Abstract
Protein splicing is a post-translational process by which an intervening protein, or intein, catalyzes its own excision from flanking polypeptides, or exteins, coupled to extein ligation. Four inteins interrupt the MCM helicase of the halophile Haloquadratum walsbyi, two of which are mini-inteins that lack a homing endonuclease. Both inteins can be over-expressed in E. coli and purified as unspliced precursors; splicing can be induced in vitro on incubation with salt. However, one intein can splice at 0.5 M NaCl in vitro, whereas the other splices efficiently only above 2 M NaCl; the organism also requires high salt to grow, with the standard growth media containing over 3 M NaCl and about 0.75 M magnesium salts. Consistent with this difference in salt-dependent activity, an intein-containing precursor protein with both inteins promotes conditional alternative protein splicing (CAPS) to yield different spliced products dependent on the salt concentration. Native Trp fluorescence of the inteins suggests that the difference in activity may be due to partial unfolding of the inteins at lower salt concentrations. This differential salt sensitivity of intein activity may provide a useful mechanism for halophiles to respond to environmental changes.
Keywords: Intein, Protein Splicing, Alternative Splicing, Halophile
Introduction
Inteins mediate the process of protein splicing, a post-translational event by which the intervening protein, or intein, catalyzes its own excision from the flanking polypeptides, or exteins, concomitant with extein ligation1 (Figure 1A). Splicing is a four-step process (Figure S1): (1) an amide to thioester rearrangement of the peptide bond linking the N-extein to the first residue of the intein (Cys for the inteins described here), (2) a transesterification reaction that results in the transfer of the N-extein from the side chain of the first residue of the intein to the side chain of the first residue of the C-extein (Ser for these inteins) to produce a branched ester intermediate, (3) cleavage of the intein from the exteins via cyclization of the C-terminal residue of the intein (Asn) coupled to peptide bond cleavage, and (4) reversion of the ester bond linking the ligated exteins to the native peptide bond.1
Figure 1. Schematic of protein splicing and the H. walsbyi cdc21 precursor.
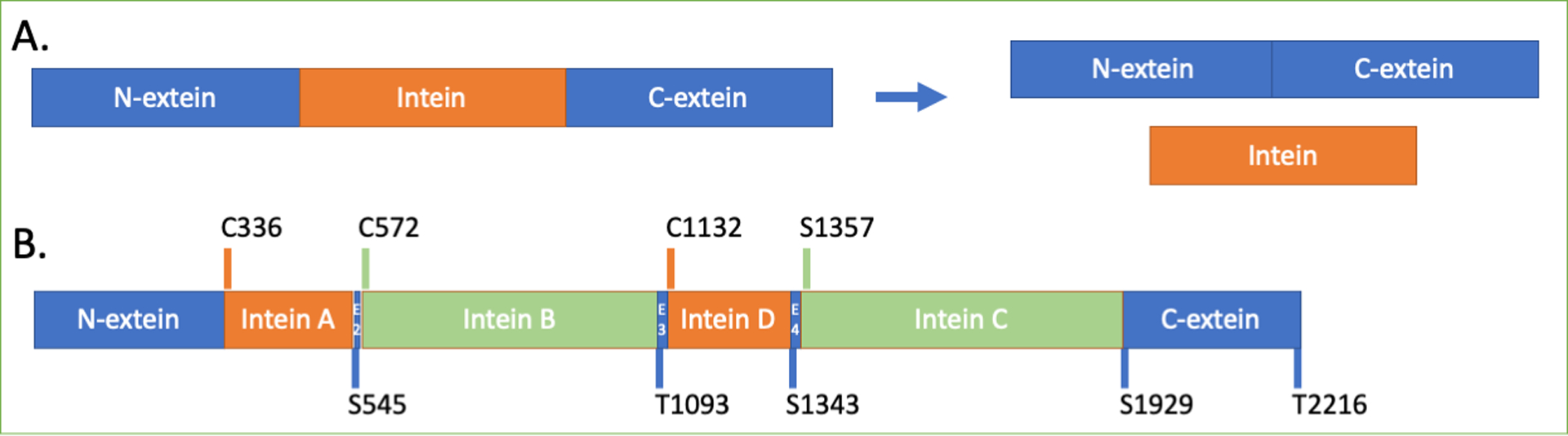
A. Protein splicing results in the excision of an intein from flanking polypeptides (the N- and C-exteins) concomitant to extein ligation to produce a functional extein protein. B. The insertion sites of the four inteins that interrupt the cdc21 protein in H. walsbyi. Mini-inteins (A and D) are in orange, and homing endonuclease-containing inteins are in blue (B and C). The codes above the schematic indicate the amino acid residue that is the first residue of the intein (orange or green bar), and those below indicate the first residue of the extein segment (blue). The insertion sites were named in the order of their discovery and are therefore out of alphabetical order, and the schematic is approximately to scale.
Improper coordination of the steps of splicing can lead to unproductive side reactions. The thioester or ester formed in steps 1 and 2 can be subject to hydrolysis if step 3 is prevented or delayed. This side reaction is called N-terminal cleavage and results in the production of the N-extein separate from the intein-C-extein fusion protein. Alternatively, if step 3 happens before step 1 or 2, C-terminal cleavage can result, with the N-extein and intein fused and the C-extein liberated.1
Many inteins are interrupted by nested homing endonuclease (HEN) domains similar to those found in mobile introns.2 Other inteins lack this domain and are called mini-inteins. The HEN domain makes a double-stranded break in an intein-less allele, facilitating intein homing by homologous recombination. This parasitic nature of inteins has been described as a homing cycle, by which an intein that splices poorly may exert a fitness cost on the host organism and loss of the intein by a random genetic event may result from this negative selective pressure. However, the HEN domain permits reintegration of the intein as long as an intein-containing allele is present either in a polyploid cell or in a population capable of mating.3, 4 Selective pressure against inefficient splicing also could result in the molecular evolution of a more efficient intein that splices co-translationally and exerts no significant burden on the host. In this case, the selective pressure to retain the HEN domain is lost, and the domain may be degraded by mutation or may be lost to form a mini-intein. Subsequent genetic deletion of these mini-inteins would result in an intein-less allele that could be subject to reinvasion by a HEN-containing intein. The genetic removal of an intein must be precise as most inteins invade near the active sites of essential enzymes involved in DNA replication, repair, and recombination.5, 6 It is likely that a dynamic equilibrium exists in the archaeal ecosystem between intein-containing and intein-lacking organisms.7
In contrast to this parasitic view of inteins, it is possible that some inteins may play a role for the host and be maintained by positive selective pressure. If so, such inteins likely would splice conditionally, subject to an environmental cue that might signal for the production of active extein segments from an inactive intein-interrupted precursor. For example, split inteins have been engineered to splice conditionally when induced with light or the binding of small molecules.8–17 Full-length inteins have been engineered to be triggered to splice by temperature, light, pH, or small molecules.18–20 Inteins from thermophiles may require elevated temperatures to splice in vitro,21–26 and inteins may require reduction of native disulfide bonds involving active site Cys residues to allow splicing.27–30 No direct evidence of conditional splicing playing a beneficial role in a native system has been shown. However, the activity of the Methanoculleus marisnigri PolII intein is regulated by the redox state of an internal disulfide bond, and its splicing activity is sensitive to redox state in E. coli.31 The splicing of the Pyrococcus horikoshii RadA intein, when interrupting the native recombinase, is activated by single-stranded DNA, which suggests that splicing may be triggered when ssDNA signals the need for recombinase activity.32–35 The Mycobacterium tuberculosis SufB intein is sensitive to oxidative and nitrosative stress that may link such stressors to Iron-Sulfur cluster assembly.36 The splicing of a Mycobacterium smegmatis DnaB intein is sensitive to oxidative stress in M. smegmatis, suggesting that pausing DNA replication may be connected to a stress response.37 These instances suggest that splicing-mediated gene regulation might serve a role for host cells.
Some genes contain multiple sites for intein insertion in different species, while in some cases multiple inteins are present in a single gene.37–40 If these inteins were to splice differently under environmentally relevant conditions, they could produce alternatively spliced products, which we will call conditional alternative protein splicing, or CAPS. For instance, the two DnaB inteins in M. smegmatis are differentially sensitive to oxidative stress and splice at different rates.37 Here, we describe another potential example in the extreme halophile Haloquadratum walsbyi,41, 42 where the differential dependence on salt concentration can lead to CAPS.
