Key Points
Question
What is the association between 3 doses of mRNA COVID-19 vaccine and symptomatic SARS-CoV-2 infection with the Omicron and Delta variants?
Findings
In this test-negative case-control analysis that included 70 155 tests from symptomatic adults, the likelihood of vaccination with 3 mRNA vaccine doses (vs unvaccinated) was significantly lower among both Omicron (odds ratio, 0.33) and Delta (odds ratio, 0.065) cases than SARS-CoV-2–negative controls; a similar pattern was observed with 3 vaccine doses vs 2 doses (Omicron odds ratio, 0.34; Delta odds ratio, 0.16).
Meaning
These findings suggest that vaccination with 3 doses of mRNA COVID-19 vaccine, compared with being unvaccinated and with receipt of 2 doses, was associated with protection against both the Omicron and Delta variants, although higher odds ratios for the association with Omicron infection suggest less protection for Omicron than for Delta.
Abstract
Importance
Assessing COVID-19 vaccine performance against the rapidly spreading SARS-CoV-2 Omicron variant is critical to inform public health guidance.
Objective
To estimate the association between receipt of 3 doses of Pfizer-BioNTech BNT162b2 or Moderna mRNA-1273 vaccine and symptomatic SARS-CoV-2 infection, stratified by variant (Omicron and Delta).
Design, Setting, and Participants
A test-negative case-control analysis among adults 18 years or older with COVID-like illness tested December 10, 2021, through January 1, 2022, by a national pharmacy-based testing program (4666 COVID-19 testing sites across 49 US states).
Exposures
Three doses of mRNA COVID-19 vaccine (third dose ≥14 days before test and ≥6 months after second dose) vs unvaccinated and vs 2 doses 6 months or more before test (ie, eligible for a booster dose).
Main Outcomes and Measures
Association between symptomatic SARS-CoV-2 infection (stratified by Omicron or Delta variants defined using S-gene target failure) and vaccination (3 doses vs unvaccinated and 3 doses vs 2 doses). Associations were measured with multivariable multinomial regression. Among cases, a secondary outcome was median cycle threshold values (inversely proportional to the amount of target nucleic acid present) for 3 viral genes, stratified by variant and vaccination status.
Results
Overall, 23 391 cases (13 098 Omicron; 10 293 Delta) and 46 764 controls were included (mean age, 40.3 [SD, 15.6] years; 42 050 [60.1%] women). Prior receipt of 3 mRNA vaccine doses was reported for 18.6% (n = 2441) of Omicron cases, 6.6% (n = 679) of Delta cases, and 39.7% (n = 18 587) of controls; prior receipt of 2 mRNA vaccine doses was reported for 55.3% (n = 7245), 44.4% (n = 4570), and 41.6% (n = 19 456), respectively; and being unvaccinated was reported for 26.0% (n = 3412), 49.0% (n = 5044), and 18.6% (n = 8721), respectively. The adjusted odds ratio for 3 doses vs unvaccinated was 0.33 (95% CI, 0.31-0.35) for Omicron and 0.065 (95% CI, 0.059-0.071) for Delta; for 3 vaccine doses vs 2 doses the adjusted odds ratio was 0.34 (95% CI, 0.32-0.36) for Omicron and 0.16 (95% CI, 0.14-0.17) for Delta. Median cycle threshold values were significantly higher in cases with 3 doses vs 2 doses for both Omicron and Delta (Omicron N gene: 19.35 vs 18.52; Omicron ORF1ab gene: 19.25 vs 18.40; Delta N gene: 19.07 vs 17.52; Delta ORF1ab gene: 18.70 vs 17.28; Delta S gene: 23.62 vs 20.24).
Conclusions and Relevance
Among individuals seeking testing for COVID-like illness in the US in December 2021, receipt of 3 doses of mRNA COVID-19 vaccine (compared with unvaccinated and with receipt of 2 doses) was less likely among cases with symptomatic SARS-CoV-2 infection compared with test-negative controls. These findings suggest that receipt of 3 doses of mRNA vaccine, relative to being unvaccinated and to receipt of 2 doses, was associated with protection against both the Omicron and Delta variants, although the higher odds ratios for Omicron suggest less protection for Omicron than for Delta.
This case-control study uses data from the Centers for Disease Control and Prevention Increasing Community Access to Testing platform to estimate the association between receipt of 3 doses of Pfizer-BioNTech BNT162b2 or Moderna mRNA-1273 vaccine, compared with 2 doses or unvaccinated, and symptomatic SARS-CoV-2 infection, stratified by Omicron and Delta variant.
Introduction
On November 24, 2021, health authorities in South Africa reported the emergence of a new SARS-CoV-2 variant, B.1.1.529 (Omicron).1 Omicron has spread rapidly, and as of January 6, 2022, was identified in 149 countries across all 6 World Health Organization regions.2 Omicron was first detected in the US on December 1, 2021, and by January 1, 2022, was estimated to be responsible for 95% of sequenced new cases.3,4
Sequencing of early Omicron strains documented more than 30 mutations in the spike protein, including in the receptor binding domain.5,6 These mutations, combined with observed exponential growth in case counts, even in settings with substantial rates of COVID-19 vaccination or previous SARS-CoV-2 infection, raised concerns about potential for increased transmissibility and immune escape.2,7,8,9,10 There is an urgent need to understand the protection provided by current vaccination regimens against Omicron, including any additional protection derived from booster doses.
In this analysis, a subset of data from the national Increasing Community Access to Testing (ICATT) platform was used to estimate the association of receipt of 3 doses of a mRNA COVID-19 vaccine (vs unvaccinated and vs 2 doses) with symptomatic infection with the Omicron and Delta variants, using an internally validated genetic proxy for variant identification.
Methods
Study Protocol Approval
The human subjects advisor for the Centers for Disease Control and Prevention (CDC) National Center for Immunization and Respiratory Diseases determined that this analysis met the requirements for public health surveillance as outlined in 45 CFR §46.102(l)(2). Because data were collected during routine operational procedures, this secondary data analysis did not require informed consent and was conducted consistent with applicable federal law and CDC policy.
Data Source
Data from the ICATT platform11—a Department of Health and Human Services (HHS) partnership facilitating no-cost, drive-through SARS-CoV-2 testing at pharmacies across all 50 states, the District of Columbia, and Puerto Rico—were analyzed. Testing sites were selected by HHS to prioritize access in racially and ethnically diverse communities and areas with moderate-to-high social vulnerability. Data for this analysis were limited to tests occurring between December 10, 2021, and January 1, 2022, at testing sites that collected booster vaccination history and sent specimens to a single laboratory chain (Aegis Sciences Corp) for processing. The laboratory used the TaqPath COVID-19 Combo Kit (Thermo Fisher Scientific), which identifies SARS-CoV-2 infections by detecting 3 targets from the viral ORF1ab, S, and N gene regions. The laboratory reported overall test results and cycle threshold (Ct) values for each of these targets for SARS-CoV-2–positive specimens.
Individuals registered online for testing at drive-through sites where nasal swabs were collected. During registration, individuals self-reported symptom status (asymptomatic or symptomatic with ≥1 COVID-like symptom), race, ethnicity, sex, age, state of residence, history of prior SARS-CoV-2 infection, and underlying conditions. Fixed categories were used to capture data for symptoms, race, ethnicity, and underlying chronic conditions including the presence of an immunocompromising condition (defined in the questionnaire as “such as from immunocompromising medications, solid organ or blood stem cell transplant, HIV, or other immunocompromising conditions”). Race and ethnicity were collected as required data elements under HHS COVID-19 laboratory reporting requirements.12 The testing program geocoded testing sites to identify their census tract Social Vulnerability Index (SVI) score.13
Patients also self-reported COVID-19 vaccination status, including the number of doses (up to 4), product, and month and year of receipt for each dose. For doses received in the same month or the month prior to testing, an additional question was asked to specify whether the dose was received 14 days or more before testing. Vaccination reporting was not mandatory, and information was not verified. Data were reported to HHS with an estimated 3-day lag and were deidentified to remove any personally identifying information.
Study Design
A retrospective test-negative case-control analysis was conducted on samples collected from December 10, 2021, to January 1, 2022, from adults 18 years or older with symptomatic COVID-like illness. The test-negative design is a commonly used observational method for evaluating the performance of vaccines in which participants are enrolled based on a clinical case definition, tested for the vaccine-preventable outcome of interest, and classified as cases or controls based on that testing; the odds of prior vaccination among cases and controls are compared as an estimate of the association between vaccination and the outcome.14,15 A strength of the test-negative design is that all participants seek care (or testing) for a common clinical case definition, which can help reduce bias resulting from confounding by differential care-seeking behavior.16,17
The unit of analysis for this study was tests; positive results were classified as cases, and negative results as controls. The short study period and restriction to symptomatic individuals limited the probability of individuals contributing more than 1 test.
