Abstract
Mounting evidence support that glia play a key role in organismal ageing. However, the mechanisms by which glia impact ageing are not understood. One of the processes that has significant impact on the rate of ageing is the unfolded protein response. The more robust the UPR, the more the organism can counteract the effect of environmental and genetic stressors. However, how decline of cellular UPR translates into organismal ageing and eventual death is not fully understood. Here we discuss recent findings highlighting that neuropeptides released by glia act long distance to regulate ageing in C. elegans. Taking advantage of the short lifespan and the genetic amenability of this organism, the endoplasmic reticulum unfolded protein responses (UPRER) can be activated in C. elegans glia. This leads to cell-nonautonomous activation of the UPRER in the intestine. Activation of intestinal UPRER requires the function of genes involved in neuropeptide processing and release, suggesting that neuropeptides signal from glia to the intestine to regulate ER stress response. Importantly, the cell-nonautonomous activation of UPRER leads to extension of lifespan. Taken together, these data suggest that environmental and genetic factors that impact the response of glia to stress have the potential to influence organismal ageing. Further research on the specific neuropeptides involved should cast new light on the mechanism of ageing and may suggest novel anti-ageing therapies.
Keywords: Glia, Neuropeptides, Ageing, Stress, Unfolded protein response, C. elegans
1. Introduction
Despite being discovered at the same time as two distinct cell types almost 200 years ago (Virchow, 1856), glia and neurons were not given the same importance and were not equally investigated. This stems primarily from the discovery of “animal electricity” by Galvani at the end of the eighteen century, which led investigators to focusing on neurons and to assuming that these were the most important cells in the nervous system (Galvani, 1791). However, in the last 70 years, we have been re-evaluating the importance of glial cells in the function of the nervous system. In particular, we now appreciate that their disfunction can significantly contribute to diseases such as Alzheimer’s disease, autism, psychiatric disorders, multiple sclerosis, and Parkinson’s disease among others (Coleman et al., 1985; Cotter et al., 2001; Damier et al., 1993; Nagele et al., 2004; Schonrock et al., 1998).
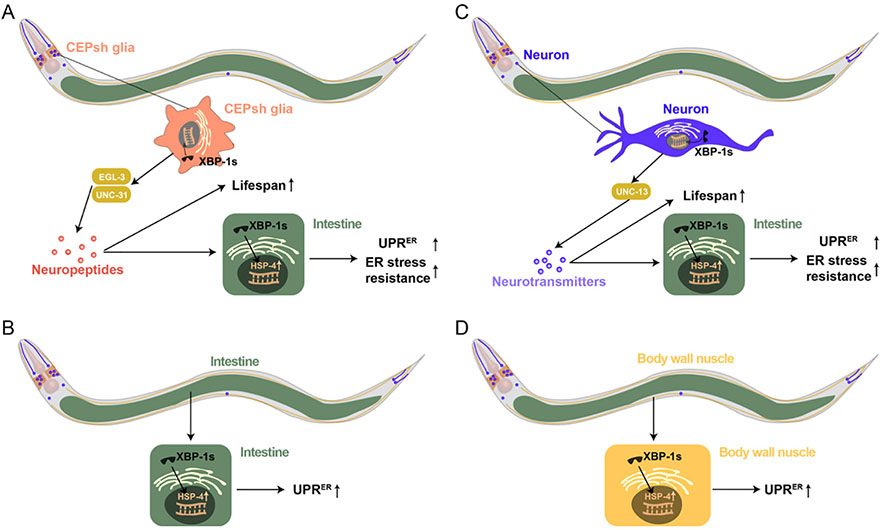
A paper recently published in Science takes this a step further and puts the importance of glia in the context of organismal pathophysiology (Frakes et al., 2020). Frakes and colleagues showed that activation of the endoplasmic reticulum unfolded protein response (UPRER) in the cephalic sheath (CEPsh) glial cells of C. elegans regulates longevity via release of neuropeptides (Fig. 1A). Activation of the UPRER in these glial cells was achieved via overexpression of spliced xbp-1 (xbp-1s), a transcription factor that is part of one of the three signaling pathways leading to UPRER activation (Yoshida et al., 2001) (Fig. 2), and was monitored using a GFP reporter for the gene hsp-4, the worm homologs of HSPA5, also known as GRP78/BiP, a molecular chaperone activated by ER stress (Nuss et al., 2008). The neuropeptides released by CEPsh glia overexpressing xbp-1s act 300 μm away (about 1/3 of the worm’s body length) on intestinal cells, where they control clearance of intestinal protein aggregation, which is known to worsen with age. The involvement of neuropeptides in this process was elucidated by the use of mutants in genes involved in neuropeptides’ processing and release. Indeed, the effect of overexpressing xbp-1s in CEPsh glia on longevity and on cell-nonautonomous UPRER activation is lost in egl-3 and unc-31 mutant C. elegans. While egl-3 encodes a peptidase involved in the processing of neuropeptides, unc-31 is a homolog of CADPS (Ca2+-dependent secretion activator) and it is involved in the fusion of dense core vesicles. Importantly, expression of xbp-1s in the intestine causes cell-autonomous activation of the UPRER, but does not activate the UPRER in any other cell and does not cause changes in lifespan (Fig. 1B).
Fig. 1.
Activation of UPRER by overexpression of xbp-1s in CEPsh glia or neurons extends the lifespan of C. elegans. (A) UPRER activated by overexpression of xbp-1s in CEPsh glia induces the release of neuropeptides that act on intestinal cells to increase ER stress resistance. These peptides also induce lifespan extension. Indeed, the effect on lifespan and on activation of intestinal UPRER of overexpressing spliced xbp-1 (xbp-1s) in CEPsh glia is lost in egl-3 and unc-31 mutant C. elegans (Frakes et al., 2020). While egl-3 encodes a peptidase involved in the processing of neuropeptides, unc-31 is a homolog of CADPS (Ca2+-dependent secretion activator) and it is involved in the fusion of dense core vesicles. HSP-4 is the worm homologs of HSPA5, also known as GRP78/BiP, the molecular chaperone for ER stress (Nuss et al., 2008). (B) Overexpressing xbp-1s in intestine activates cell-autonomous UPRER but does not affect lifespan (Frakes et al., 2020). (C) UPRER activated by overexpressing xbp-1s in neurons induces the release neurotransmitters in a unc-13-dependent manner. unc-13 is the C. elegans homolog of UNC13, a vesicular protein involved in neurotransmitter release. The released neurotransmitters activate the UPRER in the intestine, leading to an increase in ER stress resistance in this tissue. The neurotransmitters also lead to lifespan extension (Taylor and Dillin, 2013). (D) Overexpressing xbp-1s in body wall muscle activates cell-autonomous UPRER but does not affect lifespan (Taylor and Dillin, 2013).
Fig. 2.
