Abstract
In this work, we describe a multiplex PCR assay for the detection of clinically relevant antibiotic resistance genes harbored by some Staphylococcus aureus isolates and for the simultaneous identification of such isolates at the species level. Conditions were optimized for the simultaneous detection of the 310-, 456-, and 651-bp regions of the mecA (encoding high-level methicillin resistance), ileS-2 (encoding high-level mupirocin resistance), and femB (encoding a factor essential for methicillin resistance) genes, respectively, from a single colony in a single reaction tube. The femB PCR fragment allows the specific identification of S. aureus. Validation of the method was performed using 50 human isolates of methicillin-resistant S. aureus (MRSA) and the appropriate control strains. This assay offers a rapid, simple, feasible, specific, sensitive, and accurate identification of mupirocin-resistant MRSA clinical isolates and could be systematically applied as a diagnostic test in clinical microbiology laboratories, facilitating the design and use of antibiotic therapy.
The selective pressure resulting from the extensive use of antibiotics over the last 50 years has led to the emergence of bacterial resistance and to the dissemination of resistance genes among pathogenic microorganisms (2, 17, 18). The progressive emergence and rapid dissemination of antibiotic resistance in staphylococci and its association with the use and consumption of antibiotics constitute a major health concern and have been considered a global crisis (7, 11, 16, 32). Staphylococci are ubiquitous microorganisms present in the respiratory tract and on the skin of a high percentage of adults. However, several population groups are at serious risk of suffering pathogenic staphylococcal infections. Within the genus Staphylococcus, S. aureus is the causal agent of most staphylococcal infections and is associated with serious community-acquired and nosocomial diseases. Serious complications occur because of multiple-antibiotic-resistant S. aureus. The introduction of new antibiotics in the fight against staphylococcal infections has stimulated a remarkable case of bacterial evolution in the face of changing selective pressures. Thus, the use of a new drug has always been followed by the prompt appearance of new staphylococcal resistance.
The first semisynthetic penicillin, namely, methicillin, was introduced in 1959 to overcome the problems that arose from the increasing prevalence of penicillinase-producing S. aureus resistant to penicillin (15). During the 1980s, methicillin-resistant S. aureus (MRSA) started to constitute a widespread human health concern (7). Methicillin resistance in S. aureus is primarily mediated by the overproduction of PBP2a, an additional altered penicillin-binding protein with low affinity for beta-lactam antibiotics. The mecA gene, the structural determinant encoding PBP2a, is therefore considered a useful molecular marker of putative methicillin resistance in S. aureus. MRSA is one of the most important pathogens that cause nosocomial infections worldwide. Moreover, S. aureus strains have a tendency to accumulate additional resistance determinants, resulting in the formation of multiple-antibiotic-resistant MRSA strains, which are creating increasing therapeutic problems, limiting the choice of therapeutic options.
One of the few antibiotics that are still active against MRSA is mupirocin (pseudomonic acid A), a natural antibiotic derived from Pseudomonas fluorescens (1, 7). Mupirocin inhibits bacterial growth by binding isoleucyl tRNA-synthetase (encoded by the ileS gene) and is used as a topical agent to avoid MRSA spread. Moreover, during the last few years it has also been proposed as an advisable antibiotic to be applied when invasive surgeries are employed, since it could prevent MRSA colonization (1, 7, 15). Unfortunately, 2 years after introduction of this drug, high-level mupirocin resistance appeared and is slowly increasing (7, 20). Such resistance is usually mediated by a conjugative plasmid-associated ileS-2 gene, which encodes an additional isoleucyl tRNA-synthetase that is not bound by mupirocin (3, 9).
Multiple-antibiotic-resistant S. aureus strains constitute a major health care problem; therefore, the availability of sensitive and specific methods for the accurate detection of antibiotic resistance in these bacteria has become an important tool in clinical diagnosis. Since phenotypic typing methods are not discriminating enough and are highly dependent on growth conditions, it is essential to use molecular techniques to stop the spread of multiple-antibiotic-resistant S. aureus. These techniques allow a rapid, accurate identification of staphylococci and their resistance type. Thus, fast, sensitive, and specific molecular methods will be an essential diagnostic tool for microbiology laboratories. The use of PCR for the sensitive and specific detection of microorganisms and antibiotic resistance genes is increasing in clinical microbiology laboratories. There are several reports in the literature describing the use of multiplex PCR (MPCR) for detection of MRSA strains, but most of these protocols are designed to detect only one or two gene fragments from overnight liquid cultures (20, 27, 30).
