Abstract
Background
Schistosomiasis, or bilharzia, is a parasitic disease caused by trematode flatworms of the genus Schistosoma. Infection by Schistosoma mansoni in humans results when cercariae emerge into water from freshwater snails in the genus Biomphalaria and seek out and penetrate human skin. The snail Biomphalaria straminea is native to South America and is now also present in Central America and China, and represents a potential vector host for spreading schistosomiasis. To date, genomic information for the genus is restricted to the neotropical species Biomphalaria glabrata. This limits understanding of the biology and management of other schistosomiasis vectors, such as B. straminea.
Findings
Using a combination of Illumina short‐read, 10X Genomics linked‐read, and Hi‐C sequencing data, our 1.005 Gb B. straminea genome assembly is of high contiguity, with a scaffold N50 of 25.3 Mb. Transcriptomes from adults were also obtained. Developmental homeobox genes, hormonal genes, and stress-response genes were identified, and repeat content was annotated (40.68% of genomic content). Comparisons with other mollusc genomes (including Gastropoda, Bivalvia, and Cephalopoda) revealed syntenic conservation, patterns of homeobox gene linkage indicative of evolutionary changes to gene clusters, expansion of heat shock protein genes, and the presence of sesquiterpenoid and cholesterol metabolic pathway genes in Gastropoda. In addition, hormone treatment together with RT-qPCR assay reveal a sesquiterpenoid hormone responsive system in B. straminea, illustrating that this renowned insect hormonal system is also present in the lophotrochozoan lineage.
Conclusion
This study provides the first genome assembly for the snail B. straminea and offers an unprecedented opportunity to address a variety of phenomena related to snail vectors of schistosomiasis, as well as evolutionary and genomics questions related to molluscs more widely.
Background
With >240 million people worldwide estimated to require treatment, the World Health Organization considers schistosomiasis to be the second most prevalent parasitic disease after malaria [1]. As such, schistosomiasis is a global health problem that causes considerable economic and social burdens.
Infection by Schistosoma mansoni (NCBI:txid6183) in humans results when cercariae emerge into the water from their freshwater snail intermediate hosts in the genus Biomphalaria, and seek out and penetrate submerged body parts through the skin. Once inside the human body, adult worms lay eggs, which are deposited in the blood venules and will cross the intestinal wall to leave the body in the faeces. In addition, eggs that fail to cross the intestinal wall (named “reflux eggs”) circulate to the liver where they grow, emerge, and cause disease. Miracidia larvae hatch from eggs that reach water, then seek out and penetrate a new snail intermediate host. Following this, sporocysts develop in the infected snails, and subsequently free-living cercariae emerge from the snail into the water, completing the parasitic life cycle. Among the 34 described species of Biomphalaria snails, 18 species (including Biomphalaria straminea) have been demonstrated to be potential vectors for S. mansoni. Different geographical locations are dominated by different species of Biomphalaria.
The native range of Biomphalaria snails is South America and Africa [2, 3]. However, several species have been introduced to other areas, presenting a risk of schistosomiasis infection. The occurrence of B. straminea in Asia was first reported at Lam Tsuen valley in Hong Kong during the 1970s ([4]; Fig. 1A), presumably having somehow spread from its native range in South America into Central America and southern China [5]. B. straminea have since been identified at a number of locations in Hong Kong and Guangdong Province [4, 6–9]. While S. mansoni is not yet endemic in either Hong Kong or mainland China, cases of schistosomiasis caused by the parasite are currently increasing in China. According to the records from the database of the National Notifiable Disease Report System, 355 cases of imported schistosomiasis cases had been reported in 15 provinces in China between 1979 and 2019, including 78 cases infected with S. mansoni, 262 cases with Schistosoma haematobia, and 15 cases with unidentified Schistosoma. Since B. straminea had already been discovered in Guangdong province in southern China, it is believed that the imported S. mansoni increases the risk of its transmission in China [10, 11].
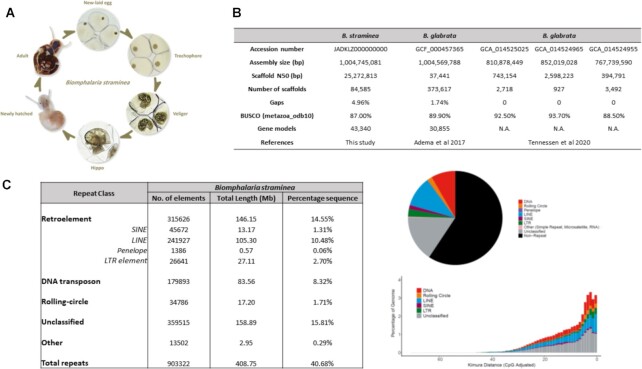
Figure 1.
: (A) Life cycle of snail Biomphalaria straminea; (B) comparison of snail Biomphalaria genome assembly quality [12, 13]; (C) transposable elements in Biomphalaria straminea.
Whole-genome sequences are valuable resources for obtaining deeper understanding of the biological characteristics of any organism. Despite the importance of the phylum Mollusca, there is a lack of genomic resources [88]. In the case of B. straminea, such a resource will affect questions of how they may interact with S. mansoni and how similar the genetic mechanisms are between different Biomphalaria species, with possible implications for how treatments and management strategies might be transferable. To date, only the genome of Biomphalaria glabrata has been sequenced and analysed ([12, 13]; Fig. 1B), and a high-quality genome of B. straminea is lacking, hindering further understanding of the species. To address this issue, we provide and analyse a high-quality genome assembly for B. straminea together with accompanying transcriptomes.
Results and Discussion
Genome quality evaluation
Genomic DNA (gDNA) was extracted from single individuals of B. straminea (Fig. 1A). Genome sequences were first assembled using short reads followed by scaffolding with Hi-C data. The genome assembly (without the mitochondrial genome) is 1.005 Gb with a scaffold N50 of 25.3 Mb (Fig. 1B). This high physical contiguity is matched by high completeness, with an 87.0% complete BUSCO score [14] (Fig. 1B). A total of 43,340 gene models, including 3,122 transfer RNA and 40,218 protein-coding genes, were generated by mapping transcriptome data to the genome assembly (Supplementary Information S1). The mean exon length is 262 bp, mean intron length is 1,603 bp, and mean deduced protein length is 377 amino acids. The genome quality generated in this study is comparable to the previously published genome assemblies of another schistosomiasis-carrying vector snail, B. glabrata ([12, 13]; Fig. 1B).
Repeat element analysis
We identified a total repeat content of 40.68% in the genome of B. straminea (Fig. 1C), demonstrating that repeats make up a large proportion of total genome size in the species. A considerable proportion of repeats were unclassified (15.81%), suggesting that many of the annotated repeats represent new repeat families (Fig. 1C), which is not unexpected given the relatively sparse attention given to the analysis of repeats in gastropod molluscs to date. Of the remaining repeats, LINE elements and DNA transposons are most abundant (LINEs: 10.48%, DNA transposons: 8.32%), whereas SINEs, LTR elements, and rolling-circle elements are present only in low proportions (LTR elements: 2.70%, rolling-circle elements: 1.71%, SINEs: 1.31%) (Fig. 1C). Consideration of a repeat landscape plot suggests that there has been a long-term ongoing expansion of repeats in B. straminea, with a recent spike in activity. The recent spike is evident from the relatively large percentage of repeats in the genome that are separated from their family consensus sequences by short distances, while the long tail of increasing divergence from the consensus is suggestive of a gradual increase in activity over a relatively long period (Fig. 1C). LINEs and DNA transposons have expanded most significantly; however, there has also been a less considerable expansion of LTR and rolling circle elements (Fig. 1C).
Homeobox-containing gene content and linkage
Hox cluster genes
Homeobox genes are transcription factors involved in regulating animal development. Not only are they highly conserved between distantly related lineages, but also many of the genes are linked or clustered in genomes. Besides the most well-known clusters such as the Hox and ParaHox clusters, many homeobox genes are linked including other ANTP class genes in NK and SuperHox clusters, and also amongst other classes of PRD, TALE, and SINE homeobox genes [15–17]. These clusters have been maintained or dispersed differently in different animal lineages. Changes to gene clustering may represent the breakdown of regulatory constraints that normally maintain clusters and are thought to be the mechanism holding together the tightly regulated Hox cluster, for instance. Genomic clustering also reflects the ancient origins of many of these homeobox genes by tandem duplication, e.g., the 4 ANTP clusters in the bilaterian ancestor that arose via subsequent expansions from a single proto-ANTP gene [18]. Among molluscs, a diverse phylum to which gastropods belong, alongside other conchiferans (monoplacophorans, bivalves, scaphopods, and cephalopods), as well as aculiferans (aplacophorans and polyplacophorans), some of the diversity of body plans may be underpinned by changes to developmental genes like homeobox genes. Hox genes have been co-opted to the development of novel morphological structures in cephalopods [89], and this corresponds to a breakdown of the Hox cluster across several large scaffolds, and the loss of a few genes [90]. Other mollusc genomes show a breakdown of homeobox clustering overall, like the Pacific oyster (Crassostrea gigas [19]), while a more recent chromosome-level assembly reveals large-scale patterns of linkage in Magallana hongkongensis [20]. This genome assembly of B. straminea improves our understanding of homeobox gene linkage in comparison to other molluscs, which are lophotrochozoans and, alongside well-studied ecdysozoans like flies, provide a more thorough protostome comparison to vertebrates, which are within the Deuterostomia.
