Abstract

Drugs targeting SARS-CoV-2 could have saved millions of lives during the COVID-19 pandemic, and it is now crucial to develop inhibitors of coronavirus replication in preparation for future outbreaks. We explored two virtual screening strategies to find inhibitors of the SARS-CoV-2 main protease in ultralarge chemical libraries. First, structure-based docking was used to screen a diverse library of 235 million virtual compounds against the active site. One hundred top-ranked compounds were tested in binding and enzymatic assays. Second, a fragment discovered by crystallographic screening was optimized guided by docking of millions of elaborated molecules and experimental testing of 93 compounds. Three inhibitors were identified in the first library screen, and five of the selected fragment elaborations showed inhibitory effects. Crystal structures of target–inhibitor complexes confirmed docking predictions and guided hit-to-lead optimization, resulting in a noncovalent main protease inhibitor with nanomolar affinity, a promising in vitro pharmacokinetic profile, and broad-spectrum antiviral effect in infected cells.
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) has caused the greatest health crisis of this generation and already led to >5 million deaths worldwide.1 Despite promising vaccination programs against COVID-19, antiviral drugs will likely be crucial to control the inevitable future outbreaks of coronaviruses. Variants of SARS-CoV-2 for which the vaccines are less effective have already emerged, which is a strong indication that antiviral drugs are needed to complement vaccines in the long term.2 Analogous to common cold viruses, SARS-CoV-2 is expected to continue to circulate and remain a major threat to our society. In this scenario, antiviral agents are needed to treat patients that have been infected as well as be given prophylactically to protect high-risk groups. Although the road to development of a drug may be long, discovery of inhibitors targeting coronavirus replication must be prioritized as such therapeutic agents can improve the quality of life of millions of patients worldwide.
In early 2020, major global efforts were initiated to develop drugs to treat coronavirus infections. Attempts to repurpose approved drugs identified several promising candidates,3 but in larger clinical studies most of these compounds (e.g., remdesivir and hydroxychloroquine) had little or no effect on mortality or the duration of hospitalization.4 Among the proteins encoded by the SARS-CoV-2 genome, the main protease (Mpro) has emerged as a promising target.5 Inhibition of Mpro blocks the processing of polyproteins produced by translation of the viral RNA, which is an essential step in SARS-CoV-2 replication. Targeting proteases has been a successful strategy for infections caused by the human immunodeficiency and hepatitis C viruses,6 but as Mpro is structurally and mechanistically different, new inhibitors need to be developed for coronaviruses. Prior to the COVID-19 pandemic, several compounds targeting Mpro of coronaviruses via covalent (e.g., GC376(7)) or noncovalent mechanisms (e.g., ML188(5,8,9)) had been identified. However, the noncovalent scaffolds were peptidomimetics, a chemotype that tends to have poor pharmacokinetic properties, and covalent modifiers typically require extensive optimization to modulate activity and selectivity.10−12 It was clear that development of safe and efficacious drugs targeting coronaviruses could benefit from the identification of novel noncovalent Mpro inhibitors with more favorable properties.
The size of commercial compound libraries is growing rapidly, and >10 billion make-on-demand molecules are currently available from chemical suppliers.13 These libraries provide opportunities to identify potential therapeutic agents that can readily be synthesized and tested for activity but require development of effective strategies for navigation in this enormous chemical space. The determination of high-resolution crystal structures of SARS-CoV-2 proteins14 has enabled virtual screening campaigns to identify hits that can be developed into antiviral drugs. Structure-based docking algorithms can sample and score binding poses in seconds, making it possible to evaluate large libraries, and this approach is not restricted to compounds that are physically available. As only a small set of top-scoring compounds are synthesized and tested, docking screens have the potential to substantially improve the efficiency of lead discovery.15 Encouragingly, molecular docking of chemical libraries also resulted in astonishingly high hit rates16 and identified novel chemotypes with high affinity.17,18 Although numerous virtual screens of large chemical libraries for SARS-CoV-2 inhibitors have recently been presented,19,20 only a small fraction of these have tested their predictions experimentally.21,22 Considering the high false-positive rate of docking, thorough validation of hits by several experimental methods is essential for progress toward developing drugs.15
We explored two different strategies to search for Mpro inhibitors in ultralarge chemical libraries using structure-based docking. In the first screen, a library with several hundred million diverse lead-like molecules was docked to the active site of Mpro, and top-ranked compounds were selected for experimental evaluation. The second screen focused on optimization of a fragment identified in a crystallographic screen23 by creating a focused library with millions of compounds. Hits emerged from both sets of compounds, and structure-guided hit-to-lead optimization identified potent inhibitors with antiviral effect in cellular models. The most promising lead compounds were compared to previously identified Mpro inhibitors, including the clinical candidate PF-07321332.24 Comparison of the two screening approaches illuminates their utility as effective strategies to navigate in ultralarge chemical space for finding starting points for drug discovery.
Results
Docking Screen of an Ultralarge Library for Mpro Inhibitors
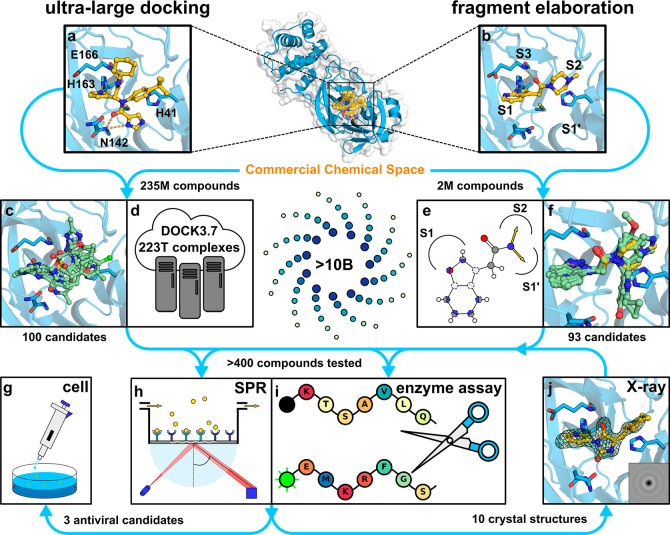
In the first virtual screen, 235 million compounds were docked to a crystal structure of Mpro determined in complex with the substrate-based inhibitor X77 (PDB code: 6W63(25)). X77 is an Mpro inhibitor that occupies all four major pockets (S1, S1′, S2, and S3) of the active site (Figure 1a). X77 (racemic) was a suitable reference compound in our experimental assays, in which it had an affinity (KD) of 1.9 μM (Table 1) and a half maximal inhibitory concentration (IC50) of 2.8 μM (Supplementary Figure 1). The screened chemical library was composed of structurally diverse compounds with physicochemical properties characteristic of good starting points for drug discovery.26 The vast majority of the compounds originated from a commercial make-on-demand library and were hence novel chemical structures that had never been synthesized before. Each compound in the library was sampled in thousands of conformations in the Mpro active site, and a total of >223 trillion complexes were evaluated using the DOCK3.7 scoring function27 (Figure 1d) The screen required 83261 core hours (corresponding to 10 CPU years) and was performed in approximately 1 day on 3500 cores. The 300000 top-ranked compounds, corresponding to 0.12% of the library, were then clustered by topological similarity to identify a diverse set of candidate molecules. From the 5000 top-ranking clusters, 82 compounds were selected for experimental evaluation based on visual inspection of their complementarity to the active site. An additional set of 18 molecules was selected among the 3000 top-ranked compounds that formed the same set of hydrogen bonds as X77, i.e., to His163, Glu166, and Gly143. In the compound selection step, we also took into account contributions to ligand binding that are not included in the docking scoring function. Examples of reasons to exclude compounds from experimental testing are ligand strain, unsatisfied polar atoms of binding site residues or the compound, and unlikely tautomeric/ionization states.28
Figure 1.
Overview of the two virtual screening approaches. (a) Crystal structure of Mpro in complex with inhibitor X77 (PDB accession code: 6W63) was used in the ultralarge docking screen. (b) Crystal structure of Mpro in complex with a fragment (compound 4, PDB accession code: 5RF7) was used in the docking screen of a focused library (fragment elaboration). (c, d) Compounds selected from molecular docking of a ZINC15 library containing 235 M lead-like molecules. (e, f) Compounds were selected from molecular docking of focused libraries with elaborated fragments. (g–j) Predicted inhibitors were evaluated in biophysical and biochemical assays, by crystallography, and in virus-infected cell models.