Here, we describe a rapid and simple MPCR assay for simultaneous detection of femB (fragment specific for S. aureus), mecA (encoding high resistance to methicillin), and ileS-2 (encoding high-level resistance to mupirocin) in a single reaction tube and using a method of extracting DNA from a single colony.
(Part of this work was presented during the 10th European Congress on Biotechnology, Madrid, Spain, July 2001.)
MATERIALS AND METHODS
Bacterial isolates.
A total of 50 staphylococcal isolates were used in this study. Included were 2 reference strains of methicillin- and mupirocin-susceptible S. aureus (ATCC 29213 and ATCC 25923), 9 clinical isolates of methicillin- and mupirocin-susceptible S. aureus, 2 clinical isolates of highly methicillin-resistant and intermediately mupirocin-resistant S. aureus, 23 clinical isolates of highly methicillin-resistant and mupirocin-susceptible S. aureus, 13 clinical isolates of highly methicillin- and highly mupirocin-resistant S. aureus, 3 clinical isolates of coagulase-negative staphylococci (1 methicillin- and mupirocin-susceptible Staphylococcus epidermidis isolate, 1 highly methicillin-resistant, mupirocin-susceptible S. epidermidis isolate, and 1 highly methicillin- and highly mupirocin-resistant S. epidermidis isolate). All of them were biochemically identified. These strains were all provided by the Microbiology Service of Nuestra Señora de Candelaria Hospital.
Identification of staphylococcal isolates and susceptibility testing.
Screening for methicillin resistance was done by 1-μg oxacillin disk diffusion testing, in which the disk was placed on Mueller-Hinton agar (Difco Laboratories, Detroit, Mich.) and incubated for 24 h at 30°C following the NCCLS guidelines (19). Intermediate resistance (disk zone diameter between 11 and 12 mm) was confirmed by the MIC determined with oxacillin E-test strips (AB Biodisk). Isolates of MRSA were screened for mupirocin resistance by the disk diffusion method (Oxoid, Basingstoke, England): 5-μg disks of mupirocin were used to detect low-level resistance, and resistant isolates were then screened against 200-μg disks in order to detect high-level resistance. Later confirmation of high resistance was performed with E-test strips (AB Biodisk). Moreover, the E-tests yielded the exact MIC for every highly mupirocin-resistant isolate, the MIC always being above 1,024 μg/ml. Biochemical identifications of staphylococci were performed according to standard laboratory criteria.
Rapid DNA extraction method.
After overnight culture on brain heart infusion (Difco Laboratories) agar plates, one colony of each sample was resuspended in 25 μl of sterile distilled water and the suspension was then placed in a 100°C heat block for 15 min. From this suspension, a 5-μl volume was directly used as a template for PCR amplification.
Oligonucleotide primers.
The oligonucleotide primers used in this study have been previously described (3, 10, 14) and were obtained from a commercial source (Roche Diagnostics): FemB1 (5′-TTA CAG AGT TAA CTG TTA CC-3′) and FemB2 (5′-ATA CAA ATC CAG CAC GCT CT-3′) (14) for femB, MecA1 (5′-GTA GAA ATG ACT GAA CGT CCG ATA A-3′) and MecA2 (5′-CCA ATT CCA CAT TGT TTC GGT CTA A-3′) (10) for mecA, and MupA (5′-TAT ATT ATG CGA TGG AAG GTT GG-3′) and MupB (5′-AAT AAA ATC AGC TGG AAA GTG TTG-3′) (3) for ileS-2 (Fig. 1).
FIG. 1.
Agarose gel electrophoresis patterns showing single PCR and MPCR amplification products for the S. aureus genes femB, ileS-2, and mecA amplified from a highly methicillin- and highly mupirocin-resistant S. aureus isolate. Lanes 1 to 3, PCR amplicons from femB, ileS-2, and mecA, respectively; lane 4, triplex PCR amplicons, i.e., femB, ileS-2, and mecA simultaneously amplified; lane M, DNA molecular size marker (100-bp ladder). No bands were present in the negative control (data not shown). A schematic representation of the fragments amplified is shown on the lefthand side of the figure.
