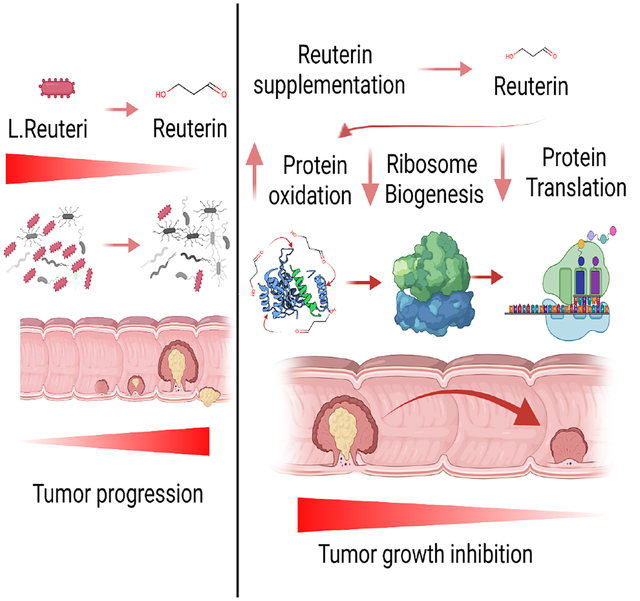
Summary
Microbial dysbiosis is a colorectal cancer (CRC) hallmark and contributes to inflammation, tumor growth, and therapy response. Gut microbes signal via metabolites, but how the metabolites impact CRC is largely unknown. We interrogated fecal metabolites associated with mouse models of colon tumorigenesis with varying mutational load. We find that microbial metabolites from healthy mice or humans are growth-repressive, and this response is attenuated in mice and patients with CRC. Microbial profiling reveals that Lactobacillus reuteri and its metabolite, reuterin are downregulated in mouse and human CRC. Reuterin alters redox balance, and reduces proliferation and survival in colon cancer cells. Reuterin induces selective protein oxidation and inhibits ribosomal biogenesis and protein translation. Exogenous Lactobacillus reuteri restricts colon tumor growth, increases tumor reactive oxygen species, and decreases protein translation in vivo. Our findings indicate that a healthy microbiome and specifically, Lactobacillus reuteri, is protective against CRC through microbial metabolite exchange.
Keywords: Microbiome, Reuterin, metabolites, protein oxidation, Lactobacillus reuteri, colorectal cancer
Graphical Abstract
eTOC Blurb:
Colon cancer has a significantly altered microbiome that promotes tumor growth. Bell et al. identified a decrease in anti-tumor bacteria Lactobacillus reuteri in colon cancer. Lactobacillus reuteri inhibits colorectal cancer by inducing oxidative stress and inhibiting protein translation. Recolonization with Lactobacillus reuteri increases survival in multiple colon tumor models.
Introduction
Colorectal cancer (CRC) is the third most common cancer and the fourth most significant cause of cancer mortality (Arnold et al., 2017). In addition to inherited mutations in a subset of patients, a combination of lifestyle, diet, inflammation, and microbiome alteration influence the development of CRC (Thanikachalam and Khan, 2019). Outcomes for CRC patients are often favorable if regional or distant metastases are not present. However, effective treatments for patients with metastatic disease are lacking, and most CRCs are not responsive to immunotherapy (Le et al., 2018; Overman et al., 2018)
Alterations of the intestinal microbiome are common in CRC patients. Germ-free or broad-spectrum antibiotic mouse models have been utilized to understand the role of commensal bacterial communities in CRC (Leystra and Clapper, 2019). The data obtained from these studies is not consistent, likely due to differences in the models employed and variations in the intestinal microbiome among different colonies. However, the vast majority of these studies indicate that commensal bacteria promote tumorigenesis (Leystra and Clapper, 2019). Germ free mice have enhanced colon tumorigenesis in a colitis-associated colon cancer model (Zhan et. al, 2013). Cumulatively, the data suggests that microbial species can either promote or inhibit colon tumor growth depending on the context. The complex role of the intestinal microbiome in colon tumorigenesis is also highlighted by work demonstrating that microbial-epithelial interactions maintain gut barrier function, sustain metabolism, and regulate the systemic and mucosal immune response (Kelly et al., 2015; Song et al., 2020). Disturbances in the resident microbial communities during the progression of CRC may lead to pathological dysbiosis which then promotes tumor growth and survival. Recent metagenomic studies have identified stage-specific shifts within the microbiota of patients with polyploid adenomas and intramucosal carcinomas (Yachida et al., 2019). The tumor microbiota is enriched for pathogenic bacteria Fusobacteria, Bacterioides fragilis and Prevotella (Pleguezuelos-Manzano et al., 2020; Tilg et al., 2018). Recent studies have advanced insights into the mechanisms by which pathogenic bacteria promote tumor growth and survival, such as by inducing inflammation, generating biofilms, and secreting genotoxins (Dejea et al., 2020, Pleguezuelos-Manzano et al., 2020, Tomkovich et al., 2019, Nougayrede J et al., 2021). However, few studies have reported on anti-tumor properties of specific host microbes.
Commensal microbes use host and dietary factors to generate metabolites via cell autonomous biosynthetic pathways. The best studied commensal microbial factors are short-chain fatty acids (SCFA), such as butyrate, which decrease growth of normal intestinal stem cells and a battery of cancer-derived cell lines (Kaiko et al., 2016). Heterocellular metabolic interactions between microbiota, tumor epithelium, and stromal cells is an emerging concept, and it is not clear how the microbiome alters tumor growth via metabolite exchange. In this work, we found that fecal metabolites from heathy humans and mice potently inhibited colon cancer growth. Mouse colon cancer mouse models with a higher mutational load were linked to dysbiosis and microbial metabolites that were growth permissive in CRC. We identified a commensal microbial species Lactobacillus reuteri (L. reuteri) and its major metabolite, reuterin, that potently reduced colon cancer progression. We found that the microbiome of wild-type mice altered the redox state of colon cancer cells and inhibited ribosomal biogenesis. Together, our findings identify that a healthy microbiome and specifically, Lactobacillus reuteri, is a potential probiotic approach to treat CRC.
Results
Fecal metabolites from healthy mice inhibit the growth of CRC cells.
To address the potential contributions of microbial metabolites in the development of CRC, we used genetically engineered mouse models of colon tumorigenesis. We used the tamoxifen-regulated CDX2p-CreERT2 transgene to target both copies of the murine Apc allele (SingleMut), which leads to the development of non-invasive adenomas in the distal colon and rectum(Hinoi et al., 2007). We generated the DoubleMut, where both Apc and Trp53 alleles are concurrently inactivated in colon epithelium, and the TripleMut combining the APC/p53 knockout with a Kras G12D knock-in allele (Figure 1A). Ten days following a single treatment of tamoxifen to initiate tumorigenesis, fecal microbial metabolites were isolated and used for the treatment in HCT116, SW480, DLD1, and RKO CRC cell lines (Figure 1B, Figure S1A). Fecal metabolites from wild-type mice were robustly inhibitory to CRC cell line proliferation. Fecal metabolites isolated from SingleMut and DoubleMut reduced proliferation to a lesser extent, whereas metabolites from the TripleMut were not growth suppressive. Fecal metabolites from wild-type mice induced cell death and decreased colony formation in all four cell lines (Figure 1C and D, Figure S1B and C). Similarly, normal human fecal metabolites, but not, metabolites isolated from CRC patients with active disease repressed the growth of CRC cell lines (Figure 1E). Microbial metabolites did not repress a non-cancerous human intestinal cell line NCM460 (Figure 1E). This fecal metabolite-based repression of the CRC cell lines was microbial in origin (Figure 1F).
Figure 1: Fecal metabolites from wild-type mice suppress colorectal cancer cell growth in vitro.
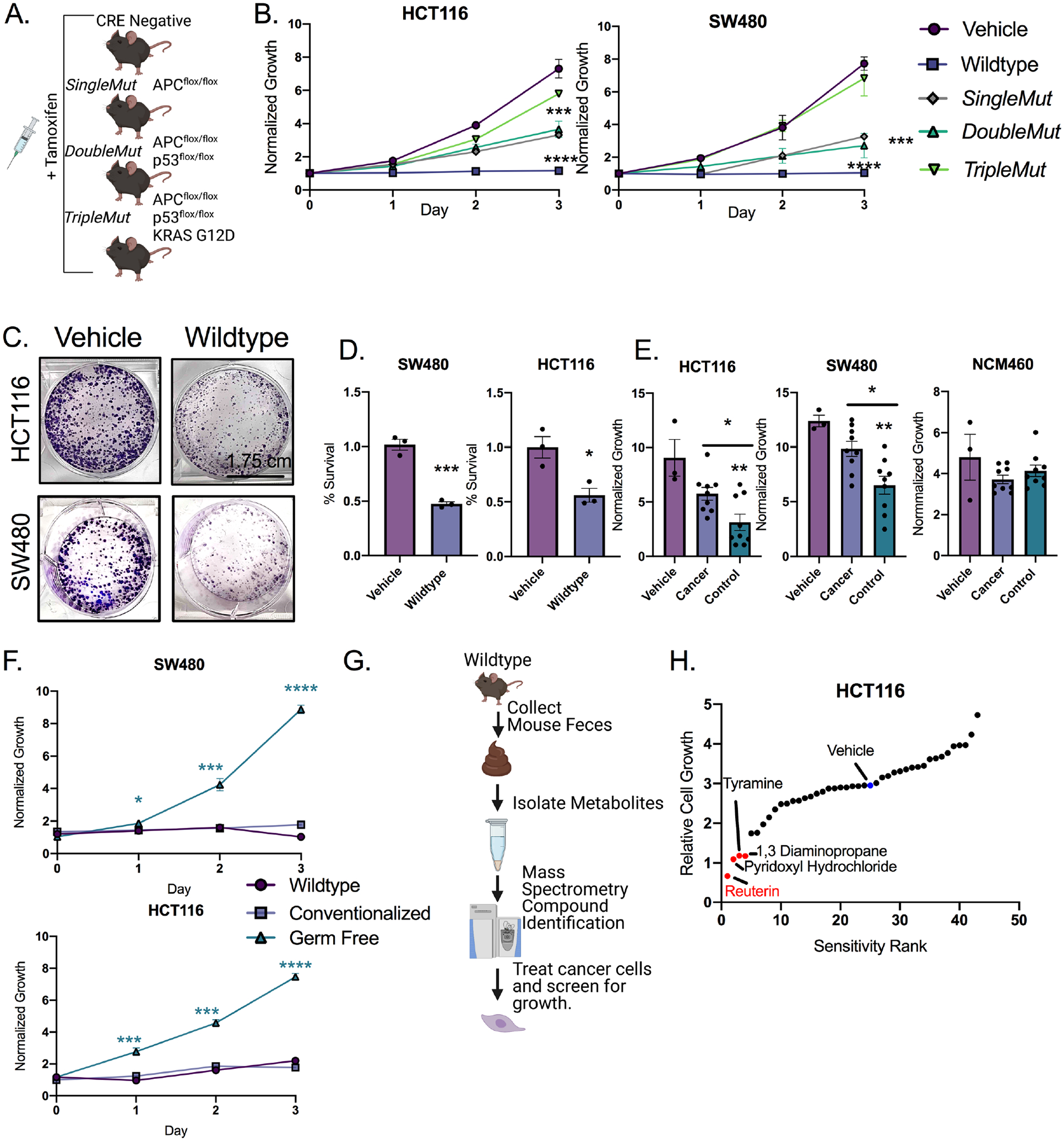
A) Mouse models and treatment strategy employed. B) Dried, organic, metabolite pellets were resuspended in DMSO and treated at 100X concentration and growth was assessed by live cell imaging. (n=9). Error bars are standard error of the mean +/− the mean C) Colony forming assay of cells treated with 100X wild-type fecal metabolites for 14 days. (n=3). D) LDH assay of cells treated with 100X fecal metabolites at 24 hours. (n=3). Mean +/− the standard error of the mean. E) Growth assays from organic fecal metabolites from age matched controls and cancer patients. Each human sample was assayed in triplicate. Mean +/− the standard error of the mean. F) Growth assays from fecal metabolites from wild-type, germ-free mice, and recolonized germfree. (n=3 per group). G) Schematic representation of the screen to identify inhibitory compounds. H) Sensitivity rank graph of cell growth after 72 hours of treatment at 1 mM for each indicated compound. (n=3). Statistical significance was measured by a one-way ANOVA (Panels B, E, and F) or T-test (Panel D) *p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. Data is presented as mean +/− the standard error of the mean. All experiments were performed in triplicates at least three times.
Untargeted metabolomics identified the most abundant microbial metabolites in wild-type mice. A small molecule library was generated from microbially derived metabolites exclusively (Figure 1G) (Wikoff et al., 2009). The fifty most abundant metabolites were screened based on cell proliferation. We focused on the most inhibitory compounds. A secondary growth screen in a second cell line was performed. This led to the identification of 1,3 diaminopropane and reuterin, both of which are present in high abundance in normal colon (Das et al., 2020)(Figure 1H). Tyramine and pyridoxyl hydrochloride inhibited CRC growth to a lesser extent (Figure 1H). The initial screen used a single dose of 1 mM, the second screen a dose of 500μm. Reuterin was confirmed as the most inhibitory compound (Figure S1D).
Reuterin is actively reduced in human and mouse colon cancer.
We performed 16s rRNA sequencing on the feces of wild-type littermates (CRE negative), DoubleMut and TripleMut mice to determine how microbial communities are altered as tumors progress. The DoubleMut and TripleMut have comparable proliferative and dysplastic indexes at early time points. There were no changes in microbiome between the uninduced mice, or cage-specific stratification (Figure S2A and B). Principal coordinate analysis (PCoA) confirmed that CRE negative mice, DoubleMut, and TripleMut all had significantly different microbiomes by Analysis of MOlecular Variance (AMOVA), and clustered separately on both PCoA axes (Figure 2A). Interestingly, the PCoA plot identified Lactobacillus as the top driver leading to differences between the groups (Figure 2B and Supplemental Table 1). This was followed by Lachnospiracaea and Muribaculaceae, both of which are associated with altered risk of inflammation and colon cancer (Flemer et al., 2018; Lang et al., 2020)
Figure 2: L. reuteri abundance and host metablotes are altered in mouse colon tumor models and human colorectal cancer.
