Abstract
A specific PCR for the detection of a variant of the gene encoding Shiga toxin 1 (stx1) called stx1OX3 (GenBank accession no. Z36901) was developed. The PCR was used to investigate 148 Stx1-producing Escherichia coli strains from human patients (n = 72), cattle (n = 27), sheep (n = 48), and a goat (n = 1) for the presence of the stx1OX3 gene. The stx1OX3 gene was present in 38 Shiga toxin-producing E. coli (STEC) strains from sheep belonging to serogroups O5, O125, O128, O146, and OX3 but was absent from Stx1-positive ovine STEC O91 strains. The stx1OX3 gene was also detected in 22 STEC strains from humans with nonbloody diarrhea and from asymptomatic excreters. Serotypes O146:H21 and O128:H2 were most frequently associated with stx1OX3-carrying STEC from sheep and humans. In contrast, Stx1-producing STEC strains from cattle and goats and 50 STEC strains from humans were all negative for the stx1OX3 gene. The stx1OX3-negative strains belonged to 13 serotypes which were different from those of the stx1OX3-positive STEC strains. Moreover, the stx1OX3 gene was not associated with STEC belonging to enterohemorrhagic E. coli (EHEC) serogroups O26, O103, O111, O118, O145, and O157. A bacteriophage carrying the stx1OX3 gene (phage 6220) was isolated from a human STEC O146:H21 strain. The phage was able to lysogenize laboratory E. coli K-12 strain C600. Phage 6220 shared a similar morphology and a high degree of DNA homology with Stx2-encoding phage 933W, which originates from EHEC O157. In contrast, few similarities were found between phage 6220 and Stx1-encoding bacteriophage H-19B from EHEC O26.
The production of Shiga toxins (verocytotoxins) Stx1 and Stx2 is associated with certain strains of Escherichia coli. Ruminant animals such as cattle, sheep, and goats are naturally colonized with Shiga-toxin-producing strains of E. coli (STEC) and shed these organisms with their feces into the environment. Humans can be infected with STEC by contaminated food and by direct transmission of bacteria, and some STEC types pathogenic for humans cause diarrhea, hemorrhagic colitis, and hemolytic-uremic syndrome (16). Over 200 serotypes of E. coli are known to be associated with the production of Shiga toxins (16). Analysis of the genes encoding Stx1 and Stx2 revealed the existence of various stx subtypes which show differences in their nucleotide sequences. In some strains, the stx genes were found to be carried by temperate bacteriophages, such as stx1 in STEC O26 and stx1 and stx2 in STEC O157 (12, 18, 20, 32, 34). A variant of the stx gene of Shigella sonnei and a variant of the stx2e gene, which is present in porcine STEC strains, were recently found to be carried by bacteriophages (7, 15). On the other hand, the stx gene of Shigella dysenteriae type 1 and some other variants of stx1 and stx2 present in E. coli were not associated with viable bacteriophages (11, 23, 35).
Paton et al. have described four variants of the stx1 gene in STEC strains isolated from humans and animals (22, 24). We found that one of these variants, called stx1OX3, is closely associated with STEC from sheep. The stx1OX3 gene was also detected in some human STEC isolates which belonged to the same serotypes as ovine STEC strains. Since the stx1OX3 gene was found in serologically diverse STEC strains, we have investigated its possible association with transmissible bacteriophages. Here, we describe the isolation and characterization of an Stx1OX3-encoding bacteriophage from an STEC O146:H21 strain. Our findings indicate that sheep could function as a source of stx1OX3-carrying STEC which might be transmitted to humans and which may cause gastrointestinal disease.
MATERIALS AND METHODS
Bacteria.
The STEC strains were fecal isolates from humans and animals and were from different locations in Germany (2, 5, 6). The strains were previously characterized for production of Stx1 and Stx2 by the verocytotoxin-producing E. coli reverse passive latex agglutination test and for the presence of the stx1 and stx2 genes by DNA hybridization and PCR (4, 6). A representative collection of 148 STEC strains which had previously tested positive for production of Stx1 and for stx1 sequences by PCR was investigated for the presence of the stx1OX3 variant (Table 1).
TABLE 1.
Presence of stx1OX3 gene in STEC strains of different serotypes and origin
| STEC serotype | Origin (no. of strains) | No. of strains with stx1 genotypea:
|
|
|---|---|---|---|
| stx1OX3 | stx1 | ||
| O5:NM | Sheep (10) | 10 | 0 |
| O26:H11/NMb | Cattle (5), human (7) | 0 | 12 |
| O103:H2 | Goat (1), human (9) | 0 | 10 |
| O111:H8/NM | Cattle (2), human (5) | 0 | 7 |
| O118:H16/NM | Cattle (5), human (5) | 0 | 10 |
| O145:NM | Cattle (1), human (5) | 0 | 6 |
| O157:H7/NM | Human (10) | 0 | 10 |
| O22:H8 | Cattle (4), human (2) | 0 | 6 |
| O22:H21 | Human (2) | 2 | 0 |
| O76:H19 | Human (2) | 2 | 0 |
| O76:H21 | Cattle (2) | 0 | 2 |
| O91:NM | Sheep (10) | 0 | 10 |
| O105:H18 | Cattle (1), human (1) | 0 | 2 |
| O125:H- | Sheep (1) | 1 | 0 |
| O146:H28 | Cattle (3) | 0 | 3 |
| O146:H21 | Sheep (10), human (10) | 20 | 0 |
| O156:Hdivc | Cattle (3), human (3) | 0 | 6 |
| O128:H2 | Sheep (11), human (8) | 19 | 0 |
| O128:H10 | Human (1) | 0 | 1 |
| OX3d:H2 | Cattle (1), human (2) | 0 | 3 |
| OX3:H8 | Sheep (6) | 6 | 0 |
All strains were positive for production of Stx1. The stx1OX3 variant was detected by PCR, as described in the text.