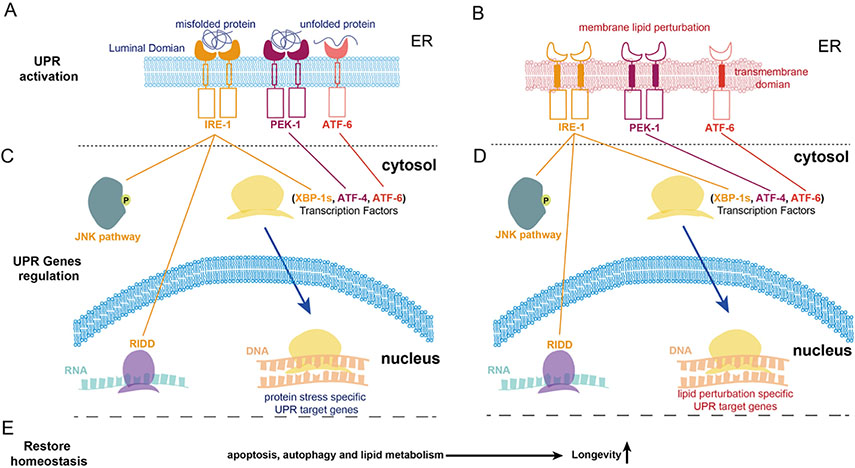
Activation of the endoplasmic reticulum UPR (UPRER) in C. elegans. There are three transducers of the UPRER on the ER membrane: IRE1, PERK, and ATF6 (Okada et al., 2002; Ye et al., 2000; Yoshida et al., 2001). These are activated by the accumulation of misfolded/unfolded proteins (A) or by membrane lipid perturbation (B). (C—D) Once the UPR is activated by lipid perturbation or unfolded protein, the IRE-1 branch regulates downstream genes through the JNK pathway, the IRE1-dependent decay of mRNA, and the spliced XBP-1. Spliced XBP-1 functions as a transcription factor. The PEK-1 branch regulates downstream genes via the transcription factor ATF-4. The ATF-6 branch regulates downstream genes through the cleaved N-term of ATF-6. (E) The downstream UPR genes include apoptosis, autophagy, and lipid metabolism pathways’ genes. Specific response genes are transcribed based on whether the UPR is activated by protein or lipid stress (Kammoun et al., 2009; Rao et al., 2001; Yorimitsu et al., 2006). Activation of spliced xbp-1 in the CEPsh glia or neurons extends lifespan in C. elegans (Frakes et al., 2020; Taylor and Dillin, 2013).
The Dillin lab had already shown that activation of the UPRER in one tissue can lead to its cell-nonautonomous activation in other tissues (Taylor and Dillin, 2013). Using similar strategies used in their recent manuscript, they showed that activation of UPRER in neurons via expression of xbp-1s leads to activation of the UPRER in the intestine (Fig. 1C). The cell-nonautonomous activation of the UPRER in the intestine requires the function of unc-13, a gene that encodes a calmodulin binding protein needed for fusion of the clear core vesicles. Thus, neuronal-mediated cell non-autonomous activation of the UPRER requires neurotransmitters, rather than neuropeptides. Similar to Frakes et al. (2020), Taylor and colleagues showed that cell-autonomous activation of the UPRER in the intestine or in muscles does not extend lifespan (Fig. 1B and D) (Taylor and Dillin, 2013). Importantly, the neuronal and glial mechanisms of cell-nonautonomous activation of UPRER and their effects on longevity can be completely separated. Indeed, the effects of neuronal UPRER activation persist in animals in which CEPsh glia have been removed (Frakes et al., 2020). It is noteworthy though to mention that C. elegans glia release at least the neurotransmitter GABA (Duan et al., 2020). However, it is currently not known whether GABA is released via unc-13-dependent vesicular release or through ion channels such as bestrophins (Lee et al., 2010).
Frakes et al. (2020) adds to the list of papers supporting that glia act long distance to regulate ageing (Apfeld and Kenyon, 1999; Cao et al., 2013; Kounatidis et al., 2017; McQuary et al., 2016; Volkenhoff et al., 2015; Walkowicz et al., 2017; Yin et al., 2017). In this review, we discuss the findings published by Frakes and colleagues (Frakes et al., 2020), while briefly reviewing what is known about the role of glia and of mechanisms that maintain proteostasis such as the UPR and the heat shock response, in organismal ageing.
2. Glia and ageing in C. elegans
C. elegans is a powerful model to determine the role of glia in organismal ageing for many reasons but in particular for three: 1) it has a simple stereotyped nervous system (simpler than the Drosophila’s nervous system) with a fixed number of neurons and glial cells, 2) in C. elegans glia are not needed for neuronal survival, and 3) it lives for just over 2 weeks (Singhvi and Shaham, 2019). The hermaphrodite C. elegans nervous system contains 302 neurons and 56 glia. Forty-six of these glial cells are ectodermally derived and ensheath the neuronal receptive endings at the tips of sensory dendrites. Six, known as GLR cells, are of mesodermal origin and guide muscle arms to appropriate territories during development. GLR cells also regulate axon specification of nearby RME neurons through gap junctions (White et al., 1986). Finally, the four CEPsh cells, have dual function via their association with both with CEP mechanosensory neurons and the nerve ring, the “brain” of the worm (Oikonomou and Shaham, 2011). The CEPsh glial cells are the ones that most closely resemble astrocytes both structurally and functionally. Structurally, they resemble astrocytes because they extend sheet-like processes that wrap around the nerve ring probably acting like a “blood-brain barrier” in worms (White et al., 1986). Functionally, the CEPsh glia, like astrocytes, express glutamate transporters that clear glutamate from the extracellular space (Katz et al., 2019).
A role of glia in ageing has been suggested in C. elegans by previous studies. The Kenyon lab showed in the late 90s that C. elegans lifespan is controlled by the worm’s ability to sense its environment. Indeed, mutations in genes that either alter the shape of the sensory neurons’ receptive endings (the cilia) or their function extend lifespan in C. elegans (Apfeld and Kenyon, 1999). Interestingly, the Kenyon lab also showed that a similar extension of lifespan could be seen in worms in which the Amphid sheath glia (AMsh), two glial cells that ensheath the cilia of sensory neurons, had been genetically ablated (Apfeld and Kenyon, 1999). The Shaham’s lab demonstrated years later that the AMsh glia indeed control the structure and function of C. elegans sensory cilia in an age dependent manner providing an explanation as to why AMsh ablation phenocopied mutations in sensory neurons’ genes (Bacaj et al., 2008; Singhvi et al., 2016). However, a more recent study demonstrating that AMsh glia function as bona fide odorant receptor cells by responding directly to odours via odorant receptors, show that the link between the environment and glia is a lot more direct than previously suspected (Bianchi, 2020; Duan et al., 2020). Thus, these recent results suggest that environmental stimuli might regulate ageing by directly acting on glial cells. Taken together, this work supports that the environment regulates lifespan in the worm via activation of sensory neurons and/or glia.
