Abstract
Background
The nuclear receptor REV-ERBα (nuclear receptor subfamily 1, Group D member 1, NR1D1) is one of the essential components of the circadian clock which modulates cell proliferation, glucose metabolism, inflammation, and many other biological processes. Modulation of these processes are also relevant to cancer development. Previous studies have suggested that activation of REV-ERBα correlates with cancer cell senescence and death, but how REV-ERBα play roles in tumor progression require further elucidation.
Methods
We investigated the expression of REV-ERBα in clinical samples by immunohistochemistry (IHC). REV-ERBα is downregulated by shorth hairpin RNA (shRNA). The gene expression level of each group was detected by Western blot analysis. The effects of REV-ERBα downregulation on apoptosis and cell cycles was assessed by flow cytometry assay. A549 cell growth curve under different treatments measured by MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay. Cell invasion ability under different treatments was measured by Transwell assay. Immunostaining analysis was also used for evaluating the effects of downregulation of REV-ERBα on nuclear factor-κB (NF-κB).
Results
Compared to 81.8% (54/66) of samples exhibiting a lower expression level of REV-ERBα in cancer tissue than in paired normal tissue, only 18.2% (12/66) were higher or equally expressed in lung cancer tissue. Furthermore, downregulation of REV-ERBα by RNA interference can significantly enhance the transcription of nuclear factor-κB (NF-κB), while the expression of p53 remained the same. Downregulation of REV-ERBα was also shown to stimulate the invasion and promote the proliferation of lung adenocarcinoma cell line A549.
Conclusions
Our findings suggest that tumorigenesis and progression of lung carcinoma is relevant to downregulation or inhibition of REV-ERBα. This pathophysiological process also correlates with regulation of the NF-κB signaling pathway, indicating that REV-ERBα is a potential target of lung cancer therapy.
Keywords: REV-ERBα, lung adenocarcinoma, nuclear factor-κB (NF-κB), cell proliferation, invasion
Introduction
Lung cancer was the malignant tumor with the highest morbidity and mortality in the world until 2019 (1). Non-small cell lung cancer (NSCLC) accounts for a more than 80% of all lung cancer diagnoses (2). A large number of lung cancer patients have local or distant metastasis at the time of diagnosis, leading to poor prognosis. Therefore, with the rapid development of lung cancer therapeutic research, one of the key motivations to improve the prognosis and the long term survival rates of lung cancer patients is to discover biomarkers for early screening and diagnosis (2,3). The two main subtypes of NSCLC are lung adenocarcinoma (LUAD) and lung squamous cell carcinoma (LUSC). The significant tumor heterogeneity of lung adenocarcinoma is an important reason for short-term recurrence and susceptibility to treatment resistance (4-6). Although a variety of therapeutic methods have been developed, including chemotherapy and targeted therapy towards certain genes, the overall survival (OS) rate still needs to be enhanced with precision medicine.
Recently, genes related to circadian regulation have been identified as playing significant roles in cancer progression (7-11). Investigations in the field of oncology have shown that the treatment effects and survival outcomes of patients are often related to the expression level of circadian genes (9-12). The autonomous circadian clock of cancer cells can coordinate a complex network of physiological processes in many ways (13). Since the circadian clock components can directly control several important pathways for tumor development, the pharmacological regulation of these components may provide a promising selective anticancer strategy.
The nuclear receptor REV-ERBα, encoded by Nrld1, is a heme-binding circadian clock component which acts as a key factor of several biological processes involved in tumorigenesis (9,13-16). REV-ERBα is a protein in charge of cell energy sources, responsible for the synthesis of lipids in cells and autophagy. When the circadian clock balance is broken, the cells will take in excess energy for proliferation and metabolism; and the balance of autophagy is also broken. It has been shown that REV-ERBα has a repressive function on cell proliferation and metabolism (17-20). The emergence of SR9009 and SR9011 as pyrrole derivatives was an important indicator for researching the regulation of REV-ERBα activity. Weakening the molecular signaling pathways related to abnormal activities by strengthening the circadian metabolic rhythm of cells can inhibit tumor cell activity (19). As we know, it has been indicated that REV-ERBα is involved in the development process of tumor, such as Breast cancer, colon cancer, melanoma, etc. But it is rarely reported in the field of non-small cell lung cancer (NSCLC), especially lung adenocarcinoma.
Accordingly, the circadian components in LUAD may be an important oncogenic factor that associates with lung cancer incidence. In this study, we aimed to investigate the role of REV-ERBα in lung adenocarcinoma progression by examining the expression of REV-ERBα in clinical samples by immunohistochemistry (IHC) and evaluating the role of REV-ERBα in the regulation of cell proliferation, apoptosis, and invasion of lung adenocarcinoma cell line A549, to further indicate that whether downregulation of REV-ERBα can promote cancer progression and influence the efficacy of chemotherapy.
