Abstract
Coronavirus disease 2019 (COVID-19) has become a new public health crisis threatening the world. Dysregulated immune responses are the most striking pathophysiological features of patients with severe COVID-19, which can result in multiple-organ failure and death. The cytochrome P450 (CYP) system is the most important drug metabolizing enzyme family, which plays a significant role in the metabolism of endogenous or exogenous substances. Endogenous CYPs participate in the biosynthesis or catabolism of endogenous substances, including steroids, vitamins, eicosanoids, and fatty acids, whilst xenobiotic CYPs are associated with the metabolism of environmental toxins, drugs, and carcinogens. CYP expression and activity are greatly affected by immune response. However, changes in CYP expression and/or function in COVID-19 and their impact on COVID-19 pathophysiology and the metabolism of therapeutic agents in COVID-19, remain unclear. In this analysis, we review current evidence predominantly in the following areas: firstly, the possible changes in CYP expression and/or function in COVID-19; secondly, the effects of CYPs on the metabolism of arachidonic acid, vitamins, and steroid hormones in COVID-19; and thirdly, the effects of CYPs on the metabolism of therapeutic COVID-19 drugs.
Keywords: COVID-19, SARS-CoV-2, CYP = cytochrome P450, inflammation, drug metabolism
1 Introduction
Coronavirus disease 2019 (COVID-19), which is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), has become a global challenge. As of December 29, 2021, there have been over 281 million confirmed cases of COVID-19, including more than five million deaths, reported to the WHO (WHO, 2021). Numerous SARS-CoV-2 variants have been detected around the world. Many SARS-CoV-2 variants are more infectious than original wild strain, which have brought new challenges to the prevention and control of COVID-19 (Tian et al., 2021). Dysregulated immune response, particularly cytokine storm, is a prominent feature of COVID-19, which can result in multiple-organ failure and death. The cytochrome P450 (CYP) enzymes form a large family of heme-containing enzymes that catalyze the metabolism of a variety of chemical compounds, and play a significant role in the metabolism of endogenous or exogenous substances. CYP expression and activity are greatly affected by immune mediators, such as interleukin (IL)-6, tumor necrosis factor (TNF)-α, IL-1, and interferon (IFN)-γ. However, changes in CYP expression and/or function in COVID-19 and their impact on its pathophysiology, and on the metabolism of therapeutic agents in COVID-19 remain unclear. This review focuses on the involvement of CYPs in the pathophysiology and pharmacotherapeutics of COVID-19.
2 Pathophysiological Characteristics of COVID-19
Dysregulated immune response is the most striking pathophysiological feature in severe COVID-19 patients; characterized by cytokine storm and lymphopenia; resulting in acute respiratory distress syndrome (ARDS), multiple-organ failure, and even death. SARS-CoV-2 may activate both innate and adaptive immune responses in patients, including lymphopenia, cytokine storm, and abnormal activation of macrophages and their complementary system (Qin et al., 2020; Tan et al., 2020; Xu et al., 2020; Jamal et al., 2021). Severe COVID-19 patients commonly exhibit a hyperinflammatory state referred to as cytokine storm, marked by elevation of IL-2, IL-4, IL-6, TNF-α, and IFN-γ (Copaescu et al., 2020; Hu et al., 2021; Paniri and Akhavan_Niaki, 2020; Qin et al., 2020; Zhang et al., 2021a). Elevated IL-6 concentration was shown to be associated with detectable serum SARS-CoV-2 RNA in patients with COVID-19 (Chen et al., 2020). A number of studies highlighted that elevation of IL-6 levels was correlated with adverse outcomes in SARS-CoV-2 infection, defined as severe COVID-19 occurrence, the requirement for mechanical ventilation, and death (Copaescu et al., 2020; Gao et al., 2020; Ruan et al., 2020; Belaid et al., 2021; Potere et al., 2021). IL-6 and IL-1 blockade may be associated with clinical improvement in patients with COVID-19 (Cavalli et al., 2020; Pinzon et al., 2021).
3 Cytochrome P450 Enzymes
The CYP system is the most important drug metabolizing enzyme family existing amongst species, and plays a role in the metabolism of endogenous and exogenous substances (Stipp and Acco, 2021). CYP enzymes are located mainly within intestinal and hepatic tissues, but are also present in the skin, lung and kidneys etc. (He and Feng, 2015) There are 18 mammalian CYP families, located in the endoplasmic reticulum, or in mitochondrial membranes, which encode 57 genes in the human genome (Nebert et al., 2013; Korobkova, 2015; Stipp and Acco, 2021). CYP nomenclature reflects the characteristic absorption spectrum of the reduced enzyme at 450 nm, and the enzyme designation consists of a number-letter-number sequence on the basis of amino acid sequence homology (Figure 1).
FIGURE 1.
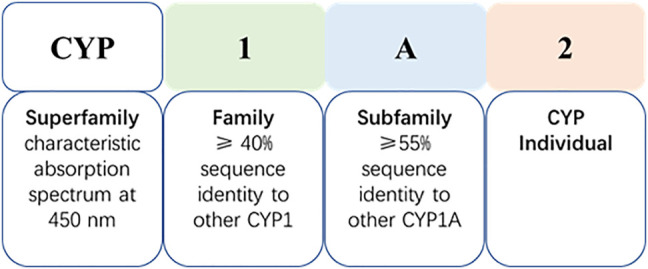
Cytochrome P450 nomenclature (take CYP1A2 as an example).
CYPs are classified into two categories: endogenous (CYP family 7–51) and xenobiotic (CYP family 1–4) (Fan et al., 2016; Stipp and Acco, 2021). Endogenous CYPs participate in the biosynthesis, or catabolism of endogenous substance, whilst xenobiotic CYPs are associated with the metabolism of environmental toxins, drugs, and carcinogens. CYP1, CYP2, and CYP3 account for ∼75% of enzymes involved in the metabolism of all clinical use drugs and other xenobiotics (Guengerich, 2008; Stipp and Acco, 2021), whilst CYP4 is involved in eicosanoid metabolism. Nevertheless, several drug-metabolizing CYPs are also involved in the metabolism of endogenous compounds, such as CYP3A4 and CYP3A5.
CYP3A4, CYP2C9, CYP2C8, CYP1A2, and CYP2E1 are highly expressed in the liver, whilst, CYP2A6, CYP2D6, CYP2B6, CYP2C19, and CYP3A5 are less abundant in the liver. CYP2J2, CYP1A1, and CYP1B1 are mainly expressed extrahepatically (Zanger and Schwab, 2013; Stipp and Acco, 2021).
4 Changes in Cytochrome P450 Expression and/or Function in COVID-19
CYP gene expression is regulated by the activation of several nuclear receptors, including constitutive androstane receptor (CAR), pregnane X receptor (PXR) and aryl hydrocarbon receptor (AhR) (Moriya et al., 2012; Stipp and Acco, 2021). CYP expression and activity are also thought to be affected by multiple factors such as hormone levels, and environment, as well as pathological conditions such as infection, inflammation, and cancer (Morgan, 2009; Zanger and Schwab, 2013; Morgan, 2017; Esteves et al., 2021). Previous studies have shown that viral infection, inflammatory mediators and hepatic injury may affect the expression and activity of some CYPs, which are prevalent in COVID-19. Therefore, we attempted to explore the changes of CYPs in expression and/or function in COVID-19 patients.
4.1 Virus Infection
Until now, no studies have focused on the effects of SARS-CoV-2 on the expression and activity of CYPs. However, previous studies have found that CYPs expression changed in several viral infections. CYP1A1 activity was suppressed by 75% in coxsackievirus B3 infected mice (Funseth et al., 2002). CYP3A4 activity was suppressed in primary hepatocytes infected with adenovirus, and adenovirus-induced modification of PXR may be responsible for changes in hepatic CYP3A4 activity (Wonganan et al., 2014). CYP2A5 and CYP3A expression increased in hepatitis B virus (HBV)-transgenic mice (Kirby et al., 1994), whilst CYP2D6 expression decreased in hepatitis C virus (HCV) infected mice (Kikuchi et al., 2010).
4.2 Cytokines
Expression of CYPs is markedly regulated during inflammatory processes. In vitro, CYPs were regulated (nearly all down-regulation) by multiple cytokine treatments, including IL-6, TNF-α, IFN-γ, TGF-β and IL-1β (Table 1).
TABLE 1.
The effect of cytokines on CYPs expression.
| Cytokine | CYPs | Effects on CYPs | mRNA or protein or activity | Condition | Studies |
|---|---|---|---|---|---|
| IL-6 | CYP1A1 | ↓ | mRNA and protein | Human HepG2 hepatoma cells | Fukuda and Sassa (1994) |
| CYP1A2 | ↓ | mRNA | Hepatoma cells (HepG2, HepG2f and Hep3B) | Fukuda et al. (1992) | |
| Turpentine-induced aseptic inflammation in IL-6-deficient mice | Siewert et al. (2000) | ||||
| Human HepaRG hepatoma cell line | Rubin et al. (2015) | ||||
| ↓ | Activity | Human HepaRG hepatoma cell line | Rubin et al. (2015) | ||
| ↑ | Activity | Clinical study in patients with active rheumatoid arthritis | Zhuang et al. (2015) | ||
| CYP2A5 | ↓ | mRNA | Turpentine-induced aseptic inflammation in IL-6-deficient mice | Siewert et al. (2000) | |
| CYP2A12 | ↓ | Activity | IL-6 knockout mice after LPS administration | Warren et al. (2001) | |
| CYP2B6 | ↓ | mRNA | Human primary hepatocytes | Aitken and Morgan (2007) | |
| Human HepaRG hepatoma cell line | Rubin et al. (2015) | ||||
| ↓ | Activity | Human HepaRG hepatoma cell line | Rubin et al. (2015) | ||
| CYP2C8 | ↓ | mRNA | Human primary hepatocytes | Aitken and Morgan (2007) | |
| CYP2C9 | ↓ | mRNA | Human primary hepatocytes | Aitken and Morgan (2007) | |
| Activity | Clinical study in patients with active rheumatoid arthritis | Zhuang et al. (2015) | |||
| CYP2C19 | ↓ | mRNA | Human primary hepatocytes | Aitken and Morgan (2007) | |
| Activity | Clinical study in patients with active rheumatoid arthritis | Zhuang et al. (2015) | |||
| CYP2E1 | ↓ | mRNA | Human primary hepatocytes | Abdel-Razzak et al. (1993) | |
| CYP3 subfamily | ↓ | Activity | Clinical study in patients with active rheumatoid arthritis | Zhuang et al. (2015) | |
| CYP3A3 | ↓ | mRNA | Hepatoma cells (HepG2, HepG2f and Hep3B) | Fukuda et al. (1992) | |
| CYP3A4 | ↓ | mRNA | Both HepG2 and Caco-2 cells | Enokiya et al. (2021) | |
| Human primary hepatocytes | Aitken and Morgan (2007) | ||||
| Activity | Clinical study in patients with rheumatoid arthritis | Lee et al. (2017) | |||
| Clinical study in patients with rheumatoid arthritis | Schmitt et al. (2011) | ||||
| Human HepaRG hepatoma cell line | Rubin et al. (2015) | ||||
| CYP3A5 | ↓ | mRNA | Both HepG2 and Caco-2 cells | Enokiya et al. (2021) | |
| CYP3A11 | ↓ | mRNA | Turpentine-induced aseptic inflammation in IL-6-deficient mice | Siewert et al. (2000) | |
| TNF-α | CYP1A1 | ↓ | mRNA and protein | Rat liver epithelial WB-F344 cells | Umannová et al. (2008) |
| CYP1A2 | ↓ | mRNA | Human primary hepatocytes | Dallas et al. (2012) | |
| CYP1B1 | ↑ | mRNA and protein | Rat liver epithelial WB-F344 cells | Umannová et al. (2008) | |
| CYP2A4/5 | ↑ | mRNA | C. rodentium mice model of infectious colitis | Nyagode et al. (2014) | |
| CYP2C8 | ↓ | mRNA | Cynomolgus hepatocytes | Uno et al. (2020) | |
| CYP2C76 | ↓ | mRNA | Cynomolgus hepatocytes | Uno et al. (2020) | |
| CYP2D6 | ↓ | mRNA | Human primary hepatocytes | Dallas et al. (2012) | |
| CYP2E1 | ↓ | mRNA | Human primary hepatocytes | Abdel-Razzak et al. (1993) | |
| CYP3A11 | ↓ | mRNA | Mouse hepatocytes | Kinloch et al. (2011) | |
| C. rodentium mice model of infectious colitis | Nyagode et al. (2014) | ||||
| CYP3A25 | ↓ | mRNA | Mouse hepatocytes | Kinloch et al. (2011) | |
| C. rodentium mice model of infectious colitis | Nyagode et al. (2014) | ||||
| CYP3A4 | ↓ | mRNA | Human primary hepatocytes | Dallas et al. (2012) | |
| IL-1 | CYP1A1 | ↓ | mRNA | Isolated rat hepatocytes | Barker et al. (1992) |
| cultured rabbit hepatocytes | Calleja et al. (1997) | ||||
| CYP1A2 | ↓ | mRNA | Isolated rat hepatocytes | Barker et al. (1992) | |
| cultured rabbit hepatocytes | Calleja et al. (1997) | ||||
| IL-1β | CYP1A1 | ↓ | mRNA | Cynomolgus hepatocytes | Uno et al. (2020) |
| CYP1A2 | ↓ | mRNA | Human primary hepatocytes | Abdel-Razzak et al. (1993) | |
| CYP2B6 | ↓ | mRNA | Human hepatocytes | Assenat et al. (2004) | |
| CYP2C8 | ↓ | mRNA | Cynomolgus hepatocytes | Uno et al. (2020) | |
| CYP2C9 | ↓ | mRNA | Human hepatocytes | Assenat et al. (2004) | |
| CYP2C11 | ↓ | mRNA | IL-1β-induced fevered rat | Kihara et al. (1998) | |
| CYP2C19 | ↓ | mRNA | Cynomolgus hepatocytes | Uno et al. (2020) | |
| CYP2C76 | ↓ | mRNA | Cynomolgus hepatocytes | Uno et al. (2020) | |
| CYP3A4 | ↓ | mRNA | Human hepatocytes | Assenat et al. (2004) | |
| CYP3A5 | ↑ | mRNA | Cynomolgus hepatocytes | Uno et al. (2020) | |
| CYP3A subfamily | ↓ | mRNA | IL-1β-induced fevered rat | Kihara et al. (1998) | |
| IFN-γ | CYP1A2 | ↓ | Protein | Human primary hepatocytes | Donato et al. (1997) |
| CYP2B9 | ↓ | mRNA | LPS-induced septic mice model | Nyagode et al. (2010) | |
| CYP2D9 | ↓ | mRNA | C. rodentium-induced colitis mice model | Nyagode et al. (2010) | |
| CYP2D22 | ↓ | mRNA | C. rodentium-induced colitis mice model; LPS-induced septic mice model | Nyagode et al. (2010) | |
| CYP2E1 | ↓ | mRNA | LPS-induced septic mice model | Nyagode et al. (2010) | |
| CYP3A1 | ↓ | mRNA | Rat primary hepatocytes | Tapner et al. (1996) | |
| CYP3A2 | ↓ | mRNA | Rat primary hepatocytes | Tapner et al. (1996) | |
| CYP3A4 | ↓ | Protein | Human primary hepatocytes | Donato et al. (1997) | |
| CYP3A11 | ↓ | mRNA | C. rodentium-induced colitis mice model | Nyagode et al. (2010) | |
| CYP3A25 | ↓ | mRNA | C. rodentium-induced colitis mice model | Nyagode et al. (2010) | |
| CYP4F18 | ↓ | mRNA | C. rodentium-induced colitis mice model | Nyagode et al. (2010) |
Abbreviations: COVID-19, coronavirus disease 2019; CYP, cytochrome P450; IL, interleukin; TNF, tumor necrosis factor; IFN, interferon; LPS, lipopolysaccharide. ↑ (Increased), ↓ (Reduced).