Table 1. Hits from the Virtual Screens, Inhibitory Potencies, Equilibrium Constants, and Structures of Complexes with Mpro.
Percent enzyme activity is expressed as mean ± SEM from two independent experiments. IC50 values are expressed as mean ± SD of 2–3 independent experiments (Supplementary Figure 1).
Uncertainties are reported as standard deviations (n = 1). KD values were determined by SPR biosensor analysis. The sensorgrams are shown in Supplementary Figure 2.
Crystal structures with (Fo – Fc) electron density difference maps (green isomesh) at +3σ carved at 1.5 Å from the ligand are shown in blue and docking models in gray. Mpro is shown as a cartoon. Selected side chains and the inhibitors are shown as sticks.
Fragment identified by crystallographic screening.23
The 100 selected compounds (Figure 1c, Supplementary Data file 1) were tested in an Mpro enzyme inhibition assay (Figure 1i) and a direct binding interaction assay (Figure 1h) using a surface plasmon resonance (SPR) based biosensor at three concentrations (5, 15–20 and 50 μM). Nineteen compounds showed measurable and dose-dependent binding in the SPR biosensor screen, and three of these (compounds 1–3) were also found to inhibit Mpro in the enzyme inhibition assay (<70% activity at 50 μM, Table 1, Supplementary Data file 1). For the three hits that were confirmed in both assays, the KD values ranged from 23 to 61 μM and the enzyme activity was reduced to 26–63% at the highest tested concentration (50 μM). In agreement with the low affinities estimated for 2 and 3 by the SPR biosensor assay (Supplementary Figure 2), these compounds only showed inhibitory effects at 50 μM in the enzyme activity assay. Compound 1 had the highest inhibitory potency with an IC50 value of approximately 40 μM. The KD determined by the SPR biosensor assay was 23 μM, corresponding to a good ligand efficiency of 0.30 kcal mol–1 heavy atom–1. Crystal structures of compounds 1 and 3 in complex with Mpro were determined at 1.6 and 1.8 Å resolution, respectively (Figure 1j, Supplementary Table 1). The crystallographic binding modes of the two inhibitors, which were based on a hydantoin scaffold, agreed remarkably well with the predicted complexes obtained by molecular docking with root-mean-square deviations (RMSDs) of 0.6 and 1.4 Å, respectively (Figure 2a,b). The hydantoin carbonyl groups formed hydrogen bonds to the backbone of residues Gly143 and Glu166, and substituents on the hydantoin core extended into the S2 and S1 pockets. Based on the docking scores, compound 1 was ranked in the top 0.002% of the chemical library, and the pyridinyl-hydantoin scaffold was strongly enriched by the virtual screen. Compound 2 was also among the high-ranking molecules and composed of a phenylpiperazine scaffold connected to an indazole-4-carbonyl group, which were predicted to extend into the S2 and S1 pockets, respectively (Table 1).
Figure 2.
Confirmation of predicted binding modes by high-resolution crystal structures. The complexes predicted by docking (PDB accession codes of the protein structures: 6W63 for (a, b) and 5RF7 for (c, d); protein and inhibitors are shown as gray cartoons and green sticks, respectively) are shown together with solved crystal structures (protein and inhibitors are shown in blue and yellow, respectively). Selected side chains are shown as sticks, and hydrogen bonds are shown as dashed lines.
Fragment-Guided Virtual Screening for Mpro Inhibitors
The second virtual screen was initiated based on a hit from a crystallographic fragment screen performed at the Diamond Light Source.23 One of the 24 identified active site bound fragments (compound 4) was selected as a starting point for further elaboration (Figure 1b). In the crystal structure (PDB code: 5RF7), compound 4 occupied the S1 and S2 pockets but did not extend into S1′ or S3 in contrast to the larger inhibitor X77. The fragment showed superstoichiometric binding to Mpro in the SPR biosensor assay (KD > 200 μM) and had no effect on enzyme activity at 50 μM (Table 1). Fragment-to-lead optimization was guided by searches in a library with >10 billion make-on-demand compounds combined with docking screens to select the best candidates. Based on visual analysis of the complex, we designed chemical patterns that could encompass the key features of the fragment’s polar interactions and also had the potential to place growth vectors into the S1′ and S2 pockets (Figure 1e,f). A five-membered aromatic heterocyclic ring and an amide that formed hydrogen bonds to His163 and Glu166, respectively, were considered to be crucial for the scaffold, but the size and topology of the heterocycle in the S1 pocket were allowed to vary. We also searched for both branched and tethered amides with potential to grow into the S2 pocket. More than two million elaborations in the make-on-demand library matched these patterns and were subsequently docked to the active site. Of these, ∼5000 compounds maintained the core-binding mode with favorable docking scores. These complexes were visually inspected, and 93 compounds were selected for experimental evaluation (Supplementary Data file 1). The selected compounds explored diverse heterocyclic rings to optimize hydrogen-bonding interactions with the S1 pocket and aliphatic as well as aromatic chemical groups in the S2 and S1′ pocket.
Twenty-one elaborated fragments showed dose-dependent binding in the SPR biosensor experiments, and five of these also inhibited Mpro activity at 50 μM (<70% activity, compounds 5–9, Table 1 and Supplementary Data file 1). KD values determined by SPR biosensor measurements ranged from 7.2 to 79 μM for the five hits confirmed in both assays (Supplementary Figure 2). Compound 5 had an IC50 of 20 μM in the enzyme assay and a KD of 7.2 μM in the SPR assay (Supplementary Figure 2), corresponding to an excellent ligand efficiency of 0.37 kcal mol–1 heavy atom–1. Crystal structures of complexes were successfully determined for compounds 5, 6, and 8 with resolutions of 1.6 Å in all three cases (Supplementary Table 1). The crystallographic binding modes confirmed the docking predictions with RMSDs of 1.3, 1.5, and 1.4 Å, respectively (Figure 2c,d).
Structure-Guided Inhibitor Optimization
Hit-to-lead optimization was pursued for three scaffolds identified from the virtual screens. Compound design was guided by docking predictions and determined crystal structures. Rapid design-make-test cycles were enabled by combining commercial make-on-demand compounds with in-house synthesis, which led to the discovery of nanomolar Mpro inhibitors in less than 4 months.
