Abstract
Enzymatic methylation catalyzed by methyltransferases has a significant impact on many human biochemical reactions. As the second most ubiquitous cofactor in humans, S-adenosyl-l-methionine (SAM or AdoMet) serves as a methyl donor for SAM-dependent methyltransferases (MTases), which transfer a methyl group to a nucleophilic acceptor such as O, As, N, S, or C as the byproduct. SAM-dependent methyltransferases can be grouped into different types based on the substrates. Here we systematically reviewed eight types of methyltransferases associated with human diseases. Catechol O-methyltransferase (COMT), As(III) S-adenosylmethionine methyltransferase (AS3MT), indolethylamine N-methyltransferase (INMT), phenylethanolamine N-methyltransferase (PNMT), histamine N-methyltransferase (HNMT), nicotinamide N-methyltransferase (NNMT), thiopurine S-methyltransferase (TPMT) and DNA methyltansferase (DNMT) are classic SAM-dependent MTases. Correlations between genotypes and disease susceptibility can be partially explained by genetic polymorphisms. The physiological function, substrate specificity, genetic variants and disease susceptibility associated with these eight SAM-dependent methyltransferases are discussed in this review.
Keywords: Methyltransferases, S-adenosyl-l-methionine, Methylation, Phenotypes, Genotypes, Single nucleotide polymorphisms
1. Introduction
Methylation reactions catalyzed by methyltransferases are found in many biochemical pathways and are associated with genetic diseases, including cancer and metabolic diseases. The SAM-dependent methyltransferase (MTase) superfamily could be classified either by substrate specificity or based on the nucleophilic atoms targeted for methylation (e.g. O, As, N, S or C,). Based on the properties and functions of the substrates and products, the methylation catalyzed by these enzymes is involved in biosynthesis, signal transduction, protein repair and other physiological processes. Aberrant methylation is closely associated with the occurrence of cancer, genetic disorders or other diseases. Polymorphisms in MTase genes can affect enzyme activity. Single nucleotide polymorphisms (SNPs) are the most common source of protein genetic variation. SNPs are found in promoters, exons or introns. They affect mRNA transcription, splicing, protein expression, structure, enzymatic activity and stability. There are millions of SNPs in the human genome when compared to a reference genome. In the last decade, genetic variations in methyltransferase proteins have been the subject of considerable research. There are now numerous MTase SNPs related to clinical disease. This review aims to systematically summarize eight SAM-dependent methyltransferases, with an emphasis on their substrate specificity, genetic polymorphisms, and crystal structures, as well as the relationships between SNPs and human disease (Table 1). It is not meant to be an exhaustive review, but simply to highlight key disease associations. This information may contribute to a more precise understanding of correlations between genotypes and disease-susceptibility phenotypes or individual risk of toxicity from drug treatment.
Table 1.
A brief summary of eight SAM-depended MTases.
| SAM-dependent methyltransferases | Gene Localization | Prototypic substrate | Catalysis of the reaction | Related diseases |
|---|---|---|---|---|
| COMT (Catechol O-methyltransferase) | 22q11.21 | Catacholamines (dopamine, epinephrine) | SAM + a catechol = SAH + a guaiacol | Schizophrenia [1,2] PD [3,4] AD [5] ADHD [6] |
| AS3MT (Arsenite methyl transferase) | 10q24.32 | Arsenic Antimony Tryptamine |
SAM + arsenite = SAH + methylarsonate SAM + methylarsonate = SAH + dimethylarsinate SAM + an amine = SAH + a methylated amine. |
Hypertension [7] Skin lesions [8] Diabetes [9] |
| INMT (Indolethylamine N-methyltransferase) | 7p14.3 | Serotonin Thioethers Selenium Tellurium |
SAM + dimethyl sulfide = SAH + trimethylsulfonium | Hirschsprung’s disease [10] |
| PNMT (Phenylethanolamine N-Methyltransferase) | 17q12 | Nonrepinephrine | SAM + phenylethanolamine = SAH + N-methylphenylethanolamine | Hypertension [11] AD [12] Sickle cell disease [13] PD [14,15,16,17] |
| HNMT (Histamine N-methyltransferase) | 2q22.1 | Histamine | SAM + histamine = SAH + N(tau)-methylhistamine | Asthma [18,19] Schizophrenia [16] Migraine [20] |
| Nicotinamide | Cancer [21] | |||
| NNMT (Nicotinamide N-methyltransferase) | 11q23.2 | Pyridine | SAM + nicotinamide = 1-methylnicotinamide + SAH | Schizophrenia [22] Obesity [23] Diabetes [24] |
| TPMT (Thiopurine S-methyltransferase) | 6p22.3 | Azathioprine 6-Mercaptopurine 6-Thioguanine |
SAM + a thiopurine = SAH + a thiopurine S-methylether | ALL [25] |
| DNMT1 | 19p13.2 | Cytosine | SAM + DNA = SAH + DNA containing | ICF syndrome [26,27] |
| DNMT3A | 2p24.1 | Cancer [28] | ||
| DNMT3B (DNA (cytosine-5)-methyltransferase) | 20q11.21 | Adenine | 5-methylcytosine | Breast cancer [29] Acute myeloid leukemia (AML) [30] |
Abbreviations: S-adenosyl l-methionine, SAM; S-adenosyl-l homocysteine, SAH or AdoHcy; Alzheimer disease, AD; Attention-deficit/hyperactivity disorder: ADHD; Parkinson’s disease: PD; Acute lymphoblastic leukaemia: ALL; Acute myeloid leukaemia: AML; Immunodeficiency, centromeric instability and facial anomalies syndrome: ICF.
To start, we searched the Web of Science database for reports on the eight types of S-adenosyl l-methionine (SAM)-dependent methyltransferases within the past 30 years (Fig. 1). Of a total of 2188 publications recovered, the prevalence of reports on MTase polymorphisms occurred in the order TPMT > COMT > DNMT, with lower numbers of publications for the other enzymes. Each of the SAM MTases was correlated with polymorphisms in the corresponding gene. Studies included in the review were selected using the following criteria: (1) Search for literature types: ‘Review’, ‘Clinical Trial’ or ‘Case Report’ in the Web of Science database; (2) Search synchronously the NCBI web site [chose SNP category, entered specific human MTase, refined by ‘PubMed Cited’ with validation status of ‘by-frequency,’ refined by clinical significance with ‘drug response/likely benign/risk factor’ to track papers]. Finally, papers from NCBI database were combined with papers from Web of Science database.
Fig. 1. Publications on S-adenosyl l-methionine (SAM)-dependent methyltransferases in the past 30 years.
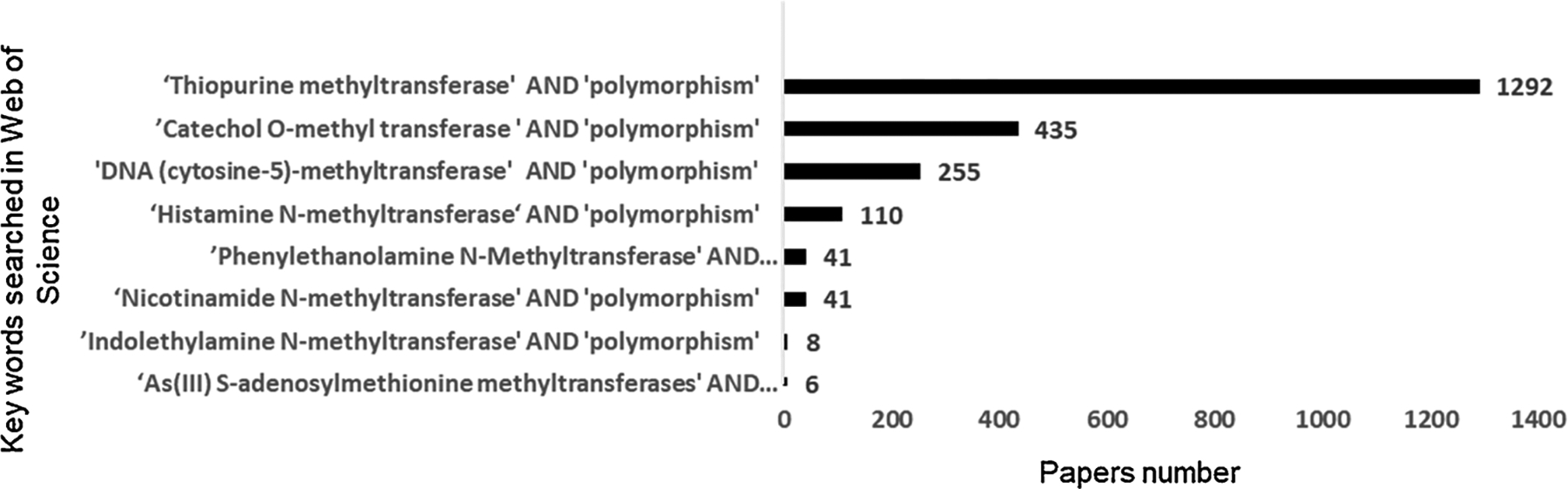
A database search (Web of Science, Thomson Reuters) was performed on 20 April 2021. Depicted is the total number of publications using the following search strategies: ‘catechol O-methyl transferase’ AND ‘polymorphism’; ‘As(III) S-adenosylmethionine methyltransferases’ AND ‘polymorphism’; ‘Histamine N-methyltransferase ‘AND ‘polymorphism’; ‘Indolethylamine N-methyltransferase’ AND ‘polymorphism’; ‘Nicotinamide N-methyltransferase’ AND ‘polymorphism’; ‘Phenylethanolamine N-Methyltransferase’ AND ‘polymorphism’; ‘Thiopurine methyltransferase’ AND ‘polymorphism’; and ‘DNA (cytosine-5)-methyltransferase’ AND ‘polymorphism’.
2. Catechol-O-methyltransferase (COMT)
Catechol-O-methyltransferases (COMT) catalyze transfer of a methyl group from SAM to the hydroxyl group on a catechol nucleus, resulting in the deactivation of catecholamine neurotransmitters such as dopamine, epinephrine, and norepinephrine [31,32] (Fig. 2). The COMT gene is located on the long (q) arm of human chromosome 22 at position 11.21, which codes for both soluble COMT (S-COMT) and membrane-bound COMT (MB-COMT). The differences between these two COMTs are that MB-COMT has 50 additional hydrophobic amino acids [33]. COMT is crucial to the function of the prefrontal cortex of the brain, specifically in organizing and coordinating information from other parts of the brain. A U-shaped relationship between hippocampal gray matter volume and presumed COMT activity has been proposed [34]. The COMT Val158Met SNP (rs4680) affects hippocampal grey matter volume and appears to be modified by P2 promoter A > G SNP (rs2097603). Thus, there are four haplotypes: (1) A-Val (P2 promoter carrying base A and rs4680 carrying animo acid Val); (2) A-Met (P2 promoter carrying base A and rs4680 carrying animo acid Met); (3) G-Val (P2 promoter carrying base G and rs4680 carrying animo acid Val); (4) G-Met (P2 promoter carrying base G and rs4680 carrying animo acid Met). When ordered by putative genetic variation of enzymatic activity, A–Val has the highest activity, followed by G–Val, A–Met and finally G–Met, corresponding to high, low, low, and high (U shape) hippocampal gray matter volumes, respectively. The enzymatic activity and hippocampal gray matter volume differences were consistent with a nonlinear effect of extracellular dopamine, extracellular dopamine levels of A-Val<G-Val<A-Vet<G-Vet. Abnormal levels of dopamine neurotransmitters result in psychiatric manifestations, including schizophrenia [35], other psychoses [36], mood disorders, schizophrenia, obsessive-compulsive disorder and substance abuse [37,38].
Fig. 2. Human biosynthetic pathway for trace amines and catecholamines.
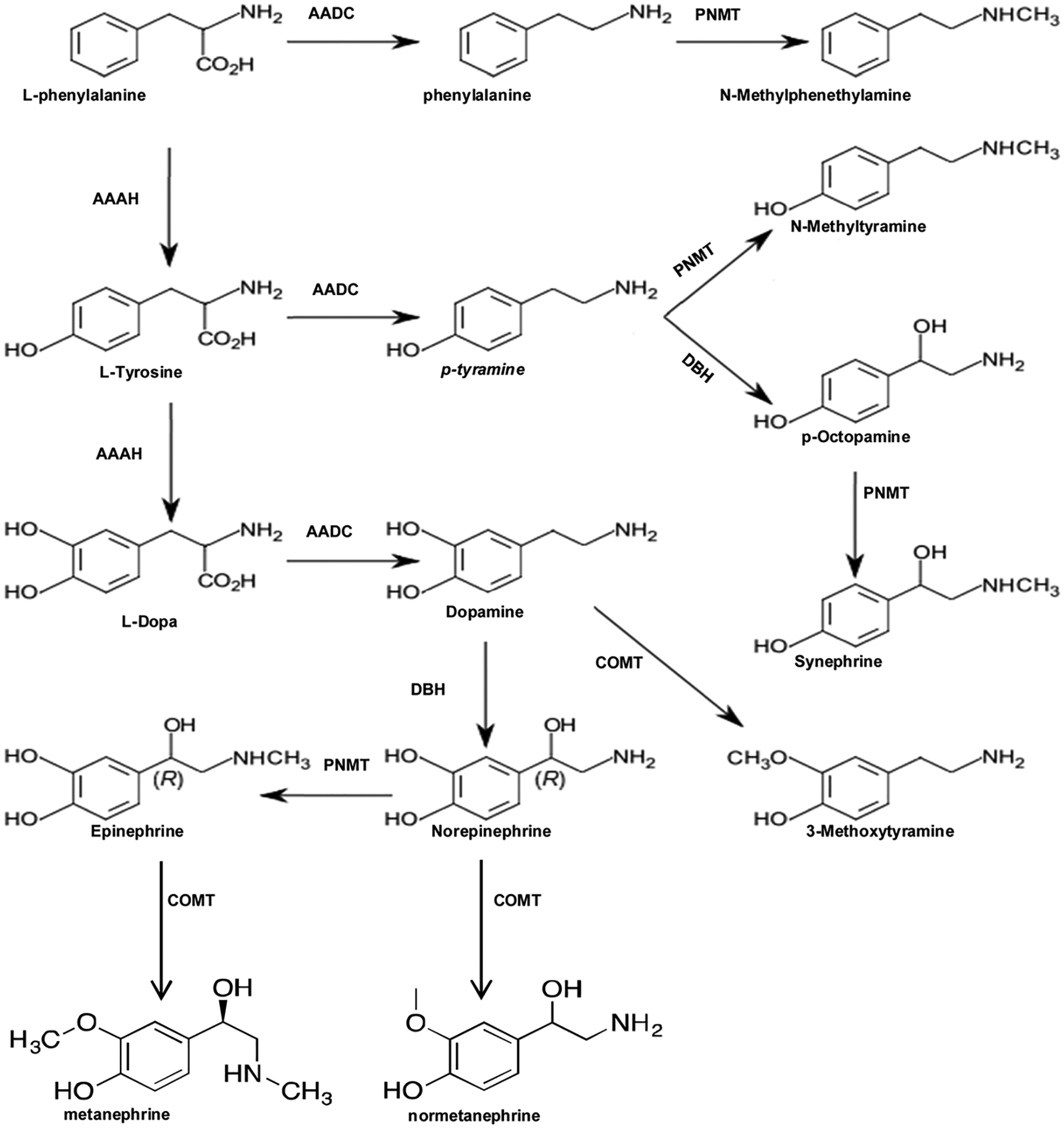
L-Phenylalanine is converted into l-tyrosine by the enzyme AAAH (aromatic amino acid hydroxylases). l-Tyrosine is converted into l-DOPA (l-dihydroxyphenylalanine) by the enzyme AAAH. l-DOPA is catalyzed to dopamine by the enzyme aromatic l-amino acid decarboxylase (AADC). Dopamine itself is also used as a precursor in the synthesis of the neuro-transmitters, norepinephrine and epinephrine. Dopamine is converted into norepinephrine by the enzyme dopamine β-hydroxylase (DBH) and inactivated by COMT Norepinephrine is converted into epinephrine by the enzyme PNMT with SAM as the cofactor. Epinephrine and norepinephrine are inactivated by COMT. [32,39,40].
Microdeletions in COMT are observed in most individuals who present with velocardiofacial syndrome or related clinical syndromes, referred to as 22q11.2 deletion syndrome [41]. People with 22q11.2 deletion have increased risk of schizophrenia, depression, anxiety and bipolar disorder compared with normal individuals [42]. Variations in the COMT gene have been identified both in the promotor region and the transcribed sequence. Promoter region variants of MB-COMT (rs2020917, rs737865) are associated with depressive symptomatology among females [43], specifically in the South African Xhosa population [44]. A frequent G > A polymorphism (rs4680) within the coding region of COMT produces a Val to Met substitution at codons 108 and 158 of S-COMT and MB-COMT, respectively [45], which leads to altered activity of both S-COMT and MB-COMT. A meta-analysis determined that the Val158Met polymorphism has a 37 % minor allele frequency (MAF) (Table 2), and Val-carriers show significantly less improvement in positive symptoms relative to Met/Met individuals in schizophrenic and schizo-affective disorder patients [odds ratio (OR) Met/Met = 1.54, 95 % confidence interval (CI): 1.11–2.14, P = 0.0098] [1]. In patients with Parkinson’s Disease (PD), a high-COMT activity haplotype (G-C-C-G for rs6269, rs4633, rs4818, and rs4680) is associated with cognitive decline (hazard ratio = 3.24; P = 0.02) [3]. Further studies demonstrated that the COMT rs4680 G/G or G/A genotypes and the COMT rs6267 G/T genotype contribute to pain in PD patients with P = 0.04 and <0.01, respectively [4]. Compared with Met/Val and Val/Val individuals, Met/Met individuals experience a cognitive deficit (n = 107, P = 0.017) [46] and a possible increased risk of bipolar disorder, panic disorder (n = 298, P = 0.0089) [47], obsessive-compulsive disorder and ADHD (n = 230, P = 0.043) [6]. These conditions may be related to inefficient processing of information in the prefrontal cortex and altered levels of muscarinic M1 receptor mRNA in the cortex [48]. It is worth noting that some associations between polymorphisms of COMT and diseases are inconsistent. Results from two meta-analyses do not support an association between COMT SNPs and ADHD [49,50]. Regarding the association between COMT SNP rs4680 and AD, one meta-analysis supported the association between the COMT G/G and G/A genotype and AD in an Asian group (OR = 0.702, 95 % CI = 0.517–0.953, P = 0.023) [5], but neither COMT genotype proved to be independently associated with AD risk [51,52]. Also, there is research support for a significant association between high alcohol consumption in patients with AD and the COMT polymorphism rs4680 [53].
