Abstract
The quest for novel broad spectrum bioactive compounds is needed continuously because of the rapid advent of pathogenic multi drug resistant organisms. Actinomycetes, isolated from unexplored habitats can be a solution of this problem. The motive of this research work was isolation of actinomycetes having potential antimicrobial activities from unexplored regions of Devbag and Tilmati beach. The isolated actinomycetes were screened against pathogenic microbes for antimicrobial activities through cross streak method. Enzyme production activity was checked for these actinomycetes for amylase, protease, cellulase and lipase enzymes. Further antimicrobial activity of ethyl acetate extract of the potent strain KS46 was performed. The strain KS46 was identified with 16S rRNA gene sequencing and secondary structure was analysed. Gas chromatography–Mass spectrometry (GC–MS) profiling was conducted to ascertain the presence of bioactive metabolites in the ethyl acetate extract. The collected samples were pre-treated and 70 actinomycetes were isolated. The Streptomyces sp. strain KS46 showed the best antimicrobial activity in primary screening. Ethyl acetate extract of the strain KS46 revealed antimicrobial activity against S. aureus, B. subtilis, B. cereus, E. faecalis, K. pneumoniae, E. coli, S. flexneri, C. albicans and C. glabrata. The 16S rRNA gene sequencing identified the strain KS46 as Streptomyces levis strain KS46. The GC–MS metabolite profiling of the ethyl acetate extract revealed the availability of 42 compounds including fatty acid esters, fatty acid anhydrides, alkanes, steroids, esters, alcohols, carboxylic ester, etc. having antibacterial, antifungal, antiproliferative, antioxidant activities. This study indicated that Devbag and Tilmati beaches being untapped habitats have enormous diversity of promising antimicrobial metabolite producing actinomycetes. Therefore, further exploration should be carried out to characterize the potential actinomycetes, which can be optimistic candidates for generation of novel antimicrobial drugs.
Keywords: Actinomycetes, Streptomyces levis, Antimicrobial, Enzymes, 16S rRNA gene sequencing, GC–MS, Devbag and Tilmati beaches
1. Introduction
Actinomycetes are spore producing, gram positive microorganisms, which has the features of both fungi and bacteria. The actinomycetes are largely distributed in soil, fresh water and marine environments. There were an impressive number of actinomycetes isolated worldwide from soil samples (Berdy, 2005). Soil is one of the major habitats of actinomycetes population, although Streptomycetes sp. remains as the most important component of population (Oskay et al., 2005). It was reported that the Streptomyces sp. are mainly found in river water and the species of the genus Micromonospora are dominant in river sediments (Rifaat, 2003). The fresh water actinomycetes include Micromonospora sp., Actinoplanes sp., Thermoactinomyces sp., Rhodococcus sp., etc. These species are common fresh water actinomycetes, where they decompose lignin, cellulose, chitin, etc. (Rajagopal et al., 2018). Marine ecosystems represent an extreme environmental condition of temperature, pressure and salinity and considered as a huge source for isolation of novel microorganisms including actinomycetes. The microorganisms survive in that environment must have unique physiological and structural characteristics to withstand the environmental conditions and have potentiality of transformation and mineralization of organic materials (Das et al., 2007, Usama et al., 2014). However, actinomycetes are also present in plants as endophytes, and other extreme habitats, such as volcanic caves, hydrothermal vents, desert, antarctica environment, etc. (Karuppiah and Mustaffa, 2013).
Actinomycetes belong to phylum Actinomycetales and constitute the biggest taxonomic unit within the bacterial kingdom. They are gram positive organisms, aerobic or facultatively anaerobic in nature and have more than 50% guanine and cytosine content in DNA (Dhakal et al., 2017). They are coccoid or rod shaped or produce branching mycelia. The mycelia are divided into aerial mycelium and substrate mycelium. The substrate mycelia branch profusely and get attached on surface of media in search of nutrition, whereas the aerial mycelia are borne on substrate mycelia and bear chains of spores or sporangia (Rajagopal et al., 2018). Around 70% of the earth’s crust is surrounded by seas and oceans, which is regarded as a less investigated ecosystem for microbial diversity. Marine actinomycetes are considered as economically priceless and treasure house of prokaryotic organisms due to ability for production of ample number of bioactive metabolites. It was documented that they are even responsible for production of around 70% of antibiotics available in treatment of pathogenic diseases (Berdy, 2005). There are wide ranges of promising secondary metabolites, which include antibiotics, antitumour compounds, antiparasitic compounds, antivirals, herbicides, pesticides, vitamins, enzymes, enzyme inhibitors, etc. (Rashad et al., 2015). The marine actinomycetes produce several classes of antibiotics, such as ansamycins, anthracyclines, macrolides, aminoglycosides, β-lactams, terpenes, tetracyclines, peptides, fatty acids, polyketides, alkaloids, sugars, etc. (Dina et al., 2020).
In today’s world the pathogenic microbes have increased their resistance against antibiotics due to the misuse and overuse of these in healthcare, agriculture and pharmaceutical industries. These organisms are causing serious threat to human lives. It was reported that around 700,000 people every year lose their lives worldwide for the reason of drug resistant microbial infections (Miethke et al., 2021). Therefore successful isolation of marine actinomycetes form unexplored regions has been proven as a strategy for discovery of potent bioactive molecules. The present study is focused on bio-prospection and metabolite profiling of actinomycetes from marine water and sediment samples.
2. Material and methods
2.1. Collection of marine samples
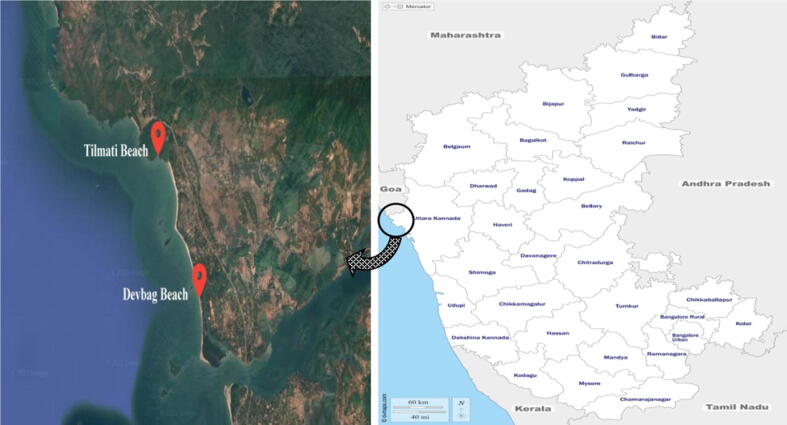
Thirty six marine sediment and water samples were collected from Devbag beach at latitude of 14°51′ 47.8548″N and longitude of 74°6′ 35.3556″E and Tilmati beach at latitude of 14°54′ 0.4824″N and longitude of 74°5′ 29.2668″E at the bank of Arabian Sea during August, 2018 (Fig. 1). The samples were obtained from 5 to 10 cm depth and placed in sterile containers. Upon return to the laboratory the samples were stored at 4 °C for future use (Gebreselema et al., 2013).
Fig. 1.
Geographical overview and sampling sites.
2.2. Pre-treatment of samples
The sediment and water samples were subjected to pre-treatment methods to facilitate the isolation of actinomycetes. The sediment samples were dried for 7 to 8 h in laboratory temperature (~26 °C). After drying the large particles were removed through sieving followed by a heat-treatment for 20min in a water bath at 70 °C to exclude the undesirable gram negative bacteria (Sengupta et al., 2015).
2.3. Isolation of marine actinomycetes
The pre-treated samples (1 g each) were thoroughly mixed with 10 ml of autoclaved physiological saline solution (0.9% NaCl) to make uniform suspension. Later, serial dilution was accomplished for all the suspensions up to 10−7 and 100 µL from each was evenly spread on surface of starch agar (SA), international streptomyces project (ISP)-2, ISP-6 and actinomycetes isolation agar (AIA) media. Each of the media was supplemented with Amphoteresin-B and Streptomycin (25 µg/ml) to constrict the growth undesirable fungi and bacteria. All the petri-dishes were incubated after inoculation at 30 °C for 7 to 14 days for the appearance of actinomycetes colonies. The number of colonies were counted, purified and stored for further use. The actinomycetes were differentiated from diverse microbial colonies by their appearance, such as leathery, tough, powdery, dry, fuzzy and pigmented colonies. The isolated colonies were preserved in SA medium at 4 °C for future studies (Njenga et al., 2017, Nayaka et al., 2020).
