Abstract
Background
The YTH domain family protein 3 (YTHDF3) is an important N6-methyladenosine (m6A) reader which is involved in multiple cancers. However, the biological role and mechanisms of action for YTHDF3 in triple-negative breast cancer (TNBC) remains to be elucidated.
Methods
The expression of YTHDF3 in TNBC tissues was evaluated using The Cancer Genome Atlas (TCGA) database, BC-GenExMiner, and immunohistochemistry (IHC) staining. Cell migration, invasion, and epithelial-mesenchymal transition (EMT) were validated by wound healing assays, transwell assays, and Western blot (WB) analyses. The association between YTHDF3 and zinc finger E-box-binding homeobox 1 (ZEB1) was confirmed by Pearson correlation analysis. RNA-binding protein immunoprecipitation (RIP) assays and mRNA actinomycin stability analyses were applied to confirm whether YTHDF3 could interact with ZEB1in an m6A-dependent manner.
Results
The expression of YTHDF3 was correlated with poorer disease-free survival (DFS) and overall survival (OS) in TNBC patients. Functional experiments indicated that YTHDF3 positively regulated cell migration, invasion, and EMT in TNBC cells. Moreover, ZEB1 was identified as a key downstream target for YTHDF3 and YTHDF3 could enhance ZEB1 mRNA stability in an m6A-dependent manner. Inhibition of YTHDF3 reduced migration, invasion, and EMT, all of which were reversed by rescue experiments overexpressing ZEB1.
Conclusions
The findings herein confirmed that the YTHDF3/ZEB1 axis plays an important role in the progression and metastasis of TNBC. YTHDF3 is a promising prognosis biomarker and potential therapeutic target for patients with TNBC.
Keywords: Triple-negative breast cancer (TNBC), metastasis, epithelial-mesenchymal transition (EMT), YTH domain family 3 (YTHDF3), zinc finger E-box-binding homeobox 1 (ZEB1)
Introduction
Breast cancer (BC) is now the most common malignant tumor for women worldwide (1). Triple-negative breast cancer (TNBC) is one of the most aggressive BC subtypes, characterized by the lack of estrogen receptor (ER), progesterone receptor (PR), and human epidermal growth factor receptor 2 (HER-2) expression (2). Although TNBC only accounts for about 15–20% of all BC cases, treatment response is generally poor, with high metastatic potential and unfavorable prognosis (3). To date, there is no effective targeted therapy for TNBC other than surgery and conventional chemotherapy (2,4). Therefore, novel prognostic biomarkers and molecular therapeutic targets for TNBC patients are urgently needed.
N6-methyladenosine (m6A) is the most abundant internal modification of RNA in eukaryotic cells and regulates almost all important biological processes by mRNA splicing, translation, and stability (5-8). There are three types of m6A regulators which includes m6A “writers” (m6A methyltransferases including METTL3, METTL14, and WTAP), “erasers” (m6A demethylases including FTO and ALKBH5), and “readers” (m6A binding proteins including YTH domain family protein 1–3, YTH domain containing 1–2 and HNRNPA2B1). Increasing evidence have indicated that the m6A regulators are involved in multiple cancers and play vital roles in tumor cell proliferation and metastasis (9,10). Several m6A regulators, such as METTL3, FTO and HNRNPA2B1, have been identified to promote the malignant progression and therapeutic response of BC (11-14). YTH domain family protein 3 (YTHDF3) is an important reader as it plays a significant role in modulating translation of m6A-modified mRNAs and affects methylated mRNA decay in an m6A methylation-dependent manner (15,16). YTHDF3 has been shown to contribute to the progression of several types of tumors, including gastric and colorectal cancer (17,18). In patients with BC, YTHDF3 has been associated with brain metastasis and poor prognosis (19,20). However, to date, there have been no studies specifically describing the function of YTHDF3 in TNBC. Our previous proteomic study indicated that YTHDF3 was highly expressed in TNBC tumor tissues with more advanced grade (21). Therefore, this study was conducted to validate the expression and function of YTHDF3 in TNBC.
The present study showed that YTHDF3 expression was closely associated with lymph node metastasis and higher histological grade in patients with TNBC. YTHDF3 was also indicated to be an independent prognostic biomarker for TNBC patients and knockdown of YTHDF3 inhibited the migration, invasion, and epithelial-mesenchymal transition (EMT) in TNBC cells. Furthermore, the EMT transcriptional factor zinc finger E-box-binding homeobox 1 (ZEB1) was identified as a key target of YTHDF3 and YTHDF3 could enhance ZEB1 mRNA stability in an m6A-dependent manner. Taken together, these findings offer insights into novel therapeutic targets in patients with TNBC.
We present the following article in accordance with the MDAR reporting checklist (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6857/rc).
Methods
Gene expression profiling analysis
The microarray datasets of TNBC patients were extracted from The Cancer Genome Atlas (TCGA) database (https://portal.gdc.cancer.gov/). BC-GenExMiner (http://bcgenex.ico.unicancer.fr/) is a statistical mining tool with published annotated BC transcriptomic data (22). The prognostic module was used to analyze the prognostic effects of YTHDF3 in TNBC.
TNBC tumor samples
A total of 224 TNBC tissue specimens were selected from female patients (aged 27–72 years) undergoing surgery in the Fujian Medical University Union Hospital between June 2013 and June 2018. All patients received radical surgery and had at least six cycles of adjuvant chemotherapy. Disease-free survival (DFS) was calculated as the time from the date of diagnosis to the date of clinical relapse (with histopathology confirmation or radiological evidence of tumor recurrence). Overall survival (OS) was calculated as the time from the date of diagnosis until death from any cause. The last follow-up time was July 1, 2021. All patients agreed to the use of their specimens and signed the informed consent forms. This study was approved by the Research Ethics Committee of Fujian Medical University Union Hospital (No. 2021KJCX039). All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Cell culture and transfection
The human TNBC cell lines MDA-MB-231, BT-549, HCC1937, and Hs578T were purchased from the Cell Bank of Type Culture Collection of The Chinese Academy of Sciences. Cell were cultured in Dulbecco's Modified Eagle Medium (DMEM; HyClone, Logan, USA), supplemented with 10% fetal bovine serum (FBS; Gibco, Carlsbad, USA) and 1% penicillin and streptomycin solution and incubated at 37 °C and 5% CO2. The short hairpin RNA (shRNA) targeting YTHDF3 was subcloned into the GV493 lentiviral shRNA vector (Genechem, Shanghai, China). For overexpression of ZEB1, the construct was generated by subcloning PCR amplified full-length human ZEB1 cDNA into the vector. The constructed lentiviral vectors were packaged into viruses in 293 T cells which were then harvested and the concentrated viruses were added to TNBC cells and cultured for 72 hours. Gene sequences are provided in Table S1.