Table 2.
Frequencies of the primary SAM-dependent methyltransferases polymorphisms1 in the 1000 Genomes Project2, ExAC3, and the BUSHMAN Population4.
| MTases | dbSNP rs# [Minor Allele Frequency (MAF)] | Amino Acid change | Base change | Allele frequency (numbers of individuals)5 | Genotype frequency (numbers of individuals)6 | Data source |
|---|---|---|---|---|---|---|
| COMT | rs4680 (0.37) | Val158Met Val108 Met |
472G > A | G: 0.631 (3159) A: 0.369 (1849) |
G|G: 0.413 (1034) A|G: 0.436 (1091) A|A: 0.151 (379) |
1000 Genomes Project |
| COMT | rs4633 (0.37) | His62His Synonymous Variant Leu136Leu |
186C > T | C: 0.628 (3147) T: 0.372 (1861) C: 0.703 (3521) |
C|C: 0.411 (1028) C|T: 0.436 (1091) T|T: 0.154 (385) C|C: 0.509(1275) |
1000 Genomes Project |
| COMT | rs4818 (0.30) | Synonymous Variant | 408C > G/T | G: 0.297 (1487) | C|G: 0.388(971) G|G: 0.103 (258) |
1000 Genomes Project |
| COMT | rs6267 (0.01) | Ala72Thr/Ser | 214G > A / T | G: 0.987 (4941) T: 0.013 (67) |
G|G: 0.973 (2437) G|T: 0.027 (67) |
1000 Genomes Project |
| COMT | rs2020917 (0.21) | intron variant (Promotor region) | −628C > T | C: 0.787 (3941) T: 0.213 (1067) |
C|C: 0.625 (1566) C|T: 0.323 (809) T|T: 0.052 (129) |
1000 Genomes Project |
| COMT | rs737865 (0.23) | intron variant (Promotor region) | 92 + 701A > G | A: 0.773 (3873) G: 0.227 (1135) |
A|A: 0.602 (1507) A|G: 0.343 (859) G|G: 0.055 (138) |
1000 Genomes Project |
| AS3MT | rs201702937 (0.01) | His51Arg | 152A > G | A: 0.9998 (5007) G: 0.000199 (1) |
A|A: 0.9996 (2503) A|G: 0.00039 (1) |
1000 Genomes Project |
| AS3MT | rs80317306 (<0.01) | Cys61Trp | 183T > G | T: 0.995 (120200) G: 0.005 (646) |
T|T 0.991 (60100) T|G 0.009 (646) |
Exome Aggregation Consortium (ExAC) |
| AS3MT | rs112056792 (< 0.01) | Ile136Thr | 407T > C | NA 1 |
NA 1 (Male) |
BUSHMAN POP |
| AS3MT | rs35232887 (0.01) | Arg173Trp | 517C > T | C: 0.999 (5003) T: 0.001 (5) |
C|C: 0.998 (2499) C|T: 0.002 (5) |
1000 Genomes Project |
| AS3MT | rs370022454 (< 0.01) | Trp203Cys | 609G > T | G: 0.999734 (3763) T: 0.000265675 (1) |
G|G: 0.999 (1881) G|T: 0.001 (1) |
African-American of NHLBI Exome Sequencing Project7 |
| AS3MT | rs139656545 (<0.01) | Arg251His | 752G > A | G: 0.998 (4997) A: 0.002 (11) T: 0.923 (4622) |
G|G: 0.996 (2493) A|G: 0.004 (11) T|T: 0.854 (2138) |
1000 Genomes Project |
| AS3MT | rs11191439 (0.08) | Met287Thr | 860 T > C/A | C: 0.077 (386) | C|T: 0.138 (346) C|C: 0.008 (20) |
1000 Genomes Project |
| AS3MT | rs34556438 (<0.01) | Thr306Ile | 917C > T | C: 0.99951 (8164) T: 0.000489716 (4) T: 0.765 (3829) |
C|C: 0.999 (4080) C|T: 0.001 (4) T|T: 0.589 (1475) |
European-American of NHLBI Exome Sequencing Project7 |
| AS3MT | rs3740392 (0.24) | Intron Variant | 610 + 63T > C | C: 0.235 (1179) A: 0.230 (1153) |
C|T: 0.351 (879) C|C: 0.060 (150) A|A: 0.082 (205) |
1000 Genomes Project |
| INMT | rs6970396 (0.23) | 3 Prime UTR Variant | 8797A > G | G: 0.770 (3855) | A|G: 0.297 (743) G|G: 0.621 (1556) |
1000 Genomes Project |
| INMT | rs77743549 (0.01) | His46Pro | 137A > C | A: 0.995 (4984) C: 0.005 (24) C: 0.744 (3728) |
A|A: 0.990 (2480) A|C: 0.010 (24) C|C: 0.558 (1397) |
1000 Genomes Project |
| NNMT | rs694539 (0.26) | Intron variant | −216–151C > T | T: 0.256 (1280) A: 0.443 (2218) |
C|T: 0.373 (934) T|T: 0.069 (173) A|A: 0.204 (510) |
1000 Genomes Project |
| NNMT | rs1941404 (0.44) | Intron variant | −224 + 122A > G | G: 0.557 (2790) G: 0.510 (2555) |
A|G: 0.478 (1198) G|G: 0.318 (796) G|G: 0.276 (691) |
1000 Genomes Project |
| NNMT | rs10891644 (0.49) | Intron variant | −129–11693G > T | T: 0.490 (2453) G: 0.587 (2940) |
G|T: 0.468 (1173) T|T: 0.256 (640) G|G: 0.368 (921) |
1000 Genomes Project |
| PNMT | rs876493 (0.41) | 5 prime UTR variant | 93 + 222G > A | A: 0.413 (2068) C: 0.940 (4710) |
A|A: 0.194 (485) A|G: 0.438 (1098) C|C: 0.886 (2219) |
1000 Genomes Project |
| HNMT | rs11558538 (0.06) rs1800460 |
Thr105Ile | 314C > T | T: 0.060 (298) C: 0.987 (4944) |
C|T: 0.109 (272) T|T: 0.005 (13) C|C: 0.975 (2441) |
1000 Genomes Project |
| TPMT | TPMT*3B (0.01) rs1142345 |
Ala154Thr | 460C > T | T: 0.013 (64) T: 0.961 (4812) |
C|T: 0.025 (62) T|T: 0.0004 (1) T|T: 0.925 (2316) |
1000 Genomes Project |
| TPMT | TPMT*3C (0.04) | Tyr240Cys Pro463Pro |
719T > C | C: 0.039 (196) T: 0.466 (2334) |
C|C: 0.003 (8) C|T: 0.072 (180) T|T: 0.228 (570) |
1000 Genomes Project |
| Dnmt1 | rs2228611 (0.47) | Synonymous Variant | 1389T > A/C | C: 0.534 (2674) T: 0.802 (4015) |
C|T: 0.477 (1194) C|C: 0.296 (740) T|T: 0.653 (1636) |
1000 Genomes Project |
| Dnmt1 | rs2228612 (0.20) | Ile327Phe | 979T > A/C /G | C: 0.198 (993) G: 0.722 (3615) |
C|T: 0.297 (743) C|C: 0.050 (125) G|G: 0.534 (1338) |
1000 Genomes Project |
| DNMT1 | rs2162560 (0.28) | Intron Variant | G > A | A: 0.278 (1393) T: 0.955 (4782) |
A|G: 0.375 (939) A|A: 0.091 (227) T|T: 0.918 (2299) |
1000 Genomes Project |
| DNMT1 | rs16999593 (0.05) | His97Arg | 290 T > C | C: 0.045 (226) | C|T: 0.073 (184) C|C: 0.008 (21) |
1000 Genomes Project |
| Dnmt3A | rs1550117 (0.11) | Upstream Transcript Variant | A > G/T | A: 0.114 (572) G: 0.886 (4436) C: 0.312 (1562) |
A|A: 0.018 (46) A|G: 0.192 (480) G|G: 0.790 (1978) C|C: 0.134 (336) |
1000 Genomes Project |
| Dnmt3B | rs2424913 (0.31) | Intron Variant | C > A/T/G | T: 0.688 (3446) G: 0.276 (1383) |
C|T: 0.355 (890) T|T: 0.510 (1278) G|G: 0.125 (312) |
1000 Genomes Project |
| Dnmt3B | rs1569686 (0.28) | Intron Variant | G > T/A/C | T: 0.724 (3625) | G|T: 0.303 (759) T|T: 0.572 (1433) |
1000 Genomes Project |
Original data were mined from the Ensembl genome browser database (http://useast.ensembl.org/index.html) accessed April 5, 2021. For the 1000 Genomes Project, all donors were over 18 and declared themselves to be healthy at the time of collection. The health conditions for other data source populations are unknown.
1000 Genomes Project population: African, American, East Asian, European, South Asian.
Exome Aggregation Consortium (ExAC) population: African/African American, Latino, East Asian, Finnish, Non-Finnish European, South Asian and others.
BUSHMAN Population: Northern Kalahari of Africa.
Presented in order as Ancestral; Variants.
Presented in order as Homozygous dominant; Heterozygous; Homozygous recessive.
African-American (AA), European-American (EA) of NHLBI Exome Sequencing Project are AA or EA population from National Heart Lung and Blood Institute (NHLBI) Exome Sequencing Project using deep whole exome resequencing of >7000 individuals to study genetic contributions to the risk of several heart, lung and blood phenotypes.
3. As(III) S-adenosylmethionine methyltransferases (AS3MT)
Arsenic (As) is one of the most common environmental contaminants. It exists in four oxidation states (+V, +III, 0, −III) [54]. In most mammalian species, inorganic As(III) can be methylated primarily to methylarsenite [MAs(III)] and dimethylarsenite [DMAs(III)], and, to a lesser extent, to trimethylarsine [TMAs(III)] by the enzyme As(III) S-adenosylmethionine methyltransferase (AS3MT in animals and ArsM in microbes) [55]. Nearly all ArsM/AS3MT enzymes have four conserved cysteine residues (Cys 32, Cys 61, Cys156 and Cys 206 in the human enzyme) that are required for As(III) binding and catalysis (Fig. 3). Biochemical analyses have focused on the activity of purified human AS3MT. Thomas and co-workers first purified AS3MT from rat liver cytosol, and the rat and human genes were subsequently cloned [56,57]. Styblo and coworkers showed that AS3MT and the most common SNP that encodes the M287 T isoform catalyze arsenic methylation with different activities [58]. We have purified seven additional polymorphic hAS3MT proteins and characterized their enzymatic properties. Each enzyme had low methylation activity due to decreased affinity for substrate [59].
Fig. 3. Multiple sequence alignment of As (III) SAM methyltransferases.
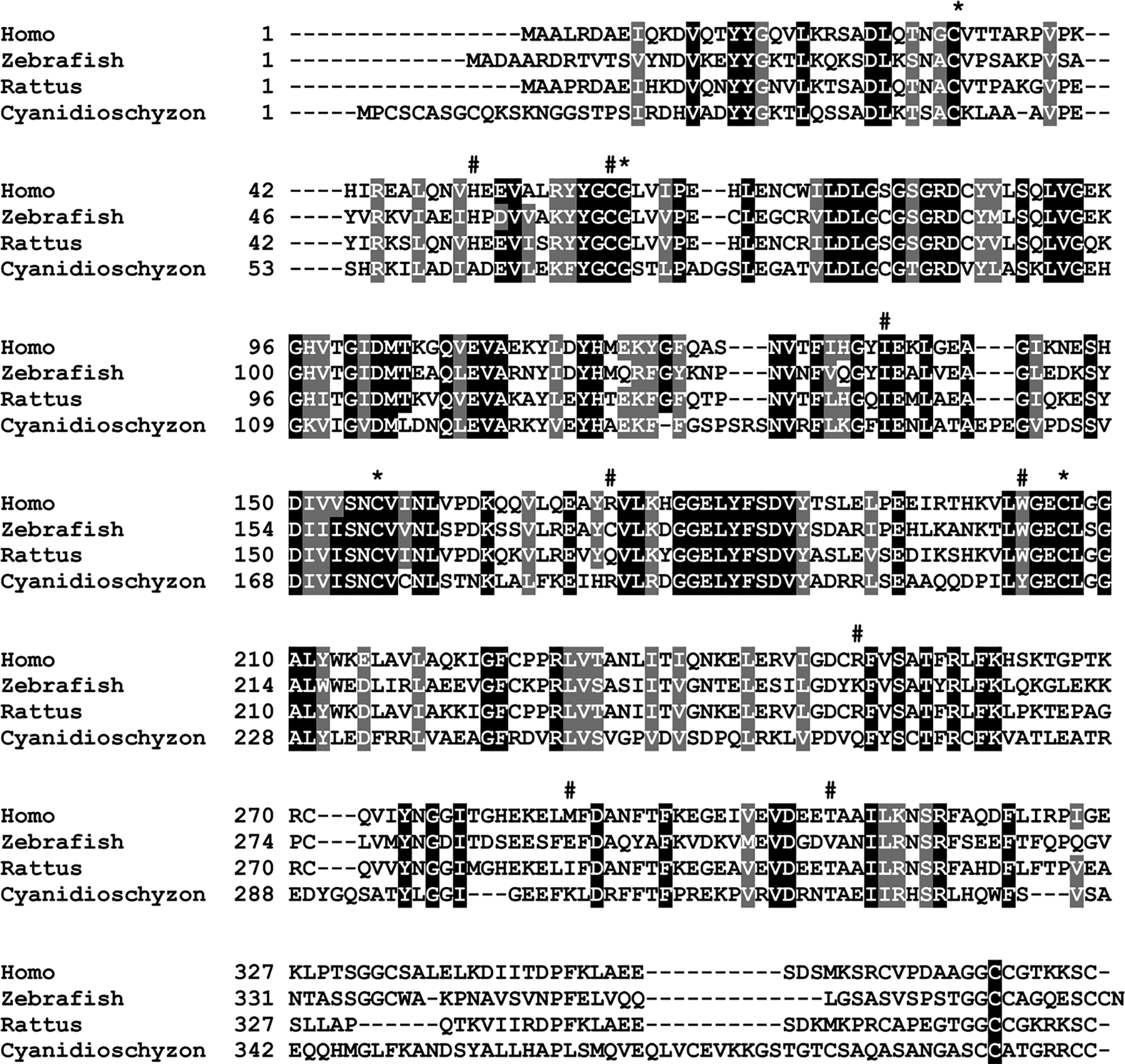
Homo sapiens As(III) S-adenosylmethionine methyltransferase (AAI19639) was aligned with eukaryotic orthologues from Danio rerio (zAS3MT, NP_001034928), Rattus norvegicus (rAS3MT, NP_543166) and Cyanidioschyzon merolae (CmArsM, FJ476310). Black shading indicates conserved residues, grey shading indicates conservative replacements, * denotes conserved cysteine residues, # denotes polymorphic residues.
The human AS3MT gene, located on chromosome 10q24 has 11 exons and is approximately 32-kb long [57]. Splicing variants of the AS3MT gene have been shown to affect the capacity for arsenic methylation. In human AS3MT, there are eight well-studied exonic SNPs with low arsenic methylation activity (Arg173Trp, Met287Thr, Thr306Ile, His51Arg, Cys61Trp, Ile136Thr, Trp203Cys, and Arg251His) (see Fig. 3 and Table 2) [59]. Inefficient arsenic methylation capacity (high MAs% and low DMAs%) is associated with increased disease susceptibility [60], including skin lesions, hypertension [7] or bladder cancer [61]. The SNP rs11191439 (Met287Thr, 860 T > C) occurred with a very high MAF (>5 %) in an arsenic endemic area of Mexico [62]. A case-control study (71 cases with skin lesions and 51 controls) showed individuals carrying the C (T/C and C/C) allele (Thr) were at increased risk of developing skin lesions [OR = 4.28; 95 % CI: 1.0–18.5)] [8].
Individuals with one or more copies of the C allele in rs11191439 (the Mer287Thr polymorphism) may have an elevated risk of bladder cancer (OR = 1.17; 95 % CI = 1.04–1.32) [63]. Moreover, AS3MT rs11191438 (C > G) G/G genotype, AS3MT rs10748835 (A > G) G/G genotype, and AS3MT rs1046778 (C > T) T/T genotype were found to be related to a decreased risk of bladder cancer, where the ORs (95 % CI) were 0.50 (0.31–0.82), 0.49 (0.30–0.79), and 0.54 (0.36–0.80), respectively [61]. Individuals with the AS3MT rs3740392 (intron variant) A/G and G/G genotype genotypes had a lower secondary methylation index, which is related to developmental delay with an OR (95 % CI) of 1.59 (1.08–2.34) [64]. Low level arsenic exposure was reported to be associated with increased cardiovascular disease mortality, and hyperlipidemia was associated with AS3MT polymorphism rs10748835 genotype A/G vs. A/A (P < 0.05) (n = 499) [65]. Wood et al. identified 26 SNPs, including 3 non-synonymous coding SNPs (cSNPs) in the AS3MT coding regions: Arg173Trp, C > T (rs35232887) in exon 6, Met287Thr, T > C (rs11191439) in exon 9 and T306I, C > T (rs34566438) in exon 10, as well as a variable number of tandem repeats in exon 1 corresponding to the 5′-untranslated region of the gene [57]. The most common exonic SNP is Met287Thr (in exon 9, SNP ID: 1445, rs11191439; T860C; Met287Thr), which is associated with several diseases as discussed previously. A genome-wide association study (GWAS) of 124 women from the Argentinean Andes and a PHIME-CROME study of a Croatian-Slovenian population indicate that arsenic is an environmental stressor that likely has driven an increase in the frequencies of protective alleles of AS3MT [66]. This supports the concept that some SNPs confer a positive selective advantage and represent human adaptation to arsenic-rich environments.