McQuary and colleagues identified another possible mechanism by which glia in C. elegans regulate lifespan (McQuary et al., 2016). In search for a potential mechanism underlying lifespan extension mediated by the loss of function of the S6 kinase, which is part of the TOR signaling pathway, the authors applied proteomics techniques to S6 mutant worms (rsks-1 mutants). They found that the most enriched protein in rsks-1 mutants, as compared to wild type, was ARGK-1, one of five C. elegans arginine kinases. Using a GFP reporter and co-labelling with glia enriched gene kcc-3, the authors showed that argk-1 is expressed in a small number of cells in the head of the worm, including the CEPsh glia. The function of arginine kinases is to maintain intracellular ATP levels by catalyzing the reversible reaction: ATP + arginine ⇌ ADP + phospho-arginine. McQuary and colleagues showed that ARGK-1 was required for lifespan extension mediated by loss of function of S6 kinase, suggesting that energy regulation in glia might play an important role in longevity, as suggested by studies on glycolytic enzymes done in Drosophila (Delgado et al., 2018; Volkenhoff et al., 2015). Thus, these previous studies highlight that glia play an important role in organismal longevity via different mechanisms that may or may not involve sensory neurons.
3. Transcriptome studies in mammals
Given that glia appear to regulate ageing, it is important to determine what glial genes might be implicated in these effects. One approach is to establish what glial genes are up or down regulated by ageing. In pursue of this goal, an interesting observation was recently made: the cell types that undergo that most change during ageing in terms of gene expression are glia not neurons. Both Soreq and colleagues, and Boisvert and colleagues, in mice and human respectively, showed that astrocytes- and oligodendrocytes-specific genes, but not neurons-specific genes, undergo the largest shifts in expression during ageing (Boisvert et al., 2018; Soreq et al., 2017). In addition, astrocytes exhibit a reactive phenotype, increased number, volume and complexity, and decreased neuronal and synaptic support as the animal ages (Baulies et al., 2014; Pertusa et al., 2007; Wilhelmsson et al., 2006). Aged astrocytes also display enhanced response to inflammation, which results in a toxic effect on neurons (Clarke et al., 2018). Collectively, these results suggest that glia may be the first cells in the nervous system to respond to “ageing stress”.
4. Protein homeostasis and ageing
As the organism ages, more and more damaged/unfolded proteins accumulate leading to cellular toxicity and eventually cellular demise. The organism though has built in mechanisms to counteract the deleterious effects of the accumulation of unfolded proteins and to prevent cellular death. The response mechanisms that help the cell deal with unfolded proteins include the unfolded protein response in the endoplasmic reticulum and in the mitochondria, and the heath shock response in the cytosol. All these three response mechanisms detect the cellular stress, communicate this stress to the nucleus, and cause change of gene expression which consequently leads to the mounting of a defense program which restores cellular homeostasis. Below we briefly summarize what it is known about these three defense mechanisms and how they relate to ageing.
4.1. The endoplasmic reticulum unfolded protein response (UPRER) and ageing
The ER unfolded protein response (UPRER), one of the major protein quality control mechanisms in the cell, is activated by stress within the endoplasmic reticulum. Accumulation of unfolded/misfolded proteins and lipid stress are the two major ways in which the UPRER is activated (Bravo et al., 2013) (Fig. 2A and B). The UPRER is so critical to organismal homeostasis that its dysregulation is linked to several age-related diseases including Type 2 diabetes, non-alcoholic fatty liver diseases, cancer, and neurodegenerative diseases such as Alzheimer, Parkinson, and Huntington’s disease (Hoozemans et al., 2005; Puri et al., 2008; Ryu et al., 2002; Wang et al., 2009; Ye et al., 2010; Zuleta et al., 2012).
ER stress is regulated by three highly conserved ER-bound membrane proteins: ATF6 (activating transcription factor 6), IRE1α (inositol requiring enzyme 1 alpha), and PERK (Okada et al., 2002; Wang et al., 2000; Ye et al., 2000; Yoshida et al., 2001). Upon ER stress activation, IRE1 and PERK become auto-phosphorylated and dimerize to regulate their own subset of genes to restore ER homeostasis (Maly and Papa, 2014). The activation of ATF6 begins with its cleavage in the Golgi (Chen et al., 2002). The N terminus of ATF6 is then transported into the nucleus where it begins serving as a transcription factor to regulate downstream UPR genes (Fig. 2C and D). Following activation of the UPRER, the responding UPR specific genes regulate protein translation, increase expression of ER chaperones, and clear misfolded proteins for degradation in an attempt to restore ER homeostasis (Brodsky and McCracken, 1999; Rao et al., 2002) (Fig. 2E). However, if the stress accumulates that beyond the UPRER can handle, the cell will die via programmed cell death, autophagy or necrosis (Iurlaro and Munoz-Pinedo, 2016).
The IRE1 branch of the UPRER is the branch that was exploited by Frakes and colleagues in their work (Frakes et al., 2020). This branch has the largest number of regulated genes as compared to the PERK and ATF6 branches. These genes are regulated through the excision by IRE1 of an intron from the XBP1 mRNA. The excision causes a translational frame shift that results in the production of the spliced/activated form of XBP1 protein, an active transcription factor responsible for the induction of the expression of downstream UPR genes (Yoshida et al., 2001) (Fig. 2C and D). Frakes and colleagues indeed expressed the spliced/activated form of xbp-1 (xbp-1s) in C. elegans glia to induce activation of the UPRER in these cells (Frakes et al., 2020). The other well-known IRE1 downstream target is c-jun NH2 terminal kinase (JNK). JNK phosphorylation stimulates pro-apoptotic factors BID and BiM that consequently inhibit anti-apoptotic factors BCL-2, BCL-XL and MCL-1 (Urano et al., 2000). On the other hand, prolonged UPRER activates the regulated IRE1-dependent decay (RIDD), a process by which IRE1 degrades ER-bound mRNAs through cleavage at both stem-loop sites and non-stem-loop sites (Hollien et al., 2009) (Fig. 2C and D). This process is thought to reduce the folding load of nascent proteins entering the ER therefore alleviating ER stress.
As mentioned above, membrane lipid perturbation activates the UPRER and this occurs via engagement of one or more of the three UPR transducers ATF6, IRE1α, and PERK (Fig. 2B). For example, disruption of the ratio between the membrane phosphatidylcholine (PC) and phosphatidylethanolamine (PE), or the depletion of the phospholipid building block inositol strongly activated IRE1 (Halbleib et al., 2017; Ling et al., 2012). Lipid perturbation activates the effectors of the UPRER via their transmembrane domains (Promlek et al., 2011; Volmer et al., 2013). Interestingly, owning to the different activation mechanism, activation of the UPRER by lipid perturbation leads to regulation of genes that are involved in lipid metabolism as opposed to genes encoding chaperones (Acosta-Alvear et al., 2007; Tam et al., 2018).