Because protection from mRNA COVID-19 vaccines is substantially lower in immunocompromised individuals18 and the recommended vaccine dosing regimen is different from that of immunocompetent individuals,19 tests from individuals reporting an immunocompromising condition were excluded. Tests from persons reporting a positive COVID-19 test result within the previous 90 days were excluded to reduce potential misclassification. Tests with unknown vaccination status or incomplete vaccination data (ie, missing vaccination dates or products), from individuals reporting prior receipt of 1 or 4 mRNA vaccine doses or of non-mRNA COVID-19 vaccines, or from persons with improbable ages (defined as >100 years) were also excluded.
Additionally, among tests with positive results, Ct values were described by variant and by vaccination status (3 doses, 2 doses, and unvaccinated). Ct values reflect the number of cycles during polymerase chain reaction amplification needed to detect viral genetic material and are inversely proportional to the amount of target nucleic acid in the tested sample.20 Ct values were examined to better understand the relative amounts of genetic material present in positive samples by variant and vaccination status.
Exposure
The exposure of interest was self-report of any 3 doses of BNT162b2 (Pfizer-BioNTech) or mRNA-1273 (Moderna) vaccine (including mixed-product regimens) vs unvaccinated and vs any 2 doses of BNT162b2 or mRNA-1273 vaccine. For individuals reporting 2 vaccine doses, tests were excluded if the second dose was received less than 6 months prior to test date to ensure eligibility for a booster dose. For those reporting 3 doses, tests were excluded if the interval between second and third doses was less than 6 months, as per recommendations during the analysis period for booster doses among immunocompetent individuals,19 or if the most recent dose was received less than 14 days before testing. Another exposure examined was any 2 doses of BNT162b2 or mRNA-1273 vaccine, with the second dose received 14 days or more before testing vs unvaccinated; assessment of this exposure did not limit to those eligible for a booster dose (ie, did not limit to those with 6 or more months elapsed between the second dose and date of testing).
Outcomes
The primary outcome was symptomatic SARS-CoV-2 infection with the Omicron or Delta variant. For this analysis, an Omicron case was defined as presence of S-gene target failure (SGTF) in the test sample and a Delta case as absence of SGTF in the test sample. All samples that were determined by the processing laboratory to be SARS-CoV-2 positive had Ct values for at least 2 of the N, ORF1ab, and S genes. SARS-CoV-2–positive samples were considered to have SGTF if they had Ct values for the N and ORF1ab genes but not for the S gene; otherwise, samples were considered not to have SGTF.
SGTF may serve as a proxy for the presence of the Omicron variant in samples tested with the TaqPath COVID-19 Combo Kit assay because of the presence of deletions in the S-gene region for Omicron that are not present in Delta2,21; the deletions lead to S-gene–negative results in Omicron lineages BA.1 and B.1.1.529 but not the majority of Delta samples. While levels of non-Delta circulating variants other than Omicron remain low, samples with SGTF may be presumed to be Omicron.22 At the time of this analysis 99.9% of sequenced samples in the US prior to the emergence of Omicron were Delta.4 To validate the use of SGTF as a proxy for Omicron in the ICATT data, the frequency of SGTF was examined in a randomly selected subset of tests with positive results that were sequenced during the same period as the analysis. The sequenced subset was drawn from the complete database of test results from the laboratory, including tests not eligible for the main analysis. Sequencing data were not available for most cases included in the main analysis, which is why SGTF was used as a proxy. The sensitivity of SGTF for detecting Omicron (B.1.1.529 or BA.1 lineage) was 83.4% and the specificity was 99.2% (eTable 1 in the Supplement).
Secondary outcome measures included Ct values for the N and ORF1ab genes among Omicron and Delta cases and S gene among Delta cases.
Statistical Analysis
The association between symptomatic infection with the Omicron or Delta variants and vaccination was estimated by comparing the odds of prior 3-dose vaccination vs unvaccinated and the odds of prior 3-dose vaccination vs 2-dose vaccination in cases vs controls using multivariable multinomial logistic regression. The odds ratio (OR) for 3 doses vs unvaccinated was used as an estimate of 3-dose vaccine effectiveness (effectiveness = [1 – OR] × 100%), with lower ORs suggesting more protection. The OR for 3 doses vs 2 doses was used as an estimate of relative vaccine effectiveness, reflecting additional protection from a booster dose relative to 2 doses. ORs were estimated for any combination of mRNA vaccine and separately for BNT162b2 and mRNA-1273.
Models included the number of days between the start of the analysis period and test date (as a continuous variable), age group, sex, race, ethnicity, testing site HHS region, testing site census tract SVI (dichotomized as 0 to <0.5 and ≥0.5-1), and number of underlying chronic conditions (0, 1, or ≥2) as covariates to adjust for potential confounding bias. Unknown race and ethnicity were coded as categories of their respective variables instead of null values to retain these records in regression models. Data with missing values for other model covariates (specifically, sex and SVI) were coded as null values and therefore dropped from adjusted regression models.
Two-sided 95% CIs were calculated for each reported OR, with 95% CIs that excluded 1 considered statistically significant. Two-sided P values for the association between vaccination and symptomatic infection with Omicron compared with Delta were corrected for false-discovery rate (FDR) using the Benjamini-Hochberg method (to account for type I error due to multiple comparisons), and results with Q values (ie, P values adjusted for the FDR) less than .001 were considered statistically significant.
To aid in the interpretation of associations of 3 vaccine doses vs unvaccinated and vs 2 vaccine doses, a secondary analysis was performed examining the association between 2 doses vs unvaccinated by time since receipt of second dose. The association between infection and 2 mRNA vaccine doses vs unvaccinated was examined separately for each product-variant combination by logistic regression incorporating the same covariates as above as well as month (0-11) since second dose using a 2-knot spline at months 3.5 and 7.5.
For comparison of Ct values among cases by variant and exposure status (3 doses vs unvaccinated, 3 doses vs 2 doses, and 2 doses vs unvaccinated), the 2-sided Mann-Whitney U test was used to identify significant differences in median Ct values. Correction for FDR was applied for each comparison of exposure status, and results with Q < .001 were considered statistically significant.
Statistical analyses were performed in RStudio and R, version 4.0.3 (R Foundation). Multinomial logistic regression was performed using the nnet R package, version 7.3-16.
Results
A total of 70 155 tests from 4666 sites on samples collected between December 10 and January 1 across 49 states met inclusion criteria (Figure 1), including 23 391 cases (13 098 Omicron; 10 293 Delta) and 46 764 controls (Table 1) (mean age, 40.3 [SD, 15.6] years; 42 050 [60.1%] women). Included tests were most frequently performed on persons aged 25 to 34 years (30.4%), followed by those aged 35 to 44 years (19.3%), and on persons who reported being of White race (75.5%). More than one-third of tests (36.4%) were from people with reported underlying health conditions, with high blood pressure most common, followed by overweight. Compared with controls, cases were more frequently tests from persons aged 25 to 34 years (Omicron 35.1% and Delta 31.0% vs controls 28.9%), of Black/African American race (Omicron 24.4% and Delta 14.9% vs controls 11.9%), and of Hispanic/Latino ethnicity (Omicron 22.5% and Delta 17.7% vs controls 17.4%) (Table 1).
Figure 1. Inclusion Criteria for a Case-Control Study of Association Between 3 Doses of COVID-19 mRNA Vaccine and Symptomatic SARS-CoV-2 Infection Caused by the Omicron or Delta Variants.
Nucleic acid amplification tests (NAATs) were from the Increasing Community Access to Testing platform. EUA indicates Emergency Use Authorization.
aNine tests were removed because of reported ages older than 100 years.
bOf all tests with positive results, 17 620 were randomly selected for whole-genome sequencing and used to perform internal validation of S-gene target failure as a marker for the Omicron variant (eTable 1 in the Supplement). This included 4905 tests also included in the main case-control analysis.
cDuring online registration for SARS-CoV-2 testing, individuals self-reported their COVID-19 vaccination status, including the number of doses (up to 4), product, and month and year of receipt for each dose. Vaccination data were considered incomplete if the number of doses reported did not match the products and dates reported.
dThese 17 177 samples were used for the main case-control analysis that compared vaccination status between Omicron and Delta cases and SARS-CoV-2–negative controls. This included 2048 tests also included in the internal validation of S-gene target failure as a marker for the Omicron variant.
eFor vaccine products, individuals could select from 4 options: Johnson & Johnson/Janssen, Pfizer-BioNTech, Moderna, and “other.”
fPersons were considered eligible for a booster if they reported 2 doses, with a testing date 6 months or more after the second dose, or if they reported 3 doses, with the third dose 6 months or more after the second dose.
gThese 52 978 samples were used for the main case-control analysis that compared vaccination status between Omicron and Delta cases and SARS-CoV-2–negative controls. This included 2857 tests also included in the internal validation of S-gene target failure as a marker for the Omicron variant.
