Abstract
The aim of this study was to assess the diversity and genomic variability of Pseudomonas aeruginosa isolates from cystic fibrosis (CF) patients being treated at a university hospital in Brazil. Ninety-seven isolates of P. aeruginosa from 43 CF patients were characterized by macrorestriction analysis of chromosomal DNA by pulsed-field gel electrophoresis (PFGE) and tested for susceptibility to 20 antimicrobial agents by broth microdilution. It was possible to evaluate single isolates from 20 patients and multiple isolates (two to seven) from 23 patients collected during a 22-month period. Among all of the unrelated patients, we detected only one pair of patients sharing a common strain. Among the 77 isolates from 23 patients who had multiple isolates analyzed, we identified 37 major types by PFGE, and five different colonization patterns were recognized. The isolates were susceptible to several antimicrobial agents, although consecutive isolates from the same patient may display differences in their susceptibilities. Mucoid isolates were more resistant (P < 0.001) than nonmucoid isolates to five antibiotics. Our results indicate that CF patients remain colonized by more than one strain of P. aeruginosa for long periods of time. In addition, the finding of several different genotypes in the same patient suggests that the colonizing strain may occasionally be replaced.
When cystic fibrosis (CF) was first described in 1938, 80% of affected babies died within the first year of life (2). The better understanding of the disease and the improvements in treatment conditions in recent years have resulted in a dramatic increase in the median survival time for patients with CF (7). Nowadays, most patients reach adulthood, but Pseudomonas aeruginosa continues to be the most prevalent cause of infections in these patients. This pathogen is one of the main causes of morbidity and mortality in CF patients (3, 8, 9, 11, 13).
It is generally accepted that once colonized with P. aeruginosa, most patients tend to harbor a conserved type (or clone) during the course of the disease (5, 21, 26, 28, 29, 30). Patients may also harbor variants of the original clone, which are characterized by minor differences in genome fingerprints. Conversely, Boukadida et al. (4) found distinct strains within the same patient, indicating that there may be a relatively high heterogeneity of strains of P. aeruginosa in the pulmonary microbiota of chronically infected CF patients. Those authors also found that the emergence of distinct strains in the same CF patient was often associated with previous courses of antibiotic therapy. It therefore appears that the epidemiology of P. aeruginosa in CF patients may vary in different medical centers.
Hospital de Clínicas de Porto Alegre (HCPA) is a 700-bed university hospital located in southern Brazil. HCPA includes a center for treatment of individuals with CF where more than 150 patients have been monitored. The present study aimed to characterize isolates of P. aeruginosa from CF patients being treated at HCPA according to their susceptibility to antimicrobial agents and their DNA macrorestriction profiles.
MATERIALS AND METHODS
Bacterial isolates.
We collected P. aeruginosa isolates from 43 CF patients attending the CF center of HCPA from March 1996 to December 1997. The clinical specimens were processed for qualitative aerobic culture according to conventional diagnostic methods (18). The identification of isolates was performed with the Vitek systems (BioMerieux, Hazelwood, Mo.) using the GNI card. All isolates were subcultured on nutrient agar slants and stored at room temperature before complementary tests (antibiogram and molecular typing).
Antibiogram.
The antimicrobial susceptibilities of isolates to 20 compounds were evaluated by broth microdilution using MicroScan Dried Gram Negative MIC/Combo panels according to NCCLS instructions (19). A standard suspension of the organism was used to inoculate the panels, which were incubated at 35°C for a minimum of 16 h. The MIC was determined as the lowest antimicrobial concentration showing inhibition of growth. The following antimicrobial agents were tested: cefoxitin, ceftizoxime, ceftazidime, cefotaxime, ceftriaxone, cefoperazone, cefpodoxime, cefepime, imipenem, meropenem, ampicillin-sulbactam, ciprofloxacin, aztreonam, amoxicillin-clavulanate, ticarcillin-clavulanate, mezlocillin, sparfloxacin, piperacillin-tazobactam, netilmicin, and azlocillin. P. aeruginosa ATCC 27853 was used as the quality control strain. The results were interpreted according to the NCCLS criteria (20).