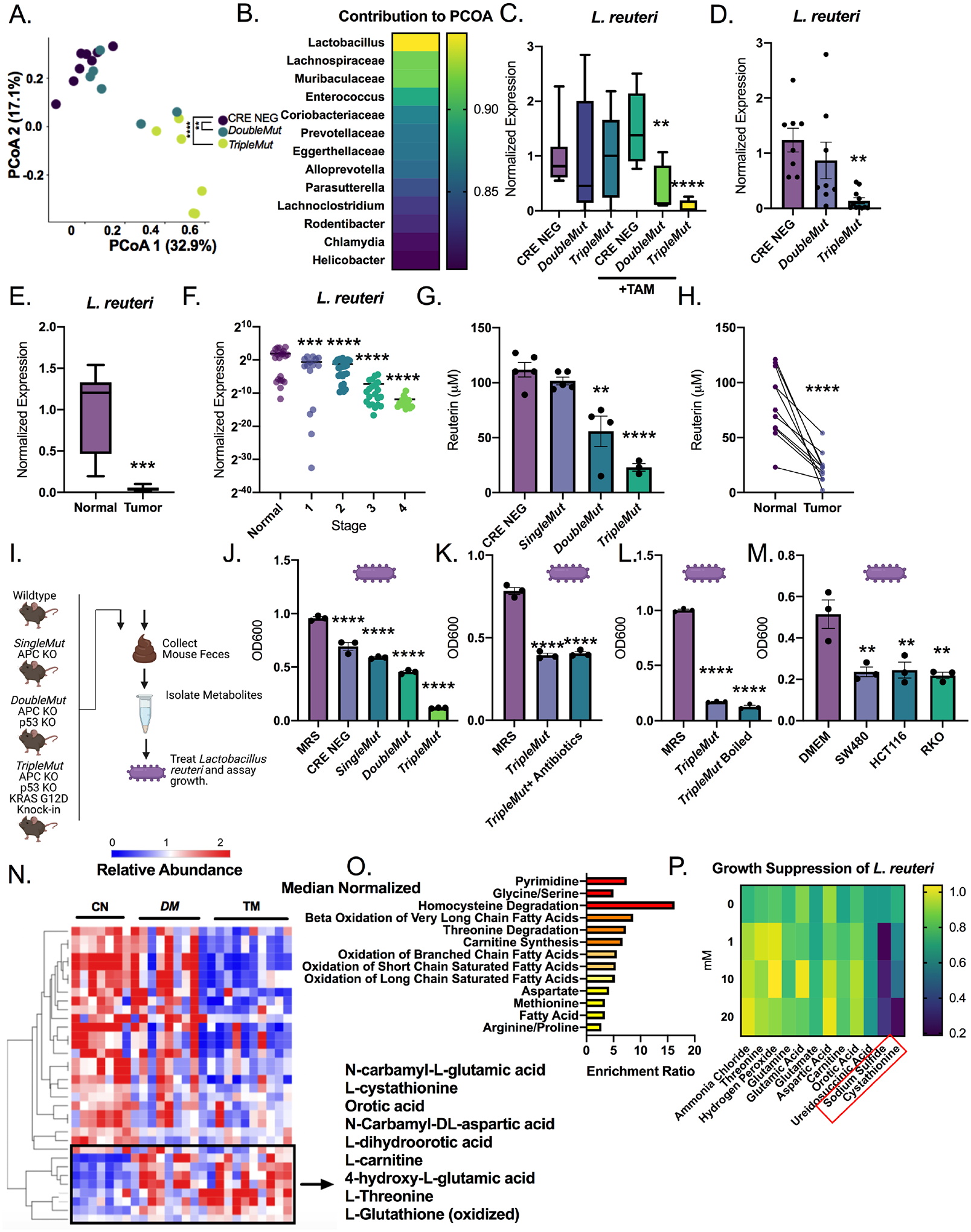
A) PCoA plots of 16S rRNA sequencing of fecal DNA from the induced (50 mg/kg tamoxifen) mice(n=6–8). Significance is compared to the Cre-negative control mice. B) Length of the vector of the PCoA plot by the indicated species. (n=6–8). C) qPCR of L. reuteri in fecal DNA of indicated mouse model induced with 50 mg/kg tamoxifen. (n=6–8). D) qPCR of indicated mouse strain from colon mucosal scrapes 14 days after induction (same animals as used for tissue 16s sequencing). (n=6–8). E) qPCR of human colon tumor and normal samples. (n=7–8). F) qPCR for L. reuteri of human colon tumor tissue isolated from patients with different stages of cancer. (n=96). One way ANOVA compared to normal human colon tissue. G) Reuterin quantification from colon samples by mass spectrometry. (n=3–5). H) Reuterin quantification from human whole colon normal and tumor paired sections. (n=9). I) Schematic representation of studies of metabolite extracts and treatment of L. reuteri. J) Treatment of L. reuteri with fecal extracts isolated from indicated mice for 24 hours at a 100X concentration. Control is grown in MRS broth. (n=3). K) Treatment of L. reuteri with fecal extracts isolated from TripleMut mice treated with a broad-spectrum antibiotic. (n=3). L) Treatment of L. reuteri with fecal extracts isolated from TripleMut mice which were boiled for 10 minutes. (n=3). M) Cells were grown in DMEM and 10% FBS for 48 hours, then media was supplemented with 20% MRS broth before incubation with L. reuteri. (n=3). N) Unsupervised clustering of metabolite extracts from colon scrapes from induced indicated mouse models (n= 7–8). Metabolites significantly different in the TripleMut compared to the CRE negative are shown, and metabolites that are upregulated in the TripleMut are boxed. O) Metaboanalyst pathway analysis of significantly upregulated metabolites listed by p value. P) L. reuteri incubated with indicated metabolite for 24 hours, then growth assessed by reading at 600 nm. Statistical significance measured by one way ANOVA (A, C, F, F, G, J, K, L, M) or t-test (E, H). Statistical significance was measured by a one-way ANOVA or T-test compared to vehicle control at Day 3, ** p < 0.01, *** p < 0.001, **** p < 0.0001. Data is presented as mean +/− the standard error of the mean. Microbial 16 sequencing experiments, metabolomics, and human tissue analysis were performed once, all other assays were performed in triplicates at least three times.
Reuterin, or 3-hydroxypropionaldehyde, was identified in our microbial metabolite growth screen. Reuterin has predominantly been studied as an antimicrobial(Asare et al., 2020). Reuterin is primarily produced by L. reuteri, a natural colonizer of the human gut, and is an intermediate in the metabolism of glycerol to 1,3-propanediol(Martín-Cabrejas et al., 2017; Zhang et al., 2020). L. reuteri was significantly decreased in the 16S-seq samples in the DoubleMut and TripleMut fecal samples (Figure 2C). Lactobacillus genus trended downward in the induced DoubleMut and TripleMut mouse models (Figure S2C). Other Lactobacillus species, such as L. acidophillus, were also downregulated (Figure S2D, E). L. reuteri was found to be robustly decreased in an additional independent cohort of TripleMut mice (Figure S2F). We next performed 16S rRNA sequencing on tumor mucosa samples. The genus Lactobacillus was not the main contributor to the PCoA vectors in tumors (Figure S2G and S2H). However, L. reuteri is strongly reduced in the TripleMut tumor mucosa (Figure 2D). No other Lactobacillus species assessed was significantly reduced in the tumor microbiome (Figure S2I). This suggests that although Lactobacillus as a genus may be globally downregulated in the feces, L. reuteri is specifically downregulated in the tumor mucosa.
We first mined publicly available datasets to assess alterations in L. reuteri between the CRC and normal human microbiome. In a metanalysis of eight colon intestinal microbiome studies, Lactobacillus was significantly reduced in both early and late-stage CRC (Figure S2J) (Wirbel, et. al, 2019). Sequenced luminal contents and tumor sections demonstrated a decrease in Lactobacillus in carcinomas (Figure S2K). Lastly, 16S rRNA sequencing on early and invasive colorectal cancers observed lower Lactobacillus in invasive compared to early cancer (Figure S2L). We next delved into a small cohort of CRC tissue samples with adjacent normal tissue to quantify L. reuteri (Figure 2E). We saw that L. reuteri levels were significantly reduced in the tumor samples. We next used a larger cohort of patient tissue samples from age matched normal controls, and different stages of CRC. L. reuteri was significantly reduced in all stages of colorectal cancer compared to normal controls in this independent cohort (Figure 2F).
We also used an inflammation-based carcinogenesis model, AOM-DSS to induce tumors. L. reuteri was reduced in tumors compared to the normal tissue, demonstrating that the reduction of L. reuteri in colon tumors was conserved (Figure S2M). Consistent with the decrease of L. reuteri in tumors, there was a graded decrease of reuterin in the sporadic and AOM-DSS colon tumor mouse models (Figure 2G and Figure S2N). Reuterin levels were also decreased in human tumor tissue compared to adjacent normal tissue (Figure 2H). This suggests that the cancerous epithelium locally effects Lactobacillus reuteri and thus reuterin levels. Diaminopropane was not altered, demonstrating that the decrease in reuterin is specific (Figure S2O and S2P, S2Q).
Interestingly, the TripleMut mouse fecal metabolites robustly inhibited L. reuteri growth in vitro (Figure 2I and J). In contrast, the CRE NEG extracts prohibited the growth of CRC cells, but have significantly less effect on the growth of L. reuteri. This effect is unlikely to be only a result of tumor burden, as the mice have the same percentage of the colon transformed. We verified that the TripleMut fecal extracts are not inhibitory to either E. coli or a complex compendium of fecal bacteria (Figure S2R, Figure S2S). We sought to identify whether the potential source of the inhibitory compound was microbial or epithelial. We treated TripleMut mice with a cocktail of broad-spectrum antibiotics for one week prior to tamoxifen induction. We discontinued the antibiotics at induction and generated fecal extracts at an endpoint of day 10. We observed that the growth inhibition on L. reuteri was sustained (Figure 2K). Moreover, our data suggests that the growth inhibition on L. reuteri from TripleMut mice is metabolitederived, as the compound remains growth-inhibitory following heating. (Figure 2L). We investigated whether conditioned media from CRC cell lines inhibits L. reuteri (Figure 2M). Conditioned media from SW480, HCT116, and RKO inhibited L. reuteri growth. Boiling the media did not alter the growth repressive functions, suggesting that the inhibitory factor is a metabolite (Figure S2T). We developed an shRNA knockdown of KRAS in the mouse CRC cell line CT26 (Figure S2U). Decreasing KRAS did not reverse the repression of L. reuteri growth (Figure S2V).
Targeted metabolomics were performed on the DoubleMut, TripleMut and CRE negative mouse models. 2D unsupervised clustering showed clear differences between the mouse models (Figure 2N). We focused on metabolites that were upregulated in the TripleMut. We analyzed the enriched pathways for increased metabolites, and found that redox balance and homocysteine degradation were the most enriched (Figure 2O). We treated L. reuteri in vitro with increasing doses of these metabolites, and found that L. reuteri growth was repressed by the homocysteine degradative metabolites hydrogen sulfide and cystathionine (Figure 2P). Supplementation with the antioxidant glutathione ethyl ester did not alter L. reuteri growth, indicating that L. reuteri is not inhibited specifically by oxidative stress (Figure S2T). Together, the data suggests a bidirectional metabolite exchange between bacterial and host metabolites.
Reuterin is cytotoxic and growth inhibitory to colon cancer cells.
Reuterin inhibited the growth of HCT116, SW480, RKO and DLD1 CRC at a dose of 25μM (Figure 3A and S3A). The CRC, PDAC, lymphoma, ovarian and melanoma cell lines were significantly growth inhibited by reuterin (Figure 3B, S3B). Breast and cervical cancer cell lines were relatively resistant to reuterin. Complementary data showing growth inhibition was also obtained using colony forming assays (Figure 3C, Figure S3C). We confirmed reuterin-induced cell death with an LDH assay (Figure 3D and S3D). Cytotoxicity of reuterin in vitro was seen around 50μM, which is below the reuterin concentration of the normal colon (around 100μM). We hypothesized that growth suppressive effects of reuterin are preferentially affecting cancer cells. Enteroid cultures were generated from Cre-negative control mice or induced TripleMut mice ten days after Cre-mediated gene targeting in vivo. TripleMut enteroids and wildtype enteroids were incubated with 100μM of reuterin and cell death was assessed. Reuterin increased cell death in TripleMut colon epithelial cells, but not the control epithelial cells (Figure 3E). Moreover, we found that a panel of normal cell lines are significantly less sensitive to reuterin than colon cancer cell lines (Figure 3F). Wild-type mouse fecal metabolites inhibited CRC cell lines HCT116 and SW480 in a dose dependent manner, whereas no change in growth was observed in a non-cancerous human colon cell line NCM460 (Figure 3G).
Figure 3: Reuterin preferentially inhibits the growth of colorectal cancer cells over normal colon epithelial cells.
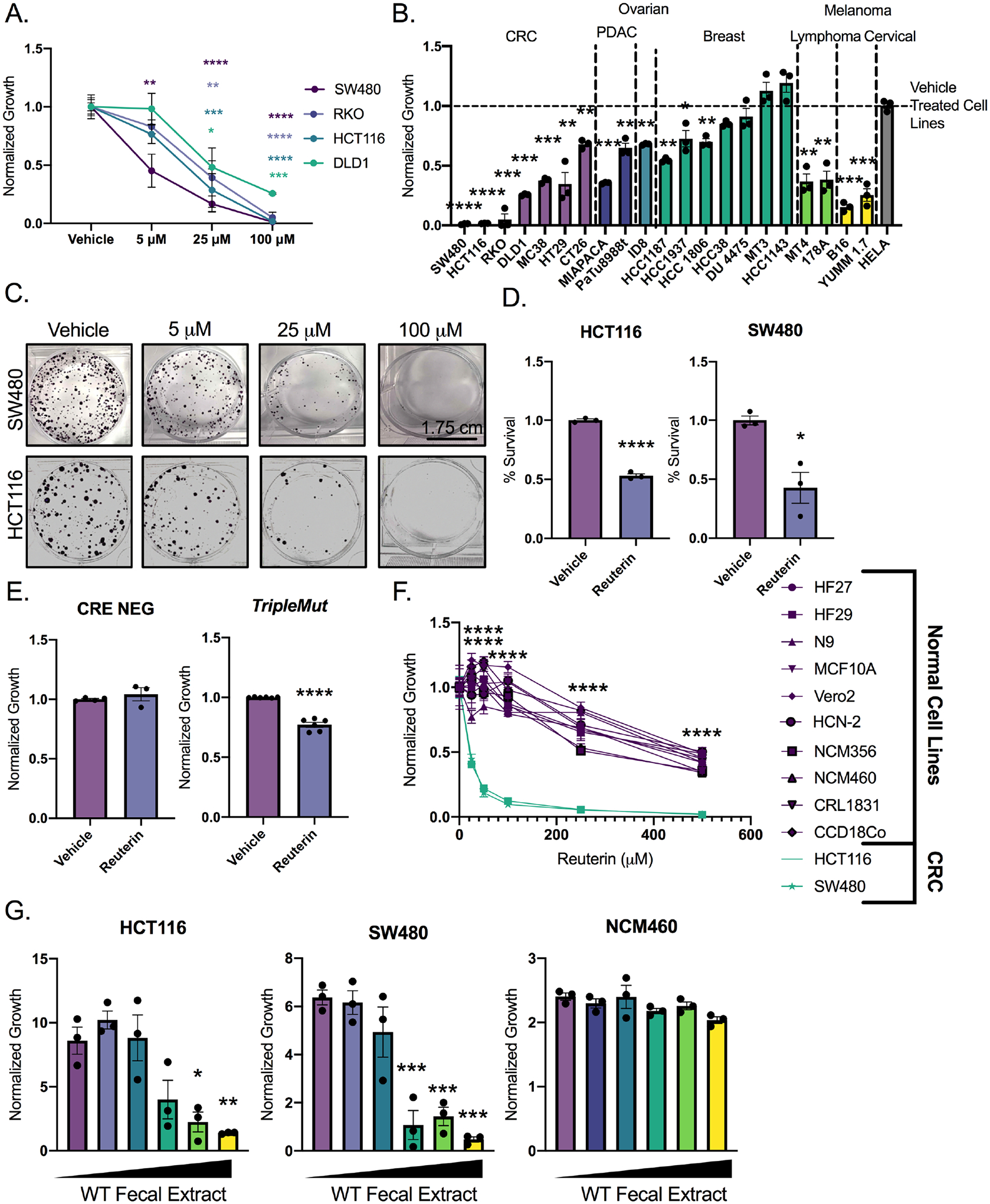
A) Dose curve of panel of cell lines treated reuterin for 72 hours. (n=3). B) Cell growth following 100μM reuterin treatment. (n=3). C) Colony forming assay of cells treated with indicated concentration of reuterin. (n=3, representative shown). D) LDH assay of cells treated with 100μM reuterin for 24 hours. (n=3). E) Epithelial cell death in indicated mouse models 14 days after induction. (n=3). F) Cell growth in normal and cancer cells. Purple: normal cell lines; green: colon cancer cell lines. Data was collected at 72 hours. (n=3). G) Dose curve of wild-type fecal extracts on indicated cell line. (n=3). Statistics were calculated with one-way ANOVA (A, B, F, and G) or t-test (D and E). *p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. Data is presented as mean +/− the standard error of the mean. All experiments were performed in triplicates at least three times with the exception of panel F which was performed in triplicate a single time.