Quantitative real time-polymerase chain reaction (qRT-PCR)
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, USA) and the complementary DNA was synthesized by PrimeScript RT Master Mix (Takara, Dalian, China) according to the manufacturer’s instructions. The SYBR Green kit (Takara, Dalian, China) was used for qRT-PCR (model 7500 Real-Time PCR System; Applied Biosystems) according to the manufacturer’s protocol. The glyceraldehyde 3-phosphate dehydrogenase (GAPDH) gene was used for normalization of data. The fold change in gene expression was calculated using the 2−ΔΔct method with three independent replicates of all biological samples. The primer sequences are listed in Table S1.
Wound healing migration assay
Cells were seeded onto 96-well plates and cultured overnight. A line wound was made by scraping a 10 μL pipette tip across the confluent cell layer. The floating cells were washed three times with phosphate buffered saline (PBS) and the serum-free medium was applied to maintain the cells. Cell images were captured at 0 and 24 hours with an inverted light microscope.
Transwell invasion assays
Cells were washed with PBS and suspended in DMEM without FBS at 1×105 cells/mL. The upper chamber of the Transwell (Corning, New York, USA) was precoated with Matrigel (BD Biosciences, San Diego, USA) and filled with 100 µL of cell suspension. The lower chamber was filled with 600 µL DMEM supplemented with 30% FBS. After incubation for 24 hours at 37 °C, cells were fixed with 4% paraformaldehyde for 30 minute and stained with 0.5% crystal violet for 5 minutes at room temperature. The chamber was then washed with PBS solution and the cells on the surface of the chamber were wiped off with cotton swabs. The images of the stained cells on the lower side were captured (×200 magnification).
WB analysis
Total protein was extracted using RIPA lysis buffer (Beyotime, Shanghai, China) and the concentration was determined with the BCA Protein Assay Kits (Beyotime, Shanghai, China). A total of 20 µg of protein was loaded for sodium dodecyl sulphate polyacrylamide gel electrophoresis (SDS-PAGE). Samples were transferred to polyvinylidene fluoride (PVDF) membranes and incubated with the following primary antibodies: anti-YTHDF3 (ab220161, Abcam, Cambridge, UK), anti-ZEB1 (21544-1-AP, Proteintech, Wuhan, China), anti-E-cadherin (20874-1-AP, Proteintech, Wuhan, China), anti-N-cadherin (22018-1-AP, Proteintech, Wuhan, China), anti-vimentin (10366-1-AP, Proteintech, Wuhan, China), and anti-GAPDH (ab181602, Abcam, Cambridge, UK). Membranes were then incubated with a secondary horseradish peroxidase-conjugated antibody and visualized with the ECL reagent (Thermo Fisher Scientific, Waltham, USA).
Immunohistochemistry (IHC) staining
IHC staining analysis was performed on paraffin-embedded tissues to assess the protein expression of YTHDF3 in TNBC tissues. Slides were incubated with anti-YTHDF3 (1:800; ab220161, Abcam, Cambridge, UK) and anti-ZEB1 (1:800, 21544-1-AP, Proteintech, Wuhan, China) antibody according to the standard procedure. The IHC staining scores of YTHDF3 were evaluated by 2 independent pathologists blinded to the corresponding patients. The percentage of stained positive cells was scored from 1 to 4 as followed: 1 point for 0–25% of cells stained; 2 points for 26–50% of cells stained; 3 points for 51–75% of cells stained; and 4 points for 75–100% of cells stained. The staining intensity score was calculated from 0 to 3 as follows: 0 points indicates no staining; 1 point indicates weak staining; 2 points indicates moderate staining; and 3 points indicates strong staining. The percentage of positive tumor cells and the staining intensity were multiplied to produce a weighted score for each patient. A score of 8–12 was defined as high expression level and a score of 0–7 was defined as low expression.
RNA-binding protein immunoprecipitation (RIP) assay
The RIP assay was conducted with Magna RIP Kits (Millipore, Billerica, USA) according to the manufacturer’s instructions. The antibodies used in this study were anti-YTHDF3 (ab220161, Abcam, Cambridge, UK), anti-m6a (202003, Synaptic Systems, Goettingen, Germany), and normal IgG. The RNA complexes were extracted by proteinase K and phenol/chloroform/isoamyl alcohol, and amplified by qRT-PCR.
RNA stability assay
To detect the ZEB1 mRNA stability, the cells were treated with actinomycin (6 mg/mL) for 0, 2, 4, 6, and 8 hours. The relative RNA level was measured by qRT-PCR.
Statistical analysis
Each experiment was repeated 3 times and presented as the means ± standard deviation (SD) in this study. The student’s t-test was used to compare variables between two groups. The Chi-square test was used to examine the clinicopathological characteristics between patients with high and low YTHDF3 expression. The correlation between the expression levels of YTHDF3 and ZEB1 was evaluated by Spearman rank correlation coefficients. Survival curves were plotted by the Kaplan-Meier method and analyzed by log-rank tests. Cox proportional hazard regression model was performed for univariate and multivariate survival analysis. A two-sided P value of less than 0.05 was considered statistically significant. All statistical analyses were performed using the SPSS 20.0 software (IBM, USA) and GraphPad Prism 7.0 software (USA).