The activation of the UPRER declines with age, which results in the accumulation of unfolded/misfolded proteins further contributing to disruption of protein homeostasis. These findings are consistent with the observation that there is a decrease in the expression level of ER proteins with ageing, including chaperones such as the protein disulfide isomerase-A1 (PDI) and the immunoglobulin heavy chain-binding protein (GRP78/BiP) (hsp-4 in C. elegans) (Nuss et al., 2008). Thus, ageing reduces the protective functions of the UPRER through a decrease in the pro-apoptotic, autophagy, and lipophagy signaling.
Consistent with data in mammals, longevity studies in C. elegans showed that knockout of the worm ire-1 or xbp-1 genes leads to shorter lifespan as compared to wild type, indicating that the UPRER has an important function in lifespan regulation in the nematode as well (Wang et al., 2018). Moreover, it was shown that worm atf-6 regulates lifespan through ER-mitochondrial calcium homeostasis (Burkewitz et al., 2020). Finally, the Dillin lab showed robust activation of cell-autonomous and cell-nonautonomous UPRER by overexpression of spliced/activated xbp-1(xbp-1s) in both neurons and glia (Frakes et al., 2020; Taylor and Dillin, 2013). These studies support conservation of these mechanisms across species and justify the use of C. elegans as a model to deepen our understanding of the link between the UPR and longevity.
4.2. The mitochondrial unfolded protein response and ageing
There are ~1500 proteins in the mitochondria, but only a handful of these are encoded by the mitochondrial genome; the rest are encoded by nuclear genes. Thus, the vast majority of the mitochondrial proteins are translated in the cytosol and then are transported into the mitochondria. The transport of these proteins into the mitochondria is mediated by the translocase of the outermembrane (TOM) complex and by the presequence pathway, both of which can transport only unfolded proteins (Chacinska et al., 2009). Thus mitochondrial proteins originated in the cytosol of the cell are folded in the mitochondria. This is achieved via the function of two major chaperones: the mtHSP70, which is part of presequence complex and thus folds proteins as they are transported, and the multimeric HSP60–HSP10 machinery in the matrix (Chacinska et al., 2009). In the mitochondria there are also proteases that recognize and degrade proteins that fail to fold (Tatsuta and Langer, 2008). As unfolded proteins accumulate due to ageing or dysfunction in the mitochondrial quality control mechanisms, a compensatory mechanism is activated to try to bring the mitochondria back to homeostasis. This is known as the mitochondrial unfolded protein response (UPRMT) and it leads to the transcription in the nucleus of mitochondrial chaperones, mitochondrial detoxifying factors, mitochondrial transporters, and genes involved in glycolysis (Nargund et al., 2012; Wu et al., 2018; Zhao et al., 2002).
While the UPRMT was first discovered in mammals, it has been investigated much more extensively in C. elegans. Not only C. elegans was instrumental in identifying stimuli that activate the UPRMT and the factors that are involved in the activation and translation of its signal (Pellegrino et al., 2013), but was also essential for discovering a link between the UPRMT and organismal ageing. First, a genome wide RNAi screen aimed at identifying genes involved in mitochondrial function found that the vast majority of these genes also influence ageing (Lee et al., 2003) and second, mutations in genes encoding components of the electron transport chain (ETC) or in genes responsible for mediating the translation of these proteins, such as mrps-5, activate the UPRMT and also extend lifespan (Dillin et al., 2002; Houtkooper et al., 2013). The reason why mutations in components of the ETC cause activation of the UPRMT is because these cause disruption of stoichiometric balance between components of the ETC which are encoded by both mitochondrial and nuclear genomes.
Interestingly, as seen for the activation of the UPRER, the UPRMT can be activated in one tissue and then spread to others via factors acting long distance. Durieux and colleagues showed that impairment of the ETC in neurons causes activation of the UPRMT in the C. elegans intestine, suggesting the existence of extracellular mitokines capable of transmitting this signal long distance (Durieux et al., 2011). A more recent publication has shown that mitokines can be released also by glia to induce the activation of the UPRMT in other tissues. Yin and colleagues recently showed that a polymorphism in the glial-expressed peptide encoding gene rgba-1 controls the rate of age-dependent decline in mating behaviour in C. elegans. Knock-out of either rgba-1 or of the gene encoding the neuropeptide receptor npr-28 slows decline in mating behaviour via a mechanism that requires the function of the histone deacetylase sirtuin gene sir-2.1 and the activation of the UPRMT (Yin et al., 2017). Importantly though, while rgba-1 peptides regulate the rate of ageing, they do not affect lifespan (Yin et al., 2017).
4.3. The heat shock response and ageing
The heat shock response (HSR) is one of the most ancient and conserved cellular survival mechanisms and it relies on the function of proteins that are sensitive to heat shock and other stressors called chaperonins (Cheng et al., 1990; Hohfeld and Hartl, 1994; Liu et al., 1997). The mechanism used by chaperonins to fold proteins is not completely understood but it is known to require ATP and the formation of large structures that function as folding chambers (Lopez-Buesa et al., 1998; Obermann et al., 1998; Wolmarans et al., 2016).
As the organism ages, the efficiency of the HSR declines. This suggests that lifespan could be extended by activation of HSR. Indeed, overexpression of the HSR chaperonins HSP-16 and HSF-1 extends the lifespan in C. elegans (Hsu et al., 2003; Walker and Lithgow, 2003). On the contrary, inactivation of the gene encoding chaperonin HSP22 decreases lifespan in flies (Morrow et al., 2004). Furthermore, life extension mediated by conditions that increase cellular stress, such as starvation, requires the activation of the HSR (Steinkraus et al., 2008; Westerheide et al., 2009).
Interestingly, there is a difference in the activation of neuronal versus glial HSR. For example, cultured astrocytes elicit faster and higher levels of Hsp70 expression after heat shock as compared to cultured cortical neurons (Nishimura et al., 1991) and oligodendrocytes show stronger induction of the HSR as compared to striatal projection neurons after treatment with the HSR inducer, NVP-HSP990 (Carnemolla et al., 2015). We suggest that the higher HSR of glial cells could be harnessed to provide protection in neurodegenerative diseases.
5. Neuropeptides and ageing
Frakes and colleagues showed that activation of the UPRER in C. elegans glia regulates longevity via release of neuropeptides (Frakes et al., 2020). However, they did not establish which neuropeptides are involved in this process. Indeed, they found that the extension of lifespan in C. elegans expressing xbp-1s in glia is blocked in egl-3 mutants; however, EGL-3 is an endopeptidase homologous to mammalian subtilisin-like proprotein convertases that can process many different pro-peptides.