NM, nonmotile.
Includes O156:H21 (n = 4), O156:H25 (n = 1), and O156:Hrough (n = 1) strains.
OX3 is a provisional name for a new O antigen in E. coli (2).
The E. coli strains which were used as reference strains for stx1 and its variants are listed in Table 2. E. coli K-12 strains harboring the Stx1-encoding bacteriophage H-19B (C600–H-19B) and the Stx2-encoding bacteriophage 933W (C600-W34) were used for production of phage H-19B and 933W lysates, respectively. Nontoxigenic laboratory E. coli K-12 strain C600 harbors no inducible bacteriophage and was used for plating and propagation of phages. Four STEC O146:H21 strains and two STEC O128:H2 strains were used for induction of Stx-encoding bacteriophages (Table 2). One of these strains, called CB6220 (O146:H21), was the source of the bacteriophage carrying the stx1OX3 gene which was investigated in the present study.
TABLE 2.
E. coli strains used for characterization of Stx1OX3-encoding bacteriophages
| Strain no. | Origin | Serotype | stx gene(s)a | Reference |
|---|---|---|---|---|
| C600 | E. coli K-12 | − | − | 28 |
| DH5α | E. coli K-12 | − | − | 28 |
| JM83 | E. coli K-12 | − | − | 28 |
| C600–H-19B | E. coli K-12 | − | stx1 | 3 |
| C600–W34 | E. coli K-12 | − | stx2 | 3 |
| C600–6220 | E. coli K-12 | stx1OX3 | C600 phage 6220 lysogen (this work) | |
| M109(pJCP526) | E. coli K-12 | − | stx1OX3 | 24 |
| M109(pJCP525) | E. coli K-12 | − | stx1CB | 24 |
| M109(pJCP524) | E. coli K-12 | − | stx1O48 | 24 |
| CB6220bc | Human | O146:H21 | stx1OX3 | This work |
| DG131/2bc | Sheep | O146:H21 | stx1OX3 | This work |
| DG137/4b | Sheep | O146:H21 | stx1OX3 + stx2d-Ount | This work |
| DG224/5b | Sheep | O146:H21 | stx1OX3 + stx2d-Ount | This work |
| CB6562b | Human | O128:H2 | stx1OX3 | This work |
| CB6223b | Human | O128:H− | stx1OX3 + stx2d-Ount | This work |
| CB7367c | Human | O128:H2 | stx1OX3 + stx2d-Ount | This work |
| DG128/1c | Sheep | O128:H2 | stx1OX3 + stx2d-Ount | This work |
| CB6595c | Human | O76:H19 | stx1OX3 | This work |
| CB7151c | Human | O22:H8 | stx1OX3 | This work |
| CDC 2950–54d | Human | O146:H21 | − | 21 |
| Ciglerisd | Human | O128:H2 | − | 21 |
| H77d | Human | O90:H− | − | 21 |
| H307bd | Human | O91:H− | − | 21 |
| F41d | Human | O26:H− | − | 21 |
Investigated by PCR and restriction fragment analysis of PCR products, as described in Materials and Methods; −, stx negative.
Strains used for induction of Stx-encoding bacteriophages.
determination of the stx1OX3 PCR product by nucleotide sequencing.
Wild-type E. coli strains used for propagation of stx1OX3 bacteriophage.
Isolation of temperate bacteriophages and preparation of phage lysates.
Bacteria were grown from single colonies in Luria broth (28) to the exponential growth phase. Mitomycin C was added to the cultures to a final concentration of 0.5 μg/ml. These cultures were then further incubated for 5 h and the bacterial culture fluid was harvested by centrifugation. The bacteria were removed from the supernatant by membrane filtration (pore size, 0.2 μm). The supernatant was incubated with 1 μg each of DNase and RNase C (Sigma-Aldrich, Steinheim, Germany) per ml for 30 min at 37°C. The presence of bacteriophages in the culture fluid was examined by titration on E. coli K-12 strain C600. Purification of high-titer phage lysates was performed on CsCl step gradients by ultracentrifugation as described previously (28).
Electron microscopy.
CsCl-purified bacteriophage particles were absorbed to carbon films and negatively stained with 1% uranyl acetate (pH 4.5) or with 2% potassium phosphowolframate (pH 7.2), as described by Steven et al. (33). The samples were examined with a Philips CM100 electron microscope.
Isolation of E. coli strains lysogenic for phage 6220.
Lysogenic E. coli strains harboring phage 6220 were selected by spotting 10 μl of phage lysate onto 0.7% Luria-Bertani (LB) top agar (28) which had been inoculated with E. coli C600. Bacteriophage lysis zones became visible after overnight growth of bacteria at 37°C. Colonies in the lysis zones were propagated on Endo agar (Merck, Darmstadt, Germany) and purified to single-colony isolates. Lysogenization of C600 was examined by mitomycin C induction, as described above. The lysogenic C600 strains were examined for the presence of the stx1OX3 toxin gene by the stx1OX3 PCR.
Bacteriophage immunity test.
Phage immunity was tested on LB plates by spotting 10 μl of phage 6220 lysate onto the top agar overlay containing 100 μl of an overnight culture of E. coli K-12 or wild-type E. coli strains (Table 2).