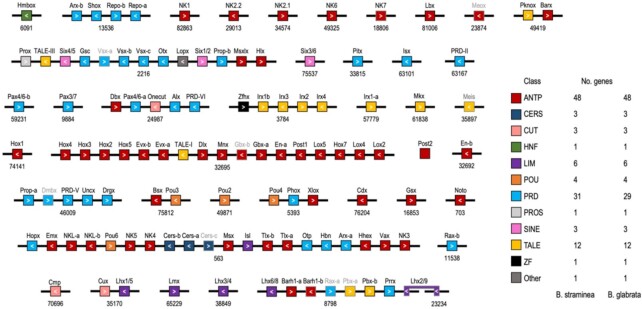
We found 114 homeobox genes in the genome of B. straminea, belonging to 11 recognized classes, and 1 lophotrochozoan-specific gene, Lopx (Supplementary information S2a; [21]). Many of these genes are clustered (situated on the same chromosome with no or very few non-homeobox genes in between) or linked (on the same chromosome, but with intervening non-homeobox genes) in the genome (Fig. 2). Nine of the 11 Hox genes are found on scaffold 32,695, in an arrangement that suggests several intrachromosomal rearrangements. In an ordered cluster as seen in the gastropod L. gigantea, for instance, the Hox genes are situated in the genome in the ancestral bilaterian order from anterior-acting Hox1 to posterior-acting Post1, and no other non-Hox genes are found amongst the Hox genes [22]. Here, however, we find that Hox2, Hox3, and Hox4 are upstream of Hox5. In addition, Hox2–Hox5 are downstream of the posterior half of the cluster, including Lox5, Hox7, Lox4, Lox2, and Post1. Hox1 is found on another scaffold, while the sequence for Post2 is not in the genomic assembly, although its sequence is found in our transcriptome data. The Hox arrangement in B. straminea provides more linkage information than the B. glabrata assembly, where the short scaffolds corroborate only fragments of the Hox cluster like the linkage of Hox4, Hox3, and Hox2 but do not confirm the rearrangements in B. straminea, such as the linkage of Hox5 to Hox2 (Supplementary Information S2b). We do see a difference in the arrangement of the posterior half of the Hox cluster, however, where in B. glabrata, Lox4, Lox2, Post2, and Post1 are linked in that order on scaffold 139, with Lox4 and Lox2 in the negative strand and Post2 and Post1 on the positive, which is slightly different from many other molluscs in which only Post1 differs in orientation relative to the remainder of the posterior end of the Hox cluster genes [20, 22]. In B. straminea, there has been a rearrangement separating Post1, placing it with Lox5 and Hox7 and in the same orientation as Lox4 and Lox2 (Fig. 2). Thus, the Hox genes of Biomphalaria seem highly rearranged relative to the ancestral order and each other. Clearly then, there are no (or minimal) long-range regulatory mechanisms operating across these genes that could have constrained their organization and prevented rearrangement. At most, there may be remains of some form of subcluster mechanisms, such as enhancer sharing, operating over the small regions (i.e., Hox2–4 and Lox2–4) whose similar arrangement may be indicative of constraints conserved across Biomphalaria species. Future expression and regulatory element analyses may help resolve this possibility.
Figure 2.
: Distribution of Homeoboxes in the genome of Biomphalaria straminea. Class is denoted by colour; arrows show orientation on each scaffold, which are represented by black lines and are numbered underneath. Post2 is not found in the genomic sequence but is found in the transcriptome so is not shown on a scaffold. Grey gene names and box outlines denote partial homeodomain sequences.
ParaHox cluster genes
The ParaHox cluster is the evolutionary sister to the Hox cluster [91]. The homeodomains of the 3 ParaHox genes (Gsx, Xlox, and Cdx) are found on 3 separate scaffolds in B. straminea (Fig. 2); however, 3 upstream exons of Cdx are on scaffold 5,393, which also has the Xlox gene (Supplementary Information S2a). This is in contrast to the genome of B. glabrata, where Gsx and Xlox are linked on scaffold 3 (Supplementary Information S2a and b). Perhaps this pattern reflects maintained linkage between all 3 ParaHox genes in Biomphalaria species and only because of the draft level of all the assemblies this is not evident. However, if this is the case, the ParaHox genes are separated by large amounts of sequence and have not retained the ancestral order of Gsx-Xlox-Cdx. B. glabrata Xlox is nearly 4 Mb from the start of its scaffold, while in B. straminea, Xlox is at a location with another homeobox-containing gene (Phox) 15 Mb away on 1 side and the first 3 Cdx exons are almost 5 Mb away on the other side of Xlox. Thus, although the Biomphalaria ParaHox genes may be linked, they cannot be considered to be clustered. This dispersal of ParaHox genes is typical for molluscs in general, with several species also showing loose linkage of some of the genes [20], which contrasts with the relatively tight clustering of these genes in many deuterostomes [23–25] and the likely pan-cluster regulation that may operate in these deuterostomes.
ANTP-class homeobox genes
Beyond Hox and ParaHox, there are other linkages among and between the classes of homeobox genes that hint at their ancient evolutionary origins and genomic arrangement in clusters. Despite the many rearrangements to the Hox cluster, many genes linked to Hox clusters in other species are also found on the same scaffold in B. straminea, including Mnx, Gbx-a and Gbx-b, En-a, Evx-a and Evx-b, and Dlx [15, 18, 20, 26, 27]. These linkages give further support for the hypothesized Super-Hox cluster of non-Hox ANTP-class genes linked to the Hox genes in bilaterians [15].
SINE homeobox genes
Another highly conserved cluster besides Hox and ParaHox is the SINE-class cluster, typically composed of the Six3/6, 1/2, and 4/5 genes or their protostome orthologues [16]. In B. straminea, Six4/5 and Six1/2 are on the same scaffold but with a number of genes between them, and Six3/6 is on a distinct scaffold (Fig. 2). In B. glabrata, Six3/6 is linked to Hlx (Supplementary Information S2b), the last homeobox gene at the end of the Six4/5-Six1/2 scaffold in B. straminea (Fig. 2). Thus, there is clearly not a SINE-class gene cluster conserved in B. straminea, but the linkage of at least some of these genes indicates that the dispersal of this cluster has not yet proceeded to the extent of these genes being separated onto different chromosomes. Also, the location of the Hlx gene relative to different Six genes indicates a certain degree of genomic rearrangement between the 2 Biomphalaria species (i.e., conserved macrosynteny but divergent microsynteny).
IRX homeobox genes
Homeobox genes in the IRX family within the TALE class are also observed to be clustered in several lineages, e.g., the 3-gene (ara, caup, and mirr) cluster in Drosophila, 2 clusters of 3 genes in vertebrates, and 4 genes in the limpet L. gigantea (irx4, irx2, irx1, and irx3) [28–30]. These clusters are thought likely to have arisen convergently by independent tandem duplications in the arthropod, vertebrate, and mollusc lineages [31, 28–30]. Both Biomphalaria species have 5 IRX-family genes, 1 pair of which seems to be a product of a more recent, possibly Biomphalaria-specific, duplication (Irx1-a and Irx1-b). Perhaps surprisingly, none of the Biomphalaria Irx genes, Irx1 (a and b), Irx2, Irx3, and Irx4, show clear orthology to specific gastropod (limpet) or bivalve (oyster) genes in a phylogenetic tree (Supplementary Information S2c). A paucity of phylogenetically informative amino acid changes is the most likely explanation for this lack of resolution. Despite this lack of resolution of Irx orthology across species the B. straminea genome assembly does provide a new example of Irx gene clustering. Irx3, Irx2, and Irx4 are closely clustered in the genome, while Irx1-b is 7 Mb away on the same scaffold, also with Zhx, a ZF-class gene another 6 Mb further. The 2 Irx1 paralogues, however, are on separate scaffolds, which may represent either a rearrangement following their duplication, convergence of the sequence of the homeodomain, or thirdly, an assembly artefact. In B. glabrata, only the linkage of Irx4 with Irx2 is corroborated owing to the shorter scaffold lengths of that assembly. Further work, perhaps using other conserved domains from these genes and with a wider breadth of lophotrochozoan species, could potentially determine whether in fact the 4 Irx gene types in Biomphalaria species are orthologous to genes in other species’ Irx clusters. A multi-gene IRX-family cluster in Biomphalaria species with evidence of ≥1 independent expansion (Irx1-a and Irx1-b) provides an interesting addition to our understanding of IRX-family clusters, and the mechanisms behind gene expansions and subsequent maintenance of clustering in general.