MPCR amplification.
MPCR assays were all directly performed from the bacterial suspension obtained after the rapid DNA extraction method described above. An aliquot of 5 μl of this suspension was added to 45 μl of PCR mixture consisting of 1× reaction buffer [16 mM (NH4)2SO4, 67 mM Tris-HCl (pH 8.8)], a 0.2 mM concentration of each of the four deoxyribonucleoside triphosphates (dATP, dCTP, dGTP, and dTTP) (Promega Corp., Madison, Wis.), 3 mM MgCl2, 75 pmol of each femB primer, 20 pmol of each ileS-2 and mecA primer, and 1.25 U of Taq DNA polymerase (Bioline). For each sample, one reaction was performed with the femB pair of primers to identify S. aureus strains and with the mecA and ileS-2 pairs of primers to detect both resistance markers. In order to reduce the formation of nonspecific extension products, a hot-start PCR protocol was used; the tubes were placed in the thermal cycler when the denaturing temperature was reached. All MPCR assays were carried out with a negative control containing all of the reagents without DNA template. DNA amplification was carried out in a GeneAmp PCR system 2400 and in a GeneAmp PCR system 9700 thermocycler (PE Applied Biosystems, Foster City, Calif.) with the following thermal cycling profile: an initial denaturation step at 94°C for 5 min was followed by 10 cycles of amplification (denaturation at 94°C for 30 s, annealing at 64°C for 30 s, and extension at 72°C for 45 s) and 25 cycles of amplification (denaturation at 94°C for 45 s, annealing at 50°C for 45 s, and extension at 72°C for 1 min) ending with a final extension step at 72°C for 10 min. After PCR amplification, 5 μl was removed and subjected to agarose gel electrophoresis (2% agarose, 1× Tris-borate-EDTA, 100 V, 100 min) to estimate the sizes of the amplification products by comparison with a 100-bp molecular size standard ladder (Roche Diagnostics). The gel was stained with ethidium bromide, and the amplicons were visualized using a UV light box.
RESULTS
Rapid DNA extraction method.
In order to accelerate the procedure of identification in clinical microbiology laboratories, it is very important to have a simple and rapid method for DNA extraction. There are several reports in the literature describing methods of extracting DNA from overnight liquid cultures (20, 27, 30). In this report, we describe a rapid method for bacterial DNA extraction directly from a single colony that gave quality DNA for PCR in as little as 15 min. This protocol yielded good-quality target DNA for PCR amplification. Amplifications using that DNA gave rise to good quantities of the expected PCR fragments. When PCR was performed using DNA obtained by this method or previously reported methods (12, 20), no differences were observed (data not shown).
MPCR for detection of selected staphylococcal genes.
The reaction conditions for the MPCR assay were optimized to ensure that all of the target gene sequences were satisfactorily amplified. The primers used in this study differ in annealing temperatures, which increased the possibility of occurrence of unwanted bands originating from nonspecific amplification. Therefore, we performed both a hot start and two rounds of amplification with different annealing temperatures. On the other hand, there is evidence that MPCR with targets that differ widely in size often favors amplification of the shorter target over the longer one, resulting in different amounts of amplified products (4, 20). For this reason, to optimize the conditions for the MPCR analysis, we assayed different primer concentrations, template DNA preparations, and MgCl2 concentrations. As described in Materials and Methods, the final quantities that we used to obtain optimal results were 3 mM MgCl2, 75 pmol of each femB primer, 20 pmol of each ileS-2 and mecA primer, and 5 μl of the DNA solution obtained with our DNA extraction method.