Inteins are broadly distributed in the haloarchaea, with at least 24 different genes invaded by inteins.40 Four inteins from halophiles have been purified as unspliced precursor fusion proteins in E. coli and induced to splice efficiently in vitro with incubation at high salt concentrations, another example of conditional protein splicing.43–45 Three of these inteins are mini-inteins (Halobacterium salinarum PolII and cdc21 and Halorhabdus utahensis MCM2), with the H. utahensis intein splicing at lower salt concentrations than the inteins from H. salinarum.43, 44 The other is a full-length intein with an intervening homing endonuclease domain (Haloferax volcanii PolB-c).45 Haloquadratum walsbyi has 15 inteins, with four separate inteins invading the cdc21 mini-chromosome maintenance (MCM) gene, a DNA helicase key to replication.39 Two of these inteins have nested homing endonuclease domains, and the other two, studied here, are mini-inteins.
Below, we show that the two mini-inteins in the H. walsbyi cdc21 protein can be expressed as unspliced fusion proteins in E. coli. The inteins splice at different salt concentrations in vitro, both when expressed separately and also when expressed as part of the same fusion protein. Analysis of their global structural stability by native Trp fluorescence shows that they fold at different salt concentrations, which likely contributes to the differential dependence of intein activity on salt concentration.
Materials and Methods
Plasmid preparation
To create E. coli expression plasmids for the H. walsbyi inteins, we obtained H. walsbyi from the Leibniz Institute DSMZ German Collection of Microorganisms and Cell Culture (DSM 16790, strain HBSQ001) and cultured the cells unshaken at 37°C in medium 1091.41 We harvested the cells by centrifugation and purified the genomic DNA using a Wizard Genomic DNA purification kit (Promega) by the manufacturer’s instructions. The plasmids described below are listed in Table S1.
To create plasmids pMIH-A and pMIH-D, which express intein precursor proteins MIH-A and MIH-D, we used PCR to amplify the intein genes from genomic DNA. For intein A, we used primers MIHAU and MIHAL, and, for intein D, we used primers MIHDU and MIHDL, digested the PCR products with StuI and EcoRI, and inserted them between those same sites in plasmid pMIHMma, described previously.31 (See Table S2 for oligonucleotide sequences.) Plasmid pMIH-A expresses a 397-residue N-extein with N-terminal MBP followed by a short linker and the nine C-terminal residues of the native N-extein, fused in-frame to the 209 residue intein and 23 residue C-extein, which has 11 native C-extein residues and a His-tag. Plasmid pMIH-D expresses a 398-residue N-extein with N-terminal MBP followed by a short linker and the 10 C-terminal residues of the native N-extein, fused in-frame to the 211-residue intein and a 23-residue C-extein, which has 11 native C-extein residues and a His-tag.
To create plasmid pMIHAD, which encodes the fusion protein M-(IA)-E-(ID)-H, we purchased a synthetic gene from Genscript in pET28-b(+) as an in-frame fusion of the gene for the E. coli maltose binding protein (MBP), a short poly-N linker, the 22 C-terminal residues of the N-extein, the 209 residue intein A, the following 66 extein residues (27 between inteins A and B and 39 between inteins B and D), the 211 residue intein D, and the 14 extein residues between inteins D and C, followed by a short linker and a His-tag. In addition, we obtained site-directed mutations of the plasmid pMIHAD from Genscript to create pMIHADInAX, for which Cys1, Asn209, and Ser+1 are all changed to Ala for intein A to make fusion protein M-(mIA)-E-(ID)-H, and pMIHADInDX, for which Cys1, Asn211, and Ser+1 are all changed to Ala for intein D to make fusion protein M-(IA)-E-(mID)-H.
To create plasmids pNIH-A and pNIH-D to express proteins for native Trp fluorescence experiments, we used Gibson assembly with linearized pET21-b(+) as the vector. We used pMIH-A or pMIH-D as templates for PCR for the insert, using the InFusion HD cloning kit per the manufacturer’s instructions (Takara Bio USA). To make pNIH-A, we used PCR primers AGibF and AGibR and for pNIH-D we used PCR primers DGibF and DGibR. Plasmid pNIH-A expresses NIH-A, a nine residue N-extein fused in-frame to the 209 residue intein and a 24-residue C-extein including a C-terminal His-tag. Plasmid pNIH-D expresses NIH-D, a 10 residue N-extein fused in-frame to the 211 residue intein and a 24-residue C-extein including a C-terminal His-tag. To prevent splicing, site-directed mutagenesis was performed by Genscript to make plasmids pNIH-AX and pNIH-DX, which have mutations to Ala for residues Cys1, Ser+1, and the final Asn residue for each intein. Plasmids pNIH-AX and pNIH-A expressed poorly. Therefore, we inserted a GB1 tag using annealed and phosphorylated oligonucleotides GBU and GBL into the NdeI site of pNIH-A and pNIH-D to make pGBAI and pGBDI. Plasmid pGBAI expresses GbA1, the 56-residue GB1 tag fused in-frame to a 6 residue linker, 7 residues of the native N-extein, the 209 residue intein and a 24-residue C-extein including a C-terminal His-tag. Plasmid pGBD1 expresses GbD1, the 56-residue GB1 tag fused in-frame to a 6-residue linker, 7 residues of the native N-extein, the 211 residue intein and a 24-residue C-extein including a C-terminal His-tag. Genscript then made site-directed mutants that resulted in the conversion of Trp43 of the GB1 tag to Phe, Cys1 and Ser+1 of each intein to Ala, and Asn209 (GBAI) or Asn211 (GBDI) to Ala to make plasmids pGBA and pGBD to express proteins GBA and GBD.
Protein expression and purification
In order to over-express intein fusion proteins, the plasmids described above were transformed into E. coli BL21(DE3). A 1:50 inoculum in LB media was grown to mid-log phase, induced with isopropyl-β-d-1-thiogalactopyranoside (IPTG) to 1 mM, and incubated with shaking at 20°C for 3 h. Cells were pelleted by centrifugation and resuspended in buffer A (20 mM HEPES, pH 7.5, 500 mM NaCl) supplemented with 0.1 mM phenylmethylsulfonyl fluoride, 10 units of benzonase nuclease, and an EDTA-free protease inhibitor cocktail. Cells were disrupted by passage through a French-pressure cell, and the lysate was clarified by centrifugation at 16 000 × g for 30 min at 4°C. Proteins were purified using the C-terminal His-tag by immobilized metal affinity chromatography using Talon metal affinity resin (Takara Bio USA) per manufacturer’s instructions. Elution fractions were exchanged into buffer A using an EMD-Millipore Amicon Centrifugal Filer with a 3000 MWCO, and the protein concentration was determined by a Bradford assay.46
Protein splicing activity was assayed by incubating about 0.2 mg/ml protein in a reaction cocktail that has 20 mM HEPES, pH 7.5, 2 mM Tris(2-carboxyethyl)phosphine (TCEP), 5 mM ethylenediaminetetraacetic acid (EDTA), and sodium chloride at the concentrations indicated in the figure legends at 20°C. The protein reaction was diluted 1:1 with water before SDS-PAGE to prevent salt precipitation on addition of the SDS-PAGE loading buffer.
The extent of protein splicing was estimated by SDS-PAGE stained with QC Colloidal Coomassie Stain (Bio-Rad). For MIH-A, the precursor protein MIH has a Mr of 68,356.42, and protein splicing would yield MH (Mr 46,176.05) and I (Mr 22,198.38). For MIH-D, the precursor protein MIH has a Mr of 69,116.90, and protein splicing would yield MH (Mr 46,432.20) and I (Mr 22,702.72).
For M-(IA)-E-(ID)-H, the precursor (Mr 100,502.63) can splice into MEH (Mr 55,637.56), IA (Mr 22,198.38) and ID (Mr 22,702.72). Thus, splicing of intein ID first would yield M-(IA)-E-H (Mr 77,817.53) and splicing of intein IA first would yield M-E-(ID)-H (Mr 78,322.26). The percentage of splicing was estimated using densitometry via ImageJ,47 using the formula % splicing = 100* (spliced product) / ((precursor) + (spliced product)), which each densitometry reading corrected for relative molar mass.
His-tagged proteins were detected using InVision His-tag in-gel stain (Life Technologies) and visualized with a BioRad Gel Doc Imager with a UV tray. For mass spectrometry, high resolution mass spectra were obtained with electrospray ionization on an Orbitrap Fusion mass spectrometer following an in-line C4 desalting column by the University of Massachusetts, Amherst.
Native Trp fluorescence
We measured native Trp fluorescence using a Perkin Elmer LS55 fluorescence spectrometer, with a constant temperature of 25°C maintained by a Peltier control system. An excitation wavelength of 295 nm was used to excite only the Trp residues in the intein rather than Tyr residues.48 Emission was monitored from 310 nm to 380 nm at a scan rate of 10 nm/min, with 2.5 nm slit widths for excitation and emission on a medium gain setting. Protein Gb1A was measured at 10 μM and protein Gb1D was measured at 8 μM in 20 mM HEPES, pH 7.5, at varying concentrations of NaCl.