We had previously found that reuterin altered the activity of hypoxia-inducible factor (HIF)2α (Das et al., 2020). HIF2α is an important transcription factor for CRC growth (Xue et. al, 2016). We generated a HIF2α-knockout HCT116 cell line (Figure S3E), but no change in sensitivity to reuterin was observed (Figure S3F). We have also previously shown that iron is central in potentiating electrophile-induced cell death (Singhal et al., 2021). We co-treated cells with reuterin and iron but observed no potentiation (Figure S3G). Taken together, these findings imply that reuterin is acting through a previously undescribed mechanism.
Reuterin induces oxidative stress in colorectal cancer cells.
In our efforts to generate molecular and mechanistic insights into the growth inhibitory effects of reuterin in intestinal cells, we used metabolomics, transcriptomics and proteomics. RNA-sequencing studies in an intestinal cell line treated with 100μM reuterin for 24 hours demonstrated definitive transcriptional changes (Figure 4A and Supplemental Table 2). Using PCA, gene expression in cells treated with reuterin were significantly altered (Figure S4A). We observed upregulation in genes essential for oxidative stress response, along with genes acting downstream of nuclear factor erythroid 2–related factor 2 (NRF2) (Figure 4B, Figure S4B). The most significantly enriched pathways include ribosomal subunit, translation initiation, and intramolecular transferase (Figure 4C). Targeted metabolomics identified glutathione metabolism as a significantly enriched pathway, with a 6-fold enrichment (Figure 4D and Supplemental Table 3). Subsequently, we created a network that integrates the transcriptomic and metabolomic data. This network shows enrichment of glutathione and glutamate metabolism, suggesting that oxidative stress is a potential mechanism (Figure S4C). We quantified how transcripts in the metabolic pathways of significantly altered metabolites were modulated. This approach also identified glutathione metabolism (Figure 4E). As a complementary approach, we quantified oxidized L-glutathione, a marker of the redox stress which confirmed that reuterin induced oxidative stress (Figure 4F). Subsequently, we observed that NAC inhibited the induction of NRF2-dependent oxidative stress genes (Figure S4D). These results provide detailed molecular confirmation that reuterin directly controls the redox balance of a cell.
Figure 4: Multi-omics approach reveals Reuterin induces oxidative stress.
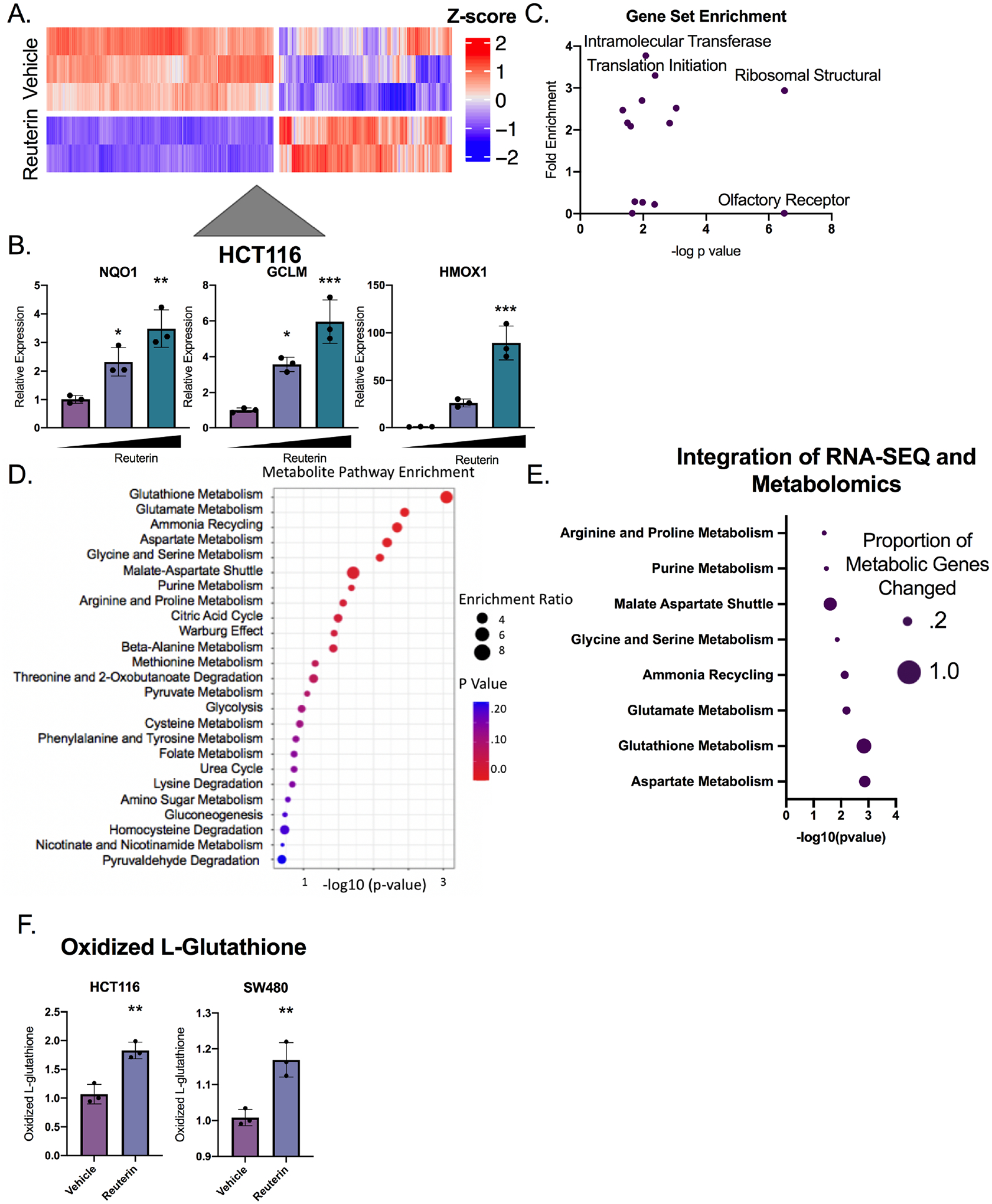
A) Heat map of altered genes treated with 100 μM reuterin for 24 hours. (n=2–3). B) qPCR of NRF2 target genes with vehicle control, 25μm and 100μm reuterin treatment for 24 hours. (n=3). C) KEGG pathway enrichment for genes differentially expressed between vehicle and reuterin treated cells. D) Metabolomics pathway enrichment of significantly altered metabolites in SW480 cells. E) Proportion of genes altered in the RNA-SEQ of metabolic pathway associated genes. The size of the dot represents the percentage of genes altered transcriptionally in the indicated metabolic pathway. The p value is the significance of the metabolic pathway enrichment. F) Quantification of oxidized L-glutathione. (n=3). Statistics were calculated with one-way ANOVA (B) or t-test (F). *p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. Data is presented as mean +/− the standard error of the mean. Metabolomics and RNA-SEQ experiments were performed a single time in triplicate, all other experiments were performed at least three times in triplicate.
Reuterin acts through Glutathione depletion.
To further address the role of reuterin in mediating oxidative stress, reactive oxygen species (ROS) were measured in SW480, HCT116, RKO and DLD1 cells. A dose-dependent increase in ROS was observed in all sensitive colon cancer cell lines studied following reuterin treatment (Figure 5A). We measured ROS in the reuterin-resistant cell line HeLa, and the normal colon, reuterin-resistant cell line NCM460. Reuterin did not induce ROS in HeLa or NCM460 cells (Figure 5A). Moreover, growth inhibition following reuterin treatment was completely rescued by NAC (Figure 5B–D, S5A). Our integrative multiomic approach identified glutathione as a key node in cellular response following reuterin treatment (Figure S4C). Directly increasing glutathione using supplementation of glutathione ethyl ester robustly protected against the cytotoxic effects of reuterin (Figure S5B). Buthionine sulfoxamine (BSO), a compound that inhibits the synthesis of glutathione, potentiated the growth inhibitory effects of reuterin. (Figure 5E, S5C) (Lee et al., 2019). We generated dose response matrices for reuterin and BSO and found that reuterin and BSO synergize (Supplemental Table 7). NAC rescued cancer cell growth inhibition by the fecal metabolites from wild-type mice (Figure 5F, S5D). This indicates that wild-type microbial metabolites inhibit colon cancer cell growth through oxidative stress similar to reuterin.
Figure 5: Reuterin induces oxidative stress in a GSH-dependent manner.
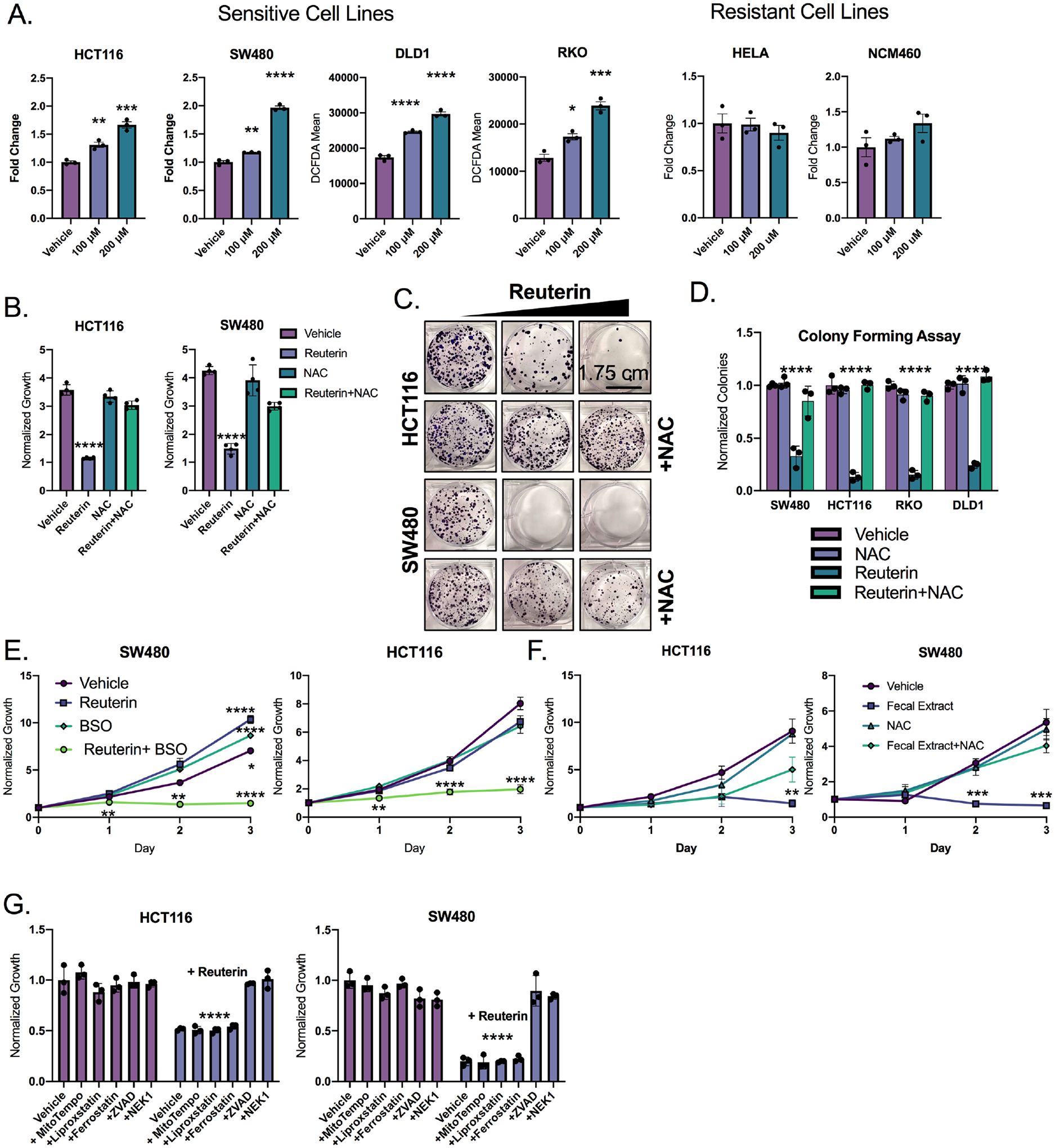
A) Cells were treated with indicated reuterin concentration for 24 hours, then stained with DCFDA ROS dye and analyzed by flow cytometry. Values are MFI. (n=3). B) Cells were pretreated with NAC for 24 hours, then treated with reuterin for 72 hours, then analyzed by live cell imaging. (n=3). C) Representative colony forming assay (CFA) at 14 days for cells pretreated with NAC for 12 hours. (n=3, representative shown). D) Quantification of CFA of cells treated as in panel C. (n=3). E) Cells were cotreated with 10μm Reuterin and 100μm BSO. (n=3). F) Cells were pretreated with NAC for 12 hours then treated with wild-type organic fecal extract. (n=3). G) Cells were pretreated with mitotempo (1 mM), liproxstatin (10μM), ferrostatin (10μM), ZVAD(50μM) or necrostatin-1 (50μM) then treated with 100μm reuterin for 3 days, growth was assessed via live cell imaging. (n=3). Statistics were calculated with one-way ANOVA (A, B, D, E,F, and G). *p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. Data is presented as mean +/− the standard error of the mean. All experiments were performed in triplicates at least three times.