4. Indolethylamine N-methyltransferase (INMT)
In 1961 Julius Axelrod discovered the enzymes indolethylamine N-methyltransferase (INMT) and arylamine N-methyltransferase, which convert serotonin and tryptamine to the psychotomimetic metabolites bufotenine and N,N-dimethyltryptamine, respectively [67]. INMT is widely distributed in mammalian tissues including lung [68], liver and brain [69]. INMT catalyzes metabolism of a group of small molecule acceptors such as tryptamine, serotonin, and other endogenous indole-containing compounds. The human INMT gene is located at chromosome 7p15.2–p15.3, has three exons, and is structurally similar to the NNMT and PNMT genes in other species. INMT utilizes methyl group acceptors as substrates and accommodates a large variety of amines, among which tryptamine is the preferred substrate with the highest apparent Kcat [70]. Tryptamine is found in trace amounts in mammal brains and is considered a neuromodulator or neurotransmitter [71] involved in mental illness [72]. INMT produces N,N-dimethyltryptamine (DMT) [73], which is a hallucinogen and has high affinity for various serotonergic, adrenergic, histaminergic and dopaminergic receptors (Sig-1R) [74]. DMT is synthesized via decarboxylation of tryptophan, followed by double-methylation utilizing AdoMet as a methyl donor (Fig. 4). DMT is found in the brain at an average concentration of 0.56 nM [69] and is considered as a monoamine neurotransmitter. Hallucinogens are psychoactive substances that alter cognition and perception by triggering neurotransmitter receptors in the brain [75]. Consequently DMT, as a product of INMT methyltransferase, is considered to be a neurotransmitter. Differences in the function or levels of INMT variant enzymes can lead to mental behavioral phenotypes generated by exogenous DMT [76].
Fig. 4.
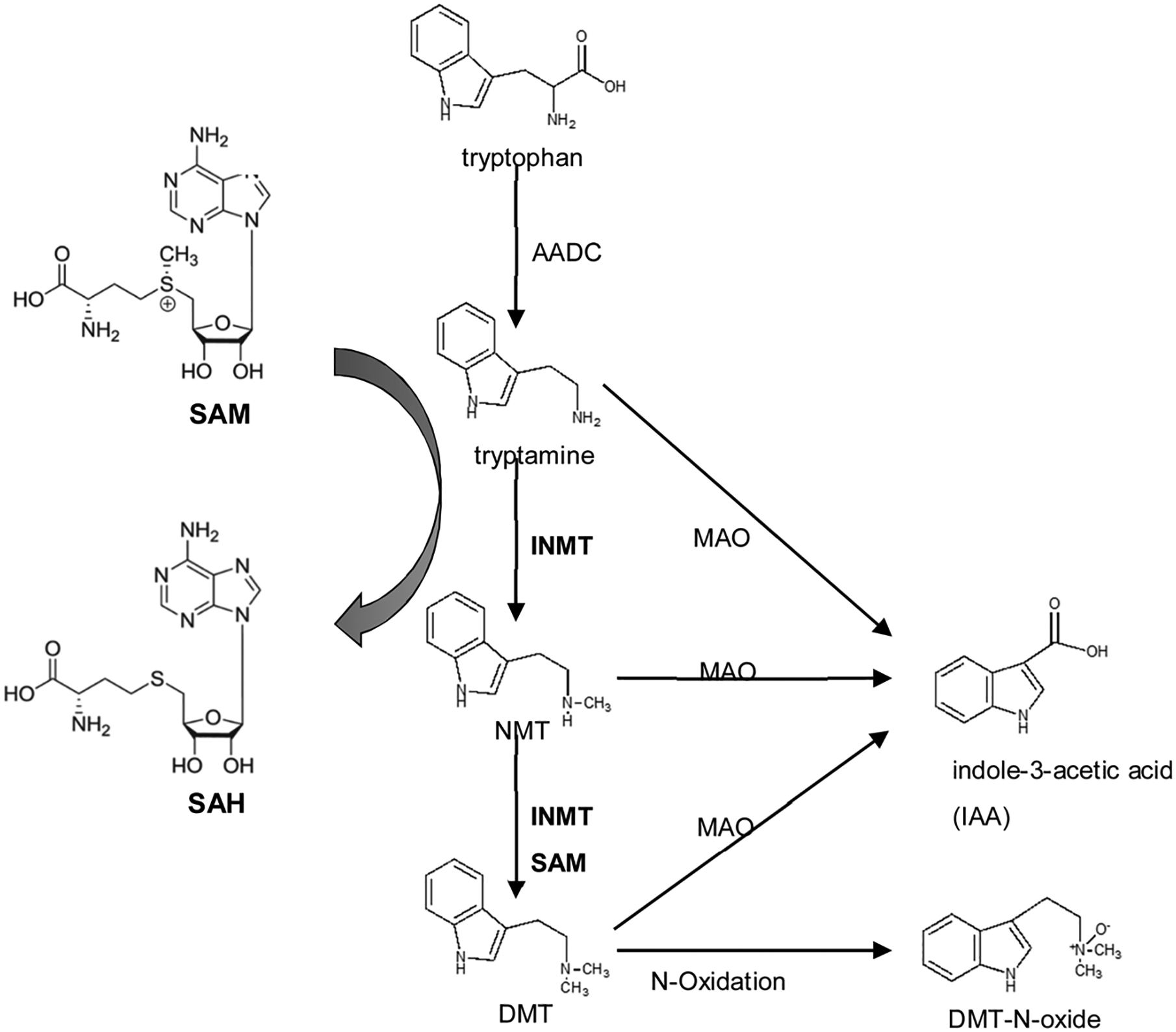
Pathways for the biosynthesis and metabolism of DMT. Tryptophan is converted to tryptamine by aromatic amino acid decarboxylase (AADC). Trytamine is dimethylated to first yield N-methyltryptamine (NMT) and then N,N-dimethyltryptamine (DMT) by indole-N-methyltransferase (INMT), using S-adenosyl-methionine (SAM) as the methyl source. Metabolism: tryptamine, NMT and DMT are all substrates for monoamine oxidase (MAO), yielding indole-3-acetic acid (IAA) as both a common precursor metabolite and the most abundant metabolite of DMT itself. DMT is also converted to DMT-N-oxide as the second-most abundant metabolite. Adapted and modified from [77].
Contributions to the understanding of the relationships between INMT, its polymorphisms and human health have been made primarily over the last decade. A study involving 187 Hirschsprung’s disease (HSCR) patients and 283 controls found a nonsynonymous SNP in the coding region of INMT (rs77743549, His46Pro) was associated with significantly increased risk of HSCR (OR = 1.77; corrected P = 0.002) [10]. INMT also methylates the essential element selenium (Se), although Se metabolism in humans is not well characterized. A cohort study involving subjects from Bangladesh and Argentina identified 3 SNPs (rs1061644; rs4270015; rs6970396) in the INMT gene that are strongly associated with metabolism of Se to the trimethylselenonium ion (TMSe) [78]. Also, samples collected from pregnant women (n = 226) in rural Bangladesh at gestational weeks 8, 14, 19, and 30 showed that urinary concentrations of TMSe were associated with the INMT rs6970396 A/G and A/A genotypes [79]. Another study demonstrated that the INMT SNP (rs6970396), and AS3MT haplotypes, including Aquaporin 4 and 9 polymorphisms, affected As and Se metabolism in the Croatian-Slovenian population (n = 509) [80]. Although there is some evidence of a relationship between maternal Se and child neurodevelopment, no solid conclusion regarding INMT polymorphism and neuropsychological development could be drawn [81].
5. Phenylethanolamine N-methyltransferase (PNMT)
Phenylethanolamine N-methyltransferase catalyzes the terminal step of catecholamine biosynthesis with norepinephrine as substrate, forming the product epinephrine, using SAM as the methyl donor (Fig. 2). PNMT plays a key step in regulating epinephrine production [82] (Fig. 1). The PNMT gene, located on chromosome 17 (17q21), has been linked to hypertension, which has both genetic and environmental components [83]. PNMT mRNA levels are significantly increased in all four chambers of the heart in hypertensive rats, with the highest expression in the right atrium [84]. Increased PNMT gene expression is a possible mechanism to explain why prenatal exposure to elevated levels of glucocorticoids leads to hypertension later in life [85].
Epinephrine accounts for 5–10 % of the catecholamine content in brain, with the exception of the adrenal medulla. Epinephrine function in the central nervous system has been implicated to control not only blood pressure and respiration, but also neurodegenerate disorders such as AD [86]. AD is a polygenic disorder, with multiple genes contributing to the disease phenotype [12]. PNMT activity is decreased in the brain of AD individuals. The decrease in the hippocampus correlates significantly with the degree of dementia [87]. Transport of the PNMT protein in advanced AD is correspondingly decreased compared with early Alzheimer’s cases [88].
Eighteen PNMT SNPs, including four non-synonymous cSNPs (Asn9Ser, Thr98Ala, Arg112Cys and Ala175Thr) were identified in 2005 [100]. The Thr98Ala allozyme displayed significantly lower levels of both activity and immunoreactivity compared to the reference enzyme, indicating that polymorphisms in PNMT could be one explanation for inherited variations in the ability to form epinephrine from norepinephrine [100]. In addition, a case-control study in 2001 reported a significant association (P ≤ 0.007) between early-onset AD and two polymorphisms, G353A and G148A, in the promoter region of the gene coding for PNMT [12]. Further, there is a connection between PNMT SNPs and hypertension. Two common PNMT promoter SNPs, G367A (rs3764351) and G161A (rs876493), are associated with decreased risk of essential hypertension in Han Chinese [11]. Another case-control study showed that PNMT + 1543 G allele was associated with the development of acute kidney injury (AKI) [OR = 2.19, 95 % CI = 1.04–4.60], and the PNMT −161A (rs876493) allele was associated with lower mortality from AKI [94]. PNMT gene polymorphisms rs876493, rs2934965 and rs2941523 have also been associated with crisis pain in sickle cell disease with P ≤ 0.005. [13]. Finally, rs876493 was shown to increase PNMT promoter activity and reduced risk of asthma [101].
6. Histamine N-methyltransferase (HNMT)
Histamine N-methylation is the major process responsible for termination of the neurotransmitter action of histamine in the brain [102]. Histamine-N-methyltransferase (HNMT) methylates histamine by transferring a methyl group to the imidazole ring, using SAM as the methyl group donor, yielding methylhistamine and SAH. As a neurotransmitter, histamine affects physiological processes including memory formation, hormonal control, cardiovascular control and thermoregulation [103].
The HNMT gene is located on chromosome 2q22.1, is 50 kb in length, and includes six exons. HNMT mRNA is found in most human tissues, with the highest mRNA levels in kidney and liver cells [104]. HNMT activity was determined in 127 clinical surgery renal biopsy samples. Those samples with the lowest thermal stability also had the lowest levels of enzyme activity [104], similar to what was found with polymorphic COMT [105]. C314 T and A939 G transitions have also been examined. A C314 T transition (rs11558538 s), which corresponds to a Thr105Ile change in the enzyme, led to a significant decrease in HNMT activity [19]. The HNMT 939A > G polymorphism lowers HNMT enzymatic activity by decreasing HNMT mRNA stability in patients with acetylsalicylic acid-intolerant chronic urticaria [106]. The observations that these polymorphisms affect the levels of enzyme activity imply that HNMT polymorphisms might be related to the pathophysiology of diseases that involve variations in histamine metabolism, such as allergy, asthma and neuropsychiatric illness.
The105lle HNMT variant has been proposed to confer a selective advantage. Higher histamine levels might be protective because histamine, IgE and mast cells played defensive roles against nematode infections [107]. In addition, elevated histamine levels in the brain have been associated with PD and schizophrenia [108]. In one case-control study, lower HNMT activity, such as that of the Ile105 allele with 30 %–50 % lower HNMT activity, was associated with the pathogenesis of PD [chi square (χ2) = 11.65, P = 0.0006] [109]. In another case-control cohort of Han Chinese patients with PD, schizophrenia, and healthy controls, the HNMT Thr105Ile polymorphism was suggested to protect against PD (OR = 0.516, P = 0.007) and schizophrenia (OR = 0.499, P = 0.011) [16]. A meta-analysis that included 4 eligible case-control association studies for the HNMT rs11558538 SNP and the risk for PD (2108 patients, 2158 controls) showed a statistically-significant association with a decreased risk for PD in Caucasian patients (OR = 0.63, 95 % CI = 0.45–0.88) [14]. The C314 T (Thr105Ile) polymorphism was reported as a risk factor for asthma [18,19]. Individuals with HNMT Thr105Ile and His645Asp mutations are more prone to developing symptoms at lower IgE levels [110]. In addition to the non-synonymous C314 T (Thr105Ile) SNP, the A939 G (rs1050891) polymorphism in the 3′-UTR of the HNMT gene was also shown to decreased histamine release due to an increased enzyme activity [106]. Histamine participates in immune responses and is involved in autoimmune disorders. The autoimmune disease myasthenia gravis has also been associated with the A939 G HNMT variant, but not the C314 T SNP [111].
7. Nicotinamide N-methyltransferase (NNMT)
Nicotinamide is the active form of vitamin B3, as well as a component of the coenzyme nicotinamide adenine dinucleotide. Nicotinamide has anti-inflammatory actions. Nicotinamide has been considered a beneficial agent for treating inflammatory skin disorders for the past 50 years, as a long-term oral therapy [112]. By enhancing DNA repair and reducing UV-induced suppression of skin immune responses, nicotinamide also acts as a chemoprevention agent [113]. Nicotinamide is methylated by nicotinamide N-methyltransferase (NNMT) [114] and yields two products: S-adenosyl-l-homocysteine and N1-methylnicotinamide, which are excreted into urine.
NNMT was first identified by cDNA cloning from the liver, where NNMT is predominantly expressed [115]. Interestingly, NNMT was highly-expressed in various cancers, such as papillary thyroid carcinoma [116], liver cancer [117], colorectal cancer (CRC) [118], lung cancer and oral carcinoma [119]. NNMT is a novel serum tumor marker for CRC [118]. Some studies demonstrated that hepatocellular carcinoma patients with higher NNMT mRNA expression levels tended to have a shorter overall survival and significantly shorter disease-free survival [117]. Also, significantly higher NNMT enzyme activities were detected in follicular cell lines [116]. The significant role of NNMT in regulation of metabolic pathways and high levels in several types of cancers indicates that it is a potential molecular target for cancer therapy. Besides being a potential tumor marker, NNMT has been associated with energy metabolism and the body mass index (BMI). NNMT was found to be upregulated in the liver of a mouse model of obesity [120]. A cross-sectional cohort study demonstrated that NNMT knockdown in adipose tissue and liver could prevent diet-induced obesity [121]. A case-control study of obese (n = 289) versus normal BMI individuals (n = 498) was carried out to explore the association between obesity and the NNMT gene polymorphism rs10891644 (intron variant, −129-11693G > T with three genotypes : homozygous dominant G/G, heterozygous G/T, and homozygous recessive T/T). The percentage of body fat difference between individuals with the heterozygous G/T and homozygous (G/G and T/T) genotypes were highly significant (P < 0.01). The ORadjusted value of the G/T versus (G/G andT/T) was 1.716, which means that the chance of G/T individuals having high body fat is 1.76 times that of the homozygous (G/G and T/T) individuals, indicating that the heterozygous individuals (G/T carriers) are susceptible to obesity [23]. In addition, a study of hyperlipidemic patients (n = 395) and controls (n = 316) showed that NNMT SNP rs1941404 is significantly associated with hyperlipidemia (P < 0.0026) [122]. rs1941404 genotype carriers in the Chinese Han population were also found to be associated with type 2 diabetes (P < 0.002, n = 558 T2D patients and 442 healthy controls) [24]. These case-control studies are listed in Table 3, which includes additional epidemiologic studies on MTase polymorphisms.
Table 3.