Results
YTHDF3 is correlated with clinicopathological features and predicts poor prognosis in TNBC patients
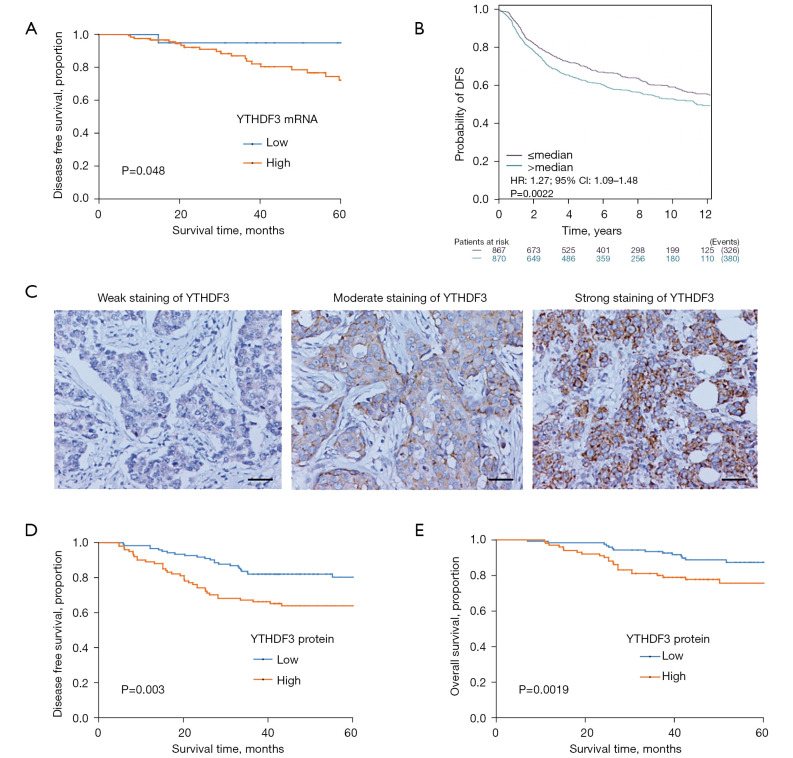
To detect YTHDF3 expression in TNBC tumor tissues, data mining was performed and the YTHDF3 mRNA profiles from the publicly available TCGA database and BC-GenExMiner were analyzed. The mRNA expression of YTHDF3 was shown to be associated with the DFS for patients with TNBC [hazard ratio (HR) =6.79, 95% confidence interval (CI): 1.13 to 7.01, P=0.048; and HR =1.27, 95% CI: 1.09 to 1.48; Figure 1A,1B]. Immunohistochemical (IHC) staining was conducted on the 224 TNBC patient specimens and the YTHDF3 protein expression level was scored based on the staining intensity and percentage of positive tumor cells (Figure 1C). Consistent with our previous proteomic study (16), high YTHDF3 expression was correlated with more advanced histological grade (P=0.014; Table 1). In addition, patients with elevated expression of YTHDF3 had a greater probability of lymph node metastasis (P=0.028; Table 1). Moreover, Kaplan-Meier survival analysis showed that high YTHDF3 expression exhibited a significantly poorer DFS and OS compared to those patients with low YTHDF3 expression (Figure 1D,1E). Multivariate Cox analysis revealed that YTHDF3 expression is an independent prognostic factor for both DFS and OS (HR=1.93, 95% CI: 1.14 to 3.28, P=0.015; and HR=1.99, 95% CI: 1.02 to 3.86, P=0.044, respectively; Table 2). In summary, these results indicated that high expression of YTHDF3 is a risk factor and YTHDF3 may be a potential prognostic biomarker for patients with TNBC.
Figure 1.
YTHDF3 expression is correlated with poor prognosis in TNBC patients. (A,B) Kaplan-Meier analysis of the association between DFS and YTHDF3 mRNA expression in TNBC patients using TCGA database and BC-GenExMiner. (C) Representative IHC images of weak, moderate, and strong staining of YTHDF3 in TNBC tumor tissues (×200 magnification, scale bar: 50 μm). (D,E) Kaplan-Meier analysis of the association between DFS and OS with YTHDF3 protein expression in TNBC patients using the IHC method. YTHDF3, the YTH domain family 3; TNBC, triple-negative breast cancer; DFS, disease-free survival; TCGA, The Cancer Genome Atlas; IHC, immunohistochemistry; OS, overall survival.
Table 1. Associations of YTHDF3 expression with clinicopathological characteristics for triple-negative breast cancer patients.
| Characteristics | All patients(n=224) | Low YTHDF3 (n=123) |
High YTHDF3 (n=101) |
P valuea | |||
|---|---|---|---|---|---|---|---|
| No. | (%) | No. | No. | ||||
| Age at diagnosis(years) | 0.570 | ||||||
| ≤50 | 104 | 46.4 | 55 | 49 | |||
| >50 | 120 | 53.6 | 68 | 52 | |||
| Tumor size | 0.706 | ||||||
| ≤2 cm | 94 | 42.0 | 53 | 41 | |||
| >2 cm | 130 | 58.0 | 70 | 60 | |||
| Lymph node metastasis | 0.028 | ||||||
| No | 131 | 58.5 | 80 | 51 | |||
| Yes | 93 | 41.5 | 43 | 50 | |||
| Tumor Grade | 0.014 | ||||||
| I + II | 84 | 37.5 | 55 | 29 | |||
| III | 140 | 62.5 | 68 | 72 | |||
| Lymphovascular invasion | 0.121 | ||||||
| No | 141 | 62.9 | 83 | 58 | |||
| Yes | 83 | 37.1 | 40 | 43 | |||
a, the P value was calculated among all groups by the Chi-square test. YTHDF3, the YTH domain family 3.
Table 2. Univariate and multivariate cox proportional hazard model for DFS and OS in TNBC patients.
| Variables | Univariate analysis | Multivariate analysis | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| DFS | OS | DFS | OS | ||||||||||
| HR (95% CI) | Pa | HR (95% CI) | Pa | HR (95% CI) | Pa | HR (95% CI) | Pa | ||||||
| Age (years) | |||||||||||||
| ≤50 | Reference | Reference | Reference | ||||||||||
| >50 | 0.56 (0.34–0.95) | 0.031 | 0.81 (0.43–1.55) | 0.526 | 0.76 (0.44–1.30) | 0.308 | |||||||
| Tumor size | |||||||||||||
| ≤2 cm | Reference | Reference | Reference | Reference | |||||||||
| >2 cm | 1.97 (1.12–3.46) | 0.019 | 2.09 (1.01–4.32) | 0.046 | 2.08 (1.17–3.71) | 0.013 | 2.07 (1.01–4.27) | 0.049 | |||||
| Lymph nodes metastasis | |||||||||||||
| No | Reference | Reference | Reference | Reference | |||||||||
| Yes | 2.25 (1.34–3.79) | 0.002 | 2.46 (1.27–4.78) | 0.008 | 1.85 (1.08–3.17) | 0.026 | 2.24 (1.15–4.36) | 0.018 | |||||
| Grade | |||||||||||||
| I + II | Reference | Reference | Reference | ||||||||||
| III | 1.94 (1.08–3.49) | 0.026 | 1.99 (0.94–4.22) | 0.072 | 1.83 (1.01–3.31) | 0.047 | |||||||
| Lymphovascular invasion | |||||||||||||
| No | Reference | Reference | Reference | ||||||||||
| Yes | 1.73(1.04–2.88) | 0.036 | 1.76 (0.92–3.36) | 0.086 | 1.59 (0.94–2.70) | 0.084 | |||||||
| YTHDF3 expression | |||||||||||||
| Low | Reference | Reference | Reference | Reference | |||||||||
| High | 2.22 (1.31–3.74) | 0.003 | 2.21 (1.14–4.30) | 0.019 | 1.93 (1.14–3.28) | 0.015 | 1.99 (1.02–3.86) | 0.044 | |||||
a, the P value was adjusted by the univariate Cox proportional hazard regression model. DFS, disease free survival; OS, overall survival; TNBC, triple-negative breast cancer; HR, hazard ratio; CI, confidence interval; YTHDF3, the YTH domain family 3.
