Abstract
Emerging clinical evidences highlight the association of Interleukin-8 (IL8) with endodontic pulpitis. Relatively higher expression of IL8 has been found in the pulp samples of pulpitis patients with moderate/severe pain. It is speculated that IL8 can be considered as a potential target for therapeutics of endodontic pulpitis. A library consisting of 3072 small molecules from the ZINC database was used to identify potential lead molecules with drug-like properties against the IL8. Based on the in-silico structure-assisted drug designing involving molecular docking, MD simulations, and MMPBSA analyses, we found a small molecule ZINC14613097 inhibits IL8. This study provides a new lead molecule than can be further validated in in-vitro, in-vivo, and ongoing clinical studies for the therapeutic management of endodontic pulpitis.
Keywords: Endodontic, Inhibitor, Interleukin-8, Pulpitis, ZINC14613097
1. Introduction
Endodontic pulpitis is a painful inflammation in the dental pulp which is characterized by, hyperalgesia, sensitivity, allodynia, and difficulty in achieving adequate local anesthesia(Galicia et al., 2016, Karapanou et al., 2008, Rechenberg et al., 2016). There are variable symptoms associated with this disease and the progression is unpredictable. It is typically caused by commensal oral microorganisms and is tightly regulated by number of vascular and cellular events arbitrated by different molecular factors (Ahlquist and Franzen, 1994, Hahn et al., 1991, Rechenberg et al., 2016). Human dental pulp consists of different types of immune-competent cells that trigger several immune responses in endodontic pulpitis(Keller et al., 2011, Smith, 2002, Staquet et al., 2011). These cascades of events are associated with release of different immune system mediators and these mediators have been reported to be correlated with different clinical conditions most importantly they exhibit pulpal or odontogenic pain, inflammation and in advanced stages, pulpal necrosis(Avellán et al., 2008, Galicia et al., 2016, Karapanou et al., 2008).
Several studies have compared and identified the physiological and molecular differences between healthy dental pulps and inflamed ones. Altered expression of cytokines, cell surface receptors and proteins have been associated with inflamed dental pulps (Caviedes-Bucheli et al., 2007, Caviedes-Bucheli et al., 2005, CAVIEDESBUCHELI et al., 2008, Esmaeili et al., 2011, Gong et al., 2010, Huang et al., 2009, Jiang et al., 2008, Keller et al., 2011, Kokkas et al., 2007, Korkmaz et al., 2011, Lundy et al., 2010, Mutoh et al., 2009) . In addition to this, there are a few studies that profiled the expression of genes in inflamed human pulps (Kaneko et al., 2010, McLachlan et al., 2005, Tete et al., 2008). Several studies have reported that differential expression of Interleukin8 (IL8) in healthy, inflamed pulp and severe pulpitis conditions ranging from basal, moderate to mild level(Galicia et al., 2016, Karapanou et al., 2008). Galicia et al., reported elevated IL8 expression in endodontic pulpitis samples and authors correlated with the moderate/severe pain in pulpitis patients as compared to pulpitis samples of patients having asymptomatic/mild pain (Galicia et al., 2016). Likewise, Karapanou et al., reported higher expression of IL8 in GCF in acute pulpitis patients (Karapanou et al., 2008). These emerging clinical evidences highlight the link between IL8 and endodontic pulpitis and reflect that IL8 can be a potential target for therapeutics and management of endodontic pulpitis. Therefore, in this current study we aimed to conduct virtual screening of molecules/compounds from ZINC data base (https://zinc12.docking.org/) and correlating the structural scaffolds of these molecules for modulatory activity against IL8 for the clinical management of endodontic pulpitis.
2. Materials and methods
2.1. Small drug and target molecules preparation
Protein Data Bank (PDB ID: 5d14) and SPDBV were used to download the crystal structure of IL8 and to minimize the energy respectively (Boreak, 2021) (Sultan et al., 2021). In the crystal structure of the protein atoms that are missing, alternate location, added waters, more than a molecule, chain breaks, etc. were examined. Addition of Polar hydrogen and assigning Kollman United Atom Charges were done (Boreak, 2021). ZINC data base was used to download the chemical structures of the small molecules (n = 3072) used in the study (https://zinc12.docking.org/) and ChemBioDraw® Ultra 12.0 was used in the process (Kerwin, 2010).