Seven of the 20 antimicrobial agents tested were chosen to establish the antibiotype of each isolate: ceftazidime, cefepime, imipenem, ciprofloxacin, netilmicin, aztreonam, and piperacilin-tazobactam. These compounds were selected because they represent the different classes of antimicrobial agents and because they are commonly used for the treatment of P. aeruginosa infections. Differences between the results from susceptible to resistant and from resistant to susceptible with at least one of these seven antimicrobial agents characterized a different antibiotype, which was represented by a capital letter. Differences in the results from susceptible to intermediate or from intermediate to susceptible characterized a variation of that antibiotype (subtype), and an arabic number was added to the capital letter. The MIC at which 50% of the isolates tested are inhibited (MIC50), MIC90, and percent susceptibility for these compounds were also calculated.
The isolates were classified according to their colonial morphology on blood agar as mucoid or nonmucoid, and the rates of susceptibility to each antimicrobial agent of these two groups were compared using the Fisher exact test (10).
Molecular typing.
All isolates were characterized by macrorestriction analysis of chromosomal DNA by pulsed-field gel electrophoresis (PFGE) (24). Genomic DNA inserts were digested at 37°C overnight (12 to 16 h) with 10 U of SpeI enzyme (New England Biolabs, Inc., Beverly, Mass.). Electrophoresis was performed in a CHEF-DRII apparatus (Bio-Rad, Richmond, Calif.) with the following conditions: 0.5 Tris-borate-EDTA, 1% agarose, 13°C, and 200 V. The electrophoresis was run for 24 h, and the switch interval was ramped from 5 to 90 s (24). Photographs of ethidium bromide-stained gels were examined visually. Isolates were considered distinct strains if there were more than six fragment (band) differences between PFGE profiles. Isolates were considered related (subtypes) if there were only two to six band differences between PFGE profiles. Isolates with the same PFGE profile were considered indistinguishable (31).
RESULTS
A total of 97 clinical isolates of P. aeruginosa from 43 CF sputum samples were obtained between March 1996 and December 1997. The carbapenens meropenem (96.9% susceptibility; MIC90, 2 μg/ml) and imipenem (93.8% susceptibility; MIC90, 4 μg/ml) were the most active compounds against the P. aeruginosa isolates evaluated in this study. Piperacillin-tazobactam (85.6% susceptibility), ticarcillin-clavulanate (83.5% susceptibility), and ceftazidime (81.4% susceptibility) also displayed reasonable in vitro antimicrobial activity. The results of MIC50, MIC90, and susceptibility rate determinations were classified according to colonial morphology (mucoid and nonmucoid) of the isolates (Table 1). The nonmucoid isolates were significantly more susceptible (P < 0.001) to cefoperazone, cefepime, ciprofloxacin, aztreonam, and sparfloxacin (Table 1). The nonmucoid isolates also displayed higher rates of susceptibility than the mucoid isolates to ceftazidime, imipenem, ticarcillin-clavulanate, piperacillin-tazobactam, and meropenem, but with no statistical significance.
TABLE 1.