4.2.1 Interleukin-6
IL-6 was considered to be the principal regulator of the hepatic acute-phase response. Previous studies have focused on investigating the effect of IL-6 on CYPs levels. When hepatoma cells were treated with IL-6, the levels of CYP1A1, CYP1A2, CYP2B6, CYP3A3, and CYP3A4 mRNAs were markedly suppressed, as well as activities of CYP1A2, CYP2B6, and CYP3A4 (Fukuda et al., 1992; Fukuda and Sassa, 1994; Rubin et al., 2015). In both HepG2 and Caco-2 cells, IL-6 also induced a significant concentration- and time-dependent decrease in CYP3A4 and CYP3A5 expression (Enokiya et al., 2021). In a rat hepatocyte and Kupffer cell co-culture (HKCC) model treated with trovafloxacin or acetaminophen, lipopolysaccharide (LPS) activation showed decreased IL-6 production with concomitant increases in CYP3A activity (Rose et al., 2016).
Additionally, CYP2A12 activity increased in IL-6 knockout mice after LPS administration compared to wild type (WT) mice (Warren et al., 2001). However, Siewert et al. showed that IL-6 was the major determinant in the down-regulation of CYP1A2, CYP2A5, and CYP3A11 in mice models of aseptic inflammation, whereas in the case of LPS-mediated septic mice models, the effects of IL-6 on CYP downregulation can be compensated by other cytokines (Siewert et al., 2000).
In human hepatocytes, IL-6 also decreases both rifampicin- and phenobarbital-mediated induction of CYP2B6, CYP2C8, CYP2C9, and CYP3A4, by negatively regulating PXR and CAR gene expression (Pascussi et al., 2000). Several other studies demonstrated that IL-6 induces drops in CYP2B6, CYP2C8, CYP2C9, CYP2C19, CYP2E1, and CYP3A4 mRNA levels in human hepatocytes, but studies on the effects of IL-6 on CYP1As expression have shown inconsistent results (Abdel-Razzak et al., 1993; Muntané-Relat et al., 1995; Aitken and Morgan, 2007; Dickmann et al., 2012). Jover et al. have explored the molecular mechanism of IL-6 regulation of CYP expression and demonstrated that IL-6 down-regulates CYP3A4 through translational induction of C/EBPβ-LIP (Jover et al., 2002).
Moreover, several clinical studies have demonstrated that IL-6 inhibitors enhance drug metabolism via CYP3A4, 2C9, and 2C19, but reduced the drug metabolism via CYP1A2 (Schmitt et al., 2011; Lee et al., 2017; White et al., 2021). Blocking IL-6 receptors, via the monoclonal antibodies tocilizumab and sarilumab has reversed CYP3A4 activity suppression in rheumatoid arthritis patients (Lee et al., 2017). Halting IL-6 signaling via the monoclonal antibody sirukumab also reversed IL-6-mediated suppression of CYP3A, CYP2C9, and CYP2C19 activity in rheumatoid arthritis patients (Zhuang et al., 2015), suggesting that IL-6 is an important regulator of CYP enzymes.
4.2.2 Tumor Necrosis Factor-α.
TNF-α enhances the induction of CYP1B1, whilst simultaneously suppressing benzo (a)pyrene-induced CYP1A1 expression in rat liver epithelial WB-F344 cells (Umannová et al., 2008). CYP3A11 and 3A25 were effectively down-regulated in mouse hepatocytes treated with TNF-α (Kinloch et al., 2011). In human hepatocytes, TNF-α down-regulated the gene expression of CYP1A1, CYP1A2, CYP2C8, CYP2D6, CYP2E1 and CYP3A4 (Abdel-Razzak et al., 1993; Aitken and Morgan, 2007; Dallas et al., 2012; Muntané-Relat et al., 1995). NF-κB was shown to play a significant role in CYP1A1 suppression caused by TNF-α and LPS (Ke et al., 2001).
A novel antagonist of soluble TNFα (XPro1595) selectively blocked the down-regulation of CYP3A11 and CYP3A25 mRNAs, as well as the induction of CYP2A4/5 in a C. rodentium model of infectious colitis (Nyagode et al., 2014). A recent study investigated the effects of TNF-α on CYP expression in hepatocytes from cynomolgus macaques, which showed significant reduction of CYP2C8 and CYP2C76 mRNA expression by TNF-α (Uno et al., 2020).
4.2.3 Interleukin-1
Previous studies demonstrated that treatment of mice with IL-1, decreased CYPs contents (Bertini et al., 1989; Sujita et al., 1990). IL-1 rapidly suppressed CYP1A1 and CYP1A2 mRNA in rat hepatocytes and rabbit hepatocytes (Barker et al., 1992; Calleja et al., 1997). And CYP1A2, CYP2C8, CYP2E1, CYP3A, and CYP4A11 mRNA levels were down-regulated in human hepatocyte after IL-1β treatment (Abdel-Razzak et al., 1993; Dickmann et al., 2012). Immunoblot analysis of the CYP isozymes indicated that CYP2C11 and CYP3A were extensively reduced in IL-1β-induced fevered rat (Kihara et al., 1998). IL-1β significantly reduced CYP1A1, CYP2C8, CYP2C19, and CYP2C76 mRNA expression, but increased CYP3A5 mRNA expression in several cynomolgus hepatocytes (Uno et al., 2020). IL-1β also decreases phenobarbital- or bilirubin-mediated induction of CYP2B6, CYP2C9, CYP3A4 mRNA expression by negatively regulating CAR expression (Assenat et al., 2004). Lee et al. showed IL-1β down-regulated CYP3A expression at post-transcriptional level in a novel dual mode: nitric oxide (NO)- and proteasome-dependent at earlier time points and NO- and proteasome independent at later times (Lee et al., 2009).
4.2.4 Interferon-γ
IFN-γ down-regulated the expression of CYP2D9, CYP2D22, CYP3A11, CYP3A25, and CYP4F18 mRNAs in a C. rodentium infection mice model and CYP2B9, CYP2D22, and CYP2E1 in a septic mice model (Nyagode et al., 2010). Furthermore, IFN-γ was shown to down-regulate CYP2E1 expression by suppressing native CYP2E1 promoter activity (Qiu et al., 2004). In human hepatocytes, the down-regulation of CYP1A2 and CYP3A4 expression by IFN-γ was observed (Donato et al., 1997). In male rat hepatocytes, IFN-γ reduced mRNA of CYP3A2 and CYP3A1, as well as CYP3A protein (Tapner et al., 1996).
4.3 Hepatic Injury Induced CYPs Alteration
Since the liver is one of the most affected organs in COVID-19 outside of the respiratory system, liver damage is common in COVID-19 patients (Fan et al., 2020). Previous study showed the expression or activity changed in hepatic injury. A reduction in CYP activity (CYP1A, 2C19 and 3A) was reported in cirrhosis (Villeneuve and Pichette, 2004). Acute experimental liver injury induced by CCl4, drastically reduced the activities of main liver CYP isoenzymes, such as CYP1A2, CYP2C6, CYP2E1 and CYP3A2 (Xie et al., 2014). Additionally, diminished expression and reduced enzymatic activity of CYP2E1, 3A11, 1A2, and 2C29 were found in drug-induced liver injury mice models (Bao et al., 2020). Consequently, COVID-19 associated haptic injury is likely to lead to changes in CYP expression and activity.
5 Altered CYPS in the Pathophysiology of COVID-19
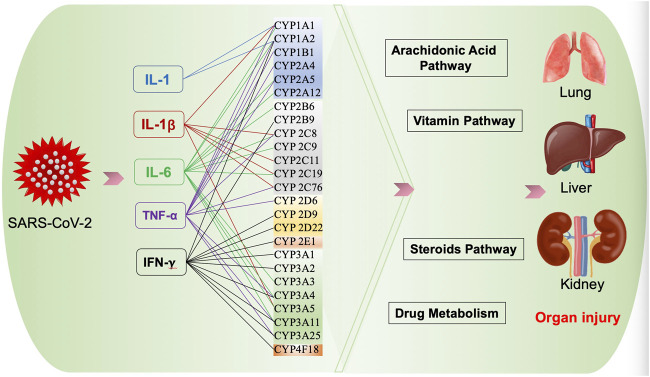
CYPs participate in the biosynthesis or catabolism of steroids, vitamins, eicosanoids, and fatty acids (Guengerich, 2008), which may be involved in the pathogenesis of COVID-19 (Figure 2).
FIGURE 2.
Possible effect of CYPs on COVID-19.
5.1 Arachidonic Acid Pathway
AA is a polyunsaturated fatty acid produced from membrane phospholipids by phospholipase-A2 (PLA2) in inflammatory condition. AA-derived lipid autacoids, including prostaglandins (PGs), thromboxanes, and leukotrienes, are critical mediators in inflammation, and tissue homeostasis (Hoxha, 2020; Ripon et al., 2021). An integrated genomic-scale metabolic model of normal human bronchial epithelial cells (NHBE) infected with SARS-CoV-2, shows that AA metabolism was one of the most affected lipid metabolic pathways in SARS-CoV-2 infection (Nanda and Ghosh, 2021). One in vitro experiment revealed that AA metabolism was markedly perturbed by human coronavirus 229E (HCoV-229E) infection, and the exogenous supplement of AA in HCoV-229E-infected cells significantly suppressed HCoV-229E virus replication (Yan et al., 2019).
Aside from cyclooxygenase (COX) and lipoxygenase pathways, the CYP pathway is another important AA metabolism pathway (Ripon et al., 2021). CYP4A1 and CYP4A2 enzymes convert AA to hydroxyeicosatetraenoic acids (HETEs), which promote the expression of inflammatory cytokines and adhesion molecules (Ishizuka et al., 2008). Additionally, the CYP epoxygenase enzymes of CYP2C and CYP2J families generate epoxyeicosatrienoic acids (EETs) from AA, resulting in anti-inflammation, vasodilation, and pro-angiogenic effects (Iliff et al., 2010; Zhang et al., 2014). Multiple studies demonstrated that both EETs and HETEs play a role in lung injury and kidney injury (Hoff et al., 2019; Zhu et al., 2020). Consequently, it is considered that therapeutic strategies related to specific CYP inhibitors or inducers that improve AA metabolism may be beneficial in COVID-19 (Shoieb et al., 2020).