2.2. Molecular dynamics simulations and Molecular docking of small molecules with IL8
Molecular Dynamics simulations and Molecular docking were studied with Ubuntu 14.04.5 LTS operating system with Intel® Xeon® CPU E5-2609 v3@1.90 GHz processor on DELL® workstation, RAM with 64 GB and hard disk of terabyte. All the small molecules were docked blindly using IL8 using AutoDock Vina and MGL tools (Sultan et al., 2021)(Vchirawongkwin et al., 2009). For X, Y, and Z coordinates the grid size was 59, 75, and 68, centralized grid 36.20, 14.51, and 62.67, respectively. The grid pacing a 1.00 Å with the exhaustiveness of 8. Molecular Dynamics simulations were done with GROMACS 5.1.1. For visualizing, evaluating and analyzing of Molecular Dyanamic trajectories PyMOL, QtGrace and Discovery Studio Visualizer were used (Vchirawongkwin et al., 2009) (Guex and Peitsch, 1997). Molecular dynamics (50 ns) of IL8 and IL8-ZINC14613097 complex were simulated with GROMOS96 43A1 force-field in GROMACS 5.1.1 at the molecular mechanics level at 300 K. Conformations sampling was done every 10 ps for IL8 and IL8-ZINC14613097 complex during 50 ns simulations. GROMACS 5.1.1 utilities gmx energy, gmx gyrate, gmxrmsf, gmx sham, gmx rms, and gmx sasa for analysing the trajectories. RMSD, RMSF, SASA, Rg, kinetic energy, density and volume, potential energy, enthalpy, of the systems (Boreak, 2021). QtGrace was used for plotting all the graphs and figures (Guex and Peitsch, 1997).
2.3. Calculating inhibition constant
The inhibition constant (Ki; nM) was calculated in AutoDock4 using the formula (Sultan et al., 2021):
In which G = docking energy;
R = 1.98719 cal K-1 mol-1;
T 298.15°k
Ki = EXP ((A*1000)/ (198719*29815))
2.4. MM-PBSA calculation
Interaction free energy was calculated with Molecular Mechanics/Poisson Boltzmann Surface Area (MM-PBSA), which is commonly aided in studying the interactions in the biomolecules (Sultan et al., 2021). The MM-PBSA was calculated with Molecular Dynamic scripts (Bhardwaj et al., 2021). The g_mmpbsa script of GROMACS was used for calculating the MM-PBSA binding free energies (Kumari et al., 2014). Equation mentioned below was used in calculating the binding energy:
where, Gbinding stands for total energy for binding the protein–ligand complex, Greceptor is free receptor binding energy, and Gligand is the unbounded ligand binding energy.
3. Results
3.1. Crystal structure of IL8protein and energy minimization
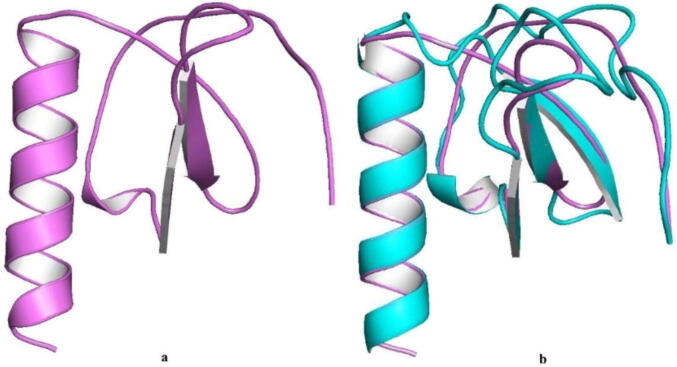
The crystal structure of IL8, was downloaded from Protein Data Bank (PDB ID: 5d14; Fig. 1a). After retrieval of protein structure, energy was minimized by SPDB viewer by applying Gromos96 43B1 force field parameters to remove unwanted energy from loop, sheets and bonds stored. After that structure was superimposed using PyMOL to observe structural deviation in terms of RMSD. Calculated RMSD was 0.041, reflecting that structure gain minimal conformational structural changes (Fig. 1b).
Fig. 1.
(a) Cartoon representation (violet color) of IL-8 (PDB ID: 5d14). (b) Superimposed structure of IL-8 after energy minimization (native in violet color and energy minimized structure in cyan color).
3.2. Analysis of interaction between IL8 and small molecules
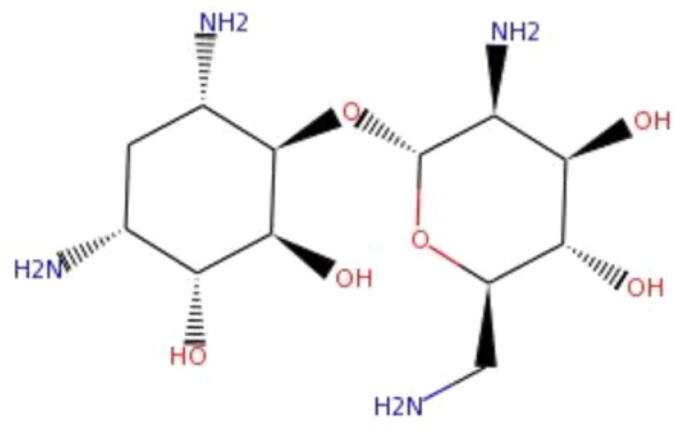
The binding affinity, energy for binding, and bound conformations of potential interacting of residual amino acid with intermolecular distances was detected using molecular docking analysis. In this analysis we docked all the listed small molecules against IL8 to check the binding affinity. Table 1 summarizes the top 10 molecules along with the parameters such as binding energy and inhibition constant. We observed high binding affinity for the small molecule ZINC14613097 (-9.7) and have significant conventional amino acid residual interactions. Our observations suggest ZINC14613097 as a potent inhibitor of IL8 (Table 1). The structure of ZINC14613097 is depicted in Fig. 2.
