Abstract
Six native isolates of Trichoderma and Bacillus having potential for biocontrol and plant growth-promoting activities in rice were isolated from different rice growing regions of India. These isolates were screened for their efficiency in both in vitro and in vivo conditions for three years. The identity of the isolates was confirmed both by morphological and molecular characterization. Three Bacillus spp. viz., Bacillus velenzensis strain BIK2, Bacillus cabrialesii strain BIK3 and Bacillus paralicheniformis strain BIK4 and Trichoderma spp. viz., Trichoderma asperellum strain TAIK1, and T. asperellum strain TAIK5, native to the Telangana state, in Southern India except for strain TAIK4 (Rewa district in the state of Madhya Pradesh in Central India). These promising isolates were subjected for whole genome sequencing using the Illumina platform and data was presented. The data was emanated for Trichoderma asperellum (TAIK1), Trichoderma asperellum (TAIK4), Trichoderma asperellum (TAIK5), Bacillus velezensis (BIK2), Bacillus cabrialesii (BIK3) and Bacillus paralicheniformis (BIK4) isolates had an average 100X coverage of 109X, 150X and 116X; 1447X, 905X and 585X respectively. Further studies on the annotation of the data obtained in correlation with the lab and field performance of these microbes would enable them to be used in metagenomics studies to compare their performance under natural conditions with different microbiota and popular rice varieties. Bioformulation of these strains would be more appropriate with the availability of this genomic data.
Keywords: Biocontrol agents, Bacillus, Trichoderma, Whole genome sequencing
Specifications Table
| Subject | Microbiology |
| Specific subject area | Biocontrol agents, antagonists |
| Type of data | Assembly (fasta files), Tables, Figures |
| How data were acquired | Whole genome sequencing conducted on Illumina HiSeq 2500 instrument platform |
| Data format | Raw data |
| Parameters of data collection | The microbes were isolated from the rhizosphere soil of rice in the farmers’ fields. Genomic DNA was extracted from pure culture of individual isolates |
| Description of data collection | Total genomic DNA was isolated from three Bacillus and three Trichoderma spp., purified and subjected to HiSeq Illumina sequencing (2*150 bp) de novo assembly. |
| Data source location |
Trichoderma asperellum (TAIK1)- Hyderabad Trichoderma asperellum (TAIK4)- Rewa Trichoderma asperellum (TAIK5)- Hyderabad Bacillus velezensis (BIK2)- Karimnagar Bacillus cabrialesii (BIK3)- Hyderabad Bacillus paralicheniformis (BIK4)- Nalgonda |
| Data accessibility | Data is publicly available at NCBI GenBank from the following links: Assembly accessions and Bio project accessions https://www.ncbi.nlm.nih.gov/assembly/GCF_019336145.1/ https://www.ncbi.nlm.nih.gov/assembly/GCF_018829645.1/ https://www.ncbi.nlm.nih.gov/assembly/GCF_019336205.1 https://www.ncbi.nlm.nih.gov/assembly/GCA_019594945.1 https://www.ncbi.nlm.nih.gov/assembly/GCA_019594925.1/ https://www.ncbi.nlm.nih.gov/assembly/GCA_019481625.1/ Bio project IDs PRJNA744701- BIK2- Bacillus velenzensis PRJNA735062- BIK3- Bacillus cabrialesii PRJNA744714- BIK4- Bacillus paralicheniformis PRJNA727916- TAIK1-Trichoderma asperellum PRJNA735060- TAIK4- Trichoderma asperellum PRJNA745529- TAIK5- Trichoderma asperellum |
| Related research article | C. Kannan, M. Divya, G. Rekha, P. Maruthi, Hajira Shaik and R. M. Sundaram, Diversity analysis of antagonistic microbes against bacterial leaf and fungal sheath blight diseases of rice. Egypt J Biol Pest Control. 31(2021) 115. doi:10.1186/s41938-021-00462-x |
Value of the Data
-
•
This whole genome sequence data of six isolates of native biocontrol agents viz., three Bacillus and three Trichoderma isolates serve as an important source towards an understanding of these bioagents which suppress the plant pathogens like Rhizoctonia solani and Xanthomonas oryzae pv. oryzae in rice and in addition induces plant growth promotion in rice.