Table 1. Characteristics of SARS-CoV-2 Tests Included in a Case-Control Analysis of Association Between Symptomatic SARS-CoV-2 Infection Caused by the Omicron or Delta Variants and mRNA COVID-19 Vaccination, by Case-Control and Vaccination Status, Increasing Community Access to Testing Platform, December 10, 2021, to January 1, 2022.
| Characteristic | No. (%) | ||||||
|---|---|---|---|---|---|---|---|
| Cases | Controls (SARS-CoV-2–negative) (n = 46 764) | Unvaccinated (n = 17 177) | Doses of mRNA COVID-19 vaccine | Overall (N = 70 155) | |||
| Omicron (n = 13 098) | Delta (n = 10 293) | 2 (n = 31 271)a | 3 (n = 21 707)b | ||||
| SARS-CoV-2 test resultc | |||||||
| Omicron variant–positive | 13 098 (100) | 0 | 0 | 3412 (19.9) | 7245 (23.2) | 2441 (11.2) | 13 098 (18.7) |
| Delta variant–positive | 0 | 10 293 (100) | 0 | 5044 (29.4) | 4570 (14.6) | 679 (3.1) | 10 293 (14.7) |
| SARS-CoV-2–negative | 0 | 0 | 46 764 (100) | 8721 (50.8) | 19 456 (62.2) | 18 587 (85.6) | 46 764 (66.7) |
| Vaccination historyd | |||||||
| Unvaccinated | 3412 (26.0) | 5044 (49.0) | 8721 (18.6) | 17 177 (100) | 0 | 0 | 17 177 (24.5) |
| 2 Doses | 7245 (55.3) | 4570 (44.4) | 19 456 (41.6) | 0 | 31 271 (100) | 0 | 31 271 (44.6) |
| 3 Doses | 2441 (18.6) | 679 (6.6) | 18 587 (39.7) | 0 | 0 | 21 707 (100) | 21 707 (30.9) |
| Age group, y | |||||||
| 18-24 | 2471 (18.9) | 1713 (16.6) | 6624 (14.2) | 3838 (22.3) | 5471 (17.5) | 1499 (6.9) | 10 808 (15.4) |
| 25-34 | 4591 (35.1) | 3191 (31.0) | 13 526 (28.9) | 6072 (35.3) | 10 082 (32.2) | 5154 (23.7) | 21 308 (30.4) |
| 35-44 | 2588 (19.8) | 2171 (21.1) | 8756 (18.7) | 3453 (20.1) | 6188 (19.8) | 3874 (17.8) | 13 515 (19.3) |
| 45-54 | 1707 (13.0) | 1515 (14.7) | 6245 (13.4) | 2046 (11.9) | 4430 (14.2) | 2991 (13.8) | 9467 (13.5) |
| 55-64 | 1153 (8.8) | 1131 (11.0) | 6259 (13.4) | 1221 (7.1) | 3449 (11.0) | 3873 (17.8) | 8543 (12.2) |
| ≥65 | 588 (4.5) | 572 (5.6) | 5354 (11.4) | 547 (3.2) | 1651 (5.3) | 4316 (19.9) | 6514 (9.3) |
| Sex | (n = 13 074) | (n = 10 273) | (n = 46 601) | (n = 17 151) | (n = 31 164) | (n = 21 633) | (n = 69 948) |
| Women | 7577 (58.0) | 5392 (52.5) | 29 081 (62.4) | 9538 (55.6) | 18 744 (60.1) | 13 768 (63.6) | 42 050 (60.1) |
| Men | 5497 (42.0) | 4881 (47.5) | 17 520 (37.6) | 7613 (44.4) | 12 420 (39.9) | 7865 (36.4) | 27 898 (39.9) |
| Racee | (n = 9861) | (n = 7355) | (n = 35 485) | (n = 12 950) | (n = 23 059) | (n = 16 692) | (n = 52 701) |
| American Indian or Alaska Native | 86 (0.9) | 92 (1.3) | 357 (1.0) | 210 (1.6) | 243 (1.1) | 82 (0.5) | 535 (1.0) |
| Asian | 851 (8.6) | 420 (5.7) | 3113 (8.8) | 305 (2.4) | 2468 (10.7) | 1611 (9.7) | 4384 (8.3) |
| Black or African American | 2405 (24.4) | 1098 (14.9) | 4226 (11.9) | 3348 (25.9) | 3089 (13.4) | 1292 (7.7) | 7729 (14.7) |
| Native Hawaiian or Other Pacific Islander | 58 (0.6) | 52 (0.7) | 143 (0.4) | 88 (0.7) | 121 (0.5) | 44 (0.3) | 253 (0.5) |
| White | 6461 (65.5) | 5693 (77.4) | 27 646 (77.9) | 8999 (69.5) | 17 138 (74.3) | 13 663 (81.9) | 39 800 (75.5) |
| Hispanic/Latino ethnicity, No./total (%)f | 2720/12 076 (22.5) | 1656/9364 (17.7) | 7516/43 308 (17.4) | 3220/15 596 (20.6) | 6217/28 798 (21.6) | 2455/20 354 (12.1) | 11 892/64 748 (18.4) |
| HHS regiong | |||||||
| East North Central | 2769 (21.1) | 3046 (29.6) | 9709 (20.8) | 4183 (24.4) | 6541 (20.9) | 4800 (22.1) | 15 524 (22.1) |
| South Atlantic | 3599 (27.5) | 1642 (16.0) | 9440 (20.2) | 3686 (21.5) | 6607 (21.1) | 4388 (20.2) | 14 681 (20.9) |
| West South Central | 1933 (14.8) | 711 (6.9) | 4440 (9.5) | 1595 (9.3) | 3420 (10.9) | 2069 (9.5) | 7084 (10.1) |
| Pacific | 924 (7.1) | 781 (7.6) | 5104 (10.9) | 1565 (9.1) | 3212 (10.3) | 2032 (9.4) | 6809 (9.7) |
| Mid-Atlantic | 925 (7.1) | 903 (8.8) | 3881 (8.3) | 1130 (6.6) | 2730 (8.7) | 1849 (8.5) | 5709 (8.1) |
| West North Central | 580 (4.4) | 966 (9.4) | 3594 (7.7) | 1308 (7.6) | 2073 (6.6) | 1759 (8.1) | 5140 (7.3) |
| East South Central | 1071 (8.2) | 706 (6.9) | 3291 (7.0) | 1548 (9.0) | 2067 (6.6) | 1453 (6.7) | 5068 (7.2) |
| Mountain | 577 (4.4) | 692 (6.7) | 3211 (6.9) | 1139 (6.6) | 1918 (6.1) | 1423 (6.6) | 4480 (6.4) |
| New England | 413 (3.2) | 784 (7.6) | 3282 (7.0) | 935 (5.4) | 1996 (6.4) | 1548 (7.1) | 4479 (6.4) |
| Puerto Rico | 307 (2.3) | 62 (0.6) | 812 (1.7) | 88 (0.5) | 707 (2.3) | 386 (1.8) | 1181 (1.7) |
| Site census tract SVIh | (n = 12 787) | (n = 10 228) | (n = 45 939) | (n = 17 089) | (n = 30 555) | (n = 21 310) | (n = 68 954) |
| 0 to <0.5 (less vulnerable) | 5940 (46.5) | 4916 (48.1) | 22 716 (49.4) | 6881 (40.3) | 15 217 (49.8) | 11 474 (53.8) | 33 572 (48.7) |
| 0.5-1.0 (more vulnerable) | 6847 (53.5) | 5312 (51.9) | 23 223 (50.6) | 10 208 (59.7) | 15 338 (50.2) | 9836 (46.2) | 35 382 (51.3) |
| Underlying chronic conditionsi | |||||||
| High blood pressure | 1661 (12.7) | 1428 (13.9) | 8348 (17.9) | 1894 (11.0) | 4467 (14.3) | 5076 (23.4) | 11 437 (16.3) |
| Overweight | 1562 (11.9) | 1265 (12.3) | 8370 (17.9) | 1664 (9.7) | 4756 (15.2) | 4777 (22.0) | 11 197 (16.0) |
| Smoking | 818 (6.2) | 1126 (10.9) | 4806 (10.3) | 2055 (12.0) | 2587 (8.3) | 2108 (9.7) | 6750 (9.6) |
| Diabetes | 623 (4.8) | 523 (5.1) | 2998 (6.4) | 687 (4.0) | 1636 (5.2) | 1821 (8.4) | 4144 (5.9) |
| Lung disease or asthma | 454 (3.5) | 399 (3.9) | 2782 (5.9) | 738 (4.3) | 1422 (4.5) | 1475 (6.8) | 3635 (5.2) |
| Heart condition | 384 (2.9) | 357 (3.5) | 2363 (5.1) | 545 (3.2) | 1065 (3.4) | 1494 (6.9) | 3104 (4.4) |
| Kidney disease | 27 (0.2) | 25 (0.2) | 165 (0.4) | 46 (0.3) | 82 (0.3) | 89 (0.4) | 217 (0.3) |
| Liver disease | 9 (0.1) | 21 (0.2) | 86 (0.2) | 25 (0.1) | 40 (0.1) | 51 (0.2) | 116 (0.2) |
| No conditions | 9328 (71.2) | 6964 (67.7) | 28 335 (60.6) | 12 025 (70.0) | 20 895 (66.8) | 11 707 (53.9) | 44 627 (63.6) |
| 1 Condition | 2471 (18.9) | 2090 (20.3) | 10 812 (23.1) | 3438 (20.0) | 6459 (20.7) | 5476 (25.2) | 15 373 (21.9) |
| ≥2 Conditions | 1299 (9.9) | 1239 (12.0) | 7617 (16.3) | 1714 (10.0) | 3917 (12.5) | 4524 (20.8) | 10 155 (14.5) |