In addition, recently experimental studies have shown that silencing or down-regulating the expression of target genes with the help of shorth hairpin RNA (shRNA) is an effective means of RNA interference technology. Using plasmid and viral vectoring systems, the transcription of shorth hairpin RNA (shRNA) precursors that are effectively processed by the RNA interference pathway can lead to potent gene knockdown. In this research, shorth hairpin RNA (shRNA) was used to achieve the effect of downregulation of REV-ERBα.
We present the following article in accordance with the MDAR checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6405/rc).
Methods
Cells culture
The A549 cell line is considerably representative in the cell experimental research of lung adenocarcinoma. Lung adenocarcinoma cell line A549 was purchased from American Type Culture Collection (ATCC; Manassas, VA, USA). The cell incubator was set to contain 5% carbon dioxide and at 37 °C. Dulbecco’s modified Eagle medium (DMEM) medium including 1% penicillin/streptomycin (Gibco, Waltham, MA, USA) and 10% fetal bovine serum (FBS; Hyclone, Logan, UT, USA) was used as the cell culture medium. We subcultured A549 in a 25 cm2 culture flask with ethylenediamine tetraacetic acid (EDTA) containing 0.25% trypsin in a ratio of 1:3. The cells in each group underwent respective experimental intervention according to the experimental scheme. Each procedure was repeated 3 times.
Human samples
Paired lung cancer tissue and normal tissues (>5 cm away from the tumor site) were collected from patients with LUAD, who had undergone lobectomy at the Tianjin Chest Hospital. Specimens are aseptically collected during surgery and stored at a low temperature (−80 °C) in liquid nitrogen. No patient had received chemotherapy or radiotherapy before surgery. The diagnostic criteria of lung cancer was made based on histopathological diagnosis. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Tianjin Chest Hospital Academic Ethics Committee (No. 2019LW-012). Informed consent was taken from all the patients.
IHC
The LUAD tissue (experimental group) and adjacent normal lung tissue (control group) were fixed in formalin, dehydrated with alcohol and embedded in paraffin. Tissues were then sliced into 5 um thick sections, dewaxed and rehydrated. They were then washed in citrate buffer (10 mm, pH=6.0) for antigen retrieval, exposed to an REV-ERBα antibody (1:100 dilution), and incubated in a refrigerator at 4 °C for 16 h. The sections were then stained with 3,3’-diaminobenzidine (DAB) and counterstained with hematoxylin dye.
At least 10 visual areas were observed randomly under a light microscope. Those with the same staining as normal lung tissue were classified as positive, and those with absent or weak staining were negative. We then calculated the percentage of positive cells among the total number of cells, according to the following: 1 point for positive cell rate <10%, 2 points for positive cell rate >10% and ≤50%, 3 points for positive cell rate >50% and ≤75%, and 4 points for positive cell rate >75%. The cells were scored according to the intensity of staining, as follows: 1 for negative staining, 2 for weak staining, 3 for medium intensity staining, and 4 for strong staining. Judging by the product of the 2 scores: ≤4 points was (−); >4 points and ≤8 points was (+); >8 points and ≤12 points was (++); >12 points and ≤16 points was (+++); and 16 points was (++++).
Reverse transcription polymerase chain reaction analysis
According to the instructions of biologics, RNAeasy (Qiagen, Hilden, Germany) was used to extract total RNA from tissues and cells and DNase was used for reverse transcription. We used qScript cDNA SuperMix (Quanta BioSciences, Beverly, MA, USA) to generate complementary DNA (cDNA). Then, the IQ SYBR-Green Supermix on the iCycler IQ Multicolor Detection System (Bio-Rad Technologies, Inc., Hercules, CA, USA) was used to perform quantitative polymerase chain reaction (qPCR). The expression level of actin was used to normalize the expression level of the target gene. The following primer sequences were used: NR1D1/REV-ERBα forward, 5'-TGCGGGAGGTGGTAGAGTTTG-3'; NR1D1/REV-ERBα reverse, 5'- GCTGTAGGTGGTGCGGCTTA-3'; Actin forward, 5'- CACCCAGCACAATGAAGATCAAGAT-3'; Actin reverse, 5'- CCAGTTTTTAAATCCTGAGTCAAGC-3'.
Shorth hairpin RNA plasmid and transfection
The oligonucleotides for REV-ERBα shRNA and Control shRNA constructs were synthesized and subsequently inserted into the pLKO.1 vector. The short hairpin RNA (shRNA) target sequence of human REV-ERBα is 5'-GCUUAAGGCUGGCACCUUUTT-3'. Transient transfections were done using Attractene™ transfection reagent (Qiagen, Germany), with an appropriate amount of DNA and serum free OPTIMEM (Gibco, Galway, Ireland). The transfection reagent was used at a 3:1 ratio with DNA. The amount of DNA, transfection reagent, and media were adjusted according to the number of cells being transfected. At 16 h after the addition of the transfection mix, the serum free media was replaced with media supplemented with an additional 10% FBS. The A549 cells were then used for experiments with the addition of 200 ng/mL of tetracycline to the media.