-
•
The data is useful in the annotation of the genes involved in the pathways of enzymes, effector proteins and metabolites/alkaloids, involved in the bioagent-host plant-pathogen interactions from the perspective of these antagonistic bioagents
-
•
The data provides valuable information on these native bioagents and enables their efficient use by all the stakeholders including the biopesticide industries to use them as biocontrol agents and as biofertilizers in sustainable eco-friendly cultivation of rice. The genomic data of these potential bioagents submitted will help in the breeding of cultivars that respond well to the bioagents when applied. For instance, TAIK1 application on 30th day of transplantation released growth promoting substances and also suppress the infection induced by R. solani and S. oryzae. It has also been reported that the bioagents application needs to be standardised for different varieties [1].
1. Data Description
Biological control is the process of using friendly bioagents or their products to suppress the pathogens leading to the sustainable integrated management of plant diseases [2]. Species belonging to the genera Trichoderma, Bacillus and Pseudomonas are more commonly found in the plant rhizosphere that helps in the growth promotion of the plants and induces resistance/tolerance against biotic and abiotic stresses. Members of the genus Bacillus, a common soil saprophytic gram-positive bacterium and Trichoderma a saprophytic fungus in rhizosphere soil, are used for their plant growth promotion and biocontrol qualities that make them a better alternative to chemical pesticides in long term use [3].
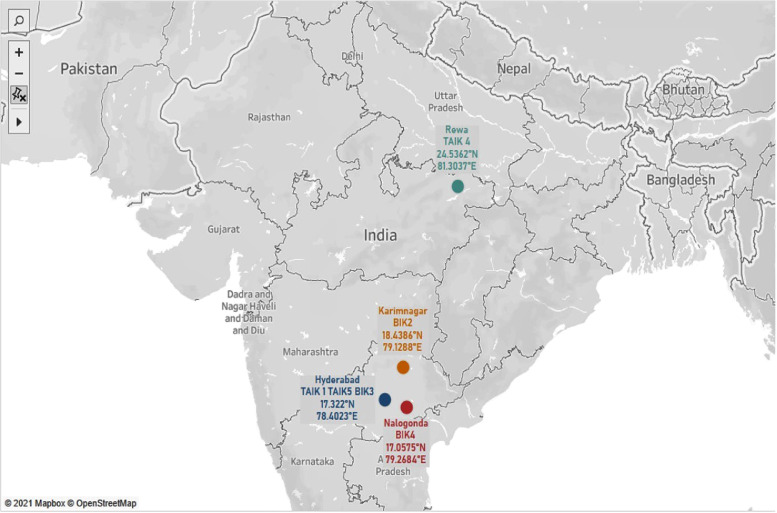
In this manuscript, we report the whole genome sequencing (WGS) data of three Bacillus isolates (BIK2, BIK3 and BIK4) and Trichoderma isolates (TAIK1, TAIK4 and TAIK5) collected from different states of India using standard dilution method [4]. The geographic data of the sampling sites and the origin of the isolates are represented as Fig. 1. Detailed statistics of three Bacillus isolates viz., BIK2, BIK3 and BIK4 and three Trichoderma isolates viz., TAIK1, TAIK4 and TAIK5 were presented in Tables 2 and 3.
Fig. 1.
Illustration of map indicating the location of the strains collected from India (Tableau public 2021.2).
Table 2.
Assembly Statistics of three Bacillus and Trichoderma isolates.
| Attributes/ Statistics | Bacillus velenzensis | Bacillus cabrialesii | Bacillus paralicheniformis | Trichoderma asperellum | Trichoderma asperellum | Trichoderma asperellum |
|---|---|---|---|---|---|---|
| Isolate | BIK2 | BIK3 | BIK4 | TAIK1 | TAIK4 | TAIK5 |
| Contigs | 26 | 28 | 30 | 702 | 473 | 449 |
| Largest contig | 10,78,503 | 5,75,880 | 10,56,155 | 10,48,585 | 6,24,435 | 7,25,734 |
| Total Length | 39,00,416 | 41,08,741 | 44,18,047 | 3,72,93,549 | 3,99,77,543 | 3,60,36,647 |
| N50 | 10,29,777 | 3,20,958 | 6,27,466 | 2,26,906 | 2,07,650 | 1,61,701 |
| N75 | 4,40,514 | 1,91,033 | 2,26,402 | 1,14,355 | 1,07,158 | 87,099 |
| L50 | 2 | 5 | 3 | 50 | 64 | 70 |
| L75 | 4 | 10 | 6 | 109 | 132 | 144 |
| GC% | 46.52 | 44.08 | 45.47 | 47 | 48 | 49 |
Table 3.