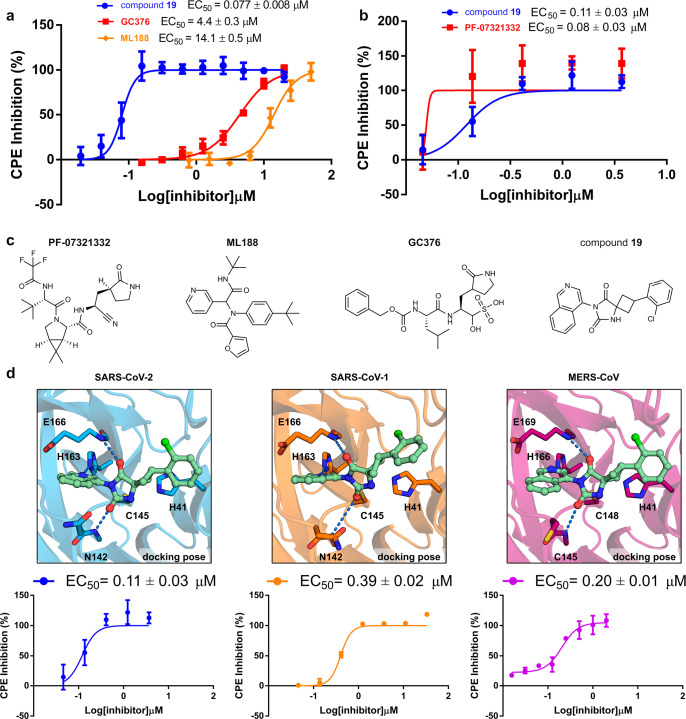
Hit optimization of the scaffold represented by compounds 1 and 3 was guided by crystal structures of these inhibitors bound to Mpro (Figure 3a). As their common hydantoin scaffold showed excellent hydrogen-bonding complementarity to the active site, this chemotype was maintained. The commercial make-on-demand library contained 43 million hydantoins, and we performed searches among these to identify promising candidates for experimental evaluation. Variations in the S1 and S2 pockets were explored systematically in several cycles to identify the optimal combination of substituents. Chemical pattern matching based on these two growth vectors identified >200000 possible elaborations. In total, 137 hydantoin-based analogues were experimentally evaluated (Supplementary Data file 1). In the S1 pocket, pyridine-based moieties based on compound 1 resulted in the greatest increase of potency (Figure 3a). Guided by structural information, substitutions at positions adjacent to the aromatic nitrogen were excluded because these were likely to clash with the protein surface. Instead, efforts were focused on the growth vector extending toward Asn142 in the crystal structure, which was also supported by the bulkier benzotriazole scaffold of compound 5 occupying S1 and by merging fragments identified by crystallographic screening.23 Pyridine elaborations of increasing size gradually improved the potency of derivatives of compound 1 from a weak effect at 50 μM to IC50 values of 18, 7.5, and 1.3 μM for compounds 10, 11, and 12, respectively. The isoquinoline moiety, which was also identified as a promising elongation of the pyridine by the COVID Moonshot Consortium,29 resulted in the highest potency in this series. In the S2 pocket, the best substituents (compounds 14 and 15) were identified from spirocyclic analogues of compound 3. Combining the spiro-indanyl moiety of compound 3 with a pyridine in the S1 pocket (compound 13) did not improve the potency compared to the virtual screening hit, but the predicted binding mode of this inhibitor was supported by crystallography (Supplementary Figure 3). Several attempts were then made to incorporate a tethered motif that would rigidify the alkyl substituent of compound 1 and increase complementarity to the S2 pocket. Spiro-cyclobutyl substitutions in compounds 14 and 15 improved IC50 values to 8.3 and 6.9 μM, respectively, and the SPR biosensor measurements confirmed that compound 15 interacted with Mpro (KD = 6.6 μM, Supplementary Figure 2). The optimal substituents in the S1 and S2 pockets were integrated into a single chemical series by in-house synthesis, resulting in a synergistic improvement of inhibitor potency and affinity. Compounds 16 and 17 had IC50 values of 0.46 and 0.33 μM, respectively (Supplementary Figure 1), and also showed high affinities for Mpro (KD values of 0.14 and 0.15 μM, respectively, Supplementary Figure 2). A crystal structure (2.2 Å resolution) supported the predicted interactions of the isoquinoline and hydantoin rings of compound 17 (Figure 3). The stereoisomeric mixtures of compounds 16 and 17 were separated and their structures were determined using NOESY experiments as compounds 16a, 16b, 17a, and 17b (Supplementary Figure 9). Of these, 16a and 17b showed the highest inhibitory potencies with IC50 values of 0.24 and 0.35 μM (Supplementary Table 4). As further elaboration of the aliphatic tails of compounds 16 and 17 was synthetically challenging, we introduced a phenyl ring at this position, and this inhibitor displayed similar activity (IC50 = 0.39 μM and KD = 0.17 μM). A crystal structure of Mpro in complex with compound 18 (2.0 Å resolution, Figure 3c) was obtained and guided further optimization. Addition of an o-chloro substituent to the phenyl resulted in compound 19, which showed improved potency (IC50 = 0.077 μM, Figure 3c,d) and affinity (KD = 0.038 μM, Supplementary Figure 2). In the same enzyme inhibition assay, GC376 and PF-07321332 had potencies of 0.073 and 0.033 μM, respectively. The potency of the latter compound may be misleading considering that it has a covalent mechanism with a reversibility that may not be accounted for in this steady-state-based assay. Compared to the virtual screening hit (compound 1), 19 had >600-fold higher affinity and an improved ligand efficiency (0.37 kcal mol–1 heavy atom–1 for the optimized compounds and 0.30 kcal mol–1 heavy atom–1 for compound 1, calculated from KD values) with physicochemical properties characteristic of promising leads for drug discovery (Supplementary Table 2).30,31
Figure 3.
Overview of hit-to-lead optimization. (a) Docking hit confirmed by crystallography revealed two potential growing vectors (toward the S1 and S2 pockets) that were explored in parallel. Crystal structures of Mpro in complex with compounds 1 and 17 are shown. (b) Crystal structures of two docking hits inspired a fragment merging, yielding the more potent inhibitor 21. Crystal structures of Mpro in complex with compounds 5 and 21 are shown. (c) Crystal structure of Mpro in complex with compound 18 led to the design of the potent inhibitor 19. (d) Mpro activity assay of compound 19. Data points represent mean ± SD from three independent experiments. IC50 values are expressed as means of three to four independent experiments. Percentage enzyme activity values are expressed as means of two independent experiments. Electron density difference maps (Fo – Fc) at +3σ carved at 1.5 Å from ligands are shown as green isomeshes.
Using the same strategy as for compounds 1 and 3, improvement of the phenylpiperazine scaffold represented by compound 2 involved testing of 67 compounds (Supplementary Data file 1). In this case, optimization of interactions in the S2 pocket was guided by the docking model of compound 2. Decorating the aromatic ring in the S2 pocket with small substituents yielded the micromolar inhibitor 20 (IC50 = 7.2 μM, Supplementary Figures 1 and 4).
The benzotriazole scaffold identified by fragment elaboration was further optimized by fine-tuning interactions in the S2 pocket, and the most successful strategy was to integrate information from several crystal structures combined with molecular docking of commercial compounds (Figure 3b). Elaborated compounds (Supplementary Data file 1) were identified by an iterative chemical pattern searching among >10 billion make-on-demand molecules. By combining features of compounds 5 and 8, the improved inhibitor 21 (IC50 = 2.1 μM) was identified after evaluation of 79 commercially available compounds, and crystal structures confirmed the predicted interactions in the S2 pocket (Figure 3b).
For compounds 20 and 21, the KD values determined by the SPR biosensor assay were 4.7 and 2.3 μM, respectively (Supplementary Figure 2). These two scaffolds have good physicochemical properties and ligand efficiencies (0.29 and 0.35 kcal mol–1 heavy atom–1 for 20 and 21, respectively) and, hence, represent favorable starting points for further optimization (Supplementary Table 2).
Counter Screens
To assess if inhibition involved covalent modification of cysteine residues, we performed assays in the presence of the reducing agent dithiothreitol (DTT) for several of the discovered compounds (Supplementary Table 3, Supplementary Figure 5). Addition of DTT ensures that accessible cysteines are in their reduced form and will indicate if inhibitors act via a false mechanism of redox-cycling.32 The IC50 values of compounds 16–21 were not altered by the addition of DTT. Additional enzymatic assays for Mpro were performed in the presence of detergent to control for promiscuous inhibition due to colloidal aggregation.33 IC50 values were not sensitive to the presence of Triton X-100, supporting that the observed inhibition was not due to colloidal aggregation. Selectivity was assessed by testing the inhibitors against human cathepsin S. Compounds 16–21 did not show any effect on cathepsin S activity (IC50 > 50 μM, Supplementary Figure 5), but the peptidomimetics GC376 and PF-07321332 inhibited this off-target (IC50 = 0.002 and 5.7 μM respectively). To further assess the selectivity profile of our scaffold, compound 19 was tested against a panel of nine other human proteases. Compound 19 showed no significant inhibitory effect against these antitargets at a concentration of 10 μM (Supplementary Table 5). Together with SPR biosensor assays, these controls indicated that the discovered compounds were noncovalent and selective Mpro inhibitors (Supplementary Figure 5, Supplementary Table 5).
Antiviral Effect in Cell Assays and In Vitro Pharmacokinetic Profiling
The antiviral effect of compounds 16, 17, and 19 was evaluated in SARS-CoV-2-infected cells (Figure 1g). The three compounds showed dose-dependent inhibition of the cytopathic effect (CPE) (Figure 4a, Supplementary Figure 6) with EC50 values of 1.7, 1.6, and 0.077 μM and no significant cytotoxicity at the highest tested concentration in Vero E6 cells (50% cytotoxicity concentration, CC50 > 20 μM, Supplementary Table 6). The antiviral activity was also confirmed in a yield reduction assay that assessed the inhibitory activity of the compounds on the viral replication using RT-qPCR. In these experiments, compounds 16, 17, and 19 inhibited SARS-CoV-2 viral replication with EC50 values of 1.0, 0.9, and 0.044 μM, respectively (Supplementary Figure 7). The previously reported Mpro inhibitors GC376, ML188, and X77 were also tested in the CPE-based assay for comparison (Figure 4c). The covalent peptidomimetic GC376 and noncovalent inhibitors ML188 and X77 (rac) showed antiviral effect (EC50 = 4.4, 14.1, and 34.3 μM, respectively, in Vero E6 cells), but their potencies were lower than for 16, 17, and 19. Pfizer’s clinical candidate (PF-07321332) and compound 19 were compared in CPE-based assays performed in Huh7 cells. These Mpro inhibitors were found to have comparable potencies, with EC50 values of 0.08 and 0.11 μM, respectively (Figure 4b).