2.4. Bacteria and fungi pathogens used
Bacterial strains, including Staphylococcus aureus (MTCC6908), Bacillus subtilis (MTCC6633), Bacillus cereus (MTCC11778), Enterococcus faecalis (MTCC6845), Escherichia coli (MTCC40), Pseudomonas aeruginosa (MTCC9027), Klebsiella pneumoniae (MTCC9238), Shigella flexneri (MTCC1457) and fungal strains, such as Candida glabrata (MTCC3019) and Candida albicans (MTCC227) were used as pathogenic organisms. All the organisms were purchased from NCIM, Pune, India.
2.5. Screening for bioactive isolates
The primary screening for evaluation of the antimicrobial potential was executed by perpendicular streak or cross streak method (Gebreselema et al., 2013). The actinomycetes strains were grown as straight lines at the centre of Muller-Hinton agar medium plates and incubated for growth at 30 °C for 7 days. Later the pathogenic microorganisms, such as S. aureus, B. cereus, B. subtilis, E. faecalis, E. coli, P. aeruginosa, K. pneumoniae, C. albicans and C. glabrata were streaked at perpendicular to actinomycetes growth. After incubation at 37 °C for 24 h the antimicrobial potential of actinomycetes were recorded.
2.6. Screening for enzymatic potential
The actinomycetes strains were explored for their potential in synthesis of various extracellular enzymes, namely amylase, cellulase, protease and lipase.
2.6.1. Amylolytic activity
The actinomycetes strains were screened for their amylolytic activity on SA medium containing 1% soluble starch by starch hydrolysis method. The actinomycetes strains were streaked on medium and incubated for 7 days at 30 °C. The surface of the agar medium was flooded with 1% freshly prepared iodine solution post incubation period. The appearance of hydrolysis zones around actinomycetes colonies implied the production of amylase enzyme.
2.6.2. Proteolytic activity
The pure strains of actinomycetes were screened for proteolytic activity on skim milk agar medium. The isolates were inoculated on the medium and incubated at 30 °C for 7 days. The opaque hydrolysis zones surrounding the colonies indicated protease producing organisms.
2.6.3. Cellulolytic activity
The isolated actinomycetes strains were grown on carboxymethylcellulose (CMC) agar medium for 7 days at 30 °C. The plates were covered with grams iodine for 5 min followed by incubation period. The clear zones around the colonies indicated cellulase producing organisms.
2.6.4. Lypolytic activity
The actinomycetes were inoculated on peptone tween agar medium containing 1% Tween 80 (V/V), an oleic acid ester. The media was autoclaved and cooled to 40–50 °C and then the previously sterilized tween 80 was added aseptically. After incubation period the plates were observed for lipase activity. The presence of clear zones around actinomycetes colonies considered as indication of enzyme activity.
2.7. Extraction of secondary metabolites and antimicrobial activity
The actinomycetes strain showing the best inhibition against growth of pathogens in primary screening was selected for extraction of secondary metabolites. The potential Streptomyces sp. strain KS46 was subjected to submerged fermentation in 500 ml of starch casein broth at 30 °C for 7 days. This was mixed with 1:1 (v/v) ethyl acetate and extracted with the help of a separating funnel. Ten mg of ethyl acetate extract was dissolved with 1 ml of di-methyl sulphoxide and used as working solution. Later, the antimicrobial activity of 100 µL of ethyl acetate extract was carried out against pathogens on nutrient agar medium at pH 7.4 through agar well diffusion method (Kadriye et al., 2013).
2.8. Morphological characterization
The colour of substrate and aerial mycelia, pigmentation and colony shape of Streptomyces sp. strain KS46 were recorded. The scanning electron microscope (SEM) analysis of the strain KS46 was done to observe spore chain and spore surface morphology with the help of a SEM instrument (JSM-IT500 InTouchScope™, Scanning Electron Microscope, JEOL, Japan) (Ajit et al., 2015).
2.9. Molecular characterization
2.9.1. Isolation of genomic DNA
The taxonomical identification of the Streptomyces sp. strain KS46 was carried out by 16S rRNA gene sequencing. The genomic DNA of the strain was extracted with the help of HipurA Streptomyces DNA purification kit (MB527) according to manufacturer’s instructions (Nayaka et al., 2020).
2.9.2. Gene sequencing
The 16S rRNA gene sequencing was carried out using a thermal cycler (Applied Biosystems 2720, Thermal Cycler, ALT, US) with the aid of 27F (forward primer) and 1492R (reverse primer). Later, the PCR amplicons were confirmed by 1% agarose gel electrophoresis with a reference DNA ladder. The 16S rRNA gene was purified and sequenced using a DNA analyser and BLAST analysis was performed at NCBI website. Finally, a phylogenetic tree was composed based on neighbour joining method using MEGA7 software (Kumar et al., 2016). The secondary structure of 16S rRNA was anticipated using RNA structure software, Version 6.3 (Mathews Lab, University of Rochester Medical Centre).
2.10. Characterizations of ethyl acetate extract
2.10.1. Ultraviolet (UV) spectroscopy
The UV absorption spectrum of ethyl acetate extract was scanned using an UV–Vis. spectrophotometer (UV-9600A, Metash Instruments Co. Ltd., Shanghai, China) in the range of 200 to 700 nm. Ethyl acetate was used as reference solvent.
2.10.2. Fourier transform infrared (FTIR) spectroscopy
The FTIR spectra of ethyl acetate extract was analysed to identify the functional groups in the range of 400 cm−1 to 4000 cm−1 using a FTIR spectrophotometer (Nicolet 6700, Thermo Fisher Scientific, Waltham, Massachusetts, USA) (Noura et al., 2017).
2.10.3. GC–MS analysis
The GC–MS exploration of Streptomyces sp. strain KS46 ethyl acetate extract was performed with the help of gas chromatograph and mass spectrometer (GC-2010 Plus, Shimadzu Europa GmbH, Germany) operating in electron ionization mode. Two µl ethyl acetate extract was injected into an EC-5 column. Helium was used an inert carrier gas and set at a constant flow rate of 2 ml/min. The temperature of the oven was increased up to 450 °C at a rate of 20 °C per minute. Electron ionization mode was utilized for detection of mass spectra (Noura et al., 2017).
3. Results
Marine actinomycetes have emerged as a potential source of novel compounds including antibiotics, enzymes and other industrially important secondary metabolites. But the studies on marine actinomycetes in Indian peninsula are scanty and needs to be explored. Marine sediments and water samples were collected from different locations of Devbagh and Tilmati beaches. Thirty six samples were collected and seventy actinomycetes strains were isolated. Among the media used; the SA medium exhibited the highest number of actinomycetes expressed in colony-forming units. However, the other media such as ISP-2, ISP-6 and AIA revealed less number of actinomycetes. The isolated actinomycetes were shown in Fig. 2.
Fig. 2.
Isolated marine actinomycetes strains.
3.1. Primary screening
Out of 70 marine actinomycetes strains isolated from samples of Devbag and Tilmati beaches, only 7 expressed good antagonistic activity against almost all pathogenic bacteria and fungi (Table 1). However, all the strains revealed antimicrobial activity against at least one tested pathogen. The isolate strain KS46 revealed strongest activity among all.
Table 1.
Antimicrobial activity of actinomycetes strains against pathogenic bacteria and yeasts.