Genome features of three Bacillus and Trichoderma isolates.
| Genome features and gene ontology | Bacillus velenzensis | Bacillus cabrialesii | Bacillus paralicheniformis | Trichoderma asperellum | Trichoderma asperellum | Trichoderma asperellum |
|---|---|---|---|---|---|---|
| Isolate | BIK2 | BIK3 | BIK4 | TAIK1 | TAIK4 | TAIK5 |
| Protein coding genes | 3751 | 4095 | 4495 | 11,592 | 14,174 | 11,589 |
| Biological processes | 1077 | 2095 | 2074 | 4051 | 5686 | 4045 |
| Molecular functions | 2111 | 4120 | 4228 | 10,717 | 14,080 | 10,692 |
| Cellular components | 863 | 1837 | 1801 | 4480 | 5568 | 4469 |
* N50 - sequence length of the shortest contig at 50% of the total genome length; L50- number of contigs length making up half of the genome size.
2. Experimental Design, Materials and Methods
2.1. Culture and DNA extraction
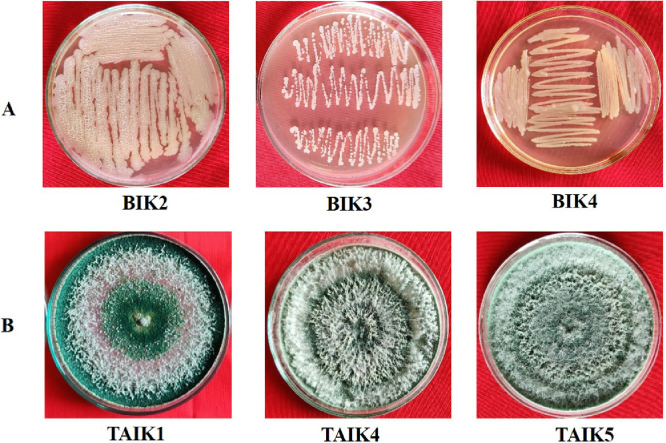
Bacillus and Trichoderma isolates were obtained from the rice rhizosphere of different regions of India, using the standard serial dilution method (Fig. 1). Trichoderma specific medium (TSM) and peptone yeast extract medium (PYEM) was used as a selective medium for the isolation and purification of fungal and bacterial antagonists, respectively [4]. Key morphological and microscopic characters were used for the identification of Trichoderma isolates [5] and Bacillus isolates [6] (Fig. 2; Table 1). For whole genome sequencing, genomic DNA from the three Bacillus and three Trichoderma strains were isolated using DNA isolation kit NucleoSpin® microbial DNA kit as per the manufacturer's protocol (Macherey-Nagel, Germany). The DNA libraries for Whole Genome Sequencing were processed using standard protocols and sequenced using the HiSeq 2500 instrumentation platform (Agri Genome Labs Private Limited, Kochi, India).
Fig. 2.
Culture plates of (A) Bacillus isolates (B) Trichoderma isolates.
Table 1.
Morphological identification of Bacillus and Trichoderma isolates.
| Colony morphology |
Sporulation |
|||||
|---|---|---|---|---|---|---|
| Isolate code |
Scientific name | colour | Area covered by radial growth of colonies in 36 h (mm) |
Texture | Colour of spores | Days for maturation |
| BIK2 | B. velezensis | Grey white | 15.0 ± 0.03 | Round, smooth and moist | - | - |
| BIK3 | B. cabriesii | Of-white | 21.0 ± 0.10 | Flat, opaque and dry | - | - |
| BIK4 | B. paralicheniformis | Pinkish whit | 18.0 ± 0.09 | Irregular and extra slimy | - | - |
| TAIK 1 | Trichoderma asperellum | Dark green | 37.0 ± 0.12 | Smooth mat with concentric rings | Yellowish Green | 4 |
| TAIK 4 | Trichoderma asperellum | Dark green | 41.0 ± 0.10 | Fluffy mat | Dark green | 3 |
| TAIK 5 | Trichoderma asperellum | Dark green | 45.0 ± 0.04 | Smooth mat | Dark green | 2 |
The table is modified from Tables 1 and 2 from the article referred - 10.1186/s41938-021-00462-x[1].