PRD- and LIM-class homeobox genes
We also observe linkages amongst PRD-class genes, with clusters on scaffolds 13,536, 2,216, 46,009, and 563 (Fig. 2). The PRD-class cluster that is widely found across various species is the so-called HRO cluster, composed of the genes Otp, Rx/Rax, and Hbn/Arx-like [16, 17], which ancestrally was likely embedded within a more extensive PRD/LIM-class mega-cluster, including the PRD-class genes Gsc and Otx and the LIM-class gene Isl [16]. In B. straminea there is a remnant of the HRO cluster, with Otp clustered with Hbn, internally on a large scaffold (563) and flanked by other homeobox genes (Fig. 2) including another PRD-class gene (Arx-a) now in this Biomphalaria PRD-class cluster, but the Rax genes are on other scaffolds. Interestingly, the Isl gene is also on this large 563 scaffold in B. straminea, consistent with descent from the hypothesized PRD/LIM-class mega-cluster [16]. B. glabrata provides an interesting contrast as the HRO cluster is now complete (with Otp, Hbn,andRax-b), in contrast to B. straminea, and again Arx-a is also in the Biomphalaria cluster (Fig. 2; Supplementary Fig. S2b). Why the PRD-class HRO cluster would remain intact in 1 species of Biomphalaria but not the other remains to be resolved. Also, whether the inclusion of the Arx-a gene in this cluster in these snails is found elsewhere in the animal kingdom and is of any functional significance also remains a topic for future work. Overall, the PRD-class gene clustering provides a mixed signal, of both conservation of remnants of ancient clustering alongside rearrangements between closely related, con-generic species.
Duplicated homeobox genes
There are several duplications shared between the 2 species, which we infer to be at least ancestral to the genus. These include paralogues of Arx, Pax4/6, Irx1, En, Evx, Abox, Barhl, Pbx, and Tlx, as well as 3 paralogues of Vsx and Cers. Notably, the 2 paralogues each of Vsx and Cers genes remain clustered in the genome, reflecting their likely origin by tandem duplication. This is also seen for En, Tlx, Evx, and Abox. B. straminea is the only species of the 2 with 2 paralogues of Gbx, although 1 has an apparently odd arrangement that would mean it is unlikely to be a functional gene, if this arrangement were real. The homeodomain is split across 2 exons, the first of which is in 1 orientation, while there are 2 copies of the second exon in the opposite orientation, indicating that the second Gbx gene may be a pseudogene or an assembly artefact (Supplementary Information S2a).
Giga-cluster homeobox genes
An overarching framework for understanding the genomic organization of homeobox-containing genes comes from hypotheses about their ancient linkage patterns following their presumed origins largely via tandem duplications. This ancestral clustering goes beyond the class-specific clusters already described above and is captured by the Giga-cluster hypothesis [16]. High-quality genome assemblies, such as that described here for B. straminea, are key resources for testing this hypothesis and potentially expanding it. Several instances of linkage of different classes of homeobox gene are present in the B. straminea assembly, most notably on scaffolds 563, 8,789, 2,216, and 24,987 (Fig. 2). Scaffold 2,216 is interesting for the linkage of the SINE-class genes Six4/5 and Six1/2 with some of the members of the ancestral PRD/LIM-class Mega-cluster (i.e., the PRD-class genes Gsc and Otx) that has undergone some dispersal in the Biomphalaria lineage (as described above). Also, some of the other members of this dispersed PRD/LIM Mega-cluster (Isl, Otp, Hbn) are on scaffold 563, which are now linked with many members of the dispersed NK-cluster (e.g., NK5, NK4, Msx, Tlx-a and -b, and NK3) as well as a member of the ancestral SuperHox cluster (i.e., Hhex) [15, 16]. Other members of the SuperHox cluster are still linked with the true Hox genes (EuHox genes) on scaffold 32,695. These linkages of genes from different homeobox classes along with the further new instances of interclass linkage on scaffold 8,798 (Fig. 2) are all consistent with the Giga-cluster hypothesis [16]. However, how much of all of these linkages represent ancestral associations (i.e., descended from primary clustering) versus instances of coming together in the genome convergently in evolution (i.e., secondary clustering) should be resolvable with comparisons to further high-quality genome sequences as well as a better understanding of the dynamics of genome evolution and rearrangements (reviewed in [16]).
Synteny analysis of B. straminea with other molluscs
The homeobox analyses described above provide instances of linkages that indicate varied synteny conservation across various mollusc and animal clades, even between the 2 Biomphalaria species now sequenced. The B. straminea genome shows considerable conserved linkage within and between classes of homeobox, and the maintenance of certain conserved clusters or linkages observed throughout wider lineages (i.e., instances of remnants of the Hox, ParaHox, SuperHox, and Giga-clusters [16]). In comparison to B. glabrata, in which less linkage can be observed because of shorter scaffold lengths, there is some conserved synteny. A few differences between the species may be due to species-specific genomic rearrangements resulting in the disruption of gene order, but the alternative possibility of assembly artefacts cannot be excluded entirely at present without further work. Of particular interest for further study is the major rearrangement of the Hox cluster in B. straminea. Perhaps more thorough sequencing of B. glabrata or assemblies of additional Biomphalaria species could determine whether this is shared in the genus or is a novelty of B. straminea. Regardless of this, the impact of this rearrangement on Hox gene expression and function is of interest. Hox cluster rearrangements could indicate the loss of shared regulatory elements that constrain Hox clusters in other lineages and may reflect changes to Hox gene expression, perhaps underpinning developmental changes in these snails. Similarly, the impacts of the dispersal of the ParaHox cluster on gene expression will be interesting to resolve. The patterns of clustering, linkage, and rearrangement of homeobox genes are good markers for genome organization, and these results show that key differences between the species may represent higher levels of genomic divergence than expected for these 2 snails. Here we observe specific cases of differences between our new B. straminea genome and that of B. glabrata within the context of ancestral linkages, and this pattern may be a good indicator of wider differences between the genetics and molecular processes operating in the 2 species.
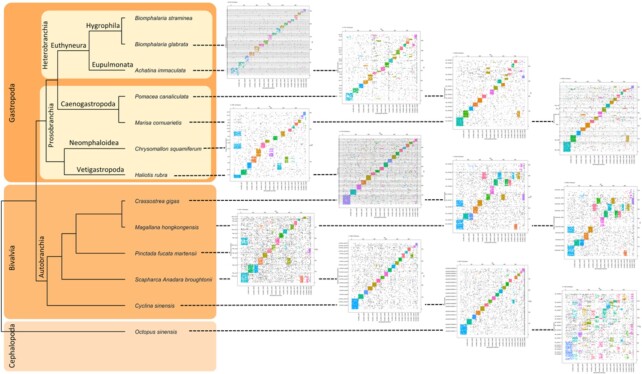
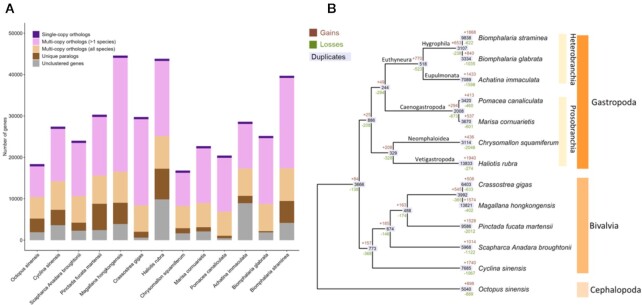
To examine the syntenic relationships more generally between Biomphalaria and mollusc genomes, we constructed Oxford dot plots, comparing the chromosomal positions of orthologous genes between published mollusc genomes, as available from GenBank for gastropod, bivalve, and cephalopod molluscs. As shown in Fig. 4, the relationship of pseudo-chromosomes (2n = 36 [12]) and scaffolds between B. straminea and molluscs of other classes were conserved in most cases. Previous phylogenetic tree constructions for different Biomphalaria species suggested a monophyletic clade of African species with the remaining lineages being neotropical species [2, 3]. Based on this phylogenetic relationship, our data show that the neotropical species have not undergone any significant interchromosomal rearrangements from their last common ancestor after separation to different geographical regions. One-to-one synteny block could be identified between B. straminea and the eupulmonata gastropod, Achatina immaculata. However, in the comparison of B. straminea to the more evolutionarily distant species, a few one-to-many blocks were found. These patterns indicated that some chromosome duplication and alteration occurred from the most recent common ancestor of B. straminea, B. glabrata, and A. immaculata (the ancestor of Hygrophila and Eupulmonata). Furthermore, species with closer evolutionary distance shared more similar synteny patterns against B. straminea (e.g., between Pomacea canaliculata and Marisa cornuarietis, as well as between C. gigas and M. hongkongensis, which share more similar synteny blocks), suggesting the dynamic changes of chromosome arrangements in different molluscs. In Octopus sinensis, the gene order and synteny blocks to B. straminea were largely lost, suggesting that more duplication, translocation, and rearrangement events occurred since the divergence of O. sinensis (Cephalopoda) and the common ancestor of Gastropoda and Bivalvia [32].
Figure 4.
: Synteny between B. straminea and 12 mollusc genomes. The species tree is constructed using 2,047 orthogroups with ≥12 of 13 mollusc genomes having single-copy genes in each orthogroup. In the Oxford dot plot, each dot represents a pair of orthologous genes between B. straminea and the specific mollusc. Horizontal and vertical dashed lines represent chromosome or scaffold boundaries. Orthologous genes are coloured according to their position in B. straminea scaffolds. Significance of synteny blocks is computed using 1-tailed Fisher exact test, and synteny blocks with Benjamini & Hochberg–corrected P > 0.05 are indicated in grey.