| Isolate code |
Gram positive |
Gram negative |
Yeast |
||||||
|---|---|---|---|---|---|---|---|---|---|
| Sa | Bc | Bs | Ef | Ec | Pa | Kp | Ca | Cg | |
| KWS-1 | + | + | ++ | − | + | ++ | − | − | − |
| KSS-2 | ++ | − | + | − | − | − | + | − | − |
| KWS-3 | − | − | + | ++ | − | + | + | − | + |
| KWS-4 | + | + | − | − | + | + | − | + | − |
| KS-5 | ++ | ++ | + | − | +++ | ++ | − | + | + |
| KSS-6 | − | − | − | − | − | − | − | − | − |
| KSS-7 | + | + | + | + | + | + | − | + | + |
| KSS-8 | + | − | − | + | + | +++ | − | − | − |
| KSS-9 | + | − | + | + | − | − | − | + | + |
| KSS-10 | + | + | − | − | − | − | − | + | + |
| KWS-11 | − | − | − | − | ++ | − | − | + | + |
| KSS-12 | + | − | − | + | − | − | + | − | − |
| KSS-13 | − | − | − | − | + | − | − | − | − |
| KWS-14 | + | + | + | ++ | − | + | − | + | ++ |
| KWS-15 | + | − | − | + | − | + | + | + | − |
| KSS-16 | − | − | + | + | − | − | − | − | + |
| KS-17 | +++ | ++ | − | ++ | − | − | + | ++ | + |
| KS-18 | + | − | − | + | − | − | ++ | − | − |
| KSS-19 | − | − | + | − | − | − | − | − | − |
| KS-20 | ++ | +++ | − | ++ | + | − | ++ | − | ++ |
| KWS-21 | − | + | ++ | − | − | − | − | + | + |
| KSS-22 | − | + | − | − | − | − | + | − | − |
| KSS-23 | − | − | − | ++ | − | + | + | − | − |
| KWS-24 | + | + | − | − | + | ++ | − | ++ | − |
| KWS-25 | + | − | + | − | − | + | − | − | + |
| KSS-26 | − | − | + | − | − | − | − | − | − |
| KSS-27 | + | − | − | + | − | − | − | + | − |
| KSS-28 | + | +++ | − | + | − | − | − | + | − |
| KSS-29 | − | + | − | − | ++ | − | + | − | + |
| KSS-30 | + | ++ | − | − | − | +++ | − | − | ++ |
| KSS-31 | − | − | + | − | − | + | − | − | − |
| KSS-32 | − | + | + | − | + | − | − | + | + |
| KWS-33 | + | − | + | + | − | − | − | + | − |
| KSS-34 | ++ | − | − | − | + | + | + | ++ | − |
| KSS-35 | − | − | + | + | − | ++ | − | − | − |
| KSS-36 | + | + | + | + | + | − | − | + | − |
| KWS-37 | + | ++ | − | − | − | − | − | + | + |
| KSS-38 | + | + | − | − | − | + | − | − | − |
| KSS-39 | ++ | − | − | − | − | − | + | + | + |
| KSS-40 | − | − | + | − | + | − | − | − | − |
| KWS-41 | − | ++ | − | − | + | − | + | + | + |
| KWS-42 | + | − | − | − | − | − | − | − | + |
| KSS-43 | − | − | − | + | + | − | − | − | − |
| KSS-44 | + | − | + | − | + | − | − | + | + |
| KSS-45 | + | − | − | + | − | − | + | + | − |
| KS-46 | ++ | ++ | − | +++ | − | ++ | ++ | +++ | ++ |
| KWS-47 | ++ | − | + | − | − | + | − | − | − |
| KSS-48 | + | − | − | − | − | − | − | − | − |
| KWS-49 | − | + | + | − | − | − | + | + | + |
| KSS-50 | − | − | − | − | ++ | − | − | − | − |
| KSS-51 | + | − | − | + | + | + | + | − | + |
| KSS-52 | − | − | + | − | − | − | + | − | − |
| KSS-53 | + | − | + | + | − | − | − | − | − |
| KWS-54 | + | + | − | − | − | − | − | + | − |
| KSS-55 | − | − | − | + | + | − | − | − | − |
| KSS-56 | + | − | ++ | − | − | + | − | − | + |
| KWS-57 | − | + | − | − | + | − | − | − | − |
| KS-58 | ++ | + | ++ | − | ++ | ++ | − | ++ | + |
| KSS-59 | − | − | − | − | + | − | − | − | − |
| KWS-60 | − | − | + | + | − | − | − | − | + |
| KWS-61 | + | ++ | + | + | − | + | + | − | − |
| KSS-62 | + | + | − | − | ++ | − | − | − | + |
| KSS-63 | − | − | + | + | − | − | − | + | + |
| KSS-64 | + | + | + | − | + | − | + | − | − |
| KSS-65 | − | + | − | − | + | − | − | − | + |
| KSS-66 | + | − | − | − | − | − | − | + | + |
| KS-67 | − | ++ | ++ | + | + | ++ | + | − | +++ |
| KWS-68 | + | − | − | − | − | − | − | − | + |
| KSS-69 | − | − | + | + | − | − | + | + | − |
| KWS-70 | + | + | − | − | − | − | − | − | − |
Staphylococcus aureus (Sa), Bacillus cereus (Bc), Bacillus subtilis (Bs), Enterococcus faecalis (Ef), Escherichia coli (Ec), Pseudomonas aeruginosa (Pa), Klebsiella pneumoniae (Kp), Candida albicans (Ca) and Candida glabrata (Cg), +++ strong activity, ++ moderate activity, + weak activity. − No activity.
3.2. Screening for enzymatic potential
Actinomycetes have been being commercially explored since decades for the production of enzymes having importance in pharmaceutical industries. In this work, all the isolated strains of actinomycetes were subjected to enzymatic screening for amylase, protease, cellulase and lipase (Table 2). The amylase producing strains showed a clear zone against dark blue plates upon covering with iodine solution. 61.42% of the total actinomycetes strains showed positive result for amylase production. Production of protease enzyme was exhibited by 42.85% of isolated actinomycetes strains. The clear zones produced by the organisms on milk agar medium indicated the production of protease enzyme. The cellulase enzyme was produced by 35.71% of actinomycetes strains on CMC agar medium and evidenced by clear hydrolysis zones around colonies. Lipase is one of the important enzymes and was produced by 41.42% of actinomycetes strains.
Table 2.
Enzymatic screening of marine actinomycetes strains.
| Isolate code | Amylase | Protease | Cellulase | Lipase | Isolate code | Amylase | Protease | Cellulase | Lipase |
|---|---|---|---|---|---|---|---|---|---|
| KWS-1 | ++ | − | − | − | KSS-36 | − | ++ | ++ | + |
| KSS-2 | + | − | + | + | KWS-37 | − | − | + | − |
| KWS-3 | − | + | − | + | KSS-38 | ++ | − | − | − |
| KWS-4 | − | − | − | ++ | KSS-39 | + | + | − | − |
| KS-5 | ++ | − | +++ | ++ | KSS-40 | − | − | − | − |
| KSS-6 | + | − | − | − | KWS-41 | + | − | + | − |
| KSS-7 | − | + | − | − | KWS-42 | ++ | − | − | − |
| KSS-8 | ++ | − | − | + | KSS-43 | − | + | − | + |
| KSS-9 | − | − | − | − | KSS-44 | +++ | + | +++ | + |
| KSS-10 | − | − | ++ | − | KSS-45 | − | + | − | − |
| KWS-11 | + | − | − | − | KS-46 | +++ | ++ | ++ | +++ |
| KSS-12 | ++ | − | +++ | − | KWS-47 | − | − | + | − |
| KSS-13 | + | − | − | + | KSS-48 | + | + | − | − |
| KWS-14 | − | + | − | − | KWS-49 | + | ++ | − | − |
| KWS-15 | + | − | − | − | KSS-50 | + | − | + | + |
| KSS-16 | + | − | + | − | KSS-51 | ++ | − | − | − |
| KS-17 | ++ | +++ | ++ | + | KSS-52 | + | − | − | − |
| KS-18 | + | + | − | − | KS-53 | − | − | − | − |
| KSS-19 | + | − | − | − | KWS-54 | − | − | − | − |
| KS-20 | + | +++ | ++ | ++ | KSS-55 | + | ++ | − | + |
| KWS-21 | − | ++ | − | − | KSS-56 | + | ++ | − | + |
| KSS-22 | + | − | + | − | KWS-57 | + | + | − | − |
| KSS-23 | − | − | − | + | KS-58 | +++ | ++ | + | ++ |
| KWS-24 | − | − | − | − | KSS-59 | − | + | + | − |
| KWS-25 | ++ | +++ | − | − | KWS-60 | + | + | − | − |
| KSS-26 | + | − | − | + | KWS-61 | + | − | − | + |
| KSS-27 | + | − | − | − | KSS-62 | − | ++ | − | − |
| KSS-28 | − | + | − | ++ | KSS-63 | + | ++ | − | + |
| KSS-29 | + | − | − | + | KSS-64 | ++ | − | + | − |
| KSS-30 | + | − | + | + | KSS-65 | +++ | ++ | − | − |
| KSS-31 | − | + | − | ++ | KSS-66 | − | +++ | + | − |
| KSS-32 | − | − | − | + | KS-67 | ++ | + | + | ++ |
| KWS-33 | ++ | +++ | − | + | KWS-68 | − | + | + | − |
| KSS-34 | + | − | + | + | KSS-69 | + | ++ | − | − |
| KSS-35 | − | − | − | − | KWS-70 | − | − | ++ | − |
+++ strong activity, ++ moderate activity, + weak activity. - No activity
3.3. Morphological characterization
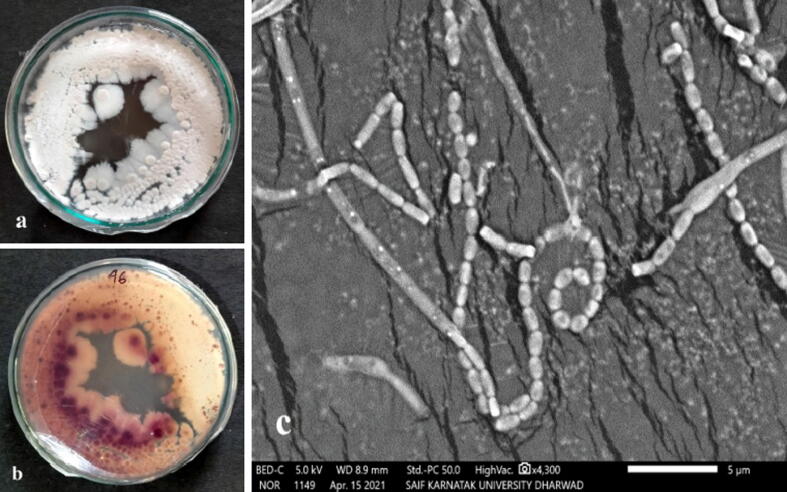
The aerial mycelium of the Streptomyces sp. strain KS46 was whitish-pink in colour (Fig. 3a) and the vegetative mycelium was pink to brown in colour (Fig. 3b). The strain produced a diffusible pigment in medium, which was brown in colour. The colonies of the strain KS46 were flat having filiform margins. The SEM analysis of Streptomyces sp. strain KS46 showed spiral spore chain having smooth spores as shown in Fig. 3c. At the maturity of the strain long spore chains were produced from aerial mycelia. The spore chains were spiral type having a closed, compact spiral structure and the spores were smooth walled and ovoid in shape.