Previous to the optimization of the triplex PCR, we ensured that our PCR protocol was adequate for the individual amplification of all three DNA fragments. Each individual amplification yielded the fragment of the expected size, i.e., 651, 310, and 456 bp, respectively (Fig. 1). Figure 2 shows an agarose gel stained with ethidium bromide to illustrate the typical results obtained with the optimized MPCR assay. Amplification of femB, ileS-2, and mecA targets produced distinct bands corresponding to their respective molecular sizes that were easily recognizable. The femB fragment was always amplified in the case of S. aureus strains and never in the case of S. epidermidis strains (Fig. 2). With respect to the mecA fragment, it was detected in all the strains exhibiting high methicillin resistance. Finally, amplification of the ileS-2 target always occurred for highly mupirocin-resistant strains but did not occur in the case of intermediate-resistance isolates, since intermediate resistance levels were not due to the ileS-2 gene. Intermediate resistance levels could be due to mutations in the endogenous ileS gene or to the presence of any additional chromosomal gene encoding an extra isoleucyl-tRNA synthetase (3, 21). This protocol, including the rapid DNA extraction method from a single colony and electrophoretic analysis of the amplified products on an agarose gel, was performed in less than 5 h.
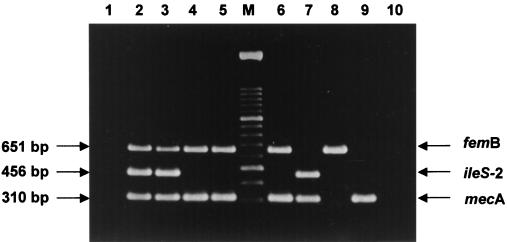
FIG. 2.
Agarose gel electrophoresis patterns showing MPCR amplification products from different S. aureus and S. epidermidis isolates. Lane 1, negative control; lanes 2 and 3, mupirocin-resistant MRSA isolates; lanes 4 and 5, mupirocin-susceptible MRSA isolates; lane 6, mupirocin-intermediate MRSA isolate; lane 7, methicillin-resistant and mupirocin-resistant S. epidermidis isolate; lane 8, methicillin-susceptible and mupirocin-sensitive S. aureus strain (ATCC 29213); lane 9, methicillin-resistant and mupirocin-susceptible S. epidermidis isolate; lane 10, methicillin-susceptible and mupirocin-sensitive S. epidermidis isolate; lane M, DNA molecular size marker (100-bp ladder).
Correlation between susceptibility testing and the multiple PCR assays.
We compared oxacillin and mupirocin susceptibility results determined by the disk diffusion method and E-test for 50 staphylococcal isolates with the results obtained by the MPCR assays for the detection of antibiotic resistance genes. All cases in which high resistance to oxacillin or mupirocin was detected, using these phenotypic assays, were later confirmed by our PCR protocol. Moreover, in the case of borderline strains that did not yield a clear phenotypic result, the MPCR described here allowed us to confirm the presence or absence of the mecA or the ileS-2 gene. Thus, for the strains included in this study, we have found a 100% concordance between microbiological and PCR results (Table 1).
TABLE 1.
Correlation between phenotypic groups and PCR results
| Species | Resistance phenotypea
|
No. of isolates | Presence of fragment:
|
|||
|---|---|---|---|---|---|---|
| Methicillin | Mupirocin | mecA | ileS-2 | femB | ||
| S. aureus | S | S | 9 | − | − | + |
| R | I | 2 | + | − | + | |
| R | S | 23 | + | − | + | |
| R | R | 13 | + | + | + | |
| S. epidermidis | S | S | 1 | − | − | − |
| R | S | 1 | + | − | − | |
| R | R | 1 | + | + | − | |
R, highly resistant; I, intermediately resistant; S, susceptible.
DISCUSSION
During the last decade, many studies have demonstrated the extremely high capacity of PCR for specifically detecting bacteria and genes of interest (22). That ability has revealed PCR as a powerful tool in clinical microbiology studies (8). Several authors have already shown the feasibility of the PCR methodology for the identification of S. aureus strains and for the detection of antibiotic resistance genes (8, 12, 31). PCR identification of S. aureus has been based on the detection of different specific target sequences such as nuc (6) and coaA (24, 25) or factors essential for methicillin resistance such as femA (5, 13, 29, 30) or femB (5, 13). On the other hand, different studies have also shown the applicability of PCR to the detection of staphylococcal antibiotic resistance genes (3, 5). Our aim was to develop an MPCR for the simultaneous identification of S. aureus strains and detection of the genes conferring high methicillin and mupirocin resistance, namely, mecA and ileS-2, respectively. For the identification of S. aureus, we employed PCR primers targeted to the femB gene. Although the femB gene encodes an important enzyme involved in the cross-linking of peptidoglycan in various Staphylococcus spp., previous studies had demonstrated the feasibility of these primers for the unequivocal identification of this bacterial species (12, 14). Indeed, Staphylococcus auricularis also would yield a PCR fragment using these primers, but this staphylococcal species is not of clinical importance and appears not to colonize body sites normally screened for MRSA (12).