5.2 Vitamin Pathway
Vitamins are essential dietary components due to their antioxidant properties and immunomodulatory effects, which are beneficial in various infectious diseases, such as COVID-19 (Kumar et al., 2021a; Shakoor et al., 2021). A recent study evaluated the nutritional status of hospitalized COVID-19 patients aged 8–18 years, and results showed vitamin D deficiency in 82%, vitamin B12 deficiency in 18%, vitamin C deficiency in 17%, vitamin A deficiency in 13%, and vitamin E deficiency in 7% of patients (Karakaya Molla et al., 2021).
Vitamin A, also called retinoic acid (RA), exhibited a protective effect on HBV and measles virus infection (Li et al., 2018). Yuan et al. revealed that a retinoid derivative, is highly effective in interrupting the life cycle of Middle East respiratory syndrome (MERS) coronavirus and influenza A virus (Yuan et al., 2019).
Vitamin Bs are important for the normal physiological functioning of body cells (Kumar et al., 2021b). A recent study revealed the potential use of vitamin B9 (Folic acid) against SARS-CoV-2, after screening hundreds of nutraceuticals compounds against known SARS-CoV-2 therapeutic targets. Results indicate that vitamin B9 could contribute to fight against the COVID-19 pandemic (Kumar et al., 2021a).
Vitamin C is well known for its antiviral, antioxidant, anti-inflammatory and immunomodulating properties, which make it a potential therapeutic candidate against COVID-19 infection. Several recent studies show that most severe, or critically ill, COVID-19 patients had hypovitaminosis C, indicating that vitamin C can potentially be used as an adjunctive therapy in the critical care of COVID-19 patients (Arvinte et al., 2020; Chiscano-Camón et al., 2020; Holford et al., 2020).
Vitamin D helps to maintain calcium–phosphorus metabolism and inhibits the overexpression of inflammatory cytokines such as IL-1α, IL-1β, TNF-α (Hughes and Norton, 2009; Kumar et al., 2021b). Serum levels of vitamin D were also low in most of the critically ill COVID-19 patients admitted into intensive care units (ICU) (Arvinte et al., 2020).
COVID-19 may predispose to venous and arterial thrombosis disease due to excessive inflammation or hypoxia. Vitamin K1 could potentially help combat thrombotic complications in COVID-19 patients, due to its ability to activate the coagulation system. A clinical study has also shown that a low vitamin K status was associated with mortality in patients with COVID-19 (Linneberg et al., 2021).
Furthermore, multiple CYPs regulate vitamin metabolism. CYP26 enzymes are involved in the metabolism and elimination of vitamin A (Ross and Zolfaghari, 2011; Stevison et al., 2015). Research in both humans and a variety of animal species have revealed that several CYPs, such as CYP2R1, CYP27A1, CYP3A4, CYP2D25, CYP24A1, CYP27B1, and CYP11A1 are involved in vitamin D metabolism (Tuckey et al., 2008; Annalora et al., 2010; Wang et al., 2012; Jones et al., 2014; Wang et al., 2018a; Maksymchuk and Kashuba, 2020). CYP4F2 and CYP4F11 are both vitamin K1 and K2 ω-hydroxylases (Edson et al., 2013). In addition, CYP4F2 is the only human enzyme shown to metabolize vitamin E (Sontag and Parker, 2002; Bardowell et al., 2012). However, how CYP is involved in the pathophysiological process of COVID-19 through the vitamin pathway still needs further exploration.
5.3 Steroids
Recent advances suggest endocrine system dysfunction in COVID-19 patients. Angiotensin-converting enzyme 2 (ACE2) and transmembrane serine protease 2 (TMPRSS2) are expressed in several endocrine tissues, including the hypothalamus, pituitary, thyroid, adrenal, gonads, and pancreatic islets (Lazartigues et al., 2020; Puig-Domingo et al., 2020), suggesting that SARS-CoV-2 may invade and affect the endocrine system. SARS studies show that 39.3% survivors had evidence of hypocortisolism 3 months after recovery, and the majority of the hypothalamic–pituitary–adrenal (HPA) axis dysfunction resolved within a year (Leow et al., 2005). In the current COVID-19 pandemic, as well as in the SARS-CoV and MERS epidemics, females have a substantially lower mortality rate than males, which can be explained by sex differences in the response to inflammation and sex steroid hormones (Al-Lami et al., 2020; Leach et al., 2021).
On the other hand, CYPs are important for catalyzing specific reactions of steroid precursors in the steridogenic pathway (Ghayee and Auchus, 2007). CYP monooxygenase systems have been found to be involved in the process of arginine vasopressin (AVP)-induced adrenocorticotropic hormone (ACTH) secretion (Okajima and Hertting, 1986). Human CYP11B2 catalyzes the 11-hydroxylation of both progesterone and androstenedione (Glass et al., 2021), whilst CYP11B1 located in the zona fasciculata catalyzes the conversion of 11-deoxycortisol to cortisol (Portrat et al., 2001). CYP17A1 is required for the production of androgen and oestrogen precursors in the zona reticularis, testes and ovaries due to its 17a-hydroxylase activity and subsequent 17, 20-lyase activity (Ghayee and Auchus, 2007; Storbeck et al., 2015). CYP3A4 was the most efficient metabolic catalyst for several of the most frequently prescribed inhaled glucocorticoids (Moore et al., 2013). CYP3A5 activity in lung cells is also related to the metabolism of inhaled glucocorticoid fluticasone propionate, which increases the effective concentration at its target site (Murai et al., 2010). Additionally, CYP3A5 catalyzes 6β-hydroxylation of endogenous cortisol, which is associated with sodium and water retention in the kidney (Rais et al., 2013). Taken together, changes in CYPs may affect endocrine system function in COVID-19, but this still needs to be confirmed by a large number of future studies.
6 Possible Mechanism of CYPS Invovled in Organ Injury in COVID-19
Some COVID-19 patients, especially those with severe diseases, suffered from lung injury, kidney injury, even multi-organ failure. Numerous previous studies showed CYPs played a role in lung injury, kidney injury and liver injury, including CYPs we mentioned above that may affected in COVID-19, suggesting these CYPs may be involved in the pathophysiological process of severe COVID-19.
6.1 Acute Lung Injury
Lung is the main target of SARS-CoV-2, and lung injury is common in severe COVID-19 patients. Increasing studies showed CYPs play a role in ALI. A recent study found that CYP1A1 knockout enhanced LPS-induced ALI, as evidenced by increased IL-6, TNF-α, IL-1β in lung (Tian et al., 2021). CYP1A1 also protects mice models against hyperoxic lung injury by decreasing oxidative stress and susceptibilities to hyperoxia (Jiang et al., 2018; Lingappan et al., 2014; Lingappan et al., 2017), while CYP1B1 enzymes increase oxidative DNA adduct under hyperoxic conditions, contributing to lung injury. Additionally, CYP2E1 and CYP2A can also contribute to hyperoxic lung injury in ethanol and nicotine metabolism through oxidative stress pathway (Stading et al., 2021).
CYPs metabolizes AA to EETs and 20-hidroxyeicosatetranoic acids (20-HETEs), which is believed to play a protective role in lung injury (Stading et al., 2021). CYP4A and CYP4F, which are downregulated by inflammatory mediators (Nyagode et al., 2010; Dickmann et al., 2012), metabolize AA to 20-HETEs, which could also impact hyperoxic lung injury via the vasodilating effects of 20-HETEs (Stading et al., 2021).
6.2 Acute Kidney Injury
The incidence of AKI was about 8.9% in COVID-19 patients, but can reach 25% in critically ill patients (Chen et al., 2020; Gabarre et al., 2020). Current evidence suggests a link between CYPs and AKI. The expression and activity of CYP3A11 was predominant reduced in sepsis-AKI mice models (Sukkummee et al., 2019). CYP2C18, 2C19 expressions were significantly lower in uremic patients (Hu et al., 2018). And CYPs were shown to play an important role in metabolizing xenobiotics and thus reduce xenobiotics-induced renal toxicity (Xiao et al., 2008; Yao et al., 2014). Additionally, AA-derived CYP metabolites, EETs and 20-HETEs, play a key role in ischemia/reperfusion-induced AKI (Hoff et al., 2019; Zhu et al., 2020).
6.3 Hepatic Injury
In the current COVID-19 pandemic, hepatic dysfunction has been observed in 14–53% of patients, particularly in severe cases (Jothimani et al., 2020). Hepatic injury interacted with CYP expression and activity. Liver diseases cause changes in the expression and activity of CYPs, while CYPs also implicated in hepatic injury. Induction of CYP2E1 enzyme is known to play a role in the pathogenesis of alcoholic liver disease and thioacetamide induced-liver injury (Ramaiah et al., 2001; Stice et al., 2015). Several studies demonstrated that CYP2E1 inhibitor protects the liver against chemical-induced hepatic injury (Choi et al., 1996; Jeong, 1999; Lin et al., 2012).
7 Pharmacokinetics in COVID-19 Therapy
Since the primary function of CYP1-3 enzymes is facilitating drug metabolism, the main concern of the dysregulation of CYP expression in COVID-19 is the direct impact on drug disposition and pharmacokinetics in humans. Currently, there is no certified medication to treat COVID-19. Several drugs that are considered as potentially effective are being used in COVID-19 treatment, many of which are metabolized by CYPs. Of the total CYPs discovered to date, six of these are responsible for 90% of drug metabolism, including CYP1A2, CYP2C9, CYP2C19, CYP2D6, CYP2E1, and CYP3A4 (Gilani and Cassagnol, 2021). In this section, we aimed to summarize the role of these CYPs in COVID-19 drug therapy from the aspect of routine treatment, symptomatic support treatment and treatment of comorbidities (Table 2).
TABLE 2.
CYPs involved in the metabolism of drugs in COVID-19 treatment.
| CYP enzymes | Anti-viral drugs | Symptomatic and supportive treatment | Pharmacological therapy for comorbidity | Traditional Chinese medicine |
|---|---|---|---|---|
| CYP1A2 | — | — | Clopidogrel clozapine, theophylline | Qingfei paidu decoction (1A family) |
| Jingyin ranules (1A family) | ||||
| CYP2B6 | — | Propofol | Clopidogrel | — |
| Diazepam | ||||
| Tramadol | ||||
| CYP2C8 | Remdesivir | Morphine | Pioglitazone | Qingfei paidu decoction |
| Loperamide | Rosiglitazone | |||
| Ibuprofen | Repaglinide | Jingyin ranules | ||
| CYP2C9 | — | Diazepam | Irbesartan | Qingfei paidu decoction |
| Losartan | ||||
| Nateglinide | ||||
| Ibuprofen | Sulfonylureas | Jingyin ranules | ||
| Clopidogrel | ||||
| Celecoxib | Carvedilol | |||
| Warfarin | ||||
| CYP2C19 | — | Diazepam | Indaparnide | Qingfei paidu decoction |
| Omeprazole | Clopidogrel | Jingyin ranules | ||
| CYP2D6 | Remdesivir | Tramadol | Propranolol | Qingfei paidu decoction |
| Carvedilol | ||||
| Chloroquine hydroxychloroquine | Loperamide | Diltiazem | ||
| Metoprolol | Jingyin ranules | |||
| Nifedipine | ||||
| CYP2E1 | — | Acetaminophen | Theophylline | Qingfei paidu decoction |
| Jingyin ranules | ||||
| CYP3A4 | Lopinavir–ritonavir | Fentanyl | Indaparnide | Qingfei paidu decoction (3A family) |
| Morphine | ||||
| Midazolam | CCBs | |||
| Remdesivir | Alprazolam | Losartan | ||
| Tramadol | Jingyin ranules (3A family) | |||
| Chloroquine hydroxychloroquine | Loperamide | Clopidogrel | ||
| Acetaminophen | Statin drugs |
Abbreviations: COVID-19, coronavirus 2019; CYP, cytochrome P450.
7.1 The Association Between Routine Drug Treatment and CYPs
7.1.1 Lopinavir–Ritonavir
Upon referring to previous antivirus activity studies, lopinavir–ritonavir was proposed as an emergency treatment in COVID-19 (Magro et al., 2021). Lopinavir and ritonavir are both CYP3A4 substrates (Cvetkovic and Goa, 2003), so there is a potential for elevated levels following infection and inflammation-related down-regulation of CYP3A4 expression. This is supported by COVID-19 clinical pharmacokinetic data. Recent studies demonstrated that lopinavir trough concentrations were 3.5-fold higher in COVID-19 patients than in HIV-infected patients (Croxtall and Perry, 2010; Marzolini et al., 2020), which positively correlates with CRP values and was significantly lower when tocilizumab (IL-6 receptor antagonist) was pre-administered (Marzolini et al., 2020; Schoergenhofer et al., 2020).