Suppressing YTHDF3 inhibits migration, invasion, and endothelial-mesenchymal transition in TNBC cells
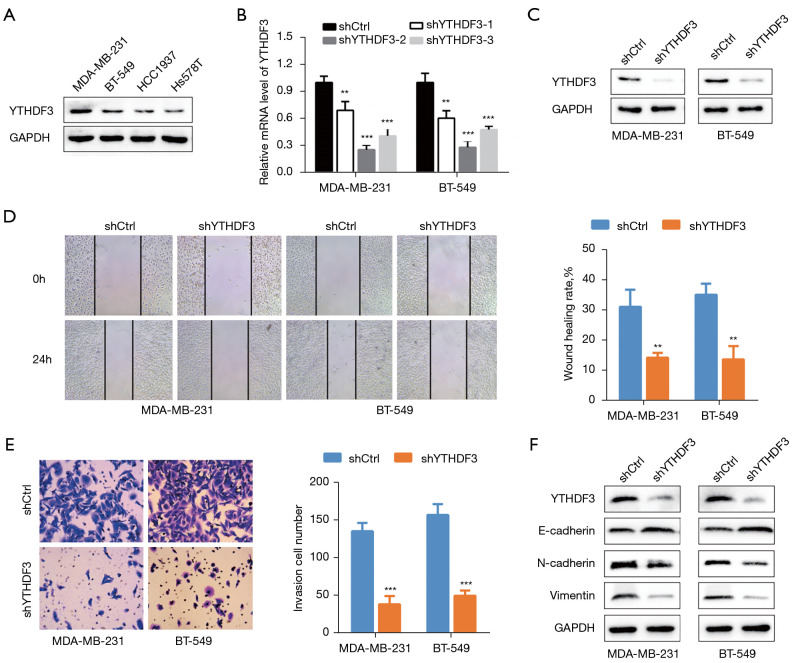
The protein expression of YTHDF3 was assessed in 4 TNBC cell lines (Figure 2A). The MDA-MB-231 and BT-549 cell lines showed higher YTHDF3 expression and were selected for further studies. Three shRNAs were designed and synthesized to suppress YTHDF3 expression in these cell lines (Figure 2B). The shRNAs that exhibited higher interference efficiency was validated by WB and chosen for subsequent experiments (Figure 2C). The wound healing assay demonstrated that YTHDF3 inhibition significantly reduced the migration potential of both cell lines (Figure 2D). In the transwell assays, a notably decrease in invasion ability was observed when YTHDF3 expression was knocked down (Figure 2E). In addition, WB ting was performed to assess the expression levels of several EMT markers related to cell migration and invasion. Knockdown of YTHDF3 upregulated the expression of E-cadherin and downregulated the expression of N-cadherin and vimentin compared with the shRNA negative control group (shCtrl) (Figure 2F). Taken together, these findings demonstrated that YTHDF3 promotes the migration, invasion, and EMT ability in TNBC cells.
Figure 2.
Knockdown of YTHDF3 suppresses cell migration, invasion, and EMT in MDA-MB-231 and BT-549 TNBC cells. (A) The protein expression of YTHDF3 in the MDA-MB-231, BT-549, HCC1937, and HS578T TNBC cell lines was detected by Western blot. (B,C) The knockdown efficiency of shYTHDF3 was analyzed by qRT-PCR and Western blot in MDA-MB-231 and BT-549 cells. (D,E) Cell migration and invasion ability were measured using the wound healing assay and transwell assay with YTHDF3 inhibition in MDA-MB-231 and BT-549 cells (transwell assay with crystal violet staining, ×200 magnification). (F) The protein levels of the EMT markers (E-cadherin, N-cadherin, and vimentin) were detected by Western blot with YTHDF3 knockdown in MDA-MB-231 and BT-549 cells. **, P<0.01; ***, P<0.001. YTHDF3, the YTH domain family 3; GAPDH, glyceraldehyde 3-phosphate dehydrogenase; EMT, endothelial mesenchymal transition; TNBC, triple-negative breast cancer; qRT-PCR, quantitative real time-polymerase chain reaction.
YTHDF3 regulates ZEB1 expression in TNBC
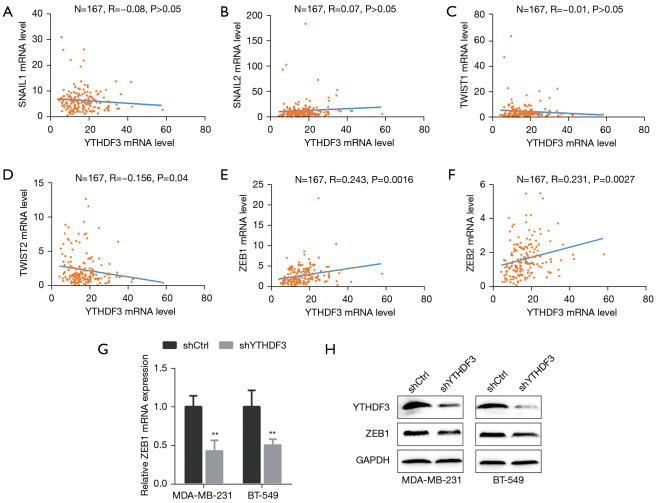
EMT is regulated by a complicated network. It is well known that several EMT transcriptional factors (EMT-TFs), namely SNAIL, TWIST, and members of the ZEB family, play vital roles in metastasis and drug resistance in cancer (23-27). To explore the underlying mechanisms of YTHDF3 in TNBC, the correlation between the mRNA expression levels of YTHDF3 and EMT-TFs in TNBC samples was examined using the TCGA database (Figure 3A-F). TWIST2, ZEB1, and ZEB2 were found to be correlated with YTHDF3 (r=–0.156, P=0.004; r=0.243, P=0.0016; and r=0.231, P=0.0027, respectively). Subsequently, YTHDF3 was stably knocked down in the MDA-MB-231 and BT-549 cell lines and the expression of TWIST2, ZEB1, and ZEB2 was examined. Indeed, knockdown of YTHDF3 markedly decreased ZEB1 mRNA and protein expression in both cell lines (Figure 3G,H). Therefore, ZEB1 was identified as a downstream target for YTHDF3 in TNBC.