Western blot analysis
Ice-cold radioimmunoprecipitation (RIPA) buffer (Solarbio, Beijing, China) to dissolve the A549 cells after the proposed experiment. The dissolved samples were placed on ice for 30 min then centrifuged (4 °C, 12,000 rpm, 10 min). The supernatant was collected for sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE). After transferring, REV-ERBα, β-actin, nuclear factor-κB (NF-κB), and p53 monoclonal antibodies (Abcam, Cambridge, UK) were used to incubate polyvinylidene fluoride (PVDF) membrane (1:1,000 dilution). The membranes were washed 3 times in 1× tris-buffered saline with Tween 20 (TBST) buffer and subsequently incubated with horseradish peroxidase (HRP)-conjugated secondary antibody at 1:5,000 dilution. An enhanced chemiluminescence (ECL) exposure kit (Millipore, Burlington, MA, USA) was used to discover the protein bands. The GBox ChemiXX9 system (Syngene, Bangalore, India) was used for imaging and photographing.
Immunostaining analysis
The A549 cells were fixed with 4% paraformaldehyde liquid for 15 min, then permeabilized with 0.25% Triton X-100 for 10 min. A primary antibody (bovine serum albumin,1%) was used to incubate A549 cells for 1 h. the cells were then washed 3 times in phosphate-buffered saline (PBS). Subsequently, a secondary antibody (bovine serum albumin, 1%) was used to incubate the A549 cells for 1 h in the dark at room temperature. The DNA was stained with Hoechst 33342 (1×PBS). A fluorescence microscope was used to observe the slides after oil-immersion.
3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide cell viability assay
The A549 cells in the logarithmic growth stage were made into cell suspension with DMEM (fetal bovine serum, 10%) and inoculated in 96 well culture plate (100 uL, 1×104 per well) with different experimental interventions, with 8 multiple wells in each group as a parallel experiment. After replacing the new culture medium, 10 uL 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (5 mg/mL, Sigma-Aldrich, St. Louis, MO, USA) was added into each well and continued to culture in an incubator (5% CO2, 37 °C) for 3 h. We then added 100 uL 10% SDS solution (0.01 mol/L HCl) to each hole and incubated it for 12 h to completely dissolve the formazan. A plate-reading spectrophotometer (Perkin-Elmer, Waltham, MA, USA) was used to detect the optical density (OD) at 595 nm wavelength.
Flow cytometry apoptosis assay by Annexin V-FITC staining
After the proposed treatments, the cells were collected into a flow cytometric tube from 6-well plates by digesting the cells using trypsin according to the subculture procedures. We then added 3 mL 4 °C PBS to resuspend the cells, followed by centrifugation at 1,500 rpm for 5 min. Then, the suspensions were discarded, the precipitates resuspended by 200 µL binding buffer, and 5 µL Annexin V-FITC and 5 µL propidium iodide (PI) were added. The tubes were mixed well and allowed to stand at room temperature for 15 min. Then, cells were sorted via FACS Caliber [Becton, Dickinson, and Co. (BD), Franklin Lakes, NJ, USA]. Analysis was performed using Flowjo software (BD, USA).
Cell cycle analysis
Cells and their suspensions were collected as described above after the proposed treatments. After washing with pre-chilled PBS, cells were fixed in 75% ethanol overnight at 4 °C. Fixed cells were then resuspended with 100 µL PBS and permeabilized using 2 µL 1 mg/mL RNaseA, followed by 37 °C water bathing for 40 min. Then, 100 µL of 100 µg/mL PI staining solution was added and rested for 20 min. Finally, the cells were sorted via a FACS Caliber (BD, USA). Analysis was performed using the Flowjo software.
Transwell cell invasion test
Transwell inserts (Corning, Inc., New York, NY, USA) of 8-µm pore size was used to detect the invasive ability of cells. The treated cells of different groups were seeded in the serum-free medium of the upper chamber plate which was overlaid with 1:8 diluted Matrigel (70 µL). We then added 500 µL the medium containing 10% FBS to the bottom chamber as a chemokine and the cells were incubated for 24 h. The cells on the upper side of the filter membrane were removed with a cotton swab and 95% ethyl alcohol was used to fix migrated cells located in the lower chamber. Subsequently, 0.5% crystal violet solution was used to stain the cells in the upper chamber. The number of invasive cells was counted at ×200 magnified field of view in 5 different visual fields of each filter membrane. Each experiment was repeated 3 times independently.