Table 1.
Top ten small molecules having high binding affinity to IL8 and the binding parameters.
| S.No. | Small molecules |
Target |
Binding |
Inhibition |
|---|---|---|---|---|
| molecule |
affinity |
constant, Ki |
||
| (kcal/Mol) | (nM) | |||
| 1. | ZINC14613097 | −9.7 | 4.19575E-07 | |
| 2. | ZINC01482077 | −8.3 | 8.24165E-07 | |
| 3. | ZINC00000061 | −7.7 | 2.26893E-06 | |
| 4. | ZINC08551464 | −8.1 | 1.15509E-06 | |
| 5. | ZINC00896717 | IL8 | −8.1 | 1.15509E-06 |
| 6. | ZINC03830793 | −7.6 | 2.6861E-06 | |
| 7. | ZINC00004351 | −7.6 | 2.6861E-06 | |
| 8. | ZINC03830326 | −7.4 | 3.76464E-06 | |
| 9. | ZINC03830332 | −7.2 | 5.27625E-06 | |
| 10. | ZINC03831537 | −7.2 | 5.27625E-06 |
Fig. 2.
Structure of small molecule ZINC14613097.
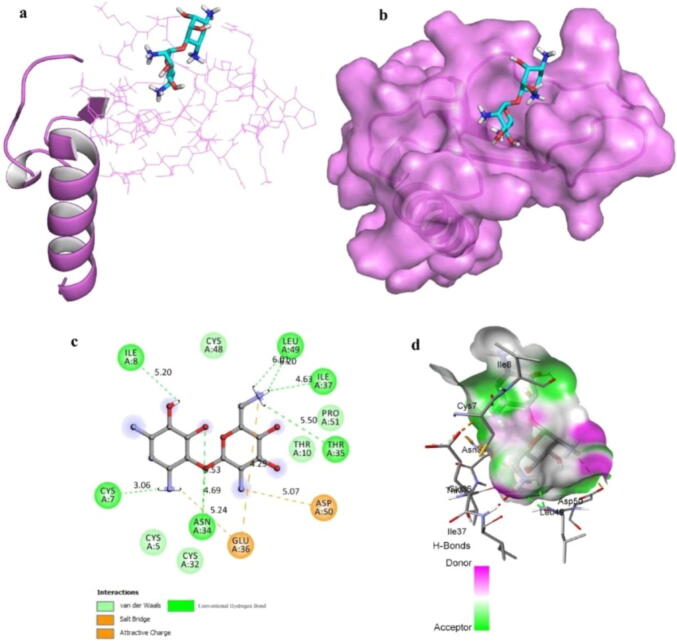
In Fig. 3a, we represent the complex formation of ZINC14613097 with the substrate binding site of IL8. Binding of inhibitor with in this cleft leads to reduced binding accessibility to ZINC14613097. We further carried out molecular docking analysis with IL8 and the small molecule to choose docking pose that was best on the entire surface of the target protein (Fig. 3b). We found that within the main groove of IL8 there is a strapping binding pattern of ZINC14613097.
Fig. 3.
a-d. The crystal structure of IL8 in complex with ZINC14613097. (a) A cartoon representation of IL8-ZINC14613097 complex. (b) Surface representation of IL8-ZINC14613097complex. ZINC14613097 is represented as coloured sticks. (c) A zoomed view of substrate binding pocket of IL8 representing the key amino acid residues forming interactions with inhibitor molecule ZINC14613097 (d) Surface representation of conserved substrate-binding pocket of IL8.
We further analyzed the bonding pattern between IL8 and ZINC14613097. Our results revealed there are many potent amino acid residues in IL8 which can form hydrogen bonds with ZINC14613097 in optimum bond length (Fig. 3c and 3d). Bond lengths are given in Fig. 3c. It was finally resulted that ZINC14613097 forms most of interactions (conventional hydrogen bonds) with CYS7, ASN34, THR35, ILE8, ILE37, and LEU49 amino acid residues of IL8. These hydrogen bonds are geometrically configured around the IL8 active site. Additionally, hydrogen bonds, IL8 also formed van der Waals with ZINC14613097 with CYS5, CYS32, CYS48, THR10, and PRO51 (Fig. 3c and 3d).