Case-control and prospective cohort studies relating SAM-dependent MTases polymorphisms to disease.
| Polymorphisms | Diseases(symptom) | Study sample | Statistical analysis | Notable observations |
|---|---|---|---|---|
| COMT rs4680 | Schizophrenia (SCZ) | meta-analysis (n = 1416); atypical antipsychotics (n = 1207) | P = .0098, OR Met/Met = 1.54, 95% CI: 1.11–2.14 | COMT Val158Met polymorphism is associated with response to antipsychotics in schizophrenia and schizo-affective disorder patients [1] |
| COMT rs6269, rs4633, rs4818, rs4680 | Parkinson’s disease (PD) | 409 PD patients | Hazard ratio = 3.24; P = 0.02 | high-COMT activity haplotype (G-C-C-G for rs6269, rs4633, rs4818, and rs4680) showed a high risk of cognitive decline in PD [3] |
| COMT rs740603 | Surgery acute pain | 241 children of African American and 277 of European Caucasian ancestry | A allele, OR: 0.69, 95 % CI: 0.48–0.99, P = 0.046 | COMT rs740603 was related to high pain in European Caucasian subjects [89] |
| COMT rs6267 | Pain in PD patient | 418 PD patients for evaluating pain severity | P < 0.01 | COMT rs4680 and COMT rs6267 contribute to pain in PD patients [4] |
| COMT rs4680 | Stress mindset on affect and cognition | Participants (n = 107) were exposed to a stress mindset manipulation | (F4,32 = 3.52, p = .017, η2 = .306 | Genetic variation at rs4680 modified the effects of stress mindset on affective and cognitive responses to stress [90] |
| COMT rs4680 | Alzheimer disease (AD) | 345 AD and 253 healthy controls | OR = 6.71, 95 %CI 3.36–13.41, P < 0.001 | OMT Val158 Met polymorphism as an associated risk factor for Alzheimer disease [52] |
| AS3MT rs11191438 rs10748835 rs1046778 |
Bladder cancer | 216 bladder cancer patient and 648 healthy controls | (OR) (95 % CI) was 0.50 (0.31–0.82), 0.49 (0.30–0.79), and 0.54 (0.36–0.80), | rs1119438 C/C, rs10748835 A/A and rs1046778 C/C, were associated with bladder cancer risk; AS3MT rs1046778 C/C + C/T genotype increase the risk of BC [61] |
| AS3MT rs3740392 | Developmental delay | 179 children with developmental delay and 88 controls | OR and 95 %o CI, 1.59 (1.08–2.34) | AS3MT rs3740392 A/G + G/G genotype had a significantly higher OR than those with AS3MT low-risk haplotypes [64] |
| AS3MT rs11191439 | Diabetes | N = 255 | OR = 11.4 (95 % CI 2.2–58.8) | Individuals with M287 T polymorphisms were more frequently diabetic than the respective wild-type carrier [9] |
| AS3MT rs11191439 | Skin lesions | 71 cases with skin lesions and 51 controls | OR = 4.28; 95 % CI (1.0–18.5) | Met287Thr influences the susceptibility to premalignant As skin lesions [8] |
| INMT rs77743549 | Hirschsprung’s disease (HSCR) | 187 HSCR patients and 283 controls | OR = 1.77; corrected P = 0.002 | rs77743549 of INMT may be associated with the risk for HSCR [10] |
| NNMT rs694539 | ALL | 245 pediatric ALL patients (cases) and from 500 blood bank donors (controls) | OR = 2.2; 95 % CI, 1.1–4.6; P = 0.04 | the NNMT IVS —151 TT genotype showed a 2.2-fold increased ALL risk [91] |
| NNMT rs694539 | SCZ | cohort of 42 SZ patients and 86 healthy controls | P = 0.0015 | NNMT rs694539 contribute to etiology of SCZ in a Han Chinese female population [92] |
| NNMT rs694539 | Epilepsy | 215 patients with epilepsy and 239 healthy controls | χ (2) = 6.682, P = 0.035 | NNMT gene rs694539 variant involve in the etiology of epilepsy in male patients [93] |
| NNMT rs1941404 | SCZ | 202 unrelated families including a schizophrenia patient and her/his parents | P = 0.033 | NNMT rs1941404 were significantly associated with schizophrenia [22] |
| NNMT rs10891644 | Obesity | 289 of high body fat group, 494 of low body fat group | Padjusted = 0.002, 95% CI = 1.240–2.235 | the variation of the tagSNP, rs10891644 is significantly associated with obesity [23] |
| PNMT rs5638 | Acute kidney injury (AKI) | 194 AKI patients and 767 controls | OR = 2.19, 95 % CI = 1.04–4.60 | The PNMT + 1543 G allele was associated with AKI [94] |
| PNMT G-353A and G-148A | AD | 131 necropsy confirmed AD cases, and 947 adult nondemented controls | P ≤ 0.007 | genetic variation in the promoter of the PNMT gene is associated with increased susceptibility to Early-Onset AD [12] |
| PNMT rs3764351 rs876493 |
Hypertension | 316 hypertension patients and 316 controls | P = 0.01; adjusted OR 0.17; 95% CI 0.05–0.58 | The 2-SNP AA haplotype in the PNMT promoter is associated with decreased risk of essential hypertension in Han Chinese [11] |
| HNMT rs11558538 | PD | 2108 patients with PD, 2158 controls | OR = 0.63; 95 % CI = 0.45–0.88 for Caucasian patients | HNMT rs11558538 minor allele reduced risk of developing PD [14]. |
| HNMT rs11558538 | Asthma | 192 asthma patients and 237 controls | OR = 1.9, P < 0.01 | Variants with low HNMT activity were associated with asthmatics [19] |
| HNMT rs11558538 | PD | 913 patients with PD and 958 controls | χ(2) = 11.65; P = 0.0006 | Lower HNMT activity plays a role in the pathogenesis of PD [15] |
| HNMT rs11558538 | PD and SCZ | 564 PD patients and 496 controls; 423 SCZ patients and 457 healthy controls | PD (OR = 0.516, p = 0.007); SCZ (OR = 0.499, P = 0.011) | Ile allele was associated with reduced risk of PD and SCZ [16] |
| DNMT1 rs2228611 rs2228612 |
Primary osteoarthritis (OA). | 244 OA patients and 244 controls | OR (95 % CI) 0.71 (0.51–0.97) | Haplotype C-T exhibited a lower risk with significant differences [OR (95% CI) 0.71 (0.51–0.97)] [95] |
| DNMT3b rs2424913 | Primary osteoarthritis (OA). | 244 patients and 244 controls | OR (95 % CI) 1.6 (1.1–2.4) | The C/C genotype of rs2424913 of DNMT3B was associated with an increased risk [95] |
| Dnmt1 rs2228612 | Outcome of melanoma patients | 123 melanoma patients | Hazard ratio = 6.620, 95 % confidence interval (CI): 2.214–19.791, P = 0.001 [15] | DNMT1 rs2228612 carriers had an increased risk for adverse outcome of nemanoma [96] |
| DNMT3B rs406193 | Outcome of melanoma patients | 123 melanoma patients | P = 0.012 | DNMT3B rs406193 polymorphism correlated significantly with the presence of tumor-infiltrating lymphocytes [96] |
| DNMT3B rs1569686 | Gastric Cancer | 3959 cases and 5992 controls | OR 0.74, 95 %CI 0.61–0.90 | rs1569686 might be a protective factor against gastric carcinogenesis [97] |
| DNMT3A rs1550117 | Colorectal cancer | 2184 cancer cases and 3420 controls | AA vs. GG (OR, 3.16; 95 % CI, 1.58–6.29; P = 0.001) | DNMT3A rs1550117 A > G polymorphism increased risk of colorectal cancer [98] |
| Dnmt1 rs2162560 | Chemotherapy-Associated Cognitive Impairment (CACI) | Prospective cohort study with 351 early-stage breast cancer patients | OR = 0.45, 95 % CI: 0.25–0.82, P = 0.01 | The allele carriers were associated with lower odds of self-reported cognitive decline [99] |
The human NNMT gene, located at 11q23.1 as determined by fluorescence in situ hybridization [115], is approximately 16.5 kb in length, with 3 exons and 2 introns. Individual differences in xenobiotic and drug toxicity may result from the variation in human hepatic NNMT activity. Human NNMT activity shows a bimodal frequency distribution [123], which was predicted to be a consequence of a genetic polymorphism [124]. We analyzed the NNMT gene mutation data in COSMIC (Catalogue of Somatic Mutations in Cancer, https://cancer.sanger.ac.uk/cosmicv), the world’s largest and most comprehensive resource for exploring the impact of somatic mutations in human cancer. There are 468 non-synonymous mutations reported in NNMT. NNMT mutations are associated with a variety of cancers, with the largest number of different mutations found in skin cancers and the greatest incidence of mutation found in large intestine cancer [21]. NNMT polymorphisms are also associated with increased incidence of ALL. A study involving 245 pediatric ALL patients (cases) and 500 blood bank donors (controls) found an NNMT SNP (rs694539; C < T, T/T genotype) conferred a 2-fold increased risk of ALL (OR = 2.2; 95 % CI: 1.1–4.6; P = 0.04) [91].
In recent years, SNPs in the human NNMT genes have been reported to be significantly associated with neurologic/mental illness, including two noncoding region SNP (rs694539 and rs1941404) [21]. NNMT has been implicated in the pathogenesis of neuropsychiatric diseases. SNP rs694539 is associated with several disorders, including bipolar disorder [125], epilepsy [93], migraine [126], and schizophrenia [22]. First, low NNMT serum levels were found to be significantly associated with bipolar disorder [127]. The rs694539 variant of NNMT was associated with bipolar disorder in a case-control study of 95 bipolar disorder patients and 201 healthy controls (χ2 = 13.382, P = 0.001). Second, the NNMT rs694539 variant was shown to be involved in the etiology of epilepsy (χ2 = 6.682, P = 0.035, n = 454) [93]; Third, the association of the rs694539 variant with migraine was reported in a case-control study of 433 patients with migraine and 229 healthy controls (χ2 = 6.076, P = 0.048) [126]; Fourth, several NNMT haplotypes were reported to be significantly associated with schizophrenia (global P values <0.05 following permutation test adjustment) [22]. Specifically, a study of a Han Chinese female population (n = 42 schizophrenia patients and 86 healthy controls) found rs694539 has a role in the etiology of schizophrenia (P = 0.0015) [92], with rs1941404 also associated with schizophrenia (P = 0.033) [22].
8. Thiopurine methyltransferase (TPMT)
In the 1960s, the function of thiopurine S-methyltransferase (TPMT) was found to be the transfer of the methyl group from SAM to the sulfur atom of thiopurines [128]. TNMT, with a molecular mass of 26 kDa is expressed in liver, kidney, intestine, erythrocytes, leukocytes and a number of other tissues [129]. Interest in TPMT emerged initially because of the important role this enzyme plays in metabolic transformation of thiopurine drugs [130]. The thiopurine drugs azathioprine and mercaptopurine are effective in the treatment of disorders of immune regulation, ALL and organ transplant recipients [131]. Thiopurine drugs are converted to toxic compounds, thioguanine nucleotide metabolites (TGNs) that are cytotoxic, immunosuppressive and kill immune system cells in the bone marrow [25]. However, the TPMT enzyme also metabolizes thiopurine drugs into inactive, nontoxic compounds. Thus, the therapeutic window between toxicity and efficacy of thiopurine drugs is narrow. The use of azathioprine in a patient with complete TPMT deficiency resulted in death from neutropenia sepsis [132].
The TPMT gene, located in 6p22.3, is approximately 34 kb in length and consists of 10 exons and 9 introns. TPMT variants are the most well-characterized genetic polymorphisms that affect drug metabolism, and variants have been studied in most populations [133]. More than 40 allelic variants of the TPMT gene have been reported to the TPMT nomenclature committee and assigned allele names (http://www.imh.liu.se/tpmtalleles) [134]. TPMT has been reported to exhibit genetic polymorphism in Caucasian populations, among which 89 % of individuals have high TPMT activity, 11 % of individuals exhibit intermediate activity, and one in 300 has extremely low or absent activity [135]. A novel TPMT allele, TPMT*45, was identified in a Korean girl whose findings suggested decreased TPMT activity [134]. Currently, approximately 20 variant alleles have been associated with low TPMT enzymatic activity in humans (Table 4). The most prominent TPMT variants include amino acid substitution variants (TPMT*2, *3A, *3B, *3C, *3D, *5, *6, *7, *8), a premature termination variant (TPMT*3D), and a variant that eliminates a splice site (TPMT*4). The predominant variant alleles, TPMT*3C, *3A and *2, account for over 95 % of inherited TPMT deficiencies [136]. TPMT*2, containing 238 G > C, was the first identified variant with decreased catalytic activity [137]. The second identified and more prevalent variant allele (TPMT*3A) contains two nucleotide transition mutations (G460A and A719 G), leading to amino acid substitutions Ala154Thr and Tyr240Cys [138,139]. A single 719A > G polymorphism in the open reading frame (ORF) of the TPMT*3C allele also results in reduced protein stability [140]. TPMT*4 is the first reported allele with low TPMT enzymatic activity as the result of a mutation within an intron [141]. TPMT*4, with a G > A transition at the intron 9–exon 10 junction, has the final nucleotide of the intron at the 3′ acceptor splice site sequence deleted, which reduces TPMT activity.
Table 4.
List of allele variants at the human TPMT locus.
| Locus | SNP | Location | Enzyme activity |
|---|---|---|---|
| TPMT*1 | Wild type: high activity | ||
| TPMT*1A | C178T | Exon 1 | Higher enzymatic activity [142] |
| TPMT*2 (the 1st identified variant allele) | G238C (Ala80Pro) | Exon 5 | This mutation led to a 100-fold reduction in TPMT activity and very low levels of immunodetectable protein [137] |
| TPMT*3A (contains two transition mutations, *3B and *3C) | G460A (Ala154Thr) and A719 G (Tyr240Cys) | Exon 7 and 10 | The most common variant allele responsible for low TPMT activity in Caucasians; 400-fold decrease in protein levels and no detectable enzyme activity [139] |
| TPMT*3B | G460A (Ala154Thr) | Exon 7 | Four-fold decrease in protein levels |
| TPMT*3C | A719 G (Tyr240Cys) | Exon 10 | More frequently identified in African and Southeast Asian populations; 1.4-fold reduction in protein level and associated with lower immunodetectable TPMT protein and catalytic activity [140] |
| TPMT*3D | G292 T/G460A (Glu-Stop) | Exon 5 and 7 | Formation of a premature stop codon [143,144] |
| TPMT*4 | G to A transition | Intron 9–exon 10 junction | Destruction of a splice site [141], with reduced TPMT activity |
| TPMT*5 | T146C (Leu49Ser) | Exon | Intermediate TPMT activity [145] |
| TPMT*6 | A539 T (Tyr180Phe) | Exon 8 | Korean subject with intermediate activity [145] |
| PMT*7 | T681 G (His227Glu) | Exon 10 | European subject with intermediate TPMT activity |
| TPMT*8 | G644A (Arg215His) | Exon 10 | African American subject with intermediate activity [146] |
| TPMT*9 | A356C (Lys119Thr) | Exon | No function [147] |
| TPMT*10 | G430C (Gly144Arg) | Exon | Deficient activity |
| TPMT*11 | G395A (Cys132Tyr) | Exon | No function |
| TPMT*12 | C374 T (Ser125Leu) | Exon | No function |
| TPMT*13 | A83 T (Glu28Val) | Exon | No function |
| TPMT*14 | A1G (Met1Val),-G1A, | Exon 3 | Splicing defect [148] |
| TPMT*15 | G488A (Arg163His), | Intron 7 | |
| TPMT*16 | C124 G (Gln42Glu), | Exon | |
| TPMT*17 | G211A (Gly71Arg) | Exon | |
| TPMT*18 | Exon | ||
| TPMT*1S | T474C | Exon | Silent mutation that does not lead to any change in enzymatic activity [149] |
| TPMT*45 | C676 T (Pro226*) | Exon | nonsense mutation that decreased TPMT activity [134] |
In childhood ALL, the most common cancer in children, the bone marrow produces too many immature lymphocytes [150]. As the number of leukemia cells increases in the blood and bone marrow of ALL patients, healthy white blood cells, red blood cells and platelets are crowded out, leading to infection, anemia and bleeding. The purine analogues mercaptopurine and thioguanine are drugs that have been used as cytotoxic agents in the treatment of leukemias for more than 50 years [151]. Because the therapeutic window between toxicity and efficacy is narrow, thiopurine drugs can potentially have life-threatening toxicity [152]. The impact of the thiopurine methyltransferase genotype on thiopurine dose intensity, myelosuppression and treatment outcome was investigated in the United Kingdom childhood ALL trial, ALL97 [25]. There were 1935 patients on this trial with 1206 patients carrying homozygous wild-type TPMT*1/*1, and 128 patients having low activity variant alleles (99 TPMT*1/*3A; 17 TPMT*1/*3C; 4 TPMT*1/*2; two children with the rare alleles TPMT*1/*9, TPMT*1/*21; three children with novel alleles TPMT*1/*32, TPMT*1/*33, TPMT*1/*34; one compound heterozygote TPMT*2/*3A; one homozygous TPMT*3A/*3A and one TPMT*3C/*3C). The findings from that trial demonstrated that TPMT polymorphism affected event-free survival (EFS) of ALL patients [25]: TPMT*1/*3A heterozygotes had a better EFS than TPMT wild-type patients (n = 1206, EFS 80 %, P = 0·05) or TPMT*1/*3C patients (n = 17, EFS 53 %, P = 0·002). In addition, the United States Food and Drug Administration states that TPMT genotype testing should be considered if a patient has clinical or laboratory evidence of severe toxicity, particularly myelosuppression. Testing for TPMT status is recommended prior to initiation of mercaptopurine therapy, so that the starting dosages can be adjusted accordingly. If the patient is homozygous with two or more functional alleles, the patient should receive the normal starting dose of mercaptopurine. If the patient is heterozygous, with one functional allele and one nonfunctional allele, the patient should receive reduced doses of mercaptopurine. If the patient carries only a low or deficient variant (homozygous phenotype), alternative agents for treatment should be considered [153].
9. DNA (cytosine-5)-methyltransferase (DNMT)
Epigenetics is defined as heritable changes in gene activity and expression that occurs without alteration in DNA sequence that guide the genome on” what to express and what not to.” DNA methylation catalyzed by DNA methyltransferase enzymes (DNMTs) is a major epigenetic modification in higher eukaryote genomes. In mammals, DNA methylation is essential for embryonic development and plays important roles in gene expression, genomic imprinting, X chromosome inactivation, and maintenance of genome integrity [154–156]. DNA methylation appears to regulate gene expression at different levels. Methylation of CpG sites within the promoters of genes can lead to their silencing, a feature found in a number of human cancers (i.e., the silencing of tumor suppressor genes). DNA methylation has been shown to be involved in silencing of an entire chromosome, as seen in the case of X inactivation [157]. DNA methylation can also affect gene expression by acting on a large chromosome domain containing multiple genes, such as some imprinted loci [158]. Furthermore, DNA methylation can act directly on transcription either by blocking transcription factor binding to the promoter sequences or by interfering with transcription elongation [159].