Results and Discussion
The cdc21 protein from H. walsbyi is interrupted by four inteins, as shown schematically in Figure 1B. Inteins A and D are mini-inteins, and inteins B and C have nested HEN domains. (The inteins are named by insertion site, which were named as they were discovered and are not in alphabetical order.) To study the splicing of the inteins separately, we elected to focus on the mini-inteins, as inteins with nested HEN domains can be challenging to express in E. coli.38 An alignment of the intein sequences is given in Figure S2. The inteins have 32% sequence identity and 50% similarity.
We created expression vectors to over-express inteins A and D separately as fusion proteins with an N-terminal maltose binding protein (M) and a C-terminal His-tag (H) serving as the exteins, creating fusion proteins MIH-A and MIH-D. To avoid any effects on splicing by non-native flanking residues, we designed the expression vectors such that the M is followed by a flexible linker and nine (intein A) or ten (intein D) native N-extein residues, and 11 native C-extein residues precede the His-tag for both inteins.
We were able to separately purify the intein fusion proteins MIH-A or MIH-D from E. coli via immobilized metal affinity chromatography. The intein fusion protein was incubated in vitro with EDTA to prevent any potential inhibition of splicing by divalent cations,49–52 with TCEP to reduce a potential disulfide bond between Cys1 and the internal Cys residues,27–29, 31 and with varying concentrations of sodium chloride.
Intein A promoted protein splicing at 20°C and 2.0 M NaCl, as seen by conversion of the precursor band MIH to the splicing products MH and I in Figure 3A. The identity of the SDS-PAGE bands were confirmed by InVision staining, which identifies bands that contain a His-tag (MIH and MH, but not I) (Figure 3B). This confirms that we are observing splicing and not N-terminal cleavage, which would produce M and IH. We also confirmed the identity of the protein products by LC-MS (Table S3). These confirmations are important as halophile proteins can have aberrant migration patterns on SDS-PAGE, likely due to their highly negative surface charge.53, 54 For the MS peaks corresponding to the excised inteins, the Mr values are about 18 Da smaller than expected, which may be due to the intein retaining the C-terminal aminosuccinimide from the third step of splicing rather than undergoing subsequent hydrolysis to either Asn or iso-Asn.
Figure 3. In vitro protein splicing activity of H. walsbyi inteins as separate fusion proteins.
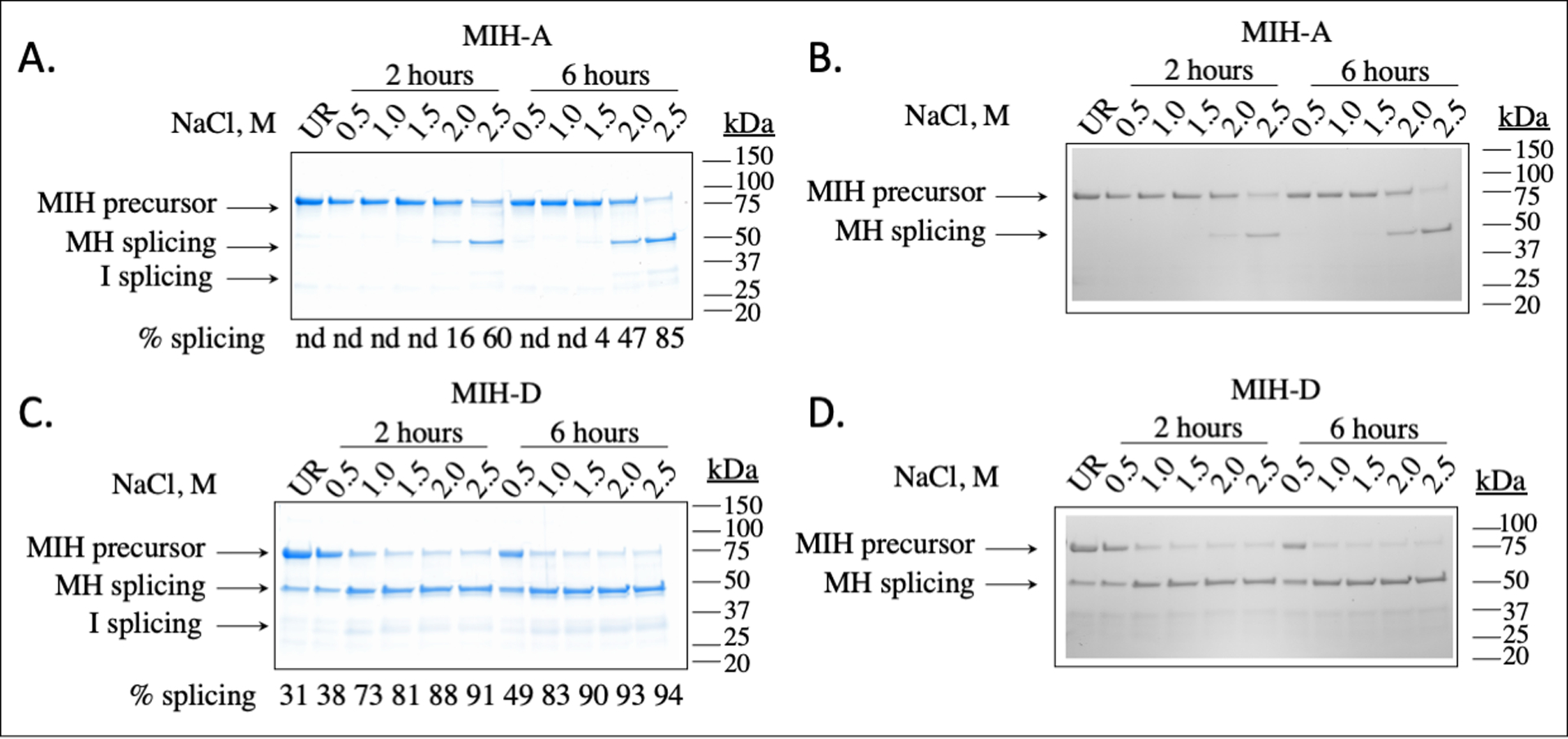
SDS-PAGE analysis of in vitro protein splicing of MIH, stained with Coomassie blue. Splicing of precursor MIH (68 kDa) results in MH (46 kDa) and I (22 kDa). In panels A and C, purified precursor fusion protein MIH-A or MIH-D (0.2 mg/mL) was incubated for 2 h or 6 h at 20°C at the total NaCl concentrations indicated above the lanes in 20 mM HEPES buffer, pH 7.5, with 2 mM TCEP and 5 mM EDTA, as described in the Materials and Methods. The percentage of splicing was estimated by densitometry using ImageJ and represents the average of two independent trials.47 In panels B and D, the gels were probed with InVision stain, which detects proteins with a His-tag. Hence, we would expect to see bands corresponding to MIH, MH, or IH, but not MI or I. UR is unreacted protein, nd is not determined.
Likewise, we were able to show that intein D also promotes splicing at 20°C (Figure 3C) and confirmed this splicing with InVision staining and LC-MS (Figure 3D and Table S3). However, efficient protein splicing occurs at lower salt concentrations. Although splicing efficiency improves at higher salt concentrations, we observe splicing at 0.5 M NaCl, and observe some splicing in E. coli by the appearance of the MH band in the purified and unreacted fraction. This suggests that intein D does not have the same requirement for high salt as intein A. At 2 M NaCl, the inteins splice over a range of temperatures from 15°C to 30°C to a similar extent (Figure S3). Splicing of intein A increases slightly over 6 h, whereas splicing of intein D is mostly complete after 2 h of incubation (Figure 3).
Although the inteins display significant differences in salt dependence, there is a high degree of similarity in their amino acid sequences (Figure S2). These two intein sequences are 32% identical and 50% similar, as measured by the EMBOSS Water tool.55 Within the conserved intein sequence blocks the similarity is even higher, with 80% or higher similarity in blocks A, F, and G, which comprise the N-terminal (A) and C-terminal (F and G) sections of the active site. The sequences are only 50% similar in conserved block B, but the highly conserved Thr and His residues are present. As calculated by the ExPasy pI/Mw tool, the isoelectric point (pI) values of the two inteins are both low and similar, at 4.48 and 4.43, respectively.56 Both inteins have similar numbers of potentially charged residues, with intein A having 33 acidic and 21 basic residues and intein D having 34 acidic and 22 basic residues.