Ecdysteroid genes
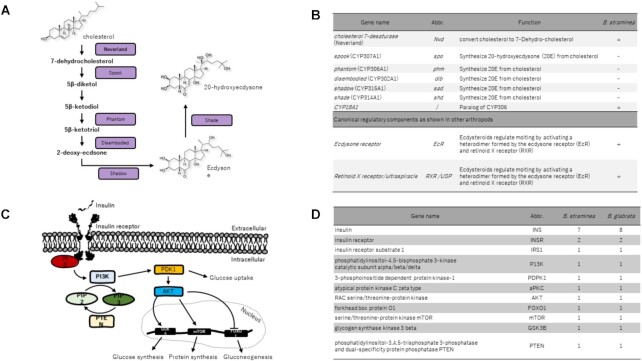
Ecdysteroids play important roles in regulating growth (in particular molting and metamorphosis) and sexual maturation of insects and other arthropods [33, 34]. Although it has long been known that gastropods contain ecdysteroids and that β-ecdysone could stimulate host location activities in S. mansoni miracidia and enhance growth and egg production in B. glabrata [35, 36], the biosynthetic pathway genes for ecdysteroids have not been systematically studied in mollusc genomes to date. As shown in Fig. 3A and B, typical genes involved in this pathway including CYP307A1, CYP306A1, CYP302A1, CYP315A1, and CYP314A1 are all absent from the B. straminea genome assembly and transcriptome data. Nevertheless, the receptors including EcR, RXR/USP, and oxygenase-like protein Nvd that are essential regulators of cholesterol metabolism are present in B. straminea and other mollusc genomes (Fig. 3A and B; Supplementary Information S3). We thus treated B. straminea with 10–6 M ecdysteroid 20-hydroxyecdysone for 24 hours but did not observe any significant expression changes in the downstream genes E74, FOXO, and Nvd. Similar hormone treatments have been shown to elicit the downstream genes in insects in previous studies [37–39]. It is unclear whether only certain forms of ecdysteroids may induce endogenous ecdysteroid pathway genes under particular conditions, and this warrants further investigation. This is the first systematic analysis of ecdysteroid pathway genes in a mollusc genome, thus providing the foundations for future work to determine how ecdysteroids have their effect in these animals.
Figure 3.
: (A) Schematic diagram of biosynthetic pathway of ecdysteroids; (B) presence and absence of ecdysteroid pathway genes in B. straminea; (C) schematic diagram of biosynthetic pathway of insulin; (E) number of gene copies of insulin pathway genes in B. straminea.
Insulin signalling pathway genes
Peptide hormones involved in growth and reproduction have been suggested as candidates for the development of novel methods of schistosomiasis control via manipulation of snail numbers [40]. Insulin is another understudied hormonal pathway in molluscs despite its potential functional roles. For instance, in the pond snail Lymnaea stagnalis, a decrease of insulin in the central nervous system correlated with better associative learning behaviour [41], while insulin-related peptides with potential roles in sexual reproduction have been identified in the oyster C. gigas [42]. In both B. straminea and B. glabrata genomes, we were able to identify all key signalling pathway genes (Fig. 3C and D, Supplementary Information S4). This establishes a foundation on which to further explore the functions of these hormones in molluscs.
Widespread gene turnover between Biomphalaria snails and other molluscs
Gene gains and losses in mollusc genomes
A phylogenomic tree was constructed using 2,047 orthogroups with ≥12 of 13 mollusc genomes having single-copy genes in each orthogroup (Supplementary Information S6). Gene family analysis among these genomes revealed the expansion of 1,868 orthogroups and contraction of 622 orthogroups in B. straminea, but in B. glabrata, the expansion of 840 orthogroups and contraction of 1,035 orthogroups (Fig. 5). These data highlight the importance of having the B. straminea genomic resource and potentially suggest that specific control strategies might be needed for B. straminea rather than treating it as identical to B. glabrata.
Figure 5.
: Summaries of gene families in B. straminea and 12 molluscs. (A) Gene family clustering; only the longest isoform for each gene was used. (B) Gene family expansion and contraction between mollusc genomes. Brown and green colour indicate the number of significantly (P < 0.05) expanded or contracted gene families at each node, respectively.
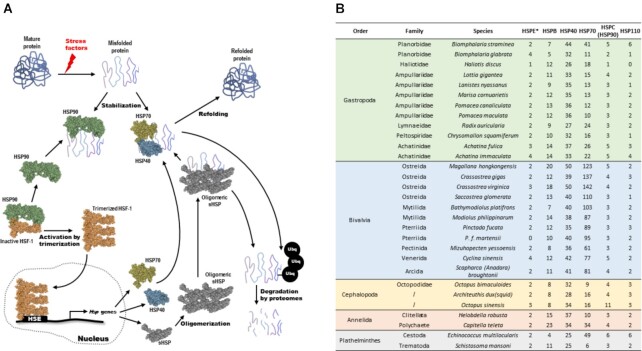
Expansion of heat shock protein family among mollusc lineages
Heat shock proteins are important stress-responsive candidates involved in protein folding for molluscs, activated in response to such things as changing pH, oxygen level, and temperature. In some mollusc genomes, such as that of the Pacific oyster C. gigas, an expansion of heat shock protein 70 (HSP70) has been observed in the genome and hypothesized to be important to the animals’ adaptation to changes in ambient environmental factors or pressures [43]. We thus identified the heat shock protein family genes in Biomphalaria and compared these to other lophotrochozoans to understand their evolution in different lineages (Fig. 6). Among the different heat shock protein families in the investigated set of gastropods, bivalves, cephalopods, annelids, and platyhelminths, a dramatic expansion is seen specifically in the HSP70 family in the bivalve molluscs (Fig. 6; Supplementary Information S7). Our data and analyses agree with previous studies (e.g., [43]), suggesting that the expansion of HSP70 is linked to the life history of molluscs having a sessile stage. This survey also provides the foundation for future work on the expression and function of particular HSP genes/proteins and their activity in these parasite vectors, which may contribute to their adaptive ability as invasive species and possibly to the recent range expansion of B. straminea.
Figure 6.
: (A) Schematic diagram showing the heat shock protein actions; (B) number of gene copies of heat shock proteins in different mollusc genomes.
Differential sesquiterpenoid and cholesterol genes in certain mollusc lineages
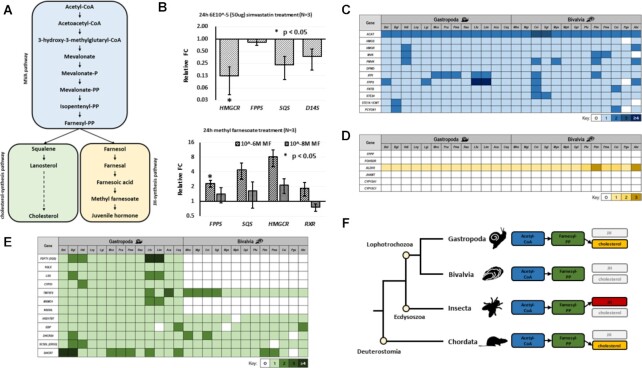
Sesquiterpenoid hormones were once considered specific to insects and crustaceans, where they control development and reproduction [33, 44, 45]. However, recent analyses have shown that the sesquiterpenoid system is also present in myriapods, annelids, and cnidarians [31, 34, 46, 47]. Conversely, vertebrates can only produce cholesterol but not sesquiterpenoids [48, 49], and a recent study revealed the canonical cholesterol biosynthesis pathway in sponges, placozoans, and deuterostomes, suggesting that cnidarians and protostomes experienced massive losses of these genes ([50]; Fig. 7A). Treatment of B. straminea with 10–6 M simvastatin and methyl farnesoate changed the expression of sesquiterpenoid pathway genes HMGCRand FPPS, suggesting a sesquiterpenoid responsive system (Fig. 7B and C). Comparison of sesquiterpenoid pathway genes in mollusc genomes further identified differential utilization of biogenesis pathways in bivalves and gastropods, where only gastropods but not the bivalves are able to produce cholesterol similar to vertebrates (Fig. 7D and F). This is the first systematic study showing the differential sesquiterpenoid and cholesterol synthesis pathways possessed by different mollusc lineages.
Figure 7.
: (A) Schematic diagram showing the mevalonate pathway, and the downstream sesquiterpenoid and de novo cholesterol synthesis pathways. (B) Expression of genes upon 6 × 10–5M simvastatin, 10–6 and 10–8 M methyl farnesoate treatment for 24 hours; *P < 0.05. FC: fold change. The bars represent the relative expression level, and the error bars are shown for independently repeated experiments. (C) Heat map of mevalonate pathway orthologues identified in gastropod and bivalve genomes. (D) Heat map of sesquiterpenoid synthesis pathway orthologues identified in gastropod and bivalve genomes. (E) Heat map of de novo cholesterol synthesis pathway orthologues identified in gastropod and bivalve genomes. (F) Schematic diagram showing the evolution of sesquiterpenoid pathway genes in bilaterians.