Isolation and characterization of bacteriophage DNA.
DNA was isolated from phages 6220, H-19B, and 933W with the Qiagen Lambda kit (Qiagen, Hilden, Germany), according to the instructions of the supplier. Purified DNA was digested with EcoRI and PvuII (MBI Fermentas, Vilnius, Lithuania), and the restriction fragments were separated on 0.8% agarose gels as described previously (28). After electrophoresis, DNA was transferred to nitrocellulose membranes (Hybond-N+; Amersham Pharmacia Biotech Limited, Little Chalfont, Buckinghamshire, United Kingdom) by capillary blotting (28). DNA probes were labeled with fluorescein-N6-ATP (NEN Life Science, Boston, Mass.), according to the manufacturer's instructions. The membranes were hybridized overnight at 65°C.
Detection of stx gene sequences and subtyping of stx genes.
The PCR conditions and primers used in the present work are listed in Table 3. Different types of stx genes were amplified by the PCR described previously (1, 14). Primer pair KS7-KS8 (29) was used for nonspecific amplification of stx1 and its genetic variants (29, 30). Primer pair LP43–44 was used for nonspecific amplification of the stx2 and stx2 gene variants (8). Analysis of the stx2 variants by PCR and restriction endonuclease digestion was performed as described by Pierard et al. (25).
TABLE 3.
PCR conditions and primers used in this study
| Primer pair | Nucleotide sequence of primers | Specificity | PCR product size (bp) | PCR conditionsa | Reference(s) |
|---|---|---|---|---|---|
| Lin-up | 5′-GAACGAAATAATTTATATGT-3′ | stx1 and stx2 (all variants, A and B subunits) | ≅900b | 94.0°C, 60 sc | 1, 14 |
| Lin-down | 5′-TTTGATTGTTACAGTCAT-3′ | 43.0°C, 90 sd | |||
| 72.0°C, 90 se | |||||
| Lin-up | 5′-GAACGAAATAATTTATATGT-3′ | stx1OX3 (A subunit) | 555 | 94.0°C, 60 sc | This study |
| 1OX3 | 5′-CTCATTAGGTACAATTCT-3′ | 48.1°C, 90 sd | |||
| 72.0°C, 90 se | |||||
| StxV1F1 | 5′-TCGCATGAGATCTGACC-3′ | stx1 (all variants, complete A and B subunits) | 1,470 | 94.0°C, 60 sc | 24 |
| StxV1B1 | 5′-AACTGACTGAATTGAGATG-3′ | 49.9°C, 90 sd | |||
| 72.0°C, 90 se | |||||
| LP43 | 5′-ATCCTATTCCCGGGAGTTTACG-3′ | stx2 (all variants, A subunit) | 584 | 94.0°C, 30 sc | 8 |
| LP44 | 5′-GCGTCATCGTATACACAGGAGC-3′ | 57.0°C, 60 sd | |||
| 72.0°C, 60 se |
All PCRs were run for 30 cycles with a final extension step of 5 min at 72°C.
The size of the amplified DNA varies by a few nucleotides, depending on the stx gene variant (1, 14).
Denaturing.
Annealing.
Extension.
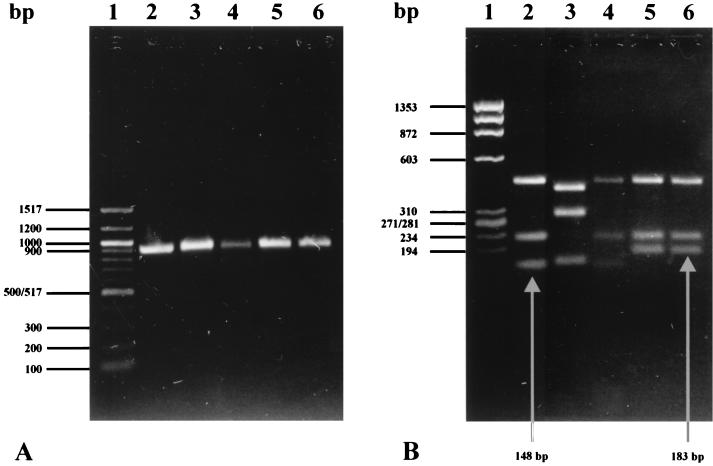
Subtyping of stx genes was performed by restriction fragment analysis of the ≅900-bp amplification product obtained by PCR with primers Lin-up and Lin-down as described previously (1, 14). The reference strains for the different stx types and the interpretation of restriction fragment patterns were described previously (1). The stx1OX3 variant could be distinguished from other stx1 types by its characteristic restriction profile when the PCR amplification product was digested with HinfI (Fig. 1B).
FIG. 1.
(A) Agarose gel showing amplified PCR products obtained with primers Lin-up and Lin-down. Lane 1, molecular mass marker (100-bp ladder; Gibco-BRL); lane 2, C600–H-19B (stx1); lane 3, C600-W34 (stx2); lane 4, CB6220 (stx1OX3); lane 5, C600-6220, (stx1OX3); lane 6, phage 6220 (stx1OX3). (B) HinfI digestion fragments of PCR products shown in panel A. Lane 1, molecular mass marker (φX174/HaeIII; MBI Fermentas); lanes 2 to 6 correspond to those in panel A.
PCR.