Interestingly, despite relatively high amino acid similarity, the inteins show differential activity as a function of salt concentration. In contrast, the three inteins that interrupt the Pyrococcus abyssi ribonucleotide reductase all splice efficiently in E. coli, despite having little sequence similarity and being flanked at their C-termini by different nucleophiles to promote step 2.38 On the other hand, the two inteins that interrupt the DnaB from M. smegmatis splice differently.37 The second M. smegmatis intein splices relatively quickly and its splicing is not sensitive to oxidative or nitrosative stress. However, the first intein splices more slowly and its splicing is sensitive to oxidative stress, both in vitro and in M. smegmatis. The first intein is also susceptible to inhibition by zinc and reactive chlorine species.57, 58 These two inteins also can facilitate conditional alternative protein splicing (CAPS) as a function of time when expressed as part of the same precursor. The M. smegmatis inteins differ considerably, as the first intein is a non-canonical class three intein;37 class three inteins splice by an alternative mechanism utilizing an internal nucleophile in place of the N-terminal Cys or Ser.59, 60 In contrast, the second intein is a traditional class one intein that splices by the canonical mechanism, so it is not surprising that the two inteins might promote CAPS from the same precursor. We were interested in whether the two more similar H. walsbyi mini-inteins can also promote CAPS.
To test whether the inteins retained their differential salt dependence when interrupting the same protein, we created the fusion protein M-(IA)-E-(ID)-H, which encodes for N-terminal MBP, followed by a flexible linker, the 22 residues that flank the N-terminus of intein A, intein A, the next 66 extein residues (Exteins-2 and -3 in Figure 1B) that would result from the splicing of intein B, then intein D, the 14 extein residues between inteins D and C, and a His-tag (Figure 1B). We anticipated that at low salt concentrations, intein D would splice, producing M-(IA)-E-H and ID. We expected both inteins to splice at higher salt concentrations, producing M-E-H, IA and ID (Figure 2). These predictions are supported by the results in Figure 4. In Figure 4A, product MIAEH is produced at 1.0 M salt, indicating that intein D has been excised. At 2.0 M salt, the fully spliced product MEH is produced, resulting from the splicing of intein A from MIAEH. The extent of splicing at 2 h and 6 h is also consistent with the extent of splicing of the individual inteins at those times (Figures 2 and 3). The identity of the bands is supported by InVision staining (Figure S4), which indicates which bands have a C-terminal His-tag. Given the similarity in size between inteins A and D, protein MIAEH (from splicing of intein D) and MEIDH (splicing of intein A) would be similar in size on SDS-PAGE. Therefore, we also analyzed the splicing via LC-MS (Table S3). For a reaction run for 6 h at 20°C at 2 M salt for a fusion protein with both active inteins, we observed a peak consistent with the MIAEH intermediate but not the MEIDH intermediate, as expected if the reaction follows the scheme in Figure 2 (Table S3).
Figure 2. Schematic of conditional alternative protein splicing (CAPS).
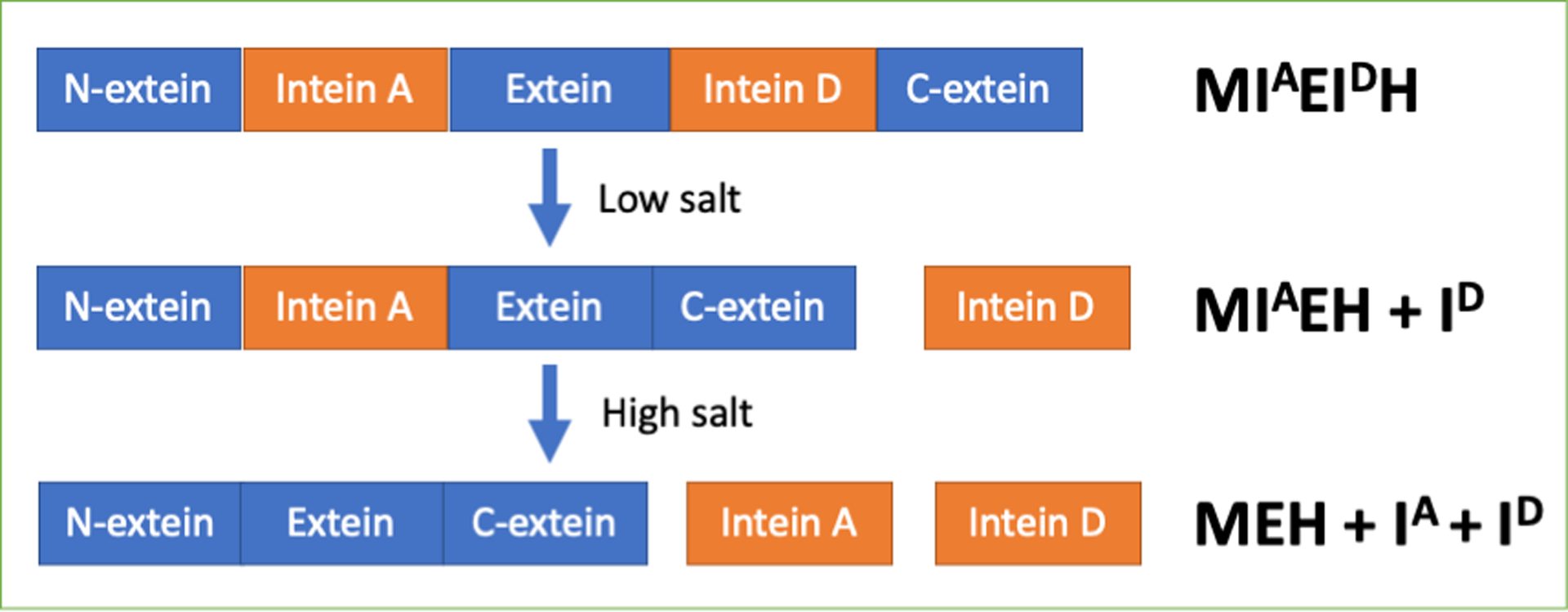
If intein D splices at lower salt concentrations than intein A, it would be removed first to produce an alternative precursor fusion protein. Intein A would then splice on incubation at a higher salt concentration. The codes to the right of the cartoon schematic correspond to the expressed protein products observed in Figure 4.
Figure 4. In vitro protein splicing activity of H. walsbyi inteins as part of one fusion protein.