MIC50s, MIC90s, and percent antimicrobial susceptibilities of 97 samples of P. aeruginosaa
| Antimicrobial agent | Breakpointsb (μg/ml)
|
Mucoid strains (n = 40)
|
Nonmucoid strains (n = 57)
|
P (Fisher exact test) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| S | I | R | MIC50 (μg/ml) | MIC90 (μg/ml) | % Susceptibility | MIC50 (μg/ml) | MIC90 (μg/ml) | % Susceptibility | ||
| Ceftizoximec | ≤8 | 16–32 | ≥64 | >32 | >32 | 7.5 | 32 | >32 | 5.3 | 0.65 |
| Ceftazidime | ≤8 | 16 | ≥32 | 4 | >32 | 70.0 | 2 | 16 | 89.5 | 0.096 |
| Cefotaximec | ≤8 | 16–32 | ≥64 | 32 | >64 | 25.0 | 32 | >64 | 14.0 | 0.173 |
| Ceftriaxonec | ≤8 | 16–32 | ≥64 | 32 | >64 | 27.5 | 64 | >64 | 21.0 | 0.464 |
| Cefoperazon | ≤16 | 32 | ≥64 | 16 | >32 | 57.5 | 3 | >32 | 79.0 | 0.00086d |
| Cefepime | ≤8 | 16 | ≥32 | 16 | >16 | 42.5 | 4 | 16 | 83.0 | 0.000047d |
| Imipenem | ≤4 | 8 | ≥16 | 1 | 4 | 90.0 | 1 | 2 | 96.5 | 0.19 |
| Ampicillin-sulbactamc | ≤8–4 | 16–8 | ≥32–16 | >32–16 | >32–16 | 5.0 | >32–16 | >32–16 | 1.7 | 0.365 |
| Ciprofloxacin | ≤1 | 2 | ≥4 | 1 | >4 | 50.0 | ≤0.25 | 2 | 86.0 | 0.00012d |
| Aztreonam | ≤8 | 16 | ≥32 | 8 | >32 | 50.0 | 4 | 32 | 84.0 | 0.00031d |
| Amoxicillin-clavulanatec | ≤8–4 | 16–8 | ≥32–16 | >32–16 | >32–16 | 2.5 | >32–16 | >32–16 | 1.7 | 0.800 |
| Ticarcillin-clavulanate | ≤64–2 | ≥128–2 | ≤16 | 128 | 87.5 | ≤16 | 128 | 80.7 | 0.376 | |
| Mezlocillin | ≤64 | ≥128 | 64 | >128 | 52.5 | 32 | >128 | 70.2 | 0.077 | |
| Sparfloxacin | ≤1 | 2 | ≥4 | 2 | >2 | 30.0 | 1 | >2 | 68.4 | 0.00020d |
| Piperacillin-tazobactam | ≤64–4 | ≥128–4 | ≤8 | >64 | 80.0 | ≤8 | 64 | 89.5 | 0.19 | |
| Netilmicin | ≤8 | 16 | ≥32 | 8 | >16 | 50.0 | 8 | >16 | 56.0 | 0.55 |
| Meropenem | ≤4 | 8 | ≥16 | ≤1 | 2 | 95.0 | ≤1 | 2 | 98.0 | 0.365 |
| Azlocillin | ≤64 | ≥128 | ≤64 | >65 | 62.5 | ≤64 | >64 | 84.2 | 0.015 | |
All samples were resistant to cefoxitin and cefpodoxime.
Based on interpretive breakpoints as indicated in NCCLS document M100-S10 (M7) (20) S. susceptible; I, intermediate; R, resistant.
Drug not indicated for treatment of P. aeruginosa infections, even with susceptibility (19).