PCRs were performed with a GeneAmp PCR system 2400 (Perkin-Elmer, PE Applied Biosystems, Norwalk, Conn.) in a total volume of 50 μl. Target DNA was diluted 1:20 with double-distilled water. Overnight cultures of E. coli were diluted 1:40 with distilled water and heated at 96°C for 10 min prior to addition of the reaction mixture (27.5 μl). The reaction mixture contained 5 μl of 10-fold-concentrated polymerase buffer, 5 μl of 25 mM MgCl2, 5 μl of each primer (30 ng/μl), and 2.5 μl of Taq polymerase (1 U/μl; MBI Fermentas).
Nucleotide sequencing.
Complete nucleotide sequencing of the stx1OX3 gene was performed with PCR products obtained with primers StxV1F1 and StxV1B1 (24). The PCR product of 1,470 bp encompasses the complete stx gene and adjacent sequences (Table 3). Both strands of the stx1OX3 toxin gene were sequenced with a cycle sequencing kit (Prism Big Dye FS terminator cycle sequencing ready reaction kit; PE Applied Biosystems) and were run on an automated ABI Prism 377 sequencer (Perkin-Elmer). Nucleotide sequences were analyzed with AutoAssembler software packages (PE Applied Biosystems).
Nucleotide sequence accession number.
The accession number for the stx1OX3 gene is EMBL AJ413986.
RESULTS
Molecular detection of stx1OX3 variant gene.
Four variants of the stx1 gene were described by Paton et al. (22, 24). One of these variants, called stx1OX3, showed the greatest deviation from the stx1 gene, with a total of 43 nucleotide differences leading to changes in 12 amino acid residues (24). This allowed us to develop a specific PCR for the stx1OX3 variant which was used for detection of stx1OX3-positive strains among STEC strains of different origins and serotypes (Table 3).
The primer pair used for specific amplification of the stx1OX3 sequence was developed by alignment of the stx1OX3 gene sequence with previously published nucleotide sequences from various stx genes (sequences with GenBank accession nos. AF125520 [stx2], AF034975 [stx1], Z36899 [stx1O46], Z36900 [stx1CB], Z36901 [stx1OX3], and L04539 [stx1O111]. The forward primer (primer Lin-up) is specific for a region in the StxA subunit which has a conserved sequence in various stx1 and stx2 genes (1, 14). Backward primer 1OX3 covers a sequence in the A subunit which is specific for the stx1OX3 gene. The specificity of the product obtained by PCR with the primers Lin-up and 1OX3 was investigated by nucleotide sequencing of the 555-bp amplification products obtained from five stx1OX3-positive strains listed in Table 2 and from purified bacteriophage 6220 (see below). Amplification of stx genes other than stx1OX3 was not observed with this primer pair (data not shown).
Amplification of the stx1OX3 gene with the primers Lin-up and Lin-down resulted in a 901-bp PCR product showing a characteristic restriction profile with HinfI. Corresponding to the nucleotide sequence, HinfI digestion generated five fragments (of 435, 222, 148, 61, and 35 bp) with the stx1 gene and four fragments (of 436, 222, 183, and 59 bp) with the stx1OX3 gene (Fig. 1B).
Prevalence of stx1OX3 variant in STEC isolates from animals and humans.
One hundred forty-eight STEC strains of human and animal origin belonging to 24 different O:H serotypes were investigated for the presence of the stx1OX3 gene by PCR. The 148 strains were chosen as representatives among Stx1-positive STEC strains which were isolated from humans (n = 72) and animals (n = 76) in Germany between 1989 and 1999 (2–6). The results are shown in Table 1.
The stx1OX3 variant was found in 60 of the 148 strains. All STEC isolates from sheep which belonged to serogroups O5 (n = 10), O125 (n = 1), O128 (n = 11), O146 (n = 10), and OX3 (n = 6) were positive for the stx1OX3 gene. However, ovine STEC O91 strains (n = 10) were all stx1OX3 negative. The stx1OX3 gene was also found in 22 STEC strains from humans which belonged to serogroups O22 (n = 2), O76 (n = 2), O146 (n = 10), and O128 (n = 8). Serotypes O146:H21 and O128:H2 were present among both human and ovine stx1OX3-positive STEC strains (Table 1). Thirteen of the 22 human STEC strains which carried the stx1OX3 gene were isolated from adults. The human STEC strains carrying the stx1OX3 gene were from patients with nonbloody diarrhea (n = 17) or were from patients who had no symptoms of gastrointestinal disease (n = 5).
The stx1OX3 variant gene was not found among STEC isolates from cattle (n = 27) and goats (n = 1), which belonged to 11 different serotypes. The remaining 50 STEC isolates from humans were also negative for the stx1OX3 gene. The Stx1OX3-negative STEC strains from cattle, goats, and humans shared eight common serotypes (serotypes O22:H8, O26:H11, O103:H2, O105:H18, O111:H8, O118:H16, O145:NM, and OX3:H2). All STEC strains which belonged to the so-called enterohemorrhagic E. coli (EHEC) serogroups (serogroups O26, O103, O111, O145, and O157) were negative for the stx1OX3 gene, independent of their human or animal origin.
Isolation of Stx1OX3-encoding bacteriophages.