DNA methylation is predominantly found in cytosine-phosphate-guanine (CpG) dinucleotide sites of the mammalian genome, called CpG Islands (CpGIs), which are 0.4–3 kb in length and relatively rich in guanine and cytosine nucleobases (>55 %) [160]. In mammalian cells, DNA methylation occurs at the 5′ position of the cytosine ring within CpG dinucleotides via addition of a methyl group to create a 5-methyl-cytosine (m5C) [161]. The modification at m5C is catalyzed by DNMTs, including DNMT1, DNMT3a, and DNMT3b in mammals [28]. These enzymes can be further classified as de novo methyltransferases (e.g., DNMT3a and DNMT3b) or maintenance methyltransferases (e.g., DNMT1). De novo methyltransferases methylate previously unmethylated CpG sequences, but maintenance methyltransferases copy pre-existing methylation marks onto new DNA strands during replication [162]. DNMT1, DNMT3a and DNMT3b map to chromosomes 19p, 2p and 20q, respectively. The DNA methyltransferase homolog DNMT2 maps to 10p and the DNMT3 family member named DNMT3L maps to 21q [163].
DNMT1 is a major maintenance methyltransferase in vivo and ensures replication of DNA methylation patterns after each round of cell division [155]. Global genome demethylation caused by mutation in the Dnmt1 gene plays an essential role in X-inactivation, genomic imprinting and genome stabilization [164]. The de novo establishment of DNA methylation patterns in early mammalian development involves DNMT3 family members, DNMT3A and DNMT3B, and the DNMT3-like non-enzymatic regulatory factor, DNMT3L [155]. Given their function in the de novo establishment of DNA methylation, both Dnmt3a and Dnmt3b, in conjunction with their nonenzymatic cofactor DNMT3L, play important roles in normal development and disease [165]. DNMT3A recognizes the unmethylated state of lysine 4 in histone H3 [166], and de novo DNA methylation by DNMT3A requires alteration of chromatin structure. DNMT3b is specifically required for methylation of centromeric minor satellite repeats. The autosomal recessive disease, Immunodeficiency, Centromeric instability and Facial anomalies syndrome, type I (ICF-1), is associated with hypomorphic mutations in the DNMT3b gene, the altered expression of which has also been correlated with the development of tumors [167,168]. The first example of naturally occurring mutations in a mammalian DNA methyltransferase gene related to ICF was reported in 1999 (two missense substitutions and a 3-aa insertion in the Dnmt3b gene) [169]. A hydrogen bond in the catalytic loop of DNMT3B causes lower CpG specificity than exhibited by DNMT3A, and site-specific epigenomic alterations seen in ICF syndrome with DNMT3B mutations can be partly explained by a shift in the substrate preference of DNMT3A and DNMT3B [170]. Homozygous missense variants in the DNMT3B gene (NM_ 006892; 2477 G > A, Arg826His) account for ICF-1 cases [27].
DNMT1, DNMT3A and DNMT3B polymorphisms are associated with increased or decreased risk of various tumors. DNMT1 (rs2228612, rs2228611, and rs2114724) and DNMT3B (rs406193 and rs2424932) polymorphisms were examined in 123 melanoma patients. DNMT1 and DNMT3B polymorphisms were shown to affect the clinical course and outcome of melanoma. The DNMT1 rs2228612 genotype was associated with poorer survival (P = 0.000), poorer recurrence-free survival, (P = 0.000), as well as greater risk of adverse outcome [hazard ratio (HR) = 6.620, 95 % CI: 2.214–19.791, P = 0.001]. The DNMT3B rs406193 polymorphism correlated significantly with the presence of tumor-infiltrating lymphocytes (P = 0.012) [96]. A prospective, longitudinal study (from 2011 to 2017, n = 351) showed a significant association between the DNMT1 rs2162560 polymorphism and chemotherapy-associated cognitive impairment of breast cancer patients [99]. In 1957 cases, the most frequent DNMT3a mutations (Arg882His and Arg882Cys) occurred at the C-terminal region outside of the MTase domain [30]. At the molecular level, DNMT3A Arg882His confers an 80 % reduction in the methyltransferase activity compared to wild-type [171]. Missense mutations in the DNMT3A gene are often observed in hematologic malignances, especially in AML, where the Arg882His mutation is prevalent [172]. A meta-analysis including 2184 cancer cases and 3420 controls concluded that the DNMT3A rs1550117 A > G polymorphism may be associated with cancer susceptibility, which was supported by a significant association between DNMT3A rs1550117 A > G polymorphism and increased risk of CRC (OR = 3.16; 95 % CI: 1.58–6.29; P = 0.001) [98]. Another meta-analysis (3959 cases and 5992 controls) investigated the effect of DNMT1 (rs16999593, rs2228611, rs8101866), DNMT3A (rs1550117, rs13420827) and DNMT3B (rs1569686, rs2424913) polymorphisms on susceptibility to gastric cancer. This meta-analysis demonstrated that rs16999593 (dominant model: OR 1.36, 95 %CI 1.15–1.60) and rs1550117 (dominant model: OR 1.20, 95 %CI 1.01–1.42) could contribute to gastric cancer risk and that rs1569686 (dominant model: OR 0.74, 95 %CI 0.61–0.90) might be a protective factor against gastric carcinogenesis [97].
10. Crystal structure of SAM-dependent MTases
SAM-dependent MTases are widely distributed among almost all organisms and often include conserved Rossmann fold, TIM barrel, and D × G × G×G motifs [173]. X-ray crystal structures of MTase provide insights into substrate recognition and reaction mechanisms. From structure-function comparisons of wild-type and variants, it is possible to infer how single amino acid changes produce phenotypic differences. The first reported structure of a SAM-dependent methyltransferase was the DNA C5-cytosine MTase reported by Hhal in 1993 [174]. The crystal structure of SAM-dependent methyltransferases (SAM-MTs) consists of a Rossmann-like fold (domain I) and a substrate-binding domain (domain II). The cofactor molecule (SAM) binds at the interface between adjacent domains, presumably near to the active site(s) of the enzyme [175]. There are five main classes (I-V) of SAM-dependent MTase categorized on the basis of their structural features. The large majority of MTases belong to the Class I or Rossmann-like fold family, catalyzing the majority of methylation reactions across all domains of life.
The crystal structure of human DNMT1, with all the structural domains (hDNMT1, residues 351–1600) in complex with SAH at 2.62 Å (Å) resolution is described in Table 5 and depicted in Fig. 5A. Fig. 5 depicts hDNMT1, composed of CXXC, tandem bromo-adjacent homology (BAH1/2), and methyltransferase domains bound to DNA-containing unmethylated CpG sites [176]. In 2018, a 2.65 Å crystal structure of the DNMT3A-DNMT3L-DNA complex was reported that showed Arg836 of the target recognition domain makes crucial contacts with the CpG dinucleotide, ensuring the DNMT3A enzymatic preference towards CpG sites [177] (Fig. 5B). DNMT3a and DNMT3b preferentially bind to nucleosomes. The structure suggests that nucleosomal DNA must be moved relatively to the nucleosome core for de novo methylation to occur. This movement allows the catalytic-like domain of the accessory protein DNMT3B to bind to the acidic patch of the nucleosome core, which orients binding of DNMT3A to the linker DNA [178]. From the structure of the human DNMT3B-3 L complex, human DNMT3b was shown to use two flexible loops to enclose DNA, specifically recognizing CpG sites in the DNA through its Asn779 and Lys777 residues [179].
Table 5.
Crystal structure information of SAM-dependent MTases extracted from the Protein Data Bank (PDB).1.
| SAM-dependent methyltransferase | PDB ID | Resolution (ångstöm /Å) | Amino acid sequence length | Organism(s) | Reference |
|---|---|---|---|---|---|
| COMT | 3BWM | 1.98 | 214 | Homo sapiens | [180] |
| COMT 108 Met mutant | 3BWY | 1.30 | 214 | Homo sapiens | [180] |
| ArsM (AS3MT) | 4FS8 | 1.60 | 383 | Cyanidioschyzon sp. 5508 | [181] |
| HNMT | 1JQD | 2.28 | 292 | Homo sapiens | [182] |
| INMT | 2A14 | 1.70 | 263 | Homo sapiens | [183] |
| NNMT | 2IIP | 2.05 | 283 | Homo sapiens | [184] |
| PNMT | 1HNN | 2.40 | 282 | Homo sapiens | [185] |
| TPMT | 2BZG | 1.58 | 232 | Homo sapiens | [186] |
| DNMT1 | 4WXX | 2.62 | 1256 | Homo sapiens | [187] |
| DNMT1-DNA | 3PTA | 3.60 | 956 | Homo sapiens | [176] |
| DNMT3A | 5YX2 | 2.65 | 285 | Homo sapiens | [177] |
| DNMT3B | 6KDL | 3.27 | 286 | Homo sapiens | [188] |
RCSB PDB: Homepage, https://www.rcsb.org/.
Fig. 5.
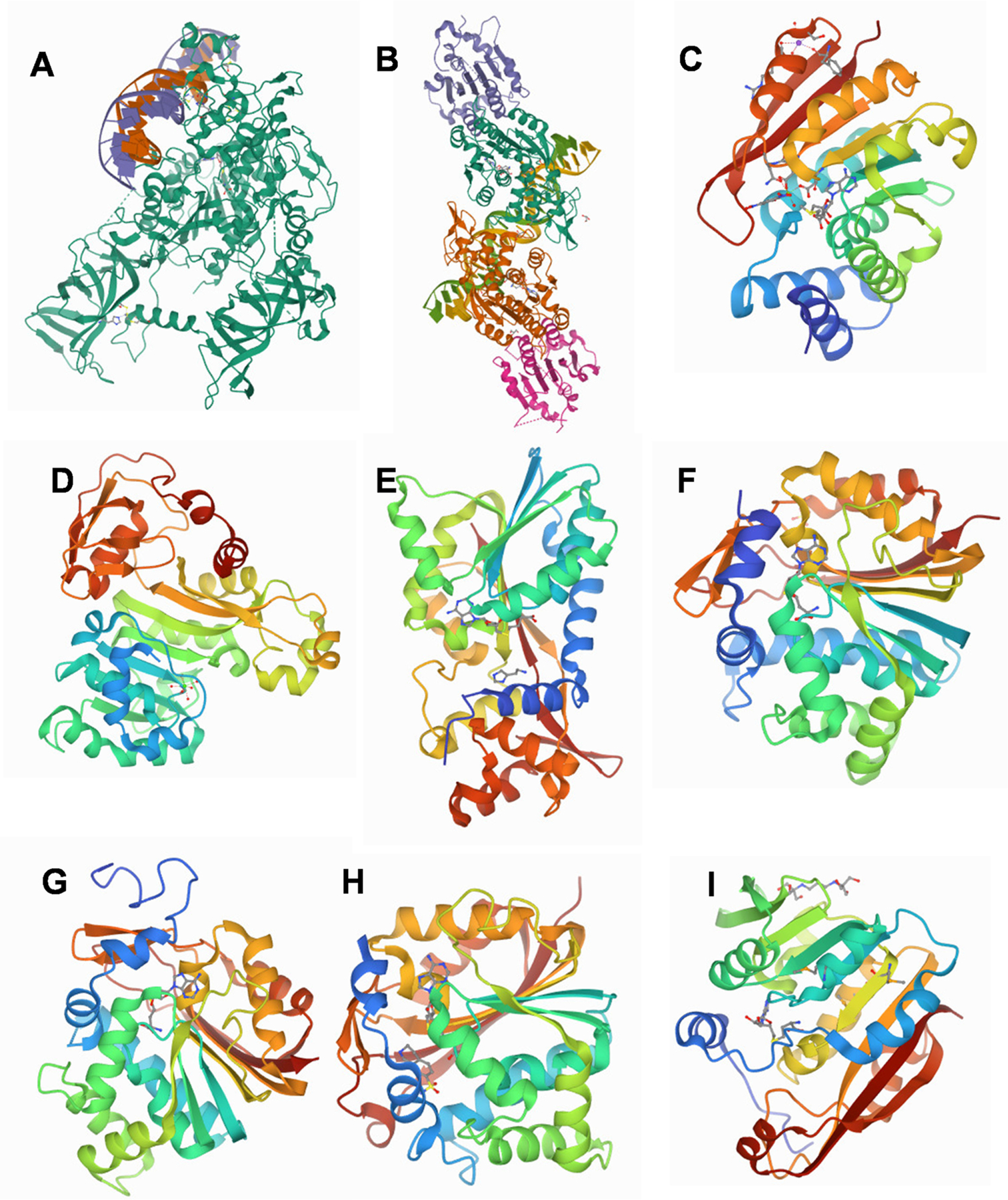
Crystal structures of SAM-dependent MTases. (A) human DNMT1(646–1600) in complex with DNA;(B)DNMT3A-DNMT3L in complex with DNA containing two CpG sites;(C) human COMT with bound SAM and DNC;(D) As3MT; (E) human HNMT (Thr105 Polymorphic Variant) complexed with SAH and histamine. DOI: 10.2210/pdb1JQD/pdb; (F) human INMT with SAH. DOI: 10.10.2210/pdb2A14/pdb; (G) human NNMT. DOI: 10.10.2210/pdb2IIP/pdb; (H) Crystal structure of human PNMT complexed with SK&F 29661 and AdoHcy; (I) TPMT. DOI: 10.10.2210/pdb2BZG/pdb.
The structure of human COMT is composed of a seven-stranded β-sheet core sandwiched between two sets of α-helices. The active site of COMT is close to the enzyme surface, which allows a variety of non-native catechol substrates to be methylated [50]. Residue 108 is located in a surface loop with SAM-binding residues at their distal end. Bounding SAM in the COMT protein might restrict the conformational changes caused by the polymorphism at residue 108. For COMT polymorphism rs4689 (Val108/158Met), the methionine variant is less stable than the valine variant, resulting in decreased enzyme activity and protein levels. The slight differences in the structures of 108 Val COMT and 108 Met COMT are attributable to differing interactions of residues Val108 and Met108 within the polymorphic site. Residues in the SAM-binding site were more exposed to the solvent in 108 Met than in the 108 Val COMT protein and the methionine side chain is packed more tightly within the polymorphic site than valine, consequently, interacts more closely with residues A22 (α2) and R78 (α4) than does valine, which may account for enzyme activity differences. [180].
The first X-ray crystallography structure of an As(III) S-adenosylmethionine methyltransferase, the CmArsM from the eukaryotic red alga Cyanidioschyzon merolae, was reported in 2012 [181]. A structural model of the human AS3MT structure was built upon the 1.78 Å ligand-free CmArsM structure [181] (Fig. 5D). From a comparison of residues 136–180 in the human AS3MT model with the rs35232887 Arg173Trp polymorphism, the Arg to Trp amino acid substitution is predicted to produce a conformational change due to a shift in the hydrogen bond between Arg173 and Glu170 (3.0 Å) to a longer hydrogen bond between Trp173 and Glu170 (4.5 Å) [181]. This conformation change can explain in part the lower catalytic activity of the Trp173 variant [59]. Trivalent arsenic(III) is methylated up to three times to form methylarsenite [MAs(III)], dimethylarsenite [DMAs(III)] and the volatile trimethylarsine [TMAs(III)] by ArsM (As3MT). Although the human As3MT crystal structure has not been reported, the catalytic mechanism of arsenic (III) SAM MTases in which a disulfide-bond (Cys44 and Cys72) cascade maintains the products in the trivalent state has been proposed based on structure of CmArsM [189] (Fig. 6A). The initial step in the As(III) SAM MTase catalytic cycle [arsenic (III) methylated to Mas(III)] was depicted by a conformational change in the N-terminal domain of CmArsM that moves a loop to allow formation of the 3-coordinate As(III) binding site. And the structure of CmArsM with bound As(III) and SAH [As(lIl)/SAH-bound CmArsM, PDB entry 6CX6] has As(III) bound coordinately in three places to Cys44, Cys174, and Cys224, and SAH in the SAM binding site [190] (Fig. 6B). The distance between the sulfur atom of SAH and bound As(III) is approximately 4.9 Å (Fig. 6C1). The distance from the S-methyl group of SAM to the As atom is 2.9 Å, when both are bound, indicating that the methyl group is poised for transfer from SAM to As [190] (Fig. 6C2).
Fig. 6.