Figure 4.
Overview of cell assays of compound 19 and reference inhibitors. (a) Inhibitory effect of compounds 19, GC376, and ML188 (rac) on CPE induced by SARS-CoV-2 infection in Vero E6 cells 72 h post infection. Assay was performed with a 3 h preincubation step for GC376 and ML188. EC50 values are expressed as mean ± SEM from at least two independent experiments. (b) Inhibitory effect of compound 19 and PF-07321332 on CPE induced by SARS-CoV-2 infection in Huh7 cells (without preincubation). EC50 values are expressed as mean ± SEM from two independent experiments. (c) Compounds tested in cellular assays. (d) Predicted binding modes of compound 19 in the active sites of Mpro from SARS-CoV-2 (blue), SARS-CoV-1 (orange), and MERS-CoV (purple) are shown. Protein–ligand hydrogen bonds are shown as dashed lines. CPE developed in infected Huh7 cells (SARS-CoV-2 and MERS-CoV) and Vero E6 cells (SARS-CoV-1) treated with compound 19. EC50 values are expressed as mean ± SEM from two independent experiments.
To further assess the potential of compound 19 as a starting point for development of broad-spectrum coronavirus drugs, we computationally docked this inhibitor to homology models of 29 reported active site mutants of Mpro and crystal structures of SARS-CoV-1 and MERS-CoV variants.34−36 The binding mode of compound 19 was maintained in all of the predicted complexes (Figure 4d, Supplementary Table 7), which indicated that our inhibitor could be active against other coronaviruses. In agreement with our computational modeling, compound 19 also inhibited the cytopathic effect of SARS-CoV-1 (EC50 = 0.39 μM, Vero E6 cells) and MERS-CoV (EC50 = 0.20 μM, Huh7 cells) (Figure 4d).
The in vitro ADME properties of compound 19 were promising with good metabolic stability in the presence of human liver microsomes (intrinsic clearance CLint = 22 μL/min/mg) and plasma protein binding in human plasma (fraction unbound fu = 3.3%). In agreement with the potent antiviral effect in cells, compound 19 was highly permeable in a Caco-2 cell assay (Papp AB = 5.9 × 10–5 cm/s and Papp BA = 5.0 × 10–5 cm/s) with no observed efflux effect (efflux ratio of 0.8).
Discussion
Three main findings emerged from our virtual screens of ultralarge chemical libraries for SARS-CoV-2 inhibitors. First, molecular docking of diverse and focused screening libraries identified eight Mpro inhibitors, and the predicted binding modes were confirmed by crystallography for five of these. Second, efficient hit-to-lead optimization was enabled by crystal structures and searches among billions of virtual compounds, resulting in three series of noncovalent inhibitors. Finally, the most promising leads have nanomolar IC50 values in an Mpro enzymatic assay, good in vitro ADME properties, and showed a potent antiviral effect against several coronaviruses in cellular assays.
The billions of compounds that are available in commercial libraries provide novel avenues for drug discovery, but navigating this vast chemical space to find leads is challenging. We compared two strategies to exploit ultralarge chemical libraries using docking screens. Mpro inhibitors were discovered by docking of an unbiased library with several hundred million molecules and a focused library based on a fragment. More diverse scaffolds were accessible in the first screen, but we anticipated that higher hit rates and potencies would be obtained from the focused library as the binding mode of the core fragment had been validated by crystallography.23 Encouragingly, both virtual screens delivered novel and nonoverlapping sets of inhibitors. However, to our surprise, the hit rates of the two strategies (i.e., the percentage of active compounds among those tested experimentally) were similar, as shown by validation both in the enzyme activity assay (3% and 5%) and the more sensitive SPR biosensor screen (19% and 21%). Although the most potent hit was discovered by the fragment elaboration (compound 5, IC50 = 20 μM), compound 1 from the diverse library only had a 2-fold higher IC50 value (∼40 μM). These observations could reflect that docking scoring functions may be more proficient in ranking diverse chemotypes rather than differentiating between closely related elaborations of the same scaffold.37 Our results suggest that docking of large and unbiased libraries should be prioritized over focused subsets because more diverse leads will be identified with a marginal loss of hit rates and activities.
The commercial libraries also facilitated hit-to-lead optimization. Optimization was primarily driven by searches among >10 billion make-on-demand compounds, which enabled rapid design-make-test cycles. Remarkably, millions of analogues with diverse substituents were available for each scaffold in the first iterations. These molecules had generally not been previously synthesized, but as they are made with well-characterized reactions and available building blocks, the chemical supplier was able to deliver optimized inhibitors within a few weeks. In only a few iterations, three scaffolds were optimized from ∼20–40 μM to single-digit micromolar potencies. However, as the size and molecular complexity of the compounds increased, the number of analogues in the make-on-demand library decreased and designs based on the active site structure were not available. As we approached the outer edge of commercial chemical space, in-house synthesis proved essential to obtain the leads 16, 17, and 19. These compounds belong to the small set of noncovalent and nonpeptidomimetic inhibitors of Mpro with nanomolar inhibitory activities.29,38,39 Compared to the noncovalent inhibitors that were available when the virtual screen was performed (ML188 and X77), compound 19 has a better ligand efficiency and physicochemical properties and an antiviral activity that is 2 orders of magnitude more potent. Subsequent to our discovery of the hydantoin-based Mpro inhibitors, the COVID Moonshot Consortium also explored this moiety in their lead series.29 These results illustrate the ability of different structure-guided design approaches to identify privileged scaffolds that can rapidly be developed to potent inhibitors.29,38 Our study represents one of the few prospective docking campaigns of several hundred million compounds17,18,40 and further supports how access to make-on-demand libraries can expedite drug discovery. The initial hit (compound 1) had good ligand efficiency and low lipophilicity, which was an excellent starting point for optimization. Only six additional heavy atoms were added in optimization of the hydantoin scaffold, leading to >600-fold increase of affinity and an improved ligand efficiency, an unusual achievement in hit-to-lead optimization.
The clinical candidate from Pfizer (PF-07321332) is the most advanced effort to develop an Mpro inhibitor to treat SARS-CoV-2 infections.24 In our enzyme inhibition assays, compound 19 and PF-07321332 both have nanomolar potencies. Encouragingly, compound 19 did not inhibit any human proteases considered to be potential off-targets, whereas PF-07321332 also inhibits human cathepsin S, indicating that promiscuity may be a liability of this compound class. Moreover, compound 19 showed comparable efficacy as PF-07321332 against SARS-CoV-2 and also displayed antiviral effect in SARS-CoV-1 and MERS-CoV infected cells. Based on the broad-spectrum antiviral effect combined with promising selectivity and in vitro pharmacokinetic profile, the scaffold represented by 19 is one of the most promising for development of an antiviral drug targeting SARS-CoV-2.