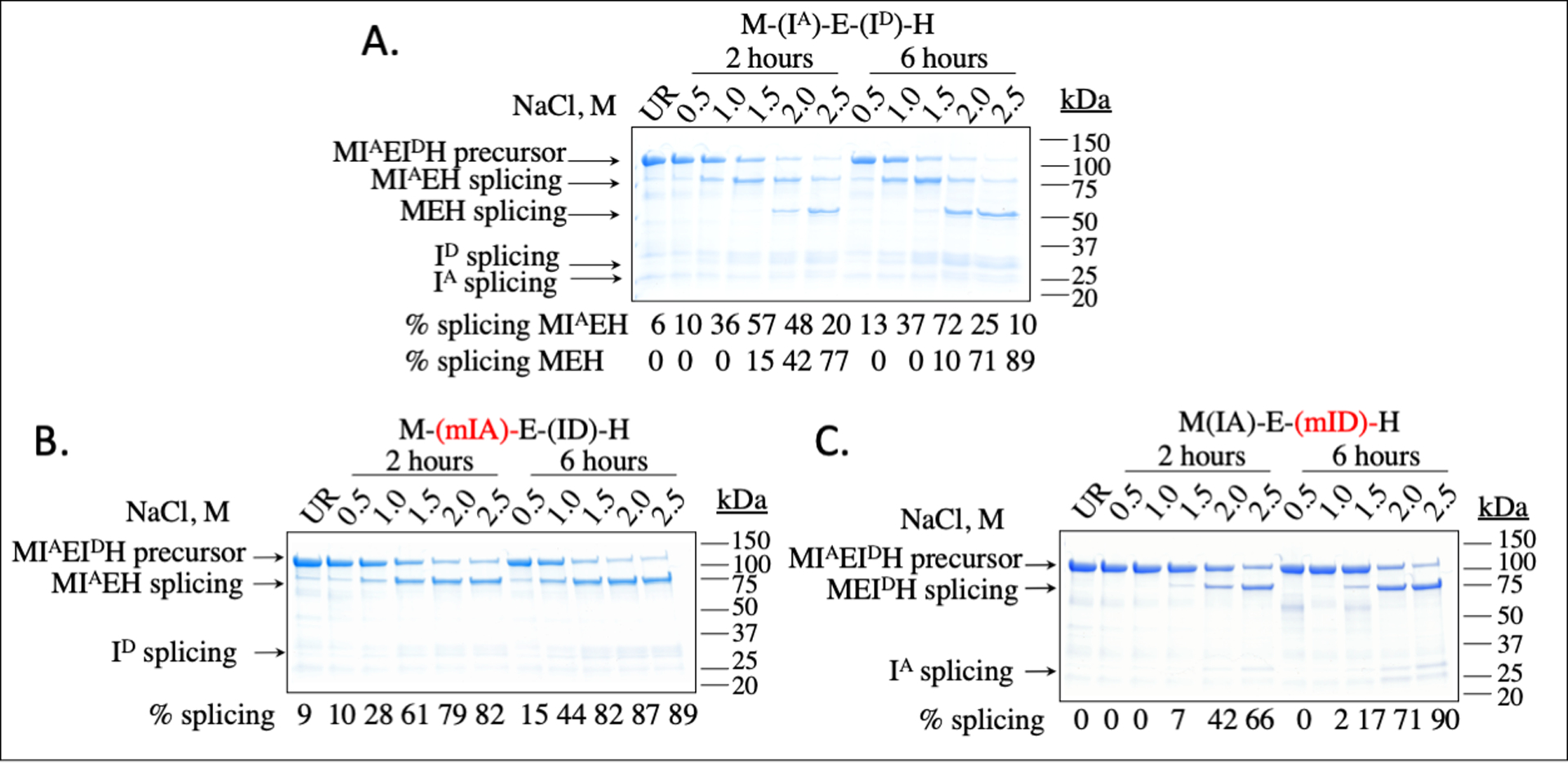
SDS-PAGE analysis of in vitro protein splicing of M-(IA)-E-(ID)-H, stained with Coomassie blue. Splicing of precursor M-(IA)-E-(ID)-H (68 kDa) results in MIAEH (78 kDa) or MEIDH after splicing of one intein, MEH (56 kDa) after splicing of both inteins, as well as inteins IA (22 kDa) or ID (23 kDa). In panel A, purified precursor fusion protein M-(IA)-E-(ID)-H (0.2 mg/mL) was incubated for 2 h or 6 h at 20°C at the total NaCl concentrations indicated above the lanes in 20 mM HEPES buffer, pH 7.5, with 2 mM TCEP and 5 mM EDTA, as described in the Materials and Methods. In panels B and C, the same reaction was run, but with inactivating mutations to intein A (M-(mIA)-E-(ID)-H) or to intein D (M-(IA)-E-(mID)-H). The percentages reflect the average of two independent trials.
Given the similarity in sizes between inteins A and D, we confirmed the splicing via mutational analysis and via LC-MS. To inactivate one intein, we made mutations to key catalytic residues, Cys1, Ser+1, and C-terminal Asn, to Ala to prevent splicing activity. We made one fusion protein with mutated intein A (M-(mIA)-E-(ID)-H), and another with mutated intein D (M-(IA)-E-(mID)-H). As seen in Figure 4B, when intein A is mutated we observe splicing at a salt concentration similar to that of intein D alone in Figure 3. When intein D is mutated as seen in Figure 4C, we observe splicing at a salt concentration similar to that of intein A in Figure 3. We confirmed the identity of the products of the splicing products for the two intein-inactivated mutants via both Invision staining (Figure S4) and LC/MS (Table S3).
In order to determine if the difference in intein activity is correlated to protein folding, we used native Trp fluorescence to measure the folding of the intein as a function of salt concentration (Figure 5). We attempted to express the inteins from separate plasmids as a fusion protein with an N-terminal His-tag rather than MBP. To prevent splicing, we mutated Cys1, the C-terminal Asn, and Ser+1 all to Ala. However, protein expression was poor, particularly with intein A. This is not surprising, given that the excised intein bands in Figures 2 and 3 are faint relative to the MBP-containing spliced product, suggesting compromised solubility for the excised intein. Therefore, we expressed the intein genes such that they were separately expressed as fusion proteins with an N-terminal Gb1 tag.61 We mutated the Trp in the Gb1 sequence to Phe such that we could monitor the three Trp residues in intein A and the two Trp residues in intein D, using an excitation wavelength of 295 nm to specifically excite the Trp fluorescence.48 The fluorescence of intein A at salt concentrations below 1.5 M NaCl is low, suggesting that the protein folds and hence buries the Trp residues as the salt concentration increases to values near those that permit splicing (Figure 5). In comparison, intein D, which can splice at lower salt concentration, also appears to be well folded at 1 M salt. The blue shift of the maximum emission wavelength also depends on salt concentration (Figure S5). These results suggest that the splicing dependence of the intein activity is due to substantive unfolding or disorder at sub-optimal ionic strength.
Figure 5. Salt-dependence of intein folding as measured by native Trp fluorescence.
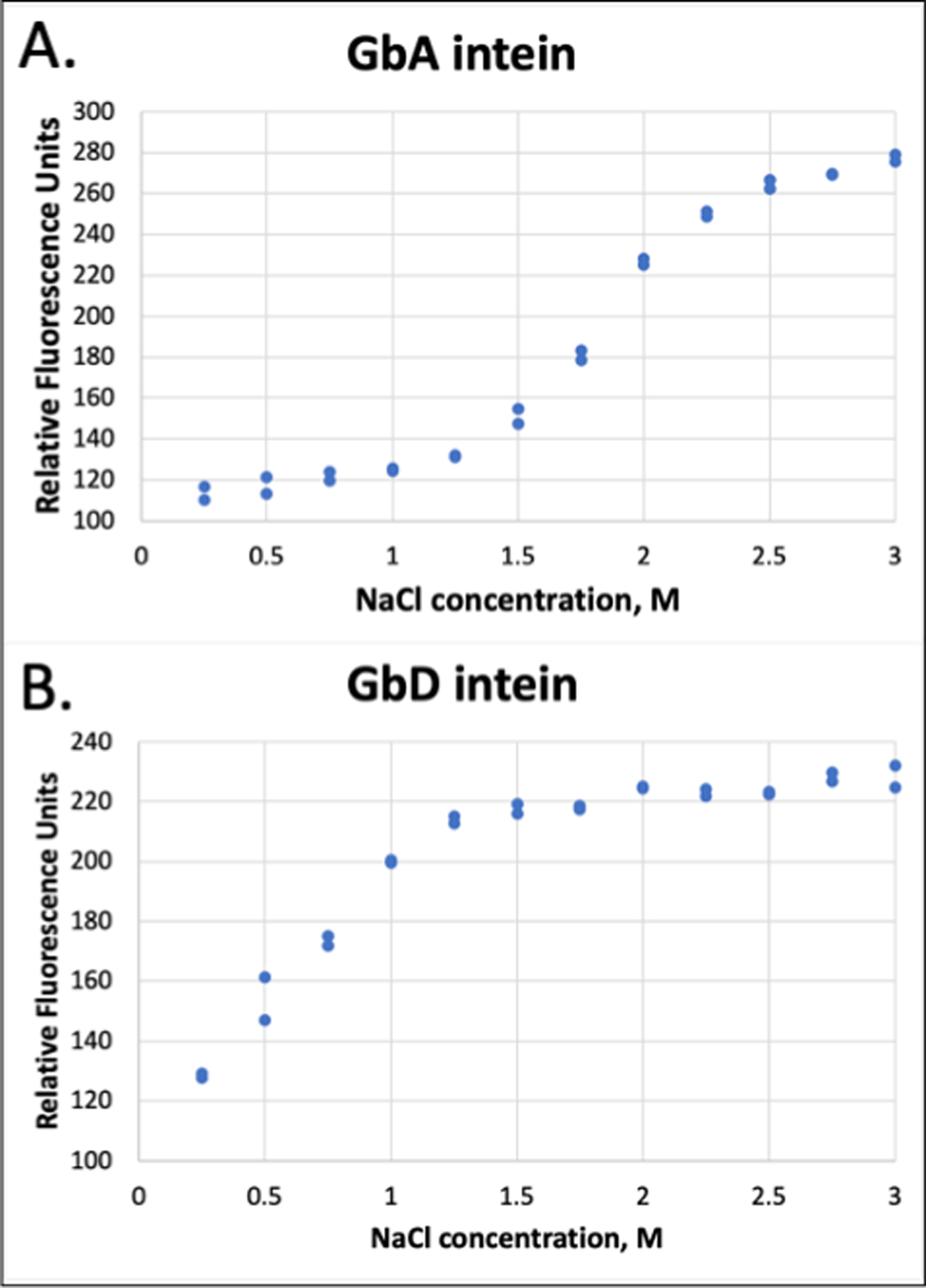
Native Trp fluorescence was measured at 340 nm via excitation at 295 nm using a Perkin Elmer LS55 fluorescence spectrometer. Gain was set to medium with emission and excitation slit widths of 2.5 nm. Temperature was maintained at 25°C with a Peltier system. Protein GBA was at 10 μM, and protein GBD at 8 μM. Samples were measured in 20 mM HEPES buffer at pH 7.5 at the indicated salt concentrations on the abscissa. Data reported are from two independent trials.