Electrophilic compounds such as reuterin can target selective macromolecules for cellular damage. We measured lipid ROS using BODIPY 581/591 and observed no increase in lipid oxidation following treatment (Figure S5E). Moreover, liproxstatin and ferrostatin, two inhibitors of lipid ROS, completely failed to rescue reuterin cytotoxicity (Figure 5G) (Angeli et al., 2014; Gaschler et al., 2018). MitoTEMPO, a mitochondrial targeted superoxide dismutase, also did not alter cell growth following reuterin (Figure 5G). However, apoptotic inhibitor ZVAD rescued reuterin induced inhibition (Figure 5G).
We found that reuterin did not synergize with the double-strand break repair inhibitor olaparib (Figure S5F and Supplementary Table 7). Moreover, gamma-H2AX, a marker of DNA damage was not increased following reuterin treatment (Figure S5G). This suggests that reuterin is not functioning primarily through damaging nucleic acids. Interestingly, reuterin did synergize in cell lines with the chemotherapeutic 5-Fluorouracil, which is known to increase ROS (Figure S5H). Together, this work suggests that reuterin is inducing oxidative stress in a GSH-dependent manner without oxidizing DNA or lipids.
Reuterin induces protein oxidation and selectively inhibiting ribosomal biogenesis.
Our work suggests that reuterin is a highly selective electrophile that does not require HIF2α, high levels of iron or targets nucleic acid and lipids. Electrophiles can interact with cysteines, causing irreversible oxidation, loss of protein function and cell death (Filipovic et al., 2018). Sodium sulfide can prevent protein oxidation by binding to cysteines leading to protective and reversible persulfidation (Figure 6A) (Filipovic et al., 2018). Sodium sulfide protected cells from reuterin-induced growth inhibition (Figure 6B). High-throughput cysteine-activity based proteomics is a recently developed approach to identify which cysteines an electrophile interacts with (Kuljanin et al., 2021). Cells were treated with reuterin, dimethyl fumarate (DMF) or vehicle for 24 hours. DMF was used as an additional control since this electrophile induces cell death mechanistically distinct from reuterin (Singhal et al., 2021) (Figure 6C and Supplemental Table 4). Cysteine proteomics revealed that reuterin selectively bound to cysteine residues in multiple biological replicates. The data was plotted as a competition ratio, where a high competition ratio indicates a higher proportion of reuterin bound protein. Several protein hits were significant as assessed by having a competition ratio three times greater than the standard deviation (Figure 6D). Similar analysis was performed with DMF to identify unique targets (Figure S6A).
Figure 6: Reuterin induces protein oxidation and inhibits ribosomal biogenesis.
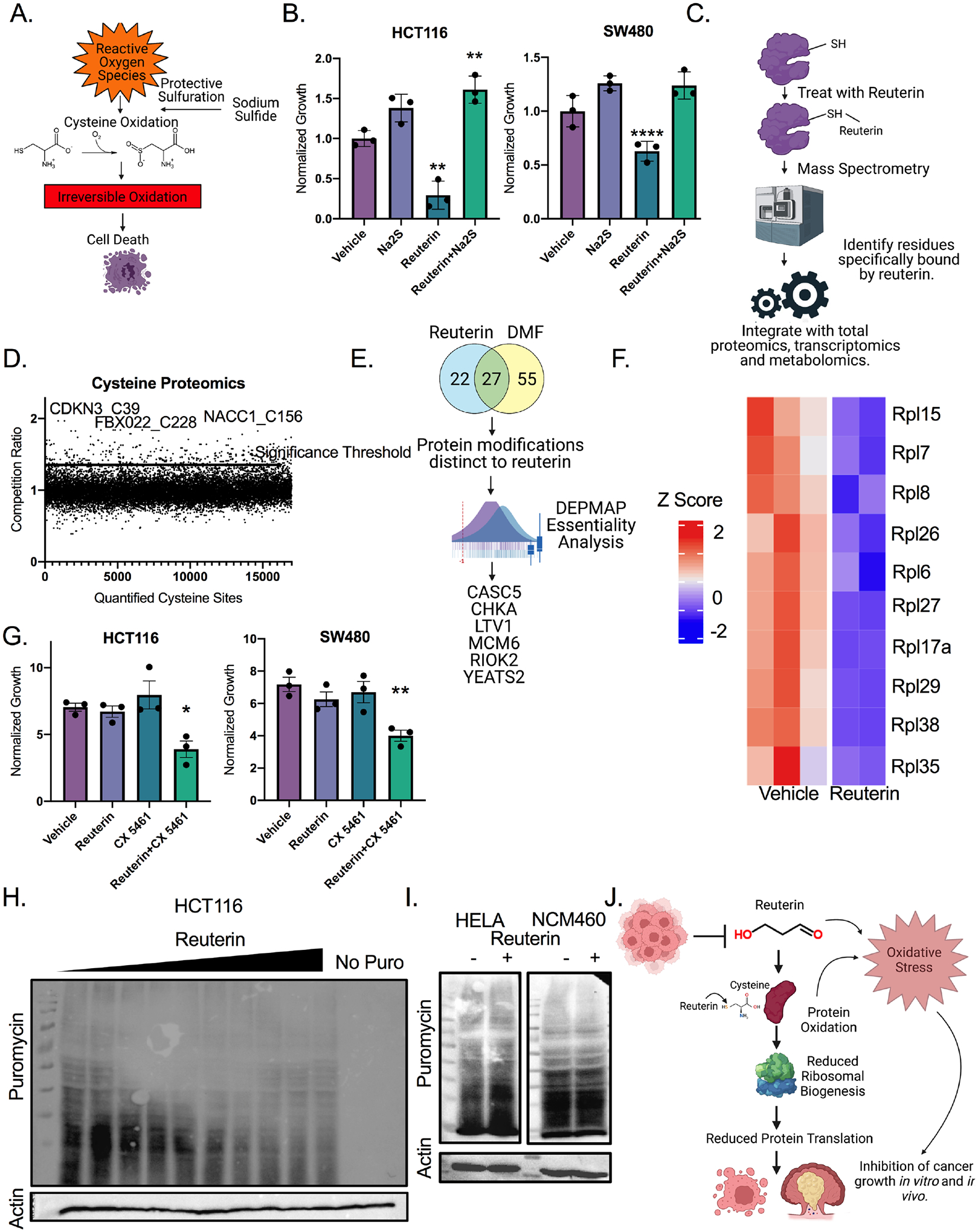
A) Schematic of protective persulfidation from protein oxidation. B). Cells were pretreated with 50μm reuterin for 12 hours, then media was washed out, and cells were treated with 200μm sodium sulfide for 24 hours before LDH assay. (n=3). C) Schematic for integrated proteomics, transcriptomics and metabolomics. D) Competition ratio of average of three separate samples for cysteine sites bound by reuterin. (n=3, repeated twice). E) Analysis of cysteine proteomics data with DepMap. F) Heat map of YEATS2 target genes in cells treated with reuterin. (n=5). G) Growth assays of cells co-treated with 2μm of CX5461 and 10μm of reuterin for 72 hours. (n=3). H) Puromycin western blot of cells treated with doses increasing from 1 to 150μm reuterin for 24 hours. I) Puromycin western blot of cells treated with 100μm reuterin for 24 hours. J) Schematic of proposed mechanism of action for reuterin. Statistics were calculated with one-way ANOVA (Panels B and G). *p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. Data is presented as mean +/− the standard error of the mean. Cysteine proteomics was performed in triplicate two times, RNA-SEQ experiment was performed in triplicate once. All other experiments were performed in triplicates at least three times.
Consistent with different mechanisms of action we found that reuterin had a significantly different cysteine proteomics profile than DMF. Pathway analysis showed enrichment for protein binding and catalysis (Figure S6B). The reuterin and DMF proteomic profiles were distinct (Figure S6C). Whole cell lysate proteomics also revealed significant differences in proteins induced by reuterin and DMF (Figure S6D and Supplemental Table 5). We utilized a network approach to integrate the proteomics with the metabolomics and transcriptomics data. We used known links from genes to metabolites to score each of our significantly changed metabolites by the number of linked genes differentially expressed. The metabolic pathways critical in NAD was the most significant hit, indicating an important role of reuterin in redox balance (Figure S6E). The network approach further implicated oxidative stress as primary mechanism of action for reuterin (Figure S6F).
We next assessed the proteins bound by reuterin in a dose-dependent manner. Proteins that were unique to reuterin were further filtered for essentiality using the Cancer Dependency Map project (Tsherniak et al., 2017) (Figure 6E). We identified 6 proteins. 3 of these 6 proteins, Ltv1 ribosome biogenesis factor (LTV1), Rio Kinase 2 (RIOK2), and YEATS domain containing protein 2 (YEATS2) are involved with ribosomal biogenesis(Cerezo et al., 2021; Collins et al., 2018; Mi et al., 2017). LTV1, CASC5, RIOK2, CHKA, MCM6 and YEATS2 were not downregulated transcriptionally in reuterin-treated HCT116 cell line (Figure S6G). This combined with our previous enrichment for ribosomal structural proteins in the RNA-sequencing analysis (Figure 4B), led us to hypothesize that inhibiting ribosomal assembly is an important cytotoxic pathway of reuterin. YEATS2 regulates a large subset of ribosomal protein-encoding genes(Mi et al., 2017). YEATS2 target genes were decreased after reuterin treatment (Figure 6F) (Mi et al., 2017). We utilized CX-5461, an established inhibitor of the initiation of ribosome biogenesis (Mars et al., 2020). We found that low dose CX-5461 and reuterin synergized but CX-5461 and DMF did not (Figure 6G and S6H, Supplemental Table 7). We utilized a puromycin incorporation assay (SUnSET) to quantify active protein translation in reuterin treated cell lines. Reuterin reduced protein translation in HCT116 and SW480 cell lines, but not reuterin resistant HeLa and NCM460 (Figure 6I and Figure S6H and Figure S6I). These results demonstrate that reuterin induces protein oxidation, and that this oxidation inhibits ribosomal biogenesis and downstream protein translation which inhibits cancer growth in vitro (Figure 6J).
Reuterin inhibits colorectal cancer growth in vivo.
We implanted HCT116 and SW480 colon cancer xenograft tumors subcutaneously in nude mice. The mice were gavaged daily with L. reuteri (reuterin-producing) or L. reuteri genetically engineered to not produce reuterin daily. Treatment with wild-type but not mutant L. reuteri reduced HCT116 and SW480 tumor growth significantly beginning two days after treatment initiation (Figure 7A). We quantified systemic reuterin in these syngeneic tumors. Reuterin was significantly increased in treated tumors (Figure 7B). Reuterin levels quantified in treated tumors are at a level that is cytotoxic in cell culture. Cleaved caspase 3 (cCaspase3) was increased in wild-type but not mutant L. reuteri treated mice in both HCT116 and SW480 tumors (Figure 7C). We quantified ROS and found that wild-type but not mutant L. reuteri significantly increased ROS (Figure 7D). We next verified wild-type L. reuteri decreased syngeneic flank tumor growth of the mouse cell line MC38. Again, L. reuteri reduced tumor size significantly and reuterin levels were increased almost three-fold (Figure 7E and 7F). The tumors in mice gavaged with wild-type L. reuteri were noticeably smaller (Figure 7G). A marked decrease in KI67 levels were observed in reuterin-treated tumors compared to vehicle control treatment (Figure S7A). Moreover, we verified mutant and wild-type L. reuteri strains have similar colonization and in vivo growth by performing QPCR in the colons of wild-type mice gavaged daily for one week (Figure S7B). We found a significant increase in ROS species in wild-type L. reuteri mice compared to mutant L. reuteri treated mice (Figure 7H).
Figure 7: Reuterin inhibits colorectal cancer growth in vivo.