7.1.2 Remdesivir
Remdesivir is one of few Food and Drug Administration (FDA)-approved treatments for severe cases of COVID-19 (Tao et al., 2021). It is metabolized by both CYPs and non-CYP enzymes, and previous studies have demonstrated that remdesivir is a substrate for CYP2C8, CYP2D6, and CYP3A4 (Deb et al., 2021). Additionally, remdesivir also acts as an inhibitor of CYP1A2, CYP2C9, CYP2C19, CYP2D6, and CYP3A4 (Aleissa et al., 2020). Since CYP3A4 is a critical enzyme responsible for about 70% of the drugs that are clinically available (Deb and Arrighi, 2021), it should be noted that the suppression of CYP3A4 expression by concomitant inflammatory conditions and simultaneous application of other drugs metabolized by CYPs, could reduce the elimination of remdesivir and lead to unpredictable dose-toxicity.
7.1.3 Chloroquine and Hydroxychloroquine
Chloroquine and hydroxychloroquine have been suggested as having an antiviral effect in COVID-19 patients, with side effects including arrythmias, cardiovascular complications, hepatological effects, and adverse vision effects (Cortegiani et al., 2020; Réa-Neto et al., 2021). Both of these drugs are metabolized by CYP3A4 and CYP2D6. Although CYP2D6 expression is not as affected by inflammatory factors as CYP3A4, the highly frequent polymorphic presence of CYP2D6 could also lead to modified elimination of these drugs and eventually life-threatening drug-adverse effects (Nefic, 2018; Deb et al., 2021). However, hydroxychloroquine plasma concentrations appear to have no correlation with CRP values in COVID-19 patients (Marzolini et al., 2020).
7.1.4 Corticosteroids
Corticosteroids are widely used in the treatment of COVID-19 due to their anti-inflammatory and immunosuppressive effects (Wang et al., 2021). Corticosteroids are substrates and inducers for CYP3A4 (Gentile et al., 1996; Mccune et al., 2000). Glucocorticoids are predominantly metabolized by CYP3A, and their plasma concentrations are influenced by CYP3A activity (Varis et al., 1998; Varis et al., 2000a; Varis et al., 2000b). Low hepatic CYP3A activity caused by hyperinflammation in COVID-19 may significantly contribute to the risk of glucocorticoid-related complications, such as steroid-induced osteonecrosis of the femoral head (Kaneshiro et al., 2006). However, glucocorticoids at doses used clinically also increased CYP3A4 activity, but with extensive intersubject variability (Mccune et al., 2000). Therefore, due to the heterogeneity of the induction effect of glucocorticoids on CYP activity, whether it can counteract the suppression effect of CYP expression and activity caused by inflammatory mediators may vary among individuals.
7.1.5 Symptomatic and Supportive Treatment
The symptoms of COVID-19 patients are varied, with the most common being fever, cough, digestive tract symptoms, sleep disorders and headaches, which are often treated by medication (Guan et al., 2020a; Bhat and Chokroverty, 2021; Fernández-De-Las-Peñas et al., 2021). Ibuprofen and acetaminophen are the most commonly used antipyretics. Ibuprofen metabolism is strongly linked to CYP2C8 and CYP2C9 (García-Martín et al., 2004), whilst CYPs (CYP3A4, CYP2E1) have some role mainly at toxic concentrations of acetaminophen. CYP3A4 is the major CYP enzyme involved in acetaminophen bioactivation. Alprazolam is a CYP3A4 substrate, often prescribed to treat COVID-19 patients with sleep disorders (Boulenc et al., 2016; Sánchez Díaz et al., 2021). Proton pump inhibitors (PPI), commonly used drugs for gastrointestinal diseases, are metabolized by CYP2C19 (Zvyaga et al., 2012). Celecoxib, a cyclooxygenase (COX)-2 inhibitor, is the substrate of CYP2C9 (Chan et al., 2009), and tramadol is a substrate of CYP2D6 (Xu et al., 2014), and therefore both can be used to treat headaches (Piovesan et al., 2002; Robbins, 2004).
ARDS is a major complication in severe COVID-19 patients, when analgesics and sedatives are routinely used for patients on mechanical ventilation. Fentanyl and morphine are also commonly used for analgesia in patients on mechanical ventilation. Fentanyl and sufentanil are metabolized by CYP3A4 (Tateishi et al., 1996; Labroo et al., 1997), and hepatic CYP3A4 and CYP2C8 are the main CYPs responsible for morphine N-demethylation (Projean et al., 2003). Propofol, benzodiazepine and dexmedetomidine are commonly used for sedation. Propofol is metabolized by CYP2B6 (Murayama et al., 2007; Oshio et al., 2019), whilst the most commonly used benzodiazepine drug Midazolam is metabolized by CYP3A4 and serves as a probe for CYP3A catalytic activity (Olkkola and Ahonen, 2008; Nassi et al., 2020). It has been reported that CYPs (mainly CYP2A6) primarily mediated aliphatic hydroxylation of dexmedetomidine, generating 3-hydroxy dexmedetomidine and other metabolites, in human liver microsomes (Weerink et al., 2017; Wang et al., 2018b). Most of these medications can be extremely harmful if plasma levels are increased following a lack of metabolism by inflammation-mediated CYP3A4 or CYP2B6 suppression.
7.2 Pharmacological Therapy for Comorbidity
Elderly patients have the highest mortality rate amongst COVID-19 patients, which is generally related to co-existing underlying diseases. Similar to most studies, our previous data showed that the most common comorbidities in this population were hypertension, coronary heart disease, and diabetes (Guan et al., 2020b; Huang et al., 2020; Wang et al., 2020). Many drugs used to treat chronic diseases are also metabolized by CYPs.
Anti-hypertensive drugs incorporate several classes: diuretics, angiotensin-converting enzyme (ACE) inhibitors, calcium channel blockers (CCBs), angiotensin II receptor blockers (ARBs), and beta-blockers (Peyriere et al., 2012). Indaparnide, a long-acting thiazide-related diuretic, is metabolized by CYP3A4 and CYP2C19 (Yan et al., 2012). Several beta-blockers, such as propranolol, are largely metabolized by CYP2D6 (Peyriere et al., 2012). All CCBs are substrates for CYP3A4. Losartan, the leading ARB, is bioactivated by CYP2C9 and subsequently metabolized by CYP3A4. Another ARB, irbesartan, is metabolized by CYP2C9 (Peyriere et al., 2012).
Anti-platelet and anti-coagulant drugs are commonly used in the treatment of cardiovascular diseases. CYP2C19, CYP1A2, and CYP2B6 catalyze clopidogrel to the immediate precursor of its pharmacologically active metabolite, whilst CYP3A4, CYP2B6, CYP2C19, and CYP2C9 contribute to the active metabolite formation (Kazui et al., 2010). CYP2C9 is responsible for warfarin metabolism (Mikheeva et al., 2008).
Several oral antidiabetic drugs are also metabolized by CYPs. For example, pioglitazone and rosiglitazone are metabolized mainly by CYP2C8, whilst sulfonylureas are mainly metabolized by CYP2C9; and to a lesser extent by CYP3A4 (Holstein and Bell, 2009). Repaglinide is metabolized mainly through CYP2C8 whereas nateglinide metabolism predominantly involves CYP2C9 (Holstein and Bell, 2009).
A recent study of 227 hospitalized COVID-19 patients showed that 38% had at least one clinically significant potential drug–drug interaction. More than half of the interactions were between lopinavir/ritonavir and regularly prescribed medications for the management of comorbidities or COVID-19 symptoms (Mahboobipour and Baniasadi, 2020). As CYPs are the most important family of drug metabolism enzymes, the interactions between drugs metabolized by CYPs need to be carefully considered by clinicians.
Additionally, traditional Chinese medicine is widely used in the treatment of COVID-19 in China (Liang et al., 2021; Wu and Zhong, 2021). Jingyin granules and Qingfei Paidu decoction, have been recommended for treating the H1N1 influenza A virus infection and COVID-19 in China, and have exhibited an inhibitory effect on CYP1A, CYP2A6, CYP2C8, CYP2C9, CYP2D6, CYP2E1, CYP2C19 and CYP3A (Zhang et al., 2021b; Zhang et al., 2021c).
8 Conclusion
The recent emergence of the COVID-19 pandemic has caused unprecedented global healthcare problems. The most striking pathophysiological feature of COVID-19 is the state of excessive inflammatory response. As the most common drug metabolizing enzyme family, CYPs are closely related to the metabolism of endogenous and exogenous substances. In this review, we analyzed and summarized current evidences regarding the possible changes and roles of CYPs in COVID-19. In COVID-19, viral infection, excessive inflammatory response, and hepatic impairment may all affect CYP expression. CYPs may influence the pathophysiological process of COVID-19 through AA, vitamins, and steroid pathways. Moreover, many of the drugs that are likely to be used in COVID-19 patients are metabolized by CYPs. Since the expression of CYPs may be greatly altered in COVID-19 patients, drug pharmacokinetics may also vary, and drug-related side effects may increase in these patients. In the case of co-administration of multiple drugs, the risk of drug interactions may even increase, and therefore, monitoring of drug concentrations and side effects is essential in this population. Overall, the information on the relationship between COVID-19 pathophysiology and CYPs status, will potentially minimize drug-related toxicity and optimize the treatment of infected individuals.
Author Contributions
GW, BX, JD, YL, LG, and YZ contributed to the review writing. YZ and JL critically edited the review. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Scientific Research Project of Hunan Provincial Health Commission (No. 202117010786), Natural Science Foundation of Hunan Province (No. 2021JJ40872), and National Natural Science Foundation of China (No. 82102283).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Glossary
- AA
Arachidonic acid
- ACE
Angiotensin-converting enzyme
- ACTH
Adrenocorticotropic hormone
- AhR
Aryl hydrocarbon receptor
- AKI
Acute kidney injury
- ARBs
Angiotensin II receptor blockers.