Materials and Methods
Molecular Docking
A crystal structure (PDB accession code: 6W63(25)) of the SARS-CoV-2 Mpro bound to an inhibitor (X77) was used in the large-scale docking screen. Crystallographic waters and other solvent molecules were first removed from the structure. The atoms of X77 were used to define the binding site by generating 45 matching spheres. DOCK3.727 uses a flexible ligand algorithm that superimposes rigid segments of a molecule’s precalculated conformational ensemble on top of the matching spheres. A total of 19 additional matching spheres were added to enhance orientational sampling in the binding site. Histidine protonation states were assigned manually based on visual inspection of hydrogen bonding networks. For example, the key residue His163 was protonated at the Nε atom because of the hydrogen-bond interaction with the pyridine of X77 (acceptor) and hydroxyl group of Tyr161 (donor). Histidines 41, 163, 164, 172, and 246 were protonated at the Nε, whereas histidines 64 and 80 were protonated at the Nδ. The remainder of the enzyme structure was protonated by REDUCE41 and assigned AMBER42 united atom charges. The dipole moments of three residues involved in recognition of X77 were increased to favor interactions with these. This technique is common practice for users of DOCK3.7 to improve docking performance43 and has been used in previous virtual screens.17,18,22,44,45 For residue His163, the partial atomic charges of the Nε and Hε atoms were increased without changing the net charge of the residue. The dipole moment of the backbone amide of residues Asn142 and Glu166 was also increased using the same technique. The atoms of the cocrystallized inhibitor were combined with atoms of bound noncovalent fragments from other crystal structures23 to create two sets of sphere layers on the protein surface (referred to as thin spheres). One set of thin spheres with radius 1.2 Å described the low protein dielectric and defines the boundary between solute and solvent. A second set of thin spheres with radius 0.3 Å was used to calibrate ligand desolvation penalties. Scoring grids were precalculated using QNIFFT46 for Poisson–Boltzmann electrostatic energies, SOLVMAP47 for ligand desolvation energies, and CHEMGRID48 for AMBER van der Waals energies. The crystal structure (PDB accession code: 5RF7(23)) of Mpro bound to compound 4 was used for fragment elaboration docking calculations. Scoring grids were generated using the same protocol as described above, and the dipole moments of residues His163 and Glu166 were increased in this case. Property-matched and property-perturbed decoys49,50 of noncovalent bound fragments identified by crystallography23 were generated using in-house scripts. The obtained control sets were used to evaluate the performance of the docking grids by means of ligands-over-decoys enrichments. Enrichment of ligands and the predicted binding poses of the fragments were used to select the final grid parameters.
In the screen of the ultralarge library, DOCK3.7 was used to dock the lead-like subset of ZINC1551 (http://zinc15.docking.org) to the Mpro active site. This subset is characterized by good physicochemical properties for screening assays (cLogP ≤ 3.5 and molecular weight ≤350 Da). The library contained more than 235 M commercially available molecules, of which 228 M molecules were successfully docked. For each compound, 5551 orientations were calculated on average, and for each orientation, an average of 475 conformations was sampled. For each ligand, the best scoring pose was optimized using a simplex rigid-body minimizer. Top-scoring molecules were filtered using a PAINS-filter to reduce the risk of encountering false positives.52 The diversity among the top-ranked compounds was increased by clustering the 300000 best scoring molecules using ECFP4-based fingerprints and a Tanimoto coefficient of 0.5.53 The best scoring member of the cluster was chosen as a representative. Out of the 33876 resulting clusters, the 5000 top-scoring were visually inspected. In addition, the top 3000 nonclustered molecules that formed hydrogen bonds with residues His163, Glu166, and Gly143 were also visually inspected. In the screens of the >2 million fragment elaborations (based on compound 4), the 50 lowest energy poses of each molecule were retained from the docking. The lowest energy pose that had an RMSD value below 2 Å from compound 4 was considered as the most relevant pose. Using an in-house script, the RMSD value was calculated between atoms that fulfilled the SMARTS definition, allowing for the exchange of heteroatoms in the core scaffold.
Crystal structures of the SARS-CoV-1 (PDB accession code: 7LMG) and MERS-CoV (PDB accession code: 4YLU) Mpro bound to inhibitors were prepared for molecular docking using the same protocols as for the SARS-CoV-2 Mpro structures.35,36 The atoms of the cocrystallized inhibitors were used to generate 45 matching spheres in the active sites. For SARS-CoV-1, dipole moments of Asn142, His163, and Glu166 were increased with the same protocol as the SARS-CoV-2 model. For MERS-CoV, the dipole moments of Cys145, His166, and Glu169 were increased.
Cheminformatics and Preparation of Chemical Libraries
Chemical SMARTS patterns were constructed using structural information and an interactive SMARTS visualizer (https://smarts.plus).54 Chemical pattern matching was performed with OpenEye’s OEToolkits and Enamine’s November 2019 REAL space library (12.3 billion compounds). Matching molecules were filtered by a PAINS-filter to reduce the risk of having false positives.52 Molecules were prepared for docking using DOCK3.7 protocols (db2 format). Conformational ensembles were capped at 400 conformations per rigid segment and an interconformer RMSD diversity threshold of 0.25 Å.
Modeling of SARS-CoV-2 Mpro Mutants
Mutation data were obtained from the GISAID SARS-CoV-2 database.34 A total of 841 unique mutant protein sequences were aligned to the reference sequence. For each sequence, a ligand-bound homology model was constructed using MODELER with a SARS-CoV-2 crystal structure (PDB code: 6W63) as template.55 Molecular docking grids were prepared using the same protocols as for the Mpro crystal structures. Symmetry-corrected ligand RMSD values were calculated after aligning the homology model to a reference crystal structure (PDB code: 7QBB).
Expression and Purification of Mpro for Protease Activity Assays and Crystallography
SARS-CoV-2 Mpro was produced adopting a previously described construct67 containing nucleotide sequences corresponding to residues S1-Q306 (Chinese isolate, NCBI accession no. YP_009725301). In this construct, Mpro is flanked by an N-terminal GST (glutathione S-transferase) tag followed by a Mpro recognition sequence for autoproteolysis and a C-terminal 6 × His-tag preceded by a HRV Mpro recognition sequence. Except for some minor adjustments, the expression and purification of SARS-CoV-2 Mpro was performed according to a previously described procedure.68 The vector (pGEX-6P-1) containing the coding sequence of the SARS-CoV-2 Mpro (obtained from Diamond Light Source, Oxford, UK) was transformed into E. coli BL21 (DE3)-T1R competent cells. L-Broth media (Formedium, Norfolk, UK) supplemented with carbenicillin (100 μg/mL) was inoculated with fresh transformants and grown at 37 °C until an OD600 of 1.5 was reached. The starter culture was then used to inoculate the main culture in Auto Induction Media (AIM) Terrific Broth base with trace elements (Formedium, Norfolk, UK) supplemented with 1% glycerol and carbenicillin (100 μg/mL). The cultures were grown at 37 °C until an OD600 of 2 was reached, and the protein expression was continued overnight at 18 °C for 13.5 h. Cells were thereafter harvested by centrifugation (10 min at 4500 gav, 4 °C), resuspended in IMAC lysis buffer (50 mM Tris, 300 mM NaCl, pH 8.0) supplemented with benzonase nuclease (10 μL/1.5 L culture, 250 U/μL, E1014, Merck, Darmstadt, Germany), and disrupted by sonication (4s/4s 3 min, 80% amplitude, Sonics Vibracell-VCX750, Sonics & Materials Inc., Newtown, CT, USA). Lysates were centrifuged at 49000 gav for 20 min at 4 °C. The supernatants were filtered (Corning bottle-top vacuum filter, 0.45 μm, Corning, NY), and imidazole was added to a final concentration of 10 mM before loading onto an HisTrap HP 5 mL (Cytiva, Uppsala, Sweden), mounted on an ÄKTAxpress system (Cytiva, Uppsala, Sweden). The column was washed with wash buffer (50 mM Tris, 300 mM NaCl, 25 mM imidazole, pH 8.0), and the bound protein was eluted with elution buffer (50 mM Tris, 300 mM NaCl, 500 mM imidazole, pH 8.0). For crystallization experiments, the protein was further purified by size-exclusion chromatography (SEC) using a HiLoad 16/60 Superdex 200 pg (Cytiva, Uppsala, Sweden) pre-equilibrated with gel filtration buffer (50 mM Tris, 300 mM NaCl, pH 8.0). To remove the His-tag, the protein containing fractions were pooled and treated with HRV 3C protease (1 μg/500 μg target protein, SAE0045, Merck, Darmstadt, Germany) overnight at 4 °C in gel filtration buffer supplemented with 0.5 mM TCEP and 0.5 mM DTT. For the Mpro protease activity assay the protein was treated with HRV 3C protease directly after the IMAC purification step, and the buffer was at the same time exchanged by dialysis (dialysis buffer 50 mM Tris, 300 mM NaCl, 0.5 mM TCEP and 0.5 mM DTT, pH 8.0) with a dialysis cassette (Slide-A-Lyzer Dialysis Cassette, 10K MWCO, 3 mL, Thermo Fisher Scientific, Waltham, MA) overnight at 4 °C. The cleaved SARS-CoV-2 Mpro samples were subsequently purified by reverse IMAC purification using a HisTrap 1 mL (Cytiva, Uppsala, Sweden). The same wash buffer described above was used, and the flow through was collected. The reverse IMAC purification was followed by a second SEC step using the same column and buffer as described earlier. Fractions containing the target protein were examined by SDS–PAGE, pooled together, and concentrated with Vivaspin 20 mL centrifugal concentrators (10 kDa MWCO, Sartorius, Göttingen, Germany) at 4000 gav, 4 °C. The protein was finally flash frozen in liquid nitrogen and stored at −80 °C.