The difference in the susceptibility rates between the mucoid and nonmucoid groups was considered statistically significant.
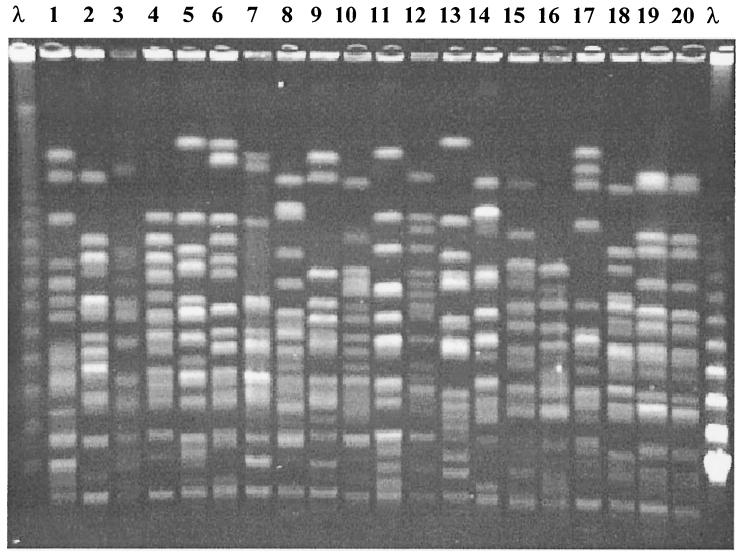
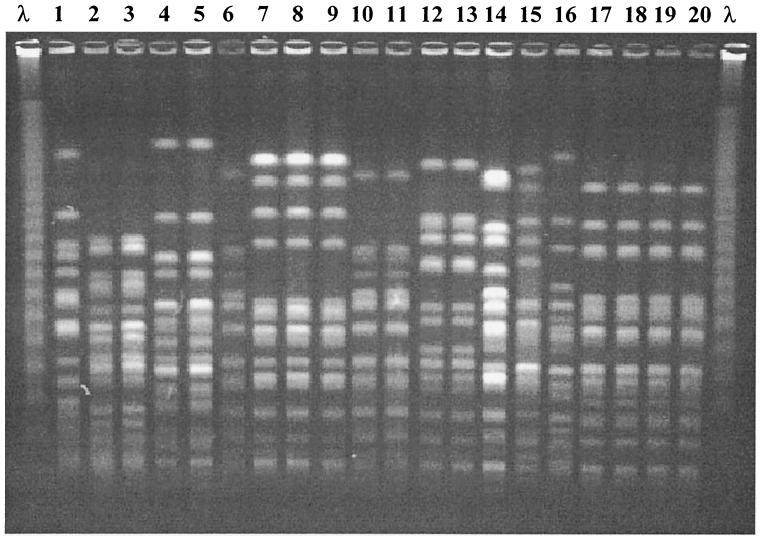
The isolates were separated into two different groups: group 1, which was composed of 20 isolates from 20 patients, and group 2, which included 77 isolates from 23 patients (two to seven isolates from each patient). A total of 19 distinct strains (major PFGE profiles) were observed in group 1 isolates. Only two unrelated patients were colonized with indistinguishable strains of P. aeruginosa in this group (Fig. 1). On the other hand, it was possible to identify 37 major PFGE profiles among the isolates of group 2, indicating a considerable genomic diversity of P. aeruginosa from different patients (Fig. 2). Two patients (siblings) of group 2 shared indistinguishable strains of P. aeruginosa. In total, 13 isolates of these two patients were typed, and 12 of them displayed the same PFGE pattern.
FIG. 1.
PFGE of 20 clinical isolates of P. aeruginosa from 20 different patients. Lane λ, lambda ladder (48.5 kb). Lanes 1 to 20, clinical isolates of P. aeruginosa from 20 different patients.
FIG. 2.
Genotypes of P. aeruginosa isolates from six CF patients. Lanes 1, 2, and 3, patient 9; lanes 4, 5, and 6, patient 10; lanes 7, 8, and 9, patient 17; lanes 10, 11, 12, and 13, patient 6; lanes 14, 15, and 16, patient 5; lanes 17, 18, 19, and 20, patient 14; lane λ, lambda ladder (48.5 kb).
It was possible to obtain multiple isolates from the 23 patients of group 2 over a period of time of up to 17 months. Thirteen patients (56%) harbored at least two distinct strains over their follow-up period, although most of these patients tended to harbor a single strain (genotype) over many months (persistence of genotype) (Table 2). P. aeruginosa of antibiotype A (susceptible to ceftazidime, ticarcillin-clavulanate, piperacillin-tazobactam, imipenem, and meropenem) or its subtypes was seen in 14 patients, but no association between genotype and antibiotype was observed, as isolates of the same genotype displayed different antibiotypes and vice versa (Table 2).
TABLE 2.