3.3. Dynamics of IL8-ZINC14613097 complex
Molecular dynamics simulations (50 ns) was used to comprehend the conformational changes, stability and interactions of both free IL8 and IL8-ZINC14613097 complex were analyzed. Preceding the Molecular Dynamics analysis, IL8-ZINC14613097 complex and free IL8 average potential energy were determined. Average potential energy of IL8 and IL8-ZINC14613097 complex were −337696 kJ mol-1 and −203177 kJ mol-1, respectively, supporting stability and equilibrium of systems. Further, system RMSD, RMSF, SASA, Rg, kinetic energy of the solvent, density and volume, enthalpy, were assigned and listed in Table 2.
Table 2.
Calculated MD parameters for IL8 and IL8-ZINC14613097 systems obtained after simulation.
| Average |
Average |
Average |
Average |
Kinetic |
Potential |
Enthalpy |
Volume |
Density |
|
|---|---|---|---|---|---|---|---|---|---|
| RMSD |
RMSF |
Rg |
SASA |
Energy |
energy |
kJ/mol |
(nm3) |
(kg/m3) |
|
| (nm) | (nm) | (nm) | (nm2) | (kJ/mol) | (kJ/mol) | ||||
| IL8 | 0.325722 | 0.165734 | 1.24268 | 57.1367 | 56160.4 | −337696 | −281522 | −220.338 | 1015.78 |
| IL8- | 0.404839 | 0.173671 | 1.27579 | 56.6026 | 39341.9 | −203177 | −163826 | −155.742 | 1005.74 |
| ZINC14613097 | |||||||||
| complex |
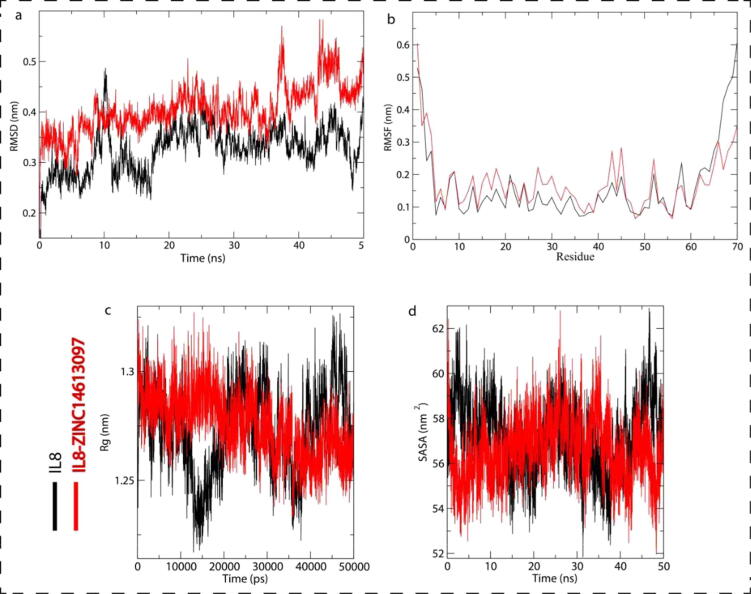
Binding of ligand to the target molecule can alter the structure, stability and can cause conformational changes (Mobley and Dill, 2009). The RMSD value conveys the structural and conformational deviations and stability (Kuzmanic and Zagrovic, 2010). The RMSD values were observed as 0.325722 nm and 0.404839 nm for IL8 and IL8-ZINC14613097, respectively (Table 2). RMSD plot revealed the ZINC14613097 attachment in IL8 binding pocket caused less deviation in structure and conformational from the native IL8 structure (Fig. 4a). However, some minor fluctuations were also observed because initially the orientation of ZINC14613097 was in binding pocket of IL8. After the initial fluctuations, we observed stable equilibrium of the system throughout the simulation. Our results suggest the stability and structure of IL8 was not altered by the binding of ZINC14613097 to IL8. Throughout the simulation RMSD was low at several parts and equilibration which suggests strongly the IL8-ZINC14613097 complex stability (Fig. 3a).
Fig. 4.
a-d: Structural dynamics of IL8 up on ZINC14613097 binding. (a) RMSD plot of IL8 and IL8-ZINC14613097 complex as a function of time. (b) RMSF plot of free IL8 and upon silmitasertib binding. (c) Time evolution of radius of gyration of IL8 and IL8-ZINC14613097 complex. (d) SASA plot of IL8 and IL8-ZINC14613097 complex as a function of time. The values were obtained from 50 ns MD simulations time scale. Black and red represents values obtained for free IL8 and IL8-ZINC14613097 complex, respectively.