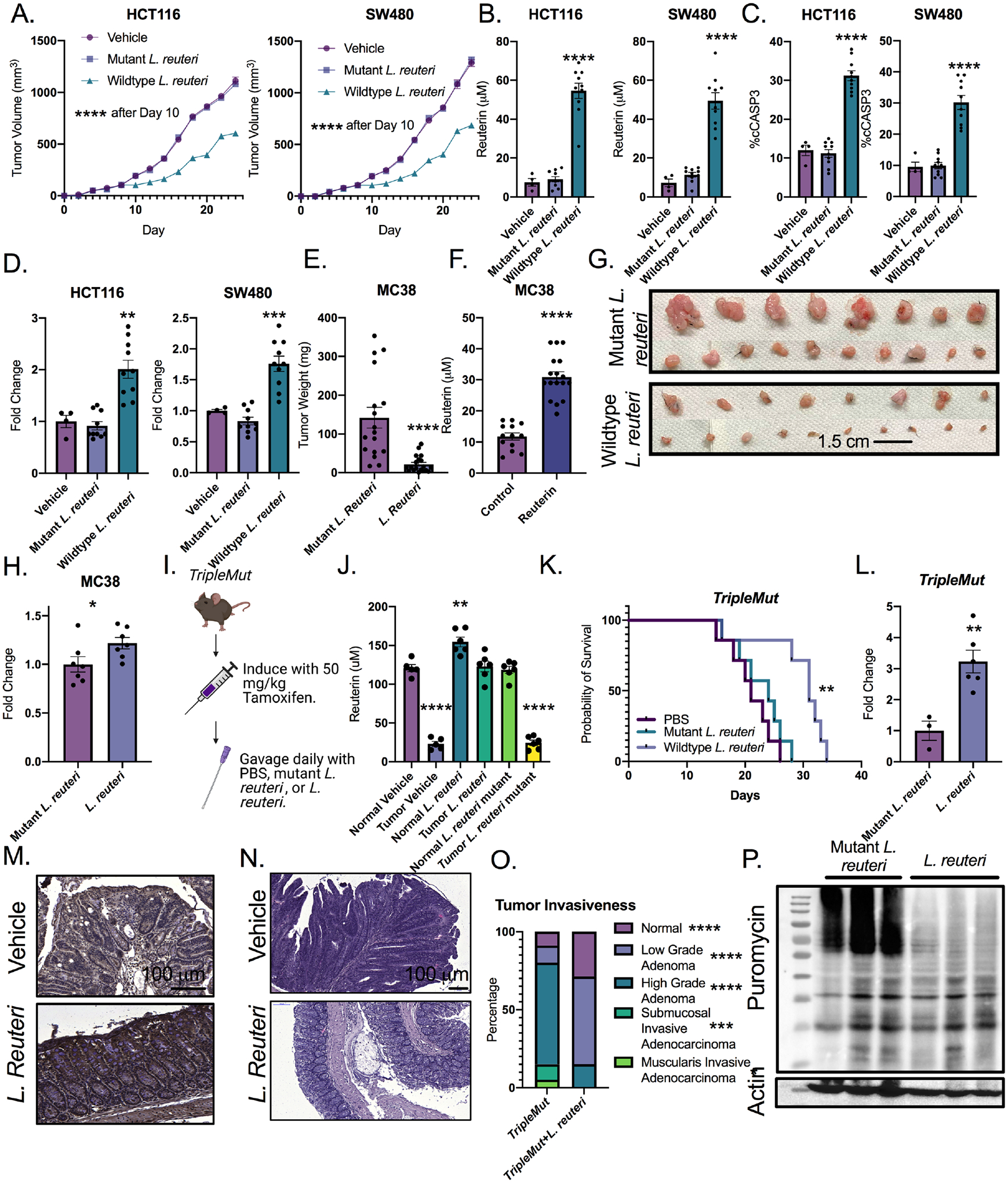
A) Quantification of tumor size of implanted HCT116 and SW480 cells in nude mice followed by daily gavage with wildtype or mutant L. reuteri or PBS (n=4–10). B) Quantification of reuterin in tumor from panel A. C) Percent cleaved caspase 3 cells (cCASP3) in tumors from panel A. D) Fold change of DCFDA ROS signal in tumors from panel A. E) End point weight of tumors of MC38 in mice gavaged daily with L. reuteri (n=8–9). F) Quantification of reuterin in MC38 tumors from E. G) Picture of representative tumors from D. H) Fold change of DCFDA ROS measurements from MC38 tumors. I) Schematic for treatment of the TripleMut. J) Quantification of reuterin in whole colon from indicated treatment groups. (n=4–6). K) Survival curve of mice gavaged daily by either wild-type or mutant L. reuteri. (n=7). L) DCFDA ROS measurements from the colons of induced TripleMut mice treated with either mutant or wildtype L. reuteri.(n=3–6). M) Representative 4HNE immunohistochemistry in induced TripleMut mice. (n=3–6, representative shown). N) H and E of PBS treated or L. reuteri treated TripleMut mice. (n=3–6, representative shown). O). Pathological score of TripleMut mice treated with wild-type or mutant L. reuteri. (n=4–5). P) Puromycin western blot of induced TripleMut mice sacrificed on Day 10 and treated with daily gavage of either wild-type or mutant L. reuteri. Colons were washed, homogenized, and stained with puromycin ex vivo for thirty minutes. (n=3–6, representative shown). Statistics were calculated with one-way ANOVA (Panels A, B, C, D, J, K, and O) or t-test (panels E, F, H and L). *p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001. Data is presented as mean +/− the standard error of the mean. Xenograft experiments were performed once. MC38 syngeneic experiments were performed two times with the indicated mouse numbers.
To extend our results to a genetic model of colon cancer, we induced a cohort of TripleMut mice with low dose tamoxifen, and gavaged daily with wild-type L. reuteri, as well as mutant L. reuteri strain (Figure 7I) (Schaefer et al., 2010). Daily gavages of wild-type L. reuteri, but not the mutant strain, increased reuterin levels in colon tissues to roughly the levels observed in healthy tumor free mice (Figure 7J). We quantified colonic diaminopropane and saw no changes in any of the treatment groups (Figure S7C). We found a robust protective effect for the wild-type L. reuteri (reuterin-producing) treatment, with a significant increase in survival compared to mouse cohorts treated with PBS or the mutant L. reuteri strain (Figure 7K). L. reuteri treatment did not alter normal tissues homeostasis as assessed by ALT and AST, body weight and H&E histological assessments (Figure S7D, S7E, and S7F). L. reuteri treated mice did not have any changes in immune infiltration and pro-inflammatory cytokines (Figure S7G and S7H). Moreover, reuterin has previously been shown to activate the aryl hydrocarbon receptor (AHR) pathway (Cervantes-Barragan et. al, 2018). Neither TripleMut mice or syngeneic tumors treated with L. reuteri have any significant, consistent changes in AHR pathways (Figure S7I and S7J). However, colons of TripleMut mice treated with wild-type L. reuteri have a more than three-fold increase in ROS species than mutant L. reuteri treated mice (Figure 7L). We also saw an increase in 4HNE, a marker of reactive oxygen species in TripleMut mice treated with wild-type L. reuteri (Figure 7M). A striking decrease in dysplastic transformation was observed in reuterin treated mouse colons (Figure 7N). Mice treated with L. reuteri had lower grade and less invasive lesions (Figure 7O). This data is consistent with a decrease in KI67 staining and an increase in cCASP3 in wild-type L. reuteri treated TripleMut mice (Figure S7K and S7L). Consistent with our in vitro analysis, an ex vivo puromycin assay in the TripleMut mice was performed. Mice gavaged with L. reuteri showed reduced translation compared to mice gavaged with mutant L. reuteri, supporting our in vitro mechanism (Figure 7P). Together, the data demonstrates a critical role of L. reutri and reuterin in modulating CRC redox balance and growth.
Discussion
Cancer metabolism is a rapidly developing field, but insight into how microbial metabolites impact colon cancer initiation and progression is limited. Recent work has highlighted that certain bacterial populations can impair or facilitate cancer growth (Chen et al., 2017, Cullin et al., 2021). Microbial byproducts like butyrate and other short chain fatty acids have been shown to have variable effects on cancer cells. While some work has shown that butyrate induces cancer cell death at high concentrations, other studies have shown that carbohydrate-derived metabolites such as butyrate induce hyper-proliferation and increased colorectal tumors in colon epithelial cells (Belcheva et al., 2014, Liu et al., 2018, Kaiko et al., 2016, Okumura et al., 2021). Moreover, other short chain fatty acids have been shown to drive hepatocellular carcinoma (Singh et al., 2018).
Studies examining interactions between the microbiome and metabolic byproducts from the tumor microenvironment (TME) are in their infancy. The TME is notably nutrient poor (Lyssiotis and Kimmelman, 2017). The rapid consumption of nutrients by cancer cells leads to the suppression of the T cell response (Bian et al., 2020; Moir et al., 2020; Nachef et al., 2021). How the TME effects dysbiosis, and how this cycle impacts colon cancer growth is unknown. We clearly show a metabolite exchange where the healthy microbiome produces several metabolites, including reuterin that decreased colon cancer growth. However, since these are endogenous metabolites produced at high levels, it was not clear how tumors bypassed this toxicity to establish growth. Our work suggests that dysbiosis is central for CRC progression through limiting the production of anti-cancer, endogenous, cytotoxic metabolites. Further, our data suggests that homocysteine and cystathionine metabolism is dysregulated in our advanced colon cancer model, and in turn this inhibits colonization of L. reuteri. Much more work is needed to understand the mechanistic basis of dysbiosis in colon cancer. Future work is needed to identify what molecular pathways facilitate production of these metabolites. We clearly show that increased mutational burden rapidly changes that microbiome. However, knockdown of KRAS in cell lines did not rescue L. reuteri growth in vitro. Moreover, cell lines with different mutational landscapes decreased L. reuteri growth in vitro. This suggests that there is a complex yet undefined interplay with tumor progression, mutations and microenvironmental stressors that are central to dysbiosis.
Electrophilic species such as reuterin can bind to thiols and alter or decrease protein function. Therefore, it is entirely possible two parallel, but essential pathways lead to cytotoxicity, (i) by inducing ROS and (ii) direct binding and altered function of ribosomal biogenesis (Figure 6J). Future work will understand if key thiols on ribosomal biogenesis proteins are essential to mediate cancer selective cell death. Our findings also demonstrate that distinct electrophiles maintain selectivity and different mechanisms of action. DMF and reuterin are cytotoxic electrophiles that induce death via protein oxidation. However, DMF and reuterin have distinct molecular targets, differential HIF2α dependency, and only reuterin targets ribosomal biogenesis (Singhal et al., 2021). Our data suggests that microbial metabolites may alter host response through modulating selective cysteine oxidation. This is a promising area for future studies.
Reuterin is an antimicrobial compound, and we demonstrate that reuterin has potent cytotoxic effects specific to cancer cells. Reuterin has been shown to be well tolerated in vivo at high doses (Asare et al., 2020). It is currently unknown how normal cells preferentially avoid reuterin induced growth inhibition. We hypothesize that a heightened state of oxidative stress due to altered metabolism in cancer cells contributes to the preferential cytotoxic effects of reuterin (Harris and DeNicola, 2020). Cells under heightened oxidative stress following 5-FU or BSO treatment are significantly more susceptible to the cytotoxic effects of reuterin. Reuterin does not decrease protein translation or induce ROS in a normal intestinal cell line. It is likely a complex integration of mutational status, proliferation, and microenvironmental stressors and waste products determines reuterin sensitivity.
L. reuteri is currently being studied in several clinical trials for colitis, colic, asthma, and H. pylori infection and has a confirmed safety profile ((Dore et al., 2019; Giudice et al., 2016; Oliva et al., 2012; Sung et al., 2017). Reuterin’s robust, cancer-specific cytotoxic effect in vivo and in vitro makes it a promising target for future clinical studies, as an adjunct to traditional therapy or as a preventative approach. Our work further highlights the utility of a precision approach to probiotics.
STAR Methods:
Resource Availability:
Lead Contact:
Further information and requests for resources and reagents should be directed to and will be responded to by Yatrik Shah, shahy@umich.edu.
Materials Availability:
16s rRNA gene sequencing is deposited at GEO publicly available as identified below. No unique plasmids or mouse lines were developed for this project. Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
Data and Code Availability:
16S RNA-seq data has been deposited at GEO and is publicly available at the date of publication. GEO accession number is GSE186880 and is also listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request. Any additional information required to reanalyzed the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| HIF-2-alpha Antibody | Bethyl Laboratories | Cat# 179483; |
| Ki67 Antibody | Cell Signaling | Cat#12202T |
| Actin Antibody | Protein Tech | Cat# 60008–1-Ig |
| Cleaved Caspase 3 | Cell Signaling | Cat#9661T |
| Puromycin Antibody | Developmental Studies Hybridoma Bank | Cat# PMY-2A4 |
| 4-HNE Antibody | Bioss Antibodies | Cat# BS-6313R |
| Anti-p-histone H2A | Sigma | Cat#ZMS05636 |
| Bacterial and Virus Strains | ||
| Wildtype L. reuteri | Robert L. Britton | N/A |
| Mutant L. reuteri | Robert L. Britton | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| FG-4592 | Cayman Chemical | Cat#15294 |
| N-acetyl-cysteine | Cayman Chemical | Cat#20261 |
| Sodium Sulfide | Cayman Chemical | Cat#10122555 |
| Cystathionine | Cayman Chemical | Cat# 16061 |
| Olaparib | Cayman Chemical | Cat#10621 |
| CX-5461 | Cayman Chemical | Cat#18392 |
| L-Threonine | Sigma | Cat# 72-19-5 |
| Ammonia Chloride | Sigma | Cat#213330 |
| 5-Fluorouracil | Sigma | Cat#F6627 |
| L-carnitine | Sigma | Cat#21489 |
| Aspartic Acid | Sigma | Cat#1043819 |
| L-Glutamic Acid | Sigma | Cat#G1251 |
| L-Glutamine | Sigma | Cat#G8540 |
| Hydrogen Peroxide | Sigma | Cat#HX0636 |
| L-Buthionine-Sulfozimine | Cayman Chemical | Cat#14484 |
| Glutathione Ethyl Ester | Cayman Chemnical | Cat#14953 |
| Orotic Acid | Sigma | Cat#O2750 |
| Ureidosuccinic Acid | Cayman Chemical | Cat#34126 |
| Deposited Data | ||
| 16S Sequencing | Yachida et. al, 2019 | PMID: 31171880 |
| 16S Sequencing | Chen et. al, 2012 | PMID: 22761885 |
| 16S Sequencing | Cheng Tao Pu, 2020 | PMID: 31609493 |
| 16S Sequencing | GEO | GSE186880 |
| Experimental Models: Cell Lines | ||
| NCM460 | ATCC | N/A |
| HCT116 | ATCC | N/A |
| MC38 | ATCC | N/A |
| CT26 | ATCC | N/A |
| DLD1 | ATCC | N/A |
| SW480 | ATCC | N/A |
| RKO | ATCC | N/A |
| HF27 | ATCC | N/A |
| HF29 | ATCC | N/A |
| MCF10A | ATCC | N/A |
| Vero2 | ATCC | N/A |
| HCN2 | ATCC | N/A |
| NCM356 | ATCC | N/A |
| CRL1831 | ATCC | N/A |
| CCD18Co | ATCC | N/A |
| Experimental Models: Organisms/Strains | ||
| Wildtype: C56Bl/6J | Jackson Labs | N/A |
| SingleMut: CDX2-ERT2Cre; Apcfl/fl | Jackson Labs | N/A |
| DoubleMut: CDX2-ERT2Cre; Apcfl/fl;Trp53fl/ | Jackson Labs | N/A |
| TripleMut: Apcfl/fl;Trp53fl/fl; KrasLSLG12D | Jackson Labs | N/A |
| Oligonucleotides | ||
| List of primers | Supplemental Table 6 | N/A |
Experimental Model and Subject Details:
Mice:
All mice used in this paper are in a predominant C57Bl/6J background. Male and female were equally mixed and littermates were randomly assigned to experimental conditions. The mice were housed in a temperature controlled, specific pathogen free environment, with a 12 hour light/dark cycle. They were fed ad libitum with standard chow diet. Studied mice were between 4–6 weeks old. For reuterin studies, L. reuteri was cultured as described below, and after 18 hours of culture, bacteria were harvested by centrifugation and diluted in PBS. Serial dilutions were prepared to assess concentration, and then mice were treated with .2 mL by gavage at a dose of 2 × 109 colony forming units per mL. There were no significant gender differences. Mouse lines used were CDX2-ERT2Cre; Apcfl/fl mice, CDX2-ERT2Cre; Apcfl/fl; Trp53fl/fl, and Apcfl/fl;Trp53fl/fl; KrasLSLG12D mice. All animal studies were carried out in accordance with Association for Assessment and Accreditation of Laboratory Animal Care International guidelines and approved by the University Committee on the Use and Care of Animals at the University of Michigan.