Distribution of P. aeruginosa isolates from the 23 patients of group 2 according to genome macrorestriction type (genotype) and antimicrobial susceptibility profile (antibiotype)
| Patient | No. of isolates | Follow-up period (mo) | Genotype (SpeI)a | No. of isolates with the same genotype | Persistence of genotype (mo) | Antibiotypeb |
|---|---|---|---|---|---|---|
| 1 | 4 | 9 | 1 | 1 | A | |
| 2 | 1 | B1 | ||||
| 3 | 1 | A | ||||
| 4 | 1 | A1 | ||||
| 2 | 3 | 9 | 5a | 2 | 1 | C1, C2 |
| 5b | 1 | C | ||||
| 3 | 3 | 10 | 6 | 1 | C3 | |
| 7 | 1 | A | ||||
| 8 | 1 | A | ||||
| 4 | 3 | 8 | 9 | 3 | 8 | A |
| 5 | 3 | 12 | 10 | 1 | A | |
| 11 | 1 | A | ||||
| 12 | 1 | A | ||||
| 6 | 4 | 14 | 13 | 2 | 14 | B |
| 14 | 2 | 2 | A, A2 | |||
| 7 | 3 | 9 | 15 | 3 | 9 | A1 |
| 8 | 3 | 11 | 16 | 1 | B2 | |
| 17 | 2 | 3 | A, D3 | |||
| 9 | 3 | 12 | 18 | 1 | A | |
| 19 | 2 | 2 | A | |||
| 10 | 3 | 9 | 20 | 2 | 9 | A1, E |
| 21 | 1 | A | ||||
| 11 | 6 | 17 | 22 | 6 | 17 | D1, D2 |
| 12 | 7 | 19 | 22 | 6 | 19 | D, D3, F, G |
| 23 | 1 | G | ||||
| 13 | 2 | 3 | 24 | 2 | 3 | H |
| 14 | 4 | 16 | 25 | 4 | 16 | I, F1 |
| 15 | 2 | 13 | 35 | 1 | J | |
| 36 | 1 | K | ||||
| 16 | 5 | 11 | 26 | 1 | A | |
| 27 | 4 | 11 | A | |||
| 17 | 3 | 3 | 28 | 3 | 3 | L, M, M1 |
| 18 | 2 | 4 | 29 | 2 | 4 | A |
| 19 | 3 | 3 | 30 | 3 | 3 | A1, A, B |
| 20 | 3 | 11 | 31a | 2 | 7 | N |
| 31b | 1 | O | ||||
| 21 | 2 | 0 | 32 | 2 | A1, A3 | |
| 22 | 2 | 2 | 33 | 1 | M2 | |
| 37 | 1 | P | ||||
| 23 | 4 | 3 | 34 | 4 | 3 | A4, Q |
Isolates of the same major PFGE pattern are represented by arabic numbers; a letter following the arabic number represents a subtype.
Isolates of the same antibiotype are represented by capital letters; a number following the capital letter represents a subantibiotype.
We identified 18 patients harboring the same strain over different periods of time, and 12 of them were colonized with P. aeruginosa displaying the same resistance profile. In the remaining six patients the subsequent isolates displayed higher rates of antimicrobial resistance than the initial isolate.
Twelve patients were colonized with both mucoid and nonmucoid isolates, which were compared by molecular typing. Five patients presented mucoid and nonmucoid isolates with distinct PFGE profiles, while seven patients harbored indistinguishable isolates regardless of colonial morphology. Among these seven patients, a change of the colonies from nonmucoid to mucoid was observed in five patients, and a change from mucoid to nonmucoid was observed in two patients.
DISCUSSION
Analysis of the antimicrobial susceptibilities of isolates showed that high rates of resistance to antimicrobial agents indicated for the treatment of CF P. aeruginosa occurred, especially among mucoid isolates. However, the carbapenens (imipenem and meropenem) remained very active against both mucoid and nonmucoid isolates, with susceptibility rates of >90%. The P. aeruginosa strains evaluated in the present study showed higher rates of susceptibility than P. aeruginosa strains from CF patients evaluated in other studies (6, 27). We also observed that mucoid isolates showed a tendency for higher rates of resistance (P < 0.001) than nonmucoid isolates to several antimicrobial agents, including cefperazone, cefepime, aztreonam, sparfloxacin, and ciprofloxacin (23).
Despite the great number of studies on the epidemiology of P. aeruginosa infection in CF (1, 4, 5, 12, 14, 15, 16, 22, 26, 30), no data have been published on the colonization or infection of Brazilian CF patients with P. aeruginosa. We evaluated 97 isolates of P. aeruginosa from 43 CF patients, and we found 39 distinct PFGE patterns among isolates from 41 patients that were not epidemiologically related. Only two nonrelated patients shared indistinguishable isolates. On the other hand, we also found that 12 out of 13 isolates obtained from two CF siblings (closely related patients) displayed the same PFGE pattern. Similarly to other reports, these results suggest that cross-infection of P. aeruginosa among CF patients would occur more frequently when contact was more intense (12).