Fig. 3.
(a) Aerial mycelium, (b) Substrate mycelium and (c) SEM micrograph of the Streptomyces sp. strain KS46.
3.4. Molecular characterization
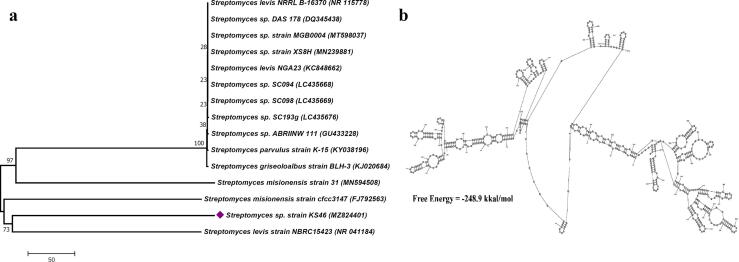
The 16S rRNA gene sequence of the Streptomyces sp. strain KS46 was 601 base pair in length and deposited in NCBI data base with a specific accession number MZ824401. Further, BLAST analysis of this sequence was done, which revealed 97.97% sequence similarity with Streptomyces levis strain NBRC 15423 (NR041184). The phylogenetic tree was constructed which revealed the evolutionary relationship of the strain KS46 and constituted a single clad with Streptomyces levis NBRC 15423 (NR041184) (Fig. 4a). Secondary structure of 16S rRNA gene sequence was predicted in Fig. 4b. The free energy of the structure was −248.9 kkal/mol.
Fig. 4.
(a) Phylogenetic tree, (b) Secondary structure of 16S rRNA gene sequence.
3.5. Antimicrobial activity
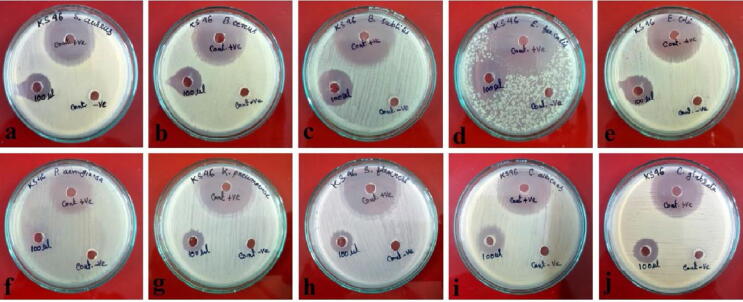
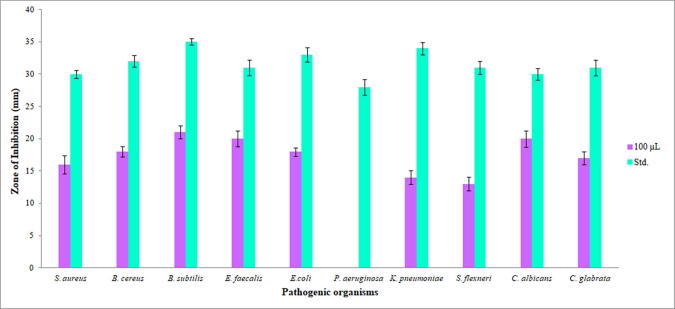
The evaporation of Streptomyces sp. strain KS46 ethyl acetate extract yielded 10 mg of reddish brown coloured semi-solid residue. The antagonistic activity of ethyl acetate extract was tested against four gram positive, three gram negative and two yeasts (Fig. 5, Fig. 6). However, the Streptomyces sp. strain KS46 showed good inhibitory activity against pathogenic organisms. The antimicrobial activity was assayed with 100 µL of ethyl acetate extract through agar well diffusion method. The Streptomyces sp. strain KS46 showed minimum activity against S. flexneri (13 ± 1.25 mm), K. pneumoniae (14 ± 1 mm), S. aureus (16 ± 1.4 mm), and C. glabrata (17 ± 1.04 mm) followed by B. cereus (18 ± 0.76 mm) and E. coli (18 ± 0.65 mm). The maximum inhibition zone was recorded against, E. faecalis (20 ± 1.20 mm), C. albicans (20 ± 1.32 mm) and B. subtilis (21 ± 1 mm). The pathogen P. aeruginosa was found resistant to Streptomyces sp. strain KS46 ethyl acetate extract.
Fig. 5.
Antimicrobial activity of ethyl acetate extract against pathogens. a. S. aureus, b. B. cereus, c. B. subtilis, d. E. faecalis, e. E. coli, f. P. aeruginosa, g. K. pneumoniae, h. S. flexneri, i. C. albicans, j. C. glabrata.
Fig. 6.
Bar diagram of Streptomyces sp. stain KS46 indicating zone of inhibition.
3.6. Characterizations of ethyl acetate extract
3.6.1. UV spectroscopy
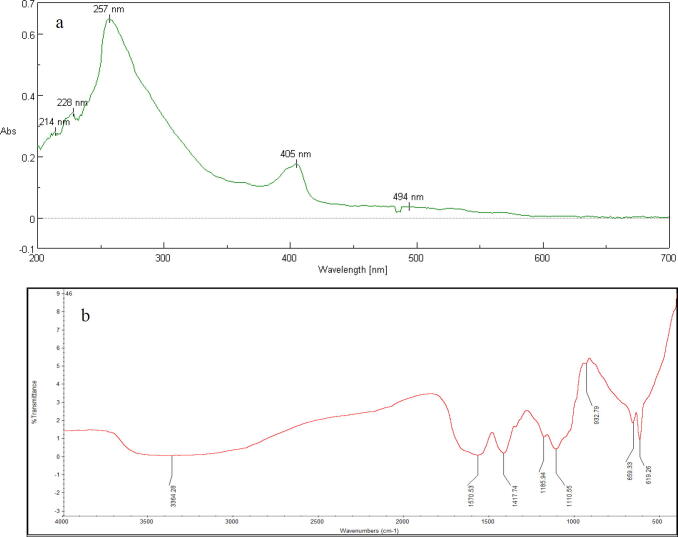
The UV spectrum of the ethyl acetate extract exhibited maximum absorption peak at 254 nm, 405 nm, 228 nm and 214 nm (Fig. 7a).
Fig. 7.
(a) UV spectrum (b) FTIR spectrum of ethyl acetate extract.
3.6.2. FTIR spectroscopy
The FTIR transmittance spectrum of ethyl acetate extract revealed eight peaks at 3364.28, 1570.53, 1417.74, 1185.94, 1110.55, 932.79, 659.33 and 619.26 cm−1 which is depicted in the Fig. 7b. The first broad and strong peak at 3364.28 cm−1 was attributed to O—H stretching of alcohol. The prominent peak at 1570.53 cm−1 indicated C C stretching of alkenes. The vibrational peak appeared at 1417.74 cm−1 indicated the presence of O—H bending of carboxylic acids. The peak positioned at 1185.94 cm−1 was emerged due to O C—O—C stretching of aliphatic esters, whereas the peak at 1110.55 cm−1 attributed to Si—O—C stretching of organic siloxanes. The vibrational peak at 932.79 cm−1 was assigned to cyclohexane ring vibrations. The peaks at 659.33 cm−1, 619.26 cm−1 were appeared due to C—Br stretching of alkyl halides and C—H bending of alkyne respectively.