2.2. Whole genome sequencing
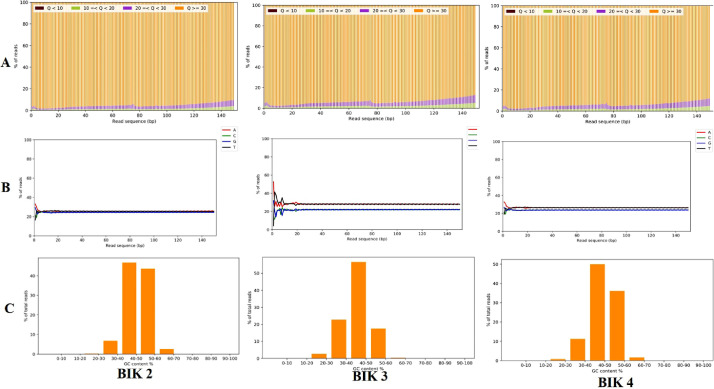
Whole Genome Sequencing (WGS) of three Bacillus isolates resulted in 20, 274, 842; 12, 674, 497 and 17, 571, 991 raw reads for BIK2, BIK3 and BIK4 respectively. The quality of raw sequence reads were assessed using Fast QC and then pre-processed using AdapterRemovalV2 version 2.3.1 tool [7] (Fig. 3) generating 20,260,548; 12,667,151 and 17,551,922 clean reads for BIK2, BIK3 and BIK4 with an average read length of 150 bp respectively, representing coverage of 1447X, 905X and 585X folds. The cleaned reads were de novo assembled using the Unicycler ver. 0.4.8 assembler [8] and CDSs in the assembled contigs were predicted using prodigal version 2.6.3 [10]. Completeness of the genome assembly was assessed by BUSCO ver. 4.0.6 [9] and quality of the genome assembly was assessed by QUAST ver. 4.6 [10]. Protein encoding genes were predicted using Prodigal ver. 2.6.3 [11].
Fig. 3.
Quality check of three Bacillus strains (A) Quality distribution (B) Base distribution (C) GC distribution.
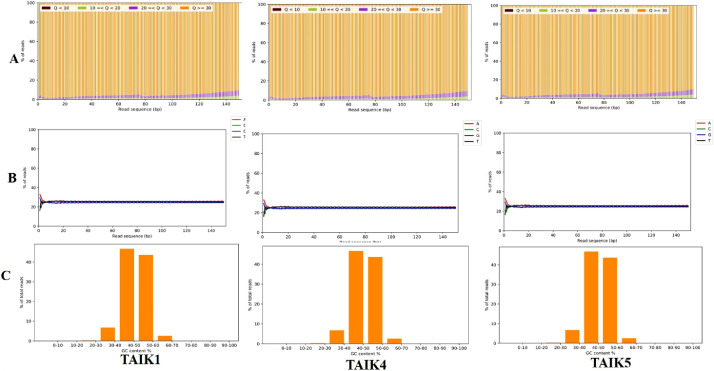
For the Trichoderma strains TAIK1, TAIK4 and TAIK5, a total of 15, 230, 394; 16, 467, 915 and 20, 615, 262 raw reads were generated and the quality of these raw sequence reads were assessed using Fast QC and then pre-processed using AdapterRemovalV2 version 2.3.1 tool [7] (Fig. 4) resulting in 11,502,933; 14,374,041; 18,498,253 clean reads respectively with an average read length of 150 bp, representing coverage of 109X, 150X and 116X folds. De novo assembly was performed using the Velvet assembler version 1.2.10 (https://angus.readthedocs.io/en/2016/week3/LN_assembly.html) and CDSs in the assembled contigs were predicted using Augustus assembler version 3.4.0 (http://bioinf.uni-greifswald.de/augustus/). Completeness of the genome assembly was assessed by BUSCO ver. 4.0.6 [8] and quality of the genome assembly was assessed by QUAST ver. 4.6 [10]. Protein encoding genes were predicted using Prodigal ver. 2.6.3 [11]. Organism annotation was determined from the predicted genes which were compared with the Uniprot database using BlastX version 2.6.0 (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/blast+/) program with E-value cut offset to 10−3 and subsequent filtering was done for the best hits based on the query coverage, identity and similarity score.
Fig. 4.
Quality check of three Trichoderma strains (A) Quality distribution (B) Base distribution (C) GC distribution.