Six stx1OX3-positive strains that were of human and ovine origin and that belonged to serotypes O128:H2 and O146:H21 were analyzed for the presence of Stx-encoding bacteriophages (Table 2). Broth cultures of the STEC strains were treated with mitomycin C and were examined for the release of phages (see Materials and Methods). Phage plaques were obtained from all strains; however, only three phage lysates from strains DG131/2, DG224/5, and CB6220 (all O146:H21) were positive for stx-specific DNA sequences by PCR. Only one of the phage isolates from human strain CB6220 produced stable, high-titer lysates (>109 phages/ml) on E. coli K-12 strain C600. The phage lysates from CB6220 were concentrated by ultracentrifugation and used for further analysis. The presence of the stx1OX3 gene on the genome of bacteriophage 6220 was indicated by PCR and was confirmed by nucleotide sequencing of the PCR amplification product of the complete stx1OX3 gene (with primers StxV1F1 and StxV1B1). The nucleotide sequence of the stx1OX3 gene on phage 6220 showed 100% homology to the previously published stx1OX3 sequence (GenBank accession no. Z36901). stx2 or stx2 variant genes were not present on the phage or in the original host strain, CB6220 (Table 2).
Morphology and host range of bacteriophage 6220.
The morphology of bacteriophage 6220 was compared with those of other well-characterized Stx-encoding bacteriophages, such as H-19B and 933W (19). Phage 6220 particles showed a head size of 69.5 to 75 nm with a tail of 25 nm. Phage 6220 and phage 933W exhibited similar morphologies. Phage H-19B is different in its morphology, with an elongated head and a longer tail (38).
The plaque-forming abilities of phage 6220 on laboratory and wild-type E. coli strains and lysogenization of laboratory and wild-type E. coli strains with phage 6220 were investigated (Materials and Methods). Three E. coli K-12 strains (strains C600, DH5α, and JM83) and five stx-negative wild-type E. coli strains belonging to different serotypes were used (Table 2). Small clear plaques were formed on the E. coli K-12 strains, but no plaques were obtained on any of the wild-type E. coli strains. Derivatives of C600 lysogenic for phage 6220 were isolated from bacterial colonies that grew in the lysis zones of phage 6220 on C600. Bacteriophage release was observed after mitomycin C induction of C600 lysogens, and the presence of the stx1OX3 gene was demonstrated by PCR and by a positive result by the Vero cell toxicity test.
DNA homologies between Stx-encoding bacteriophages.
The genome size of phage 6220 was calculated to be 60 ± 2 kb from the sum of the EcoRI restriction fragments of phage DNA. This is similar to the size of the genome of phage 933W, which is reported to be 61.67 kb (26). In contrast, phage H-19B has a smaller genome size, 41 kb (10).
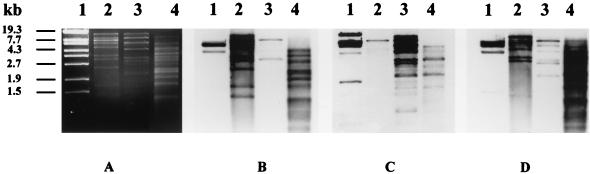
Cross hybridization of EcoRI- and PvuII-digested DNAs of phages 6220, H-19B, and 933W revealed a high degree of DNA homology between phages 6220 and 933W (Fig. 2B and D, lanes 2 and 4). In contrast, only very little DNA homology was found between phages 6220 and H-19B (Fig. 2B and C, lanes 2 and 3). Phages 933W and H-19B showed more DNA homology to each other than phages H-19B and 6220 did to each other (Fig. 2B to D, lanes 2 to 4). All three phages showed a minor degree of DNA homology with bacteriophage lambda (Fig. 2B to D, lanes 1).
FIG. 2.
(A to D) Agarose gel electrophoresis and Southern hybridization of phage DNAs. (A) Agarose gel electrophoresis of restriction endonuclease-digested DNA of phages 6220, H-19B, and 933W. Lane 1, molecular mass marker (bacteriophage λEco130I; MBI Fermentas); lane 2, EcoRI-digested DNA of phage 6220; lane 3, EcoRI-digested DNA of phage H-19B; lane 4, PvuII-digested DNA of phage 933W. (B) Hybridization of gel shown in panel A with EcoRI-digested DNA of phage 6220 as the gene probe. (C) Hybridization of gel shown in panel A with EcoRI-digested DNA of phage H-19B as the gene probe. (D) Hybridization of gel shown in panel A with PvuII-digested DNA of phage 933 as the gene probe.
DISCUSSION
Since the first detection of Shiga toxin production in E. coli, increasing numbers of genetic variants of the major types of toxin genes, stx1 and stx2, have been described. The stx1 and stx2 toxin genes were found to be carried by lysogenic bacteriophages in the chromosomes of STEC strains of different serotypes (19). The stx gene of S. dysenteriae type 1, which is highly similiar to the stx1 gene of E. coli, was recently found to be associated with bacteriophage DNA sequences, although viable phages could not be isolated from these strains (35, 36). More recently, some of the genetic variants of stx, stx1, and stx2 were shown to be encoded by bacteriophages in strains of S. sonnei and E. coli, respectively (7, 15; this work).
In the present study, we examined the association of the stx1OX3 gene with STEC strains from different sources and serotypes. The stx1OX3 variant was first described by Paton et al. (24) and was cloned from an ovine STEC OX3:H8 strain. Among the different variants of the stx1 gene, stx1OX3 is the only one which shows sufficient differences in its nucleotide sequence to be specifically detected by PCR. All other genetic variants of stx and stx1 show very few differences from each other and are thus detectable only by nucleotide sequence analysis of the toxin genes. In the present study, we have developed a specific PCR for identification of the stx1OX3 gene. The specificity of the stx1OX3 PCR was confirmed by nucleotide sequencing of PCR products obtained from different STEC strains and from bacteriophage 6220. No amplification of DNA was obtained with STEC strains carrying stx genes other than stx1OX3.