Proteins from archaeal extremophiles have various strategies to prevent unfolding or a transition to disorder under extreme conditions.62 Many archaeal halophiles use a “salt-in” strategy to avoid osmotic stress and have proteins with very low pI values and hence very negatively charged surfaces, with a slightly lower number of bulky hydrophobic residues.63–65 Although both inteins A and D have similarly low pI values and hence are likely to be similarly negatively charged, it has been suggested that more subtle structural differences at the surface, such as the choice of less bulky charged side chains, may affect the interaction surface area of halophilic proteins and hence their structural stability in the low water activity conditions of a high salt environment.66 Inteins are highly mobile genetic elements, and the inconsistency of inteins being present at the same insertion site in proteins across the haloarchaea suggests transfer and loss of inteins within the family.40 It is possible that the salt dependence of halophile inteins may be influenced by the salt requirements of the species they have invaded during their mobile evolutionary history.
H. walsbyi is the dominant archaeal species at very high aqueous salinity, despite its slow growth in laboratory-based culture.41, 67 The degree of species representation of H. walsbyi in saline pools can differ based on salinity.68 On extreme dilution from over 30% salt to about 12% salt, it has been shown that although the ecotype of H. walsbyi changes and the percentage abundance of the population declines, that the dominant ecotype of H. walsbyi is still present and recovers after evaporation results in an increase in salinity.69 H. walsbyi abundance can also vary seasonally, likely due to the balance of ions, particularly magnesium.70 These observations of variable growth under physiological cues of ionic strength and/or water activity are consistent with the organism needing biochemical triggers that induce cell division under permissive environmental conditions. It is possible that CAPS activity could serve this purpose by linking post-translational activation of intein-containing genes related to DNA replication and cell-cycle progression to successful protein splicing of an interrupted precursor. One could speculate that in times of flooding, H. walsbyi may express its MCM in an intein-interrupted, inactivated format, ready for immediate activation on a return to salt concentrations conducive to growth and cell division.
For the inteins described here, the ligated exteins constitute the cdc21 (MCM) protein, the DNA helicase important in initiation of replication.71 Comparing the sequence of the H. walsbyi MCM to the well characterized MCM from the archaea Sulfolobus solfataricus,72, 73 the core AAA+ sequences are highly similar. The four inteins in H. walsbyi interrupt this core AAA+ region. Figure S6 shows a sequence alignment between the ligated H. walsbyi exteins and the uninterrupted S. solfataricus MCM using BLAST.74 The sequences have 39% identity and 60% sequence similarity, with more conservation in the AAA+ region. The conserved regions of the MCM are noted under the S. solfataricus sequence in Figure S6.75 The Ser+1 residue of intein A is at the C-terminus of the Walker A motif, and the Ser+1 of intein D (an Asp residue in S. solfataricus) immediately follows the Walker B motif. This suggests that the inteins interrupt the motifs responsible for nucleotide and magnesium ion binding. Mutation of the Lys that would precede intein A to Ala has been shown to compromise both helicase and ATPase activity in the S. solfataricus MCM, suggesting this region is vital to MCM activity.73 Mutations of residues Ala+6 and Ala+10 for intein D to Arg result in a S. solfataricus MCM that cannot form hexamer and lacks helicase activity,76 suggesting that a region immediately C-terminal of intein D may be at an interface, and that intein D might therefore interfere with hexamer formation. Interestingly, the change of Asp for Ser at the end of the Walker B motif in the H. walsbyi might compromise the conserved Walker B sequence to allow for intein D insertion, although a Glu residue that follows could serve in the role of the conserved Asp. Although not studied here, intein C is at a region that may be trans-acting with adjacent monomers in the MCM hexamer, suggesting that intein C could interfere with hexamer formation.75 The residues six and eight positions downstream of intein B have been shown to be essential for DNA binding in the S. solfataricus MCM,77 suggesting the importance of these region. Figure S7 uses the 296 C-terminal residues of the S. solfataricus MCM crystal structure (PDB code 3F9V)76 to represent the MCM in the same orientation as the structure of the Natronomonas moolapensis MCM used by Belfort and coworkers to compare the intein insertion sites of bacterial DnaB and archaeal MCM exteins.5
Interestingly, in the S. solfataricus MCM, the AAA+ region can function as an ATPase and helicase on its own, and the helicase activity can be enhanced by addition of the N-terminal domain in trans. This suggests that the MCM protein may have different properties when expressed in fragments. One could speculate that differential protein splicing (or intein-mediated cleavage at splicing boundaries) could result in H. walsbyi MCM variants that also have different properties. Perhaps an intein remaining in the precursor might disrupt helicase activity by interfering with an interface between two monomers while still permitting ATPase activity.
It is interesting that two examples of CAPS are from M. smegmatis DnaB and H. walsbyi MCM. DnaB is the most common known intein host in bacteria, and MCM is the most common known host in the archaea.5 Both serve as replicative helicases, but are not closely related in sequence or structure.5 Given that these proteins are essential to the initiation of DNA replication and cell division, an intein-mediated trigger in these proteins could be useful to the host organism by linking cell division to appropriate environmental conditions for growth.
In addition, halophile inteins are useful in biotechnology applications including protein cyclization and semi-synthesis.43 Adding these two inteins that are easily purified as unspliced precursors to the intein toolkit could allow for further development of interesting applications, including biosensors that could detect salt concentration differences by facilitating the splicing of fluorescent proteins under selective conditions.78–80
Supplementary Material
ACKNOWLEDGMENT
We thank Alvin Gomez, Shirley Amunya, Angel Chavez, Deirdre Reidy, Lily Russo, and the late Julie Reitter for experimental assistance, and we thank Chunyu Wang for a critical reading of the manuscript. Mass spectral data were acquired by the University of Massachusetts mass spectrometry core facility. We thank Dr. Steve Eyles for assistance with mass spectrometry.
Funding Sources
This work was supported by the US National Institutes of Health NIGMS Grant 1R15GM132817-01 (KVM) and the US National Science Foundation (NSF) grant MCB-1517138 (KVM). KVM also is supported by a Henry Dreyfus Teacher-Scholar Award. The Orbitrap Fusion was funded by NIH SIG S10OD010645. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of NIGMS or NIH.
ABBREVIATIONS
- CAPS
Conditional Alternative Protein Splicing
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- HEN
homing endonuclease
- H. walsbyi
Haloquadratum walsbyi
- IPTG
isopropyl-1-β-d-thiogalactopyranoside
- MBP
E. coli maltose binding protein
- MCM
mini-chromosome maintenance
- TCEP
tris(2-carboxyethyl)phosphine
Footnotes
ACCESSION CODE
The UniProt accession ID for the intein-containing H. walsbyi cdc21 is Q18E84
Supporting Information: Schematic of the mechanism of protein splicing, amino acid sequence alignment of the two inteins, SDS-PAGE analysis of temperature dependence, Invision stain to support Figure 4, Fluorescence spectra data, annotated sequence alignment of exteins from H. walsbyi and S. solfataricus, C-terminal structure of S. solfataricus MCM with H. walsbyi intein insertion sites indicated, tables of plasmids and oligonucleotides used in cloning, and LC-MS data.
All authors have given approval to the final version of the manuscript.