Figure 3.
ZEB1 is a key downstream target for YTHDF3 in triple-negative breast cancer. (A-F) The correlation between YTHDF3 and EMT-TFs, SNAIL1, SNAIL2, TWIST1, TWIST2, ZEB1, and ZEB2, was evaluated using Spearman’s correlation analysis based on TCGA database. (G) The mRNA expression of ZEB1 was identified by qRT-PCR after YTHDF3 knockdown in MDA-MB-231 and BT-549 cells. (H) The protein expression of ZEB1 was identified by Western blot after YTHDF3 knockdown in MDA-MB-231 and BT-549 cells. **, P<0.01. YTHDF3, the YTH domain family 3; SNAIL1/2, snail family transcriptional repressor 1/2; TWIST1/2, twist family transcription factor 1/2; ZEB1/2, zinc finger E-box-binding homeobox 1/2; EMT-TF, endothelial mesenchymal transition transcription factor. TCGA, The Cancer Genome Atlas; qRT-PCR, quantitative real time-polymerase chain reaction.
YTHDF3 enhances ZEB1 mRNA stability by m6A modification
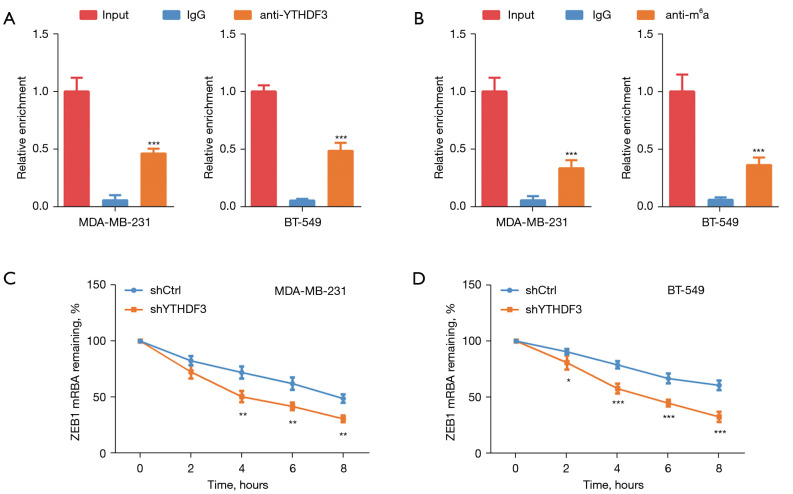
Since YTHDF3 is an important m6A reader, we investigated whether YTHDF3 could interact with ZEB1 mRNA in an m6A-dependent manner. The results of the RIP assay revealed that YTHDF3 could bind to ZEB1 mRNA in MDA-MB-231 and BT-549 TNBC cells (Figure 4A). In addition, the ZEB1 mRNA in TNBC was modified with a m6A modification (Figure 4B). The mRNA actinomycin stability analysis showed that YTHDF3 inhibition impaired the stability of the ZEB1 mRNA in both MDA-MB-231 and BT-549 cells (Figure 4C,4D). The above data confirmed that YTHDF3 could enhance ZEB1 mRNA stability in an m6A-dependent manner.
Figure 4.
YTHDF3 enhances ZEB1 mRNA stability in an m6A-dependent manner. (A) RNA-binding protein immunoprecipitation (RIP) assay confirmed that YTHDF3 can bind to ZEB1 mRNA in MDA-MB-231 and BT-549 cells. (B) RIP assay validated that ZEB1 mRNA was modified with m6A modification in MDA-MB-231 and BT-549 cells. (C,D) The mRNA stability analysis verified that ZEB1 mRNA was drastically decreased after YTHDF3 knockdown in MDA-MB-231 and BT-549 cells. *, P<0.05; **, P<0.01; ***, P<0.001. YTHDF3, the YTH domain family 3; ZEB1, zinc finger E-box-binding homeobox 1; m6A, N6-methyladenosine.
The YTHDF3/ZEB1 axis is involved in the progression of TNBC
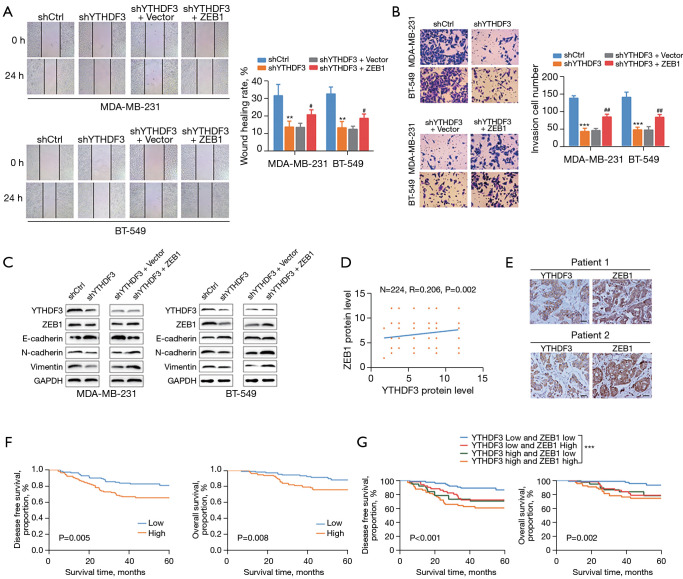
To further evaluate the effects of the YTHDF3/ZEB1 axis on TNBC, functional rescue experiments were conducted with YTHDF3 shRNA and ZEB1 overexpression vectors in TNBC cells. Wound healing and transwell assays revealed that the reduced migration and invasion ability induced by YTHDF3 inhibition could be recovered by ZEB1 overexpression (Figure 5A,5B). Moreover, the overexpression of ZEB1 also caused the downregulation of E-cadherin, as well as the upregulation of N-cadherin and vimentin in YTHDF3 knockdown cells (Figure 5C). IHC staining was performed to assess the relationship between ZEB1 and YTHDF3 through Spearman’s correlation analysis. As shown in Figure 5D,5E, the protein levels of ZEB1 were positively correlated with that of YTHDF3 in TNBC tumor tissues. Survival curve analysis also identified that patients with high ZEB1 expression were associated with poorer DFS and OS compared with patients with low ZEB1 expression (Figure 5F). Moreover, a significantly poorer prognosis was found in TNBC patients with high expression levels of both YTHDF3 and ZEB1 (Figure 5G). Collectively, these findings suggested that ZEB1 is crucial for YTHDF3-mediated TNBC migration, invasion, and EMT. Indeed, the YTHDF3/ZEB1 axis is important in the progression of TNBC.