The investigation of 148 STEC strains from humans and animals revealed that the stx1OX3 gene is associated with certain serotypes of STEC and with healthy sheep as an animal host. The only exceptions were ovine STEC O91:NM strains that carried all of the stx1 gene and that were positive for Stx1 production. STEC strains belonging to serogroups O5, O91, and O128 have frequently been isolated from healthy sheep in Europe, Australia, and North America (3, 5, 9, 13, 27). The ovine STEC strains investigated in our study originated from different flocks of sheep in Germany (3, 5). The association of the stx1OX3 gene with larger numbers of ovine STEC strains from other geographical locations needs to be further investigated.
In contrast, none of the bovine STEC strains produced Stx1, were positive for an stx1 gene, or carried the stx1OX3 variant. Similar observations were made previously for the stx2e gene, which is associated with certain STEC strains and serotypes that cause edema disease in pigs but which are not isolated or which are rarely isolated from other animals or humans (37).
Our finding that stx1OX3-carrying strains were also present in humans with diarrhea indicates that sheep may be the source of infection. Some of the human stx1OX3 STEC isolates belonged to the same serotypes (serotypes O146:H21 and O128:H2) as the ovine strains. STEC O146:H21 and O128:H2 strains are frequently found in healthy sheep in Germany, and both serotypes could be assigned to distinct clonal types of STEC (2, 5). However, it is not yet known how humans become infected with STEC stx1OX3 strains. Direct contact with STEC-excreting sheep and consumption of STEC-contaminated food of ovine origin may contribute to the spread of ovine STEC to humans.
Interestingly, none of the 55 Stx1-producing strains which belonged to the classical EHEC types were found to carry the stx1OX3 gene. Typical EHEC strains carry the genes for the attaching-and-effacing phenotype and for a plasmid-encoded hemolysin and are frequently associated with hemorrhagic colitis and hemolytic-uremic syndrome (16, 31). In contrast, all STEC strains from our study which carried the stx1OX3 gene were negative for the eae gene (unpublished results), and most of these were from adults with nonbloody diarrhea or from asymptomatic excreters. We previously reported that eae-negative STEC strains are more frequently isolated from adults in Germany and are generally associated with milder illness (6). Further studies are necessary to assess the role of the stx1OX3 gene in the pathogenicities of STEC strains for humans.
Like phages H-19B and 933W, phage 6220 is able to infect and lysogenize E. coli K-12 strains, which shows that the phage can integrate into the genomes of genetically heterogeneous E. coli types. This could explain why the stx1OX3 gene is found in serologically heterogeneous STEC strains which belong to different clonal lineages. The host range and the spread of Stx1OX3-encoding bacteriophages in E. coli need to be explored further. Only STEC O146:H21 strains produced plaque-forming Stx1OX3-encoding bacteriophages after mitomycin C induction. In contrast, stx1OX3 gene-carrying bacteriophages could not be isolated from STEC OX3:H8 and O128:H2 strains (24; this work). The phages which were isolated from STEC O128:H2 strains after mitomycin C induction may have lost their stx genes upon subculture or may be stx-negative phages carried by these strains. Further investigations with phage 6220-specific DNA probes could be used to investigate the presence of this bacteriophage in other STEC strains which carry the stx1OX3 gene.
None of the wild-type E. coli strains tested in the present study showed plaque formation with bacteriophage 6220. This finding is similar to those described for other Stx-encoding bacteriophages, which nevertheless were able to lysogenize different wild-type E. coli strains (30). Recent investigations have shown that stx1, stx2, and stx2c genes are genetically linked to bacteriophage-specific sequences in serologically different STEC strains (36). These data and the finding that genetically highly similar stx genes are present in genetically unrelated STEC strains indicate that bacteriophage transmission is the major vehicle in the spread of stx genes in E. coli.
Comparison of bacteriophage 6220 with other Stx-encoding bacteriophages revealed that phage 6220 is morphologically and genetically more related to Stx2-encoding phage 933W (26) than to stx1-carrying bacteriophage H-19B (17). Nucleotide sequence analysis of the flanking regions downstream of the stx2 or stx2c genes in E. coli O26, O145, and O157 strains revealed 83 to 86% DNA homology to bacteriophage 933W sequences (36), indicating that Stx2- and Stx2c-encoding bacteriophages are closely related to each other. The high degree of nucleotide sequence homology which we found between the genomes of phage 6220 and 933W could indicate that the stx1OX3 gene is acquired by a 933W-like bacteriophage by recombination. It appears to be unlikely that phage 6220 is a derivative of phage H-19B since both phages are morphologically different and the genome of phage H-19B is significantly smaller than that of phage 6220. Like the other Stx-encoding bacteriophages, phage 6220 showed some homology to bacteriophage lambda, which indicates that it belongs to the family of lambdoid bacteriophages (17, 26).
Our findings contribute to those of other investigators who have shown that lambdoid bacteriophages play a major role in the inheritance and transmission of stx genes in E. coli. Sheep appear to constitute a natural reservoir for STEC strains which carry the stx1OX3 gene, and the occurrence of such STEC strains in humans could indicate that these were acquired from sheep. The possible biological significance of the association of the stx1OX3 gene with STEC strains from sheep is unknown at present but needs to be further explored.
ACKNOWLEDGMENTS
We are grateful to Henry Smith, Central Public Health Laboratory, London, United Kingdom, for critical reading of the manuscript and for helpful suggestions.
This work was supported by the German Ministry of Health (BMG).