Currently, multiple-antibiotic-resistant S. aureus strains constitute a major health care problem, since they are the etiologic agent of several nosocomial and community-acquired pathological infections. For that reason, accurate and fast detection of resistant isolates constitutes a critical goal of clinical microbiology, and therefore, PCR assays have become an essential tool in laboratory programs. Although previous reports have evidenced the utility of PCR for the accurate detection of the mecA gene (23, 27) and the possibility of simultaneous identification of S. aureus and detection of mecA (10, 12, 22, 26, 28, 31), few studies have also included the detection of the ileS-2 gene (3, 20). Only one previous report describes the simultaneous detection of ileS-2 and mecA in one tube and femA in another tube (20). Furthermore, fast DNA extraction methods have also been reported, but normally they are not performed from a single colony and most of them need the use of lytic enzymes, e.g., lysostaphin, and organic solvents, e.g., phenol-chloroform (3, 30, 31). Since the convenience of performing all three PCR amplifications in a single tube and from a single colony is obvious, we focused on optimizing the triplex PCR herein described. Thus, we firstly optimized a quick method of extracting DNA from a single colony which yielded enough DNA for optimal PCR amplification without the need of overnight liquid culture, a lytic enzyme, or an organic solvent and without PCR bias due to inhibitory substances. When PCR was performed using DNA obtained by this method or by previously reported ones (3, 12, 20), no differences were observed (data not shown).
For the strains included in this study, we have found a 100% concordance between microbiological susceptibility testing and PCR results (Table 1). However, other authors have detected isolates presenting the ileS-2 fragment but not high mupirocin resistance (2, 3). In other cases, discrepancies between the PCR results and the mupirocin MICs appeared to be due to the selection of bacterial colonies with mixed mupirocin susceptibilities derived from lack of expression of the ileS-2 gene in a proportion of the cells (3). For these reasons, we believe that only a combination of both approaches should be used for a reliable identification of mupirocin-resistant MRSA isolates.
Nowadays, with only a few antibiotics such as mupirocin constituting the last defense against MRSA, and due to the increasing incidence and spread of MRSA, it is absolutely necessary that fast and sensitive laboratory methods be available for the immediate detection of multiple-antibiotic-resistant MRSA. As our results showed, the method herein described is highly sensitive, specific, fast, and feasible. Hence, considering that it represents a rapid, simple, and cost-effective method, it could be systematically applied in clinical microbiology laboratories for the identification of mupirocin-resistant MRSA, bringing insights into antibiotic therapy design and helping treatment to be initiated without delay.
ACKNOWLEDGMENTS
We thank Ninivé Batista from the Microbiology Service of Nuestra Señora de La Candelaria Hospital for the bacterial isolates.
This work was supported by grants 1999/074 and 2001/020 from the Consejería de Educación, Cultura y Deportes, Canarian Autonomous Government, and FUNCIS PI 40/00, Canarian Autonomous Government, to F.C.-M. and S.M.-A. S.M.-A. was supported by FIS contract 99/3060 (Fondo de Investigación Sanitaria, Spain).