References
- [1].Mills KV, Johnson MA, and Perler FB (2014) Protein splicing: how inteins escape from precursor proteins, J Biol Chem 289, 14498–14505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Belfort M, and Roberts RJ (1997) Homing endonucleases: keeping the house in order, Nucleic Acids Res 25, 3379–3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Barzel A, Obolski U, Gogarten JP, Kupiec M, and Hadany L (2011) Home and away- the evolutionary dynamics of homing endonucleases, BMC Evol Biol 11, 324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Barzel A, Naor A, Privman E, Kupiec M, and Gophna U (2011) Homing endonucleases residing within inteins: evolutionary puzzles awaiting genetic solutions, Biochem Soc Trans 39, 169–173. [DOI] [PubMed] [Google Scholar]
- [5].Novikova O, Jayachandran P, Kelley DS, Morton Z, Merwin S, Topilina NI, and Belfort M (2016) Intein Clustering Suggests Functional Importance in Different Domains of Life, Mol Biol Evol 33, 783–799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Swithers KS, Senejani AG, Fournier GP, and Gogarten JP (2009) Conservation of intron and intein insertion sites: implications for life histories of parasitic genetic elements, BMC Evol Biol 9, 303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Naor A, Altman-Price N, Soucy SM, Green AG, Mitiagin Y, Turgeman-Grott I, Davidovich N, Gogarten JP, and Gophna U (2016) Impact of a homing intein on recombination frequency and organismal fitness, Proc Natl Acad Sci U S A 113, E4654–4661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Berrade L, Kwon Y, and Camarero JA (2010) Photomodulation of protein trans-splicing through backbone photocaging of the DnaE split intein, Chembiochem 11, 1368–1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Binschik J, Zettler J, and Mootz HD (2011) Photocontrol of protein activity mediated by the cleavage reaction of a split intein, Angew Chem Int Ed Engl 50, 3249–3252. [DOI] [PubMed] [Google Scholar]
- [10].Brenzel S, Kurpiers T, and Mootz HD (2006) Engineering artificially split inteins for applications in protein chemistry: biochemical characterization of the split Ssp DnaB intein and comparison to the split Sce VMA intein, Biochemistry 45, 1571–1578. [DOI] [PubMed] [Google Scholar]
- [11].Brenzel S, and Mootz HD (2005) Design of an intein that can be inhibited with a small molecule ligand, J Am Chem Soc 127, 4176–4177. [DOI] [PubMed] [Google Scholar]
- [12].Mootz HD, Blum ES, Tyszkiewicz AB, and Muir TW (2003) Conditional protein splicing: a new tool to control protein structure and function in vitro and in vivo, J Am Chem Soc 125, 10561–10569. [DOI] [PubMed] [Google Scholar]
- [13].Mootz HD, and Muir TW (2002) Protein splicing triggered by a small molecule, J Am Chem Soc 124, 9044–9045. [DOI] [PubMed] [Google Scholar]
- [14].Schwartz EC, Saez L, Young MW, and Muir TW (2007) Post-translational enzyme activation in an animal via optimized conditional protein splicing, Nat Chem Biol 3, 50–54. [DOI] [PubMed] [Google Scholar]
- [15].Tyszkiewicz AB, and Muir TW (2008) Activation of protein splicing with light in yeast, Nat Methods 5, 303–305. [DOI] [PubMed] [Google Scholar]
- [16].Vila-Perello M, Hori Y, Ribo M, and Muir TW (2008) Activation of protein splicing by protease- or light-triggered O to N acyl migration, Angew Chem Int Ed Engl 47, 7764–7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Vila-Perello M, and Muir TW (2010) Biological applications of protein splicing, Cell 143, 191–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Buskirk AR, Ong YC, Gartner ZJ, and Liu DR (2004) Directed evolution of ligand dependence: small-molecule-activated protein splicing, Proc Natl Acad Sci U S A 101, 10505–10510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Skretas G, and Wood DW (2005) Regulation of protein activity with small-molecule-controlled inteins, Protein Sci 14, 523–532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Zeidler MP, Tan C, Bellaiche Y, Cherry S, Hader S, Gayko U, and Perrimon N (2004) Temperature-sensitive control of protein activity by conditionally splicing inteins, Nat Biotechnol 22, 871–876. [DOI] [PubMed] [Google Scholar]
- [21].Cambon-Bonavita MA, Schmitt P, Zieger M, Flaman JM, Lesongeur F, Raguenes G, Bindel D, Frisch N, Lakkis Z, Dupret D, Barbier G, and Querellou J (2000) Cloning, expression, and characterization of DNA polymerase I from the hyperthermophilic archaea Thermococcus fumicolans, Extremophiles 4, 215–225. [DOI] [PubMed] [Google Scholar]
- [22].Mills KV, Manning JS, Garcia AM, and Wuerdeman LA (2004) Protein splicing of a Pyrococcus abyssi intein with a C-terminal glutamine, J Biol Chem 279, 20685–20691. [DOI] [PubMed] [Google Scholar]
- [23].Shao Y, Xu MQ, and Paulus H (1995) Protein splicing: characterization of the aminosuccinimide residue at the carboxyl terminus of the excised intervening sequence, Biochemistry 34, 10844–10850. [DOI] [PubMed] [Google Scholar]
- [24].Shao Y, Xu MQ, and Paulus H (1996) Protein splicing: evidence for an N-O acyl rearrangement as the initial step in the splicing process, Biochemistry 35, 3810–3815. [DOI] [PubMed] [Google Scholar]
- [25].Xu MQ, Comb DG, Paulus H, Noren CJ, Shao Y, and Perler FB (1994) Protein splicing: an analysis of the branched intermediate and its resolution by succinimide formation, Embo J 13, 5517–5522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Xu MQ, and Perler FB (1996) The mechanism of protein splicing and its modulation by mutation, Embo J 15, 5146–5153. [PMC free article] [PubMed] [Google Scholar]
- [27].Callahan BP, Stanger M, and Belfort M (2013) A redox trap to augment the intein toolbox, Biotechnol Bioeng 110, 1565–1573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Callahan BP, Topilina NI, Stanger MJ, Van Roey P, and Belfort M (2011) Structure of catalytically competent intein caught in a redox trap with functional and evolutionary implications, Nat Struct Mol Biol 18, 630–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Chen W, Li L, Du Z, Liu J, Reitter JN, Mills KV, Linhardt RJ, and Wang C (2012) Intramolecular disulfide bond between catalytic cysteines in an intein precursor, J Am Chem Soc 134, 2500–2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Mills KV, Dorval DM, and Lewandowski KT (2005) Kinetic analysis of the individual steps of protein splicing for the Pyrococcus abyssi PolII intein, J Biol Chem 280, 2714–2720. [DOI] [PubMed] [Google Scholar]
- [31].Nicastri MC, Xega K, Li L, Xie J, Wang C, Linhardt RJ, Reitter JN, and Mills KV (2013) Internal disulfide bond acts as a switch for intein activity, Biochemistry 52, 5920–5927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Lennon CW, Stanger M, Banavali NK, and Belfort M (2018) Conditional Protein Splicing Switch in Hyperthermophiles through an Intein-Extein Partnership, MBio 9, e02304–e02317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Lennon CW, Stanger M, and Belfort M (2016) Protein splicing of a recombinase intein induced by ssDNA and DNA damage, Genes Dev 30, 2663–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Lennon CW, Stanger MJ, and Belfort M (2019) Mechanism of Single-Stranded DNA Activation of Recombinase Intein Splicing, Biochemistry 58, 3335–3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Topilina NI, Novikova O, Stanger M, Banavali NK, and Belfort M (2015) Post-translational environmental switch of RadA activity by extein-intein interactions in protein splicing, Nucleic Acids Res 43, 6631–6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Topilina NI, Green CM, Jayachandran P, Kelley DS, Stanger MJ, Piazza CL, Nayak S, and Belfort M (2015) SufB intein of Mycobacterium tuberculosis as a sensor for oxidative and nitrosative stresses, Proc Natl Acad Sci U S A 112, 10348–10353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Kelley DS, Lennon CW, Li Z, Miller MR, Banavali NK, Li H, and Belfort M (2018) Mycobacterial DnaB helicase intein as oxidative stress sensor, Nat Commun 9, 4363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Kerrigan AM, Powers TL, Dorval DM, Reitter JN, and Mills KV (2009) Protein splicing of the three Pyrococcus abyssi ribonucleotide reductase inteins, Biochem Biophys Res Commun 387, 153–157. [DOI] [PubMed] [Google Scholar]
- [39].Perler FB (2002) InBase: the Intein Database, Nucleic Acids Res 30, 383–384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Soucy SM, Fullmer MS, Papke RT, and Gogarten JP (2014) Inteins as indicators of gene flow in the halobacteria, Frontiers in microbiology 5, 299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Burns DG, Janssen PH, Itoh T, Kamekura M, Li Z, Jensen G, Rodriguez-Valera F, Bolhuis H, and Dyall-Smith ML (2007) Haloquadratum walsbyi gen. nov., sp. nov., the square haloarchaeon of Walsby, isolated from saltern crystallizers in Australia and Spain, Int J Syst Evol Microbiol 57, 387–392. [DOI] [PubMed] [Google Scholar]