Conclusion
This study presents the first high-quality genome assembly for a schistosomiasis-transmitting snail in China and Asia. The snail Biomphalaria straminea is important scientifically, as well as having considerable medical relevance. Our work provides gene and transposable element annotations, and detailed analyses of a variety of gene families, including the homeobox, ecdysteroid, insulin, heat shock protein, and sesquiterpenoid pathway genes, suggesting extensive molecular differences between B. straminea and B. glabrata, as well as among other molluscan taxa. More generally, our high-quality B. straminea genome provides a useful reference point for further understanding of the biology, ecology, and evolution of molluscs.
Methods
Sample collection and genome sequencing
One week prior to the experiment, ∼100 ramshorn snails were collected in a freshwater stream in Tai Po New Territories, Hong Kong (GPS: 22.50206300747975, 114.15354682258841). The collected animals were maintained in a laboratory aquarium and fed with lettuce 3 days a week. Samples for genome sequencing originate from single individuals for each sequencing method (Fig. 1A). gDNA was extracted using the PureLink Genomic DNA Mini Kit (Invitrogen, Waltham, Massachusetts, United States) following the manufacturer's protocol. Extracted gDNA was subjected to quality control using a Nanodrop spectrophotometer (Thermo Scientific, Waltham, Massachusetts, United States) and gel electrophoresis. Qualifying samples were sent to Novogene and Dovetail Genomics for library preparation and sequencing. The resulting library was sequenced on an Illumina HiSeq X platform (Illumina HiSeq X, RRID:SCR_016385) to produce 2 × 150 paired-end sequences. The length-weighted mean molecule length is 22.2 kb, and the raw data can be found at NCBI's SRA (SRR12963913).
Dovetail Omni-C library preparation and sequencing
For each Dovetail Omni-C library, chromatin was fixed with formaldehyde and extracted. Fixed chromatin was digested with DNAse I, and chromatin ends were repaired and ligated to a biotinylated bridge adapter followed by proximity ligation of adapter-containing ends. After proximity ligation, crosslinks were reversed and the DNA was purified. Purified DNA was treated to remove biotin that was not internal to ligated fragments. Sequencing libraries were generated using NEBNext Ultra enzymes and Illumina-compatible adapters. Biotin-containing fragments were isolated using streptavidin beads before PCR enrichment of each library. The library was sequenced on an Illumina HiSeqX platform to produce 128 million 150-bp read pairs, and the raw data can be found at NCBI's SRA (SRR12963914).
Transcriptome sequencing
Total RNA from different tissues was isolated using a combination method of cetyltrimethylammonium bromide (CTAB) pre-treatment [51] and mirVana™ miRNA Isolation Kit (Ambion, Austin, Texas, United States) following the manufacturer's protocol. The extracted total RNA was subjected to quality control using a Nanodrop spectrophotometer (Thermo Scientific, Waltham, Massachusetts, United States), gel electrophoresis, and an Agilent 2100 Bioanalyzer (Agilent RNA 6000 Nano Kit). Qualifying samples underwent library construction and sequencing at Novogene; polyA-selected RNA-Sequencing libraries were prepared using the TruSeq RNA Sample Prep Kit v2. Insert sizes and library concentrations of final libraries were determined using an Agilent 2100 bioanalyzer instrument (Agilent DNA 1000 Reagents) and real-time quantitative PCR (TaqMan Probe), respectively. Details of the sequencing data can be found in Supplementary Information S1.
Genome assembly
Chromium whole-genome sequencing reads were used to construct a de novo assembly using Supernova (v 2.1.1) with default parameters (raw coverage = 68.32×). The Supernova output pseudohap assembly and Dovetail OmniC library reads were used as input data for HiRise, a software pipeline designed specifically for using proximity ligation data to scaffold genome assemblies [52]. Dovetail OmniC library sequences were aligned to the draft input assembly using BWA [53]. The separations of Dovetail OmniC read pairs mapped within draft scaffolds were analysed by HiRise to produce a likelihood model for genomic distance between read pairs, and the model was used to identify and break putative misjoins, to score prospective joins, and make joins above a threshold.
Gene model prediction
Gene models were predicted as described in the Hong Kong oyster (M. hongkongensis) genome [20]. Briefly, the gene models were trained and predicted using funannotate (v1.7.4 [54]) [55] with the following parameters: “–repeats2evm –protein_evidence uniprot_sprot.fasta –genemark_mode ET –busco_seed_species metazoa –optimize_augustus –busco_db metazoa –organism other –max_intronlen 350 000.” The gene models from several prediction sources including GeneMark, high-quality Augustus predictions (HiQ), PASA, Augustus, GlimmerHM, and SNAP were passed to EvidenceModeler and generated the gene model annotation files, followed by PASA to update the EVM consensus predictions, and add untranslated region annotations and models for alternatively spliced isoforms. Protein-coding genes were searched with BLASTp (BLASTp, RRID:SCR_001010) against the nr and swissprot databases by diamond (v0.9.24) [56] with parameters “–more-sensitive –evalue 1e-3,” and mapped by HISAT2 version 2.1.0 (HISAT2, RRID:SCR_015530) with transcriptome reads [57]. Gene models with no similarity to any known protein and no messenger RNA support were removed from the final version.
Repetitive element annotation
Repetitive elements were identified using the transposable element annotation pipeline Earl Grey [58] as follows. First, elements were identified using RepeatMasker v.4.1 (RepeatMasker, RRID:SCR_012954) [84], using a sensitive (-s) search and ignoring low-complexity repeats (-nolow). Subsequently, a de novo repeat library was constructed using RepeatModeler v.1.0.11 (RepeatModeler, RRID:SCR_015027) [59], including RECON v.1.08 (RECON, RRID:SCR_021170) [60] and RepeatScout v.1.0.5 (RepeatScout, RRID:SCR_014653) [61]. Identified novel repeats were analysed using a “BLAST, Extract, Extend” process to characterize elements along their entire length [62]; consensus sequences and classifications for each repeat family were generated, and the resulting de novo repeat library was used to identify repetitive elements in RepeatMasker. All plots were generated using Rstudio ver. 1.2.1335 with R ver. 3.5.1 [63] and ggplot2 ver. 3.2.1 (ggplot2, RRID:SCR_014601) [85].
Gene family annotation and gene tree building
Gene family sequences were first obtained from NCBI for selected species, including B. glabrata and other lophotrochozoans. The sequences were then used to retrieve the corresponding genes from the B. straminea genome using the tBLASTn algorithm on a local server, with an E-value of <10–3. The identity of each retrieved gene was then checked by reciprocal searches against the Genbank nr database at NCBI with BLASTx. For phylogenetic analyses of gene families, DNA sequences were first translated into amino acid sequences and aligned to other reference sequences (extracted from NCBI) using Clustal W. Gapped sites were removed from alignments using MEGA 7.0 (MEGA, RRID:SCR_000667), and phylogenetic trees (neighbour-joining) were constructed using MEGA 7.0, where each phylogenetic node was analysed using 1,000 bootstrap replicates. For homeobox-containing genes, homeodomains were annotated using tBLASTn searches with HomeoDB sequences, and sequences from representative lophotrochozoan families, including the expanded Spiralia TALEs [21]. We also removed redundant hits based on their unique locations in the genome sequence, and manually detected any likely artefactual duplicates that were not carried forward into the protein sequence alignments (Supplementary Table S2). Alignments of each class were made using MUSCLE (MUSCLE, RRID:SCR_011812) [64], with homeodomain sequences from human (Homo sapiens, deuterostome), amphioxus (Branchiostoma floridae, a cephalochordate deuterostome), the ecdysozoans fruitfly (Drosophila melanogaster) and red flour beetle (Tribolium castaneum), and the lophotrochozoans oyster (Crassostrea gigas, bivalve), limpet (Lottia gigantea, gastropod), brachiopod (Lingula anatina), and annelids Platynereis dumerilii and Capitella teleta, where available from other studies [19, 21] and HomeoDB [86], [87]. The best substitution models were tested with ModelFinder, and maximum likelihood phylogenies were constructed with IQ-TREE (IQ-TREE, RRID:SCR_017254) with 1,000 bootstrap replicates [65].
Identification of orthologous genes and gene families
Orthologues and orthogroups in B. straminea and 12 other animal proteomes were inferred using OrthoFinder v. 2.5.2 (OrthoFinder, RRID:SCR_017118) [66] with default values and “-M msa” activated. To cover the gene families, the longest protein of each gene was taken as the representative in OrthoFinder analysis. Gene duplication events were then identified. Duplication ratios per node/tip were calculated by dividing the number of duplications observed in each node/tip by the total number of gene trees containing that node. CAFE5 was used to infer gene gain and loss rates [80]. Orthogroups from the output of OrthoFinder were regarded as gene families and fed to CAFE5. A divergence tree was inferred using r8s [82] from the species tree generated by OrthoFinder. We tested several gamma rate categories (-k), and k = 1 showed the best likelihood.