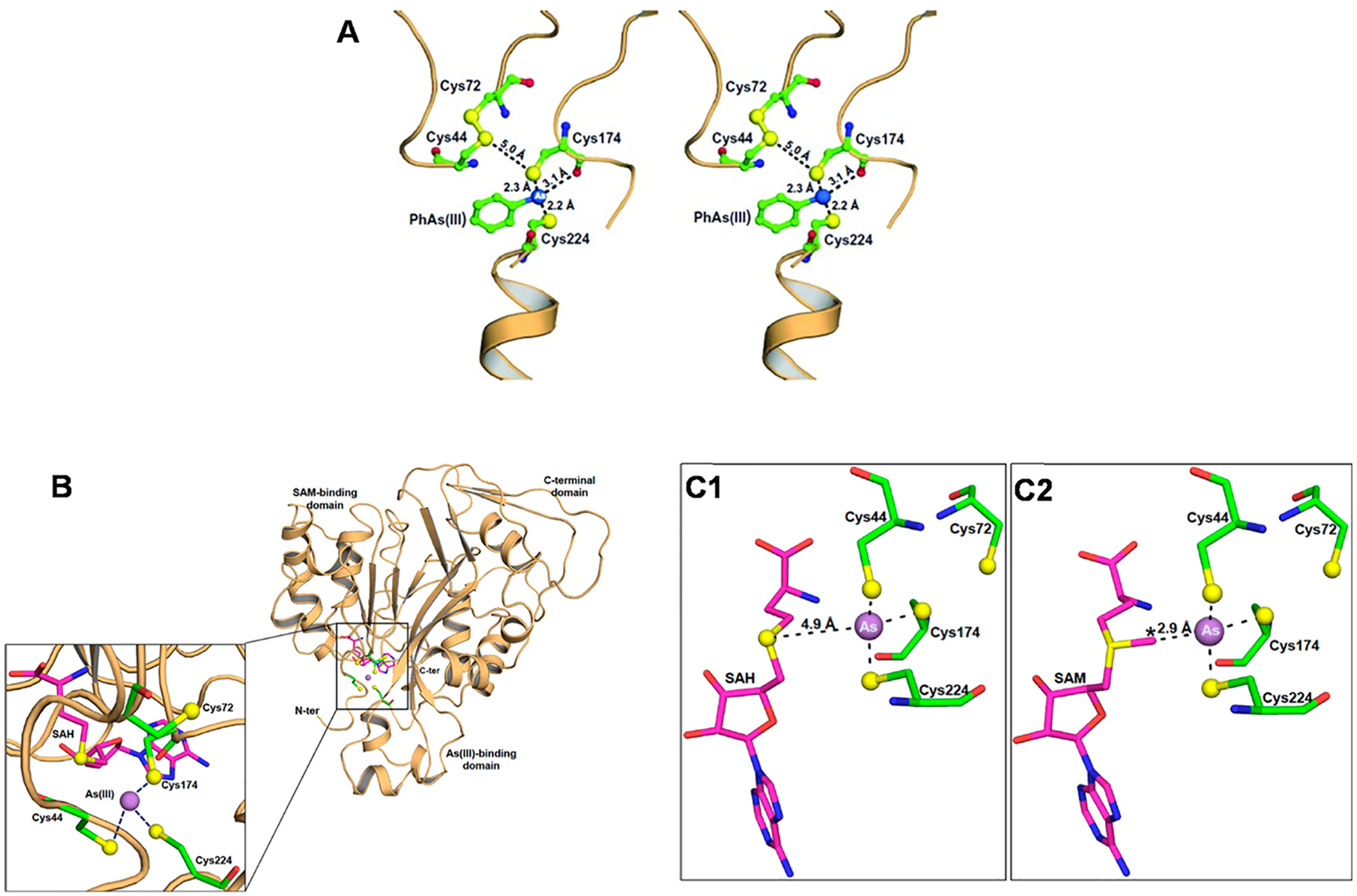
(A) A disulfide bond between Cys44 and Cys72 in the crystal structure of CmArsM with bound aromatic arsenical, phenylarsenite PhAs (III) (PDB entry 4kw7). The conserved cysteine residues are shown in ball-and-stick representation with atoms colored green (carbon), blue (nitrogen) or yellow (sulfur). The dark blue sphere is the As atom. The length of the disulfide bond is approximately 2.1 Å in the PhAs (III)-bound structure. (B) The overall structure of CmArsM consists of an N-terminal domain, an As(III) binding domain, and a C-terminal domain. The inset shows a close-up of the active site showing the four conserved cysteine residues represented by balls and sticks with atoms colored green (carbon), blue (nitrogen), red (oxygen), and yellow (sulfur). The purple sphere is the arsenic atom, and the SAH in the SAM binding site is represented by balls and sticks and with carbon colored magenta. As(III) is bound among conserved residues Cys44, Cys174, and Cys224. (C1) The As atom in the binding site consisting of Cys44, Cys174, and Cys224 is positioned near SAH in the SAM binding site. The distance between the sulfur atom of SAH and the As atom is 4.9 Å. (C2) Distances between the As atom and the sulfur atoms of SAM is 2.9 Å. The S-methyl group is poised for electron transfer from SAM to As(III).
Human HNMT is a 2-domain protein, with the MTase fold in the larger domain that contains a seven-stranded β-sheets flanked on each side by three α-helices [182]. The MTase domain is structurally most similar to COMT (Fig. 5C and E). In the AdoMet binding domain of HNMT, residue 105 is located on the surface. The surface location of residue 105 suggests that this polymorphism is unlikely to affect overall structural stability. However, Ile105 affects the stability of AdoMet binding, with lower Km values for AdoMet and histamine [104]. The HNMT common polymorphism, rs11558538 (Thr105IIe, 314C > T) exhibits high- (Thr) and low- (Ile) activity phenotypes, similar to COMT SNP rs4680 (Met158Val) [182]. In wild-type HNMT, there is a stabilizing hydrogen bond between Thr105 and main chain residue Leu101. In the Ile105 variant, this hydrogen bond is lost, resulting in an increase in the flexibility of the loop between αB and β3. This slightly destabilizes the carboxyl end of helix αB, lowering the affinity for SAM, consistent with the effect of COMT SNP rs4680 [182].
NNMT catalyzes the transfer of a methyl group from SAM onto the substrate (nicotinamide) to form 1-methyl-nicotinamide. NNMT possesses a class I SAM-dependent MTases core fold, comprising a central seven-stranded β-sheet, flanked on both sides by α-helices (Fig. 5G). In addition to the MTase core, there are two extra α-helices at the N-terminus with a β-hairpin (Y203-S212) that forms a cap over the active site [191]. Dr. Sven Ruf’s research group described their approach to identify potent small molecule inhibitors of NNMT. One kind of inhibitor (a series of imine compounds was tested) makes a hydrogen bond with the hydroxyl group of Tyr20, forming a van der Waals contact (d = 3.3 Å) with the sulfur of SAH. This is the position wherethe methyl group of the co-substrate SAM would be transferred by NNMT to the substrate nicotinamide [191]. Another inhibitor occupies both the nicotinamide and SAM pockets of the catalytic center [191]. There is a large body of crystallography research on NNMT interaction with inhibitors, aimed at identifying selective small molecule inhibitors for treating metabolic disorders [192].
PNMT uses the methyl donor AdoMet to catalyze the formation of adrenaline from noradrenaline. PNMT and COMT are both part of the catecholamine pathway and they both interact with adrenaline and noradrenaline (Fig. 2). TPMT is a single domain protein with a classic Class-I MTase fold. The crystal structure of human PNMT was solved at a resolution of 2.4 Å in complex with a cofactor product and a sub-micromolar inhibitor. The structure revealed a MTase fold with an active site protected from solvent by an extensive cover formed from several discrete structural motifs [185]. When compared with COMT, PNMT is a much more complicated structure (Fig. 5H). By comparing Fig. 5C and H, it can be seen that COMT is a relatively streamlined structure, with no inserts in the fold. However, there are four insertions in the MTase fold of PNMT, providing the motifs that enclose the active site and contribute a significant number of the ligand binding residues.
TPMT catalyzes thiopurine and thiopyrimidine S-methylation, an important metabolic pathway for thiopurine drugs. Considerable interindividual variation in thiopurine toxicity and therapeutic efficacy occurs, mainly due to genetic polymorphisms. TPMT is a single domain protein with a classic Class-I MTase fold (Fig. 5I). From the structure of TPMT mutant proteins containing variant alleles of TPMT*2, *3A, *3B, and *3C, it appears that forming intra-molecular stabilizing interactions (mainly van der Waals contacts) have the most influence on function [186]. Due to extremely low solubility of thiopurine drugs (6-MP, 6-TG, or 2-thiopurin) in water. No co-crystals of TPMT in complex with drug have been reported to date.
11. Summary and conclusions
MTases are classified based on their methyl-accepting substrates, which include drugs, xenobiotics, neurotransmitters, hormones, proteins, DNA and RNA. SAM-dependent MTases transfer a methyl group from SAM to a nucleophilic acceptor (e.g. O, As, N, S or C). In this review we systematically described eight SAM-MTases and their relationship with human health and disease. COMT is an O-methyl SAM-MTase that uses dopamine as substrate. The physiological role of COMT is to maintain appropriate levels of dopamine and norepinephrine at the front of the brain. Some symptoms in individuals with 22q11.2 deletion syndrome may be related to the deletion of COMT. AS3MT is an As(III)-SAM-MTase that uses the environmental contaminant arsenic as substrate. As(III) SAM-MTases are found in members of every kingdom from bacteria to humans. The most common AS3MT polymorphism, M287 T, is related to susceptibility to diabetes. INMT is an N-methyl SAM-MTase that catalyzes methylation of tryptamine to form the hallucinogen N,N-dimethyltryptamine (DMT). INMT gene variants are associated with Hirschsprung’s disease. Another N-methyl SAM-MTase is PNMT, which catalyzes the final step in catecholamine biosynthesis by N-methylation of norepinephrine to epinephrine. PNMT genetic polymorphisms G (−353)A and G(−148)A, in the 5′-flanking region of the PNMT gene are associated with increased risk for sporadic early onset AD, multiple sclerosis and essential hypertension. Another N-methyl MTase, HNMT, uses the neurotransmitter histamine as substrate. The HNMT polymorphism T105 l is associated with PD and schizophrenia. The N-methyl MTase, NNMT is the only enzyme known to utilize nicotinamide, which is used in synthesis of NADH and ATP as a methyl acceptor. NNMT is a potential tumor marker. One physiological function of the S-methyl MTase TPMT is metabolism of thiopurine drugs that are used for treatment of ALL, Crohn’s disease and rheumatoid arthritis. Variant alleles of TPMT are associated with low TPMT enzyme activity in humans. DNA methyltransferase plays a crucial role in the maintenance of genomic methylation patterns. Mutations in DNMT1, DNMT3A and DNMT3B are associated with cancers. These are some of the most well-characterized genetic polymorphisms of drug metabolism. The various physiological roles of the substrates and products of these eight SAM-MTases (COMT, AS3MT, INMT, DNMT, HNMT, NNMT, TPMT, DNMT) demonstrate their importance to human physiology. Polymorphisms in their genes are related to individual variations in metabolism, disease susceptibility and therapeutic effects of their substrates and products. Although we focused on eight representative SAM MTases, there are many others of considerable importance for human health. Crystal structures of SAM-MTs not only provide important information regarding the catalytic function of the enzymes, but also pave the way for developing more potent and selective inhibitors in the future.
Methyl conjugation is a critical reaction in the metabolism of many drugs (e.g., nicotinamide), thiopurine neurotransmitters (e.g., dopamine, tryptamine, histamine, norepinephrine), and xenobiotic compounds (e.g., arsenic). Individual variation in SAM-dependent MTases are responsible for individual differences in the metabolism of neurotransmitters, therapeutic effects and toxicity of xenobiotics. Most of the SNPs in the eight MTases described in this review lead to lower enzyme activity. Elucidation of the relationship between mutations and the risk of disease is essential for personalized medicine that can provide the correct drug at the appropriate dose for each individual based on the patient’s genetic background. Importantly, knowledge about these variants will contribute to a more precise understanding of correlations between genotypes and disease-susceptibility phenotypes or the risk of side effects from drugs.
Acknowledgements
This research was supported in part by the National Natural Science Foundation of China grant 32101361, Applied Basic Research Foundation of Yunnan Province202101AU070078, Yunnan Provincial Education Department grant 2021J0003 to JJ.L, in part by the National Natural Science Foundation of China grant 41967023 to J.C, and in part by US National Institutes of Health grants ES023779, GM136211 and GM055425 to B.P.R.
Footnotes
Declaration of Competing Interest
The authors state that they have no competing interests.
References
- [1].Huang E, et al. , Catechol-O-Methyltransferase Val158Met polymorphism and clinical response to antipsychotic treatment in schizophrenia and schizo-affective disorder patients: a meta-analysis, Int. J. Neuropsychopharmacol 19 (5) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Nikolac Perkovic M, et al. , Catechol-O-methyltransferase rs4680 and rs4818 haplotype association with treatment response to olanzapine in patients with schizophrenia, Sci. Rep 10 (1) (2020) 10049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Lin CH, et al. , Catechol-O-methyltransferase (COMT) genetic variants are associated with cognitive decline in patients with Parkinson’s disease, Parkinsonism Relat. Disord 50 (2018) 48–53. [DOI] [PubMed] [Google Scholar]
- [4].Lin CH, et al. , Depression and Catechol-O-methyltransferase (COMT) genetic variants are associated with pain in Parkinson’s disease, Sci. Rep 7 (1) (2017) 6306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Lee YH, Song GG, COMT Val158Met and PPARγ Pro12Ala polymorphisms and susceptibility to Alzheimer’s disease: a meta-analysis, Neurol. Sci 35 (5) (2014) 643–651. [DOI] [PubMed] [Google Scholar]
- [6].Fageera W, et al. , Association between COMT methylation and response to treatment in children with ADHD, J. Psychiatr. Res 135 (2021) 86–93. [DOI] [PubMed] [Google Scholar]
- [7].Huang YK, et al. , Arsenic methylation capability and hypertension risk in subjects living in arseniasis-hyperendemic areas in southwestern Taiwan, Toxicol. Appl. Pharmacol 218 (2) (2007) 135–142. [DOI] [PubMed] [Google Scholar]
- [8].Valenzuela OL, et al. , Association of AS3MT polymorphisms and the risk of premalignant arsenic skin lesions, Toxicol. Appl. Pharmacol 239 (2) (2009) 200–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Drobná Z, et al. , Environmental exposure to arsenic, AS3MT polymorphism and prevalence of diabetes in Mexico, J. Expo. Sci. Environ. Epidemiol 23 (2) (2013) 151–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Kim JY, et al. , Potential association of INMT nonsynonymous variant (His46Pro) with Hirschsprung’s disease, Neonatology 108 (3) (2015) 164–171. [DOI] [PubMed] [Google Scholar]
- [11].Huang C, et al. , Phenylethanolamine N-methyltransferase gene promoter haplotypes and risk of essential hypertension, Am. J. Hypertens 24 (11) (2011) 1222–1226. [DOI] [PubMed] [Google Scholar]
- [12].Mann MB, et al. , Phenylethanolamine N-methyltransferase (PNMT) gene and early-onset Alzheimer disease, Am. J. Med. Genet 105 (4) (2001) 312–316. [DOI] [PubMed] [Google Scholar]
- [13].Sadhu N, et al. , Phenylethanolamine N-methyltransferase gene polymorphisms associate with crisis pain in sickle cell disease patients, Pharmacogenomics 21 (4) (2020) 269–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Jiménez-Jiménez FJ, et al. , Thr105Ile (rs11558538) polymorphism in the histamine N-methyltransferase (HNMT) gene and risk for Parkinson disease: a PRISMA-compliant systematic review and meta-analysis, Medicine (Baltimore) 95 (27) (2016) e4147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Palada V, et al. , Histamine N-methyltransferase Thr105Ile polymorphism is associated with Parkinson’s disease, Neurobiol. Aging 33 (4) (2012) 836, e1–3. [DOI] [PubMed] [Google Scholar]
- [16].Yang X, et al. , Association of histamine N-methyltransferase Thr105Ile polymorphism with Parkinson’s disease and schizophrenia in Han Chinese: a case-control study, PLoS One 10 (3) (2015) e0119692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Agúndez JA, et al. , Nonsynonymous polymorphisms of histamine-metabolising enzymes in patients with Parkinson’s disease, Neuromolecular Med. 10 (1) (2008) 10–16. [DOI] [PubMed] [Google Scholar]
- [18].Szczepankiewicz A, et al. , Polymorphisms of two histamine-metabolizing enzymes genes and childhood allergic asthma: a case control study, Clin. Mol. Allergy 8 (2010) 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Yan L, et al. , Histamine N-methyltransferase pharmacogenetics: association of a common functional polymorphism with asthma, Pharmacogenetics 10 (3) (2000) 261–266. [DOI] [PubMed] [Google Scholar]
- [20].García-Martín E, et al. , Histamine-N-methyl transferase polymorphism and risk for migraine, Headache 48 (9) (2008) 1343–1348. [DOI] [PubMed] [Google Scholar]
- [21].Ramsden DB, et al. , Nicotinamide N-Methyltransferase: genomic connection to disease, Int. J. Tryptophan Res 13 (2020), p. 1178646920919770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Bromberg A, et al. , Nicotinamide-N-methyltransferase (NNMT) in schizophrenia: genetic association and decreased frontal cortex mRNA levels, Int. J. Neuropsychopharmacol 15 (6) (2012) 727–737. [DOI] [PubMed] [Google Scholar]
- [23].Zhou Q, Zhu XJ, Li JH, Association between nicotinamide N-methyltransferase gene polymorphisms and obesity in Chinese Han male college students, Biomed Res. Int 2017 (2017) 2984826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Li J-H, et al. , Metabolomics study on the association between nicotinamide N-methyltransferase gene polymorphisms and type 2 diabetes, Int. J. Diabetes Dev 38 (4) (2018) 409–416. [Google Scholar]
- [25].Lennard L, et al. , Thiopurine dose intensity and treatment outcome in childhood lymphoblastic leukaemia: the influence of thiopurine methyltransferase pharmacogenetics, Br. J. Haematol 169 (2) (2015) 228–240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Vukic M, Daxinger L, DNA methylation in disease: immunodeficiency, centromeric instability, facial anomalies syndrome, in: Blewitt M (Ed.), DNA Methylation, 2019, pp. 773–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Mehawej C, et al. , DNMT3B deficiency presenting as severe combined immune deficiency: a case report, Clin. Immunol 215 (2020) 108453. [DOI] [PubMed] [Google Scholar]
- [28].Zhang J, et al. , DNA methyltransferases in cancer: biology, paradox, aberrations, and targeted therapy, Cancers 12 (8) (2020) 2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Hegde M, Joshi MB, Comprehensive analysis of regulation of DNA methyltransferase isoforms in human breast tumors, J. Cancer Res. Clin. Oncol 147 (4) (2021) 937–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Hoang N-M, Rui L, DNA methyltransferases in hematological malignancies, J. Genet. Genom 47 (7) (2020) 361–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Chen J, et al. , Functional analysis of genetic variation in catechol-O-methyltransferase (COMT): effects on mRNA, protein, and enzyme activity in postmortem human brain, Am. J. Hum. Genet 75 (5) (2004) 807–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Eisenhofer G, Finberg JP, Different metabolism of norepinephrine and epinephrine by catechol-O-methyltransferase and monoamine oxidase in rats, J. Pharmacol. Exp. Ther 268 (3) (1994) 1242–1251. [PubMed] [Google Scholar]