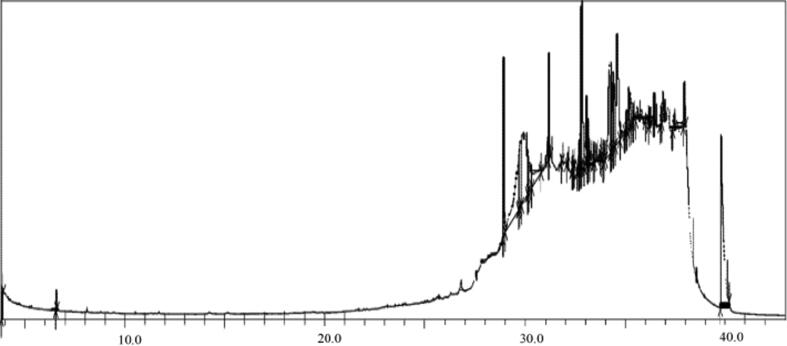
3.6.3. GC–MS analysis
The GC–MS analysis of Streptomyces sp. strain KS46 showed a detailed account on the compounds present in ethyl acetate extract. The chromatogram is shown in the Fig. 8 and associated compounds were listed in Table 3 along with their molecular formulae, molecular weight and biological activities. However, there was no biological activity reported for few compounds. The chromatogram revealed 42 compounds in the span of 43 min. The GC–MS chromatogram broadcasted that E,E,Z-1,3,12-Nonadecatriene-5,14-diol, Octadecane, 1-iodo and 22-Tricosenoic acid as major metabolic compounds present in strain KS46 extract covering a peak area of 7.17%, 7.10% and 5.05% respectively.
Fig. 8.
GC–MS chromatogram of Streptomyces sp. strain KS46 ethyl acetate extract.
Table 3.
Compounds present in ethyl acetate extract of Streptomyces sp. strain KS46.
| No. | Compound name | R. time | Area% | Chemical formula | Molecular weight | Biological activity | References |
|---|---|---|---|---|---|---|---|
| 1 | 2-Pentanone, 4-hydroxy | 3.884 | 0.81 | C5H10O2 | 102.13 | Not reported | |
| 2 | Cyclotetrasiloxane, octamethyl- | 6.564 | 0.10 | C8H24O4Si4 | 296.61 | Antioxidant, antimicrobial | Gehan et al., 2020 |
| 3 | n-Pentadecanol | 28.920 | 4.01 | C15H32O | 228.4 | Antibacterial activity | Umdale et al., 2021 |
| 4 | Octadecane, 1-iodo- | 29.565 | 7.10 | C18H37I | 380.39 | Not reported | |
| 5 | 2-methylhexacosane | 29.675 | 1.62 | C27H56 | 380.73 | Antimicrobial activity | Ryu et al., 2020 |
| 6 | Hexatriacontane | 29.740 | 3.84 | C36H74 | 506.97 | Antimicrobial, antioxidant, antiproliferative, activity | Arun et al., 2017 |
| 7 | Eicosyl trifluoroacetate | 30.132 | 1.39 | C22H41F3O2 | 394.55 | Not reported | |
| 8 | Eicosyl acetate | 30.310 | 1.95 | C22H44O2 | 340.58 | Not reported | |
| 9 | Palmitic acid vinyl ester | 30.742 | 2.20 | C18H34O2 | 282.46 | Antipruritic, antiasthmatics,anticonvulsant, antipsoriatic, antiepileptic, antimigraine | Ramya et al., 2019 |
| 10 | Nonadecyl pentafluoropropionate | 31.110 | 1.34 | C22H39F5O2 | 430.53 | Not reported | |
| 11 | Docosanoic anhydride | 31.198 | 2.36 | C44H86O3 | 663.2 | Not reported | |
| 12 | 1-Heptacosanol | 31.805 | 0.15 | C27H56O | 396.73 | Antioxidant, antimicrobial activity | Imada, 2005 |
| 13 | 9,12-Octadecadienoic acid (Z,Z)- | 32.373 | 0.22 | C18H32O2 | 280.44 | Antioxidant Antibacterial | Kim et al., 2020, Sunita and Sonam, 2017 |
| 14 | Oleic anhydride | 32.410 | 0.22 | C36H66O3 | 546.9 | Not reported | |
| 15 | Stearic acid chloride | 32.630 | 0.24 | C18H35ClO | 302.92 | Not reported | |
| 16 | 6,9-Octadecadienoic acid, methyl ester | 32.735 | 1.30 | C19H34O2 | 294.5 | Not reported | |
| 17 | Bicyclo[10.1.0]tridec-1-ene | 32.794 | 3.27 | C13H22 | 178.31 | Insecticidal activity | Nadi et al., 2021 |
| 18 | 22-Tricosenoic acid | 32.831 | 5.05 | C23H44O2 | 352.6 | Cytotoxic activity | Andre et al., 2020 |
| 19 | Glycidyl stearate | 33.037 | 2.45 | C21H40O3 | 340.5 | Not reported | |
| 20 | Hexadecanoic acid, 2-hydroxy-1-(hydroxymethyl)ethyl ester | 33.130 | 1.65 | C19H38O4 | 330.50 | Antimicrobial, antioxidant, pesticide | Tulika and Mala, 2017 |
| 21 | Docosyl ethyl carbonate | 33.310 | 0.42 | C25H50O3 | 398.66 | Not reported | |
| 22 | Phthalic acid, di(2-propylpentyl) ester | 33.447 | 0.15 | C24H38O4 | 390.55 | Orally toxic during pregnancy, sucking in long-evans rat | Krishnamoorthy and Subramaniam, 2014 |
| 23 | 9,12-Octadecadienoyl chloride, (9Z,12Z)- | 33.564 | 0.42 | C18H31ClO | 298.9 | Not reported | |
| 24 | 2-methylhexacosane | 33.820 | 0.15 | C27H56 | 380.73 | Antimicrobial, hypocholesterolemic activity | Ryu et al., 2020 |
| 25 | Urs-12-en-28-al | 33.946 | 0.44 | C30H48O | 424.7 | Not reported | |
| 26 | Phytol, acetate | 34.212 | 5.59 | C22H42O2 | 338.56 | Antileishmanial, antiinflammatory, | Priyanka et al., 2019 |
| 27 | 1-Cyclohexyldimethylsilyloxybutane | 34.255 | 2.13 | C12H26OSi | 224.42 | Not reported | |
| 28 | Hexadecanoic acid, hexadecyl ester | 34.371 | 4.52 | C32H64O2 | 480.84 | Not reported | |
| 29 | Adipic acid, β-citronellyl tetradecyl ester | 34.460 | 2.37 | C30H56O4 | 480.76 | Not reported | |
| 30 | E,E,Z-1,3,12-Nonadecatriene-5,14-diol | 34.585 | 7.17 | C19H34O2 | 294.5 | Antimicrobial | Mohammed et al., 2019 |
| 31 | Octadecanoic acid, 2,3-dihydroxypropy ester | 34.765 | 0.72 | C21H42O4 | 358.55 | Antimicrobial, anticancer | Sunita and Ganesh, 2018 |
| 32 | 3.beta.-Hydroxy-5-cholen-24-oic acid | 35.040 | 0.72 | C24H38O3 | 374.56 | Not reported | |
| 33 | 2,4,7,14-Tetramethyl-4-vinyl-tricyclo[5.4.3.0(1,8)]tetradecan-6-ol | 35.247 | 0.99 | C20H34O | 290.5 | Not reported | |
| 34 | Heneicosane | 35.320 | 0.87 | C21H44 | 296.57 | Antimicrobial | Vanitha et al., 2020 |
| 35 | Acetic acid, 13-hydroxy-4,4,6a,6b,8a,11 11,14b-octamethyldocosahydropicen-3-yl ester | 35.425 | 0.20 | C32H54O3 | 486.8 | Not reported | |
| 36 | Stigmastane-3,6-dione, (5.alpha.)- | 35.595 | 0.56 | C29H48O2 | 428.69 | Antimicrobial, antiinflammatory | Rajiv et al., 2018 |
| 37 | Hexatriacontane | 36.025 | 0.58 | C36H74 | 506.97 | Antioxidant | |
| 38 | 1-Hexadecanesulfonyl chloride | 36.192 | 0.50 | C16H33ClO2S | 324.95 | Not reported | |
| 39 | Hexadecanoic acid, tetradecyl ester | 36.422 | 0.68 | C30H60O2 | 452.79 | Xenobiotic | |
| 40 | Octatriacontyl trifluoroacetate | 36.800 | 0.47 | C40H77F3O2 | 647.03 | Insecticidal | Usha et al., 2019 |
| 41 | Stigmasta-5,22-dien-3-ol, acetate, (3.β)- | 37.415 | 0.36 | C31H50O2 | 454.72 | Antimicrobial | Achika et al., 2016 |
| 42 | Stigmast-5-en-3-ol, oleate | 37.962 | 1.16 | C47H82O2 | 679.2 | Not reported |
4. Discussions
In todays world, there is an emergence new diseases, multidrug resistant human pathogenic organisms, re-emergences of previously controlled diseases and inadequacy of therapeutic measures to control such emerging diseases have a become a global health concern. Actinomycetes due to its potentiality to produce a number of antibiotics have become the centre of interest to counteract these pathogenic diseases. Therefore the isolation of actinomycetes from underexplored habitats has become popularized to replenish the need of new antibiotics (Sengupta et al., 2015, Olga, 2018). An extensive literature survey indicated that there is no report regarding the isolation and screening of actinomycetes from Devbag and Tilmati beaches. Therefore, the isolation of marine actinomycetes from these regions was attempted to find out potential strains of actinomycetes. Total 30 marine water and sediment samples were gathered from both the beaches and 70 marine actinomycetes were isolated from these samples. During the isolation of actinomycetes, bacterial contaminants were found to grow faster and inhibit the colonization of actinomycetes. Hence the growth of these unwanted microbes needed to be inhibited through heating the samples at 70 °C for 20 min. Singh et al., (2016) reported that the spores of actinomycetes can normally withstand desiccation and show slightly better resistance to dry or moist heat than other microorganisms. A similar result was published where the heat treatment and serial dilutions of samples reduced growth of unwanted microorganisms (Mahejbin et al., 2019).