REFERENCES
- 1.Alarcón T, Sanz J C, Blanco F, Domingo D, López-Brea M. High-level mupirocin resistance among Spanish methicillin-resistant Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 1998;17:877–879. doi: 10.1007/s100960050212. [DOI] [PubMed] [Google Scholar]
- 2.Amábile-Cuevas C F. Molecular Biology Intelligence Unit. Origin, evolution and spread of antibiotic resistance genes. Austin, Tex: R. G. Landes Company; 1993. [Google Scholar]
- 3.Anthony R M, Connor A M, Power E G M, French G L. Use of the polymerase chain reaction for rapid detection of high-level mupirocin resistance in staphylococci. Eur J Clin Microbiol Infect Dis. 1999;18:30–34. doi: 10.1007/s100960050222. [DOI] [PubMed] [Google Scholar]
- 4.Bej A K, Mahbubani M H, Miller R, Dicesare J L, Haff L, Atlas R M. Multiplex PCR amplification and immobilized capture probes for detection of bacterial pathogens and indicators in water. Mol Cell Probes. 1990;4:353–365. doi: 10.1016/0890-8508(90)90026-v. [DOI] [PubMed] [Google Scholar]
- 5.Berger-Bächi B, Strassle A, Gustafson J E, Kayser F H. Mapping and characterization of multiple chromosomal factors involved in methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1992;36:1367–1373. doi: 10.1128/aac.36.7.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brakstad O G, Aasbakk K, Maeland J A. Detection of Staphylococcus aureus by polymerase chain reaction amplification of the nuc gene. J Clin Microbiol. 1992;30:1654–1660. doi: 10.1128/jcm.30.7.1654-1660.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chambers H F. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin Microbiol Rev. 1997;10:781–791. doi: 10.1128/cmr.10.4.781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cockerill F R. Genetic methods for assessing antimicrobial resistance. Antimicrob Agents Chemother. 1999;43:199–212. doi: 10.1128/aac.43.2.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dyke K G, Curnock S P, Golding M, Noble W C. Cloning of the gene conferring resistance to mupirocin in Staphylococcus aureus. FEMS Microbiol Lett. 1991;61:195–198. doi: 10.1016/0378-1097(91)90550-t. [DOI] [PubMed] [Google Scholar]
- 10.Geha D J, Uhl J R, Gustaferro C A, Persing D H. Multiplex PCR for identification of methicillin-resistant staphylococci in the clinical laboratory. J Clin Microbiol. 1994;32:1768–1772. doi: 10.1128/jcm.32.7.1768-1772.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hashimoto H, Inouye M, Hayashi I. A survey of Staphylococcus aureus by typing and drug-resistance in various areas of Japan during 1992 and 1993. Jpn J Antibiot. 1994;47:618–626. [PubMed] [Google Scholar]
- 12.Jonas D, Grundmann H, Hartung D, Daschner F D, Towner K J. Evaluation of the mecA femB duplex polymerase chain reaction for detection of methicillin-resistant Staphylococcus aureus. Eur J Clin Microbiol Infect Dis. 1999;18:643–647. doi: 10.1007/s100960050365. [DOI] [PubMed] [Google Scholar]
- 13.Kizaki M, Kobayashi Y, Ikeda Y. Rapid and sensitive detection of the femA gene in staphylococci by enzymatic detection of polymerase chain reaction (ED-PCR): comparison with standard PCR analysis. J Hosp Infect. 1994;28:287–295. doi: 10.1016/0195-6701(94)90092-2. [DOI] [PubMed] [Google Scholar]
- 14.Kobayashi N, Wu H, Kojima K, Taniguchi K, Urasawa S, Uehara N, Omizu Y, Kishi Y, Yagihashi A, Kurokawa I. Detection of mecA, femA and femB genes in clinical strains of staphylococci using polymerase chain reaction. Epidemiol Infect. 1994;113:259–266. doi: 10.1017/s0950268800051682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Livermore D M. Antibiotic resistance in staphylococci. Int J Antimicrob Agents. 2000;16:3–10. doi: 10.1016/s0924-8579(00)00299-5. [DOI] [PubMed] [Google Scholar]
- 16.Martínez J L, Baquero F. Mutation frequencies and antibiotic resistance. Antimicrob Agents Chemother. 2000;44:1771–1777. doi: 10.1128/aac.44.7.1771-1777.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Méndez-Álvarez S, Pérez-Hernández X, Claverie-Martín F. Glycopeptide resistance in enterococci. Int Microbiol. 2000;3:71–80. [PubMed] [Google Scholar]