The local structure flexibility of the IL8 and IL8-ZINC14613097 complex was determined using the RMSF value and is plotted in Fig. 4b. Table 2 summarizes the RMSF values. The average fluctuation of all residues is represented by the RMSF. It shows many residual fluctuations of IL8 structure at distinct regions, with ZINC14613097 binding were minimal during simulation bSut, binding of ZINC14613097 binding with IL8 resulted in several random residual fluctuations (Fig. 4b).
To understand further tertiary structure volume and overall conformational shape of a protein, we sought to compute the Rg, which reflects protein stability in a biological system. The flexible packing of the protein confers a higher Rg value. 1.24268 nm and 1.27579 nm were the average Rg values observed for IL8 and IL8-ZINC14613097 complex, respectively (Table 2). Insignificant deviations were observed in the Rg plot in the IL8 packingwith ZINC14613097 as a complex ensemble. Rg plot also presented high compactness initially, which can be due to tight packaging of IL8; but stable Rg equilibrium was achieved by IL8 in the subsequent simulation (Fig. 4c). There was a minimal structural deviation and no conformation changes on IL8 upon ZINC14613097 binding in the Rg plot. Throughout the simulation, the IL8 remained in tightly packed state even after the binding of ZINC14613097.
Next, we calculated the protein interaction surface area with surrounding solvent using SASA 50 ns Molecular Dynamic simulations (Mazola et al., 2015). SASA and the Rg of a protein are directly related. We observed an average SASA values of 57.1 367 nm2 and 58.6026 nm2 for IL8 and IL8-ZINC14613097 complex, respectively (Fig. 4d). Higher SASA values of IL8-ZINC14613097 complex might be due to the conformational change which leads to few residues of IL8 residues exposing to solvent.
3.4. MM-PBSA calculation
Table 3 presents the average free binding energy values along with standard deviations. Results of the MM-PBSA analysis showed that all forms of energy aid the interaction between IL8 and ZINC14613097.
Table 3.
MM-PBSA calculations of binding free energy for IL8-ZINC14613097 complex.
| Complex | Ebinding (kj/mol) | SASA (kj/mol) | Epolar solvation (kj/mol) | EElectrostatic (kj/mol) | EVan der Waal (kj/mol) |
|---|---|---|---|---|---|
| IL8-ZINC14613097 | 215.223 ± 14.124 | 24.921 ± 1.383 | 141.215 ± 15.418 | 53.281 ± 12.329 | 271.827 ± 19.215 |
| complex. |
4. Discussion
Despite the advances in molecular as well as computational techniques the diagnosis and treatment planning of endodontic pulpitis is not well explored. Treatments based on the molecular profiling of the disease is the first step towards precision treatment and provide better clinical outcome. Non-surgical endodontic therapies could be more prevalent among patients due to the minimal invasive medical procedure. In this study we explored the application of computational algorithms in drug discovery as an effective strategy in medical research, as it accelerates the discovery of drug molecules as well as their characterization. (Ou-Yang et al., 2012, Sultan et al., 2021, Wade and Goodford, 1989). Computational drug discovery is applied in all stages of drug researches such as in the discovery and development of particular drug, to identify the target of the drug, validation of the drug- target interaction, discovery of lead and the optimization process and also in preclinical studies(Ou-Yang et al., 2012, Sultan et al., 2021). Due to the availability of small molecules and biological macromolecules in larger scale, the biological experiment-based study for identifying the drugs for the target molecules will be hectic and time consuming. Currently, structure assisted screening method is the most widely implemented in discovering small molecules that exhibit drug like properties (Ou-Yang et al., 2012).
In this study, we carried out Molecular Docking analysis to identify the drug targeting IL8. We examined binding affinity, binding energy, and bound conformations of potential interaction of residual amino acid with intermolecular distances of the small molecules of IL8 (Guex and Peitsch, 1997, Vchirawongkwin et al., 2009). From our results, based upon the high binding energy value (-9.7Kcal/mol) and significant residual amino acid interactions, it was identified IL8. Was inhibited by ZINC14613097.