The primary screening of actinomycetes showed a significant result against pathogenic bacteria and fungi. It was evident from the result that all of the pathogens were susceptible to at least one isolated actinomycetes strains. In case of gram positive organisms; 60% of the isolates showed antibacterial activity against S. aureus, 44.28% of the isolates displayed antagonistic activity against B. subtilis, 42.85% and 40% of the actinomycetes indicated activity against B.cereus and E. faecalis respectively. But in case of gram negative pathogens; 37.14% of the isolates showed activity against E. coli, 32.85% expressed activity against P. aeruginosa and 31.42% of the strains suppressed the growth of K. pneumoniae. 41.42% and 45.71% of the actinomycetes strains exhibited antifungal activity against C. albicans and C. glabrata respectively.
The antagonistic activity of ethyl acetate extract of Streptomyces sp. strain KS46 showed different levels of results against tested pathogens. The maximum zone of inhibition was exhibited by B. subtilis. All the four gram positive pathogens and three out of 4 gram negative pathogens were susceptible to ethyl acetate extract. It was noticed that gram negative pathogens were more resistant when compared to gram positive pathogens. This could be attributed to the presence of lipopolysaccharide in outer membrane of gram negative bacteria as a major structural unit, whereas the gram positive bacteria are devoid of lipopolysaccharide as a protective barrier and become susceptible to the metabolites. The antimicrobial mechanisms can be grouped into 4 categories: (i) modification of drug target, (ii) limitation of drug uptake, (iii) activating drug efflux and (iv) drugs inactivation. The gram negative bacteria are capable of using all the four mechanisms. The gram positive bacteria due to the structural differences can less commonly use limiting the drug uptake and cannot induce certain types of drug efflux mechanism (Wanda, 2018). It has been reported that higher efficiency in antimicrobial activity could be achieved when extraction of bioactive compounds was performed using organic solvents compared to water-based methods (Jose et al., 2002). A similar report was represented by Noura et al. (2017), where the solvent ethyl acetate was found as the best for the extraction of antimicrobial metabolites.
The actinomycetes isolated from marine samples exhibited potentiality for the production of enzymes like amylase, protease, cellulase and lipase. Amylase is an industrially important enzyme and 61.42% of the isolated marine actinomycetes strains showed the production of this enzyme. This enzyme amylase helps microorganisms to carry out extracellular digestion when secreted outside the cell (Ranjani et al., 2016). This result was consistent with the report of Praveen et al., (2015). In case protease enzyme 42.85% of the actinomycetes strains displayed the production of this enzyme. Protease enzyme is used for several decades in industries like animal feed, detergents, leather and breweries. The protease enzymes produced by actinomycetes are cost-effective, consume less time and space for production and can be yielded in large quantity (Razzaq et al., 2019). A similar result was reported by Praveen et al., (2015), where out of 82 actinomycetes strains 14% exhibited protease activity. 35.71% of the total isolated strains showed positive result for cellulase production. The production of cellulase enzyme has a very important role in cellulolytic microbes for hydrolysis of lignocellulosic polymer materials in the natural environment. The result obtained can be confirmed with the report of Pinky et al., (2013). 41.42% of actinomycetes strains showed positive result for lipase production. The microbial lipases are commercially substantial due to their superior stability, low manufacturing cost and more availability than plant and animal lipases. The lipase enzyme from actinomycetes provides extraordinary potential because of their adaptation abilities in uninhabitable environments, such as alkaline lakes, volcanic vents, contaminated soils, hot springs, etc. (Chandra et al., 2020). The enzymes produced by some halophilic actinomycetes have optimal activity at high salinities and therefore could be used in several harsh industrial processes where the concentrated salt solutions used would otherwise inhibit many enzymatic conversions. This is in accordance with the result of Praveen et al., (2015), where 57% of actinomycetes strains revealed lipase enzyme activity.
Morphologically the organism Streptomyces sp. strain KS46 produced whitish-pink aerial mycelium and pink to brown substrate mycelium. The colony of the strain KS46 was flat with filiform margins. The SEM analysis of Streptomyces sp. strain KS46 showed spiral spore chain having smooth spores. This result is in compliance with the report of Nayaka et al. (2020), where the Streptomyces sp. NS-33 produced flat and irregular shaped colonies. The SEM analysis of the strain revealed smooth surface spores and retinaculiapreti structure of the spore chain.
The molecular characterization of Streptomyces sp. strain KS46 disclosed 97.97% sequence similarity with Streptomyces levis (NR041184). This can be compared with the report of Sreenivasa et al. (2020), where the Streptomyces sp. KAS-1 showed 98.97% similarity with Streptomyces thermocarboxydus (KJ018992) through 16S rRNA gene sequencing.
The UV spectrum of ethyl acetate extract displayed peaks at 254 nm, 405 nm, 228 nm and 214 nm. This is why the strain exhibited a broad spectrum of antimicrobial activities. The FTIR spectrum expressed various functional groups in the ethyl acetate extract, such as alcohols, alkenes, alkyne, alkyl halides, carboxylic acids, aliphatic esters, organic siloxanes, cyclohexane. This result was compared with the report of Hadi and Omid, (2011).
The ethyl acetate extract of the Streptomyces sp. stain KS46 presented 42 bioactive compounds. Out of this 42 compounds seven were fatty acid esters (9,12-Octadecadienoic acid (Z,Z)-, 6,9-Octadecadienoic acid methyl ester, Hexadecanoic acid 2-hydroxy-1-(hydroxymethyl)ethyl ester, Hexadecanoic acid hexadecyl ester, Adipic acid β-citronellyl tetradecyl ester, Octadecanoic acid 2,3-dihydroxypropyl ester and Hexadecanoic acid tetradecyl ester), four alkanes (2-methylhexacosane, Hexatriacontane, 1-Cyclohexyldimethylsilyloxybutane and Heneicosane), four steroids (Stigmastane-3,6-dione(5.alpha.)-, Stigmasta-5,22-dien-3-ol acetate(3.β)-, 3.beta.-Hydroxy-5-cholen-24-oic acid and Stigmast-5-en-3-ol oleate), four esters (Eicosyl trifluoroacetate, Eicosyl acetate, Palmitic acid vinyl ester and Acetic acid, 13-hydroxy-4,4,6a,6b,8a,11,11,14b-octamethyldocosahydropicen-3-yl ester), three alcohols (Phytol, acetate, E,E,Z-1,3,12-Nonadecatriene-5,14-diol and 2,4,7,14-Tetramethyl-4-vinyl-tricyclo[5.4.3.0(1,8)]tetradecan-6-ol), two halogenated alkanes (Octadecane 1-iodo- and Octatriacontyl trifluoroacetate), two fatty alcohols (n-Pentadecanol and 1-Heptacosanol), two carboxylic ester (Glycidyl stearate and Phthalic acid, di(2-propylpentyl) ester), two fatty acid anhydride (Docosanoic anhydride and Oleic anhydride), one ketone (2-Pentanone, 4-hydroxy), one organosilicon compound (Cyclotetrasiloxane, octamethyl-), one fatty acid chloride (Stearic acid chloride), one poly unsaturated fatty acid chloride (9,12-Octadecadienoyl chloride, (9Z,12Z)-), one carbonic acid ester (Docosyl ethyl carbonate), one sulphonyl chloride (1-Hexadecanesulfonyl chloride), one acetate surfactant (Nonadecyl pentafluoropropionate), one cyclic olefins (Bicyclo[10.1.0]tridec-1-ene), and one pentacyclic triterpene (Urs-12-en-28-al). The majority of these secondary metabolites produced by the Streptomyces sp. stain KS46 have antibacterial, antifungal, insectidal, etc. activities. The result obtained here can be compared to the result of Noura et al. (2017), where the GCMS spectrum of ethyl acetate extract of a Streptomyces sp. displayed the occurrence of 22 bioactive compounds including saturated fatty acids, alkanes, unsaturated fatty acid, alkenes, fatty acid esters and triterpene.