| Vaccination historyj | |||||||
| BNT162b2/BNT162b2/no third dose | 4714 (36.0) | 3062 (29.7) | 12 063 (25.8) | 0 | 19 839 (63.4) | 0 | 19 839 (28.3) |
| Unvaccinated | 3412 (26.0) | 5044 (49.0) | 8721 (18.6) | 17 177 (100) | 0 | 0 | 17 177 (24.5) |
| BNT162b2/BNT162b2/BNT162b2 | 1494 (11.4) | 464 (4.5) | 10 518 (22.5) | 0 | 0 | 12 476 (57.5) | 12 476 (17.8) |
| mRNA-1273/mRNA-1273/no third dose | 2520 (19.2) | 1498 (14.6) | 7365 (15.7) | 0 | 11 383 (36.4) | 0 | 11 383 (16.2) |
| mRNA-1273/mRNA-1273/mRNA-1273 | 731 (5.6) | 172 (1.7) | 6674 (14.3) | 0 | 0 | 7577 (34.9) | 7577 (10.8) |
| BNT162b2/BNT162b2/mRNA-1273 | 114 (0.9) | 26 (0.3) | 813 (1.7) | 0 | 0 | 953 (4.4) | 953 (1.4) |
| mRNA-1273/mRNA-1273/BNT162b2 | 99 (0.8) | 17 (0.2) | 571 (1.2) | 0 | 0 | 687 (3.2) | 687 (1.0) |
| mRNA-1273/BNT162b2/no third dose | 7 (0.1) | 4 (<0.1) | 15 (<0.1) | 0 | 26 (0.1) | 0 | 26 (<0.1) |
| BNT162b2/mRNA-1273/no third dose | 4 (<0.1) | 6 (0.1) | 13 (<0.1) | 0 | 23 (0.1) | 0 | 23 (<0.1) |
| mRNA-1273/BNT162b2/BNT162b2 | 1 (<0.1) | 0 | 3 (<0.1) | 0 | 0 | 4 (<0.1) | 4 (<0.1) |
| BNT162b2/mRNA-1273/BNT162b2 | 0 | 0 | 4 (<0.1) | 0 | 0 | 4 (<0.1) | 4 (<0.1) |
| mRNA-1273/BNT162b2/mRNA-1273 | 1 (<0.1) | 0 | 2 (<0.1) | 0 | 0 | 3 (<0.1) | 3 (<0.1) |
| BNT162b2/mRNA-1273/mRNA-1273 | 1 (<0.1) | 0 | 2 (<0.1) | 0 | 0 | 3 (<0.1) | 3 (<0.1) |
| Months between second dose and testingk | (n = 9686) | (n = 5249) | (n = 38 043) | (n = 0) | (n = 52 978) | ||
| Median (IQR) | 8.0 (1.0) | 8.0 (1.0) | 8.0 (2.0) | NA | 8.0 (1.0) | 8.0 (1.0) | 8.0 (2.0) |
| Range | 6.0-11.0 | 6.0-12.0 | 6.0-12.0 | NA | 6.0-12.0 | 6.0-12.0 | 6.0-12.0 |
| Months between third dose and testingk,l | (n = 2441) | (n = 679) | (n = 18 587) | (n = 0) | (n = 0) | (n = 21 707) | |
| Median (IQR) | 1.0 (1.0) | 1.0 (1.0) | 1.0 (1.0) | NA | NA | 1.0 (1.0) | 1.0 (1.0) |
| Range | 0.0-3.0 | 0.0-3.0 | 0.0-3.0 | NA | NA | 0.0-3.0 | 0.0-3.0 |
| Months between second and third dosesk | (n = 2441) | (n = 679) | (n = 18 587) | (n = 0) | (n = 0) | (n = 21 707) | |
| Median (IQR) | 7.0 (1.0) | 7.0 (1.0) | 7.0 (1.0) | NA | NA | 7.0 (1.0) | 7.0 (1.0) |
| Range | 6.0-11.0 | 6.0-11.0 | 6.0-11.0 | NA | NA | 6.0-11.0 | 6.0-11.0 |
Abbreviations: HHS, US Department of Health and Human Services; NA, not applicable; SVI, Social Vulnerability Index.
Tests were from individuals 6 months or more after dose 2.
Tests were from individuals 14 days or more after dose 3, with 6 months or more between doses 2 and 3.
Samples positive for SARS-CoV-2 were considered to have S-gene target failure (SGTF) if they had cycle threshold (Ct) values for the N and ORF1ab genes but not for the S gene; otherwise, samples were considered not to have SGTF. SGTF was used as a proxy for the Omicron variant. Samples without SGTF were presumed to be Delta.
Any 2 or 3 doses of BNT162b2 or mRNA-1273.
Race was self-reported. Test registrants could select from predefined categories American Indian/Alaska Native, Asian, Black/African American, Native Hawaiian/Pacific Islander, White, or decline to answer.
Ethnicity was self-reported. Test registrants could select from predefined categories Hispanic/Latino, Non-Hispanic/Latino, or decline to answer.
HHS regions defined as New England (Connecticut, Maine, Massachusetts, New Hampshire, Rhode Island, and Vermont), Mid-Atlantic (New Jersey, New York, and Pennsylvania), South Atlantic (Delaware, Florida, Georgia, Maryland, North Carolina, South Carolina, Virginia, District of Columbia, and West Virginia), East North Central (Illinois, Indiana, Michigan, Ohio, and Wisconsin), East South Central (Alabama, Kentucky, Mississippi, and Tennessee), West North Central (Iowa, Kansas, Minnesota, Missouri, Nebraska, North Dakota, and South Dakota), West South Central (Arkansas, Louisiana, Oklahoma, and Texas), Mountain (Arizona, Colorado, Idaho, Montana, Nevada, New Mexico, Utah, and Wyoming), Pacific (Alaska, California, Hawaii, Oregon, and Washington), and Puerto Rico.
SVI is a tool that uses census data on 15 social factors to rank social vulnerability by census tract. The scale is from 0 to 1; higher SVIs represent more vulnerable communities. Missing SVI data were included in unadjusted analyses but not included in adjusted analyses.
Test registrants were asked to report whether a health care worker had ever diagnosed them with heart conditions; high blood pressure; overweight or obesity; diabetes; current or former smoker; kidney failure or end-stage kidney disease; cirrhosis of the liver; or chronic lung disease, such as chronic obstructive pulmonary disease, moderate to severe asthma, cystic fibrosis, or pulmonary embolism.
Vaccine doses listed in sequential order.
Month and year of receipt were reported for each vaccination dose; therefore, the number of months between a vaccine dose and testing is a whole number calculated as the difference between the month and year of testing and the month and year of the vaccine dose. Similarly, the number of months between 2 vaccine doses is a whole number calculated as the difference between the month and year of receipt for each dose.
Tests from individuals reporting receipt of a vaccine dose less than 14 days before the date of testing were excluded from the main analysis.
For vaccination history, the most common combination of 2 doses was BNT162b2/BNT162b2 (63.4% of 2-dose regimens), followed by mRNA-1273/mRNA-1273 (36.4%); the most common 3-dose combination was BNT162b2/BNT162b2/BNT162b2 (57.5% of 3-dose regimens) (Table 1). Among individuals receiving 2 doses only, the median time between the second dose and test date was 8 months. Among those receiving 3 doses, the median time between the second dose and test date was 8 months, between the second and third dose was 7 months, and between the third dose and test date was 1 month. Prior receipt of 3 vaccine doses was reported for 18.6% (2441/13 098) of Omicron cases, 6.6% (679/10 293) of Delta cases, and 39.7% (18 587/46 764) of controls (Table 1).