Figure 5.
The YBX1/CTPS1 axis is involved in the progression of TNBC. (A,B) Cell migration and invasion ability were determined by wound healing and transwell assays after downregulation of YTHDF3 and overexpression of ZEB1 (transwell assay with crystal violet staining, ×200 magnification). (C) The protein levels of the EMT markers (E-cadherin, N-cadherin, and vimentin) after downregulation of YTHDF3 and overexpression of ZEB1were detected by Western blot. (D) The correlation between YTHDF3 and ZEB1 protein expression was evaluated by Spearman’s correlation analysis based on immunohistochemistry analysis. (E) Representative staining images of YTHDF3 and ZEB1 in TNBC tumor tissues (×200 magnification). (F) Kaplan-Meier analysis of the disease-free survival and overall survival with ZEB1 protein expression in TNBC patients. (G) Kaplan-Meier analysis of the disease-free survival and overall survival in TNBC patients expressing both YTHDF3 and ZEB1. Scale bar: 50 μm. **, P<0.01; ***, P<0.001 (shCtrl vs. shYTHDF3); and #, P<0.05, ##, P<0.01 (shYTHDF3 + Vector vs. shYTHDF3 + ZEB1). YTHDF3, the YTH domain family 3; ZEB1, zinc finger E-box-binding homeobox 1; TNBC, triple-negative breast cancer; EMT, epithelial-mesenchymal transition.
Discussion
TNBC is the most invasive subtype of BC with unfavorable outcomes. The lack of ER, PR, and HER2 expression renders TNBC patients inaccessible to endocrine or anti-HER2 target therapy. Currently, with no effective and clinically applicable therapeutic targets, the most common treatment strategy for TNBC is a combination of surgery and adjuvant/neoadjuvant chemotherapy (28-30). Therefore, identification of novel regulatory molecules and promising therapeutic targets is beneficial to developing new treatment strategies for patients with TNBC.
m6A is the most common post-transcriptional modification of RNA in eukaryotes, and has been identified to play key roles in various types of tumors (9,10). YTHDF3 is a crucial m6A reader and is associated with the progression of gastric and colorectal cancer (17,18). Previous studies have also demonstrated that YTHDF3 can induce brain metastasis in BC patients by enhancing the translation of m6A-enriched transcripts for ST6GALNAC5, gap junction alpha-1 protein (GJA1), and EGFR (19). However, there is a paucity of data regarding the function of YTHDF3 in TNBC. In this current study, the association of YTHDF3 expression with patient clinicopathological parameters and outcomes was evaluated using the TCGA database, the BC-GenExMiner tool, and IHC staining. The increased protein expression of YTHDF3 was closely related to more advanced histological grade and greater probability of lymph node metastasis. In addition, higher mRNA and protein expression of YTHDF3 was correlated with shortened survival for patients with TNBC. The above results collectively indicated that YTHDF3 may act as a promising biomarker for the prognosis of TNBC patients.
To further determine the function role of YTHDF3 in TNBC progression, a series of in vitro experiments were performed. Knockdown of YTHDF3 dramatically reduced the migration and invasion abilities of TNBC cells compared with the shRNA control group. It has been well demonstrated that EMT plays an important role in tumor progression and metastasis (31,32). The downregulation of epithelial markers such as E-cadherin and the upregulation of mesenchymal markers such as N-cadherin and vimentin are common hallmarks of EMT (24). Numerous studies have indicated that several m6A regulators, including METTL3, METTL14, FTO, YTHDF1, and HNRNPA2B1, can promote tumor metastasis with an EMT-feature (33-38). Our study also revealed that YTHDF3 plays a critical role in EMT for TNBC. The expression levels of E-cadherin were upregulated, while N-cadherin and vimentin expression were downregulated following the inhibition of YTHDF3. The correlation between YTHDF3 and EMT-TFs (SNAIL, TWIST, and ZEB families) was examined using TNBC samples in TCGA database. Interestingly, TWIST2, ZEB1, and ZEB2 were the 3 EMT-TFs found to be correlated with YTHDF3. Subsequent qRT-PCR and WB experiments confirmed that only ZEB1 mRNA and protein expression were markedly decreased in MDA-MB-231 and BT-549 cell lines. All these data confirmed that ZEB1 may be a downstream target for YTHDF3 in TNBC.
ZEB1 protein is a key transcriptional factor that promotes cellular plasticity and EMT in tumor cells (39,40). It has been reported to induce BC chemotherapy resistance and can transform non-invasive BC cells into highly malignant cancer stem cells (CSCs) (41-44). Since YTHDF3 is an important m6A reader and previous studies have demonstrated that it can enhance ZEB1 mRNA stability via an m6A-dependent manner in hepatocellular carcinoma (45), the YTDF3-ZEB1 interaction was further investigated using RIP assays. The relevant results indicated that YTHDF3 can bind to ZEB1 mRNA in TNBC cells and ZEB1 mRNA in TNBC can be modified with m6A modification. In addition, mRNA actinomycin stability analysis also demonstrated that inhibition of YTHDF3 can significantly impaired the stability of ZEB1 mRNA in MDA-MB-231 and BT-549 cells, further confirming that YTHDF3 can enhance ZEB1 mRNA stability in an m6A-dependent manner.