5. Conclusion
The marine ecosystem is a tremendous source for isolation of valuable species of actinomycetes. In this research work 70 marine actinomycetes strains were isolated from Devbagh and Tilmati beaches. The results suggested that the marine ecological community is a potential source of bioactive actinomycetes. Therefore, such studies are recommended for exploring the bioactive potentiality of actinomycetes associated with marine ecosystem and achieving their conservation. The outcome of this research work is encouraging and hence further work will be carried out for purification of ethyl acetate extract, characterization and identification of bioactive metabolites.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
The authors are thankful to the P.G. Department of Studies in Botany, Karnatak University, Dharwad, Karnataka, India for extending lab facilities.
Funding
The project was supported by Researchers Supporting Project number (RSP-2021/142), King Saud University, Riyadh, Saudi Arabia.
Footnotes
Peer review under responsibility of King Saud University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.sjbs.2021.11.055.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- Achika, J.I., Ndukwe, G.I., Ayo, R.G., 2016. Isolation, characterization and antimicrobial activity of 3β, 22E-Stigmasta-5, 22-dien-3-ol from the aerial part of Aeschynomene uniflora E. Mey. British J. Pharm. Res. 11(5), BJPR.23506.
- Ajit K.P., Vineet K.M., Ratul S., Vijai K.G., Bhim P.S. Isolation, abundance and phylogenetic affiliation of endophytic actinomycetes associated with medicinal plants and screening for their in vitro antimicrobial biosynthetic potential. Front. Microbiol. 2015;6:273. doi: 10.3389/fmicb.2015.00273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andre L.G., Valcineide O.A.T., Gilberto V.M.P., Oranys M., Sandro J.R.B., Suzany S., Ivan R.B., Carlos R.S. Phytochemical analysis and biological activities of in vitro cultured Nidularium procerum, a bromeliad vulnerable to extinction. Sci. Rep. 2020;10(1):7008. doi: 10.1038/s41598-020-64026-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arun K.M., Umamaheswari, Rajesh K.G., Preethy P.R., Elumalai S., Sangeetha T. Studies on the extraction and analysis of polyunsaturated fatty acids (pufa) from marine macroalgae. Int. J. Pharm. Technol. 2017;9(3):30715–30741. [Google Scholar]
- Berdy J. Bioactive microbial metabolites. J. Antibiot. 2005;58:1–26. doi: 10.1038/ja.2005.1. [DOI] [PubMed] [Google Scholar]
- Chandra P., Enespa R.S., Pankaj K.A. Microbial lipases and their industrial applications: a comprehensive review. Microb. Cell. Fact. 2020;19(1) doi: 10.1186/s12934-020-01428-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Das S., Lyla P.S., Ajmalkhan S. Biogeochemical processes in the continental slope of Bay of Bengal: I. Bacterial solubilization of inorganic phosphate. Rev. Biol. Trop. 2007;55(1):1–9. doi: 10.15517/rbt.v55i1.6052. [DOI] [PubMed] [Google Scholar]
- Dhakal D., Pokhrel A.R., Shrestha B., Sohng J.K. Marine rare Actinobacteria: isolation, characterization, and strategies for harnessing bioactive compounds. Front. Microbiol. 2017;8:1106. doi: 10.3389/fmicb.2017.01106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dina H.A., Nagwa A.A., Assem A., Sahar T., Elizabeth M.H.W. Microbiological and molecular insights on rare Actinobacteria harbouring bioactive prospective. Bull. Natl. Res. Cent. 2020;44:5. doi: 10.1186/s42269-019-0266-8. [DOI] [Google Scholar]
- Gebreselema G., Feleke M., Samuel S., Nagappan R. Isolation and characterization of potential antibiotic producing actinomycetes from water and sediments of Lake Tana, Ethiopia. Asian Pac. J. Trop. Biomed. 2013;3(6):426–435. doi: 10.1016/S2221-1691(13)60092-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gehan A.I., Saly F.G., Atef M.A.S., Omnia H.A.K. In vitro potential activity of some seaweeds as antioxidants and inhibitors of diabetic enzymes. Food Sci. Technol. 2020;40(3):681–691. doi: 10.1590/fst.15619. [DOI] [Google Scholar]
- Hadi M., Omid M. Characterization of Streptomyces Isolates with UV, FTIR Spectroscopy and HPLC Analyses. Bioimpacts. 2011;1(1):47–52. doi: 10.5681/bi.2011.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imada C. Enzyme inhibitors and other bioactive compounds from marine actinomycetes. Antonie Van Leeuwenhoek. 2005;87(1):59–63. doi: 10.1007/s10482-004-6544-x. [DOI] [PubMed] [Google Scholar]
- Jose V.M.L.F., Ana F.F.U.C., Sissi M.F., Vania M.M.M. Antibacterial activity of extracts of six macroalgae from the northeastern brazilian coast. Braz. J. Microbiol. 2002;33(4):311–313. doi: 10.1590/S1517-83822002000400006. [DOI] [Google Scholar]
- Kadriye O., Semiha C.A., Orcun K., Atac U., Esin H.K., Erdal B. Diversity and antibiotic-producing potential of cultivable marine-derived actinomycetes from coastal sediments of Turkey. J. Soils Sediments. 2013;13(8):1493–1501. doi: 10.1007/s11368-013-0734-y. [DOI] [Google Scholar]
- Karuppiah P., Mustaffa M. Antibacterial and antioxidant activities of Musa sp. Leaf extracts against multidrug resistant clinical pathogens causing nosocomial infection. Asian Pac. J. Trop. Biomed. 2013;3(9):737–742. doi: 10.1016/S2221-1691(13)60148-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim B.R., Kim H.M., Jin C.H., Kang S.Y., Kim J.B., Jeon Y.G., Park K.Y., Lee I.S., Han A.R. Composition and Antioxidant Activities of Volatile Organic Compounds in Radiation-Bred Coreopsis Cultivars. Plants. 2020;9(6):717. doi: 10.3390/plants9060717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamoorthy, K., Subramaniam, P., 2014. Phytochemical profiling of leaf, stem, and tuber parts of Solena amplexicaulis (Lam.) Gandhi using GC-MS. Int. Sch. Res. Notices. 2014, 567409. https://doi.org/10.1155/2014/567409. [DOI] [PMC free article] [PubMed]
- Kumar S., Glen S., Koichiro T. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016;33(7):1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahejbin S., Dalip S.R., Sangeeta G., Satya P.S. Cultivation and characteristics of the Marine Actinobacteria from the Sea water of Alang. Bhavnagar. Indian J. Mar. Sci. 2019;48(12):1896–1901. [Google Scholar]
- Miethke M., Pieroni M., Weber T., et al. Towards the sustainable discovery and development of new antibiotics. Nat. Rev. Chem. 2021;5:726–749. doi: 10.1038/s41570-021-00313-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohammed, Y.H., Ghaidaa, J.M., Imad, H.H., 2019. Analysis of bioactive chemical compounds of Nigella sativa using gas chromatography-mass spectrometry 8(2), 18–24. https://doi.org/10.5897/JPP2015.0364.