There is a total of 119 neuropeptide precursor genes in C. elegans. Thirty-one of them contain the conserved sequence Phe-Met-Arg-Pheamide, 40 are insulin-like peptides, and 48 are neuropeptide-like protein (nlp) genes (Li and Kim, 2010). Intriguingly, Frakes and colleagues found that the expression of ins-7 and ins-31 was significantly increased in animals over-expressing xbp-1s in CEPsh glia (Frakes et al., 2020). These data may suggest that ins-7 and ins-31 are involved in signaling from the CEP glia to other tissues to extend lifespan. However, another report shows that over-expression of insulin-like peptides ins-7 or ins-31 significantly shortens lifespan (Zheng et al., 2018). This last finding is in line with a report showing that XBP-1 functions with DAF-16, a FOXO-transcription factor that is activated in daf-2 (ortholog of human IGF1R) mutants, to enhance ER stress resistance and to activate new genes that promote longevity (Henis-Korenblit et al., 2010). Indeed, of note is that mutants of daf-2, which encodes the insulin-like peptide ligands receptor, are among the longest-lived mutant strains in C. elegans. Overall, these results suggest that peptides other than ins-7 and ins-31 mediate the effects of xbp-1s overexpression in CEPsh glia on longevity.
No flp genes were found to have differential expression in wild type animals versus animals over-expressing xbp-1s in CEPsh glia. However, of the 48 nlp genes, nlp-31 and nlp-51 were increased in expression (whereas nlp-17, nlp-24, and nlp-77 were decreased) (Frakes et al., 2020). Consistent with these data, single cell sequencing from CEPsh glia revealed that nlp-17 shows low level of expression as compared to the rest of the C. elegans cells (Taylor et al., 2019). Lifespan studies on nlp-31, nlp-51, and on nlp-17 nlp-24, and nlp-77 genes using knockout and over-expression mutants should determine whether CEPsh glia cells regulate lifespan by releasing any of these nlp type peptides. Furthermore, studies on other glial genes (in addition to peptidase gene egl-3 and vesicular release protein gene unc-31) that regulate the release of neuropeptides, both directly and indirectly, should help shed more light on how glial cells, regulate ageing of the entire organism via neuropeptide release. For example, studies on genes encoding ion channels and transporters that directly and indirectly regulate intracellular Ca2+ concentration and therefore membrane fusion, should be of interest.
6. Conclusions and future directions
Ageing is a multidimensional process that involves multiple tissues and cell types. C. elegans, one of the most powerful models to study conserved mechanisms of ageing, is starting to shed new light on exactly how these different cell types communicate to regulate the lifespan of an organism. In particular, glial cells are found capable of sending long range signals to other tissues thereby regulating their stress response and organismal ageing. These are exciting new findings that not only advance our understanding of ageing but also cement our appreciation of glia as cells capable of communicating with other cell types both locally and long distance.
However, more remains to be learned. For example, Frakes and colleagues showed that overexpression of xbp-1s in the AMsh glia is also capable of inducing UPRER in the intestine, to an extent similar to what seen in animals overexpressing xbp-1 in the CEPsh glia. However, in animals overexpressing xbp-1s in the AMsh glia there is no extension of lifespan (Frakes et al., 2020). Does this mean that CEPsh glia release two different types of neuropeptides, one needed for activation of the UPRER in the intestine and one needed for the extension of lifespan? Perhaps AMsh release only the first type of peptide? If this is the case, comparing the AMsh and CEPsh transcriptomes, especially in animals expressing xbp-1s in these cells, should help in the identification of these peptides. If AMsh glia can activate cell-nonautonomous UPRER in the intestine, can they perhaps increase lifespan in conditions not explored in Frakes and colleagues’ work and via a mechanism distinct from sensory neurons’ function (Apfeld and Kenyon, 1999; Bacaj et al., 2008; Singhvi et al., 2016)? Future studies in which the UPRER is activated via the engagement of the other UPRER pathways (PERK and ATF6) or via lipid perturbation in AMsh glia may help address these questions. In line with what discussed above, cell-autonomous activation of the UPRER in the intestine does not extend lifespan, again underscoring that activation of the UPRER per se is not sufficient to extend lifespan (Frakes et al., 2020; Taylor and Dillin, 2013).
Another intriguing result is that while rescue of unc-31 and egl-3 in CEPsh glia fully rescues lifespan extension, it has only a modest effect on rescuing the activation of UPRER in the intestine. While this might be due to lower level of expression of unc-31 and egl-3 cDNAs, as compared to genomic DNA, impacting more the cell-nonautonomous activation of the UPRER than lifespan extension, it also suggests that the intestine might require signaling from other tissues to mount a robust UPRER. Conversely, these data may suggest that activation of the UPRER in tissues other than the intestine, that perhaps poorly express the hsp-4::GFP reporter, is important to extend lifespan (Frakes et al., 2020). Finally, an entire array of environmental and genetic factors that influence glia development and function should be investigated in the context of ageing. We believe that C. elegans will be again instrumental in these studies given its amenability and given that in this organism neurons do not need glia to survive (Singhvi and Shaham, 2019). A good starting point will be to investigate the genes that Frakes and colleagues have found using RNA sequencing to be enriched in C. elegans overexpressing xbp-1s in CEPsh glia (Frakes et al., 2020).
Acknowledgments
Work in the Bianchi’s laboratory has been supported by the National Institute of Health (NS105616, NS106951, NS081259, NS070969 and NS049511) and the American Cancer Society (RGS-09-043-01-DDC).