- 18.Murray P R, Baron E J, Pfaller M A, Tenover F C, Yolken R H, editors. Manual of clinical microbiology. 7th ed. Washington, D.C.: ASM Press; 1999. [Google Scholar]
- 19.NCCLS. Performance standards for antimicrobial disk susceptibility tests. Approved standard M2–A7. 7th ed. Wayne, Pa: NCCLS; 2001. [Google Scholar]
- 20.Nunes E L, dos Santos K R, Mondino P J, Bastos M D, Giambiagi-de Marval M. Detection of ileS-2 gene encoding mupirocin resistance in methicillin-resistant Staphylococcus aureus by multiplex PCR. Diagn Microbiol Infect Dis. 1999;34:77–81. doi: 10.1016/s0732-8893(99)00021-8. [DOI] [PubMed] [Google Scholar]
- 21.Ramsey M A, Bradley S F, Kauffman C A, Morton T M. Identification of chromosomal location of mupA gene, encoding low-level mupirocin resistance in staphylococcal isolates. Antimicrob Agents Chemother. 1996;40:2820–2823. doi: 10.1128/aac.40.12.2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salisbury S M, Sabatini L M, Spiegel C A. Identification of methicillin-resistant staphylococci by multiplex polymerase chain reaction assay. Microbiol Infect Dis. 1996;107:368–373. doi: 10.1093/ajcp/107.3.368. [DOI] [PubMed] [Google Scholar]
- 23.Schmitz F J, Mackenzie C R, Hofmann B, Verhoef J, Finken-Eigen M, Heinz H P, Kohrer K. Specific information concerning taxonomy, pathogenicity and methicillin resistance of staphylococci obtained by multiplex PCR. J Med Microbiol. 1997;46:773–778. doi: 10.1099/00222615-46-9-773. [DOI] [PubMed] [Google Scholar]
- 24.Shopsin B, Gomez M, Waddington M, Riehman M, Kreiswirth B N. Use of coagulase gene (coa) repeat region nucleotide sequences for typing of methicillin-resistant Staphylococcus aureus strains. J Clin Microbiol. 2000;38:3453–3456. doi: 10.1128/jcm.38.9.3453-3456.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang Y W, Procop G W, Persing D H. Molecular diagnostics of infectious diseases. Clin Chem. 1997;43:2021–2038. [PubMed] [Google Scholar]
- 26.Tenover F C, Jones R N, Swenson J M, Zimmer B, McAllister S, Jorgensen J H the NCCLS Staphylococcus Working Group. Methods for improved detection of oxacillin resistance in coagulase-negative staphylococci: results of a multicenter study. J Clin Microbiol. 1999;37:4051–4058. doi: 10.1128/jcm.37.12.4051-4058.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tokue Y, Shoji S, Satoh K, Watanabe A, Motomiya M. Comparison of a polymerase chain reaction assay and conventional microbiologic method for detection of methicillin-resistant Staphylococcus aureus. Antimicrob Agents Chemother. 1992;36:6–9. doi: 10.1128/aac.36.1.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Towner K J, Tabot D C S, Curran R, Webster C A, Humphreys H. Development and evaluation of a PCR-based immunoassay for the rapid detection of methicillin-resistant Staphylococcus aureus. J Med Microbiol. 1998;47:1–7. doi: 10.1099/00222615-47-7-607. [DOI] [PubMed] [Google Scholar]
- 29.Unal S, Hoskins J, Flokowitsch J E, Wu C Y, Preston D A, Skatrud P L. Detection of methicillin-resistant staphylococci by using the polymerase chain reaction. J Clin Microbiol. 1992;30:1685–1691. doi: 10.1128/jcm.30.7.1685-1691.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vannuffel P, Gigi J, Ezzedine H, Vandercam B, Delmee M, Wauters G, Gala J L. Specific detection of methicillin-resistant Staphylococcus species by multiplex PCR. J Clin Microbiol. 1995;33:2864–2867. doi: 10.1128/jcm.33.11.2864-2867.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vannuffel P, Laterre P F, Bouyer M, Gigi J, Vandercam B, Reynaert M, Gala J L. Rapid and specific molecular identification of methicillin-resistant Staphylococcus aureus in endotracheal aspirates from mechanically ventilated patients. J Clin Microbiol. 1998;36:2366–2368. doi: 10.1128/jcm.36.8.2366-2368.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Voss A, Milatovic D, et al. Methicillin-resistant Staphylococcus aureus in Europe. Eur J Clin Microbiol Infect Dis. 1994;13:50–55. doi: 10.1007/BF02026127. [DOI] [PubMed] [Google Scholar]