- ARDS
Acute respiratory distress syndrome
- AVP
Arginine vasopressin
- CAR
Constitutive androstane receptor
- CCBs
Calcium channel blockers
- COVID-19
Coronavirus disease 2019
- COX
Cyclooxygenase
- CYP
Cytochrome P450
- EETs
Epoxyeicosatrienoic acids
- HCV
Hepatitis C virus
- HETEs
Hydroxyeicosatetraenoic acids
- HPA
Hypothalamic–pituitary–adrenal axis
- ICU
Intensive care units
- IFN
Interferon
- IL
Interleukin
- LPS
Lipopolysaccharide
- MERS
Middle East respiratory syndrome
- NHBE
Human bronchial epithelial cells
- PGs
Prostaglandins
- PLA2
Phospholipase-A2
- PXR
Pregnane X receptor
- RA
Retinoic acid
- SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2
- TMPRSS2
Transmembrane serine protease 2
- TNF
Tumor necrosis factor
- WHO
World Health Organization
References
- Abdel-Razzak Z., Loyer P., Fautrel A., Gautier J. C., Corcos L., Turlin B., et al. (1993). Cytokines Down-Regulate Expression of Major Cytochrome P-450 Enzymes in Adult Human Hepatocytes in Primary Culture. Mol. Pharmacol. 44, 707–715. [PubMed] [Google Scholar]
- Aitken A. E., Morgan E. T. (2007). Gene-specific Effects of Inflammatory Cytokines on Cytochrome P450 2C, 2B6 and 3A4 mRNA Levels in Human Hepatocytes. Drug Metab. Dispos 35, 1687–1693. 10.1124/dmd.107.015511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Lami R. A., Urban R. J., Volpi E., Algburi A. M. A., Baillargeon J. (2020). Sex Hormones and Novel Corona Virus Infectious Disease (COVID-19). Mayo Clin. Proc. 95, 1710–1714. 10.1016/j.mayocp.2020.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aleissa M. M., Silverman E. A., Paredes Acosta L. M., Nutt C. T., Richterman A. G., Marty F. M. (2020). New Perspectives on Antimicrobial Agents: Remdesivir Treatment for COVID-19. Antimicrob. Agents Chemother. 65, e01814–01820. 10.1128/AAC.01814-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Annalora A. J., Goodin D. B., Hong W. X., Zhang Q., Johnson E. F., Stout C. D. (2010). Crystal Structure of CYP24A1, a Mitochondrial Cytochrome P450 Involved in Vitamin D Metabolism. J. Mol. Biol. 396, 441–451. 10.1016/j.jmb.2009.11.057 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arvinte C., Singh M., Marik P. E. (2020). Serum Levels of Vitamin C and Vitamin D in a Cohort of Critically Ill COVID-19 Patients of a North American Community Hospital Intensive Care Unit in May 2020: A Pilot Study. Med. Drug Discov. 8, 100064. 10.1016/j.medidd.2020.100064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assenat E., Gerbal-Chaloin S., Larrey D., Saric J., Fabre J. M., Maurel P., et al. (2004). Interleukin 1beta Inhibits CAR-Induced Expression of Hepatic Genes Involved in Drug and Bilirubin Clearance. Hepatology 40, 951–960. 10.1002/hep.20387 [DOI] [PubMed] [Google Scholar]
- Bao Y., Wang P., Shao X., Zhu J., Xiao J., Shi J., et al. (2020). Acetaminophen-Induced Liver Injury Alters Expression and Activities of Cytochrome P450 Enzymes in an Age-dependent Manner in Mouse Liver. Drug Metab. Dispos 48, 326–336. 10.1124/dmd.119.089557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardowell S. A., Duan F., Manor D., Swanson J. E., Parker R. S. (2012). Disruption of Mouse Cytochrome P450 4f14 (Cyp4f14 Gene) Causes Severe Perturbations in Vitamin E Metabolism. J. Biol. Chem. 287, 26077–26086. 10.1074/jbc.M112.373597 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker C. W., Fagan J. B., Pasco D. S. (1992). Interleukin-1 Beta Suppresses the Induction of P4501A1 and P4501A2 mRNAs in Isolated Hepatocytes. J. Biol. Chem. 267, 8050–8055. 10.1016/s0021-9258(18)42406-4 [DOI] [PubMed] [Google Scholar]
- Belaid B., Lamara Mahammad L., Mihi B., Rahali S. Y., Djidjeli A., Larab Z., et al. (2021). T Cell Counts and IL-6 Concentration in Blood of North African COVID-19 Patients Are Two Independent Prognostic Factors for Severe Disease and Death. J. Leukoc. Bio 111, 269–281. 10.1002/jlb.4cova1020-703r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertini R., Bianchi M., Erroi A., Villa P., Ghezzi P. (1989). Dexamethasone Modulation of In Vivo Effects of Endotoxin, Tumor Necrosis Factor, and Interleukin-1 on Liver Cytochrome P-450, Plasma Fibrinogen, and Serum Iron. J. Leukoc. Biol. 46, 254–262. 10.1002/jlb.46.3.254 [DOI] [PubMed] [Google Scholar]
- Bhat S., Chokroverty S. (2021). Sleep Disorders and COVID-19. Amsterdam: Sleep Medicine. 10.1016/j.sleep.2021.07.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulenc X., Nicolas O., Hermabessière S., Zobouyan I., Martin V., Donazzolo Y., et al. (2016). CYP3A4-based Drug-Drug Interaction: CYP3A4 Substrates' Pharmacokinetic Properties and Ketoconazole Dose Regimen Effect. Eur. J. Drug Metab. Pharmacokinet. 41, 45–54. 10.1007/s13318-014-0235-4 [DOI] [PubMed] [Google Scholar]
- Calleja C., Eeckhoutte C., Larrieu G., Dupuy J., Pineau T., Galtier P. (1997). Differential Effects of Interleukin-1 Beta, Interleukin-2, and Interferon-Gamma on the Inducible Expression of CYP 1A1 and CYP 1A2 in Cultured Rabbit Hepatocytes. Biochem. Biophys. Res. Commun. 239, 273–278. 10.1006/bbrc.1997.7468 [DOI] [PubMed] [Google Scholar]
- Cavalli G., De Luca G., Campochiaro C., Della-Torre E., Ripa M., Canetti D., et al. (2020). Interleukin-1 Blockade with High-Dose Anakinra in Patients with COVID-19, Acute Respiratory Distress Syndrome, and Hyperinflammation: a Retrospective Cohort Study. Lancet Rheumatol. 2, e325–e331. 10.1016/s2665-9913(20)30127-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan A. T., Zauber A. G., Hsu M., Breazna A., Hunter D. J., Rosenstein R. B., et al. (2009). Cytochrome P450 2C9 Variants Influence Response to Celecoxib for Prevention of Colorectal Adenoma. Gastroenterology 136, 2127–e1. e2121. 10.1053/j.gastro.2009.02.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Zhao B., Qu Y., Chen Y., Xiong J., Feng Y., et al. (2020). Detectable Serum Severe Acute Respiratory Syndrome Coronavirus 2 Viral Load (RNAemia) Is Closely Correlated with Drastically Elevated Interleukin 6 Level in Critically Ill Patients with Coronavirus Disease 2019. Clin. Infect. Dis. : official Publ. Infect. Dis. Soc. America 71, 1937–1942. 10.1093/cid/ciaa449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiscano-Camón L., Ruiz-Rodriguez J. C., Ruiz-Sanmartin A., Roca O., Ferrer R. (2020). Vitamin C Levels in Patients with SARS-CoV-2-Associated Acute Respiratory Distress Syndrome. Crit. Care 24, 522. 10.1186/s13054-020-03249-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copaescu A., Smibert O., Gibson A., Phillips E. J., Trubiano J. A. (2020). The Role of IL-6 and Other Mediators in the Cytokine Storm Associated with SARS-CoV-2 Infection. J. Allergy Clin. Immunol. 146, 518–e1. e511. 10.1016/j.jaci.2020.07.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cortegiani A., Ingoglia G., Ippolito M., Giarratano A., Einav S. (2020). A Systematic Review on the Efficacy and Safety of Chloroquine for the Treatment of COVID-19. J. Crit. Care 57, 279–283. 10.1016/j.jcrc.2020.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Croxtall J. D., Perry C. M. (2010). Lopinavir/Ritonavir: a Review of its Use in the Management of HIV-1 Infection. Drugs 70, 1885–1915. 10.2165/11204950-000000000-00000 [DOI] [PubMed] [Google Scholar]
- Cvetkovic R. S., Goa K. L. (2003). Lopinavir/ritonavir: a Review of its Use in the Management of HIV Infection. Drugs 63, 769–802. 10.2165/00003495-200363080-00004 [DOI] [PubMed] [Google Scholar]
- Dallas S., Sensenhauser C., Batheja A., Singer M., Markowska M., Zakszewski C., et al. (2012). De-risking Bio-Therapeutics for Possible Drug Interactions Using Cryopreserved Human Hepatocytes. Curr. Drug Metab. 13, 923–929. 10.2174/138920012802138589 [DOI] [PubMed] [Google Scholar]
- Deb S., Arrighi S. (2021). Potential Effects of COVID-19 on Cytochrome P450-Mediated Drug Metabolism and Disposition in Infected Patients. Eur. J. Drug Metab. Pharmacokinet. 46, 185–203. 10.1007/s13318-020-00668-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deb S., Reeves A. A., Hopefl R., Bejusca R. (2021). ADME and Pharmacokinetic Properties of Remdesivir: Its Drug Interaction Potential. Pharmaceuticals 14, 655. 10.3390/ph14070655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickmann L. J., Patel S. K., Wienkers L. C., Slatter J. G. (2012). Effects of Interleukin 1β (IL-1β) and IL-1β/interleukin 6 (IL-6) Combinations on Drug Metabolizing Enzymes in Human Hepatocyte Culture. Curr. Drug Metab. 13, 930–937. 10.2174/138920012802138642 [DOI] [PubMed] [Google Scholar]
- Donato M. T., Guillén M. I., Jover R., Castell J. V., Gómez-Lechón M. J. (1997). Nitric Oxide-Mediated Inhibition of Cytochrome P450 by Interferon-Gamma in Human Hepatocytes. J. Pharmacol. Exp. Ther. 281, 484–490. [PubMed] [Google Scholar]
- Edson K. Z., Prasad B., Unadkat J. D., Suhara Y., Okano T., Guengerich F. P., et al. (2013). Cytochrome P450-dependent Catabolism of Vitamin K: ω-hydroxylation Catalyzed by Human CYP4F2 and CYP4F11. Biochemistry 52, 8276–8285. 10.1021/bi401208m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enokiya T., Nishikawa K., Hamada Y., Ikemura K., Sugimura Y., Okuda M. (2021). Temporary Decrease in Tacrolimus Clearance in Cytochrome P450 3A5 Non-expressors Early after Living Donor Kidney Transplantation: Effect of Interleukin 6-induced Suppression of the Cytochrome P450 3A Gene. Basic Clin. Pharmacol. Toxicol. 128, 525–533. 10.1111/bcpt.13539 [DOI] [PubMed] [Google Scholar]
- Esteves F., Rueff J., Kranendonk M. (2021). The Central Role of Cytochrome P450 in Xenobiotic Metabolism-A Brief Review on a Fascinating Enzyme Family. J. Xenobiot 11, 94–114. 10.3390/jox11030007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan H., Cai J., Tian A., Li Y., Yuan H., Jiang Z., et al. (2020). Comparison of Liver Biomarkers in 288 COVID-19 Patients: A Mono-Centric Study in the Early Phase of Pandemic. Front. Med. (Lausanne) 7, 584888. 10.3389/fmed.2020.584888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z., Wang Z., Chen W., Cao Z., Li Y. (2016). Association between the CYP11 Family and Six Cancer Types. Oncol. Lett. 12, 35–40. 10.3892/ol.2016.4567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández-de-las-Peñas C., Navarro-Santana M., Gómez-Mayordomo V., Cuadrado M. L., García-Azorín D., Arendt-Nielsen L., et al. (2021). Headache as an Acute and post-COVID-19 Symptom in COVID-19 Survivors: A Meta-Analysis of the Current Literature. Euro J. Neurol. 28, 3820–3825. 10.1111/ene.15040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda Y., Ishida N., Noguchi T., Kappas A., Sassa S. (1992). Interleukin-6 Down Regulates the Expression of Transcripts Encoding Cytochrome P450 IA1, IA2 and IIIA3 in Human Hepatoma Cells. Biochem. Biophys. Res. Commun. 184, 960–965. 10.1016/0006-291x(92)90684-d [DOI] [PubMed] [Google Scholar]
- Fukuda Y., Sassa S. (1994). Suppression of Cytochrome P450IA1 by Interleukin-6 in Human HepG2 Hepatoma Cells. Biochem. Pharmacol. 47, 1187–1195. 10.1016/0006-2952(94)90391-3 [DOI] [PubMed] [Google Scholar]
- Funseth E., Påhlman M., Eloranta M. L., Friman G., Ilbäck N. G. (2002). Effects of Coxsackievirus B3 Infection on the Acute-phase Protein Metallothionein and on Cytochrome P-4501A1 Involved in the Detoxification Processes of TCDD in the Mouse. Sci. Total Environ. 284, 37–47. 10.1016/s0048-9697(01)00864-6 [DOI] [PubMed] [Google Scholar]