Statistical analysis
The software SPSS 24.0 (IBM Corp., Armonk, NY, USA) was used for statistical analysis. All values were expressed as mean ± standard deviation (). One-factor analysis of variance (ANOVA) was used to compare the mean of multiple groups. Least significant difference (LSD) test was used for pairwise comparison of mean between groups. A P value <0.05 was considered statistically significant.
Results
REV-ERBα was less expressed in lung cancer tissues compared to normal tissues
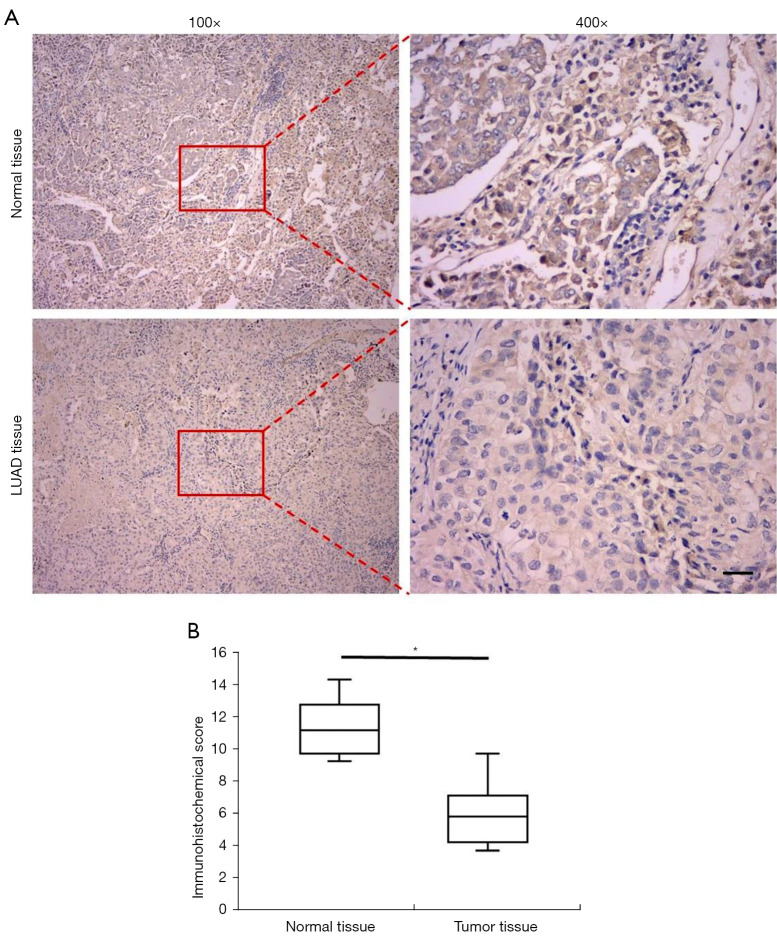
In order to elucidate the expression levels of REV-ERBα in the samples from patients with LUAD, we firstly collected 66 paired samples for IHC analysis which consisted of LUAD specimens and corresponding normal tissues. Representative images are shown in Figure 1A. In the LUAD tissue samples, less REV-ERBα staining concentrated in the cell nuclei was identified compared to normal tissue samples. Among all the samples, 54 LUAD tissues revealed lower IHC classification of REV-ERBα than paired normal tissues (81.8%), and the other 12 samples exhibited equal or higher expression levels in LUAD tissues compared to paired normal tissues (18.2%). These results indicated that downregulation of REV-ERBα may relate to tumorigenesis. The average IHC score of the total 66 LUAD specimens was 5.93, compared to the score of paired normal tissues at 11.32 (Figure 1B).
Figure 1.
IHC detection of REV-ERBα in normal tissue and tumor tissue in patients with lung adenocarcinoma. (A) Representative IHC staining images of REV-ERBα in normal tissue and tumor tissue at the magnification of ×100 and ×400 respectively (scale bar, 50 µm). (B) Statistics of IHC scores of expression of REV-ERBα in lung cancer tissue and paired non-cancerous tissues (n=66). (*P<0.05 compared with paired non-cancerous tissues). The slids were stained with 3,3’-diaminobenzidine (DAB) and counterstained with hematoxylin dye. IHC, immunohistochemical.
The clinical characteristics of 66 patients are summarized in Table 1. In terms of gender of the patients, 24 out of 26 female patients developed REV-ERBα downregulation. The downregulation of REV-ERBα was more common in patients younger than 50 years old. Besides, T stage was significantly associated with REV-ERBα downregulation, as almost all the patients under T3 and T4 displayed REV-ERBα downregulation in their tumor tissues (31 out of 33). Furthermore, patients with distant metastasis had all developed REV-ERBα downregulation. Patients with epidermal growth factor receptor (EGFR) mutations were more likely to have REV-ERBα downregulation in their tumor tissues. Taken together, these results indicated that REV-ERBα downregulation was significantly associated with the progression and malignancy of LUAD, especially the properties that related to cancer cell proliferation and migration.