Comparison of 3 Doses vs Unvaccinated
Among Omicron cases and controls, the adjusted OR for prior receipt of 3 mRNA vaccine doses vs unvaccinated was 0.33 (95% CI, 0.31-0.35); among Delta cases and controls, the adjusted OR was 0.065 (95% CI, 0.059-0.071; Q < .001 for comparison of ORs for Omicron and Delta) (Table 2). When models were stratified by mRNA product, the adjusted ORs for Omicron were 0.35 (95% CI, 0.32-0.38) for 3 doses of BNT162b2 vs unvaccinated and 0.28 (95% CI, 0.26-0.31) for 3 doses of mRNA-1273 vs unvaccinated. For Delta, the adjusted ORs were 0.077 (95% CI, 0.070-0.086) for 3 doses of BNT162b2 vs unvaccinated and 0.045 (95% CI, 0.038-0.053) for 3 doses of mRNA-1273 vs unvaccinated. Q values for all comparisons (Omicron vs Delta) of product-specific ORs were less than .001 (Table 2). An adjusted OR less than 1 indicates that relatively fewer test-positive cases had prior receipt of 3 doses (vs unvaccinated), with values closer to 0 representing a stronger magnitude of association.
Table 2. Association Between Omicron or Delta Symptomatic SARS-CoV-2 Infection and Prior mRNA COVID-19 Vaccination Among Adults 18 Years or Older Tested in the Increasing Community Access to Testing Platform, December 10, 2021, to January 1, 2022.
| Vaccine type evaluated | SARS-CoV-2 variant | Total test-positive cases | Total test-negative controls | OR (95% CI) | Q valueb | |
|---|---|---|---|---|---|---|
| Crude | Adjusteda | |||||
| 3 Doses vs unvaccinatedc | ||||||
| Any 3 doses of mRNA vaccined | Delta | 5723 | 27 308 | 0.063 (0.058-0.069) | 0.065 (0.059-0.071) | <.001 |
| Omicron | 5853 | 27 308 | 0.34 (0.32-0.36) | 0.33 (0.31-0.35) | ||
| 3 Doses of BNT-162b2e | Delta | 5508 | 19 239 | 0.076 (0.069-0.084) | 0.077 (0.070-0.086) | <.001 |
| Omicron | 4906 | 19 239 | 0.36 (0.34-0.39) | 0.35 (0.32-0.38) | ||
| 3 Doses of mRNA-1273f | Delta | 5216 | 15 395 | 0.045 (0.038-0.052) | 0.045 (0.038-0.053) | <.001 |
| Omicron | 4143 | 15 395 | 0.28 (0.26-0.31) | 0.28 (0.26-0.31) | ||
| 3 vs 2 Doses c , g | ||||||
| Any 3 doses of mRNA vaccined | Delta | 5249 | 38 043 | 0.16 (0.14-0.17) | 0.16 (0.14-0.17) | <.001 |
| Omicron | 9686 | 38 043 | 0.35 (0.34-0.37) | 0.34 (0.32-0.36) | ||
| 3 Doses of BNT-162b2e | Delta | 3526 | 22 581 | 0.17 (0.16-0.19) | 0.17 (0.16-0.19) | <.001 |
| Omicron | 6208 | 22 581 | 0.36 (0.34-0.39) | 0.35 (0.32-0.37) | ||
| 3 Doses of mRNA-1273f | Delta | 1670 | 14 039 | 0.13 (0.11-0.15) | 0.13 (0.11-0.15) | <.001 |
| Omicron | 3251 | 14 039 | 0.32 (0.29-0.35) | 0.31 (0.28-0.34) | ||
Abbreviation: OR, odds ratio.
An adjusted OR less than 1 indicated that receipt of 3 doses (vs unvaccinated and vs 2 doses) was less likely among test-positive cases vs test-negative controls. Models included the number of days between the start of the analysis period and test date (as a continuous variable), age group, sex, race, ethnicity, testing site US Department of Health and Human Services region, testing site census tract Social Vulnerability Index (dichotomized as 0 to <0.5 and ≥0.5 to 1), and number of underlying chronic conditions (0, 1, or ≥2) to adjust for potential confounding. All data for Puerto Rico were missing Social Vulnerability Index; therefore, records from Puerto Rico were not included in the adjusted analysis. Unknown race and ethnicity were coded as categorical levels within the variable to retain those tests in regression models.
Q value for the comparison of the adjusted OR for Delta vs Omicron.
For 3 doses, tests were from those 14 days or more after dose 3, with 6 months or more between doses 2 and 3.
Three doses of mRNA vaccine (n = 21 707) included vaccination histories BNT162b2/BNT162b2/BNT162b2, mRNA-1273/mRNA-1273/mRNA-1273, BNT162b2/BNT162b2/mRNA-1273, mRNA-1273/mRNA-1273/BNT162b2, mRNA-1273/BNT162b2/BNT162b2, BNT162b2/mRNA-1273/BNT162b2, mRNA-1273/BNT162b2/mRNA-1273, and BNT162b2/mRNA-1273/mRNA-1273. Two doses of mRNA vaccine (n = 31 271) included vaccination histories BNT162b2/BNT162b2/no third dose, mRNA-1273/mRNA-1273/no third dose, mRNA-1273/BNT162b2/no third dose, and BNT162b2/mRNA-1273/no third dose.
Three doses of BNT162b2 vaccine (n = 12 476) included vaccination history BNT162b2/BNT162b2/BNT162b2. Two doses of BNT162b2 vaccine (n = 19 839) included vaccination history BNT162b2/BNT162b2/no third dose.
Three doses of mRNA-1273 vaccine (n = 7577) included vaccination history mRNA-1273/mRNA-1273/mRNA-1273. Two doses of mRNA-1273 vaccine (n = 11 383) included vaccination history mRNA-1273/mRNA-1273/no third dose.
For 2 doses, tests were from individuals 6 months or more after dose 2 (ie, eligible for a booster dose).
Comparison of 3 Doses vs 2 Doses
Among Omicron cases and controls, the adjusted OR for prior receipt of 3 mRNA vaccine doses vs 2 doses was 0.34 (95% CI, 0.32-0.36); among Delta cases and controls, the adjusted OR was 0.16 (95% CI, 0.14-0.17; Q < .001) (Table 2). When models were stratified by mRNA product, the adjusted ORs for Omicron were 0.35 (95% CI, 0.32-0.37) for 3 doses of BNT162b2 vs 2 doses and 0.31 (95% CI, 0.28-0.34) for 3 doses of mRNA-1273 vs 2 doses. For Delta, the adjusted ORs were 0.17 (95% CI, 0.16-0.19) for 3 doses of BNT162b2 vs 2 doses and 0.13 (95% CI, 0.11-0.15) for 3 doses of mRNA-1273 vs 2 doses. Q values for all comparisons (Omicron vs Delta) of product-specific ORs were less than .001 (Table 2). An adjusted OR less than 1 indicates that relatively fewer test-positive cases had prior receipt of 3 doses (vs 2 doses), with values closer to 0 representing a stronger magnitude of association.