Mpro Protease Activity Assay
An internally quenched fluorogenic substrate for SARS-CoV-2 Mpro (DABCYL-Lys-HCoV-SARS Replicase Polyprotein 1ab (3235–3246)-Glu-EDANS trifluoroacetate salt, > 95% pure) was custom synthesized and obtained from Bachem AG, Switzerland.
The Mpro protein used for catalytic activity assays was obtained from the Protein Science Facility (PSF, Karolinska Institutet, Stockholm, Sweden) and is described in a prior section. All test compounds were dissolved to 10 mM stocks in 100% DMSO (Merck KGaA, Darmstadt, Germany) and transferred to Echo LDV source plates (Labcyte, Inc., CA). Mpro activity was analyzed by detection of hydrolysis of an internally quenched SARS-CoV-2 Mpro substrate. The assay was performed in 20 mM Tris, 50 mM NaCl, and 0.1 mM EDTA (Merck KGaA, Darmstadt, Germany) at room temperature (pH 7.5). Compounds were transferred with an Echo 550 noncontact dispenser (Labcyte, Inc., USA) to Corning 3575 nonbinding 384 well assay plates. Mpro (75 nM final concentration) was added to the assay plate using a 16-channel pipet (Integra ViaFlo, BergmanLabora AB, Sweden) and shaken for 15 min at 1000 rpm in an Eppendorf Mixmate. After a pulse centrifugation, the Mpro fluorogenic substrate (stock solution at 5 mM in 100% DMSO) was added to the assay plate to a final concentration of 10 μM, thus contributing with 0.2% DMSO in final assay, with a Labcyte Echo 550 noncontact dispenser. After 10 min incubation with shaking at 1000 rpm in an Eppendorf Mixmate and a pulse centrifugation, fluorescence was measured in a PerkinElmer Envision plate reader at ambient temperature with excitation at 340 nm and emission at 490 nm.
Compound Screening
Screening compounds (Supplementary Data file 1) were purchased from Enamine Ltd. (compound purity >90%, which was confirmed in-house by LC–MS). GC376 was obtained from Carbosynth, and a freshly synthesized reference sample of PF-07321332 was kindly provided by the Drugs for Neglected Diseases initiative (DNDi, Switzerland). Using the activity assay described above, compounds were screened against Mpro at three concentrations, 5, 15, and 50 μM, respectively, and hits were retested in an 11-point concentration series (1:3 dilutions, starting concentration 50 μM). The dose–response curve was generated using Echo 550 noncontact dispensing from 10 mM compound stocks. Screening results were calculated as percent of Mpro activity in each data point (100 × (RFU sample − RFU Blank control)/(RFU DMSO control − RFU Blank control)) using Microsoft Excel with XLfit 5.5, IDBS, Guildford, U.K. A nonlinear fit of 11-point dose−response curves (log(inhibitor) vs response − variable slope (four parameters) and IC50 calculations were performed using GraphPad Prism version 9.1.0 for Windows (GraphPad Software, San Diego, CA). Outliers identified by GraphPad Prism were excluded from graphs and analysis in 11-point dose response data.
Counter Assay Screening
Compounds were evaluated in an 11-point concentration series in the standard Mpro activity assay with the addition of 0.01% Triton X-100 to identify aggregating compounds. The compounds were also run with the addition of reducing agent (1 mM DTT). The effect of selected compounds on the activity on human cathepsin S was determined with the SensoLyte 440 cathepsin S Assay Kit (Anaspec, Inc.) used according to the manufacturer’s recommendations. The screen against a panel of nine human proteases (cathepsin K, cathepsin D, cathepsin B, cathepsin L, thrombin, caspase-2, elastase, calpain 1, and trypsin) was performed by Cerep/Panlabs/Eurofins. Compound 19 was tested in duplicate at 10 μM against each enzyme. Reference control compounds were included in each assay.
Surface Plasmon Resonance (SPR) Biosensor Assays
The SPR experiments were performed using a Biacore S200 instrument and Sensor Chip CM5 (Cytiva, Uppsala, Sweden) at 25 °C. Streptavidin (Sigma) was immobilized by a standard amine coupling procedure. The Sensor Chip CM5 surface was activated by an injection of a 1:1 mixture of EDC and NHS for 7 min at a flow rate 10 μL/min. Streptavidin was diluted to 250 μg/mL in sodium acetate buffer (pH 5.0) and injected over the activated surface at a flow rate 2 μL/min for 10 min. The surface was then deactivated by the injection of 1 M ethanolamine for 7 min. Subsequently, the biosensor chip was conditioned with four pulse injections of 1 M NaCl/50 mM NaOH solution. Mpro with a C-terminal Avi-tag (obtained from Diamond Light Source, Oxford, UK) was diluted to 100 μg/mL in 1.02 × running buffer (50 mM TrisHCl, pH 7.5, 0.05% Tween-20) and injected at the flow rate of 2 μL/min, reaching a typical immobilization level of 8000–9000 RU.
After immobilization, compounds were injected over the surface at 5, 20, and 50 μM, at a flow rate 30 μL/min in 50 mM TrisHCl, pH 7.5, 0.05% Tween-20. An association phase was monitored for 60 s and a dissociation phase for 120 s. A solvent correction accounting for 2% DMSO was performed. The data was analyzed using Biacore S200 Evaluation Software, v. 1.1 (Cytiva, Uppsala, Sweden). Selected compounds were further analyzed using a 10-point concentration series. Sensorgrams were double-referenced by subtracting the signals from a reference surface and the signal from one blank injection. For determination of KD values, an equation corresponding to a reversible, one-step, 1:1 interaction model was fitted by nonlinear regression analysis to report points taken at the end of the injection, representing steady-state signals. GC376 and PF-07321332 were analyzed using a single-cycle kinetics experiment due to no/slow dissociations.
Protein Crystallization
Mpro was crystallized at 20 °C in 96-Well-3-Drop MRC plates (SWISSCI AG, Switzerland) using sitting drop vapor diffusion method and Mosquito pipetting robot (TTP Labtech, UK). Initial crystallization hits (clusters of plate-like crystals) were obtained from 300 nl 1:1 protein to reservoir (100 mM MES pH 6–6.5, 9–11% (m/v) PEG8K) drops and were used to prepare crystallization seeds. After optimization, the final crystallization conditions were as follows: 100 nL of Mpro (8.3 mg mL–1 in 50 mM Tris pH 8.0, 300 mM NaCl), 50 nL seeds, and 450 nL reservoir solution (100 mM Tris pH 8.25 or 200 mM HEPES pH 7.75, 5% (v/v) DMSO, 12.5% PEG4K). Individual crystals nucleated within 24 h and grew to maximal dimensions within 2 days. Soaking was performed in 100 mM Tris pH 8.25 or 200 mM HEPES pH 7.75, 6.25–15 mM ligand, 5% DMSO, and 20% PEG4K soaking buffers, supplemented with PEG300 to 10% (v/v) for in situ cryo-protection. After 2 h of soaking at 20 °C, crystals were harvested and cryo-cooled in liquid nitrogen.
All diffraction data was collected at cryogenic temperature at the BioMAX beamline (MAX IV Laboratory, Sweden).56 Data collection parameters are given in Supplementary Table 1. Data sets were indexed, merged, and scaled on-site and analyzed with the DIMPLE pipeline, implemented within the CCP4 software suite,57 using 7K3T as a search model and an optional molecular replacement with PHASER MR.58 The data sets of interest were reprocessed with XDS59 and AIMLESS, ligand dictionaries were created using AceDRG,60 model building and ligand fitting were carried out using Coot,61 and structure refinement was performed using either Refmac5 or phenix.refine.62 Validation of the structures was performed using MolProbity.63 Data collection and refinement statistics are shown in Supplementary Table 1. Crystallographic structures and the corresponding structural factors were deposited in the Protein Data Bank (PDB IDs: 7B2U, 7AU4, 7B2J, 7B5Z, 7B77, 7BIJ, 7NEO, 7O46, 7QBB, and 7NBT). Images were generated using PyMOL (v.2.0.6). Omit difference maps were calculated by removing ligand atoms from the structural models, followed by 10 cycles of refinement in Refmac5.