2.2. Assembly statistics
Bacillus isolates viz., BIK2, BIK3 and BIK4 and Trichoderma isolates viz., TAIK1, TAIK4 and TAIK5 consisted of 26, 28 and 30 contigs; 702, 473 and 449 with a maximum size of 1078,503; 575,880 and 1056,15 bp; 1048,585; 624,435 and 725,734 respectively. The sequence length of the shortest contig (N50) of Bacillus isolates viz., BIK2, BIK3 and BIK4 and Trichoderma isolates viz., TAIK1, TAIK4 and TAIK5 were 1029,777; 320,958 and 627,466; 226,906 and 161,701 respectively. While the length of the contig (L50) were two, five and three; 50, 64 and 70 for Bacillus and Trichoderma strains respectively. The sequencing data were deposited in the Sequence Read Archive (SRA) with accession numbers JAHWRC01, JAHKKH01 and JAHWRD01 for Bacillus strains BIK2, BIK3 and BIK4 respectively and JA1AZZ01, JA1CDU01 and JAHYXG01 accessions for Trichoderma strains TAIK1, TAIK4 and TAIK5 respectively. The closest associated strains to the isolates include Bacillus velenzensis (DSM23117) for BIK2, Bacillus paralicheniformis (ATCC 9945a) for BIK4 and Trichoderma asperellum (CBS443.97) for all the three Trichoderma strains viz., TAIK1, TAIK4 and TAIK5. The Bio-project accession numbers are presented in the specifications table.
Funding Information
This work was supported by ICAR- Indian Institute of Rice Research, Hyderabad, India.
Ethical Statement
Not applicable.
CRediT authorship contribution statement
C. Kannan: Conceptualization, Supervision, Writing – review & editing. M. Divya: Methodology, Writing – review & editing. G. Rekha: Methodology, Formal analysis, Writing – review & editing. Kalyani M. Barbadikar: Validation, Investigation, Writing – review & editing. P. Maruthi: Methodology. S.K. Hajira: Methodology, Project administration. R.M. Sundaram: Supervision.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships which have or could be perceived to have influenced the work reported in this article.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.dib.2022.107923.
Appendix. Supplementary materials
References
- 1.Kannan C., Divya M., Rekha G., Maruthi P., Shaik Hajira, Sundaram R.M. Diversity analysis of antagonistic microbes against bacterial leaf and fungal sheath blight diseases of rice. Egypt. J. Biol. Pest Control. 2021;31:115. doi: 10.1186/s41938-021-00462-x. [DOI] [Google Scholar]
- 2.Gnanamanickam S.S. Vol. 8. Springer; Berlin: 2009. (Biological Control of Rice Diseases). [Google Scholar]
- 3.Mukherjee P.K., Horwitz B.A., Herrera-Estrella A., Schmoll M., Kenerley C.M. Trichoderma research in the genome era. Ann. Rev. Phytopathol. 2013;51:105–129. doi: 10.1146/annurev-phyto-082712-102353. [DOI] [PubMed] [Google Scholar]
- 4.Cavaglieri L., Passone A., Etcheverry M. Screening procedures to select rhizobacteria with biocontrol activity upon fusarium verticillioides growth and fumonisin B1 production. Res. Microbiol. 2004;155:747–754. doi: 10.1016/j.resmic.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 5.Gams W., Bissett J., Kubicek C.P., Harman G.E. Vol. 1. Taylor and Francis; London: 1998. Morphology and identification of Trichoderma; pp. 1–34. (Trichoderma and Gliocladium). [Google Scholar]
- 6.Sneath P.H.A., Holt J.G. Williams & Wilkins; Baltimore: 1998. Berger’s Manual of Determinative Bacteriology; pp. 1105–1139. 1288–1301. [Google Scholar]
- 7.Wick R.R., Judd L.M., Gorrie C.L., Holt K.E. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 2017;13(6):1–22. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simao F.A., Waterhouse R.M., Ioannidis P., Kriventseva E.V., Zdobnov E.M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31:3210–3212. doi: 10.1093/bioinformatics/btv351. [DOI] [PubMed] [Google Scholar]
- 9.Gurevich A., Saveliev V., Vyahhi N., Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29:1072–1075. doi: 10.1093/bioinformatics/btt086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hyatt D., Chen G.L., Locascio P.F, Land M.L., Larimer F.W., Hauser L.J. Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics. 2010;11:119. doi: 10.1186/1471-2105-11-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schubert M., Lindgreen S., Orlando L. Adapter removal v2: rapid adapter trimming, identification, and read merging. BMC Res. Notes. 2016;9(1):1–7. doi: 10.1186/s13104-016-1900-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.