Functional terms enrichment analysis
Orthogroups were assigned Gene Ontology (GO), EuKaryotic Orthologous Groups (KOG), KEGG, and KEGG Orthology (KO) terms by inheriting the terms from genes found within the groups. The functional term annotations were performed using eggNOG (eggNOG, RRID:SCR_002456) [67]. Functional enrichment was tested for using the function “compareCluster()” in R package “clusterProfiler” v.3.16.1 [68] under the environment of R 4.0.4 [81]. Significantly enriched terms were determined with pvalueCutoff = 0.05, pAdjustMethod = “BH,” and qvalueCutoff = 0.2. Data were visualized using R packages ggplot2 (Wickham 2016), ggtree [69], and “pathview” [70].
Macrosynteny analysis
Single-copy orthologues anchored by mutual best Diamond blastp v0.9.14.115 [76] hits (–evalue 0.001) between B. straminea and 12 other animals with chromosome-level or near chromosome-level assemblies were used in macrosynteny analysis. Oxford synteny plots were generated following previously described methods [83] using the R package ggplot2 (Wickham 2016).
Drug and hormone treatment and RT-qPCR
Experimental adult animals of ∼1 cm with reproductive capability were isolated from the culture and were rinsed in double-distilled water to remove any contaminants. Three individuals per set were placed in a glass container, with a well of 3.5 cm in diameter and 0.8 cm in depth, filled with 2 mL of double-distilled water with either 10–6 or 10–8 M of methyl farnesoate (MF) (Sigma), 6 × 10–5 M of simvastatin (Sigma), or 10–6 M of 20-hydroxyecdysone (Abcam Biochemicals) in separate set-ups. The chemicals were first dissolved in acetone and diluted to the target concentration in the treatment container. The control set-up contained the same number of individuals and was treated with the same concentration of acetone in corresponding experiments. Each replicate of snails was exposed for 24 hours to these treatments without any feeding. Post-treated animals were rinsed with double-distilled water and shells were removed prior to whole-body total RNA extraction. The RNA from each experiment was isolated using TRIzol reagent following the manufacturer's protocol. Purified RNA was dissolved in nuclease-free water. The complementary DNA (cDNA) synthesis was performed using the iScript gDNA Clear cDNA Synthesis Kit (BioRad) following the manufacturer's protocol. The cDNA was used in subsequent quantitative real-time PCR. The amplification conditions were as follows: initial denaturation at 95°C for 30 s, followed by 40 cycles of 95°C denaturation for 15 s, 57°C primer annealing for 15 s, and 72°C extension for 15 s. Primer details are listed in Supplementary File S8. The primers were tested by conventional PCR with B. straminea cDNA prior to experiments to ensure their specificity. Each sample was analysed in replicates. The expression of each target gene transcript was normalized to the housekeeping gene, myoglobin (Myo), as adopted in previous studies [71–74]. The subsequent fold induction analyses were calculated using the ΔΔCt method.
Data Availability
The raw genome and RNA sequencing data have been deposited in the SRA under Bioproject No. PRJNA673593. The final chromosome assembly was submitted to NCBI Assembly under accession No. JADKLZ000000000. All data can also be found in the GigaScience Database [75].
Additional Files
Supplementary Information S1. Sequencing data.
Supplementary Information S2. (a) Tables of homeobox gene sequences in B. straminea, B. glabrata, a synteny comparison of homeobox genes, and comparison of ParaHox gene linkage. (b) Distribution of homeoboxes in the genome of Biomphalaria glabrata. (c) Alignments and phylogenies of each class of homeobox sequences.
Supplementary Information S3. Ecdysteroid genes.
Supplementary Information S4. Insulin pathway genes.
Supplementary Information S5. Synteny information.
Supplementary Information S6. Gene expansion and contraction.
Supplementary Information S7. Heat shock protein family genes.
Supplementary Information S8. Cholesterol genes and primers.
Supplementary Information S9. Phylogenetic trees.
Supplementary Information S10. Tables.
Jacob Tennessen -- 8/28/2021 Reviewed
Coen Adema -- 9/5/2021 Reviewed
Coen Adema -- 1/5/2022 Reviewed
Abbreviations
BLAST: Basic Local Alignment Search Tool; BUSCO: Benchmarking Universal Single-Copy Orthologs; BWA: Burrows-Wheeler Aligner; cDNA: complementary DNA; gDNA: genomic DNA; kb: kilobase pairs; KEGG: Kyoto Encyclopedia of Genes and Genomes; LINE: long interspersed nuclear element; LTR: long terminal repeat; Mb: megabase pairs; NCBI: National Center for Biotechnology Information; PASA: Program to Assemble Spliced Alignments; SINE: short interspersed nuclear element; SRA: Sequence Read Archive.
Competing Interests
The authors declare that they have no competing interests.
Funding
This work was supported by the Hong Kong Research Grant Council Collaborative Research Fund(C4015-20EF), General Research Fund (14100919), NSFC/RGC Joint Research Scheme (N_CUHK401/21), and The Chinese University of Hong Kong Direct Grant (4053433, 4053489). Y.Y., W.L.S., C.F.W., S.T.S.L., and Y.L. were supported by the Ph.D. studentships of The Chinese University of Hong Kong. A.H. is supported by a Biotechnology and Biological Sciences Research Council (BBSRC) David Phillips Fellowship (BB/N020146/1). T.B. is supported by a studentship from the Biotechnology and Biological Sciences Research Council-funded South West Biosciences Doctoral Training Partnership (BB/M009122/1). M.E.A.R. is supported by a Ph.D. studentship from the School of Biology and St Andrews University.
Conflict of interest
The authors declare no conflict of interests.
Authors’ Contributions
J.H.L.H., D.E.K.F., A.H., Z.W., S.X., Z.P.K., and S.S.C. conceived the study. J.H.L.H., D.E.K.F., and A.H. supervised the study. W.N., J.H., and T.S. assembled the genome. W.N. carried out the gene model prediction and comparison. Y.Y. carried out the heat shock protein analyses. Y.X. carried out the gene gain and loss and synteny analyses. W.L.S. and C.F.W. carried out the sesquiterpenoid analyses. Y.Y., W.L.S., and S.Y.L. carried out the ecdysteroid analyses. M.E.A.R. and Y.L. carried out the homeobox gene analyses. T.B. carried out the transposable element analyses. S.T.S.L. carried out the insulin analyses. W.N., Y.Y., Y.X., W.L.S., M.E.A.R., T.B., A.H., D.E.K.F., and J.H.L.H. wrote the first draft of manuscript. All authors approved the final version of the manuscript.
ACKNOWLEDGEMENTS
The authors would like to thank Elaine Huang and Ho Yin Yip for collection and maintenance of snails. We thank Thomas Barton-Owen for help and advice on homeobox searches. The authors would also like to thank the anonymous reviewers, whose comments improved the manuscript.
Contributor Information
Wenyan Nong, School of Life Science, Simon F.S. Li Marine Science Laboratory, State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Hong Kong, China.
Yifei Yu, School of Life Science, Simon F.S. Li Marine Science Laboratory, State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Hong Kong, China.
Madeleine E Aase-Remedios, The Scottish Oceans Institute, Gatty Marine Laboratory, School of Biology, University of St. Andrews, St. Andrews, UK.
Yichun Xie, School of Life Science, Simon F.S. Li Marine Science Laboratory, State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Hong Kong, China.
Wai Lok So, School of Life Science, Simon F.S. Li Marine Science Laboratory, State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Hong Kong, China.
Yiqian Li, School of Life Science, Simon F.S. Li Marine Science Laboratory, State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Hong Kong, China.
Cheuk Fung Wong, School of Life Science, Simon F.S. Li Marine Science Laboratory, State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Hong Kong, China.
Toby Baril, University of Exeter, Exeter, UK.
Sean T S Law, School of Life Science, Simon F.S. Li Marine Science Laboratory, State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Hong Kong, China.
Sheung Yee Lai, School of Life Science, Simon F.S. Li Marine Science Laboratory, State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Hong Kong, China.
Jasmine Haimovitz, Dovetail Genomics, California, USA.
Thomas Swale, Dovetail Genomics, California, USA.
Shan-shan Chen, Institute of Agro-food Standard and Testing Technology, Shanghai Academy of Agricultural Sciences, Shanghai, China.
Zhen-peng Kai, School of Chemical and Environmental Engineering, Shanghai Institute of Technology, Shanghai, China.
Xi Sun, Sun Yat-sen University, Guangdong, China.
Zhongdao Wu, Sun Yat-sen University, Guangdong, China.
Alexander Hayward, University of Exeter, Exeter, UK.
David E K Ferrier, The Scottish Oceans Institute, Gatty Marine Laboratory, School of Biology, University of St. Andrews, St. Andrews, UK.
Jerome H L Hui, School of Life Science, Simon F.S. Li Marine Science Laboratory, State Key Laboratory of Agrobiotechnology, The Chinese University of Hong Kong, Hong Kong, China.