SARS-CoV-2 Antiviral Activity in Huh7 Cell Culture
The human hepatoma cell line Huh7 was maintained in DMEM (Gibco catalog no. 41965-039) supplemented with 10% fetal bovine serum (FBS), 2% HEPES 1 M (Gibco catalog no. 15630106), 5 mL of sodium bicarbonate 7.5% (Gibco catalog no. 25080-060) 1% nonessential amino acids (NEAA Gibco catalog no. 11140050), and 1% penicillin-streptomycin 10000 U mL–1 (Gibco catalog no. 15140148) in a humidified 5% CO2 incubator at 37 °C. Assay medium, used for producing virus stocks and antiviral testing, was prepared by supplementing DMEM with 4% FBS, 2% HEPES 1 M, 5 mL of sodium bicarbonate 7.5, and 1% NEAA. To quantify antiviral activity on Huh7 cells, a SARS-CoV-2 virus strain that produces a sufficient cytopathogenic effect (CPE) on this cell line was selected. Passage 6 of the SARS-CoV-2 strain BetaCov/Belgium/GHB-03021/2020 (EPI ISL 407976, 3 February 2020)64 was passaged three times on Huh7 cells while selecting those cultures that showed the most CPE. This resulted in a virus stock (passage 9) that confers full CPE on Huh7 (5.6 × 104 median tissue culture infectious dose (TCID50) per mL) as well as on Vero E6 cells (1.8 × 107 TCID50 per mL). The genotype of this virus stock shows four nucleotide changes as compared with the mother virus stock (P6) and these are currently being analyzed. None of the nucleotide changes occur in the part of the genome that encodes the 3C-like protease, validating this virus stock for testing protease inhibitors. For antiviral testing, Huh7 cells were seeded in 96-well plates (Corning CellBIND 96-well Microplate catalog no. 3300) at a density of 6000 cells per well in assay medium. After overnight growth, cells were treated with the indicated compound concentrations and infected with a multiplicity of infection of 0.005 TCID50 per cell of the P9 virus (final volume of 200 μL per well in assay medium). On day 4 post-infection, differences in cell viability caused by virus-induced CPE or by compound-specific side effects were analyzed using MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt). For this, an MTS:phenazine methosulfate (PMS) stock solution (2 mg mL–1 MTS (Promega) and 46 μg mL–1 PMS (Sigma-Aldrich) in PBS at pH 6–6.5) was diluted 1:20 in MEM without phenol red (Gibco catalog no. 51200038). Medium was aspirated from wells of the test plates, and 70 μL of MTS/PMS solution was added. After 0.5–1 h incubation at 37 °C, absorbance was measured at 498 nm. Cytotoxic effects caused by compound treatment alone were monitored in parallel plates containing mock-infected cells. These experiments were performed in the high-containment BSL3+ facilities of the KU Leuven Rega Institute (3CAPS) under licenses AMV 30112018 SBB 219 2018 0892 and AMV 23102017 SBB 219 2017 0589 according to institutional guidelines.
SARS-CoV-2 Antiviral Activity in Vero E6 Cell Culture
Vero E6 cells were grown in DMEM (Gibco, 41966029) supplemented with 10% FBS (Gibco, 10500064) and 1× penicillin-streptomycin (Sigma-Aldrich, PA333) and incubated at 37 °C, 5% CO2 atmosphere. SARS-CoV-2 was isolated at the Zoonosis Science Center (Uppsala University) from naso-oropharyngeal swab collected from a Swedish patient as described in Nissen et al.65 All infection experiments were performed in a biosafety level 3 laboratory (BSL3) at the Zoonosis Science Center (Uppsala University).
SARS-CoV-2 CPE-Based Antiviral Assay in Vero E6 Cell Culture
Compounds 16, 17, and GC376 were tested at final concentrations ranging from 20 to 0.156 μM with and without a 3 h preincubation step. Compound ML188 was tested at final concentrations ranging from 100 to 0.78 μM with a 3 h preincubation step. Compound 19 was tested at final concentrations ranging from 20 to 0.019 μM without a preincubation step. Each concentration was tested with triplicates in at least two independent experiments. Vero E6 cells were seeded in 96-well plates at a density of 104 cells/well in a final volume of 100 μL of DMEM (2% FBS, 1× penicillin–streptomycin) and incubated overnight at 37 °C, 5% CO2 atmosphere. On the day of the assay, aliquots of the compounds in DMSO (stored at −20 °C) were thawed, serially diluted (1:2) in DMSO, and then further diluted in DMEM (2% FBS, 1× penicillin–streptomycin) to 4× working solutions of the desired final concentrations. Cell media was removed and substituted with 50 μL of fresh cell media (2% FBS, 1× penicillin–streptomycin). Cells were infected at a MOI of 0.01 by adding 25 μL of SARS-CoV-2 solution and treated by adding 25 μL of the compounds’ 4× working solutions giving a total volume of 100 μL (1:4 dilution). Compounds 16, 17, GC376, and ML188 were also tested with a 3 h pretreatment step. Vero E6 cells were pretreated with the compounds by removing 25 μL of cell media and adding 25 μL of 4x working solutions of the compounds’ desired final concentrations to the remaining 75 μL of cell media (1:4 dilution). After 3 h of pretreatments, the cell media was removed, cells were washed with 100 μL of PBS, 50 μL of fresh DMEM (2% FBS, 1× penicillin–streptomycin) was added to each well, and cells were retreated and infected as previously explained. Treated–uninfected, infected–untreated, and untreated and infected control wells were also included in triplicates. DMSO concentration in each well was kept to 0.2% (v/v).
After 72 h, the cell media in each well was replaced with 100 μL of fresh DMEM (2% FBS, 1x Penicillin-Streptomycin) to which 10 μL of a 5 mg/mL MTT (Sigma-Aldrich, M2128) solution in PBS was added. Following 4 h incubation, 100 μL of a 10% SDS, 0.01 M HCl solution was added to solubilize the formed formazan crystal. After overnight incubation optical density (OD) at 570 and 690 nm was read using a Tecan infinite M200 PRO plate reader (Tecan Trading AG, Switzerland). OD readings at different wavelengths were subtracted, the resulting values were normalized to the controls, and EC50 were determined by nonlinear regression analysis using GraphPad Prism (v.6.0) (GraphPad Software, La Jolla California, USA).
SARS-CoV-2 Virus Yield Reduction Assay with RT-qPCR
Vero E6 cells were seeded in 96-well plates at a density of 104 cells/well in a final volume of 100 μL of DMEM (2% FBS, 1× penicillin–streptomycin). After overnight incubation at 37 °C, 5% CO2, cells were infected with SARS-CoV-2 at a MOI of 0.01 for 1 h, after which the virus inoculum was removed, cells were washed with 100 μL of PBS and 75 μL of fresh DMEM (2% FBS, 1× penicillin–streptomycin) was added to each well. Cells were then treated as explained in the CPE based assay paragraph by adding 25 μL of 4× working solutions of the compounds’ desired final concentrations. Treated–uninfected, infected–untreated, and untreated and infected control wells were also included in triplicates. DMSO concentration in each well was kept to 0.2% (v/v). After 72 h, 50 μL of supernatant from each well was collected and mixed with 150 μL of TRIzol LS (Invitrogen, ThermoFisher Scientific, Waltham, MA) for viral RNA extraction and quantification by RT-qPCR. The remaining supernatant was removed and substituted with 100 μL of fresh DMEM (2% FBS, 1× penicillin–streptomycin). Quantified viral RNA from infected wells treated with different concentrations of the compounds were normalized to the controls and EC50 values were determined by nonlinear regression analysis using GraphPad Prism v.6.0 (GraphPad Software, La Jolla CA).