- [33].Magarkar A, et al. , Membrane bound COMT isoform is an interfacial enzyme: general mechanism and new drug design paradigm, Chem. Commun 54 (28) (2018) 3440–3443. [DOI] [PubMed] [Google Scholar]
- [34].Honea R, et al. , Impact of interacting functional variants in COMT on regional gray matter volume in human brain, Neuroimage 45 (1) (2009) 44–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Kang Y, et al. , Genetic contribution of catechol-O-methyltransferase in dorsolateral prefrontal cortex functional changes in the first episode schizophrenia, Behav. Brain Res 364 (2019) 225–232. [DOI] [PubMed] [Google Scholar]
- [36].Williams HJ, Owen MJ, O’Donovan MC, Is COMT a susceptibility gene for schizophrenia? Schizophr. Bull 33 (3) (2007) 635–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Taylor S, Association between COMT Val158Met and psychiatric disorders: a comprehensive meta-analysis, Am. J. Med. Genet. B Neuropsychiatr. Genet 177 (2) (2018) 199–210. [DOI] [PubMed] [Google Scholar]
- [38].Wang MQ, et al. , Meta-analysis of the COMT Val158Met polymorphism in major depressive disorder: effect of ethnicity, J. Neuroimmune Pharmacol 11 (3) (2016) 434–445. [DOI] [PubMed] [Google Scholar]
- [39].Broadley KJ, The vascular effects of trace amines and amphetamines, Pharmacol. Ther 125 (3) (2010) 363–375. [DOI] [PubMed] [Google Scholar]
- [40].Lindemann L, Hoener MC, A renaissance in trace amines inspired by a novel GPCR family, Trends Pharmacol. Sci 26 (5) (2005) 274–281. [DOI] [PubMed] [Google Scholar]
- [41].Scambler PJ, The 22q11 deletion syndromes, Hum. Mol. Genet 9 (16) (2000) 2421–2426. [DOI] [PubMed] [Google Scholar]
- [42].Gothelf D, et al. , Biological effects of COMT haplotypes and psychosis risk in 22q11.2 deletion syndrome, Biol. Psychiatry 75 (5) (2014) 406–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Hatzimanolis A, et al. , Potential role of membrane-bound COMT gene polymorphisms in female depression vulnerability, J. Affect. Disord 148 (2–3) (2013) 316–322. [DOI] [PubMed] [Google Scholar]
- [44].Wright GEB, et al. , Association of MB-COMT polymorphisms with schizophrenia-susceptibility and symptom severity in an African cohort, Prog. Neuropsychopharmacol. Biol. Psychiatry 39 (1) (2012) 163–169. [DOI] [PubMed] [Google Scholar]
- [45].Gonzalez-Castro TB, et al. , The role of COMT gene Val108/158Met polymorphism in suicidal behavior: systematic review and updated meta-analysis, Neuropsychiatr. Dis. Treat 14 (2018) 2485–2496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Crum AJ, et al. , Correction: Catechol-O-Methyltransferase moderates effect of stress mindset on affect and cognition, PLoS One 14 (5) (2019) e0216305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hooten WM, et al. , Associations of catechol-O-methyltransferase (rs4680) single nucleotide polymorphisms with opioid use and dose among adults with chronic pain, Pain 160 (1) (2019) 263–268. [DOI] [PubMed] [Google Scholar]
- [48].Parkin GM, et al. , Catechol-O-methyltransferase (COMT) genotypes are associated with varying soluble, but not membrane-bound COMT protein in the human prefrontal cortex, J. Hum. Genet 63 (12) (2018) 1251–1258. [DOI] [PubMed] [Google Scholar]
- [49].Cheuk DK, Wong V, Meta-analysis of association between a catechol-O-methyltransferase gene polymorphism and attention deficit hyperactivity disorder, Behav. Genet 36 (5) (2006) 651–659. [DOI] [PubMed] [Google Scholar]
- [50].Kang P, et al. , Association of Val158Met polymorphism in COMT gene with attention-deficit hyperactive disorder: an updated meta-analysis, Medicine (Baltimore) 99 (48) (2020) e23400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Lanni C, et al. , Influence of COMT Val158Met polymorphism on Alzheimer’s disease and mild cognitive impairment in Italian patients, J. Alzheimers Dis 32 (4) (2012) 919–926. [DOI] [PubMed] [Google Scholar]
- [52].Martínez MF, et al. , The COMT Val158 Met polymorphism as an associated risk factor for Alzheimer disease and mild cognitive impairment in APOE 4 carriers, BMC Neurosci. 10 (2009) 125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Shibata N, et al. , Association between the catechol-O-methyltransferase polymorphism Val158Met and Alzheimer’s disease in a Japanese population, Int. J. Geriatr. Psychiatry 30 (9) (2015) 927–933. [DOI] [PubMed] [Google Scholar]
- [54].Ajees AA, Rosen BP, As(III) S-adenosylmethionine methyltransferases and other arsenic binding proteins, Geomicrobiol. J 32 (7) (2015) 570–576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Li J, Pawitwar SS, Rosen BP, The organoarsenical biocycle and the primordial antibiotic methylarsenite, Metallomics 8 (10) (2016) 1047–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Lin S, et al. , A novel S-adenosyl-L-methionine:arsenic(III) methyltransferase from rat liver cytosol, J. Biol. Chem 277 (13) (2002) 10795–10803. [DOI] [PubMed] [Google Scholar]
- [57].Wood TC, et al. , Human arsenic methyltransferase (AS3MT) pharmacogenetics: gene resequencing and functional genomics studies, J. Biol. Chem 281 (11) (2006) 7364–7373. [DOI] [PubMed] [Google Scholar]
- [58].Ding L, et al. , Methylation of arsenic by recombinant human wild-type arsenic (+ 3 oxidation state) methyltransferase and its methionine 287 threonine (M287T) polymorph: role of glutathione, Toxicol. Appl. Pharmacol 264 (1) (2012) 121–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Li J, et al. , Nonsynonymous polymorphisms in the human AS3MT arsenic methylation gene: implications for arsenic toxicity, Chem. Res. Toxicol 30 (7) (2017) 1481–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Antonelli R, et al. , AS3MT, GSTO, and PNP polymorphisms: impact on arsenic methylation and implications for disease susceptibility, Environ. Res 132 (2014) 156–167. [DOI] [PubMed] [Google Scholar]
- [61].Lin YC, et al. , Polymorphisms of arsenic (+3 oxidation state) methyltransferase and arsenic methylation capacity affect the risk of bladder cancer, Toxicol. Sci 164 (1) (2018) 328–338. [DOI] [PubMed] [Google Scholar]
- [62].Farhid F, et al. , Frequency of M287T/AS3MT single nucleotide polymorphism in an Iranian population, Int. J. Hematol. Stem Cell Res 11 (1) (2017) 19–23. [PMC free article] [PubMed] [Google Scholar]
- [63].Beebe-Dimmer JL, et al. , Genetic variation in glutathione S-transferase omega-1, arsenic methyltransferase and methylene-tetrahydrofolate reductase, arsenic exposure and bladder cancer: a case-control study, Environ. Health 11 (2012) 43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Hsieh RL, et al. , Relation of polymorphism of arsenic metabolism genes to arsenic methylation capacity and developmental delay in preschool children in Taiwan, Toxicol. Appl. Pharmacol 321 (2017) 37–47. [DOI] [PubMed] [Google Scholar]
- [65].Gong G, O’Bryant SE, Low-level arsenic exposure, AS3MT gene polymorphism and cardiovascular diseases in rural Texas counties, Environ. Res 113 (2012) 52–57. [DOI] [PubMed] [Google Scholar]
- [66].Stajnko A, et al. , Arsenic metabolites; selenium; and AS3MT, MTHFR, AQP4, AQP9, SELENOP, INMT, and MT2A polymorphisms in Croatian-Slovenian population from PHIME-CROME study, Environ. Res 170 (2019) 301–319. [DOI] [PubMed] [Google Scholar]
- [67].Axelrod J, Enzymatic formation of psychotomimetic metabolites from normally occurring compounds, Science 134 (3475) (1961) 343. [DOI] [PubMed] [Google Scholar]
- [68].Thompson MA, Weinshilboum RM, Rabbit lung indolethylamine N-methyltransferase. cDNA and gene cloning and characterization, J. Biol. Chem 273 (51) (1998) 34502–34510. [DOI] [PubMed] [Google Scholar]
- [69].Dean JG, et al. , Biosynthesis and extracellular concentrations of N,N-dimethyltryptamine (DMT) in mammalian brain, Sci. Rep 9 (1) (2019) 9333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Ansher SS, Jakoby WB, Amine N-methyltransferases from rabbit liver, J. Biol. Chem 261 (9) (1986) 3996–4001. [PubMed] [Google Scholar]
- [71].Shimazu S, Miklya I, Pharmacological studies with endogenous enhancer substances: beta-phenylethylamine, tryptamine, and their synthetic derivatives, Prog. Neuropsychopharmacol. Biol. Psychiatry 28 (3) (2004) 421–427. [DOI] [PubMed] [Google Scholar]
- [72].Barker SA, McIlhenny EH, Strassman R, A critical review of reports of endogenous psychedelic N, N-dimethyltryptamines in humans: 1955–2010, Drug Test. Anal 4 (7–8) (2012) 617–635. [DOI] [PubMed] [Google Scholar]
- [73].Chu UB, et al. , Noncompetitive inhibition of indolethylamine-N-methyltransferase by N,N-dimethyltryptamine and N,N-dimethylaminopropyltryptamine, Biochemistry 53 (18) (2014) 2956–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Fontanilla D, et al. , The hallucinogen N,N-dimethyltryptamine (DMT) is an endogenous sigma-1 receptor regulator, Science 323 (5916) (2009) 934–937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [75].Aghajanian GK, Marek GJ, Serotonin and hallucinogens, Neuropsychopharmacology 21 (Suppl. 2) (1999) 16S–23S. [DOI] [PubMed] [Google Scholar]
- [76].Dean JG, Indolethylamine-N-methyltransferase polymorphisms: genetic and biochemical approaches for study of endogenous N,N,-dimethyltryptamine, Front. Neurosci 12 (2018) 232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Barker SA, N, N-dimethyltryptamine (DMT), an endogenous hallucinogen: past, present, and future research to determine its role and function, Front. Neurosci 12 (536) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Kuehnelt D, et al. , Selenium metabolism to the trimethylselenonium ion (TMSe) varies markedly because of polymorphisms in the indolethylamine N-methyltransferase gene, Am. J. Clin. Nutr 102 (6) (2015) 1406–1415. [DOI] [PubMed] [Google Scholar]
- [79].Skröder H, et al. , Associations between methylated metabolites of arsenic and selenium in urine of pregnant bangladeshi women and interactions between the main genes involved, Environ. Health Perspect 126 (2) (2018) 027001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Stajnko A, et al. , Arsenic metabolites; selenium; and AS3MT, MTHFR, AQP4, AQP9, SELENOP, INMT, and MT2A polymorphisms in Croatian-Slovenian population from PHIME-CROME study, Environ. Res 170 (2019) 301–319. [DOI] [PubMed] [Google Scholar]
- [81].Amóros R, et al. , Selenium status during pregnancy: influential factors and effects on neuropsychological development among Spanish infants, Sci. Total Environ 610–611 (2018) 741–749. [DOI] [PubMed] [Google Scholar]
- [82].Drinkwater N, et al. , Molecular recognition of physiological substrate noradrenaline by the adrenaline-synthesizing enzyme PNMT and factors influencing its methyltransferase activity, Biochem. J 422 (3) (2009) 463–471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Baima J, et al. , Evidence for linkage between essential hypertension and a putative locus on human chromosome 17, Hypertension 34 (1) (1999) 4–7. [DOI] [PubMed] [Google Scholar]
- [84].Peltsch H, et al. , Cardiac phenylethanolamine N-methyltransferase: localization and regulation of gene expression in the spontaneously hypertensive rat, Can. J. Physiol. Pharmacol (2015) 1–10. [DOI] [PubMed] [Google Scholar]
- [85].Nguyen P, et al. , Prenatal glucocorticoid exposure programs adrenal PNMT expression and adult hypertension, J. Endocrinol 227 (2) (2015) 117–127. [DOI] [PubMed] [Google Scholar]
- [86].Burke WJ, et al. , Degenerative changes in epinephrine tonic vasomotor neurons in Alzheimer’s disease, Brain Res. 661 (1–2) (1994) 35–42. [DOI] [PubMed] [Google Scholar]
- [87].Burke WJ, et al. , Phenylethanolamine N-methyltransferase activity is decreased in Alzheimer’s disease brains, Ann. Neurol 22 (2) (1987) 278–280. [DOI] [PubMed] [Google Scholar]
- [88].Burke WJ, et al. , Evidence for decreased transport of PNMT protein in advanced Alzheimer’s disease, J. Am. Geriatr. Soc 38 (12) (1990) 1275–1282. [DOI] [PubMed] [Google Scholar]
- [89].Li J, et al. , Candidate gene analyses for acute pain and morphine analgesia after pediatric day surgery: African American versus European Caucasian ancestry and dose prediction limits, Pharmacogenomics J. 19 (6) (2019) 570–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Crum AJ, et al. , Catechol-O-Methyltransferase moderates effect of stress mindset on affect and cognition, PLoS One 13 (4) (2018) e0195883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].de Jonge R, et al. , Polymorphisms in folate-related genes and risk of pediatric acute lymphoblastic leukemia, Blood 113 (10) (2009) 2284–2289. [DOI] [PubMed] [Google Scholar]
- [92].Wang GX, et al. , Female specific association between NNMT gene and schizophrenia in a Han Chinese population, Int. J. Med. Sci 11 (12) (2014) 1234–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Sazci G, et al. , Association of Nicotinamide-N-Methyltransferase gene rs694539 variant with epilepsy, Mol. Neurobiol 53 (6) (2016) 4197–4200. [DOI] [PubMed] [Google Scholar]
- [94].Alam A, et al. , Phenylethanolamine N-methyltransferase gene polymorphisms and adverse outcomes in acute kidney injury, Nephron Clin. Pract 114 (4) (2010) c253–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Miranda-Duarte A, et al. , DNA methyltransferase genes polymorphisms are associated with primary knee osteoarthritis: a matched case-control study, Rheumatol. Int 40 (4) (2020) 573–581. [DOI] [PubMed] [Google Scholar]
- [96].Maric H, et al. , DNMT1 and DNMT3B genetic polymorphisms affect the clinical course and outcome of melanoma patients, Melanoma Res. 29 (6) (2019) 596–602. [DOI] [PubMed] [Google Scholar]
- [97].Li H, et al. , DNMT1, DNMT3A and DNMT3B polymorphisms associated with gastric Cancer risk: a systematic review and meta-analysis, EBioMedicine 13 (2016) 125–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Zhang W, et al. , Genetic polymorphism of DNA methyltransferase 3A rs1550117 a>G and risk of cancer: a meta-analysis, J. Invest. Surg 28 (6) (2015) 346–353. [DOI] [PubMed] [Google Scholar]
- [99].Chan A, et al. , An evaluation of DNA methyltransferase 1 (DNMT1) single nucleotide polymorphisms and chemotherapy-associated cognitive impairment: a prospective, longitudinal study, Sci. Rep 9 (1) (2019) 14570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Ji Y, et al. , Human phenylethanolamine N-methyltransferase pharmacogenomics: gene re-sequencing and functional genomics, J. Neurochem 95 (6) (2005) 1766–1776. [DOI] [PubMed] [Google Scholar]
- [101].Sio YY, et al. , Epistasis between phenylethanolamine N-methyltransferase and beta 2-adrenergic receptor influences extracellular epinephrine level and associates with the susceptibility to allergic asthma, Clin. Exp. Allergy 50 (3) (2020) 352–363. [DOI] [PubMed] [Google Scholar]
- [102].Schwartz JC, et al. , Histaminergic transmission in the mammalian brain, Physiol. Rev 71 (1) (1991) 1–51. [DOI] [PubMed] [Google Scholar]
- [103].Brown RE, Stevens DR, Haas HL, The physiology of brain histamine, Prog. Neurobiol 63 (6) (2001) 637–672. [DOI] [PubMed] [Google Scholar]
- [104].Preuss CV, et al. , Human histamine N-methyltransferase pharmacogenetics: common genetic polymorphisms that alter activity, Mol. Pharmacol 53 (4) (1998) 708–717. [DOI] [PubMed] [Google Scholar]
- [105].Lachman HM, et al. , Human catechol-O-methyltransferase pharmacogenetics: description of a functional polymorphism and its potential application to neuropsychiatric disorders, Pharmacogenetics 6 (3) (1996) 243–250. [DOI] [PubMed] [Google Scholar]
- [106].Kim SH, et al. , Histamine N-methyltransferase 939A>G polymorphism affects mRNA stability in patients with acetylsalicylic acid-intolerant chronic urticaria, Allergy 64 (2) (2009) 213–221. [DOI] [PubMed] [Google Scholar]
- [107].King CL, et al. , Mice with a targeted deletion of the IgE gene have increased worm burdens and reduced granulomatous inflammation following primary infection with Schistosoma mansoni, J. Immunol 158 (1) (1997) 294–300. [PubMed] [Google Scholar]
- [108].Haas HL, Sergeeva OA, Selbach O, Histamine in the nervous system, Physiol. Rev 88 (3) (2008) 1183–1241. [DOI] [PubMed] [Google Scholar]
- [109].Palada V, et al. , Histamine N-methyltransferase Thr105Ile polymorphism is associated with Parkinson’s disease, Neurobiol. Aging 33 (4) (2012) 836, e1–3. [DOI] [PubMed] [Google Scholar]
- [110].Garcia-Martin E, et al. , Polymorphisms of histamine-metabolizing enzymes and clinical manifestations of asthma and allergic rhinitis, Clin. Exp. Allergy 37 (8) (2007) 1175–1182. [DOI] [PubMed] [Google Scholar]
- [111].Kellermayer B, et al. , Association of myasthenia gravis with polymorphisms in the gene of histamine N-methyltransferase, Hum. Immunol 74 (12) (2013) 1701–1704. [DOI] [PubMed] [Google Scholar]