Footnotes
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
References
- Acosta-Alvear D, Zhou Y, Blais A, Tsikitis M, Lents NH, Arias C, Lennon CJ, Kluger Y, Dynlacht BD, 2007. XBP1 controls diverse cell type- and condition-specific transcriptional regulatory networks. Mol. Cell 27, 53–66. [DOI] [PubMed] [Google Scholar]
- Apfeld J, Kenyon C, 1999. Regulation of lifespan by sensory perception in Caenorhabditis elegans. Nature 402, 804–809. [DOI] [PubMed] [Google Scholar]
- Bacaj T, Tevlin M, Lu Y, Shaham S, 2008. Glia are essential for sensory organ function in C. elegans. Science 322, 744–747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baulies S, Cusido M, Tresserra F, Rodriguez I, Ubeda B, Ara C, Fabregas R, 2014. Pregnancy-associated breast cancer: an analytical observational study. Med. Clin 142, 200–204. [DOI] [PubMed] [Google Scholar]
- Bianchi L, 2020. C. elegans glia are bona fide odorant receptor cells. Neuron 108, 588–589. [DOI] [PubMed] [Google Scholar]
- Boisvert MM, Erikson GA, Shokhirev MN, Allen NJ, 2018. The aging astrocyte transcriptome from multiple regions of the mouse brain. Cell Rep. 22, 269–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo R, Parra V, Gatica D, Rodriguez AE, Torrealba N, Paredes F, Wang ZV, Zorzano A, Hill JA, Jaimovich E, Quest AFG, Lavandero S, 2013. Endoplasmic reticulum and the unfolded protein response: dynamics and metabolic integration. Int. Rev. Cell Mol. Biol 301, 215–290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brodsky JL, McCracken AA, 1999. ER protein quality control and proteasome-mediated protein degradation. Semin. Cell Dev. Biol 10, 507–513. [DOI] [PubMed] [Google Scholar]
- Burkewitz K, Feng GM, Dutta S, Kelley CA, Steinbaugh M, Cram EJ, Mair WB, 2020. Atf-6 regulates lifespan through ER-mitochondrial calcium homeostasis. Cell Rep. 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao Y, Chtarbanova S, Petersen AJ, Ganetzky B, 2013. Dnr1 mutations cause neurodegeneration in Drosophila by activating the innate immune response in the brain. Proc. Natl. Acad. Sci. U. S. A 110, E1752–E1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carnemolla A, Lazell H, Moussaoui S, Bates GP, 2015. In vivo profiling reveals a competent heat shock response in adult neurons: implications for neurodegenerative disorders. PLoS One 10, e0131985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chacinska A, Koehler CM, Milenkovic D, Lithgow T, Pfanner N, 2009. Importing mitochondrial proteins: machineries and mechanisms. Cell 138, 628–644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Shen J, Prywes R, 2002. The luminal domain of ATF6 senses endoplasmic reticulum (ER) stress and causes translocation of ATF6 from the ER to the Golgi. J. Biol. Chem 277, 13045–13052. [DOI] [PubMed] [Google Scholar]
- Cheng MY, Hartl FU, Horwich AL, 1990. The mitochondrial chaperonin hsp60 is required for its own assembly. Nature 348, 455–458. [DOI] [PubMed] [Google Scholar]
- Clarke LE, Liddelow SA, Chakraborty C, Munch AE, Heiman M, Barres B, 2018. Normal aging induces A1-like astrocyte reactivity. Proc. Natl. Acad. Sci. U. S. A 115, E1896–E1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman PD, Romano J, Lapham L, Simon W, 1985. Cell counts in cerebral-cortex of an autistic patient. J. Autism Dev. Disord 15, 245–255. [DOI] [PubMed] [Google Scholar]
- Cotter DR, Pariante CM, Everall IP, 2001. Glial cell abnormalities in major psychiatric disorders: the evidence and implications. Brain Res. Bull 55, 585–595. [DOI] [PubMed] [Google Scholar]
- Damier P, Hirsch EC, Zhang P, Agid Y, Javoy-Agid F, 1993. Glutathione peroxidase, glial cells and Parkinson’s disease. Neuroscience 52, 1–6. [DOI] [PubMed] [Google Scholar]
- Delgado MG, Oliva C, Lopez E, Ibacache A, Galaz A, Delgado R, Barros LF, Sierralta J, 2018. Chaski, a novel Drosophila lactate/pyruvate transporter required in glia cells for survival under nutritional stress. Sci. Rep 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dillin A, Hsu AL, Arantes-Oliveira N, Lehrer-Graiwer J, Hsin H, Fraser AG, Kamath RS, Ahringer J, Kenyon C, 2002. Rates of behavior and aging specified by mitochondrial function during development. Science 298, 2398–2401. [DOI] [PubMed] [Google Scholar]
- Duan D, Zhang H, Yue X, Fan Y, Xue Y, Shao J, Ding G, Chen D, Li S, Cheng H, Zhang X, Zou W, Liu J, Zhao J, Wang L, Zhao B, Wang Z, Xu S, Wen Q, Liu J, Duan S, Kang L, 2020. Nov 25. Sensory glia detect repulsive odorants and drive olfactory adaptation. Neuron 108 (4), 707–721.e8. 10.1016/j.neuron.2020.08.026. Epub 2020 Sep 23.PMID: 32970991. [DOI] [PubMed] [Google Scholar]
- Durieux J, Wolff S, Dillin A, 2011. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144, 79–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frakes AE, Metcalf MG, Tronnes SU, Bar-Ziv R, Durieux J, Gildea HK, Kandahari N, Monshietehadi S, Dillin A, 2020. Four glial cells regulate ER stress resistance and longevity via neuropeptide signaling in C. elegans. Science 367, 436–440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galvani L, 1791. Bon. Sci. Art. Inst. Acad. Comm, Vol. 56. [Google Scholar]
- Halbleib K, Pesek K, Covino R, Hofbauer HF, Wunnicke D, Hanelt I, Hummer G, Ernst R, 2017. Activation of the unfolded protein response by lipid bilayer stress. Mol. Cell 67, 673–684 e678. [DOI] [PubMed] [Google Scholar]
- Henis-Korenblit S, Zhang PC, Hansen M, McCormick M, Lee SJ, Cary M, Kenyon C, 2010. Insulin/IGF-1 signaling mutants reprogram ER stress response regulators to promote longevity. Proc. Natl. Acad. Sci. U. S. A 107, 9730–9735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hohfeld J, Hartl FU, 1994. Role of the chaperonin cofactor Hsp10 in protein folding and sorting in yeast mitochondria. J. Cell Biol 126, 305–315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollien J, Lin JH, Li H, Stevens N, Walter P, Weissman JS, 2009. Regulated Ire1-dependent decay of messenger RNAs in mammalian cells. J. Cell Biol 186, 323–331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoozemans JJM, Veerhuis R, Van Haastert ES, Rozemuller JM, Baas F, Eikelenboom P, Scheper W, 2005. The unfolded protein response is activated in Alzheimer’s disease. Acta Neuropathol. 110, 165–172. [DOI] [PubMed] [Google Scholar]
- Houtkooper RH, Mouchiroud L, Ryu D, Moullan N, Katsyuba E, Knott G, Williams RW, Auwerx J, 2013. Mitonuclear protein imbalance as a conserved longevity mechanism. Nature 497, 451–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu AL, Murphy CT, Kenyon C, 2003. Regulation of aging and age-related disease by DAF-16 and heat-shock factor. Science 300, 1142–1145. [DOI] [PubMed] [Google Scholar]