Our study also documented the permanence of a single genotype in a patient over time. We observed the presence of the original strain isolated two to six times over 22 months in 18 patients. Ten of these 18 patients harbored a single genotype with a conserved macrorestriction pattern over time (3 to 17 months). These results suggest that even during periods of antibiotic therapy, the original strain is not eradicated from these patients. Another interesting finding was the presence of variations of the same genotype (subtypes) of P. aeruginosa in two single patients, as published by other authors (30).
The remaining six patients carried two totally different genotypes, and usually each one of these strains was isolated from different specimens and on different days (Table 2). A mixed colonization of different clones of P. aeruginosa can explain this finding. The clone that predominates would vary over time, with one clone predominating in one period and another clone predominating in another period. This event suggests that P. aeruginosa infection and colonization of the CF patient's lung is a very dynamic process, as proposed by Renders et al. (25).
The most relevant result of our study was the finding of several different genotypes in five patients of group 2 (21.7%). These patients had two to four distinct strains (distinct major PFGE patterns) isolated during the follow-up period (2 to 13 months). All PFGE patterns were detected only once in these subgroups of patients. In contrast to other studies (1, 5, 12, 14, 15, 16, 26, 30), these results suggest that the colonizing strain may occasionally be replaced. Boukadida et al. (4) also found different strains in the same patient and observed that the emergence of distinct strains in the same CF patient was often associated with periods of antibiotic therapy.
Among the 97 clinical isolates of P. aeruginosa included in our study, 40 presented the mucoid appearance, confirming that this phenotypic alteration is also very common among P. aeruginosa strains from Brazilian CF patients (13). Moreover, it was possible to determine that mucoid and nonmucoid isolates were indistinguishable by PFGE in 7 out of 12 patients colonized with these colonial variants. This finding confirmed the clonal relationship between mucoid and nonmucoid isolates of P. aeruginosa from the same CF patient, as previously demonstrated (17, 26, 32).
In conclusion, the results of this study indicate that in spite of the fact that the CF patients had received courses of antibiotic therapy periodically, nonmucoid P. aeruginosa strains isolated from their lung showed relatively low rates of antimicrobial resistance. Among unrelated patients, we detected only one pair of patients sharing isolates with identical PFGE patterns, and we concluded that cross-infection is not common in our population. Our results also indicate that CF patients remain colonized by more than one strain of P. aeruginosa for long periods of time. In addition, the finding of several isolates with distinct genotypes in the same patient suggests that the colonizing strain may occasionally be replaced.
ACKNOWLEDGMENTS
We thank the Unidade de Microbiologia and the Cystic Fibrosis Division, HCPA, for assistance in collecting and processing the specimens for our study. We also thank the Molecular Epidemiology and Fungus Testing Lab (Pathology Department, University of Iowa) for assistance with the molecular techniques.
This work was supported by Fundação de Amparo a Pesquisa do Estado de São Paulo (FAPESP) (grant 97/02795-6) and Fundo de Incentivo a Pesquisa e Ensino from HCPA.