The best docking pose of small molecule on the surface of the protein opened ZINC14613097 in a strapping binding pattern within IL8. ZINC14613097 binds to IL8 main groove via the hydrogen bonds of the CYS7, ASN34, THR35, ILE8, ILE37, and LEU49 amino acid residues. The role of hydrogen-bonds in determining the ligand specific binding is previously reported (Wade and Goodford, 1989). We report the involvement of other type of interactions such as van der Waals in IL8 and ZINC14613097. The interaction of inhibitor molecule with IL8 via the hydrogen bonds and van der Waals collectively stabilize the complex (Wade and Goodford, 1989). The conformational changes, interaction and stability of free IL8 and IL8-ZINC14613097 complex were analysed by MD simulations (50 ns). Currently, MD simulations have advanced to comprehend macromolecular structure-to-function relationships. From the characterization of the MD parameters such as RMSD, RMSF, Rg, and SASA proved ZINC14613097 as potent inhibitor of IL8. The effect of ligand binding with the target proteins causes structural deviations, conformational changes and can also fluctuate the targeted macromolecules stability (Mobley and Dill, 2009). Calculation of RMSD values gives an insight into the structural and confirmational deviations and stability (Guex and Peitsch, 1997). Comparative RMSD values implicated minimum deviation from the native structure of IL8 upon ZINC14613097 binding strongly suggests the IL8- ZINC14613097 complex stablity. RMSF represents the local structure flexibility (Guex and Peitsch, 1997). Initially, many residual fluctuations of IL8 structure at distinct regions were found, which was reduced during the simulation process with ZINC14613097 binding, reflecting the structure flexibility of IL8-ZINC14613097 complex. The tertiary structure volume and overall conformational shape of a protein correlates with Rg (Guex and Peitsch, 1997). Rg plot showed structural deviation was minimum and there was no IL8 conformation change upon ZINC14613097 binding. Again, Rg plot revealed that throughout the simulation IL8 remained tightly packed. SASA describes the outer surface area of the protein which interacts with the solvents (Mazola et al., 2015). We found higher SASA for the IL8-ZINC14613097 complex than the IL8 alone. We hypothesis the binding of ZINC14613097 to the IL8 caused the conformational change which then led some of the internal residues of IL8 expose to solvent. The stability of the ZINC14613097-IL8 complex can be confirmed by using RMSD, RMSF, Rg, and SASA computations for molecular screening.
We also calculated IL8-ZINC14613097 complex the free binding energy with MM-PBSA analysis. Energy released during the interaction or bond formation process of ligand and target molecule is presented as binding energy (Sultan et al., 2021). It is inversely proportional to the ligand and protein binding. The sum of electrostatic, polar solvation, SASA energy and van der Wall is the final binding energy (Bhardwaj et al., 2021, Sultan et al., 2021). The MM-PBSA analysis showed all forms of energy contributed in the interaction between IL8 and ZINC14613097.
5. Conclusions
Relatively higher levels of IL8 have been found associated with inflamed pulps and moderate to severe pulpitis pain conditions as compared to normal zero to mild pulpitis pain conditions. It is speculated that inhibition of IL8 might be useful for the drug development and therapeutic management of endodontic pulpitis. Our structure-assisted drug designing approaches, simulation analysis and molecular docking revealed a small molecule ZINC14613097 could be a potential inhibitor of the proteinIL8. Our study is the first step to identify a potential inhibitor of IL8, which later can be validated and implemented in clinics for treating endodontic pulpitis.
Funding
This research received no external funding
Data Availability Statement
All the data has been included in the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
College of Dentistry, Jezan University, Kingdom of Saudi Arabia is highly acknowledged for providing research facilities.
Footnotes
Peer review under responsibility of King Saud University.
Contributor Information
Nezar Boreak, Email: nboraak@jazanu.edu.sa.
Shilpa Bhandi, Email: shilpa.bhandi@gmail.com.