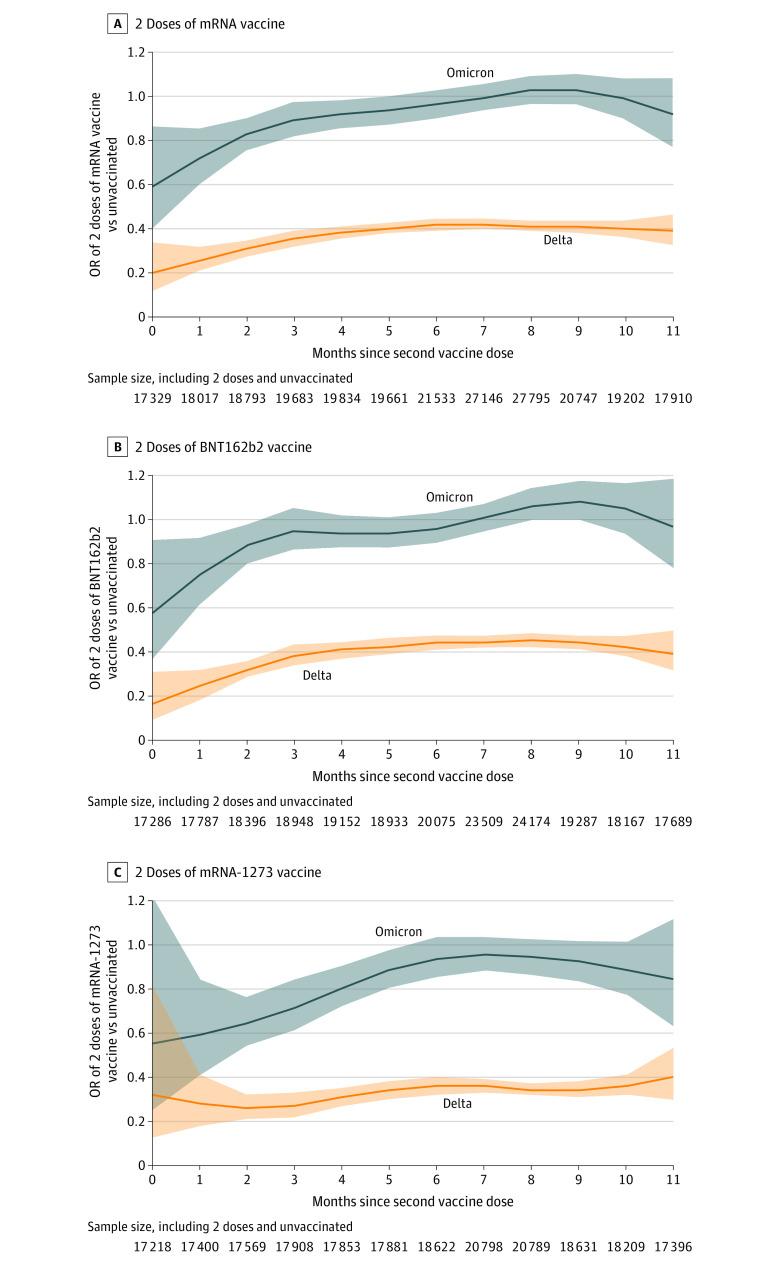
Comparison of 2 Doses vs Unvaccinated by Time Since Vaccination
Among Omicron cases and controls, the adjusted OR for prior receipt of any 2 mRNA vaccine doses vs unvaccinated was lowest soon after the second dose and generally increased over time since vaccination (Figure 2). The upper bound of the 95% CI was consistently greater than 1 starting at 3 months after second dose for BNT162b2 and at 6 months after second dose for mRNA-1273. For Delta, the OR for any 2 doses vs unvaccinated was also lowest soon after receipt of second dose; however, the ORs remained less than 0.46 for BNT162b2 and less than 0.40 for mRNA-1273, with 95% CIs that did not include 1 (Figure 2).
Figure 2. Odds Ratios for the Association of 2 Doses of mRNA Vaccine by Months Since Second Dose and Symptomatic SARS-CoV-2 Infection Caused by the Omicron or Delta Variants Among Adults 18 Years or Older Tested in the Increasing Community Access to Testing Platform, December 10, 2021, to January 1, 2022.
Shaded regions indicate 95% CIs. Months since second dose reflect 30-day increments since the second dose except for month 0, which only includes days 14 to 30 because tests from persons with second mRNA COVID-19 doses received less than 14 days before testing were excluded from the analysis. The total number of cases and controls included at each time point varied by product (total Ns reported below plots). OR indicates odds ratio.
Comparison of Ct Values
Among Omicron cases, median Ct values were significantly higher in samples from persons reporting 3 mRNA vaccine doses vs unvaccinated for the ORF1ab gene (19.25 vs 18.58; Q < .001) but not for the N gene (19.35 vs 18.71; Q = .002) (eTable 2 in the Supplement). Among Delta cases, median Ct values of the N, ORF1ab, and S genes were significantly higher in samples from persons receiving 3 doses vs unvaccinated (N gene: 19.07 vs 18.28; ORF1ab gene: 18.70 vs 17.84; S gene: 23.62 vs 19.58; Q < .001 for all comparisons) (Figure 3; eTable 2 in the Supplement). In comparing median Ct values in samples from persons reporting 3 mRNA vaccine doses vs 2 doses, all were significantly higher (Omicron N gene, 19.35 vs 18.52; Omicron ORF1ab gene, 19.25 vs 18.40; Delta N gene, 19.07 vs 17.52; Delta ORF1ab gene, 18.70 vs 17.28; Delta S gene, 23.62 vs 20.24; Q < .001 for all comparisons) (Figure 3; eTable 2 in the Supplement).
Figure 3. Cycle Threshold Values for the N, ORF1ab, and S Genes by Variant and Vaccination Status Among SARS-CoV-2–Positive Cases Tested by the TaqPath COVID-19 Combo Kit Assay in the Increasing Community Access to Testing Platform, December 10, 2021, to January 1, 2022.
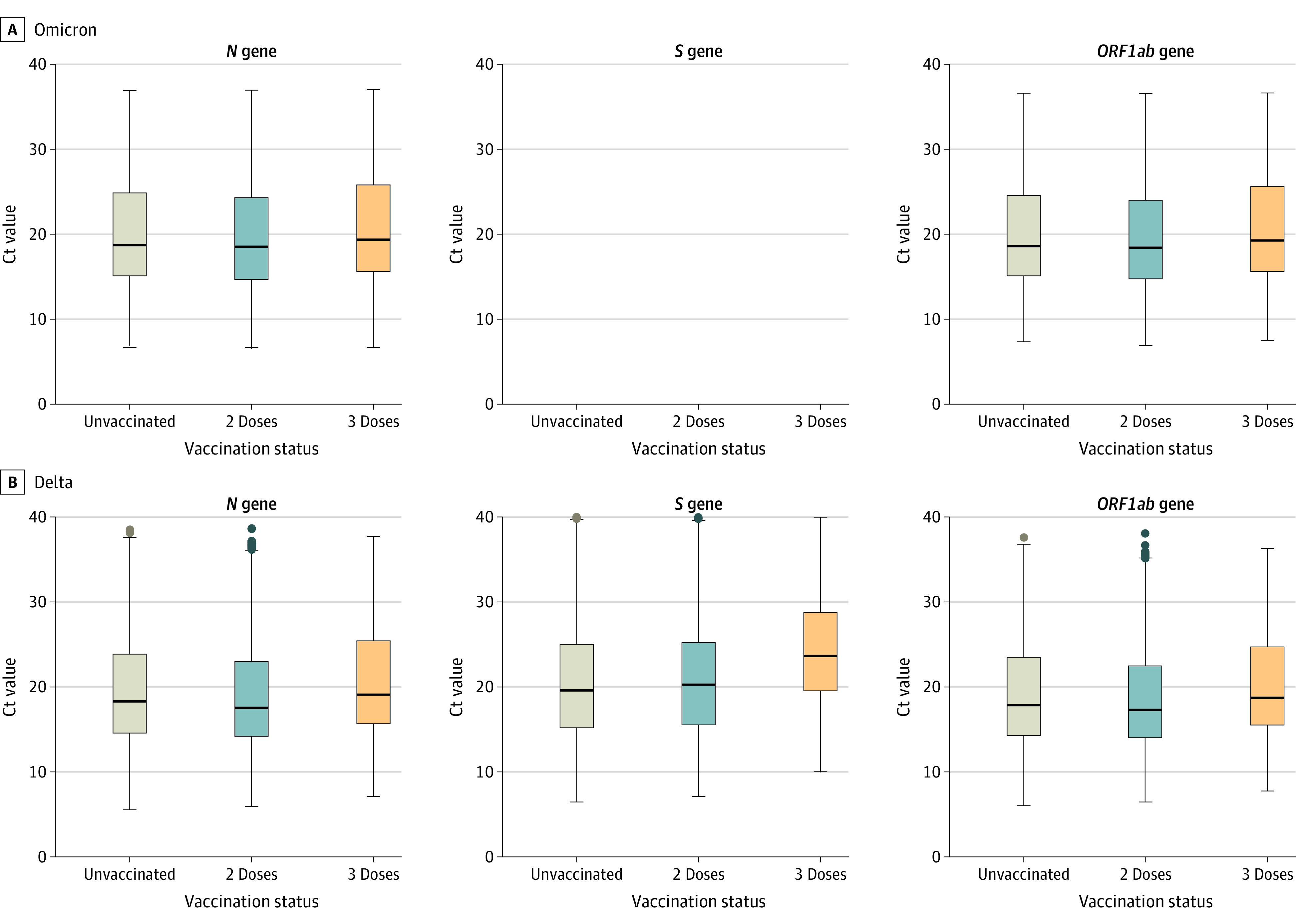
For each plot, from top to bottom, lines in the box represent the 75th percentile, median, and 25th percentile. The whiskers extend to the largest and smallest values up to 1.5 times the interquartile range from the 75th and 25th percentiles, respectively. Data beyond the ends of the whiskers are plotted individually. Unvaccinated (Omicron n = 3412; Delta n = 5044; total n = 8456) were individuals who received zero vaccine doses. Two doses (tested ≥6 months after dose 2) of mRNA vaccine (Omicron n = 7245; Delta n = 4570; total n = 11 815) included vaccination histories BNT162b2/BNT162b2/no third dose, mRNA-1273/mRNA-1273/no third dose, mRNA-1273/BNT162b2/no third dose, and BNT162b2/mRNA-1273/no third dose. Three doses (tested ≥14 days after dose 3, with ≥6 months between doses 2 and 3) of mRNA vaccine (Omicron n = 2441; Delta n = 679; total n = 3120) included vaccination histories BNT162b2/BNT162b2/BNT162b2, mRNA-1273/mRNA-1273/mRNA-1273, BNT162b2/BNT162b2/mRNA-1273, mRNA-1273/mRNA-1273/BNT162b2, mRNA-1273/BNT162b2/BNT162b2, BNT162b2/mRNA-1273/BNT162b2, mRNA-1273/BNT162b2/mRNA-1273, and BNT162b2/mRNA-1273/mRNA-1273. Cycle threshold (Ct) values were compared between 3 doses and unvaccinated, 3 doses and 2 doses, and 2 doses and unvaccinated. Significance testing was performed with a 2-sided Mann-Whitney U test and false-discovery rate–corrected for multiple comparisons. Q < .001 for all comparisons, except for comparing 3 doses vs unvaccinated for N gene for Omicron and for comparing 2 doses vs unvaccinated for the N and ORF1ab genes for Omicron and the S gene for Delta (eTable 2 in the Supplement).