REFERENCES
- 1.Agodi A, Sciacca A, Campanile F, Messina C, Barchitta M, Sciacca S, Stefani S. Molecular epidemiology of Pseudomonas aeruginosa from cystic fibrosis in Sicily: genome macrorestriction analysis and rapid PCR-ribotyping. New Microbiol. 2000;23:319–327. [PubMed] [Google Scholar]
- 2.Andersen D H. Cystic fibrosis of the pancreas and its relation to celiac disease. A clinical and pathological study. Am J Dis Child. 1938;56:344–399. [Google Scholar]
- 3.Barth A L, Pitt T L. Microbial pathogens associated with cystic fibrosis: special focus on Pseudomonas aeruginosa. Braz J Infect Dis. 1998;2:43–61. [PubMed] [Google Scholar]
- 4.Boukadida J, De Montalembert M, Lenoir G, Scheinmann P, Véron M, Berche P. Molecular epidemiology of chronic pulmonary colonization by Pseudomonas aeruginosa in cystic fibrosis. J Med Microbiol. 1993;38:29–33. doi: 10.1099/00222615-38-1-29. [DOI] [PubMed] [Google Scholar]
- 5.Burns J L, Gibson R L, McNamara S, Yim D, Emerson J, Rosenfeld M, Hiatt P, McCoy K, Castile R, Smith A L, Ramsey B W. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J Infect Dis. 2001;183:444–452. doi: 10.1086/318075. [DOI] [PubMed] [Google Scholar]
- 6.Byrne S, Maddison J, Connor P, Doughty I, Dodd M, Jenney M. Clinical evaluation of meropenem versus ceftazidime for the treatment of Pseudomonas spp. infections in cystic fibrosis patients. J Antimicrob Chemother. 1995;36:135–143. doi: 10.1093/jac/36.suppl_a.135. [DOI] [PubMed] [Google Scholar]
- 7.Fiel S B, Fitzsimmons S C, Shidlow D. Evolving demographics of cystic fibrosis. Semin Respir Crit Care Med. 1994;15:349–355. [Google Scholar]
- 8.Fitzsimmons S C. The changing epidemiology of cystic fibrosis. J Pediatr. 1995;122:1–9. doi: 10.1016/s0022-3476(05)83478-x. [DOI] [PubMed] [Google Scholar]
- 9.Gilligan P H. Microbiology of airway disease in patients with cystic fibrosis. Clin Microbiol Rev. 1991;4:35–51. doi: 10.1128/cmr.4.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Glantz S A. Primer of biostatistics. San Francisco, Calif: McGraw-Hill; 1997. pp. 108–150. [Google Scholar]
- 11.Govan J R W, Nelson J W. Microbiology of cystic fibrosis lung infections: themes and issues. J R Soc Med. 1993;86:11–18. [PMC free article] [PubMed] [Google Scholar]
- 12.Grothues D, Koopmann U, Von Der Hardt H, Tümmler B. Genome fingerprinting of Pseudomonas aeruginosa indicates colonization of cystic fibrosis siblings with closely related strains. J Clin Microbiol. 1988;26:1973–1977. doi: 10.1128/jcm.26.10.1973-1977.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Høiby N. Prevalence of mucoid strains of Pseudomonas aeruginosa in bacteriological specimens from patients with cystic fibrosis and patients with other diseases. Acta Pathol Microbiol Scand Sect B. 1975;83:549–552. [PubMed] [Google Scholar]
- 14.Høiby N, Pedersen S S. Estimated risk of cross-infection with Pseudomonas aeruginosa in Danish cystic fibrosis patients. Acta Paediatr Scand. 1989;78:395–404. doi: 10.1111/j.1651-2227.1989.tb11099.x. [DOI] [PubMed] [Google Scholar]
- 15.Hoogkamp-Korstanje J A A, Meis J F G, Kissing J, Van Der Laag J, Melchers W J G. Risk of cross-colonization and infection by Pseudomonas aeruginosa in a holiday camp for cystic fibrosis patients. J Clin Microbiol. 1995;33:572–575. doi: 10.1128/jcm.33.3.572-575.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunfeld K P, Schmidt C, Krackhardt B, Posselt H G, Bargon J, Yahaf Y, Schafer V, Brade V, Wichelhaus T A. Risk of Pseudomonas aeruginosa cross-colonisation in patients with cystic fibrosis within a holiday camp—a molecular epidemiological study. Wien Klin Wochenschr. 2000;112:329–333. [PubMed] [Google Scholar]
- 17.Loutit J S, Tompkins L C. Restriction enzyme and Southern hybridization analysis of Pseudomonas aeruginosa strains from patients with cystic fibrosis. J Clin Microbiol. 1991;29:2897–2900. doi: 10.1128/jcm.29.12.2897-2900.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: American Society for Microbiology; 1999. [Google Scholar]