References
- Ahlquist M.L., Franzen O.G. Inflammation and dental pain in man. Dent. Traumatol. 1994;10(5):201–209. doi: 10.1111/j.1600-9657.1994.tb00070.x. [DOI] [PubMed] [Google Scholar]
- Avellán N.-L., Sorsa T., Tervahartiala T., Forster C., Kemppainen P. Experimental tooth pain elevates substance P and matrix metalloproteinase-8 levels in human gingival crevice fluid. Acta Odontol. Scand. 2008;66(1):18–22. doi: 10.1080/00016350701810658. [DOI] [PubMed] [Google Scholar]
- Bhardwaj V.K., Singh R., Sharma J., Rajendran V., Purohit R., Kumar S. Identification of bioactive molecules from tea plant as SARS-CoV-2 main protease inhibitors. J. Biomol. Struct. Dyn. 2021;39(10):3449–3458. doi: 10.1080/07391102.2020.1766572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boreak N., Vozza I. Small Molecule “Silmitasertib” Repurposed as Inhibitor of Transforming Growth Factor Beta 1 for the Development of Therapeutics for Oral Submucous Fibrosis. Biomed Res. Int. 2021;2021:1–8. doi: 10.1155/2021/6631848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caviedes-Bucheli J., Arenas N., Guiza O., Moncada N.A., Moreno G.C., Diaz E., Munoz H.R. Calcitonin gene-related peptide receptor expression in healthy and inflamed human pulp tissue. Int. Endod. J. 2005;38(10):712–717. doi: 10.1111/j.1365-2591.2005.01006.x. [DOI] [PubMed] [Google Scholar]
- Caviedes-Bucheli J., Gutierrez-Guerra J.E., Salazar F., Pichardo D., Moreno G.C., Munoz H.R. Substance P receptor expression in healthy and inflamed human pulp tissue. Int. Endod. J. 2007;40(2):106–111. doi: 10.1111/j.1365-2591.2006.01189.x. [DOI] [PubMed] [Google Scholar]
- CAVIEDESBUCHELI J., MUNOZ H., AZUEROHOLGUIN M., ULATE E. Neuropeptides in Dental Pulp: The Silent Protagonists. J. Endod. 2008;34(7):773–788. doi: 10.1016/j.joen.2008.03.010. [DOI] [PubMed] [Google Scholar]
- Esmaeili A., Akhavan A., Bouzari M., Mousavi S.B., Torabinia N., Adibi S. Temporal expression pattern of sodium channel Nav 1.8 messenger RNA in pulpitis. Int. Endod. J. 2011;44:499–504. doi: 10.1111/j.1365-2591.2011.01853.x. [DOI] [PubMed] [Google Scholar]
- Galicia J.C., Henson B.R., Parker J.S., Khan A.A. Gene expression profile of pulpitis. Genes Immun. 2016;17(4):239–243. doi: 10.1038/gene.2016.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong Q., Jiang H., Wei X.i., Ling J., Wang J. Expression of Erythropoietin and Erythropoietin Receptor in Human Dental Pulp. J. Endod. 2010;36(12):1972–1977. doi: 10.1016/j.joen.2010.08.041. [DOI] [PubMed] [Google Scholar]
- Guex N., Peitsch M.C. SWISS-MODEL and the Swiss-Pdb Viewer: An environment for comparative protein modeling. Electrophoresis. 1997;18(15):2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- Hahn C.-L., Falkler W.A., Minah G.E. Microbiological studies of carious dentine from human teeth with irreversible pulpitis. Arch. Oral Biol. 1991;36(2):147–153. doi: 10.1016/0003-9969(91)90077-8. [DOI] [PubMed] [Google Scholar]
- Huang F.-M., Tsai C.-H., Yang S.-F., Chang Y.-C. The upregulation of oncostatin M in inflamed human dental pulps. Int. Endod. J. 2009;42:627–631. doi: 10.1111/j.1365-2591.2009.01567.x. [DOI] [PubMed] [Google Scholar]
- Jiang H.-W., Ling J.-q., Gong Q.-M. The Expression of Stromal Cell–derived Factor 1 (SDF-1) in Inflamed Human Dental Pulp. J. Endod. 2008;34(11):1351–1354. doi: 10.1016/j.joen.2008.07.023. [DOI] [PubMed] [Google Scholar]
- Kaneko T., Okiji T., Kaneko R., Sunakawa M., Kaneko M., Suda H. Gene Expression Analysis of Acutely Traumatized Pulps. J. Endod. 2010;36(1):78–82. doi: 10.1016/j.joen.2009.09.011. [DOI] [PubMed] [Google Scholar]
- Karapanou V., Kempuraj D., Theoharides T.C. Interleukin-8 Is Increased in Gingival Crevicular Fluid from Patients with Acute Pulpitis. J. Endod. 2008;34(2):148–151. doi: 10.1016/j.joen.2007.10.022. [DOI] [PubMed] [Google Scholar]
- Keller J.-F., Carrouel F., Staquet M.-J., Kufer T.A., Baudouin C., Msika P., Bleicher F., Farges J.-C. Expression of NOD2 is increased in inflamed human dental pulps and lipoteichoic acid-stimulated odontoblast-like cells. Innate Immun. 2011;17(1):29–34. doi: 10.1177/1753425909348527. [DOI] [PubMed] [Google Scholar]
- Kerwin S.M. ChemBioOffice Ultra 2010 Suite. J. Am. Chem. Soc. 2010;132(7):2466–2467. doi: 10.1021/ja1005306. [DOI] [PubMed] [Google Scholar]