References
- 1. Schistosomiasis (Bilharzia). https://www.who.int/health-topics/schistosomiasis. [Google Scholar]
- 2. Campbell G, Jones CS, Lockyer AE, et al. Molecular evidence supports an African affinity of the Neotropical freshwater gastropod, Biomphalaria glabrata, Say 1818, an intermediate host for Schistosoma mansoni. Proc R Soc Lond B Biol Sci. 2000;267(1460):2351–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. DeJong RJ, Morgan JA, Paraense WL, et al. Evolutionary relationships and biogeography of Biomphalaria (Gastropoda: Planorbidae) with implications regarding its role as host of the human bloodfluke, Schistosoma mansoni. Mol Biol Evol. 2001;18(12):2225–39. [DOI] [PubMed] [Google Scholar]
- 4. Meier-Brook C. A snail intermediate host of Schistosoma mansoni introduced into Hong Kong. Bull World Health Organ. 1974;51:661. [PMC free article] [PubMed] [Google Scholar]
- 5. Yang Y, Cheng W, Wu X, et al. Prediction of the potential global distribution for Biomphalaria straminea, an intermediate host for Schistosoma mansoni. PLoS Negl Trop Dis. 2018;12(5):e0006548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Attwood SW, Huo GN, Qiu JW. Update on the distribution and phylogenetics of Biomphalaria (Gastropoda: Planorbidae) populations in Guangdong Province, China. Acta Trop. 2015;141:258–70. [DOI] [PubMed] [Google Scholar]
- 7. Dudgeon D, Yipp MW. A report on the gastropod fauna of aquarium fish farms in Hong Kong, with special reference to an introduced human schistosome host species, Biomphalaria straminea (Pulmonata: Planorbidae). Malacol Rev. 1983;16:93–94. [Google Scholar]
- 8. Woodruff DS, Mulvey M, Yipp MW. The continued introduction of intermediate host snails to Schistosoma mansoni into Hong Kong. Bull World Heal Organ. 1985;63:621. [PMC free article] [PubMed] [Google Scholar]
- 9. Zeng X, Yiu WC, Cheung KH, et al. Distribution and current infection status of Biomphalaria straminea in Hong Kong. Parasit Vectors. 2017;10(1):351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang L, Wu X, Li X, et al. Imported schistosomiasis: a new public health challenge for China. Front Med (Lausanne). 2020;7:553487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu R, Xu J. [Epidemic situation of oversea imported schistosomiasis in China and thinking about its prevention and control]. Zhongguo Xue Xi Chong Bing Fang Zhi Za Zhi. 2014;26:111–4. [PubMed] [Google Scholar]
- 12. Adema CM, Hillier LDW, Jones CS, et al. Whole genome analysis of a schistosomiasis-transmitting freshwater snail. Nat Commun. 2017;8:15451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Tennessen JA, Bollmann SR, Peremyslova E, et al. Clusters of polymorphic transmembrane genes control resistance to schistosomes in snail vectors. Elife. 2020;9:e59395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Simão FA, Waterhouse RM, Ioannidis P, et al. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–2. [DOI] [PubMed] [Google Scholar]
- 15. Butts T, Holland PWH, Ferrier DEK. The Urbilaterian Super-Hox cluster. Trends Genet. 2008;24(6):259–62. [DOI] [PubMed] [Google Scholar]
- 16. Ferrier DEK. Evolution of homeobox gene clusters in animals: the giga-cluster and primary vs. secondary clustering. Front Ecol Evol. 2016;4:doi: 10.3389/fevo.2016.00036. [DOI] [Google Scholar]
- 17. Mazza ME, Pang K, Reitzel AM, et al. A conserved cluster of three PRD-class homeobox genes (homeobrain, rx and orthopedia) in the Cnidaria and Protostomia. Evodevo. 2010;1(1):3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hui JHL, McDougall C, Monteiro AS, et al. Extensive chordate and annelid macrosynteny reveals ancestral homeobox gene organization. Mol Biol Evol. 2012;29(1):157–65. [DOI] [PubMed] [Google Scholar]
- 19. Paps J, Xu F, Zhang G, et al. Reinforcing the egg-timer: recruitment of novel Lophotrochozoa homeobox genes to early and late development in the Pacific oyster. Genome Biol Evol. 2015;7(3):677–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li Y, Nong W, Baril T, et al. Reconstruction of ancient homeobox gene linkages inferred from a new high-quality assembly of the Hong Kong oyster (Magallana hongkongensis) genome. BMC Genomics. 2020;21(1):713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Barton-Owen TB, Szabó R, Somorjai IML, et al. A revised spiralian homeobox gene classification incorporating new polychaete transcriptomes reveals a diverse TALE class and a divergent hox gene. Genome Biol Evol. 2018;10:2151–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Simakov O, Marlétaz F, Cho S-J, et al. Insights into bilaterian evolution from three spiralian genomes. Nature. 2013;493(7433):526–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ikuta T, Chen YC, Annunziata R, et al. Identification of an intact ParaHox cluster with temporal colinearity but altered spatial colinearity in the hemichordate Ptychodera flava. BMC Evol Biol. 2013;13:129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Osborne PW, Benoit G, Laudet V, et al. Differential regulation of ParaHox genes by retinoic acid in the invertebrate chordate amphioxus (Branchiostoma floridae). Dev Biol. 2009;327(1):252–62. [DOI] [PubMed] [Google Scholar]
- 25. Zhang H, Ravi V, Tay BH, et al. Lampreys, the jawless vertebrates, contain only two ParaHox gene clusters. Proc Natl Acad Sci U S A. 2017;114(34):9146–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Castro LFC, Holland PWH. Chromosomal mapping of ANTP class homeobox genes in amphioxus: piecing together ancestral genomes. Evol Dev. 2003;5(5):459–65. [DOI] [PubMed] [Google Scholar]
- 27. Chourrout D, Delsuc F, Chourrout P, et al. Minimal ProtoHox cluster inferred from bilaterian and cnidarian Hox complements. Nature. 2006;442(7103):684–7. [DOI] [PubMed] [Google Scholar]
- 28. Irimia M, Maeso I, Garcia-Fernandez J. Convergent evolution of clustering of Iroquois homeobox genes across metazoans. Mol Biol Evol. 2008;25(8):1521–5. [DOI] [PubMed] [Google Scholar]
- 29. Kerner P, Ikmi A, Coen D, et al. Evolutionary history of the iroquois/Irx genes in metazoans. BMC Evol Biol. 2009;9:74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Takatori N, Butts T, Candiani S, et al. Comprehensive survey and classification of homeobox genes in the genome of amphioxus, Branchiostoma floridae. Dev Genes Evol. 2008;218(11-12):579–90. [DOI] [PubMed] [Google Scholar]
- 31. Chipman AD, Ferrier DEK, Brena C, et al. The first myriapod genome sequence reveals conservative arthropod gene content and genome organisation in the centipede Strigamia maritima. PLoS Biol. 2014;12:(11):e1002005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Smith SA, Wilson NG, Goetz FE, et al. Resolving the evolutionary relationships of molluscs with phylogenomic tools. Nature. 2011;480(7377):364–7. [DOI] [PubMed] [Google Scholar]
- 33. Cheong SPS, Huang J, Bendena WG, et al. Evolution of ecdysis and metamorphosis in arthropods: the rise of regulation of juvenile hormone. Integr Comp Biol. 2015;55(5):878–90. [DOI] [PubMed] [Google Scholar]
- 34. Qu Z, Kenny NJ, Lam HM, et al. How did arthropod sesquiterpenoids and ecdysteroids arise? Comparison of hormonal pathway genes in noninsect arthropod genomes. Genome Biol Evol. 2015;7:1951–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Bayne CJ. On the reported occurrence of an ecdysone-like steroid in the freshwater snail, Biomphalaria glabrata (Pulmonata; Basommatophora), intermediate host of Schistosoma mansoni. Parasitology. 1972;64(3):501–9. [DOI] [PubMed] [Google Scholar]
- 36. Shiff CJ, Dossaji SF. Ecdysteroids as regulators of host and parasite interactions: a study of interrelationships between Schistosoma mansoni and the host snail, Biomphalaria glabrata. Trop Med Parasitol. 1991;42:11–6. [PubMed] [Google Scholar]
- 37. Hossain MS, Liu Y, Zhou S, et al. 20-Hydroxyecdysone-induced transcriptional activity of FoxO upregulates brummer and acid lipase-1 and promotes lipolysis in Bombyx fat body. Insect Biochem Mol Biol. 2013;43(9):829–38. [DOI] [PubMed] [Google Scholar]
- 38. Ji C, Zhang N, Jiang H, et al. 20-hydroxyecdysone regulates expression of methioninesulfoxide reductases through transcription factor FOXO in the red flour beetle, Tribolium castaneum. Insect Biochem Mol Biol. 2021;131:103546. [DOI] [PubMed] [Google Scholar]
- 39. Sekimoto T, Iwami M, Sakurai S. 20-Hydroxyecdysone regulation of two isoforms of the Ets transcription factor E74 gene in programmed cell death in the silkworm anterior silk gland. Insect Mol Biol. 2007;16(5):581–90. [DOI] [PubMed] [Google Scholar]
- 40. Acker MJ, Habib MR, Beach GA, et al. An immunohistochemical analysis of peptidergic neurons apparently associated with reproduction and growth in Biomphalaria alexandrina. Gen Comp Endocrinol. 2019;280:1–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Totani Y, Aonuma H, Oike A, et al. Monoamines, insulin and the roles they play in associative learning in pond snails. Front Behav Neurosci. 2019;13:65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Cherif—Feildel M, Heude Berthelin C, Adeline B, et al. Molecular evolution and functional characterisation of insulin related peptides in molluscs: contributions of Crassostrea gigas genomic and transcriptomic-wide screening. Gen Comp Endocrinol. 2019;271:15–29. [DOI] [PubMed] [Google Scholar]