Discussion
In this analysis of SARS-CoV-2 tests performed at sites across the US during a 23-day period when incidence of Omicron was rapidly increasing, vaccination with a third dose of an mRNA COVID-19 vaccine was significantly less common among individuals infected with either the Omicron or Delta variants compared with uninfected individuals. Because of the timing of booster recommendations in the US, most booster recipients had recent vaccination (median of 1 month since booster).
The magnitude of the association between vaccination and infection depended on the referent group and variant. For 3 doses vs unvaccinated, the ORs corresponded to an estimated effectiveness (1 – OR) of 67.3% (95% CI, 65.0%-69.4%) for Omicron and 93.5% (95% CI, 92.9%-94.1%) for Delta. For 3 doses vs 2 doses, the ORs corresponded to an estimated relative effectiveness of 66.3% (95% CI, 64.3%-68.1%) for Omicron and 84.5% (95% CI, 83.1%-85.7%) for Delta. For Omicron, the similarity between ORs for 3 doses using the unvaccinated referent group and the 2-dose referent group is consistent with the attenuation of the OR for 2 doses vs unvaccinated with time since second dose, which reflected no significant association by 6 months after second dose for both products. For Delta, the association between infection and 2 doses vs unvaccinated also attenuated over time since second dose, which is consistent with previous reports23,24,25,26; however, the ORs were statistically significant even up to 11 months after the second dose.
Although these findings provide evidence supporting that 3-dose schedules are protective and that booster doses are more protective than primary series alone, the significantly higher OR for Omicron suggests that booster doses are less protective against Omicron than against Delta. These results are consistent with in-vitro neutralization assays that suggested the potential for immune evasion with Omicron.27,28,29,30 They also highlight that, in the setting of Omicron, higher booster coverage rates may be needed to achieve the same public health benefit as during Delta predominance. Additionally, nonpharmaceutical interventions may provide an important adjunct to slow the spread of Omicron.
Among both Omicron and Delta variant cases, Ct values were generally higher (reflecting less genetic material detected) among those with 3 vaccine doses compared with unvaccinated or to 2 doses; with 1 exception (N gene for Omicron, 3 doses vs unvaccinated), all comparisons were statistically significant. Ct values are not a direct measure of viral load or infectiousness and can vary for a range of reasons including timing of sample collection relative to infection onset, specimen transport times, and laboratory assays and conditions. However, they have previously been used as a crude indicator of transmission potential, with higher values representing decreased likelihood of a case being infectious.31,32,33,34,35,36,37 In this analysis, the Ct values were based on 2 targets from a single assay, performed at a single laboratory chain, increasing their comparability. The significantly higher Ct values found in individuals reporting receipt of 3 doses vs unvaccinated or 2 doses, for both the Omicron and Delta variants, may suggest decreased infectiousness in those receiving an mRNA booster dose. However, caution should be taken in interpreting these differences between groups as epidemiologically meaningful because all differences were less than 1 unit on the log scale.
Limitations
This study has several limitations. First, vaccination status and symptoms were based on patient self-reported data, potentially leading to misclassification. Second, because the testing data do not include identifiers, tests rather than persons were used as the unit of analysis, and individuals may have been included more than once. However, the analysis was restricted to symptomatic individuals to reduce inclusion of individuals serially testing for reasons other than symptomatic disease, and the short study period (23 days) reduces the probability of individuals contributing multiple test results.
Third, individuals remaining unvaccinated or unboosted may differ from individuals with 3 doses in ways that cannot be adjusted for with the variables in this data set. Fourth, some factors that could potentially be associated with both vaccination and risk of infection and thus confound the observed associations (eg, masking and social distancing) were not measured. Fifth, US adults were recommended to receive booster doses at different dates depending on age, underlying conditions, and occupation; therefore, some subgroups may have had greater access to boosters before broader recommendations were made.
Sixth, sequencing data were limited for tests included in this analysis so SGTF was used as a proxy for Omicron infection; however, sensitivity of 83.4% and specificity of 99.2% in internal validation suggest that samples classified as Omicron by SGTF were almost always Omicron. Although some Omicron samples may have been misclassified as Delta, this would not affect the association between Omicron and vaccination status and would bias the association of Delta and vaccination toward that of Omicron such that the reported differences between Omicron and Delta are conservative.
Seventh, the analysis of the association between symptomatic infection and 3 vs 2 doses did not directly account for waning of the primary series, although all tests were performed at least 6 months after receipt of a primary series, at which point protection from 2 doses was quite reduced, particularly for the Omicron variant. Eighth, associations between infection and vaccination are not constant and will likely continue to change with time since last dose.
Conclusions
Among individuals seeking testing for COVID-like illness in the US in December 2021, receipt of 3 doses of mRNA COVID-19 vaccine (compared with unvaccinated and with receipt of 2 doses) was less likely among cases with symptomatic SARS-CoV-2 infection compared with test-negative controls. These findings suggest that receipt of 3 doses of mRNA vaccine, relative to being unvaccinated and to receipt of 2 doses, was associated with protection against both the Omicron and Delta variants, although the higher odds ratios for Omicron suggest less protection for Omicron than for Delta.
