Incident Coronary Heart Disease. JAMA. 2020;323:627–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Aragam KG, Dobbyn A, Judy R, Chaffin M, Chaudhary K, Hindy G, Cagan A, Finneran P, Weng LC, Loos RJF, et al. Limitations of Contemporary Guidelines for Managing Patients at High Genetic Risk of Coronary Artery Disease. J Am Coll Cardiol. 2020;75:2769–2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Timmerman N, de Kleijn DPV, de Borst GJ, den Ruijter HM, Asselbergs FW, Pasterkamp G, Haitjema S and van der Laan SW. Family history and polygenic risk of cardiovascular disease: Independent factors associated with secondary cardiovascular events in patients undergoing carotid endarterectomy. Atherosclerosis. 2020;307:121–129. [DOI] [PubMed] [Google Scholar]
- 11.Gladding PA, Legget M, Fatkin D, Larsen P and Doughty R. Polygenic Risk Scores in Coronary Artery Disease and Atrial Fibrillation. Heart Lung Circ. 2020;29:634–640. [DOI] [PubMed] [Google Scholar]
- 12.Dalton JE, Rothberg MB, Dawson NV, Krieger NI, Zidar DA and Perzynski AT. Failure of Traditional Risk Factors to Adequately Predict Cardiovascular Events in Older Populations. J Am Geriatr Soc. 2020;68:754–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Conroy RM, Pyorala K, Fitzgerald AP, Sans S, Menotti A, De Backer G, De Bacquer D, Ducimetiere P, Jousilahti P, Keil U, et al. Estimation of ten-year risk of fatal cardiovascular disease in Europe: the SCORE project. Eur Heart J. 2003;24:987–1003. [DOI] [PubMed] [Google Scholar]
- 14.Piepoli MF, Hoes AW, Agewall S, Albus C, Brotons C, Catapano AL, Cooney MT, Corra U, Cosyns B, Deaton C, et al. 2016 European Guidelines on cardiovascular disease prevention in clinical practice: The Sixth Joint Task Force of the European Society of Cardiology and Other Societies on Cardiovascular Disease Prevention in Clinical Practice (constituted by representatives of 10 societies and by invited experts)Developed with the special contribution of the European Association for Cardiovascular Prevention & Rehabilitation (EACPR). Eur Heart J. 2016;37:2315–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.group SOw and collaboration ESCCr. SCORE2-OP risk prediction algorithms: estimating incident cardiovascular event risk in older persons in four geographical risk regions. Eur Heart J. 2021;42:2455–2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu T, Forgetta V, Yu OHY, Mokry L, Gregory M, Thanassoulis G, Greenwood CMT and Richards JB. Polygenic risk for coronary heart disease acts through atherosclerosis in type 2 diabetes. Cardiovasc Diabetol. 2020;19:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