Table 1. Clinical characteristics of 66 patients with lung adenocarcinoma.
| Variable | Total cases | REV-ERBα downregulation in LUAD (%) |
|---|---|---|
| Gender | ||
| Male | 40 | 30 (75.0) |
| Female | 26 | 24 (92.3) |
| T stage | ||
| Tx | 1 | 0 (0.0) |
| T1 | 12 | 7 (58.3) |
| T2 | 20 | 16 (80.0) |
| T3 | 31 | 29 (93.5) |
| T4 | 2 | 2 (100.0) |
| Age | ||
| ≤50 | 16 | 14 (87.5) |
| >50 | 50 | 40 (80.0) |
| M stage | ||
| Mx | 3 | 1 (33.3) |
| M0 | 58 | 48 (82.8) |
| M1 | 5 | 5 (100.0) |
| EGFR mutation | ||
| No | 30 | 22 (73.3) |
| Yes | 36 | 32 (88.9) |
LUAD, lung adenocarcinoma; EGFR, epidermal growth factor receptor.
Downregulation of REV-ERBα is associated with progression of LUAD
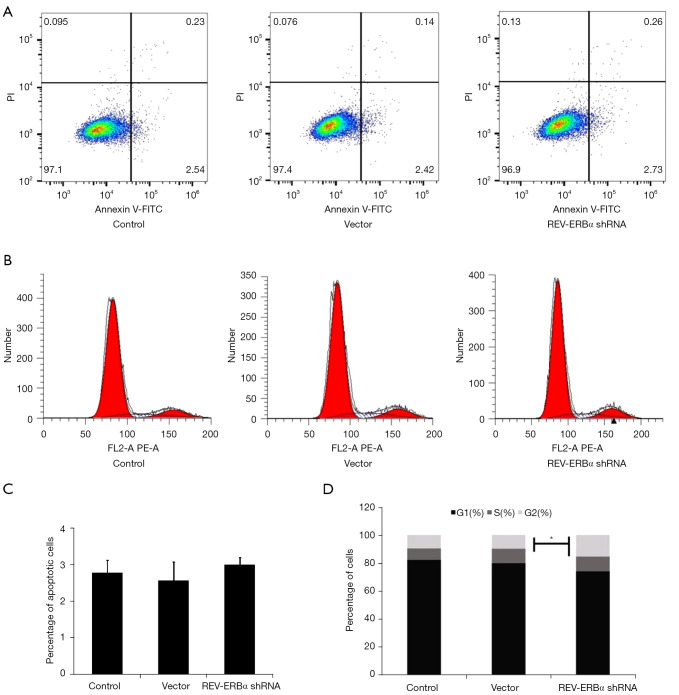
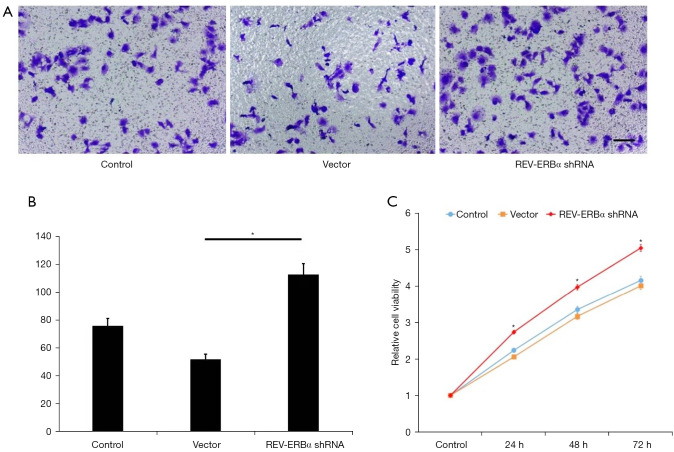
To study the functional roles of REV-ERBα in lung cancer progression, REV-ERBα was targeted by shRNA-mediated downregulation in the LUAD A549 cell line. We designed 4 shRNA and reverse transcription PCR (RT-PCR) was conducted to select the one with the best silencing efficacy. An shRNA sequence with no targeting effect served as the negative control for the experiments. We firstly evaluated whether downregulation of REV-ERBα influenced cancer cell survival. As shown in Figure 2 apoptotic cells achieved similar percentages according to the Annexin V-FITC sorting after the transfection of REV-ERBα shRNA, indicating downregulation of REV-ERBα may not affect cell death. However, cell cycle assay revealed that more cells were at the G2 phase (Figure 2B,2D), resulting in a more rapid growth rate of lung cancer cells examined by MTT assay (Figure 3). These results elucidated that downregulation of REV-ERBα can promote the proliferation of lung cancer cells.
Figure 2.