eTable 1. S-gene target failure and variant lineages among 17,620 positive SARS-CoV-2 tests whole genome sequenced in the Increasing Community Access to Testing platform between December 10th, 2021 and January 1st, 2022
eTable 2. Cycle threshold (Ct) values among 23,391 positive SARS-CoV-2 tests performed in the Increasing Community Access to Testing platform between December 10th, 2021 and January 1st, 2022
References
- 1.World Health Organization . Classification of Omicron (B.1.1.529): SARS-CoV-2 variant of concern. Updated November 26, 2021. Accessed December 23, 2021. https://www.who.int/news/item/26-11-2021-classification-of-omicron-(b.1.1.529)-sars-cov-2-variant-of-concern
- 2.World Health Organization . Enhancing response to OMICRON (COVID-19 variant B.1.1.529): Technical brief and priority actions for Member States. January 7, 2022. Accessed January 11, 2022. https://www.who.int/publications/m/item/enhancing-readiness-for-omicron-(b.1.1.529)-technical-brief-and-priority-actions-for-member-states
- 3.CDC COVID-19 Response Team . SARS-CoV-2 B.1.1.529 (Omicron) variant—United States, December 1-8, 2021. MMWR Morb Mortal Wkly Rep. 2021;70(50):1731-1734. doi: 10.15585/mmwr.mm7050e1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention . COVID Data Tracker. Accessed January 7, 2021. https://covid.cdc.gov/covid-data-tracker/#variant-proportions
- 5.Wang L, Cheng G. Sequence analysis of the emerging SARS-CoV-2 variant Omicron in South Africa. J Med Virol. 2021;1-6. doi: 10.1002/jmv.27516 [DOI] [PubMed] [Google Scholar]
- 6.GISAID . Tracking of variants. Accessed December 23, 2021. https://www.gisaid.org/hcov19-variants/
- 7.National Institute for Communicable Diseases . COVID-19 weekly epidemiology brief, South Africa: week 47 2021. November 27, 2021. https://www.nicd.ac.za/wp-content/uploads/2021/12/COVID-19-Weekly-Epidemiology-Brief-week-47-2021.pdf
- 8.UK Health Security Agency . SARS-CoV-2 variants of concern and variants under investigation in England. Technical briefing 33. December 23, 2021. Accessed December 23, 2021. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1043807/technical-briefing-33.pdf
- 9.European Centre for Disease Prevention and Control . Implications of the further emergence and spread of the SARS-CoV-2 B.1.1.529 variant of concern (Omicron) for the EU/EEA—first update. Updated December 2, 2021. https://www.ecdc.europa.eu/sites/default/files/documents/threat-assessment-covid-19-emergence-sars-cov-2-variant-omicron-december-2021.pdf
- 10.Centers for Disease Control and Prevention . Potential Rapid increase of Omicron variant infections in the United States. Updated December 20, 2021. Accessed December 23, 2021. https://www.cdc.gov/coronavirus/2019-ncov/science/forecasting/mathematical-modeling-outbreak.html
- 11.Miller MF, Shi M, Motsinger-Reif A, Weinberg CR, Miller JD, Nichols E. Community-based testing sites for SARS-CoV-2—United States, March 2020-November 2021. MMWR Morb Mortal Wkly Rep. 2021;70(49):1706-1711. doi: 10.15585/mmwr.mm7049a3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.US Department of Health and Human Services . COVID-19 Pandemic Response, Laboratory Data Reporting: CARES Act Section 18115. Updated January 8, 2021. Accessed January 11, 2022. https://www.hhs.gov/sites/default/files/covid-19-laboratory-data-reporting-guidance.pdf
- 13.Centers for Disease Control and Prevention . CDC/ATSDR SVI fact sheet. Updated April 28, 2021. Accessed December 23, 2021. https://www.atsdr.cdc.gov/placeandhealth/svi/index.html
- 14.Vandenbroucke JP, Pearce N. Test-negative designs: differences and commonalities with other case-control studies with “other patient” controls. Epidemiology. 2019;30(6):838-844. doi: 10.1097/EDE.0000000000001088 [DOI] [PubMed] [Google Scholar]
- 15.Chua H, Feng S, Lewnard JA, et al. The use of test-negative controls to monitor vaccine effectiveness: a systematic review of methodology. Epidemiology. 2020;31(1):43-64. doi: 10.1097/EDE.0000000000001116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sullivan SG, Tchetgen Tchetgen EJ, Cowling BJ. Theoretical basis of the test-negative study design for assessment of influenza vaccine effectiveness. Am J Epidemiol. 2016;184(5):345-353. doi: 10.1093/aje/kww064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jackson ML, Phillips CH, Benoit J, et al. The impact of selection bias on vaccine effectiveness estimates from test-negative studies. Vaccine. 2018;36(5):751-757. doi: 10.1016/j.vaccine.2017.12.022 [DOI] [PubMed] [Google Scholar]
- 18.Embi PJ, Levy ME, Naleway AL, et al. Effectiveness of 2-dose vaccination with mRNA COVID-19 vaccines against COVID-19–associated hospitalizations among immunocompromised adults—nine states, January-September 2021. MMWR Morb Mortal Wkly Rep. 2021;70(44):1553-1559. doi: 10.15585/mmwr.mm7044e3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mbaeyi S, Oliver SE, Collins JPet al. The Advisory Committee on Immunization Practices’ interim recommendations for additional primary and booster doses of COVID-19 vaccines—United States, 2021. MMWR Morb Mortal Wkly Rep. 2021;70:1545-1552. doi: 10.15585/mmwr.mm7044e2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Infectious Diseases Society of America and Association for Molecular Pathology . IDSA and AMP joint statement on the use of SARS-CoV-2 PCR cycle threshold (Ct) values for clinical decision-making. Updated March 12, 2021. https://www.idsociety.org/globalassets/idsa/public-health/covid-19/idsa-amp-statement.pdf
- 21.Thermo Fisher Scientific confirms detection of SARS-CoV-2 in samples containing the Omicron variant with its TaqPath COVID-19 tests. Updated November 29, 2021. Accessed December 23, 2021. https://thermofisher.mediaroom.com/2021-11-29-Thermo-Fisher-Scientific-Confirms-Detection-of-SARS-CoV-2-in-Samples-Containing-the-Omicron-Variant-with-its-TaqPath-COVID-19-Tests
- 22.Scott L, Hsiao NY, Moyo S, et al. Track Omicron’s spread with molecular data. Science. 2021;374(6574):1454-1455. doi: 10.1126/science.abn4543 [DOI] [PubMed] [Google Scholar]
- 23.Chemaitelly H, Tang P, Hasan MR, et al. Waning of BNT162b2 vaccine protection against SARS-CoV-2 infection in Qatar. N Engl J Med. 2021;385(24):e83. doi: 10.1056/NEJMoa2114114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tartof SY, Slezak JM, Fischer H, et al. Effectiveness of mRNA BNT162b2 COVID-19 vaccine up to 6 months in a large integrated health system in the USA: a retrospective cohort study. Lancet. 2021;398(10309):1407-1416. doi: 10.1016/S0140-6736(21)02183-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tabak YP, Sun X, Brennan TA, Chaguturu SK. Incidence and estimated vaccine effectiveness against symptomatic SARS-CoV-2 infection among persons tested in US retail locations, May 1 to August 7, 2021. JAMA Netw Open. 2021;4(12):e2143346. doi: 10.1001/jamanetworkopen.2021.43346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bruxvoort KJ, Sy LS, Qian L, et al. Effectiveness of mRNA-1273 against delta, mu, and other emerging variants of SARS-CoV-2: test negative case-control study. BMJ. 2021;375:e068848. doi: 10.1136/bmj-2021-068848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cao Y, Wang J, Jian F, et al. Omicron escapes the majority of existing SARS-CoV-2 neutralizing antibodies. Nature. Published online December 23, 2021. doi: 10.1038/d41586-021-03796-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cele S, Jackson L, Khoury DS, et al. Omicron extensively but incompletely escapes Pfizer BNT162b2 neutralization. Nature. Published online December 23, 2021. doi: 10.1038/d41586-021-03824-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu L, Iketani S, Guo Y, et al. Striking antibody evasion manifested by the Omicron variant of SARS-CoV-2. Nature. Published online December 23, 2021. doi: 10.1038/s41586-021-04388-0 [DOI] [PubMed] [Google Scholar]
- 30.Dejnirattisai W, Shaw RH, Supasa Pet al. Reduced neutralization of SARS-CoV-2 omicron B.1.1.529 variant by post-immunisation serum. Lancet. 2021;399(10321):234-236. doi: 10.1016/S0140-6736(21)02844-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kissler SM, Fauver JR, Mack C, et al. Viral dynamics of SARS-CoV-2 variants in vaccinated and unvaccinated persons. N Engl J Med. 2021;385(26):2489-2491. doi: 10.1056/NEJMc2102507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bullard J, Dust K, Funk D, et al. Predicting infectious severe acute respiratory syndrome coronavirus 2 from diagnostic samples. Clin Infect Dis. 2020;71(10):2663-2666. doi: 10.1093/cid/ciaa63 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Al Bayat S, Mundodan J, Hasnain S, et al. Can the cycle threshold (Ct) value of RT-PCR test for SARS CoV2 predict infectivity among close contacts? J Infect Public Health. 2021;14(9):1201-1205. doi: 10.1016/j.jiph.2021.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Singanayagam A, Patel M, Charlett A, et al. Duration of infectiousness and correlation with RT-PCR cycle threshold values in cases of COVID-19, England, January to May 2020. Euro Surveill. 2020;25(32):2001483. doi: 10.2807/1560-7917.ES.2020.25.32.2001483 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Salvatore PP, Dawson P, Wadhwa A, et al. Epidemiological correlates of polymerase chain reaction cycle threshold values in the detection of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Clin Infect Dis. 2021;72(11):e761-e767. doi: 10.1093/cid/ciaa1469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Marcus JE, Frankel DN, Pawlak MT, et al. Risk factors associated with COVID-19 transmission among US Air Force trainees in a congregant setting. JAMA Netw Open. 2021;4(2):e210202-e210202. doi: 10.1001/jamanetworkopen.2021.0202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Regev-Yochay G, Amit S, Bergwerk M, et al. Decreased infectivity following BNT162b2 vaccination: a prospective cohort study in Israel. Lancet Reg Health Eur. 2021;7:100150-100150. doi: 10.1016/j.lanepe.2021.100150 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
eTable 1. S-gene target failure and variant lineages among 17,620 positive SARS-CoV-2 tests whole genome sequenced in the Increasing Community Access to Testing platform between December 10th, 2021 and January 1st, 2022
eTable 2. Cycle threshold (Ct) values among 23,391 positive SARS-CoV-2 tests performed in the Increasing Community Access to Testing platform between December 10th, 2021 and January 1st, 2022