REFERENCES
- 1.Bastian S N, Carle I, Grimont F. Comparision of 14 PCR systems for the detection and subtyping of stx genes in Shiga-toxin-producing Escherichia coli. Res Microbiol. 1998;149:457–472. doi: 10.1016/s0923-2508(98)80001-6. [DOI] [PubMed] [Google Scholar]
- 2.Beutin L, Geier D, Steinrueck H, Zimmermann S, Scheutz F. Prevalence and some properties of verotoxin (Shiga-like toxin)-producing Escherichia coli in seven different species of healthy domestic animals. J Clin Microbiol. 1993;31:2483–2488. doi: 10.1128/jcm.31.9.2483-2488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beutin L, Geier D, Zimmermann S, Karch H. Virulence markers of Shiga-like toxin-producing Escherichia coli strains originating from healthy domestic animals of different species. J Clin Microbiol. 1995;33:631–635. doi: 10.1128/jcm.33.3.631-635.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beutin L, Zimmermann S, Gleier K. Rapid detection and isolation of Shiga-like toxin (verocytotoxin)-producing Escherichia coli by direct testing of individual enterohemolytic colonies from washed sheep blood agar plates in the VTEC-RPLA assay. J Clin Microbiol. 1996;34:2812–2814. doi: 10.1128/jcm.34.11.2812-2814.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beutin, L., D. Geier, S. Zimmermann, S. Aleksic, H. A. Gillespie, and T. S. Whittam. 1997. Epidemiological relatedness and clonal types of natural populations of Escherichia coli strains producing Shiga toxins in separate populations of cattle and sheep. 63:2175–2180. [DOI] [PMC free article] [PubMed]
- 6.Beutin L, Zimmermann S, Gleier K. Human infections with Shiga toxin-producing Escherichia coli other than serogroup O157 in Germany. Emerg Infect Dis. 1998;4:635–639. doi: 10.3201/eid0404.980415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beutin L, Strauch E, Fischer I. Isolation of Shigella sonnei lysogenic for a bacteriophage encoding gene for production of Shiga toxin. Lancet. 1999;353:1498. doi: 10.1016/S0140-6736(99)00961-7. [DOI] [PubMed] [Google Scholar]
- 8.Cebula T A, Payne W L, Feng P. Simultaneous identification of strains of Escherichia coli serotype O157 and their Shiga-like toxin type by mismatch amplification mutation assay-multiplex PCR. J Clin Microbiol. 1995;33:248–250. doi: 10.1128/jcm.33.1.248-250.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Djordjevic S P, Hornitzky M A, Bailey G, Gill P, Vanselow B, Walker K, Bettelheim K A. Virulence properties and serotypes of Shiga toxin-producing Escherichia coli from healthy Australian slaughter-age sheep. J Clin Microbiol. 2001;39:2017–2021. doi: 10.1128/JCM.39.5.2017-2021.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Huang A, Friesen J, Brunton J L. Characterization of a bacteriophage that carries the genes for production of Shiga-like toxin 1 in Escherichia coli. J Bacteriol. 1987;169:4308–4312. doi: 10.1128/jb.169.9.4308-4312.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito H, Terai A, Kurazono H, Takeda Y, Nishibuchi M. Cloning and nucleotide sequencing of Vero toxin 2 variant genes from Escherichia coli O91:H21 isolated from a patient with the hemolytic uremic syndrome. Microb Pathog. 1990;8:47–60. doi: 10.1016/0882-4010(90)90007-d. [DOI] [PubMed] [Google Scholar]
- 12.Jackson M P, Neill R J, O'Brien A D, Holmes R K, Newland J W. Nucleotide sequence analysis and comparison of the structural genes for Shiga-like toxin I and Shiga-like toxin II encoded by bacteriophages from Escherichia coli 933. FEMS Microbiol Lett. 1987;44:109–114. doi: 10.1016/0882-4010(87)90106-9. [DOI] [PubMed] [Google Scholar]
- 13.Kudva I T, Hatfield P G, Hovde C J. Characterization of Escherichia coli O157:H7 and other Shiga toxin-producing E. coli serotypes isolated from sheep. J Clin Microbiol. 1997;35:892–899. doi: 10.1128/jcm.35.4.892-899.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lin Z, Kurazono H, Yamasaki S, Takeda Y. Detection of various variant Verotoxin genes in Escherichia coli by polymerase chain reaction. Microbiol Immunol. 1993;37:543–548. doi: 10.1111/j.1348-0421.1993.tb01675.x. [DOI] [PubMed] [Google Scholar]
- 15.Muniesa M, Recktenwald J, Bielaszewska M, Karch H, Schmidt H. Characterization of a Shiga toxin 2e-converting bacteriophage from an Escherichia coli strain of human origin. Infect Immun. 2000;68:4850–4855. doi: 10.1128/iai.68.9.4850-4855.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neely M N, Friedman D I. Functional and genetic analysis of regulatory regions of coliphage H-19B: location of Shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol Microbiol. 1998;28:1255–1267. doi: 10.1046/j.1365-2958.1998.00890.x. [DOI] [PubMed] [Google Scholar]
- 18.Newland J W, Strockbine N A, Miller S F, O'Brien A D, Holmes R K. Cloning of Shiga-like toxin structural genes from a toxin converting phage of Escherichia coli. Science. 1985;230:179–181. doi: 10.1126/science.2994228. [DOI] [PubMed] [Google Scholar]