- Kokkas A.B., Goulas A., Varsamidis K., Mirtsou V., Tziafas D. Irreversible but not reversible pulpitis is associated with up-regulation of tumour necrosis factor-alpha gene expression in human pulp. Int. Endod. J. 2007;40(3):198–203. doi: 10.1111/j.1365-2591.2007.01215.x. [DOI] [PubMed] [Google Scholar]
- Korkmaz Y., Lang H., Beikler T., Cho B., Behrends S., Bloch W., Addicks K., Raab W.-H.-M. Irreversible Inflammation is Associated with Decreased Levels of the α 1 -, β 1 -, and α 2 -Subunits of sGC in Human Odontoblasts. J. Dent. Res. 2011;90(4):517–522. doi: 10.1177/0022034510390808. [DOI] [PubMed] [Google Scholar]
- Kumari R., Kumar R., Lynn A. g_mmpbsa —A GROMACS Tool for High-Throughput MM-PBSA Calculations. J. Chem. Inf. Model. 2014;54(7):1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- Kuzmanic A., Zagrovic B. Determination of Ensemble-Average Pairwise Root Mean-Square Deviation from Experimental B-Factors. Biophys. J. 2010;98(5):861–871. doi: 10.1016/j.bpj.2009.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lundy F.T., About I., Curtis T.M., McGahon M.K., Linden G.J., Irwin C.R., El Karim I.A. PAR-2 Regulates Dental Pulp Inflammation Associated with Caries. J. Dent. Res. 2010;89(7):684–688. doi: 10.1177/0022034510365652. [DOI] [PubMed] [Google Scholar]
- Mazola Y., Guirola O., Palomares S., Chinea G., Menéndez C., Hernández L., Musacchio A. A comparative molecular dynamics study of thermophilic and mesophilic β-fructosidase enzymes. J. Mol. Model. 2015;21:228. doi: 10.1007/s00894-015-2772-4. [DOI] [PubMed] [Google Scholar]
- McLachlan J.L., Smith A.J., Bujalska I.J., Cooper P.R. Gene expression profiling of pulpal tissue reveals the molecular complexity of dental caries. Biochim. Biophys. Acta - Mol. Basis Dis. 2005;1741(3):271–281. doi: 10.1016/j.bbadis.2005.03.007. [DOI] [PubMed] [Google Scholar]
- Mobley D.L., Dill K.A. Binding of Small-Molecule Ligands to Proteins: “What You See” Is Not Always “What You Get”. Structure. 2009;17(4):489–498. doi: 10.1016/j.str.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mutoh N., Watabe H., Chieda K., Tani-Ishii N. Expression of Toll-Like Receptor 2 and 4 in Inflamed Pulp in Severe Combined Immunodeficiency Mice. J. Endod. 2009;35(7):975–980. doi: 10.1016/j.joen.2009.04.005. [DOI] [PubMed] [Google Scholar]
- Ou-Yang S.-S., Lu J.-Y., Kong X.-Q., Liang Z.-J., Luo C., Jiang H. Computational drug discovery. Acta Pharmacol. Sin. 2012;33(9):1131–1140. doi: 10.1038/aps.2012.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rechenberg D.-K., Galicia J.C., Peters O.A., Kerkis I. Biological Markers for Pulpal Inflammation: A Systematic Review. PLoS One. 2016;11(11):e0167289. doi: 10.1371/journal.pone.0167289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith A.J. Pulpal Responses to Caries and Dental Repair. Caries Res. 2002;36:223–232. doi: 10.1159/000063930. [DOI] [PubMed] [Google Scholar]
- Staquet M.-J., Carrouel F., Keller J.-F., Baudouin C., Msika P., Bleicher F., Kufer T.A., Farges J.-C. Pattern-recognition Receptors in Pulp Defense. Adv. Dent. Res. 2011;23(3):296–301. doi: 10.1177/0022034511405390. [DOI] [PubMed] [Google Scholar]
- Sultan A., Ali R., Sultan T., Ali S., Khan N.J., Parganiha A. Circadian clock modulating small molecules repurposing as inhibitors of SARS-CoV-2 M pro for pharmacological interventions in COVID-19 pandemic. Chronobiol. Int. 2021;38(7):971–985. doi: 10.1080/07420528.2021.1903027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tete S., Mastrangelo F., Scioletti A.P., Tranasi M., Raicu F., Paolantonio M., Stuppia L., Vinci R., Gherlone E., Ciampoli C., Sberna M.T., Conti P. Microarray expression profiling of human dental pulp from single subject. Clin. Investig. Med. 2008;31:55. doi: 10.25011/cim.v31i2.3364. [DOI] [PubMed] [Google Scholar]
- Vchirawongkwin V., Pribil A.B., Rode B.M. Ab Initio quantum mechanical charge field study of hydrated bicarbonate ion: Structural and dynamical properties. J. Comput. Chem. NA-NA. 2009:NA–NA. doi: 10.1002/jcc.21308. [DOI] [PubMed] [Google Scholar]
- Wade R.C., Goodford P.J. The role of hydrogen-bonds in drug binding. Prog. Clin. Biol. Res. 1989;289:433–444. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
All the data has been included in the manuscript.