- Iurlaro R, Munoz-Pinedo C, 2016. Cell death induced by endoplasmic reticulum stress. FEBS J. 283, 2640–2652. [DOI] [PubMed] [Google Scholar]
- Kammoun HL, Chabanon H, Hainault I, Luquet S, Magnan C, Koike T, Ferre P, Foufelle F, 2009. GRP78 expression inhibits insulin and ER stress-induced SREBP-1c activation and reduces hepatic steatosis in mice. J. Clin. Invest 119, 1201–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz M, Corson F, Keil W, Singhal A, Bae A, Lu Y, Liang Y, Shaham S, 2019. Glutamate spillover in C. elegans triggers repetitive behavior through presynaptic activation of MGL-2/mGluR5. Nat. Commun 10, 1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kounatidis I, Chtarbanova S, Cao Y, Hayne M, Jayanth D, Ganetzky B, Ligoxygakis P, 2017. NF-kappa B immunity in the brain determines Fly lifespan in healthy aging and age-related Neurodegeneration. Cell Rep. 19, 836–848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SS, Lee RY, Fraser AG, Kamath RS, Ahringer J, Ruvkun G, 2003. A systematic RNAi screen identifies a critical role for mitochondria in C. elegans longevity. Nat. Genet 33, 40–48. [DOI] [PubMed] [Google Scholar]
- Lee S, Yoon BE, Berglund K, Oh SJ, Park H, Shin HS, Augustine GJ, Lee CJ, 2010. Channel-mediated tonic GABA release from glia. Science 330, 790–796. [DOI] [PubMed] [Google Scholar]
- Li C, Kim K, 2010. Neuropeptide gene families in Caenorhabditis elegans. In: Neuropeptide Systems as Targets for Parasite and Pest Control, Vol. 692, pp. 98–137. [DOI] [PubMed] [Google Scholar]
- Ling J, Chaba T, Zhu LF, Jacobs RL, Vance DE, 2012. Hepatic ratio of phosphatidylcholine to phosphatidylethanolamine predicts survival after partial hepatectomy in mice. Hepatology 55, 1094–1102. [DOI] [PubMed] [Google Scholar]
- Liu XD, Liu PC, Santoro N, Thiele DJ, 1997. Conservation of a stress response: human heat shock transcription factors functionally substitute for yeast HSF. EMBO J. 16, 6466–6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Buesa P, Pfund C, Craig EA, 1998. The biochemical properties of the ATPase activity of a 70-kDa heat shock protein (Hsp70) are governed by the C-terminal domains. Proc. Natl. Acad. Sci. U. S. A 95, 15253–15258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maly DJ, Papa FR, 2014. Druggable sensors of the unfolded protein response. Nat. Chem. Biol 10, 892–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuary PR, Liao CY, Chang JT, Kumsta C, She X, Davis A, Chu CC, Gelino S, Gomez-Amaro RL, Petrascheck M, Brill LM, Ladiges WC, Kennedy BK, Hansen M, 2016. C. elegans S6K mutants require a creatine-kinase-like effector for lifespan extension. Cell Rep. 14, 2059–2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrow G, Samson M, Michaud S, Tanguay RM, 2004. Overexpression of the small mitochondrial Hsp22 extends Drosophila lifespan and increases resistance to oxidative stress. FASEB J. 18, 598–599. [DOI] [PubMed] [Google Scholar]
- Nagele RG, Wegiel J, Venkataraman V, Imaki H, Wang KC, Wegiel J, 2004. Contribution of glial cells to the development of amyloid plaques in Alzheimer’s disease. Neurobiol. Aging 25, 663–674. [DOI] [PubMed] [Google Scholar]
- Nargund AM, Pellegrino MW, Fiorese CJ, Baker BM, Haynes CM, 2012. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337, 587–590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura RN, Dwyer BE, Clegg K, Cole R, de Vellis J, 1991. Comparison of the heat shock response in cultured cortical neurons and astrocytes. Brain Res. Mol. Brain Res 9, 39–45. [DOI] [PubMed] [Google Scholar]
- Nuss JE, Choksi KB, DeFord JH, Papaconstantinou J, 2008. Decreased enzyme activities of chaperones PD1 and BiP in aged mouse livers. Biochem. Biophys. Res. Commun 365, 355–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obermann WM, Sondermann H, Russo AA, Pavletich NP, Hartl FU, 1998. In vivo function of Hsp90 is dependent on ATP binding and ATP hydrolysis. J. Cell Biol 143, 901–910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oikonomou G, Shaham S, 2011. The glia of Caenorhabditis elegans. Glia 59, 1253–1263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada T, Yoshida H, Akazawa R, Negishi M, Mori K, 2002. Distinct roles of activating transcription factor 6 (ATF6) and double-stranded RNA-activated protein kinase-like endoplasmic reticulum kinase (PERK) in transcription during the mammalian unfolded protein response. Biochem. J 366, 585–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pellegrino MW, Nargund AM, Haynes CM, 2013. Signaling the mitochondrial unfolded protein response. Biochim. Biophys. Acta 1833, 410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pertusa M, Garcia-Matas S, Rodriguez-Farre E, Sanfeliu C, Cristofol R, 2007. Astrocytes aged in vitro show a decreased neuroprotective capacity. J. Neurochem 101, 794–805. [DOI] [PubMed] [Google Scholar]
- Promlek T, Ishiwata-Kimata Y, Shido M, Sakuramoto M, Kohno K, Kimata Y, 2011. Membrane aberrancy and unfolded proteins activate the endoplasmic reticulum stress sensor Ire1 in different ways. Mol. Biol. Cell 22, 3520–3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puri P, Mirshahi F, Cheung O, Natarajan R, Maher JW, Kellum JM, Sanyal AJ, 2008. Activation and dysregulation of the unfolded protein response in nonalcoholic fatty liver disease. Gastroenterology 134, 568–576. [DOI] [PubMed] [Google Scholar]
- Rao RV, Hermel E, Castro-Obregon S, del Rio G, Ellerby LM, Ellerby HM, Bredesen DE, 2001. Coupling endoplasmic reticulum stress to the cell death program – mechanism of caspase activation. J. Biol. Chem 276, 33869–33874. [DOI] [PubMed] [Google Scholar]
- Rao RV, Peel A, Logvinova A, del Rio G, Hermel E, Yokota T, Goldsmith PC, Ellerby LM, Ellerby HM, Bredesen DE, 2002. Coupling endoplasmic reticulum stress to the cell death program: role of the ER chaperone GRP78. FEBS Lett. 514, 122–128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu EJ, Harding HP, Angelastro JM, Vitolo OV, Ron D, Greene LA, 2002. Endoplasmic reticulum stress and the unfolded protein response in cellular models of Parkinson’s disease. J. Neurosci 22, 10690–10698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schonrock LM, Kuhlmann T, Adler S, Bitsch A, Bruck W, 1998. Identification of glial cell proliferation in early multiple sclerosis lesions. Neuropathol. Appl. Neurobiol 24, 320–330. [DOI] [PubMed] [Google Scholar]
- Singhvi A, Shaham S, 2019. Glia-neuron interactions in Caenorhabditis elegans. Annu. Rev. Neurosci 42, 149–168. [DOI] [PubMed] [Google Scholar]