The effects of REV-ERBα downregulation on apoptosis and cell cycles. (A) Apoptosis of A549 cells with different treatments was assessed by Annexin V-FITC/PI staining flow cytometry assay. (B) Cell cycle assay of A549 cells under different treatment by PI staining flow cytometry. (C) Statistical results of percentage of apoptotic cell under the effect of REV-ERBα shRNA. (D) Statistical results of cell cycle distributions of A549 under the effect of REV-ERBα shRNA (*P<0.05). PI, propidium iodide; shRNA, short hairpin RNA.
Figure 3.
The effects of REV-ERBα downregulation on cell migration and proliferation. (A) Representative images of Transwell assay evaluating cell invasion ability under different treatments (scale bar, 50 µm). (B) Statistical results of invaded cells in 5 observation fields in each sample (*P<0.05). (C) A549 cell growth curve under different treatments measured by MTT assay (*P<0.05). 0.5% crystal violet solution was used to stain the cells in the upper chamber. MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide.
Furthermore, we evaluated changes in cell migration ability after downregulating REV-ERBα. The results showed that migration was significantly increased by downregulation of REV-ERBα compared to the control groups (Figure 3A,3B).
Exogenous disruption of REV-ERBα contributes to upregulation of NF-κB
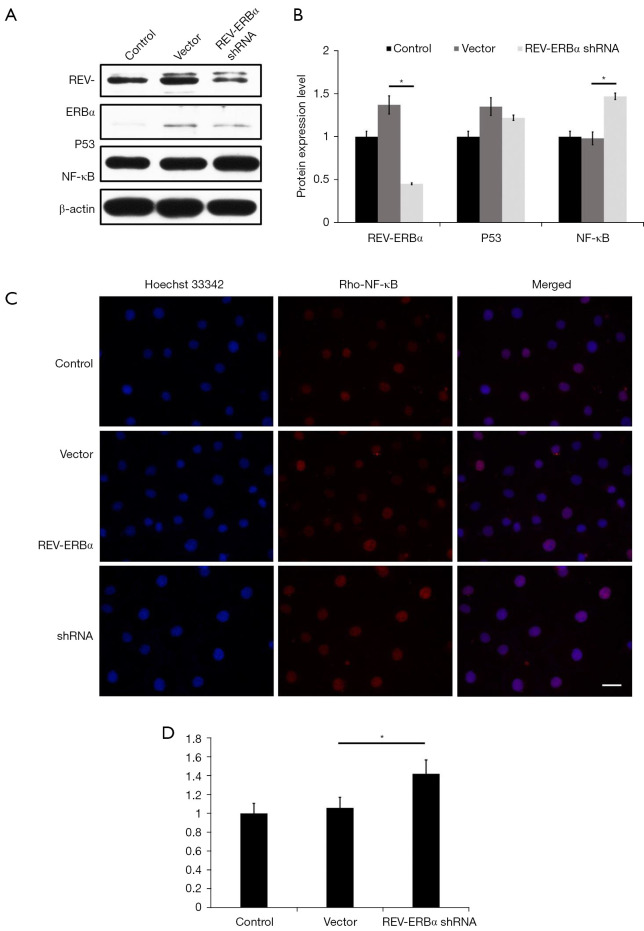
To further study the role of circadian component in LUAD, we monitored the changes in the expression levels of cancer driver gene p53 and NF-κB. Compared with the control group and the vector group, the expression level of NF-κB increased significantly (P<0.05), and the expression of p53 gene had no significant difference (P>0.05) (Figure 4A,4B). Immunostaining results also revealed an enhancement in NF-κB expression as shown in Figure 4C,4D. These results elucidated that downregulation of REV-ERBα can lead to the increase of NF-κB expression levels in LUAD, which indicates a potential molecular pathway that downregulation of REV-ERBα can promote lung cancer progression through NF-κB related pathways.
Figure 4.
Inhibition of REV-ERBα can elevate the expression of NF-κB in lung cancer cells. (A) Western blot analysis showed the expression of REV-ERBα, p53, and NF-κB under different treatments. (B) Statistics of western blot analysis of the expression level of REV-ERBα, p53, and NF-κB (*P<0.05). (C) Representative images of immunostaining analysis on the effects of downregulation of REV-ERBα on NF-κB (scale bar, 25 µm). (D) Statistical results of immunostaining analysis on the effects of downregulation of REV-ERBα on NF-κB (*P<0.05). Cell nuclei were stained with Hoechst 33342 showing a blue fluorescence. NF-κB, nuclear factor-κB.
Discussion
As one of the key components of the circadian clock, the nuclear hormone receptor REV-ERBα coordinates a variety of cellular processes including cell proliferation, metabolism, and inflammation. Several studies have indicated that disrupting circadian rhythm elevates the risk of cancer (9-11,16), suggesting that upregulation or pharmacologic activation of REV-ERBα may serve as a promising anticancer therapeutic method. Besides, research findings have also shown that maintenance or upregulation of REV-ERBα is beneficial to many physiological processes, including energy balance, anti-inflammation, bone metabolism, and chemoresistance of cancer (21-24).