Human Subjects:
The study was approved by the institutional review boards at the University of Michigan (IRBMED, protocol number HUM00085066) and the Department of Health and Human Services, Food and Drug Administration (Research Involving Human Subjects Committee/RIHSC, protocol number14–029D). The study was carried out in accordance with the protocol and applicable local regulatory requirements. All subjects (both male and female, age 18–55 years) provided written informed consents and participated after completion of a physical examination and review of their previous medical history. For L. reuteri quantitation, tissue was collected during either an open surgical procedure or a colonoscopy for both normal and tumor tissue. Tissue was immediately flash frozen until time of extraction as described below. All qPCR was normalized to a universal bacteria control and normalized to the average of the normal tissue or cohort. Feces were collected from patients and then flash frozen.
Cell Lines:
Human intestinal cell lines HCT116, RKO, SW480, and DLD1 were used for most experiments. Other cell lines used are MC38, HT29, CT26, MIAPACA, PaTu8988t, ID8, HCC1187, HCC1937, HCC 1806, HCC 38, DU 4475, MT3, HCC1143, MT4, 178A, B16F10, YUMM 5.1, HELA, HF27, HF29, N9, MCF10A, Vero2, HCN-2, NCM356, NCM460, CRL1831, and CCD18Co. Cell lines have been STR-authenticated. All cells were maintained in complete DMEM medium (supplemented with 10% fetal bovine serum and 1% antibiotic/antimycotic agent) at 37°C in 5% CO2 and 21% O2.
Method Details:
Proliferation Assay:
Growth assays were performed using MTT reagent (Thiazolyl Blue Tetrazolium Bromide). Briefly, cells were plated down and 24-hours following plating a Day 0 reading was taken. Cells were incubated for 45 minutes with MTT solution (5X concentrate stock: 5 mg/ml, in 1XPBS, pH 7.4). Media and MTT solution were then carefully aspirated followed by solubilization with dimethyl sulfoxide. Absorbance was read at 570nm. Following the Day 0 read, the cells were treated with indicated doses and readings were taken after 24-hours. Cell growth assays were also measured using the Cytation 5 Imaging Multi-Mode reader. Cells were plated down, treated 24 hours later with indicated treatments, and immediately imaged and analyzed for cell number. Images were then taken every 24 hours.
Bacterial Strains:
L. reuteri strain ATCC PTA6475 (originally isolated from human breast milk, gift of Dr. Robert Britton, Baylor College of Medicine) and L. reuteri strain PRB94 (deltapduC mutant in ATCC PTA6475 background, gift of Dr. Robert Britton, Baylor College of Medicine) was used as described previously(Das et al., 2020). The bacteria were cultured in de Man, Rogosa and Sharpe (MRS) broth at 37 degrees Celsius in a sealed tube under atmospheric oxygen concentration. The bacteria were cultured in Luria broth (LB) media at 37°C under atmospheric oxygen concentration. Fecal bacteria were grown from homogenized wild-type mouse feces inoculum, grown at 37 degrees Celsius in MRS broth.
Antibiotics Treatment
Antibiotics treatment (Abx) was performed as described previously (Ramakrishnan et al., 2019). Briefly, animals were given water containing antibiotic cocktail (Ampicillin 1 g/L, neomycin 1 g/L, gentamycin 500 mg/L; penicillin 100U/L) in the drinking water, ad libitum. In addition, oral gavage of vancomycin (1 mg/mL) and metronidazole (0.5 mg/mL) was given on alternate days for 1 week. Mice were then induced and metabolite extracts were prepared 10 days after induction and discontinuation of antibiotics.
Real time Quantitative PCR:
1μg of total RNA extracted using Trizol reagent from mouse tissues (intestinal epithelial scrapes), human intestinal cell lines, and mouse feces. RNA was reverse transcribed to cDNA using SuperScriptTM III First-Strand Synthesis System (Invitrogen). Real time PCR reactions were set up in three technical replicates for each sample. cDNA gene specific primers, SYBR green master mix was combined, and then run in QuantStudio 5 Real-Time PCR System (Applied BioSystems). The fold-change of the genes were calculated using the ΔΔCt method using β-actin as the housekeeping gene. Primers are listed (Supplemental Table 6). Bacterial qPCR was performed on extracted ribosomal DNA. Primers were designed to specifically amplify the indicated bacteria and normalized to an all bacteria primer control.
Western Blotting:
Whole-cell lysate preparations were described previously(Anderson et al., 2013). Whole cell lysates were prepared from cell lines by RIPA buffer. Homogenates were incubated in RIPA buffer for 15 minutes on ice followed by 13,000 rpm centrifugation for 15 minutes. Supernatants were transferred to a new tube and mixed with 5X Laemmli buffer and boiled for 5 minutes. Lysates containing 30–40μg of protein per well were separated by SDS-PAGE, transferred onto nitrocellulose membranes, and immunoblotted overnight at 4°C with indicated antibodies. All the primary antibodies were used at a dilution of 1:1000. HRP-conjugated secondary antibodies used were anti-rabbit and anti-mouse at a dilution of 1: 2000 and immunoblots were developed using Chemidoc imaging system (ChemiDoc, BioRad).
Immunostaining:
Tissues were fixed with formalin for 24 hours, and then placed in ethanol for 24 hours. Paraffin embedded tissues were sectioned (6μm). Antigen retrieval was performed by boiling slides for 10 minutes in citrate buffer. Sections were blocked in 5% (vol/vol) normal goat serum in TBST for 30 min at room temperature and probed with polyclonal rabbit anti–Ki67 or 4-HNE primary antibody (1:250) overnight at 4°C. The samples were then incubated with rabbit IgG Alexa Fluor 594 for Ki67 (1:500) or HRP coupled secondary for 1 hour in dark at room temperature, followed by mounting and counterstaining by Prolong™ Gold Antifade Mountant with DAPI. For Gamma H2AX ataining positive control was treated with 2 Gy radiation for 2 minutes. Cells were fixed in 10% formaldehyde and permeabilized in 0.01% Triton X 100. Primary antibody was stained overnight, then secondary for one hour in the dark in 0.01% Triton X 100 and 3% BSA. Confocal microscopy was performed.
Metabolite Extraction:
Metabolites from feces were extracted by 2:1 methanol: chloroform (v/v) in water as described previously(Sridharan et al., 2014). Pre-weighed freshly collected feces were homogenized in 3 mL of dry ice cold 80% methanol and incubated on dry ice for 10 minutes followed by centrifugation at 4,000 rpm for 10 minutes at 4 degrees Celsius. Supernatant was passed through a 70-μm cell strainer and centrifuged at 13,000 rpm for 10 minutes. 0.6 mL of ice-cold water was added per 1.5 mL of supernatant, then the sample tube was vortexed vigorously and centrifuged at 13,000 rpm for 10 minutes to obtain phase separation. The upper and lower phases were separately collected into fresh sample tubes without disturbing the interface. Samples were dried in a speed vac (Eppendorf Vacufuge) to and were resuspended in DMSO for in vitro experiments.
16S RRNA Gene Sequencing and Bacterial Community Analysis:
Bacterial sequencing analysis was done as described previously at the Microbial Systems Molecular Biology Lab, a part of the University of Michigan Host Microbiome Initiative(Ramakrishnan et al., 2019). Briefly, the V4 region of the 16S rRNA was amplified from freshly collected mouse feces or tumor mucosal scrapes using the Dual-indexing sequencing strategy. Sequencing was done on the Illumina MiSeq platform, using a MiSeq reagent kit V2 500 cycles, according to the manufacturer’s instructions. Bacterial community analysis was performed using Mothur (v1.42.3) aligning to the full SILVA reference alignment (release 132) provided by Mothur(Schloss et al., 2009). Sample quality was determined by assuring samples contained more aligned reads than the DNA extraction control and that samples clustered within their genotypes via principal component analysis in PC1/PC2.
LDH Assay:
Cell death was measured using the LDH Cytotoxicity Assay Kit from Takara Bio. 10,000 cells were seeded in a 96 well plate overnight. The next day, cells were treated with the indicated condition. Supernatants were harvested after 24 hours of treatment and a media only control and Triton X-100 control were included. The supernatant was incubated with LDH detection reagent for 30 minutes per the manufacturer’s instructions. Absorbance was taken at 490 nm in technical triplicate. Cytotoxicity was calculated using the manufacturer’s protocol.
Colony Forming Assay:
Cells were plated in biological triplicates in a 6 well plate at 600 cells per well in 2 mL of media. After 48 hours, cells were treated with different reagents as mentioned in the legends. Assays were concluded at 15 days by fixing in cold 10% buffered formalin for 10 minutes and staining with 0.5% crystal violet, 20% methanol solution for 30 minutes. Colonies were manually counted via a study blinded observer.
C11-BODIPY lipid ROS measurement:
1 million colorectal cancer cells were seeded in a 12 well plate overnight. Cells were then treated with the indicated concentration of Reuterin and incubated for 12 hours. Cells were harvested in HBSS, washed, and stained with 5μM C11-BODIPY (ThermoFisher) at 37 degrees C for 30 minutes. Fluorescent intensity was measured on the FITC channel on the Beckman Coulter MoFlo Astrios. A minimum of 20,000 cells were analyzed per condition, data was analyzed using FlowJo software (Tree Star). Values are MFI.
ROS Detection Assay:
Cell permeable free radical sensor carboxy-H2DCFDA (Invitrogen) was used. Cells were treated with indicated reuterin concentration for 24 hours, then incubated with 10μM carboxy-H2DCFDA in PBS at 37 degrees C for 45 minutes. Cells were then washed and resuspended in PBS. Mean fluorescent intensity was obtained from the Cytation 5 Imaging Multimode Reader. For in vivo ROS assay, epithelial cells were isolated from either induced TripleMut mouse colon or from syngeneic tumors using Collagenase IV incubation. Cells were then strained and then stained as above with 10μM carboxy-H2DCFDA. Mean fluorescent intensity and percent ROS positive cells was obtained from flow cytometry.
Metabolomics:
Cells were plated at 1 million cells per well in 6 well plates. After 24 hours, cells were treated with 100μM Reuterin for 24 hours. Cells were then washed once with ice cold PBS, then incubated in dry-ice cold 80% methanol on dry ice for 10 minutes. Cells were scraped and the polar metabolite supernatant was collected on dry ice. Tissues were bead homogenized in the polar metabolite methanol. Samples were clarified via high speed centrifugation. Protein concentration was determined by processing a parallel well and used for normalization. Cell or tissue solutions were lyophilized using a SpeedVac concentrator. Metabolite pellets were resuspended in 50:50 methanol/water mixture for LC-MS analysis. Data were collected using previously published parameters(Bennett et al., 2008; Chae et al., 2015). QQ data were preprocessed with Agilent MassHunter Workstation Quantitative Analysis Software (B0700). Additional analyses were postprocessed for further quality control. Each metabolite abundance level in each sample was divided by the median of all abundance levels across samples for comparison and statistics. Statistical significance was determined by a one-way ANOVA with a significance threshold of 0.05. Further pathway analysis took place on Metaboanalyst. For microbial metabolites, identification was performed as described in Das, 2019. Briefly, samples were homogenized, centrifuged, and incubated with 50 mM sodium hydroxide and propanol/pyridine. Propylchloroformate was added on ice and derivatized for 30 minutes. The samples were extracted with hexane twice and stored for GC-MS analysis. For DAP a headspace sampler was utilized, for Reuterin a standard GC/MS sampling. A HP5 ms 5% phenylmethylpolysiloxane capillary GC column (Agilent) was employed with helium as the carrier gas. The initial temperature was 70 degrees C. increased to 230 degrees C. The total run time was 20 minutes and the mass spectral data ranges were from 30–400 m/x. Reuterin was quantified by peak area of extraction m/z 219 as described previously(Talarico and Dobrogosz, 1989). L-glutathione was measured from lyophilized cell pellets. Pellets were resuspended in 50:50 methanol/water mixture for LC-MS analysis. QQ data was preprocessed with Agilent MassHunter Workstation Quantitative Analysis Software (B0700).
Histology:
Tumor tissues were rolled and fixed in formalin for 24 hours, then embedded in paraffin. Sections of 5μm were stained for H&E and mounted with Permount Mounting Medium (Thermo Fisher Scientific). For immunohistochemistry, paraffin tissue sections underwent antigen retrieval, blocking in 5% goat serum in PBS, and probed with Ki67 antibody (Cell Signaling, 1:250). Sections were washed 2 times with PBST and incubated with HRP conjugated anti-rabbit IgG (1:500, catalog 7074S, Cell Signaling Technology) for 1 hour. Sections were then washed with PBST and incubated with DAB substrate. After the sample turned brown, the reaction was stopped with distilled water and dehydration steps were completed. The slides were mounted with Permount Mounting Medium. Histological scoring of dysplasia was done by a pathologist as previously described (Triner et, al, 2019).
Immune Cell Isolation:
Colons were removed, washed thoroughly in PBS, and then incubated with 10 mM EDTA for 45 minutes. Supernatant was removed, and then cells were isolated with 1 mg/mL collagenase Type IV for 45 minutes. Immune compartment was obtained through a Percoll gradient, cells were stained using indicated antibody, and then acquired using flow cytometry.
RNA-SEQ analysis: RNA-Seq analysis:
IEC6 cells were seeded in triplicate overnight, then treated with 100μM Reuterin for 16 hours. RNA was isolated from culture tissue as described above. Quality of fastq files was assessed using FastQC v0.11.8 and MultiQC v1.7(Ewels et al., 2016). Reads were then aligned to GENCODE’s GRCm38.vM24 assembly using STAR v2.6.1a_08–27(Dobin et al., 2013; Frankish et al., 2018). Aligned reads were counted using featureCounts v1.6.3(Liao et al., 2014). PCA-based clustering was used to identify outliers and then differential expression was performed using DESeq2 v1.30.1(Ren and Kuan, 2020). Genes with an adjusted p-value < 0.05 were considered differentially expressed. All analysis was carried out on the University of Michigan Great Lakes HPC cluster.
Pathway analysis:
Significant metabolites obtained from metabolomic analysis (see above) were converted to HMDB identifiers using MetaboAnalyst v5.0(Pang et al., 2021). HMDB identifiers were then mapped to known protein interactors using MetaBridge v1.2 via KEGG and MetaCyc(Hinshaw et al., 2018). These protein-metabolite interactions were then overlaid with differentially expressed genes from RNA-sequencing analysis (see above). Metabolites with links to 3 or more differentially expressed genes were plotted in a network using cytoscape v3.8.0(Shannon et al., 2003).
Syngeneic and Xenograft Studies:
Wild-type or immunocompromised mice of both sexes were inoculated with 2 million MC38, HCT116, or SW480 cells. Cells were implanted into lower flanks, and treatment began at day 10 after visible tumors as described above. Tumor size was measured with digital calipers. At the endpoint, mice were sacrificed and tumors were excised. Tumor volume and weight was measured and tissues were prepared for histology, IHC, and flow cytometry.