- Singhvi A, Liu B, Friedman CJ, Fong J, Lu Y, Huang XY, Shaham S, 2016. A glial K/cl transporter controls neuronal receptive ending shape by chloride inhibition of an rGC. Cell 165, 936–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soreq L, Rose J, Soreq E, Hardy J, Trabzuni D, Cookson MR, Smith C, Ryten M, Patani R, Ule J, Consortium, U.B.E., Consor, N.A.B.E., 2017. Major shifts in glial regional identity are a transcriptional hallmark of human brain aging. Cell Rep. 18, 557–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steinkraus KA, Smith ED, Davis C, Carr D, Pendergrass WR, Sutphin GL, Kennedy BK, Kaeberlein M, 2008. Dietary restriction suppresses proteotoxicity and enhances longevity by an hsf-1-dependent mechanism in Caenorhabditis elegans. Aging Cell 7, 394–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tam AB, Roberts LS, Chandra V, Rivera IG, Nomura DK, Forbes DJ, Niwa M, 2018. The UPR activator ATF6 responds to proteotoxic and lipotoxic stress by distinct mechanisms. Dev. Cell 46 (327–343), e327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tatsuta T, Langer T, 2008. Quality control of mitochondria: protection against neurodegeneration and ageing. EMBO J. 27, 306–314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor RC, Dillin A, 2013. XBP-1 is a cell-nonautonomous regulator of stress resistance and longevity. Cell 153, 1435–1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor SR, Santpere G, Reilly M, Glenwinkel L, Poff A, McWhirter R, Xu C, Weinreb A, Basavaraju M, Cook SJ, Barrett A, Abrams A, Vidal B, Cros C, Rafi I, Sestan N, Hammarlund M, Hobert O, Miller DMI, 2019. Expression profiling of the mature C. elegans nervous system by single-cell RNA-sequencing. BioRxiv. 10.1101/737577. [DOI] [Google Scholar]
- Urano F, Wang XZ, Bertolotti A, Zhang YH, Chung P, Harding HP, Ron D, 2000. Coupling of stress in the ER to activation of JNK protein kinases by transmembrane protein kinase IRE1. Science 287, 664–666. [DOI] [PubMed] [Google Scholar]
- Virchow R, 1856. Cellularpathologie. [Google Scholar]
- Volkenhoff A, Weiler A, Letzel M, Stehling M, Klambt C, Schirmeier S, 2015. Glial glycolysis is essential for neuronal survival in Drosophila. Cell Metab. 22, 437–447. [DOI] [PubMed] [Google Scholar]
- Volmer R, van der Ploeg K, Ron D, 2013. Membrane lipid saturation activates endoplasmic reticulum unfolded protein response transducers through their transmembrane domains. Proc. Natl. Acad. Sci. U. S. A 110, 4628–4633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker GA, Lithgow GJ, 2003. Lifespan extension in C. elegans by a molecular chaperone dependent upon insulin-like signals. Aging Cell 2, 131–139. [DOI] [PubMed] [Google Scholar]
- Walkowicz L, Kijak E, Krzeptowski W, Gorska-Andrzejak J, Stratoulias V, Woznicka O, Chwastek E, Heino TI, Pyza EM, 2017. Downregulation of DmMANF in glial cells results in neurodegeneration and affects sleep and lifespan in Drosophila melanogaster. Front. Neurosci 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Shen JS, Arenzana N, Tirasophon W, Kaufman RJ, Prywes R, 2000. Activation of ATF6 and an ATF6 DNA binding site by the endoplasmic reticulum stress response. J. Biol. Chem 275, 27013–27020. [DOI] [PubMed] [Google Scholar]
- Wang M, Wey S, Zhang Y, Ye R, Lee AS, 2009. Role of the unfolded protein response regulator GRP78/BiP in development, cancer, and neurological disorders. Antioxid. Redox Signal 11, 2307–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Beaudoin-Chabot C, Thibault G, 2018. Glucose increases the lifespan of post-reproductive C. elegans independently of FOXO. BioRvix. 10.1101/347435. [DOI] [Google Scholar]
- Westerheide SD, Anckar J, Stevens SM Jr., Sistonen L, Morimoto RI, 2009. Stress-inducible regulation of heat shock factor 1 by the deacetylase SIRT1. Science 323, 1063–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White JG, Southgate E, Thomson JN, Brenner S, 1986. The structure of the nervous system of the nematode Caenorhabditis elegans. Philos. Trans. R. Soc. B Biol. Sci 314, 1–340. [DOI] [PubMed] [Google Scholar]
- Wilhelmsson U, Bushongt EA, Price DL, Smarr BL, Phung V, Terada M, Ellisman MH, Pekny M, 2006. Redefining the concept of reactive astrocytes as cells that remain within their unique domains upon reaction to injury. Proc. Natl. Acad. Sci. U. S. A 103, 17513–17518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolmarans A, Lee B, Spyracopoulos L, LaPointe P, 2016. The mechanism of Hsp90 ATPase stimulation by Aha1. Sci. Rep 6, 33179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Z, Senchuk MM, Dues DJ, Johnson BK, Cooper JF, Lew L, Machiela E, Schaar CE, DeJonge H, Blackwell TK, Van Raamsdonk JM, 2018. Mitochondrial unfolded protein response transcription factor ATFS-1 promotes longevity in a long-lived mitochondrial mutant through activation of stress response pathways. BMC Biol. 16, 147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye J, Rawson RB, Komuro R, Chen X, Dave UP, Prywes R, Brown MS, Goldstein JL, 2000. ER stress induces cleavage of membrane-bound ATF6 by the same proteases that process SREBPs. Mol. Cell 6, 1355–1364. [DOI] [PubMed] [Google Scholar]
- Ye R, Jung DY, Jun JY, Li J, Luo S, Ko HJ, Kim JK, Lee AS, 2010. Grp78 heterozygosity promotes adaptive unfolded protein response and attenuates diet-induced obesity and insulin resistance. Diabetes 59, 6–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin JA, Gao G, Liu XJ, Hao ZQ, Li K, Kang XL, Li H, Shan YH, Hu WL, Li HP, Cai SQ, 2017. Genetic variation in glia-neuron signalling modulates ageing rate. Nature 551, 198–203. [DOI] [PubMed] [Google Scholar]
- Yorimitsu T, Nair U, Yang ZF, Klionsky DJ, 2006. Endoplasmic reticulum stress triggers autophagy. J. Biol. Chem 281, 30299–30304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida H, Matsui T, Yamamoto A, Okada T, Mori K, 2001. XBP1 mRNA is induced by ATF6 and spliced by IRE1 in response to ER stress to produce a highly active transcription factor. Cell 107, 881–891. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Wang J, Levichkin IV, Stasinopoulos S, Ryan MT, Hoogenraad NJ, 2002. A mitochondrial specific stress response in mammalian cells. EMBO J. 21, 4411–4419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng SQ, Chiu H, Boudreau J, Papanicolaou T, Bendena W, Ian CS, 2018. A functional study of all 40 Caenorhabditis elegans insulin-like peptides. J. Biol. Chem 293, 16912–16922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuleta A, Vidal RL, Armentano D, Parsons G, Hetz C, 2012. MV-mediated delivery of the transcription factor XBP1s into the striatum reduces mutant Huntingtin aggregation in a mouse model of Huntington’s disease. Biochem. Biophys. Res. Commun 420, 558–563. [DOI] [PubMed] [Google Scholar]