- Gao Y., Li T., Han M., Li X., Wu D., Xu Y., et al. (2020). Diagnostic Utility of Clinical Laboratory Data Determinations for Patients with the Severe COVID-19. J. Med. Virol. 92, 791–796. 10.1002/jmv.25770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garciamartin E., Martínez C., Tabarés B., Frías J., Agúndez J. A. (2004). Interindividual Variability in Ibuprofen Pharmacokinetics Is Related to Interaction of Cytochrome P450 2C8 and 2C9 Amino Acid Polymorphisms*1. Clin. Pharmacol. Ther. 76, 119–127. 10.1016/j.clpt.2004.04.006 [DOI] [PubMed] [Google Scholar]
- Gentile D. M., Tomlinson E. S., Maggs J. L., Park B. K., Back D. J. (1996). Dexamethasone Metabolism by Human Liver In Vitro. Metabolite Identification and Inhibition of 6-hydroxylation. J. Pharmacol. Exp. Ther. 277, 105–112. [PubMed] [Google Scholar]
- Ghayee H. K., Auchus R. J. (2007). Basic Concepts and Recent Developments in Human Steroid Hormone Biosynthesis. Rev. Endocr. Metab. Disord. 8, 289–300. 10.1007/s11154-007-9052-2 [DOI] [PubMed] [Google Scholar]
- Gilani B., Cassagnol M. (2021). “Biochemistry, Cytochrome P450,” in StatPearls. Treasure Island (FL): StatPearls Publishing Copyright © 2021 (StatPearls Publishing LLC.). [PubMed] [Google Scholar]
- Glass S. M., Reddish M. J., Child S. A., Wilkey C. J., Stec D. F., Guengerich F. P. (2021). Characterization of Human Adrenal Cytochrome P450 11B2 Products of Progesterone and Androstenedione Oxidation. J. Steroid Biochem. Mol. Biol. 208, 105787. 10.1016/j.jsbmb.2020.105787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W. J., Ni Z. Y., Hu Y., Liang W. H., Ou C. Q., He J. X., et al. (2020b). Clinical Characteristics of Coronavirus Disease 2019 in China. N. Engl. J. Med. 382, 1708–1720. 10.1056/NEJMoa2002032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.-J., Liang W.-H., Zhao Y., Liang H.-R., Chen Z.-S., Li Y.-M., et al. (2020a). Comorbidity and its Impact on 1590 Patients with Covid-19 in China: A Nationwide Analysis. Eur. Respir. J. 55, 2000547. 10.1183/13993003.00547-2020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guengerich F. P. (2008). Cytochrome P450 and Chemical Toxicology. Chem. Res. Toxicol. 21, 70–83. 10.1021/tx700079z [DOI] [PubMed] [Google Scholar]
- He X., Feng S. (2015). Role of Metabolic Enzymes P450 (CYP) on Activating Procarcinogen and Their Polymorphisms on the Risk of Cancers. Curr. Drug Metab. 16, 850–863. 10.2174/138920021610151210164501 [DOI] [PubMed] [Google Scholar]
- Hoff U., Bubalo G., Fechner M., Blum M., Zhu Y., Pohlmann A., et al. (2019). A Synthetic Epoxyeicosatrienoic Acid Analogue Prevents the Initiation of Ischemic Acute Kidney Injury. Acta Physiol. (Oxf) 227, e13297. 10.1111/apha.13297 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holford P., Carr A. C., Jovic T. H., Ali S. R., Whitaker I. S., Marik P. E., et al. (2020). Vitamin C-An Adjunctive Therapy for Respiratory Infection, Sepsis and COVID-19. Nutrients 12, 3760. 10.3390/nu12123760 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holstein A., Beil W. (2009). Oral Antidiabetic Drug Metabolism: Pharmacogenomics and Drug Interactions. Expert Opin. Drug Metab. Toxicol. 5, 225–241. 10.1517/17425250902806424 [DOI] [PubMed] [Google Scholar]
- Hoxha M. (2020). What about COVID-19 and Arachidonic Acid Pathway? Eur. J. Clin. Pharmacol. 76, 1501–1504. 10.1007/s00228-020-02941-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Huang S., Yin L. (2021). The Cytokine Storm and COVID-19. J. Med. Virol. 93, 250–256. 10.1002/jmv.26232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Wang Y., Li X., Ren L., Zhao J., Hu Y., et al. (2020). Clinical Features of Patients Infected with 2019 Novel Coronavirus in Wuhan, China. Lancet 395, 497–506. 10.1016/s0140-6736(20)30183-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes D. A., Norton R. (2009). Vitamin D and Respiratory Health. Clin. Exp. Immunol. 158, 20–25. 10.1111/j.1365-2249.2009.04001.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iliff J. J., Jia J., Nelson J., Goyagi T., Klaus J., Alkayed N. J. (2010). Epoxyeicosanoid Signaling in CNS Function and Disease. Prostaglandins Other Lipid Mediat 91, 68–84. 10.1016/j.prostaglandins.2009.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishizuka T., Cheng J., Singh H., Vitto M. D., Manthati V. L., Falck J. R., et al. (2008). 20-Hydroxyeicosatetraenoic Acid Stimulates Nuclear Factor-kappaB Activation and the Production of Inflammatory Cytokines in Human Endothelial Cells. J. Pharmacol. Exp. Ther. 324, 103–110. 10.1124/jpet.107.130336 [DOI] [PubMed] [Google Scholar]
- Jamal M., Bangash H. I., Habiba M., Lei Y., Xie T., Sun J., et al. (2021). Immune Dysregulation and System Pathology in COVID-19. Virulence 12, 918–936. 10.1080/21505594.2021.1898790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G., Prosser D. E., Kaufmann M. (2014). Cytochrome P450-Mediated Metabolism of Vitamin D. J. Lipid Res. 55, 13–31. 10.1194/jlr.R031534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jover R., Bort R., Gómez-Lechón M. J., Castell J. V. (2002). Down-regulation of Human CYP3A4 by the Inflammatory Signal Interleukin-6: Molecular Mechanism and Transcription Factors Involved. FASEB J. 16, 1799–1801. 10.1096/fj.02-0195fje [DOI] [PubMed] [Google Scholar]
- Kaneshiro Y., Oda Y., Iwakiri K., Masada T., Iwaki H., Hirota Y., et al. (2006). Low Hepatic Cytochrome P450 3A Activity Is a Risk for Corticosteroid-Induced Osteonecrosis. Clin. Pharmacol. Ther. 80, 396–402. 10.1016/j.clpt.2006.07.004 [DOI] [PubMed] [Google Scholar]
- Karakaya Molla G., Ünal Uzun Ö., Koç N., Özen Yeşil B., Bayhan G. İ. (2021). Evaluation of Nutritional Status in Pediatric Patients Diagnosed with Covid-19 Infection. Clin. Nutr. ESPEN 44, 424–428. 10.1016/j.clnesp.2021.04.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazui M., Nishiya Y., Ishizuka T., Hagihara K., Farid N. A., Okazaki O., et al. (2010). Identification of the Human Cytochrome P450 Enzymes Involved in the Two Oxidative Steps in the Bioactivation of Clopidogrel to its Pharmacologically Active Metabolite. Drug Metab. Dispos 38, 92–99. 10.1124/dmd.109.029132 [DOI] [PubMed] [Google Scholar]
- Kihara T., Toda A., Umesue I., Ono N., Shigematsu H., Soeda S., et al. (1998). Effect of Interleukin 1 Beta-Induced Fever on Hepatic Drug Metabolism in Rat. Xenobiotica 28, 559–569. 10.1080/004982598239317 [DOI] [PubMed] [Google Scholar]
- Kikuchi R., Mccown M., Olson P., Tateno C., Morikawa Y., Katoh Y., et al. (2010). Effect of Hepatitis C Virus Infection on the mRNA Expression of Drug Transporters and Cytochrome P450 Enzymes in Chimeric Mice with Humanized Liver. Drug Metab. Dispos 38, 1954–1961. 10.1124/dmd.109.031732 [DOI] [PubMed] [Google Scholar]
- Kinloch R. D., Lee C. M., Van Rooijen N., Morgan E. T. (2011). Selective Role for Tumor Necrosis Factor-α, but Not Interleukin-1 or Kupffer Cells, in Down-Regulation of CYP3A11 and CYP3A25 in Livers of Mice Infected with a Noninvasive Intestinal Pathogen. Biochem. Pharmacol. 82, 312–321. 10.1016/j.bcp.2011.04.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirby G. M., Chemin I., Montesano R., Chisari F. V., Lang M. A., Wild C. P. (1994). Induction of Specific Cytochrome P450s Involved in Aflatoxin B1 Metabolism in Hepatitis B Virus Transgenic Mice. Mol. Carcinog 11, 74–80. 10.1002/mc.2940110204 [DOI] [PubMed] [Google Scholar]
- Korobkova E. A. (2015). Effect of Natural Polyphenols on CYP Metabolism: Implications for Diseases. Chem. Res. Toxicol. 28, 1359–1390. 10.1021/acs.chemrestox.5b00121 [DOI] [PubMed] [Google Scholar]
- Kumar P., Kumar M., Bedi O., Gupta M., Kumar S., Jaiswal G., et al. (2021a). Role of Vitamins and Minerals as Immunity Boosters in COVID-19. Inflammopharmacology 29, 1001–1016. 10.1007/s10787-021-00826-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V., Kancharla S., Jena M. K. (2021b). In Silico virtual Screening-Based Study of Nutraceuticals Predicts the Therapeutic Potentials of Folic Acid and its Derivatives against COVID-19. VirusDis. 32, 29–37. 10.1007/s13337-020-00643-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labroo R. B., Paine M. F., Thummel K. E., Kharasch E. D. (1997). Fentanyl Metabolism by Human Hepatic and Intestinal Cytochrome P450 3A4: Implications for Interindividual Variability in Disposition, Efficacy, and Drug Interactions. Drug Metab. Dispos 25, 1072–1080. [PubMed] [Google Scholar]
- Lazartigues E., Qadir M. M. F., Mauvais-Jarvis F. (2020). Endocrine Significance of SARS-CoV-2's Reliance on ACE2. Endocrinology 161, bqaa108. 10.1210/endocr/bqaa108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leach D. A., Brooke G. N., Bevan C. L. (2021). Roles of Steroid Receptors in the Lung and COVID-19. Essays Biochem. 65, 1025–1038. 10.1042/ebc20210005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee C. M., Pohl J., Morgan E. T. (2009). Dual Mechanisms of CYP3A Protein Regulation by Proinflammatory Cytokine Stimulation in Primary Hepatocyte Cultures. Drug Metab. Dispos 37, 865–872. 10.1124/dmd.108.026187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee E. B., Daskalakis N., Xu C., Paccaly A., Miller B., Fleischmann R., et al. (2017). Disease-Drug Interaction of Sarilumab and Simvastatin in Patients with Rheumatoid Arthritis. Clin. Pharmacokinet. 56, 607–615. 10.1007/s40262-016-0462-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leow M. K., Kwek D. S., Ng A. W., Ong K. C., Kaw G. J., Lee L. S. (2005). Hypocortisolism in Survivors of Severe Acute Respiratory Syndrome (SARS). Clin. Endocrinol. (Oxf) 63, 197–202. 10.1111/j.1365-2265.2005.02325.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li B., Wang Y., Shen F., Wu M., Li Y., Fang Z., et al. (2018). Identification of Retinoic Acid Receptor Agonists as Potent Hepatitis B Virus Inhibitors via a Drug Repurposing Screen. Antimicrob. Agents Chemother. 62, e00465–18. 10.1128/aac.00465-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S. B., Fang M., Liang C. H., Lan H. D., Shen C., Yan L. J., et al. (2021). Therapeutic Effects and Safety of Oral Chinese Patent Medicine for COVID-19: A Rapid Systematic Review and Meta-Analysis of Randomized Controlled Trials. Complement. Ther. Med. 60, 102744. 10.1016/j.ctim.2021.102744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linneberg A., Kampmann F. B., Israelsen S. B., Andersen L. R., Jørgensen H. L., Sandholt H., et al. (2021). The Association of Low Vitamin K Status with Mortality in a Cohort of 138 Hospitalized Patients with COVID-19. Nutrients 13, 1985. 10.3390/nu13061985 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magro P., Zanella I., Pescarolo M., Castelli F., Quiros-Roldan E. (2021). Lopinavir/ritonavir: Repurposing an Old Drug for HIV Infection in COVID-19 Treatment. Biomed. J. 44, 43–53. 10.1016/j.bj.2020.11.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahboobipour A. A., Baniasadi S. (2020). Clinically Important Drug-Drug Interactions in Patients Admitted to Hospital with COVID-19: Drug Pairs, Risk Factors, and Management. Drug Metab. personalized Ther. 10.1515/dmpt-2020-0145 [DOI] [PubMed] [Google Scholar]
- Maksymchuk O. V., Kashuba V. I. (2020). Altered Expression of Cytochrome P450 Enzymes Involved in Metabolism of Androgens and Vitamin D in the Prostate as a Risk Factor for Prostate Cancer. Pharmacol. Rep. 72, 1161–1172. 10.1007/s43440-020-00133-y [DOI] [PubMed] [Google Scholar]
- Marzolini C., Stader F., Stoeckle M., Franzeck F., Egli A., Bassetti S., et al. (2020). Effect of Systemic Inflammatory Response to SARS-CoV-2 on Lopinavir and Hydroxychloroquine Plasma Concentrations. Antimicrob. Agents Chemother. 64, e01177–20. 10.1128/aac.01177-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mccune J. S., Hawke R. L., Lecluyse E. L., Gillenwater H. H., Hamilton G., Ritchie J., et al. (2000). In Vivo and In Vitro Induction of Human Cytochrome P4503A4 by Dexamethasone. Clin. Pharmacol. Ther. 68, 356–366. 10.1067/mcp.2000.110215 [DOI] [PubMed] [Google Scholar]