- [42].Bolhuis H, Palm P, Wende A, Falb M, Rampp M, Rodriguez-Valera F, Pfeiffer F, and Oesterhelt D (2006) The genome of the square archaeon Haloquadratum walsbyi: life at the limits of water activity, BMC Genomics 7, 169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Ciragan A, Aranko AS, Tascon I, and Iwai H (2016) Salt-inducible Protein Splicing in cis and trans by Inteins from Extremely Halophilic Archaea as a Novel Protein-Engineering Tool, J Mol Biol 428, 4573–4588. [DOI] [PubMed] [Google Scholar]
- [44].Reitter JN, Cousin CE, Nicastri MC, Jaramillo MV, and Mills KV (2016) Salt-Dependent Conditional Protein Splicing of an Intein from Halobacterium salinarum, Biochemistry 55, 1279–1282. [DOI] [PubMed] [Google Scholar]
- [45].Robinzon S, Cawood AR, Ruiz MA, Gophna U, Altman-Price N, and Mills KV (2020) Protein Splicing Activity of the Haloferax volcanii PolB-c Intein Is Sensitive to Homing Endonuclease Domain Mutations, Biochemistry 59, 3359–3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding, Anal Biochem 72, 248–254. [DOI] [PubMed] [Google Scholar]
- [47].Abramoff MD, Magalhaes PJ, and Ram SJ (2004) Image Processing with ImageJ, Biophotonics International 11, 36–42. [Google Scholar]
- [48].Royer CA (2006) Probing protein folding and conformational transitions with fluorescence, Chem Rev 106, 1769–1784. [DOI] [PubMed] [Google Scholar]
- [49].Ghosh I, Sun L, and Xu MQ (2001) Zinc inhibition of protein trans-splicing and identification of regions essential for splicing and association of a split intein*, J Biol Chem 276, 24051–24058. [DOI] [PubMed] [Google Scholar]
- [50].Mills KV, and Paulus H (2001) Reversible inhibition of protein splicing by zinc ion, J Biol Chem 276, 10832–10838. [DOI] [PubMed] [Google Scholar]
- [51].Nichols NM, Benner JS, Martin DD, and Evans TC Jr. (2003) Zinc ion effects on individual Ssp DnaE intein splicing steps: regulating pathway progression, Biochemistry 42, 5301–5311. [DOI] [PubMed] [Google Scholar]
- [52].Sun P, Ye S, Ferrandon S, Evans TC, Xu MQ, and Rao Z (2005) Crystal structures of an intein from the split dnaE gene of Synechocystis sp. PCC6803 reveal the catalytic model without the penultimate histidine and the mechanism of zinc ion inhibition of protein splicing, J Mol Biol 353, 1093–1105. [DOI] [PubMed] [Google Scholar]
- [53].Iakoucheva LM, Kimzey AL, Masselon CD, Smith RD, Dunker AK, and Ackerman EJ (2001) Aberrant mobility phenomena of the DNA repair protein XPA, Protein Sci 10, 1353–1362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Reed CJ, Bushnell S, and Evilia C (2014) Circular dichroism and fluorescence spectroscopy of cysteinyl-tRNA synthetase from Halobacterium salinarum ssp. NRC-1 demonstrates that group I cations are particularly effective in providing structure and stability to this halophilic protein, PLoS One 9, e89452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rice P, Longden I, and Bleasby A (2000) EMBOSS: the European Molecular Biology Open Software Suite, Trends Genet 16, 276–277. [DOI] [PubMed] [Google Scholar]
- [56].Bjellqvist B, Hughes GJ, Pasquali C, Paquet N, Ravier F, Sanchez JC, Frutiger S, and Hochstrasser D (1993) The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences, Electrophoresis 14, 1023–1031. [DOI] [PubMed] [Google Scholar]
- [57].Lennon CW, Wahl D, Goetz JR, and Joel Weinberger II J (2021) Reactive chlorine species reversibly inhibit DnaB protein splicing in mycobacteria., Microbiology spectrum 9, e0030121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Woods D, Vangaveti S, Egbanum I, Sweeney AM, Li Z, Bacot-Davis V, LeSassier DS, Stanger M, Hardison GE, Li H, Belfort M, and Lennon CW (2020) Conditional DnaB Protein Splicing Is Reversibly Inhibited by Zinc in Mycobacteria, mBio 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Tori K, Dassa B, Johnson MA, Southworth MW, Brace LE, Ishino Y, Pietrokovski S, and Perler FB (2010) Splicing of the mycobacteriophage Bethlehem DnaB intein: identification of a new mechanistic class of inteins that contain an obligate block F nucleophile, J Biol Chem 285, 2515–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Tori K, and Perler FB (2011) Expanding the definition of class 3 inteins and their proposed phage origin, J Bacteriol 193, 2035–2041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zhou P, Lugovskoy AA, and Wagner G (2001) A solubility-enhancement tag (SET) for NMR studies of poorly behaving proteins, J Biomol NMR 20, 11–14. [DOI] [PubMed] [Google Scholar]
- [62].Reed CJ, Lewis H, Trejo E, Winston V, and Evilia C (2013) Protein adaptations in archaeal extremophiles, Archaea 2013, 373275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].DasSarma S, and DasSarma P (2015) Halophiles and their enzymes: negativity put to good use, Curr Opin Microbiol 25, 120–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Karan R, Capes MD, and Dassarma S (2012) Function and biotechnology of extremophilic enzymes in low water activity, Aquat Biosyst 8, 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Zhang G, Huihua G, and Yi L (2013) Stability of halophilic proteins: from dipeptide attributes to discrimination classifier, Int J Biol Macromol 53, 1–6. [DOI] [PubMed] [Google Scholar]
- [66].Tadeo X, Lopez-Mendez B, Trigueros T, Lain A, Castano D, and Millet O (2009) Structural basis for the aminoacid composition of proteins from halophilic archea, PLoS Biol 7, e1000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Burns DG, Camakaris HM, Janssen PH, and Dyall-Smith ML (2004) Cultivation of Walsby’s square haloarchaeon, FEMS Microbiol Lett 238, 469–473. [DOI] [PubMed] [Google Scholar]
- [68].Ventosa A, de la Haba RR, Sanchez-Porro C, and Papke RT (2015) Microbial diversity of hypersaline environments: a metagenomic approach, Curr Opin Microbiol 25, 80–87. [DOI] [PubMed] [Google Scholar]
- [69].Viver T, Conrad RE, Orellana LH, Urdiain M, Gonzalez-Pastor JE, Hatt JK, Amann R, Anton J, Konstantinidis KT, and Rossello-Mora R (2021) Distinct ecotypes within a natural haloarchaeal population enable adaptation to changing environmental conditions without causing population sweeps, Isme J 15, 1178–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Podell S, Emerson JB, Jones CM, Ugalde JA, Welch S, Heidelberg KB, Banfield JF, and Allen EE (2014) Seasonal fluctuations in ionic concentrations drive microbial succession in a hypersaline lake community, Isme J 8, 979–990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Barry ER, and Bell SD (2006) DNA replication in the archaea, Microbiol Mol Biol Rev 70, 876–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].McGeoch AT, Trakselis MA, Laskey RA, and Bell SD (2005) Organization of the archaeal MCM complex on DNA and implications for the helicase mechanism, Nat Struct Mol Biol 12, 756–762. [DOI] [PubMed] [Google Scholar]
- [73].Moreau MJ, McGeoch AT, Lowe AR, Itzhaki LS, and Bell SD (2007) ATPase site architecture and helicase mechanism of an archaeal MCM, Mol Cell 28, 304–314. [DOI] [PubMed] [Google Scholar]
- [74].Altschul SF, Gish W, Miller W, Myers EW, and Lipman DJ (1990) Basic local alignment search tool, J Mol Biol 215, 403–410. [DOI] [PubMed] [Google Scholar]
- [75].Bochman ML, and Schwacha A (2009) The Mcm complex: unwinding the mechanism of a replicative helicase, Microbiol Mol Biol Rev 73, 652–683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Brewster AS, Wang G, Yu X, Greenleaf WB, Carazo JM, Tjajadi M, Klein MG, and Chen XS (2008) Crystal structure of a near-full-length archaeal MCM: functional insights for an AAA+ hexameric helicase, Proc Natl Acad Sci U S A 105, 20191–20196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Slaymaker IM, Fu Y, Toso DB, Ranatunga N, Brewster A, Forsburg SL, Zhou ZH, and Chen XS (2013) Mini-chromosome maintenance complexes form a filament to remodel DNA structure and topology, Nucleic Acids Res 41, 3446–3456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Cheriyan M, and Perler FB (2009) Protein splicing: A versatile tool for drug discovery, Adv Drug Deliv Rev 61, 899–907. [DOI] [PubMed] [Google Scholar]
- [79].Nanda A, Nasker SS, Mehra A, Panda S, and Nayak S (2020) Inteins in Science: Evolution to Application, Microorganisms 8, 1–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Topilina NI, and Mills KV (2014) Recent advances in in vivo applications of intein-mediated protein splicing, Mobile DNA 5, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