Functional rescue experiments were performed to verify the effects of the YTHDF3/ZEB1 axis on TNBC. The reduced migration and invasion ability induced by YTHDF3 inhibition could be recovered by ZEB1 overexpression. Moreover, the overexpression of ZEB1 also caused the downregulation of E-cadherin, and the upregulation of N-cadherin and vimentin in YTHDF3 knockdown cells. The correlations between YTHDF3 and ZEB1 in TNBC patients was assessed by IHC staining of surgical specimens. The protein expression of YTHDF3 was positively correlated with the expression of ZEB1. Patients with high ZEB1 expression endured a relatively poorer DFS and OS compared with patients with low ZEB1 expression. In addition, a significantly poorer survival was observed in patients with high expression levels of both YTHDF3 and ZEB1. Together, these results may represent a novel therapeutic target in the treatment of TNBC.
Conclusions
In summary, this investigation demonstrated that YTHDF3 is associated with poor prognosis for patients with TNBC and promotes the migration, invasion, and EMT of TNBC cells. ZEB1 was identified as a key downstream target for YTHDF3 and YTHDF3 can enhance ZEB1 mRNA stability in an m6A-dependent manner. Taken together, these findings suggest that YTHDF3 may be a critical prognostic factor and potential therapeutic target in TNBC.
Supplementary
The article’s supplementary files as
Acknowledgments
Funding: This study was supported by grants from the Joint Funds for the Innovation of Science and Technology, Fujian Province (2018Y9205, 2019Y9054, 2019Y9103, 2020Y9053) and the Natural Science Foundation of Fujian Province (2021J01737).
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study was approved by the Research Ethics Committee of Fujian Medical University Union Hospital (No. 2021KJCX039). All patients agreed to the use of their specimens and signed the informed consent forms. All procedures performed in this study involving human participants were in accordance with the Declaration of Helsinki (as revised in 2013).
Reporting Checklist: The authors have completed the MDAR reporting checklist. Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6857/rc
Data Sharing Statement: Available at https://atm.amegroups.com/article/view/10.21037/atm-21-6857/dss
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://atm.amegroups.com/article/view/10.21037/atm-21-6857/coif). The authors have no conflicts of interest to declare.
(English Language Editor: J. Teoh)
References
- 1.Sung H, Ferlay J, Siegel RL, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin 2021;71:209-49. 10.3322/caac.21660 [DOI] [PubMed] [Google Scholar]
- 2.Foulkes WD, Smith IE, Reis-Filho JS. Triple-negative breast cancer. N Engl J Med 2010;363:1938-48. 10.1056/NEJMra1001389 [DOI] [PubMed] [Google Scholar]
- 3.Zhao S, Zuo WJ, Shao ZM, et al. Molecular subtypes and precision treatment of triple-negative breast cancer. Ann Transl Med 2020;8:499. 10.21037/atm.2020.03.194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bianchini G, Balko JM, Mayer IA, et al. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol 2016;13:674-90. 10.1038/nrclinonc.2016.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roundtree IA, Evans ME, Pan T, et al. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017;169:1187-200. 10.1016/j.cell.2017.05.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin S, Choe J, Du P, et al. The m(6)A Methyltransferase METTL3 Promotes Translation in Human Cancer Cells. Mol Cell 2016;62:335-45. 10.1016/j.molcel.2016.03.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang X, Zhao BS, Roundtree IA, et al. N(6)-methyladenosine Modulates Messenger RNA Translation Efficiency. Cell 2015;161:1388-99. 10.1016/j.cell.2015.05.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang X, Lu Z, Gomez A, et al. N6-methyladenosine-dependent regulation of messenger RNA stability. Nature 2014;505:117-20. 10.1038/nature12730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lan Q, Liu PY, Haase J, et al. The Critical Role of RNA m6A Methylation in Cancer. Cancer Res 2019;79:1285-92. 10.1158/0008-5472.CAN-18-2965 [DOI] [PubMed] [Google Scholar]
- 10.Chen XY, Zhang J, Zhu JS. The role of m6A RNA methylation in human cancer. Mol Cancer 2019;18:103. 10.1186/s12943-019-1033-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie J, Ba J, Zhang M, et al. The m6A methyltransferase METTL3 promotes the stemness and malignant progression of breast cancer by mediating m6A modification on SOX2. J BUON 2021;26:444-9. [PubMed] [Google Scholar]
- 12.Chen F, Chen Z, Guan T, et al. N6 -Methyladenosine Regulates mRNA Stability and Translation Efficiency of KRT7 to Promote Breast Cancer Lung Metastasis. Cancer Res 2021;81:2847-60. 10.1158/0008-5472.CAN-20-3779 [DOI] [PubMed] [Google Scholar]
- 13.Niu Y, Lin Z, Wan A, et al. RNA N6-methyladenosine demethylase FTO promotes breast tumor progression through inhibiting BNIP3. Mol Cancer 2019;18:46. 10.1186/s12943-019-1004-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Petri BJ, Piell KM, South Whitt GC, et al. HNRNPA2B1 regulates tamoxifen- and fulvestrant-sensitivity and hallmarks of endocrine resistance in breast cancer cells. Cancer Lett 2021;518:152-68. 10.1016/j.canlet.2021.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shi H, Wang X, Lu Z, et al. YTHDF3 facilitates translation and decay of N6-methyladenosine-modified RNA. Cell Res 2017;27:315-28. 10.1038/cr.2017.15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li A, Chen YS, Ping XL, et al. Cytoplasmic m6A reader YTHDF3 promotes mRNA translation. Cell Res 2017;27:444-7. 10.1038/cr.2017.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jing JJ, Zhao X, Li H, et al. Expression profiles and prognostic roles of m6A writers, erasers and readers in gastric cancer. Future Oncol 2021;17:2605-20. 10.2217/fon-2020-0630 [DOI] [PubMed] [Google Scholar]