In our study, we have shown that REV-ERBα was downregulated in human lung cancer tissues, which was closely associated with T stage, M stage, and some other clinical characteristics of patients with LUAD. Taken together with the analysis of clinical characteristics of patients with LUAD, we can conclude that the abnormal expression of REV-ERBα is relevant to the malignancy of lung cancer. The prognosis of these patients should be considered and traced to further show the link between circadian components and survival of cancer patients. We also found that downregulation of REV-ERBα can promote the migration and proliferation of LUAD cells, simultaneously elevating the expression level of NF-κB. The illustration of pathway overview is shown in Figure 5. These findings can provide potential therapeutic methods by upregulating or pharmacologically activating REV-ERBα to inhibit the proliferation of lung cancer cells.
Figure 5.
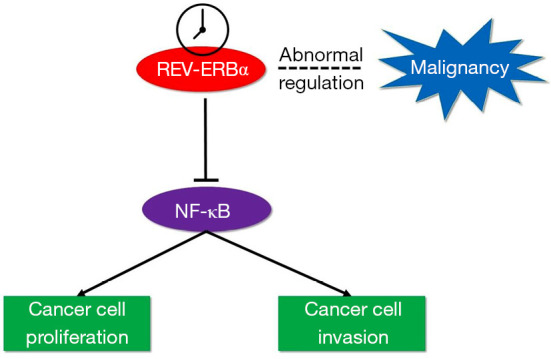
Illustration of molecular pathway of REV-ERBα’s association with cancer progression. Abnormal regulation of REV-ERBα is relevant to cancer malignancy. Downregulation of REV-ERBα can promote cancer cell proliferation and invasion as NF-κB can be upregulated when REV-ERBα was inhibited. NF-κB, nuclear factor-κB.
Based on our previous study, progression of lung adenocarcinoma is associated with downregulation or inhibition of REV-ERBα. Future work will be focused on molecular signaling pathways regarding circadian regulation of LUAD. It is needed to study the signal pathway by pathway inhibitor. The agonist of REV-ERBα may also be used to up-regulate its expression. we will next explore whether it will inhibit NF-kB signaling pathway, so as to provide a new perspective for clinical application in this field. Due to the fact that the therapeutic effect and long-term survival rate of LUAD are not satisfactory, there is an urgent need to innovate the treatment strategy. Taking the circadian rhythm mechanism as the research direction, the multidisciplinary individualized comprehensive therapy which integrates conventional therapy, radiotherapy, chemotherapy, and molecular targeted therapy will play an important role in the field of lung cancer treatment. Besides, if the upregulation of REV-ERBα is detected in samples from patients with LUAD, it should be considered to reduce the frequency and extend the interval of reexamination for the patients after receiving proper therapy.
Conclusions
In summary, our findings suggest that tumorigenesis and progression of lung carcinoma is associated with downregulation or inhibition of REV-ERBα. This pathophysiological process also correlates with regulation of the NF-κB signaling pathway, indicating that REV-ERBα is a potential target of lung cancer therapy. Further investigation at molecular levels should place emphasis on the organelles, such as the signaling pathways underlying the stimulation of mitochondria which leads to apoptosis
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This work was supported by grants from Scientific Research Project of Tianjin Chest Hospital (2018XKZ26); the project of Tianjin Science and Technology Innovation Bureau (20JCYBJC01350); the Project of Tianjin Health Commission (ZD20023); the Project of Tianjin key Clinical Subject.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013). The study was approved by Tianjin Chest Hospital Academic Ethics Committee (No. 2019LW-012). Informed consent was taken from all the patients.