- [112].Niren NM, Pharmacologic doses of nicotinamide in the treatment of inflammatory skin conditions: a review, Cutis 77 (Suppl. 1) (2006) 11–16. [PubMed] [Google Scholar]
- [113].Zhao Y, Nicotinamide for skin-cancer chemoprevention, N. Engl. J. Med 374 (8) (2016) 789. [DOI] [PubMed] [Google Scholar]
- [114].Aksoy S, Szumlanski CL, Weinshilboum RM, Human liver nicotinamide N-methyltransferase. cDNA cloning, expression, and biochemical characterization, J. Biol. Chem 269 (20) (1994) 14835–14840. [PubMed] [Google Scholar]
- [115].Aksoy S, et al. , Human nicotinamide N-methyltransferase gene: molecular cloning, structural characterization and chromosomal localization, Genomics 29 (3) (1995) 555–561. [DOI] [PubMed] [Google Scholar]
- [116].Xu J, et al. , Enhanced expression of nicotinamide N-methyltransferase in human papillary thyroid carcinoma cells, J. Clin. Endocrinol. Metab 88 (10) (2003) 4990–4996. [DOI] [PubMed] [Google Scholar]
- [117].Kim J, et al. , Expression of nicotinamide N-methyltransferase in hepatocellular carcinoma is associated with poor prognosis, J. Exp. Clin. Cancer Res 28 (2009) 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Roessler M, et al. , Identification of nicotinamide N-methyltransferase as a novel serum tumor marker for colorectal cancer, Clin. Cancer Res 11 (18) (2005) 6550–6557. [DOI] [PubMed] [Google Scholar]
- [119].Sartini D, et al. , Nicotinamide N-methyltransferase upregulation inversely correlates with lymph node metastasis in oral squamous cell carcinoma, Mol Med 13 (7–8) (2007) 415–421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [120].Kraus D, et al. , Nicotinamide N-methyltransferase knockdown protects against diet-induced obesity, Nature 508 (7495) (2014) 258–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Kannt A, et al. , Association of nicotinamide-N-methyltransferase mRNA expression in human adipose tissue and the plasma concentration of its product, 1-methylnicotinamide, with insulin resistance, Diabetologia 58 (4) (2015) 799–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [122].Zhu XJ, et al. , Physiological study on association between nicotinamide N-Methyltransferase gene polymorphisms and hyperlipidemia, Biomed Res. Int 2016 (2016) 7521942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Yan L, Otterness DM, Weinshilboum RM, Human nicotinamide N-methyltransferase pharmacogenetics: gene sequence analysis and promoter characterization, Pharmacogenetics 9 (3) (1999) 307–316. [DOI] [PubMed] [Google Scholar]
- [124].Saito S, et al. , Identification of 197 genetic variations in six human methyltranferase genes in the Japanese population, J. Hum. Genet 46 (9) (2001) 529–537. [DOI] [PubMed] [Google Scholar]
- [125].Sazci A, et al. , Association of nicotinamide-N-methyltransferase (NNMT) gene rs694539 variant with bipolar disorder, Gene 532 (2) (2013) 272–275. [DOI] [PubMed] [Google Scholar]
- [126].Sazci A, et al. , Nicotinamide-N-Methyltransferase gene rs694539 variant and migraine risk, J. Headache Pain 17 (1) (2016) 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [127].Hu Q, et al. , Lower serum nicotinamide N-methyltransferase levels in patients with bipolar disorder during acute episodes compared to healthy controls: a cross-sectional study, BMC Psychiatry 20 (1) (2020) 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Van Loon JA, Weinshilboum RM, Thiopurine methyltransferase biochemical genetics: human lymphocyte activity, Biochem. Genet 20 (7–8) (1982) 637–658. [DOI] [PubMed] [Google Scholar]
- [129].Coulthard S, Hogarth L, The thiopurines: an update, Invest. New Drugs 23 (6) (2005) 523–532. [DOI] [PubMed] [Google Scholar]
- [130].Rudin S, Marable M, Huang RS, The promise of pharmacogenomics in reducing toxicity during acute lymphoblastic leukemia maintenance treatment, Genom. Proteom. Bioinform 15 (2) (2017) 82–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Marinaki AM, Arenas-Hernandez M, Reducing risk in thiopurine therapy, Xenobiotica 50 (1) (2020) 101–109. [DOI] [PubMed] [Google Scholar]
- [132].Azimi F, et al. , Assessment of Thiopurine-based drugs according to Thiopurine S-methyltransferase genotype in patients with Acute Lymphoblastic Leukemia, Iran. J. Ped. Hematol. Oncol 4 (1) (2014) 32–38. [PMC free article] [PubMed] [Google Scholar]
- [133].Katara P, Kuntal H, TPMT polymorphism: when shield becomes weakness, Interdiscip. Sci 8 (2) (2016) 150–155. [DOI] [PubMed] [Google Scholar]
- [134].Ha C, et al. , The identification of a novel thiopurine S-Methyltransferase allele, TPMT*45, in Korean patient with Crohn’s disease, Pers. Med 13 (2020) 665–671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].McLeod HL, et al. , Thiopurine methyltransferase activity in American white subjects and black subjects, Clin. Pharmacol. Ther 55 (1) (1994) 15–20. [DOI] [PubMed] [Google Scholar]
- [136].Tamm R, et al. , Polymorphic variation in TPMT is the principal determinant of TPMT phenotype: a meta-analysis of three genome-wide association studies, Clin. Pharmacol. Ther 101 (5) (2017) 684–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Krynetski EY, et al. , A single point mutation leading to loss of catalytic activity in human thiopurine S-methyltransferase, Proc. Natl. Acad. Sci. U. S. A 92 (4) (1995) 949–953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Szumlanski C, et al. , Thiopurine methyltransferase pharmacogenetics: human gene cloning and characterization of a common polymorphism, DNA Cell Biol. 15 (1) (1996) 17–30. [DOI] [PubMed] [Google Scholar]
- [139].Tai HL, et al. , Thiopurine S-methyltransferase deficiency: two nucleotide transitions define the most prevalent mutant allele associated with loss of catalytic activity in Caucasians, Am. J. Hum. Genet 58 (4) (1996) 694–702. [PMC free article] [PubMed] [Google Scholar]
- [140].Loennechen T, et al. , Isolation of a human thiopurine S-methyltransferase (TPMT) complementary DNA with a single nucleotide transition A719G (TPMT*3C) and its association with loss of TPMT protein and catalytic activity in humans, Clin. Pharmacol. Ther 64 (1) (1998) 46–51. [DOI] [PubMed] [Google Scholar]
- [141].Otterness DM, et al. , Human thiopurine methyltransferase pharmacogenetics. Kindred with a terminal exon splice junction mutation that results in loss of activity, J. Clin. Invest 101 (5) (1998) 1036–1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Spire-Vayron de la Moureyre C, et al. , Detection of known and new mutations in the thiopurine S-methyltransferase gene by single-strand conformation polymorphism analysis, Hum. Mutat 12 (3) (1998) 177–185. [DOI] [PubMed] [Google Scholar]
- [143].McLeod HL, et al. , Genetic polymorphism of thiopurine methyltransferase and its clinical relevance for childhood acute lymphoblastic leukemia, Leukemia 14 (4) (2000) 567–572. [DOI] [PubMed] [Google Scholar]
- [144].Krynetski E, Evans WE, Drug methylation in cancer therapy: lessons from the TPMT polymorphism, Oncogene 22 (47) (2003) 7403–7413. [DOI] [PubMed] [Google Scholar]
- [145].Otterness D, et al. , Human thiopurine methyltransferase pharmacogenetics: gene sequence polymorphisms, Clin. Pharmacol. Ther 62 (1) (1997) 60–73. [DOI] [PubMed] [Google Scholar]
- [146].Hon YY, et al. , Polymorphism of the thiopurine S-methyltransferase gene in African-Americans, Hum. Mol. Genet 8 (2) (1999) 371–376. [DOI] [PubMed] [Google Scholar]
- [147].Hamdan-Khalil R, et al. , In vitro characterization of four novel non-functional variants of the thiopurine S-methyltransferase, Biochem. Biophys. Res. Commun 309 (4) (2003) 1005–1010. [DOI] [PubMed] [Google Scholar]
- [148].Lindqvist M, et al. , Identification of two novel sequence variants affecting thiopurine methyltransferase enzyme activity, Pharmacogenetics 14 (4) (2004) 261–265. [DOI] [PubMed] [Google Scholar]
- [149].Spire-Vayron de la Moureyre C, et al. , Genotypic and phenotypic analysis of the polymorphic thiopurine S-methyltransferase gene (TPMT) in a European population, Br. J. Pharmacol 125 (4) (1998) 879–887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [150].Thomas A, How can we improve on the already impressive results in pediatric ALL? Hematology Am. Soc. Hematol. Educ. Program 2015 (1) (2015) 414–419. [DOI] [PubMed] [Google Scholar]
- [151].Lee SHR, Yang JJ, Pharmacogenomics in acute lymphoblastic leukemia, Best Pract. Res. Clin. Haematol 30 (3) (2017) 229–236. [DOI] [PubMed] [Google Scholar]
- [152].Wojtuszkiewicz A, et al. , Assessment of mercaptopurine (6MP) metabolites and 6MP metabolic key-enzymes in childhood acute lymphoblastic leukemia, Nucleosides Nucleotides Nucleic Acids 33 (4–6) (2014) 422–433. [DOI] [PubMed] [Google Scholar]
- [153].Azimi F, et al. , Assessment of Thiopurine-based drugs according to Thiopurine S-methyltransferase genotype in patients with Acute Lymphoblastic Leukemia, Iran. J. Ped. Hematol. Oncol 4 (1) (2014) 32–38. [PMC free article] [PubMed] [Google Scholar]
- [154].Holliday R, Pugh JE, DNA modification mechanisms and gene activity during development, Science 187 (4173) (1975) 226–232. [PubMed] [Google Scholar]
- [155].Goll MG, Bestor TH, Eukaryotic cytosine methyltransferases, Annu. Rev. Biochem 74 (2005) 481–514. [DOI] [PubMed] [Google Scholar]
- [156].Chang SC, et al. , Mechanisms of X-chromosome inactivation, Front Biosci 11 (2006) 852–866. [DOI] [PubMed] [Google Scholar]
- [157].Avner P, Heard E, X-chromosome inactivation: counting, choice and initiation, Nat. Rev. Genet 2 (1) (2001) 59–67. [DOI] [PubMed] [Google Scholar]
- [158].Smith ZD, et al. , DNA methylation dynamics of the human preimplantation embryo, Nature 511 (7511) (2014) 611–615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Petryk N, et al. , Staying true to yourself: mechanisms of DNA methylation maintenance in mammals, Nucleic Acids Res. 49 (6) (2021) 3020–3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [160].Mendizabal I, Yi SV, Whole-genome bisulfite sequencing maps from multiple human tissues reveal novel CpG islands associated with tissue-specific regulation, Hum. Mol. Genet 25 (1) (2016) 69–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [161].Ginder GD, Gnanapragasam MN, Mian OY, The role of the epigenetic signal, DNA methylation, in gene regulation during erythroid development, Curr. Top. Dev. Biol 82 (2008) 85–116. [DOI] [PubMed] [Google Scholar]
- [162].Jeltsch A, Molecular enzymology of mammalian DNA methyltransferases, Curr. Top. Microbiol. Immunol 301 (2006) 203–225. [DOI] [PubMed] [Google Scholar]
- [163].Bestor TH, The DNA methyltransferases of mammals, Hum. Mol. Genet 9 (16) (2000) 2395–2402. [DOI] [PubMed] [Google Scholar]
- [164].Newell-Price J, Clark AJ, King P, DNA methylation and silencing of gene expression, Trends Endocrinol. Metab 11 (4) (2000) 142–148. [DOI] [PubMed] [Google Scholar]
- [165].Okano M, et al. , DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development, Cell 99 (3) (1999) 247–257. [DOI] [PubMed] [Google Scholar]
- [166].Otani J, et al. , Structural basis for recognition of H3K4 methylation status by the DNA methyltransferase 3A ATRX-DNMT3-DNMT3L domain, EMBO Rep. 10 (11) (2009) 1235–1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [167].Gagliardi M, Strazzullo M, Matarazzo MR, DNMT3B functions: novel insights from human disease, Front. Cell Dev. Biol 6 (2018) 140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [168].Vukic M, Daxinger L, DNA methylation in disease: immunodeficiency, Centromeric instability, Facial anomalies syndrome, Essays Biochem. 63 (6) (2019) 773–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [169].Hansen RS, et al. , The DNMT3B DNA methyltransferase gene is mutated in the ICF immunodeficiency syndrome, Proc. Natl. Acad. Sci. U. S. A 96 (25) (1999) 14412–14417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [170].Gao LF, et al. , Comprehensive structure-function characterization of DNMT3B and DNMT3A reveals distinctive de novo DNA methylation mechanisms, Nat. Commun 11 (1) (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [171].Russler-Germain DA, et al. , The R882H DNMT3A mutation associated with AML dominantly inhibits wild-type DNMT3A by blocking its ability to form active tetramers, Cancer Cell 25 (4) (2014) 442–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [172].Khrabrova DA, Yakubovskaya MG, Gromova ES, AML-associated mutations in DNA methyltransferase DNMT3A, Biochemistry Mosc. 86 (3) (2021) 307–318. [DOI] [PubMed] [Google Scholar]
- [173].Liao L, et al. , Crystal structure of a S-adenosyl-L-methionine-dependent O-methyltransferase-like enzyme from Aspergillus flavus, Proteins 89 (2) (2021) 185–192. [DOI] [PubMed] [Google Scholar]
- [174].Cheng X, et al. , Crystal structure of the HhaI DNA methyltransferase complexed with S-adenosyl-L-methionine, Cell 74 (2) (1993) 299–307. [DOI] [PubMed] [Google Scholar]
- [175].Pampa KJ, et al. , Crystal structure of SAM-dependent methyltransferase from Pyrococcus horikoshii, Acta Crystallogr. F Struct. Biol. Commun 73 (Pt 12) (2017) 706–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [176].Song J, et al. , Structure of DNMT1-DNA complex reveals a role for autoinhibition in maintenance DNA methylation, Science 331 (6020) (2011) 1036–1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [177].Zhang ZM, et al. , Structural basis for DNMT3A-mediated de novo DNA methylation, Nature 554 (7692) (2018) 387–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [178].Xu TH, et al. , Structure of nucleosome-bound DNA methyltransferases DNMT3A and DNMT3B, Nature 586 (7827) (2020) 151–+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [179].Lin CC, et al. , Structural insights into CpG-specific DNA methylation by human DNA methyltransferase 3B, Nucleic Acids Res. 48 (7) (2020) 3949–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [180].Rutherford K, et al. , Crystal structures of human 108V and 108M catechol O-methyltransferase, J. Mol. Biol 380 (1) (2008) 120–130. [DOI] [PubMed] [Google Scholar]
- [181].Ajees AA, et al. , Structure of an As(III) S-adenosylmethionine methyltransferase: insights into the mechanism of arsenic biotransformation, Biochemistry 51 (27) (2012) 5476–5485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [182].Horton JR, et al. , Two polymorphic forms of human histamine methyltransferase: structural, thermal, and kinetic comparisons, Structure 9 (9) (2001) 837–849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [183].Wu H, Dong A, Zeng H, Loppnau P, Sundstrom M, Arrowsmith CH, Edwards AM, Bochkarev A, Plotnikov AN, Crystal Structure of Human Indolethylamine N-methyltransferase With SAH, Structural Genomics Consortium (SGC), 2005, 10.2210/pdb2A14/pdb). [DOI] [Google Scholar]
- [184].Bernstein G, Min J, Wu H, Tempel W, Zeng H, Loppnau P, Avvakumov GV, Wasney G, Weigelt J, Sundstrom M, Arrowsmith CH, Edwards AM, Bochkarev A, Plotnikov AN, The Crystal Structure of Human Nicotinamide N-methyltransferase in Complex With SAH, Structural Genomics Consortium (SGC), 2006, 10.2210/pdb2IIP/pdb). [DOI] [Google Scholar]
- [185].Martin JL, et al. , Getting the adrenaline going: crystal structure of the adrenaline-synthesizing enzyme PNMT, Structure 9 (10) (2001) 977–985. [DOI] [PubMed] [Google Scholar]
- [186].Wu H, et al. , Structural basis of allele variation of human thiopurine-S-methyltransferase, Proteins 67 (1) (2007) 198–208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [187].Zhang ZM, et al. , Crystal structure of human DNA methyltransferase 1, J. Mol. Biol 427 (15) (2015) 2520–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [188].Lin CC, et al. , Structural insights into CpG-specific DNA methylation by human DNA methyltransferase 3B, Nucleic Acids Res. 48 (7) (2020) 3949–3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [189].Marapakala K, et al. , A disulfide-bond cascade mechanism for arsenic(III) S-adenosylmethionine methyltransferase, Acta Crystallogr. D Biol. Crystallogr 71 (Pt 3) (2015) 505–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [190].Packianathan C, Kandavelu P, Rosen BP, The structure of an As(III) S-adenosylmethionine methyltransferase with 3-coordinately bound As(III) depicts the first step in catalysis, Biochemistry 57 (28) (2018) 4083–4092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [191].Kannt A, et al. , Novel inhibitors of Nicotinamide-N-Methyltransferase for the treatment of metabolic disorders, Molecules 26 (4) (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [192].Kannt A, et al. , A small molecule inhibitor of Nicotinamide N-methyltransferase for the treatment of metabolic disorders, Sci. Rep 8 (1) (2018) 3660. [DOI] [PMC free article] [PubMed] [Google Scholar]