- Mikheeva I. A., Kropacheva E. S., Ignat'ev I. V., Bulytova I. M., Ramenskaia G. V., Sychev D. A., et al. (2008). Cytochrome P4502C9(CYP2C9) Gene Polymorphism and Safety of Therapy with Warfarin]. Kardiologiia 48, 52–57. [PubMed] [Google Scholar]
- Moore C. D., Roberts J. K., Orton C. R., Murai T., Fidler T. P., Reilly C. A., et al. (2013). Metabolic Pathways of Inhaled Glucocorticoids by the CYP3A Enzymes. Drug Metab. Dispos 41, 379–389. 10.1124/dmd.112.046318 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan E. T. (2009). Impact of Infectious and Inflammatory Disease on Cytochrome P450-Mediated Drug Metabolism and Pharmacokinetics. Clin. Pharmacol. Ther. 85, 434–438. 10.1038/clpt.2008.302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morgan E. T. (2017). “Regulation of Drug-Metabolizing Enzymes and Drug Metabolism by Inflammatory Responses,” in Drug Metabolism in Diseases. Editor Xie W. (Boston: Academic Press; ), 21–58. 10.1016/b978-0-12-802949-7.00002-x [DOI] [Google Scholar]
- Moriya N., Kataoka H., Fujino H., Nishikawa J., Kugawa F. (2012). Effect of Lipopolysaccharide on the Xenobiotic-Induced Expression and Activity of Hepatic Cytochrome P450 in Mice. Biol. Pharm. Bull. 35, 473–480. 10.1248/bpb.35.473 [DOI] [PubMed] [Google Scholar]
- Muntané-Relat J., Ourlin J. C., Domergue J., Maurel P. (1995). Differential Effects of Cytokines on the Inducible Expression of CYP1A1, CYP1A2, and CYP3A4 in Human Hepatocytes in Primary Culture. Hepatology 22, 1143–1153. [PubMed] [Google Scholar]
- Murai T., Reilly C. A., Ward R. M., Yost G. S. (2010). The Inhaled Glucocorticoid Fluticasone Propionate Efficiently Inactivates Cytochrome P450 3A5, a Predominant Lung P450 Enzyme. Chem. Res. Toxicol. 23, 1356–1364. 10.1021/tx100124k [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murayama N., Minoshima M., Shimizu M., Guengerich F. P., Yamazaki H. (2007). Involvement of Human Cytochrome P450 2B6 in the omega- and 4-hydroxylation of the Anesthetic Agent Propofol. Xenobiotica 37, 717–724. 10.1080/00498250701449431 [DOI] [PubMed] [Google Scholar]
- Nanda P., Ghosh A. (2021). Genome Scale-Differential Flux Analysis Reveals Deregulation of Lung Cell Metabolism on SARS-CoV-2 Infection. Plos Comput. Biol. 17, e1008860. 10.1371/journal.pcbi.1008860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nassi A., Quintieri L., Merlanti R., Pezzato F., Capolongo F., Pauletto M., et al. (2020). Midazolam Oxidation in Cattle Liver Microsomes: The Role of Cytochrome P450 3A. J. Vet. Pharmacol. Ther. 43, 608–613. 10.1111/jvp.12906 [DOI] [PubMed] [Google Scholar]
- Nebert D. W., Wikvall K., Miller W. L. (2013). Human Cytochromes P450 in Health and Disease. Philos. Trans. R. Soc. Lond. B Biol. Sci. 368, 20120431. 10.1098/rstb.2012.0431 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nefic H. (2018). The Genetic Variation of CYP2D6 Gene in the Bosnian Population. Med. Arch. 72, 396–400. 10.5455/medarh.2018.72.396-400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyagode B. A., Jahangardi R., Merrell M. D., Tansey M. G., Morgan E. T. (2014). Selective Effects of a Therapeutic Protein Targeting Tumor Necrosis Factor-Alpha on Cytochrome P450 Regulation during Infectious Colitis: Implications for Disease-dependent Drug-Drug Interactions. Pharmacol. Res. Perspect. 2, e00027. 10.1002/prp2.27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nyagode B. A., Lee C. M., Morgan E. T. (2010). Modulation of Hepatic Cytochrome P450s by Citrobacter Rodentium Infection in Interleukin-6- and Interferon-{gamma}-Null Mice. J. Pharmacol. Exp. Ther. 335, 480–488. 10.1124/jpet.110.171488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okajima T., Hertting G. (1986). The Possible Involvement of Cytochrome P-450 Monooxygenase in AVP-Induced ACTH Secretion. Horm. Metab. Res. 18, 281–282. 10.1055/s-2007-1012296 [DOI] [PubMed] [Google Scholar]
- Olkkola K. T., Ahonen J. (2008). Midazolam and Other Benzodiazepines. Handb Exp. Pharmacol., 335–360. 10.1007/978-3-540-74806-9_16 [DOI] [PubMed] [Google Scholar]
- Oshio T., Uehara S., Uno Y., Inoue T., Sasaki E., Yamazaki H. (2019). Marmoset Cytochrome P450 2B6, a Propofol Hydroxylase Expressed in Liver. Xenobiotica 49, 265–269. 10.1080/00498254.2018.1439204 [DOI] [PubMed] [Google Scholar]
- Paniri A., Akhavan-Niaki H. (2020). Emerging Role of IL-6 and NLRP3 Inflammasome as Potential Therapeutic Targets to Combat COVID-19: Role of lncRNAs in Cytokine Storm Modulation. Life Sci. 257, 118114. 10.1016/j.lfs.2020.118114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascussi J. M., Gerbal-Chaloin S., Pichard-Garcia L., Daujat M., Fabre J. M., Maurel P., et al. (2000). Interleukin-6 Negatively Regulates the Expression of Pregnane X Receptor and Constitutively Activated Receptor in Primary Human Hepatocytes. Biochem. Biophys. Res. Commun. 274, 707–713. 10.1006/bbrc.2000.3219 [DOI] [PubMed] [Google Scholar]
- Peyriere H., Eiden C., Macia J. C., Reynes J. (2012). Antihypertensive Drugs in Patients Treated with Antiretrovirals. Ann. Pharmacother. 46, 703–709. 10.1345/aph.1Q546 [DOI] [PubMed] [Google Scholar]
- Pinzon R. T., Wijaya V. O., Buana R. B. (2021). Interleukin-6 (IL-6) Inhibitors as Therapeutic Agents for Coronavirus Disease 2019 (COVID-19): A Systematic Review and Meta-Analysis. J. Infect. Public Health 14, 1001–1009. 10.1016/j.jiph.2021.06.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piovesan E. J., Zukerman E., Kowacs P. A., Werneck L. C. (2002). COX-2 Inhibitor for the Treatment of Idiopathic Stabbing Headache Secondary to Cerebrovascular Diseases. Cephalalgia 22, 197–200. 10.1046/j.1468-2982.2002.00346.x [DOI] [PubMed] [Google Scholar]
- Portrat S., Mulatero P., Curnow K. M., Chaussain J. L., Morel Y., Pascoe L. (2001). Deletion Hybrid Genes, Due to Unequal Crossing over between CYP11B1 (11beta-Hydroxylase) and CYP11B2(aldosterone Synthase) Cause Steroid 11beta-Hydroxylase Deficiency and Congenital Adrenal Hyperplasia. J. Clin. Endocrinol. Metab. 86, 3197–3201. 10.1210/jcem.86.7.7671 [DOI] [PubMed] [Google Scholar]
- Potere N., Batticciotto A., Vecchié A., Porreca E., Cappelli A., Abbate A., et al. (2021). The Role of IL-6 and IL-6 Blockade in COVID-19. Expert Rev. Clin. Immunol. 17, 601–618. 10.1080/1744666x.2021.1919086 [DOI] [PubMed] [Google Scholar]
- Projean D., Morin P. E., Tu T. M., Ducharme J. (2003). Identification of CYP3A4 and CYP2C8 as the Major Cytochrome P450 S Responsible for Morphine N-Demethylation in Human Liver Microsomes. Xenobiotica 33, 841–854. 10.1080/0049825031000121608 [DOI] [PubMed] [Google Scholar]
- Puig-Domingo M., Marazuela M., Giustina A. (2020). COVID-19 and Endocrine Diseases. A Statement from the European Society of Endocrinology. Endocrine 68, 2–5. 10.1007/s12020-020-02294-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin C., Zhou L., Hu Z., Zhang S., Yang S., Tao Y., et al. (2020). Dysregulation of Immune Response in Patients with Coronavirus 2019 (COVID-19) in Wuhan, China. Clin. Infect. Dis. : official Publ. Infect. Dis. Soc. America 71, 762–768. 10.1093/cid/ciaa248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qiu L. O., Linder M. W., Antonino-Green D. M., Valdes R., Jr. (2004). Suppression of Cytochrome P450 2E1 Promoter Activity by Interferon-Gamma and Loss of Response Due to the -71G>T Nucleotide Polymorphism of the CYP2E1*7B Allele. J. Pharmacol. Exp. Ther. 308, 284–288. 10.1124/jpet.103.057208 [DOI] [PubMed] [Google Scholar]
- Rais N., Hussain A., Chawla Y. K., Kohli K. K. (2013). Association between Urinary 6β-Hydroxycortisol/cortisol Ratio and CYP3A5 Genotypes in a Normotensive Population. Exp. Ther. Med. 5, 527–532. 10.3892/etm.2012.842 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Réa-Neto Á., Bernardelli R. S., Câmara B. M. D., Reese F. B., Queiroga M. V. O., Oliveira M. C. (2021). An Open-Label Randomized Controlled Trial Evaluating the Efficacy of Chloroquine/hydroxychloroquine in Severe COVID-19 Patients. Sci. Rep. 11, 9023. 10.1038/s41598-021-88509-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ripon M. A. R., Bhowmik D. R., Amin M. T., Hossain M. S. (2021). Role of Arachidonic cascade in COVID-19 Infection: A Review. Prostaglandins & Other Lipid Mediators 154, 106539. 10.1016/j.prostaglandins.2021.106539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robbins L. (2004). Tramadol for Tension-type Headache. Headache 44, 192–193. 10.1111/j.1526-4610.2004.t01-3-04041.x [DOI] [PubMed] [Google Scholar]
- Rose K. A., Holman N. S., Green A. M., Andersen M. E., Lecluyse E. L. (2016). Co-culture of Hepatocytes and Kupffer Cells as an In Vitro Model of Inflammation and Drug-Induced Hepatotoxicity. J. Pharm. Sci. 105, 950–964. 10.1016/s0022-3549(15)00192-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross A. C., Zolfaghari R. (2011). Cytochrome P450s in the Regulation of Cellular Retinoic Acid Metabolism. Annu. Rev. Nutr. 31, 65–87. 10.1146/annurev-nutr-072610-145127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruan Q., Yang K., Wang W., Jiang L., Song J. (2020). Clinical Predictors of Mortality Due to COVID-19 Based on an Analysis of Data of 150 Patients from Wuhan, China. Intensive Care Med. 46, 846–848. 10.1007/s00134-020-05991-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubin K., Janefeldt A., Andersson L., Berke Z., Grime K., Andersson T. B. (2015). HepaRG Cells as Human-Relevant In Vitro Model to Study the Effects of Inflammatory Stimuli on Cytochrome P450 Isoenzymes. Drug Metab. Dispos 43, 119–125. 10.1124/dmd.114.059246 [DOI] [PubMed] [Google Scholar]
- Sánchez Díaz M., Martín-Calvo M. L., Mateos-Campos R. (2021). Trends in the Use of Anxiolytics in Castile and Leon, Spain, between 2015-2020: Evaluating the Impact of COVID-19. Int. J. Environ. Res. Public Health 18, 5944. 10.3390/ijerph18115944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt C., Kuhn B., Zhang X., Kivitz A. J., Grange S. (2011). Disease-drug-drug Interaction Involving Tocilizumab and Simvastatin in Patients with Rheumatoid Arthritis. Clin. Pharmacol. Ther. 89, 735–740. 10.1038/clpt.2011.35 [DOI] [PubMed] [Google Scholar]
- Schoergenhofer C., Jilma B., Stimpfl T., Karolyi M., Zoufaly A. (2020). Pharmacokinetics of Lopinavir and Ritonavir in Patients Hospitalized with Coronavirus Disease 2019 (COVID-19). Ann. Intern. Med. 173, 670–672. 10.7326/m20-1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shakoor H., Feehan J., Al Dhaheri A. S., Ali H. I., Platat C., Ismail L. C., et al. (2021). Immune-boosting Role of Vitamins D, C, E, Zinc, Selenium and omega-3 Fatty Acids: Could They Help against COVID-19? Maturitas 143, 1–9. 10.1016/j.maturitas.2020.08.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoieb S. M., El-Ghiaty M. A., El-Kadi A. O. S. (2020). Targeting Arachidonic Acid-Related Metabolites in COVID-19 Patients: Potential Use of Drug-Loaded Nanoparticles. Emergent Mater., 1–13. 10.1007/s42247-020-00136-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siewert E., Bort R., Kluge R., Heinrich P. C., Castell J., Jover R. (2000). Hepatic Cytochrome P450 Down-Regulation during Aseptic Inflammation in the Mouse Is Interleukin 6 Dependent. Hepatology 32, 49–55. 10.1053/jhep.2000.8532 [DOI] [PubMed] [Google Scholar]
- Sontag T. J., Parker R. S. (2002). Cytochrome P450 omega-hydroxylase Pathway of Tocopherol Catabolism. Novel Mechanism of Regulation of Vitamin E Status. J. Biol. Chem. 277, 25290–25296. 10.1074/jbc.M201466200 [DOI] [PubMed] [Google Scholar]