Footnotes
Reporting Checklist: The authors have completed the MDAR checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6405/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6405/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6405/coif). The authors report that this work was supported by grants from Scientific Research Project of Tianjin Chest Hospital (2018XKZ26); the Project of Tianjin Science and Technology Innovation Bureau (20JCYBJC01350); the Project of Tianjin Health Commission (ZD20023); and the Project of Tianjin key Clinical Subject. The authors have no other conflicts of interest to declare.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin 2020;70:7-30. 10.3322/caac.21590 [DOI] [PubMed] [Google Scholar]
- 2.Blandin Knight S, Crosbie PA, Balata H, et al. Progress and prospects of early detection in lung cancer. Open Biol 2017;7:170070. 10.1098/rsob.170070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yang CF, Chan DY, Speicher PJ, et al. Role of Adjuvant Therapy in a Population-Based Cohort of Patients With Early-Stage Small-Cell Lung Cancer. J Clin Oncol 2016;34:1057-64. 10.1200/JCO.2015.63.8171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brambilla E. Lung adenocarcinoma expression profile: one more layer of heterogeneity. Eur Respir J 2013;42:1180-2. 10.1183/09031936.00087213 [DOI] [PubMed] [Google Scholar]
- 5.Tomonaga N, Nakamura Y, Yamaguchi H, et al. Analysis of intratumor heterogeneity of EGFR mutations in mixed type lung adenocarcinoma. Clin Lung Cancer 2013;14:521-6. 10.1016/j.cllc.2013.04.005 [DOI] [PubMed] [Google Scholar]
- 6.Bria E, Pilotto S, Amato E, et al. Molecular heterogeneity assessment by next-generation sequencing and response to gefitinib of EGFR mutant advanced lung adenocarcinoma. Oncotarget 2015;6:12783-95. 10.18632/oncotarget.3727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dierickx P, Emmett MJ, Jiang C, et al. SR9009 has REV-ERB-independent effects on cell proliferation and metabolism. Proc Natl Acad Sci U S A 2019;116:12147-52. 10.1073/pnas.1904226116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Puram RV, Kowalczyk MS, de Boer CG, et al. Core Circadian Clock Genes Regulate Leukemia Stem Cells in AML. Cell 2016;165:303-16. 10.1016/j.cell.2016.03.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sulli G, Rommel A, Wang X, et al. Pharmacological activation of REV-ERBs is lethal in cancer and oncogene-induced senescence. Nature 2018;553:351-5. 10.1038/nature25170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wagner PM, Monjes NM, Guido ME. Chemotherapeutic Effect of SR9009, a REV-ERB Agonist, on the Human Glioblastoma T98G Cells. ASN Neuro 2019;11:1759091419892713. 10.1177/1759091419892713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tao L, Yu H, Liang R, et al. Rev-erbα inhibits proliferation by reducing glycolytic flux and pentose phosphate pathway in human gastric cancer cells. Oncogenesis 2019;8:57. 10.1038/s41389-019-0168-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dong Z, Zhang G, Qu M, et al. Targeting Glioblastoma Stem Cells through Disruption of the Circadian Clock. Cancer Discov 2019;9:1556-73. 10.1158/2159-8290.CD-19-0215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fu L, Lee CC. The circadian clock: pacemaker and tumour suppressor. Nat Rev Cancer 2003;3:350-61. 10.1038/nrc1072 [DOI] [PubMed] [Google Scholar]
- 14.Bass J, Lazar MA. Circadian time signatures of fitness and disease. Science 2016;354:994-9. 10.1126/science.aah4965 [DOI] [PubMed] [Google Scholar]
- 15.Zhang Y, Fang B, Emmett MJ, et al. GENE REGULATION. Discrete functions of nuclear receptor Rev-erbα couple metabolism to the clock. Science 2015;348:1488-92. 10.1126/science.aab3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yu EA, Weaver DR. Disrupting the circadian clock: gene-specific effects on aging, cancer, and other phenotypes. Aging (Albany NY) 2011;3:479-93. 10.18632/aging.100323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Vieira E, Marroquí L, Batista TM, et al. The clock gene Rev-erbα regulates pancreatic β-cell function: modulation by leptin and high-fat diet. Endocrinology 2012;153:592-601. 10.1210/en.2011-1595 [DOI] [PubMed] [Google Scholar]
- 18.Cho H, Zhao X, Hatori M, et al. Regulation of circadian behaviour and metabolism by REV-ERB-α and REV-ERB-β. Nature 2012;485:123-7. 10.1038/nature11048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Solt LA, Wang Y, Banerjee S, et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 2012;485:62-8. 10.1038/nature11030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Woldt E, Sebti Y, Solt LA, et al. Rev-erb-α modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med 2013;19:1039-46. 10.1038/nm.3213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Borba TKF, Toscano AE, Costa de Santana BJR, et al. Central administration of REV-ERBα agonist promotes opposite responses on energy balance in fasted and fed states. J Neuroendocrinol 2020;32:e12833. 10.1111/jne.12833 [DOI] [PubMed] [Google Scholar]
- 22.Yue J, He J, Wei Y, et al. Decreased expression of Rev-Erbα in the epileptic foci of temporal lobe epilepsy and activation of Rev-Erbα have anti-inflammatory and neuroprotective effects in the pilocarpine model. J Neuroinflammation 2020;17:43. 10.1186/s12974-020-1718-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kim K, Kim JH, Kim I, et al. Rev-erbα Negatively Regulates Osteoclast and Osteoblast Differentiation through p38 MAPK Signaling Pathway. Mol Cells 2020;43:34-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Teixeira AAS, Biondo LA, Silveira LS, et al. Doxorubicin modulated clock genes and cytokines in macrophages extracted from tumor-bearing mice. Cancer Biol Ther 2020;21:344-53. 10.1080/15384047.2019.1702400 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as