Proteomics and Cysteine Proteomics:
Proteomics analysis was performed as described (Kuljanin et al., 2021). Briefly, cells were treated for 24 hours, then washed twice with PBS. Cell pellet was homogenized and sonicated, and total protein was determined. Each sample was treated with DBIA for 1 hour, and then proteins were precipitated by chloroform/methanol. Protein pellets were solubilized in 4-(2-hydroxyethyl)-1-piperazinepropanesulfonic acid at pH 8.5 and digested using LysC and trypsin overnight. For whole proteome analysis cells were lysed in 8 M urea and 200 mM EPPS at pH 8.5 Samples were homogenized and reduced. Cysteine peptides were enriched using streptavidin beads as described in Kuljanin, 2021. Mass spectrometry data were acquired using an Orbitrap Fusion Lumos mass spectrometer in line with a Proxeon NanoLC-1200 UPLC system. Intelligent data acquisition was performed using orbiter and the Comet search algorithm. Spectral searches were performed eliminating common contaminants. Cysteine modified peptides were filtered for site localization using the AScore algorithm with a cutoff of 13. A total sum signal to noise of all reporter ions of 100 was required. Cysteine engagement was assessed by the blocking of the labeling of the probe. Peptides showing a 95% reduction were assigned a maximum ratio of 20. Multiple cysteines with confident localization scores were separated. Competition ratios were found by dividing the control channel by the electrophile channel. Replicate measurements were averaged.
Puromycin Assay:
Cells were treated for 24 hours with reuterin, then incubated with 1 mg/ml puromycin for 10 minutes. Lysates were flash frozen then western blot was performed for puromycin antibody. Small pieces of tumor or colon were homogenized thoroughly for the in vivo assay, then incubated with 1 mg/mL puromycin for 30 minutes. Lysates were then generated using bead homogenization before western blotting for anti-puromycin.
Epithelial Cell Isolation:
Colons were removed and washed into PBS. Colon was incubated at 150 RPM for 30 minutes in 25 mL of 10 mM EDTA in RPMI. Colons were vortexed twice, and the liquid collected. RPMI was added, tissues were vortexed and liquid collected, repeated twice. Cells were counted, plated, and imaged on the Cytation 5 Imaging MultiMode Reader at 0 and 12 hours.
Dose Response Matrices:
Cells were plated in triplicate for each indicated dose. The average percent inhibition was calculated, and then the coefficient of drug interaction (CDI) was calculated from the matrix by dividing the 2-drug combination inhibition by the two single drug concentrations multiplied together. CDI<1 indicates synergism, CDI=1 indicates additivity and CD1>1 indicates antagonism.
Quantification and Statistical Analysis:
In vitro experiments were validated in 4 cell lines. Each cell line experiment was performed in technical replicates for each condition and repeated at least three times with biological triplicates to ensure reproducibility. Figures show a representative biological replicate unless otherwise indicated. Blinding was performed whenever appropriate. Sample description and identification was unavailable to the core personnel during data collection and analysis. Statistical details of all experiments can be found in the figure legends. The sample numbers are mentioned in each figure legend and denote biological replicates. Statistical details are reported in figure legends. Results are expressed as the mean plus or minus the standard error of the mean for all figures unless otherwise noted. Significance between 2 groups was tested using a 2 tailed unpaired t test. Significance among multiple groups was tested using a one-way ANOVA. GraphPad Prism 7.0 was used for the statistical analysis. Statistical significance is described in the figure legends as: * p < 0.05, ** p < 0.01, *** p < 0.001, **** p < 0.0001.
Supplementary Material
Supplemental Table 1: 16S PCoA: Related to Figure 2. PCoA data from 16S sequencing. Includes pcoa1, pcoa2, vector length and size and taxonomy.
Supplemental Table 2: Reuterin RNA-SEQ: Related to Figure 4. Differentially regulated genes from RNA Seq. Log fold change, p value and FDR for reuterin compared to vehicle and DAP compared to vehicle.
Supplemental Table 3: Reuterin Metabolomics: Related to Figure 4. Metabolomics data in HCCT116 and SW480. Shows all assayed compounds, relative amounts, t-test, averages between samples, and the log2 for reuterin over control.
Supplemental Table 4: Reuterin Cysteine Proteomics: Related to Figure 6. Raw cysteine proteomics data including the gene, protein, peptide identified, site position, expression in all vehicle, DMF and reuterin samples, averages in all conditions and competition ratios in all conditions.
Supplemental Table 5: Reuterin Proteomics: Related to Figure 6. Raw proteomics data including t test for significance, the unique peptide number, the p value, the q value, log2fold change, the uniprot ID and the gene symbol.
Supplemental Table 6: Primer Table: Related to Figures 2, 4, 6, and 7. Primer sequences used in the paper.
Supplemental Table 7: Reuterin Synergy: Related to Figures 5 and 6. Coefficients of drug interactions at various doses with reuterin.
Highlights:
Microbial metabolites from the healthy colon inhibit colon tumorigenesis
L. reuteri and reuterin levels are reduced in mouse and human colon cancer
Reuterin induces protein oxidation and inhibits ribosomal biogenesis
L. reuteri and reuterin decrease tumor growth and prolong survival in mice
Acknowledgements:
This work was supported by NIH grants; R01CA148828, R01CA245546, and R01DK095201 to YMS. R37CA237421, R01CA248160, R01CA244931 and R01CA215607 to CAL. R01NS099280 and R01NA086810 to PKT. VA grant BLRD BX004842 to PKT. The work was also supported by the University of Michigan Comprehensive Cancer Center Core Grant P30CA046592; the GI SPORE Molecular Pathology and Biosample Core (P50CA130810); the Center for Gastrointestinal Research (DK034933); and the Department of Defense (CA171086 to YMS). HNB was supported by T32 training grant (T32- of Michigan Systems and Integrative Biology grant. SAK was supported by an NIH F31 fellowship (F31CA247457). SLM was supported by CMB Graduate Program T32GM007315. Metabolomics studies were supported by NIH grant DK097153, and the Charles Woodson Research Fund. Proteomics studies were supported by the Hale Family Center for Pancreatic Cancer Research. The authors thank Robert L. Britton for providing the strains of Lactobacillus reuteri.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interests:
The authors declare no competing interests.
References:
- Anderson ER, Taylor M, Xue X, Ramakrishnan SK, Martin A, Xie L, Bredell BX, Gardenghi S, Rivella S, and Shah YM (2013). Intestinal HIF2α promotes tissue-iron accumulation in disorders of iron overload with anemia. Proc National Acad Sci 110, E4922–E4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Angeli JPF, Schneider M, Proneth B, Tyurina YY, Tyurin VA, Hammond VJ, Herbach N, Aichler M, Walch A, Eggenhofer E, et al. (2014). Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol 16, 1180–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, and Bray F (2017). Global patterns and trends in colorectal cancer incidence and mortality. Gut 66, 683. [DOI] [PubMed] [Google Scholar]
- Asare PT, Zurfluh K, Greppi A, Lynch D, Schwab C, Stephan R, and Lacroix C (2020). Reuterin Demonstrates Potent Antimicrobial Activity Against a Broad Panel of Human and Poultry Meat Campylobacter spp. Isolates. Microorg 8, 78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcheva A, Irrazabal T, Robertson SJ, Streutker C, Maughan H, Rubino S, Moryama EH, Copeland JK, Surendra A, Kumar S, et al. (2014). Gut microbial metabolism drives transformation of MSH2-deficient colon epithelial cells. Cell 158(2):288–299. [DOI] [PubMed] [Google Scholar]
- Bennett BD, Yuan J, Kimball EH, and Rabinowitz JD (2008). Absolute quantitation of intracellular metabolite concentrations by an isotope ratio-based approach. Nat Protoc 3, 1299–1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bian Y, Li W, Kremer DM, Sajjakulnukit P, Li S, Crespo J, Nwosu ZC, Zhang L, Czerwonka A, Pawłowska A, et al. (2020). Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. Nature 585, 277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cerezo EL, Houles T, Lié O, Sarthou M-K, Audoynaud C, Lavoie G, Halladjian M, Cantaloube S, Froment C, Burlet-Schiltz O, et al. (2021). RIOK2 phosphorylation by RSK promotes synthesis of the human small ribosomal subunit. Plos Genet 17, e1009583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes-Barragan L, Chai JN, Tianero MD, Luccia BD, Ahem PP, Merriman J, Cortez VS, Caparon MG, Sonia MS, Gilgillan S, Cella M, Gordon JI, Hsieh C, Colonna M (2018). Lactobacillus reuteri induces gut intraepithelial CD4+CD8alpha+ T cells. Science, 357, 806810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chae TU, Kim WJ, Choi S, Park SJ, and Lee SY (2015). Metabolic engineering of Escherichia coli for the production of 1,3-diaminopropane, a three carbon diamine. Sci Rep-Uk 5, 13040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J, Pitmon E, and Wang K (2017). Microbiome, inflammation and colorectal cancer. Semin Immunol 32, 43–53. [DOI] [PubMed] [Google Scholar]
- Collins JC, Ghalei H, Doherty JR, Huang H, Culver RN, and Karbstein K (2018). Ribosome biogenesis factor Ltv1 chaperones the assembly of the small subunit head. J Cell Biol 217, 4141–4154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullin N, Antunes CA, Straussman R, Stein-Thoeringer CK, Elinav E (2021). Microbiome and Cancer. Cancer Cell. 39(10):1317–1341. [DOI] [PubMed] [Google Scholar]
- Dame MK, Jiang Y, Appelman HD, Copley KD, McClintock SD, Aslam MN, Attili D, Elmunzer BJ, Brenner DE, Varani J, et al. (2014). Human colonic crypts in culture: segregation of immunochemical markers in normal versus adenoma-derived. Lab Invest 94, 222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das NK, Schwartz AJ, Barthel G, Inohara N, Liu Q, Sankar A, Hill DR, Ma X, Lamberg O, Schnizlein MK, et al. (2020). Microbial Metabolite Signaling Is Required for Systemic Iron Homeostasis. Cell Metab 31, 115–130.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejea CM, Fathi P, Craig JM, Boleij A, Taddese R, Geis AL, Wu X, Shields CE, Hechenbleikner EM, Huso DL, Anders RA, Giardiello FM, Wicks EC, Wang H, Wu S, Pardoll DM, Housseau F, and Sears CL (2018). Patients with familial adenomatous polyposis harbor colonic biofilms containing tumorigenic bacteria. Science 359(6375):592–597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dore MP, Bibbò S, Loria M, Salis R, Manca A, Pes GM, and Graham DY (2019). Twice- a-day PPI, tetracycline, metronidazole quadruple therapy with Pylera® or Lactobacillus reuteri for treatment naïve or for retreatment of Helicobacter pylori. Two randomized pilot studies. Helicobacter 24, e12659. [DOI] [PubMed] [Google Scholar]
- Ewels P, Magnusson M, Lundin S, and Käller M (2016). MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32, 3047–3048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filipovic MR, Zivanovic J, Alvarez B, and Banerjee R (2018). Chemical Biology of H2S Signaling through Persulfidation. Chem Rev 118, 1253–1337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flemer B, Warren RD, Barrett MP, Cisek K, Das A, Jeffery IB, Hurley E, O’Riordain M, Shanahan F, and O’Toole PW (2018). The oral microbiota in colorectal cancer is distinctive and predictive. Gut 67, 1454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A, Diekhans M, Ferreira A-M, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J, et al. (2018). GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 47, gky955-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaschler MM, Hu F, Feng H, Linkermann A, Min W, and Stockwell BR (2018). Determination of the Subcellular Localization and Mechanism of Action of Ferrostatins in Suppressing Ferroptosis. Acs Chem Biol 13, 1013–1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- del Giudice MM, Maiello N, Allegorico A, Iavarazzo L, Capasso M, Capristo C, and Ciprandi G (2016). Lactobacillus reuteri DSM 17938 plus vitamin D3 as ancillary treatment in allergic children with asthma. Ann Allergy Asthma Immunol 117, 710–712. [DOI] [PubMed] [Google Scholar]
- Grivennikov SI, Greten FR, and Karin M (2010). Immunity, Inflammation, and Cancer. Cell 140, 883–899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grivennikov SI, Wang K, Mucida D, Stewart CA, Schnabl B, Jauch D, Taniguchi K, Yu G-Y, Österreicher CH, Hung KE, et al. (2012). Adenoma-linked barrier defects and microbial products drive IL-23/IL-17-mediated tumour growth. Nature 491, 254–258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris IS, and DeNicola GM (2020). The Complex Interplay between Antioxidants and ROS in Cancer. Trends Cell Biol 30, 440–451. [DOI] [PubMed] [Google Scholar]
- Hinoi T, Akyol A, Theisen BK, Ferguson DO, Greenson JK, Williams BO, Cho KR, and Fearon ER (2007). Mouse Model of Colonic Adenoma-Carcinoma Progression Based on Somatic Apc Inactivation. Cancer Res 67, 9721–9730. [DOI] [PubMed] [Google Scholar]
- Hinshaw SJ, Lee AHY, Gill EE, and Hancock REW (2018). MetaBridge: enabling network-based integrative analysis via direct protein interactors of metabolites. Bioinformatics 34, 3225–3227. [DOI] [PubMed] [Google Scholar]
- Kaiko GE, Ryu SH, Koues OI, Collins PL, Solnica-Krezel L, Pearce EJ, Pearce EL, Oltz EM, and Stappenbeck TS (2016). The Colonic Crypt Protects Stem Cells from Microbiota-Derived Metabolites. Cell 167, 1137. [DOI] [PubMed] [Google Scholar]