- 19.O'Brien A D, Newland J W, Miller S F, Holmes R K. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science. 1984;226:694–696. doi: 10.1126/science.6387911. [DOI] [PubMed] [Google Scholar]
- 20.O'Brien A D, Marques L R M, Kerry C F, Newland J W, Holmes R K. Shiga-like toxin converting phage of enterohemorrhagic Escherichia coli strain 933. Microb Pathog. 1989;6:381–390. doi: 10.1016/0882-4010(89)90080-6. [DOI] [PubMed] [Google Scholar]
- 21.Orskov I, Orskov F, Jann B, Jann K. Serology, chemistry, and genetics of O and K antigens of Escherichia coli. Bacteriol Rev. 1977;41:667–710. doi: 10.1128/br.41.3.667-710.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Paton A W, Paton J C, Goldwater P N, Heuzenroeder M W, Manning P A. Sequence of a variant Shiga-like toxin type 1 operon of Escherichia coli O111:H−. Gene. 1993;129:87–92. doi: 10.1016/0378-1119(93)90700-d. [DOI] [PubMed] [Google Scholar]
- 23.Paton A W, Paton J C, Manning P A. Polymerase chain reaction amplification, cloning and sequencing of variant Escherichia coli Shiga-like toxin type II operons. Microb Pathog. 1993;15:77–82. doi: 10.1006/mpat.1993.1058. [DOI] [PubMed] [Google Scholar]
- 24.Paton A W, Beutin L, Paton J C. Heterogeneity of the amino-acid sequences of Escherichia coli Shiga-like toxin type-I operons. Gene. 1995;153:71–74. doi: 10.1016/0378-1119(94)00777-p. [DOI] [PubMed] [Google Scholar]
- 25.Pierard D, Muyldermans G, Moriau L, Stevens D, Lauwers S. Identification of new Verocytotoxin type 2 variant B-subunit genes in human and animal Escherichia coli isolates. J Clin Microbiol. 1998;36:3317–3322. doi: 10.1128/jcm.36.11.3317-3322.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plunkett G, Rose D J, Durfee T J, Blattner F R. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J Bacteriol. 1999;181:1767–1778. doi: 10.1128/jb.181.6.1767-1778.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramachandran V, Hornitzky M A, Bettelheim K A, Walker M J, Djordjevic S P. The common ovine Shiga toxin 2-containing Escherichia coli serotypes and human isolates of the same serotypes possess a Stx2d toxin type. J Clin Microbiol. 2001;39:1932–1937. doi: 10.1128/JCM.39.5.1932-1937.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 29.Schmidt H, Russmann H, Schwarzkopf A, Aleksic S, Heesemann J, Karch H. Prevalence of attaching and effacing Escherichia coli in stool samples from patients and controls. Zentbl Bakteriol Parasitenkd Infektkrankh Hyg Abt 1 Orig. 1994;281:201–213. doi: 10.1016/s0934-8840(11)80571-2. [DOI] [PubMed] [Google Scholar]
- 30.Schmidt H, Bielaszewska M, Karch H. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage φ3538 isolated from Escherichia coli O157:H7. Appl Environ Microbiol. 1999;65:3855–3861. doi: 10.1128/aem.65.9.3855-3861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schmidt H, Geitz C, Tarr P I, Frosch M, Karch H. Non-O157:H7 pathogenic Shiga toxin-producing Escherichia coli: phenotypic and genetic profiling of virulence traits and evidence of clonality. J Infect Dis. 1999;179:115–123. doi: 10.1086/314537. [DOI] [PubMed] [Google Scholar]
- 32.Smith H W, Green P, Parsell Z. Vero cell toxins in Escherichia coli and related bacteria: transfer by phage and conjugation and toxic action in laboratory animals, chickens and pigs. J Gen Microbiol. 1983;129:3121–3137. doi: 10.1099/00221287-129-10-3121. [DOI] [PubMed] [Google Scholar]
- 33.Steven A C, Trus B L, Maizel J V, Unser M, Parry D A D, Wall J S, Hainfeld J F, Studier F W. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J Mol Biol. 1988;200:351–365. doi: 10.1016/0022-2836(88)90246-x. [DOI] [PubMed] [Google Scholar]
- 34.Strockbine N A, Marques L R M, Newland J W, Smith H W, Holmes R K, O'Brien A D. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities. Infect Immun. 1986;53:135–140. doi: 10.1128/iai.53.1.135-140.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Strockbine N A, Jackson M P, Sung L M, Holmes R K, O'Brien A D. Cloning and sequencing of the genes for Shiga toxin from Shigella dysenteriae type 1. J Bacteriol. 1988;170:1116–1122. doi: 10.1128/jb.170.3.1116-1122.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Unkmeir A, Schmidt H. Structural analysis of phage-born stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect Immun. 2000;68:4856–4864. doi: 10.1128/iai.68.9.4856-4864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weinstein D L, Jackson M P, Samuel J E, Holmes R K, O'Brien A D. Cloning and sequencing of a Shiga-like toxin type II variant from Escherichia coli strain responsible for edema disease of swine. J Bacteriol. 1988;170:4223–4230. doi: 10.1128/jb.170.9.4223-4230.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willshaw G A, Smith H R, Scotland S M, Field A M, Rowe B. Heterogeneity of Escherichia coli phages encoding Vero cytotoxins: comparison of cloned sequences determining VT1 and VT2 and development of specific gene probes. J Gen Microbiol. 1987;133:1309–1317. doi: 10.1099/00221287-133-5-1309. [DOI] [PubMed] [Google Scholar]