- Nadi A.H., Nagy M.A., Dalia M.H., Salwa E.M., Arafat A.H.A.L., Amira A.I., Mohamed A.A. Evaluation of insecticidal effects of plants essential oils extracted from basil, black seeds and lavender against Sitophilus oryzae. Plants. 2021;10:829. doi: 10.3390/plants10050829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nayaka S., Chakraborty B., Bhat M.P., Shashiraj K.N., Airodagi D., Pallavi S.S., Muthuraj R., Halaswamy H., Dhanyakumara S.B., Bharati K. Biosynthesis, characterization, and in vitro assessment on cytotoxicity of actinomycete-synthesized silver nanoparticles on Allium cepa root tip cells. Beni-Suef Univ. J. Basic Appl. Sci. 2020;9:51. doi: 10.1186/s43088-020-00074-8. [DOI] [Google Scholar]
- Njenga W.P., Mwaura F.B., Wagacha J.M., Gathuru E.M. Methods of Isolating Actinomycetes from the Soils of Menengai Crater in Kenya. Arch. Clin. Microbiol. 2017;8(3):45. doi: 10.21767/1989-8436.100045. [DOI] [Google Scholar]
- Noura A.N., Ashraf A.A.B., Mamdouh A.M., Noura S.N. In vitro activity, extraction, separation and structure elucidation of antibiotic produced by Streptomyces anulatus NEAE-94 active against multidrug-resistant Staphylococcus aureus. Biotechnol. Biotechnol. Equip. 2017;31(2):418–430. doi: 10.1080/13102818.2016.1276412. [DOI] [Google Scholar]
- Olga G. Mining Actinomycetes for Novel Antibiotics in the Omics Era: Are We Ready to Exploit This New Paradigm? Antibiotics. 2018;7:85. doi: 10.3390/antibiotics7040085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oskay A.M., Usame T., Cem A. Antibacterial activity of some actinomycetes isolated from farming soils of Turkey. Afr. J. Biotechnol. 2005;3(9):441–446. doi: 10.5897/AJB10.5897/AJB2004.000-2087. [DOI] [Google Scholar]
- Pinky P., Tanuja S., Sheila B. Characterization of the cellulolytic enzyme produced by Streptomyces griseorubens (Accession No. AB184139) isolated from Indian soil. J. King Saud Univ.-Sci. 2013;25(3):245–250. doi: 10.1016/j.jksus.2013.03.003. [DOI] [Google Scholar]
- Praveen K.P., Preetam R.J.P., Nimal C.I.V.S., Sagaya J.R., Murugan N., Agastian P., Arunachalam C., Ali A.S. Screening of Actinomycetes for Enzyme and Antimicrobial Activities from the Soil Sediments of Northern Tamil Nadu, South India. J. Biol. Act. Prod. Nat. 2015;5(1):58–70. doi: 10.1080/22311866.2015.1009385. [DOI] [Google Scholar]
- Priyanka G., Bunty K.D., Neelam B., Navneet S.C. Comparative GC-MS analysis of bioactive phytochemicals from different plant parts and callus of Leptadenia reticulata Wight and Arn. Pharmacogn. J. 2019;11(1):129–140. doi: 10.5530/pj10.5530/pj.2019.110.5530/pj.2019.1.22. [DOI] [Google Scholar]
- Rajagopal G., Palaniappan S., Kannan S. Marine actinobacteria: A concise account for young researchers. Indian J. Mar. Sci. 2018;47(3):541–547. [Google Scholar]
- Rajiv R., Nor S.H.Z., Nurul N.R., Nik R.N.Y., Mohd S.M.R., Muhammad I.A., Zulhazman H., Intan H.I., Mohamad F.M.A. Evaluation of two different solvents for Azolla pinnata extracts on chemical compositions and larvicidal activity against Aedes albopictus (Diptera: Culicidae) J. Chem. 2018;2018:1–8. doi: 10.1155/2018/7453816. [DOI] [Google Scholar]
- Ramya K.S., Kanimathi P., Radha A. GC-MS analysis and antimicrobial activity of various solvent extracts from Simarouba glauca leaves. J. Pharmacogn. Phytochem. 2019;8(2):166–171. [Google Scholar]
- Ranjani, A., Dhanasekaran, D. Gopinath, P.M., 2016. An Introduction to Actinobacteria, Actinobacteria. In: Dharumadurai, D., Yi, J. (Eds.), Actinobacteria-Basics and Biotechnological Applications. IntechOpen, pp. 1-37. https://doi.org/10.5772/62329.
- Rashad F.M., Hayam M.F., Ayatollah S.Z., Ahlam M.E. Isolation and characterization of multifunctional Streptomyces species with antimicrobial, nematicidal and phytohormone activities from marine environments in Egypt. Microbiol. Res. 2015;175:34–47. doi: 10.1016/j.micres.2015.03.002. [DOI] [PubMed] [Google Scholar]
- Razzaq A., Shamsi S., Ali A., Ali Q., Sajjad M., Malik A., Ashraf M. Microbial Proteases Applications. Front. Bioeng. Biotechnol. 2019;7:110. doi: 10.3389/fbioe.2019.00110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rifaat H.M. The biodiversity of actinomycetes in the River Nile exhibiting antifungal activity. J. Mediterr. Ecol. 2003;4(3–4):5–7. [Google Scholar]
- Ryu J., Lyu J., Kim D.G., Kim J.M., Jo Y.D., Kang S.Y., Kim J.B., Ahn J.W., Kim S.H. Comparative Analysis of Volatile Compounds of Gamma-Irradiated Mutants of Rose (Rosa hybrida) Plants. 2020;9(9):1221. doi: 10.3390/plants9091221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta S., Pramanik A., Ghosh A., Bhattacharyya M. Antimicrobial activities of actinomycetes isolated from unexplored regions of Sundarbans mangrove ecosystem. BMC Microbiol. 2015;15:170. doi: 10.1186/s12866-015-0495-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh V., Haque S., Singh H., Verma J., Vibha K., Singh R., Jawed A., Tripathi C.K.M. Isolation, Screening, and Identification of Novel Isolates of Actinomycetes from India for Antimicrobial Applications. Front. Microbiol. 2016;7:1921. doi: 10.3389/fmicb.2016.01921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sreenivasa N., Shashiraj K.N., Bidhayak C., Meghashyama P.B., Pallavi S.S., Dattatraya A., Halaswamy H., Muthuraj R., Dhanyakumara S.B., Anusha R.P. Antimicrobial and Enzymatic potential of Streptomyces sp. KAS-1 isolated from the microbiologically unexplored estuary of Kali river ecosystem. Int. J. Res. Pharm. Sci. 2020;11(2):1655–1666. doi: 10.26452/ijrps.v11i2.2048. [DOI] [Google Scholar]
- Sunita A., Ganesh K. Phytochemical screening of root, stem and leaves of Cenchrus biflorus Roxb. J. Pharmacogn. Phytochem. 2018;7(1):1445–1450. [Google Scholar]
- Sunita A., Sonam M. GC-MS profiling of Ceropegia bulbosa Roxb. var. bulbosa, an endangered plant from Thar Desert, Rajasthan. Pharm. Innov. 2017;6(11):568–573. [Google Scholar]
- Tulika T., Mala A. GC-MS analysis of invasive aquatic weed, Pistia stratiotes L. and Eichhornia crassipes (mart.) solms. Int. J. Curr. Pharm. Res. 2017;9(3):111–117. [Google Scholar]
- Umdale S., Mahadik R., Otari P., Gore N., Mundada P., Ahire M. Phytochemical composition, and antioxidant potential of Frerea indica Dalz.: a critically endangered, endemic and monotypic genus of the Western Ghats of India. Biocatal. Agric. Biotechnol. 2021;35:102080. doi: 10.1016/j.bcab.2021.102080. [DOI] [Google Scholar]
- Usama R.A., Kristina B., Ute H. Diversity, abundance and natural products of marine sponge-associated actinomycetes. Nat. Prod. Rep. 2014;31(3):381–399. doi: 10.1039/C3NP70111E. [DOI] [PubMed] [Google Scholar]
- Usha N.S., Sabari A.J.V., Mangaiyarkarasi Preliminary phytochemical screening and GC-MS analysis of Cladophora glomerata: green marine algae. Int. J. Basic Clin. Pharmacol. 2019;8(4):732–737. doi: 10.18203/2319-2003.ijbcp20191108. [DOI] [Google Scholar]
- Vanitha V., Vijayakumar S., Nilavukkarasi M., Punitha V.N., Vidhya E., Praseetha P.K. Heneicosane -A novel microbicidal bioactive alkane identified from Plumbago zeylanica L. Ind. Crop. Prod. 2020;154:112748. doi: 10.1016/j.indcrop.2020.112748. [DOI] [Google Scholar]
- Wanda C.R. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018;4(3):482–501. doi: 10.3934/microbiol.2018.3.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.