- 19.National Committee for Clinical Laboratory Standards. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standard M7–A4. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1997. [Google Scholar]
- 20.National Committee for Clinical Laboratory Standards. MIC testing—supplemental tables. Approved standard M100–S10 (M7). Wayne, Pa: National Committee for Clinical Laboratory Standards; 2000. [Google Scholar]
- 21.Ogle J W, Janda J M, Woods D E, Vasil M L. Characterization and use of a DNA probe as an epidemiological marker for Pseudomonas aeruginosa. J Infect Dis. 1987;155:119–126. doi: 10.1093/infdis/155.1.119. [DOI] [PubMed] [Google Scholar]
- 22.Ojeniyi B, Frederiksen B, Hoiby N. Pseudomonas aeruginosa cross-infection among patients with cystic fibrosis during a winter camp. Pediatr Pulmonol. 2000;29:177–181. doi: 10.1002/(sici)1099-0496(200003)29:3<177::aid-ppul4>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 23.Pedersen S S, Kharazmi A, Espersen F, Høiby N. Pseudomonas aeruginosa alginate in cystic fibrosis sputum and the inflammatory response. Infect Immun. 1990;58:3363–3368. doi: 10.1128/iai.58.10.3363-3368.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pfaller M A, Hollis R J, Sader H S. Molecular biology—PFGE analysis of chromosomal restriction fragments. In: Isenberg H D, editor. Clinical microbiology procedures handbook. American Society for Microbiology, Washington, D.C. 1992. pp. 10.5.c.1–10.5.c.11. [Google Scholar]
- 25.Renders N H M, Sijmons M A F, Van Belkum A, Overbeek S E, Mouton J W, Verbrugh H A. Exchange of Pseudomonas aeruginosa strains among cystic fibrosis siblings. Res Microbiol. 1997;148:447–454. doi: 10.1016/s0923-2508(97)83875-2. [DOI] [PubMed] [Google Scholar]
- 26.Römling U, Fiedler B, Bobhammer J. Epidemiology of chronic Pseudomonas aeruginosa infections in cystic fibrosis. J Infect Dis. 1994;170:1616–1621. doi: 10.1093/infdis/170.6.1616. [DOI] [PubMed] [Google Scholar]
- 27.Saiman L, Mehar F, Niu W W, Neu H C, Shaw K J, Miller G, Prince A. Antibiotic susceptibility of multiply resistant Pseudomonas aeruginosa isolated from patients with cystic fibrosis, including candidates for transplantation. Clin Infect Dis. 1996;23:532–537. doi: 10.1093/clinids/23.3.532. [DOI] [PubMed] [Google Scholar]
- 28.Speert D P, Campbell S W, Farmer K, Volpel K, Joffe A M, Paranchych W. Use of a pilin gene probe to study molecular epidemiology of Pseudomonas aeruginosa. J Clin Microbiol. 1989;27:2589–2593. doi: 10.1128/jcm.27.11.2589-2593.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Speert D P. Pseudomonas aeruginosa infections in patients with cystic fibrosis. In: Baltch A L, Smith R P, editors. Pseudomonas aeruginosa: infections and treatment. New York, N.Y: Marcel Dekker Inc.; 1994. pp. 183–215. [Google Scholar]
- 30.Struelens M J, Schwam V, Deplano A, Baran D. Genome macrorestriction analysis of diversity and variability of Pseudomonas aeruginosa strains infecting cystic fibrosis patients. J Clin Microbiol. 1993;31:2320–2326. doi: 10.1128/jcm.31.9.2320-2326.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tenover F C, Arbeit R D, Goering R V, Mickelsen P A, Murray B E, Persing D H, Swaminathan B. Interpreting chromosomal DNA restriction patterns produced by pulsed-field gel electrophoresis: criteria for bacterial strain typing. J Clin Microbiol. 1995;33:2233–2239. doi: 10.1128/jcm.33.9.2233-2239.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolz C, Kiosz G, Ogle J W. Pseudomonas aeruginosa cross-colonisation and persistence in patients with cystic fibrosis. Use of a DNA probe. Epidemiol Infect. 1989;102:205–214. doi: 10.1017/s0950268800029873. [DOI] [PMC free article] [PubMed] [Google Scholar]