- Stading R., Couroucli X., Lingappan K., Moorthy B. (2021). The Role of Cytochrome P450 (CYP) Enzymes in Hyperoxic Lung Injury. Expert Opin. Drug Metab. Toxicol. 17, 171–178. 10.1080/17425255.2021.1853705 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevison F., Jing J., Tripathy S., Isoherranen N. (2015). Role of Retinoic Acid-Metabolizing Cytochrome P450s, CYP26, in Inflammation and Cancer. Adv. Pharmacol. 74, 373–412. 10.1016/bs.apha.2015.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stipp M. C., Acco A. (2021). Involvement of Cytochrome P450 Enzymes in Inflammation and Cancer: a Review. Cancer Chemother. Pharmacol. 87, 295–309. 10.1007/s00280-020-04181-2 [DOI] [PubMed] [Google Scholar]
- Storbeck K. H., Swart A. C., Fox C. L., Swart P. (2015). Cytochrome B5 Modulates Multiple Reactions in Steroidogenesis by Diverse Mechanisms. J. Steroid Biochem. Mol. Biol. 151, 66–73. 10.1016/j.jsbmb.2014.11.024 [DOI] [PubMed] [Google Scholar]
- Sujita K., Okuno F., Tanaka Y., Hirano Y., Inamoto Y., Eto S., et al. (1990). Effect of Interleukin 1 (IL-1) on the Levels of Cytochrome P-450 Involving IL-1 Receptor on the Isolated Hepatocytes of Rat. Biochem. Biophys. Res. Commun. 168, 1217–1222. 10.1016/0006-291x(90)91158-o [DOI] [PubMed] [Google Scholar]
- Tan M., Liu Y., Zhou R., Deng X., Li F., Liang K., et al. (2020). Immunopathological Characteristics of Coronavirus Disease 2019 Cases in Guangzhou, China. Immunology 160, 261–268. 10.1111/imm.13223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao J., Aristotelidis R., Zanowick-Marr A., Chambers L. C., Mcdonald J., Mylonakis E. E., et al. (2021). Evaluation of the Treatment Efficacy and Safety of Remdesivir for COVID-19: a Meta-Analysis. SN Compr. Clin. Med. 3, 2443–2454. 10.1007/s42399-021-01014-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tapner M., Liddle C., Goodwin B., George J., Farrell G. C. (1996). Interferon Gamma Down-Regulates Cytochrome P450 3A Genes in Primary Cultures of Well-Differentiated Rat Hepatocytes. Hepatology 24, 367–373. 10.1002/hep.510240213 [DOI] [PubMed] [Google Scholar]
- Tateishi T., Krivoruk Y., Ueng Y. F., Wood A. J., Guengerich F. P., Wood M. (1996). Identification of Human Liver Cytochrome P-450 3A4 as the Enzyme Responsible for Fentanyl and Sufentanil N-Dealkylation. Anesth. Analg 82, 167–172. 10.1097/00000539-199601000-00031 [DOI] [PubMed] [Google Scholar]
- Tian D., Sun Y., Zhou J., Ye Q. (2021). The Global Epidemic of SARS-CoV-2 Variants and Their Mutational Immune Escape. J. Med. Virol. 10.1002/jmv.27376 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuckey R. C., Li W., Zjawiony J. K., Zmijewski M. A., Nguyen M. N., Sweatman T., et al. (2008). Pathways and Products for the Metabolism of Vitamin D3 by Cytochrome P450scc. FEBS J. 275, 2585–2596. 10.1111/j.1742-4658.2008.06406.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umannová L., Machala M., Topinka J., Nováková Z., Milcová A., Kozubík A., et al. (2008). Tumor Necrosis Factor-α Potentiates Genotoxic Effects of Benzo[a]pyrene in Rat Liver Epithelial Cells through Upregulation of Cytochrome P450 1B1 Expression. Mutat. Research/Fundamental Mol. Mech. Mutagenesis 640, 162–169. 10.1016/j.mrfmmm.2008.02.001 [DOI] [PubMed] [Google Scholar]
- Uno Y., Murayama N., Yamazaki H. (2020). Interleukin-1β and Tumor Necrosis Factor-α Affect Cytochrome P450 Expression in Cynomolgus Macaque Hepatocytes. Drug Metab. Pharmacokinet. 35, 341–343. 10.1016/j.dmpk.2020.02.001 [DOI] [PubMed] [Google Scholar]
- Varis T., Kaukonen K. M., Kivistö K. T., Neuvonen P. J. (1998). Plasma Concentrations and Effects of Oral Methylprednisolone Are Considerably Increased by Itraconazole. Clin. Pharmacol. Ther. 64, 363–368. 10.1016/s0009-9236(98)90066-2 [DOI] [PubMed] [Google Scholar]
- Varis T., Kivistö K. T., Backman J. T., Neuvonen P. J. (2000a). The Cytochrome P450 3A4 Inhibitor Itraconazole Markedly Increases the Plasma Concentrations of Dexamethasone and Enhances its Adrenal-Suppressant Effect. Clin. Pharmacol. Ther. 68, 487–494. 10.1067/mcp.2000.110772 [DOI] [PubMed] [Google Scholar]
- Varis T., Kivistö K. T., Neuvonen P. J. (2000b). The Effect of Itraconazole on the Pharmacokinetics and Pharmacodynamics of Oral Prednisolone. Eur. J. Clin. Pharmacol. 56, 57–60. 10.1007/s002280050720 [DOI] [PubMed] [Google Scholar]
- Villeneuve J.-P., Pichette V. (2004). Cytochrome P450 and Liver Diseases. Curr. Drug Metab. 5, 273–282. 10.2174/1389200043335531 [DOI] [PubMed] [Google Scholar]
- Wang G., Wu C., Zhang Q., Yu B., Lü J., Zhang S., et al. (2020). Clinical Characteristics and the Risk Factors for Severe Events of Elderly Coronavirus Disease 2019 Patients. Zhong Nan Da Xue Xue Bao Yi Xue Ban 45, 542–548. 10.11817/j.issn.1672-7347.2020.200292 [DOI] [PubMed] [Google Scholar]
- Wang J., Yang W., Chen P., Guo J., Liu R., Wen P., et al. (2021). The Proportion and Effect of Corticosteroid Therapy in Patients with COVID-19 Infection: A Systematic Review and Meta-Analysis. PloS one 16, e0249481. 10.1371/journal.pone.0249481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Wang S., Qi J., Yu R., Zhuang J., Zhuang B., et al. (2018a). Impact of CYP2A6 Gene Polymorphism on the Pharmacokinetics of Dexmedetomidine for Premedication. Expert Rev. Clin. Pharmacol. 11, 917–922. 10.1080/17512433.2018.1510312 [DOI] [PubMed] [Google Scholar]
- Wang P., Qin X., Liu M., Wang X. (2018b). The Burgeoning Role of Cytochrome P450-Mediated Vitamin D Metabolites against Colorectal Cancer. Pharmacol. Res. 133, 9–20. 10.1016/j.phrs.2018.04.022 [DOI] [PubMed] [Google Scholar]
- Wang Z., Lin Y. S., Zheng X. E., Senn T., Hashizume T., Scian M., et al. (2012). An Inducible Cytochrome P450 3A4-dependent Vitamin D Catabolic Pathway. Mol. Pharmacol. 81, 498–509. 10.1124/mol.111.076356 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G. W., Van Ess P. J., Watson A. M., Mattson M. P., Blouin R. A. (2001). Cytochrome P450 and Antioxidant Activity in Interleukin-6 Knockout Mice after Induction of the Acute-phase Response. J. Interferon Cytokine Res. 21, 821–826. 10.1089/107999001753238060 [DOI] [PubMed] [Google Scholar]
- Weerink M. A. S., Struys M. M. R. F., Hannivoort L. N., Barends C. R. M., Absalom A. R., Colin P. (2017). Clinical Pharmacokinetics and Pharmacodynamics of Dexmedetomidine. Clin. Pharmacokinet. 56, 893–913. 10.1007/s40262-017-0507-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White C. M., Sicignano D. J., Smith K. (2021). Impact of Interferons and Biological Drug Inhibitors of IL-2 and IL-6 on Small-Molecule Drug Metabolism through the Cytochrome P450 System. Ann. Pharmacother. 56, 170–180. 10.1177/10600280211022281 [DOI] [PubMed] [Google Scholar]
- Who (2021). Coronavirus (COVID-19) Overview. Available from: https://covid19.who.int/December 29, 2021). [Google Scholar]
- Wonganan P., Jonsson-Schmunk K., Callahan S. M., Choi J. H., Croyle M. A. (2014). Evaluation of the HC-04 Cell Line as an In Vitro Model for Mechanistic Assessment of Changes in Hepatic Cytochrome P450 3A during Adenovirus Infection. Drug Metab. Dispos 42, 1191–1201. 10.1124/dmd.113.056663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Y., Zhong P. (2021). Clinical Progress on Management of Pneumonia Due to COVID-19 with Chinese Traditional Patent Medicines. Front. Pharmacol. 12, 655063. 10.3389/fphar.2021.655063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Y., Hao H., Wang H., Guo C., Kang A., Wang G. (2014). Reversing Effects of Lignans on CCl4-Induced Hepatic CYP450 Down Regulation by Attenuating Oxidative Stress. J. Ethnopharmacol 155, 213–221. 10.1016/j.jep.2014.05.016 [DOI] [PubMed] [Google Scholar]
- Xu J., Zhang X. C., Lv X. Q., Xu Y. Y., Wang G. X., Jiang B., et al. (2014). Effect of the Cytochrome P450 2D6*10 Genotype on the Pharmacokinetics of Tramadol in post-operative Patients. Pharmazie 69, 138–141. [PubMed] [Google Scholar]
- Xu Z., Shi L., Wang Y., Zhang J., Huang L., Zhang C., et al. (2020). Pathological Findings of COVID-19 Associated with Acute Respiratory Distress Syndrome. Lancet Respir. Med. 8, 420–422. 10.1016/s2213-2600(20)30076-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan B., Chu H., Yang D., Sze K. H., Lai P. M., Yuan S., et al. (2019). Characterization of the Lipidomic Profile of Human Coronavirus-Infected Cells: Implications for Lipid Metabolism Remodeling upon Coronavirus Replication. Viruses 11, 73. 10.3390/v11010073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan F., Hu Y., Di B., He P. L., Sun G. (2012). Effects of Some Antihypertensive Drugs on the Metabolism and Pharmacokinetics of Indapamide in Rats. J. Pharm. Pharm. Sci. 15, 208–220. a publication of the Canadian Society for Pharmaceutical Sciences, Societe canadienne des sciences pharmaceutiques. 10.18433/j3sk5v [DOI] [PubMed] [Google Scholar]
- Yuan S., Chu H., Chan J. F., Ye Z. W., Wen L., Yan B., et al. (2019). SREBP-dependent Lipidomic Reprogramming as a Broad-Spectrum Antiviral Target. Nat. Commun. 10, 120. 10.1038/s41467-018-08015-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanger U. M., Schwab M. (2013). Cytochrome P450 Enzymes in Drug Metabolism: Regulation of Gene Expression, Enzyme Activities, and Impact of Genetic Variation. Pharmacol. Ther. 138, 103–141. 10.1016/j.pharmthera.2012.12.007 [DOI] [PubMed] [Google Scholar]
- Zhang D., Guo R., Lei L., Liu H., Wang Y., Wang Y., et al. (2021a). Frontline Science: COVID-19 Infection Induces Readily Detectable Morphologic and Inflammation-Related Phenotypic Changes in Peripheral Blood Monocytes. J. Leukoc. Biol. 109, 13–22. 10.1002/jlb.4hi0720-470r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Liu W., Huang J., Chen Q. L., Wang D. D., Zou L. W., et al. (2021c). Inhibition of Drug-Metabolizing Enzymes by Jingyin Granules: Implications of Herb-Drug Interactions in Antiviral Therapy. Acta Pharmacol. Sin 1–10. 10.1038/s41401-021-00697-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang F., Huang J., Liu W., Wang C.-R., Liu Y.-F., Tu D.-Z., et al. (2021b). Inhibition of Drug-Metabolizing Enzymes by Qingfei Paidu Decoction: Implication of Herb-Drug Interactions in COVID-19 Pharmacotherapy. Food Chem. Toxicol. 149, 111998. 10.1016/j.fct.2021.111998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Kodani S., Hammock B. D. (2014). Stabilized Epoxygenated Fatty Acids Regulate Inflammation, Pain, Angiogenesis and Cancer. Prog. Lipid Res. 53, 108–123. 10.1016/j.plipres.2013.11.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Ding A., Yang D., Cui T., Yang H., Zhang H., et al. (2020). CYP2J2-produced Epoxyeicosatrienoic Acids Attenuate Ischemia/reperfusion-Induced Acute Kidney Injury by Activating the SIRT1-FoxO3a Pathway. Life Sci. 246, 117327. 10.1016/j.lfs.2020.117327 [DOI] [PubMed] [Google Scholar]
- Zhuang Y., De Vries D. E., Xu Z., Marciniak S. J., Jr., Chen D., Leon F., et al. (2015). Evaluation of Disease-Mediated Therapeutic Protein-Drug Interactions between an Anti-interleukin-6 Monoclonal Antibody (Sirukumab) and Cytochrome P450 Activities in a Phase 1 Study in Patients with Rheumatoid Arthritis Using a Cocktail Approach. J. Clin. Pharmacol. 55, 1386–1394. 10.1002/jcph.561 [DOI] [PubMed] [Google Scholar]
- Zvyaga T., Chang S. Y., Chen C., Yang Z., Vuppugalla R., Hurley J., et al. (2012). Evaluation of Six Proton Pump Inhibitors as Inhibitors of Various Human Cytochromes P450: Focus on Cytochrome P450 2C19. Drug Metab. Dispos 40, 1698–1711. 10.1124/dmd.112.045575 [DOI] [PubMed] [Google Scholar]