- Kelly CJ, Zheng L, Campbell EL, Saeedi B, Scholz CC, Bayless AJ, Wilson KE, Glover LE, Kominsky DJ, Magnuson A, et al. (2015). Crosstalk between Microbiota-Derived Short-Chain Fatty Acids and Intestinal Epithelial HIF Augments Tissue Barrier Function. Cell Host Microbe 17, 662–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuljanin M, Mitchell DC, Schweppe DK, Gikandi AS, Nusinow DP, Bulloch NJ, Vinogradova EV, Wilson DL, Kool ET, Mancias JD, et al. (2021). Reimagining high-throughput profiling of reactive cysteines for cell-based screening of large electrophile libraries. Nat Biotechnol 39, 630–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lang M, Baumgartner M, Rożalska A, Frick A, Riva A, Jarek M, Berry D, and Gasche C (2020). Crypt residing bacteria and proximal colonic carcinogenesis in a mouse model of Lynch syndrome. Int J Cancer 147, 2316–2326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le DT, Kavan P, Kim TW, Burge ME, Cutsem EV, Hara H, Boland PM, Laethem J-LV, Geva R, Taniguchi H, et al. (2018). KEYNOTE-164: Pembrolizumab for patients with advanced microsatellite instability high (MSI-H) colorectal cancer. J Clin Oncol 36, 3514–3514. [Google Scholar]
- Lee HM, Kim DH, Lee HL, Cha B, Kang DH, and Jeong Y-I (2019). Synergistic effect of buthionine sulfoximine on the chlorin e6-based photodynamic treatment of cancer cells. Arch Pharm Res 42, 990–999. [DOI] [PubMed] [Google Scholar]
- Leystra AA, and Clapper ML (2019). Gut Microbiota Influences Experimental Outcomes in Mouse Models of Colorectal Cancer. Genes-Basel 10, 900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, and Shi W (2014). featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. [DOI] [PubMed] [Google Scholar]
- Liu H, Wang J, He T, Becker S, Zhang G, Li D, and Ma X (2018). Butyrate: A Double-Edged Sword for Health? Adv Nutr 9, 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyssiotis CA, and Kimmelman AC (2017). Metabolic Interactions in the Tumor Microenvironment. Trends Cell Biol 27, 863–875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mars J-C, Tremblay MG, Valere M, Sibai DS, Sabourin-Felix M, Lessard F, and Moss T (2020). The chemotherapeutic agent CX-5461 irreversibly blocks RNA polymerase I initiation and promoter release to cause nucleolar disruption, DNA damage and cell inviability. Nar Cancer 2, zcaa032-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martín-Cabrejas I, Langa S, Gaya P, Rodríguez E, Landete JM, Medina M, and Arqués JL (2017). Optimization of reuterin production in cheese by Lactobacillus reuteri. J Food Sci Technology 54, 1346–1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mi W, Guan H, Lyu J, Zhao D, Xi Y, Jiang S, Andrews FH, Wang X, Gagea M, Wen H, et al. (2017). YEATS2 links histone acetylation to tumorigenesis of non-small cell lung cancer. Nat Commun 8, 1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moir JAG, Long A, Haugk B, French JJ, Charnley RM, Manas DM, Wedge SR, Mann J, Robinson SM, and White SA (2020). Therapeutic Strategies Toward Lactate Dehydrogenase Within the Tumor Microenvironment of Pancreatic Cancer. Pancreas 49, 1364–1371. [DOI] [PubMed] [Google Scholar]
- Nachef M, Ali AK, Almutairi SM, and Lee S-H (2021). Targeting SLC1A5 and SLC3A2/SLC7A5 as a Potential Strategy to Strengthen Anti-Tumor Immunity in the Tumor Microenvironment. Front Immunol 12, 624324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nougayrede J, Chagneau CV, Motta J, Bossuet-Greif N, Belloy M, Taieb F, Gratadoux J, Thomas M, Langella P and Oswald E (2021). A Toxic Friend: Genotoxic and Mutagenic Activity of the Probiotic Strain Escherichia coli Nissle 1917. mSphere 6(4):e0062421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura S, Konishi Y, Narukawa M, Sugiura Y, Yoshimoto S, Arai Y, Sato S, Yoshida Y, Tsuju S, Uemura K, et al. (2021). Gut bacteria identified in colorectal cancer patients promote tumorigenesis via butyrate secretion. Nat Commun 12(1):5674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva S, Nardo GD, Ferrari F, Mallardo S, Rossi P, Patrizi G, Cucchiara S, and Stronati L (2012). Randomised clinical trial: the effectiveness of Lactobacillus reuteri ATCC 55730 rectal enema in children with active distal ulcerative colitis. Aliment Pharm Therap 35, 327–334. [DOI] [PubMed] [Google Scholar]
- Overman MJ, Lonardi S, Wong KYM, Lenz H-J, Gelsomino F, Aglietta M, Morse MA, Cutsem EV, McDermott R, Hill A, et al. (2018). Durable Clinical Benefit With Nivolumab Plus Ipilimumab in DNA Mismatch Repair–Deficient/Microsatellite Instability–High Metastatic Colorectal Cancer. J Clin Oncol 36, JCO.2017.76.990. [DOI] [PubMed] [Google Scholar]
- Pang Z, Chong J, Zhou G, de Lima Morais DA, Chang L, Barrette M, Gauthier C, Jacques P-É, Li S, and Xia J (2021). MetaboAnalyst 5.0: narrowing the gap between raw spectra and functional insights. Nucleic Acids Res 49, gkab382-. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleguezuelos-Manzano C, Puschhof J, Huber AR, van Hoeck A, Wood HM, Nomburg J, Gurjao C, Manders F, Dalmasso G, Stege PB, et al. (2020). Mutational signature in colorectal cancer caused by genotoxic pks+E. coli. Nature 580, 269–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramakrishnan SK, Zhang H, Ma X, Jung I, Schwartz AJ, Triner D, Devenport SN, Das NK, Xue X, Zeng MY, et al. (2019). Intestinal non-canonical NFκB signaling shapes the local and systemic immune response. Nat Commun 10, 660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren X, and Kuan P-F (2020). Negative binomial additive model for RNA-Seq data analysis. Bmc Bioinformatics 21, 171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer L, Auchtung TA, Hermans KE, Whitehead D, Borhan B, and Britton RA (2010). The antimicrobial compound reuterin (3-hydroxypropionaldehyde) induces oxidative stress via interaction with thiol groups. Microbiology+ 156, 1589–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, et al. (2009). Introducing mothur: Open-Source, Platform-Independent, Community-Supported Software for Describing and Comparing Microbial Communities▿. Appl Environ Microb 75, 7537–7541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, and Ideker T (2003). Cytoscape: A Software Environment for Integrated Models of Biomolecular Interaction Networks. Genome Res 13, 2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V, Yeoh BS, Chassaing B, Xiao X, Saha P, Olvera RA, Lapek JD, Zhang L, Wang W, Hao S, et al. (2018). Dysregulated Microbial Fermentation of Soluble Fiber Induces Cholestatic Liver Cancer. Cell 175(3): 679–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singhal R, Mitta SR, Das NK, Kerk SA, Sajjakulnukit P, Solanki S, Andren A, Kumar R, Olive KP, Banerjee R, et al. (2021). HIF-2α activation potentiates oxidative cell death in colorectal cancers by increasing cellular iron. J Clin Invest 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M, Chan AT, and Sun J (2020). Influence of the Gut Microbiome, Diet, and Environment on Risk of Colorectal Cancer. Gastroenterology 158, 322–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sridharan GV, Choi K, Klemashevich C, Wu C, Prabakaran D, Pan LB, Steinmeyer S, Mueller C, Yousofshahi M, Alaniz RC, et al. (2014). Prediction and quantification of bioactive microbiota metabolites in the mouse gut. Nat Commun 5, 5492. [DOI] [PubMed] [Google Scholar]
- Sung V, D’Amico F, Cabana MD, Chau K, Koren G, Savino F, Szajewska H, Deshpande G, Dupont C, Indrio F, et al. (2017). Lactobacillus reuteri to Treat Infant Colic: A Meta-analysis. Pediatrics 141, e20171811. [DOI] [PubMed] [Google Scholar]
- Talarico TL, and Dobrogosz WJ (1989). Chemical characterization of an antimicrobial substance produced by Lactobacillus reuteri. Antimicrob Agents Ch 33, 674–679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomkovich S, Dejea CM, Winglee K, Drewes JL, Chung L, Housseau F, Pope JL, Gauthier J, Sun X, Muhlbauer M, et al. (2019). Human colon mucosal biofilms from healthy or colon cancer hosts are carcinogenic. J Clin Invest 129(4):1699–1712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thanikachalam K, and Khan G (2019). Colorectal Cancer and Nutrition. Nutrients 11, 164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilg H, Adolph TE, Gerner RR, and Moschen AR (2018). The Intestinal Microbiota in Colorectal Cancer. Cancer Cell 33, 954–964. [DOI] [PubMed] [Google Scholar]
- Tsherniak A, Vazquez F, Montgomery PG, Weir BA, Kryukov G, Cowley GS, Gill S, Harrington WF, Pantel S, Krill-Burger JM, et al. (2017). Defining a Cancer Dependency Map. Cell 170, 564–576.e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Triner D, Devenport SN, Ramakrishnan SK, Ma X, Frieler RA, Greenson JK, Inohara N, Nunez G, Colacino JA, Mortenson RM, Shah YM (2018) Neutrophils Restrict Tumor-Associate Microbiota to Reduce Growth and Invasion of Colon Tumors in Mice. Gastroenterology 156(5), 1467–1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veiga P, Suez J, Derrien M, and Elinav E (2020). Moving from probiotics to precision probiotics. Nat Microbiol 5, 878–880. [DOI] [PubMed] [Google Scholar]
- Wikoff WR, Anfora AT, Liu J, Schultz PG, Lesley SA, Peters EC, and Siuzdak G (2009). Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc National Acad Sci 106, 3698–3703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirbel J, Pyl PT, Kartal E, Zych K, Kashani A, Milanese A, Fleck JS, Voigt AY, Palleja A, Ponnudurai R, et al. (2019). Meta-analysis of fecal metagenomes reveals global microbial signatures that are specific for colorectal cancer. Nat Med 25, 679–689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yachida S, Mizutani S, Shiroma H, Shiba S, Nakajima T, Sakamoto T, Watanabe H, Masuda K, Nishimoto Y, Kubo M, et al. (2019). Metagenomic and metabolomic analyses reveal distinct stage-specific phenotypes of the gut microbiota in colorectal cancer. Nat Med 25, 968–976. [DOI] [PubMed] [Google Scholar]
- Zhan Y, Chen P-J, Sadler WD, Wang F, Poe S, Núñez G, Eaton KA, and Chen GY (2013). Gut Microbiota Protects against Gastrointestinal Tumorigenesis Caused by Epithelial Injury. Cancer Res 73, 7199–7210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Wang K, Oh J-H, Zhang S, van Pijkeren J-P, Cheng CC, Ren D, Wei H, Gänzle MG, and Walter J (2020). A Phylogenetic View on the Role of Glycerol for Growth Enhancement and Reuterin Formation in Limosilactobacillus reuteri. Front Microbiol 11, 601422. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Table 1: 16S PCoA: Related to Figure 2. PCoA data from 16S sequencing. Includes pcoa1, pcoa2, vector length and size and taxonomy.
Supplemental Table 2: Reuterin RNA-SEQ: Related to Figure 4. Differentially regulated genes from RNA Seq. Log fold change, p value and FDR for reuterin compared to vehicle and DAP compared to vehicle.
Supplemental Table 3: Reuterin Metabolomics: Related to Figure 4. Metabolomics data in HCCT116 and SW480. Shows all assayed compounds, relative amounts, t-test, averages between samples, and the log2 for reuterin over control.
Supplemental Table 4: Reuterin Cysteine Proteomics: Related to Figure 6. Raw cysteine proteomics data including the gene, protein, peptide identified, site position, expression in all vehicle, DMF and reuterin samples, averages in all conditions and competition ratios in all conditions.
Supplemental Table 5: Reuterin Proteomics: Related to Figure 6. Raw proteomics data including t test for significance, the unique peptide number, the p value, the q value, log2fold change, the uniprot ID and the gene symbol.
Supplemental Table 6: Primer Table: Related to Figures 2, 4, 6, and 7. Primer sequences used in the paper.
Supplemental Table 7: Reuterin Synergy: Related to Figures 5 and 6. Coefficients of drug interactions at various doses with reuterin.
Data Availability Statement
16S RNA-seq data has been deposited at GEO and is publicly available at the date of publication. GEO accession number is GSE186880 and is also listed in the key resources table. Microscopy data reported in this paper will be shared by the lead contact upon request. Any additional information required to reanalyzed the data reported in this paper is available from the lead contact upon request.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| HIF-2-alpha Antibody | Bethyl Laboratories | Cat# 179483; |
| Ki67 Antibody | Cell Signaling | Cat#12202T |
| Actin Antibody | Protein Tech | Cat# 60008–1-Ig |
| Cleaved Caspase 3 | Cell Signaling | Cat#9661T |
| Puromycin Antibody | Developmental Studies Hybridoma Bank | Cat# PMY-2A4 |
| 4-HNE Antibody | Bioss Antibodies | Cat# BS-6313R |
| Anti-p-histone H2A | Sigma | Cat#ZMS05636 |
| Bacterial and Virus Strains | ||
| Wildtype L. reuteri | Robert L. Britton | N/A |
| Mutant L. reuteri | Robert L. Britton | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| FG-4592 | Cayman Chemical | Cat#15294 |
| N-acetyl-cysteine | Cayman Chemical | Cat#20261 |
| Sodium Sulfide | Cayman Chemical | Cat#10122555 |
| Cystathionine | Cayman Chemical | Cat# 16061 |
| Olaparib | Cayman Chemical | Cat#10621 |
| CX-5461 | Cayman Chemical | Cat#18392 |
| L-Threonine | Sigma | Cat# 72-19-5 |
| Ammonia Chloride | Sigma | Cat#213330 |
| 5-Fluorouracil | Sigma | Cat#F6627 |
| L-carnitine | Sigma | Cat#21489 |
| Aspartic Acid | Sigma | Cat#1043819 |
| L-Glutamic Acid | Sigma | Cat#G1251 |
| L-Glutamine | Sigma | Cat#G8540 |
| Hydrogen Peroxide | Sigma | Cat#HX0636 |
| L-Buthionine-Sulfozimine | Cayman Chemical | Cat#14484 |
| Glutathione Ethyl Ester | Cayman Chemnical | Cat#14953 |
| Orotic Acid | Sigma | Cat#O2750 |
| Ureidosuccinic Acid | Cayman Chemical | Cat#34126 |
| Deposited Data | ||
| 16S Sequencing | Yachida et. al, 2019 | PMID: 31171880 |
| 16S Sequencing | Chen et. al, 2012 | PMID: 22761885 |
| 16S Sequencing | Cheng Tao Pu, 2020 | PMID: 31609493 |
| 16S Sequencing | GEO | GSE186880 |
| Experimental Models: Cell Lines | ||
| NCM460 | ATCC | N/A |
| HCT116 | ATCC | N/A |
| MC38 | ATCC | N/A |
| CT26 | ATCC | N/A |
| DLD1 | ATCC | N/A |
| SW480 | ATCC | N/A |
| RKO | ATCC | N/A |
| HF27 | ATCC | N/A |
| HF29 | ATCC | N/A |
| MCF10A | ATCC | N/A |
| Vero2 | ATCC | N/A |
| HCN2 | ATCC | N/A |
| NCM356 | ATCC | N/A |
| CRL1831 | ATCC | N/A |
| CCD18Co | ATCC | N/A |
| Experimental Models: Organisms/Strains | ||
| Wildtype: C56Bl/6J | Jackson Labs | N/A |
| SingleMut: CDX2-ERT2Cre; Apcfl/fl | Jackson Labs | N/A |
| DoubleMut: CDX2-ERT2Cre; Apcfl/fl;Trp53fl/ | Jackson Labs | N/A |
| TripleMut: Apcfl/fl;Trp53fl/fl; KrasLSLG12D | Jackson Labs | N/A |
| Oligonucleotides | ||
| List of primers | Supplemental Table 6 | N/A |