- 43. Zhang G, Fang X, Guo X, et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature. 2012;490(7418):49–54. [DOI] [PubMed] [Google Scholar]
- 44. Qu Z, Bendena WG, Tobe SS, et al. Juvenile hormone and sesquiterpenoids in arthropods: biosynthesis, signaling, and role of MicroRNA. J Steroid Biochem Mol Biol. 2018;184:69–76. [DOI] [PubMed] [Google Scholar]
- 45. Tsang SSK, Law STS, Li C, et al. Diversity of insect sesquiterpenoid regulation. Front Genet. 2020;11:1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Nong W, Cao J, Li Y, et al. Jellyfish genomes reveal distinct homeobox gene clusters and conservation of small RNA processing. Nat Commun. 2020;11(1):3051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Schenk S, Krauditsch C, Frühauf P, et al. Discovery of methylfarnesoate as the annelid brain hormone reveals an ancient role of sesquiterpenoids in reproduction. Elife. 2016;5:e17126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hui JHL, Bendena WG, Tobe SS. Future perspectives for research on the biosynthesis of juvenile hormones and related sesquiterpenoids in arthropod endocrinology and ecotoxicology. In: Devillers J, ed. Juvenile Hormones and Juvenoids. Modeling Biological Effects and Environmental Fate. Boca Raton: CRC Press; 2013:15–30. [Google Scholar]
- 49. Tobe SS, Bendena WG. The regulation of juvenile hormone production in arthropods. functional and evolutionary perspectives. Ann NY Acad Sci. 1999;897(1):300–10. [DOI] [PubMed] [Google Scholar]
- 50. Zhang T, Yuan D, Xie J, et al. Evolution of the cholesterol biosynthesis pathway in animals. Mol Biol Evol. 2019;36(11):2548–56. [DOI] [PubMed] [Google Scholar]
- 51. Jordon-Thaden IE, Chanderbali AS, Gitzendanner MA, et al. Modified CTAB and TRIzol protocols improve RNA extraction from chemically complex embryophyta. Appl Plant Sci. 2015;3(5):1400105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Putnam NH, Connell BO, Stites JC, et al. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 2016;26(3):342–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. https://github.com/lh3/bwa . [Google Scholar]
- 54. https://github.com/nextgenusfs/funannotate . [Google Scholar]
- 55. Palmer JM, Stajich J. nextgenusfs/funannotate: funannotate v1.7.4 (1.7.4). Zenodo 2020. 10.5281/zenodo.3679386. [DOI]
- 56. Buchfink B, Xie C, Huson DH. Fast and sensitive protein alignment using DIAMOND. Nat Methods. 2015;12(1):59–60. [DOI] [PubMed] [Google Scholar]
- 57. Kim D, Paggi JM, Park C, et al. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat Biotechnol. 2019;37(8):907–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Baril T, Imrie R, Hayward A. Earl Grey. Zenodo 2021. 10.5281/zenodo.5654616. [DOI]
- 59. Smit AFA, Hubley RR, Green PR. RepeatMasker Open-4.0. http://repeatmasker.org. 2015. [Google Scholar]
- 60. Bao Z, Eddy SR. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002;12(8):1269–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Price AL, Jones NC, Pevzner PA. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21(Suppl 1):i351–8. [DOI] [PubMed] [Google Scholar]
- 62. Platt RN, Blanco-Berdugo L, Ray DA. Accurate transposable element annotation is vital when analyzing new genome assemblies. Genome Biol Evol. 2016;8(2):403–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. R Core Team . R: A language and environment for statistical computing. Vienna, Austria; 2013. [Google Scholar]
- 64. Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32(5):1792–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Nguyen L-T, Schmidt HA, von Haeseler A, et al. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 2015;32(1):268–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Emms DM, Kelly S. OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 2019;20(1):238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Jensen LJ, Julien P, Kuhn M, et al. eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 2008;36(Database issue):D250–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Yu G, Wang L-G, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yu G, Smith DK, Zhu H, et al. ggtree: an R package for visualization and annotation of phylogenetic trees with their covariates and other associated data. Methods Ecol Evol. 2017;8(1):28–36. [Google Scholar]
- 70. Luo W, Brouwer C. Pathview: an R/Bioconductor package for pathway-based data integration and visualization. Bioinformatics. 2013;29(14):1830–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Arican-Goktas HD, Ittiprasert W, Bridger JM, et al. Differential spatial repositioning of activated genes in Biomphalaria glabrata snails infected with Schistosoma mansoni. PLoS Negl Trop Dis. 2014;8(9):e3013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jiang Y, Loker ES, Zhang SM. In vivo and in vitro knockdown of FREP2 gene expression in the snail Biomphalaria glabrata using RNA interference. Dev Compar Immunol. 2006;30:855–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Pinaud S, Tetreau G, Poteaux P, et al. New insights into biomphalysin gene family diversification in the vector snail Biomphalaria glabrata. Front Immunol. 2021;12:918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Queiroz FR, Silva LM, Jeremias WDJ, et al. Differential expression of small RNA pathway genes associated with the Biomphalaria glabrata/Schistosoma mansoni interaction. PLoS One. 2017;12(7):e0181483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Nong W, Yu Y, Aase-Remedios ME, et al. Supporting data for “Genome of the ramshorn snail Biomphalaria straminea—an obligate intermediate host of schistosomiasis.”. GigaScience Database 2022. 10.5524/100985. [DOI] [PMC free article] [PubMed]
- 76. Altschul SF, Gish W, Miller W, et al. Basic Local Alignment Search Tool. J Mol Biol. 1990;215(3):403–10. [DOI] [PubMed] [Google Scholar]
- 77. Huerta-Cepas J, Szklarczyk D, Heller D, et al. eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 2019;47(D1):D309–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for bigger datasets. Mol Biol Evol. 2016;33(7):1870–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mendes FK, Vanderpool D, Fulton B, et al. CAFE 5 models variation in evolutionary rates among gene families. Bioinformatics. 2020:doi: 10.1093/bioinformatics/btaa1022. [DOI] [PubMed] [Google Scholar]
- 81. R Core Team . R: a language and environment for statistical computing. Vienna, Austria; 2013. [Google Scholar]
- 82. Sanderson MJ. r8s: Inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19(2):301–2. [DOI] [PubMed] [Google Scholar]
- 83. Simakov O, Marlétaz F, Yue JX, et al. Deeply conserved synteny resolves early events in vertebrate evolution. Nat Ecol Evol. 2020;4(6):820–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Smit A, Hubley R. RepeatModeler Open-1.0. http://repeatmasker.org. 2015. [Google Scholar]
- 85. Wickham H. ggplot2: Elegant graphics for data analysis. Springer-Verlag: Springer; 2016. http://ggplot2.org. [Google Scholar]
- 86. Zhong Y, Holland PWH. HomeoDB2: functional expansion of a comparative homeobox gene database for evolutionary developmental biology. Evol Dev. 2011;13(6):567–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zhong Y-F, Butts T, Holland PWH. HomeoDB: a database of homeobox gene diversity. Evol Dev. 2008;10(5):516–8. [DOI] [PubMed] [Google Scholar]
- 88. Davison A, Neiman M, Mobilizing molluscan models and genomes in biology. Phil Trans R Soc. 2021; 10.1098/rstb.2020.0163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Lee P, Callaerts P, de Couet H, et al. Cephalopod Hox genes and the origin of morphological novelties. Nature. 2003;424:1061–5. [DOI] [PubMed] [Google Scholar]
- 90. Albertin C, Simakov O, Mitros T, et al. The octopus genome and the evolution of cephalopod neural and morphological novelties Nature. Nature. 2015;524:220–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Brooke N, Garcia-Fernàndez J, Holland P, The ParaHox gene cluster is an evolutionary sister of the Hox gene cluster. Nature. 1998;392:920–2. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Palmer JM, Stajich J. nextgenusfs/funannotate: funannotate v1.7.4 (1.7.4). Zenodo 2020. 10.5281/zenodo.3679386. [DOI]
- Baril T, Imrie R, Hayward A. Earl Grey. Zenodo 2021. 10.5281/zenodo.5654616. [DOI]
- Nong W, Yu Y, Aase-Remedios ME, et al. Supporting data for “Genome of the ramshorn snail Biomphalaria straminea—an obligate intermediate host of schistosomiasis.”. GigaScience Database 2022. 10.5524/100985. [DOI] [PMC free article] [PubMed]
Supplementary Materials
Jacob Tennessen -- 8/28/2021 Reviewed
Coen Adema -- 9/5/2021 Reviewed
Coen Adema -- 1/5/2022 Reviewed
Data Availability Statement
The raw genome and RNA sequencing data have been deposited in the SRA under Bioproject No. PRJNA673593. The final chromosome assembly was submitted to NCBI Assembly under accession No. JADKLZ000000000. All data can also be found in the GigaScience Database [75].