- 18.Ni W, Yao S, Zhou Y, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m6A reader YTHDF3. Mol Cancer 2019;18:143. 10.1186/s12943-019-1079-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chang G, Shi L, Ye Y, et al. YTHDF3 Induces the Translation of m6A-Enriched Gene Transcripts to Promote Breast Cancer Brain Metastasis. Cancer Cell 2020;38:857-871.e7. 10.1016/j.ccell.2020.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anita R, Paramasivam A, Priyadharsini JV, et al. The m6A readers YTHDF1 and YTHDF3 aberrations associated with metastasis and predict poor prognosis in breast cancer patients. Am J Cancer Res 2020;10:2546-54. [PMC free article] [PubMed] [Google Scholar]
- 21.Lin Y, Lin L, Fu F, et al. Quantitative proteomics reveals stage-specific protein regulation of triple negative breast cancer. Breast Cancer Res Treat 2021;185:39-52. 10.1007/s10549-020-05916-8 [DOI] [PubMed] [Google Scholar]
- 22.Jézéquel P, Campone M, Gouraud W, et al. bc-GenExMiner: an easy-to-use online platform for gene prognostic analyses in breast cancer. Breast Cancer Res Treat 2012;131:765-75. 10.1007/s10549-011-1457-7 [DOI] [PubMed] [Google Scholar]
- 23.van Staalduinen J, Baker D, Ten Dijke P, et al. Epithelial-mesenchymal-transition-inducing transcription factors: new targets for tackling chemoresistance in cancer? Oncogene 2018;37:6195-211. 10.1038/s41388-018-0378-x [DOI] [PubMed] [Google Scholar]
- 24.Wang Y, Shi J, Chai K, et al. The Role of Snail in EMT and Tumorigenesis. Curr Cancer Drug Targets 2013;13:963-72. 10.2174/15680096113136660102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Caramel J, Ligier M, Puisieux A. Pleiotropic Roles for ZEB1 in Cancer. Cancer Res 2018;78:30-5. 10.1158/0008-5472.CAN-17-2476 [DOI] [PubMed] [Google Scholar]
- 26.Fardi M, Alivand M, Baradaran B, et al. The crucial role of ZEB2: From development to epithelial-to-mesenchymal transition and cancer complexity. J Cell Physiol 2019. [Epub ahead of print]. 10.1002/jcp.28277 [DOI] [PubMed] [Google Scholar]
- 27.Budd GT, Barlow WE, Moore HC, et al. SWOG S0221: a phase III trial comparing chemotherapy schedules in high-risk early-stage breast cancer. J Clin Oncol 2015;33:58-64. 10.1200/JCO.2014.56.3296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Early Breast Cancer Trialists' Collaborative Group (EBCTCG) . Increasing the dose intensity of chemotherapy by more frequent administration or sequential scheduling: a patient-level meta-analysis of 37 298 women with early breast cancer in 26 randomised trials. Lancet 2019;393:1440-52. 10.1016/S0140-6736(18)33137-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loibl S, Poortmans P, Morrow M, et al. Breast cancer. Lancet 2021;397:1750-69. 10.1016/S0140-6736(20)32381-3 [DOI] [PubMed] [Google Scholar]
- 30.Pastushenko I, Blanpain C. EMT Transition States during Tumor Progression and Metastasis. Trends Cell Biol 2019;29:212-26. 10.1016/j.tcb.2018.12.001 [DOI] [PubMed] [Google Scholar]
- 31.Lambert AW, Pattabiraman DR, Weinberg RA. Emerging Biological Principles of Metastasis. Cell 2017;168:670-91. 10.1016/j.cell.2016.11.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Serrano-Gomez SJ, Maziveyi M, Alahari SK. Regulation of epithelial-mesenchymal transition through epigenetic and post-translational modifications. Mol Cancer 2016;15:18. 10.1186/s12943-016-0502-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao C, Ling X, Xia Y, et al. The m6A methyltransferase METTL3 controls epithelial-mesenchymal transition, migration and invasion of breast cancer through the MALAT1/miR-26b/HMGA2 axis. Cancer Cell Int 2021;21:441. 10.1186/s12935-021-02113-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liang X, Zhang Z, Wang L, et al. Mechanism of methyltransferase like 3 in epithelial-mesenchymal transition process, invasion, and metastasis in esophageal cancer. Bioengineered 2021;12:10023-36. 10.1080/21655979.2021.1994721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shi Y, Zhuang Y, Zhang J, et al. METTL14 Inhibits Hepatocellular Carcinoma Metastasis Through Regulating EGFR/PI3K/AKT Signaling Pathway in an m6A-Dependent Manner. Cancer Manag Res 2020;12:13173-84. 10.2147/CMAR.S286275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimura T, Kandimalla R, Okugawa Y, et al. Novel evidence for m6A methylation regulators as prognostic biomarkers and FTO as a potential therapeutic target in gastric cancer. Br J Cancer 2022;126:228-37. 10.1038/s41416-021-01581-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bian S, Ni W, Zhu M, et al. Identification and Validation of the N6-Methyladenosine RNA Methylation Regulator YTHDF1 as a Novel Prognostic Marker and Potential Target for Hepatocellular Carcinoma. Front Mol Biosci 2020;7:604766. 10.3389/fmolb.2020.604766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhu F, Yang T, Yao M, et al. HNRNPA2B1, as a m6A Reader, Promotes Tumorigenesis and Metastasis of Oral Squamous Cell Carcinoma. Front Oncol 2021;11:716921. 10.3389/fonc.2021.716921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Krebs AM, Mitschke J, Lasierra Losada M, et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat Cell Biol 2017;19:518-29. 10.1038/ncb3513 [DOI] [PubMed] [Google Scholar]
- 40.Chaffer CL, Marjanovic ND, Lee T, et al. Poised chromatin at the ZEB1 promoter enables breast cancer cell plasticity and enhances tumorigenicity. Cell 2013;154:61-74. 10.1016/j.cell.2013.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang X, Zhang Z, Zhang Q, et al. ZEB1 confers chemotherapeutic resistance to breast cancer by activating ATM. Cell Death Dis 2018;9:57. 10.1038/s41419-017-0087-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wellner U, Schubert J, Burk UC, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol 2009;11:1487-95. 10.1038/ncb1998 [DOI] [PubMed] [Google Scholar]
- 43.Luo H, Zhou Z, Huang S, et al. CHFR regulates chemoresistance in triple-negative breast cancer through destabilizing ZEB1. Cell Death Dis 2021;12:820. 10.1038/s41419-021-04114-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang G, Dong Y, Liu H, et al. Loss of miR-873 contributes to gemcitabine resistance in triple-negative breast cancer via targeting ZEB1. Oncol Lett 2019;18:3837-44. 10.3892/ol.2019.10697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wang M, Yang Y, Yang J, et al. circ_KIAA1429 accelerates hepatocellular carcinoma advancement through the mechanism of m6A-YTHDF3-Zeb1. Life Sci 2020;257:118082. 10.1016/j.lfs.2020.118082 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The article’s supplementary files as