Viral RNA was extracted using the Direct-zol-96 RNA Kit (Zymo Research, Irvine, CA) from the collected supernatant with a sample volume of 200 μL (50 μL of supernatant + 150 μL of TRIzo LS) according to the manufacturer’s protocol. Portions of the SARS-CoV-2 envelope small membrane protein (E) gene was amplified by RT-qPCR, using primers (Thermo Fisher Scientific, Waltham, MA) previously described and the SuperScript III OneStep RT-PCR System with Platinum Taq DNA Polymerase kit (Invitrogen, Thermo Fisher Scientific, Waltham, MA). Target E:66 forward primer 5′-ACAGGTACGTTAATAGTTAATAGCGT-3′; reverse primer 5′-TGTGTGCGTACTGCTGCAATAT-3′; and the probe 5′-FAM-ACACTAGCCATCCTTACTGCGCTTCG-TAMRA-3′. The reaction mixture contained 12.5 μL of reaction buffer (a buffer containing 0.4 mM of each dNTP, 3.2 mM MgSO4), 0.5 μL of SuperScript III RT/Platinum Taq Mix, 0.5 μL of each primer (10 μM (μM) stock concentrations), 0.25 μL probe (10 μM stock concentration), 2.4 μL of 25 mM magnesium sulfate, 3.35 μL of nuclease-free water, and 5 μL of RNA template. The RT-qPCR assay was performed on a CFX96 Touch Real-Time PCR Detection System (Bio-Rad Laboratories, Hercules CA) under the following conditions: reverse transcription at 55 °C for 30 min and 95 °C for 3 min, followed by 45 cycles of denaturation at 95 °C for 15 s, extension at 57 °C for 30 s, and collecting the fluorescence signal at 68 °C for 30 s.
All samples were run in triplicate. The corresponding number of copies for each Ct was calculated from a standard curve prepared with synthetic DNA gene fragments (gBLOCKs; IDT, San Jose, CA) with a five-base-pair deletion in the amplified regions of the viral genome diluted in deionized, nuclease-free water to concentrations of 103–105 copies per μL. The five-base-pairs were deleted to be able to distinguish between viral RNA and gBLOCKs during sequencing. The LODs for both genes were 101 copies per μL. The relative fluorescence unit (RFU) data were obtained from the CFX Maestro Software (Bio-Rad CFX Maestro for Mac 1.1 Version 4.1.2434.0214, Bio-Rad Laboratories, Hercules, CA).
SARS-CoV-1 Antiviral Activity in Vero E6 Cell Culture
The cell line Vero E6 was maintained in DMEM (Gibco catalog no. 41965-039) supplemented with 10% fetal bovine serum (FBS), 2% HEPES 1 M (Gibco catalog no. 15630106), 5 mL of sodium bicarbonate 7.5% (Gibco catalog no. 25080-060), 1% nonessential amino acids (NEAA Gibco catalog no. 11140050), and 1% penicillin–streptomycin 10000 U mL–1 (Gibco catalog no. 15140148) in a humidified 5% CO2 incubator at 37 °C. Assay medium, used for producing virus stocks and antiviral testing, was prepared by supplementing DMEM with 4% FBS, 2% HEPES 1 M, 5 mL of sodium bicarbonate 7.5, and 1% NEAA. To quantify antiviral activity on Vero E6 cells, a SARS-CoV-1 virus strain that produces sufficient cytopathogenic effect (CPE) on this cell line was selected. SARS-CoV-1 strain 200300592 was obtained from the Centers for Disease Control and Prevention (CDC, Atlanta, GA) and passaged three times on Vero E6 cells while selecting those cultures that showed the most CPE. This resulted in a virus stock (passage 3) that confers full CPE on Vero E6 cells (3.4 × 107 TCID50 per mL). For antiviral testing, Vero E6 cells were seeded in 96-well plates (Corning CellBIND 96-well Microplate catalog no. 3300) at a density of 8000 cells per well in assay medium. After overnight growth, cells were treated with the indicated compound concentrations and infected with a multiplicity of infection of 0.02 TCID50 per cell of the P3 virus (final volume of 200 μL per well in assay medium). On day 4 post infection, differences in cell viability caused by virus-induced CPE or by compound-specific side effects were analyzed using MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium, inner salt). For this, an MTS:phenazine methosulfate (PMS) stock solution (2 mg mL–1 MTS (Promega) and 46 μg mL–1 PMS (Sigma-Aldrich) in PBS at pH 6–6.5) was diluted 1:20 in MEM without phenol red (Gibco catalog no. 51200038). Medium was aspirated from wells of the test plates and 70 μL of MTS/PMS solution was added. After 0.5–1 h incubation at 37 °C, absorbance was measured at 498 nm. Cytotoxic effects caused by compound treatment alone were monitored in parallel plates containing mock-infected cells. These experiments were performed in the high-containment BSL3+ facilities of the KU Leuven Rega Institute (3CAPS) under licenses AMV 30112018 SBB 219 2018 0892 and AMV 23102017 SBB 219 2017 0589 according to institutional guidelines.
MERS-CoV Antiviral Activity in Huh7 Cell Culture
Huh7 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Lonza) with 8% fetal calf serum (Bodinco), 2 mM l-glutamine (Sigma-Aldrich), 100 IU/mL penicillin, and 100 g/mL streptomycin (Sigma-Aldrich). After infection, cells were kept in Eagle’s minimal essential medium (EMEM; Lonza) with 25 mM HEPES (Lonza), 2% FCS, l-glutamine, and antibiotics. MERS-CoV-Jordan-N3/2012 (Genbank accession no. KC776174) stocks were grown on Vero E6 cells, and titers were determined by plaque assay on the same cells. All experiments with infectious MERS-CoV were performed at the LUMC biosafety level 3 facilities.
MERS-CoV CPE-Based Antiviral Assay
Huh7 cells (1.5 × 104 cells/well) were seeded in 96-well plates 1 day before infection. Cells were incubated with 100 μL volumes of 2-fold serial dilutions of compound in infection medium, followed by infection with 225 PFU of MERS-CoV in 50 μL, yielding a total assay volume of 150 μL (highest compound concentration 2 μM). In parallel, noninfected cells in the same plate were treated with the same dilution series of compound to assess cytotoxicity. Plates were incubated for 42 h at 37 °C, after which cell viability was quantified with the CellTiter-96 aqueous nonradioactive cell proliferation kit (Promega). After incubation for ∼1.5 h, absorption at 495 nm was measured with an EnVision multilabel plate reader (PerkinElmer). Cell viability was normalized against the readings for noninfected untreated cells (100%). EC50 (compound concentration that reduces virus-induced cell death by 50%) values were determined by nonlinear regression using GraphPad Prism v8.0. Experiments, consisting of biological quadruplicates, were repeated twice.
Metabolic Stability in the Presence of Human Liver Microsomes
Metabolic stability was determined in 0.5 mg/mL human liver microsomes at a compound concentration of 1 μM in 100 mM KPO4 buffer pH 7.4 in a total incubation volume of 500 μL. The reaction was initiated by addition of 1 mM NADPH. At various incubation times, i.e., at 0, 5, 10, 20, 40, and 60 min, a sample was withdrawn from the incubation, and the reaction was terminated by addition of cold acetonitrile with warfarin as an internal standard. The amount of parent compound remaining was analyzed by liquid chromatography coupled to triple quadrupole mass spectrometry (LC–MS/MS).
Plasma Protein Binding and Plasma Stability in Human Plasma
Pooled human plasma was provided by Uppsala Academic Hospital and was collected from two donors (nonsmoking) (citric acid). In brief, 0.2 mL of the plasma (50% plasma, 50% isotonic buffer) test solution (typically 10 μM final compound concentration) was transferred to the membrane tube in the RED insert (ThermoFisher Scientific). Then 0.35 mL of isotonic phosphate buffer pH 7.4 was added to the other side of the membrane. The 96-well base plate was then sealed with an adhesive plastic film (Scotch Pad) to prevent evaporation. The sample was incubated with rapid rotation (≫900 rpm) on a Kisker rotational incubator at 37 °C for 4 h to achieve equilibrium. Prior to LC–MS/MS analysis, the plasma and buffer sample were treated by the addition of methanol (1:3) containing warfarin as the internal standard to precipitate proteins. The standard curve was created using the plasma standard. The plate was then sealed and centrifuged, and the supernatant was analyzed by LC−MS/MS.
Caco-2 Cell Permeability Assay
Caco-2 cell monolayers (passage 94–105) were grown on permeable filter support and used for transport study on day 21 after seeding. Prior to the experiment a drug solution of 10 μM was prepared and warmed to 37 °C. The Caco-2 filters were washed with prewarmed HBSS prior to the experiment, and thereafter, the experiment was started by applying the donor solution on the apical or basolateral side. The transport experiments were carried out at pH 7.4 in both the apical and basolateral chamber. The experiments were performed at 37 °C and with a stirring rate of 500 rpm. The receiver compartment was sampled at 15, 30, and 60 min and at 60 min also a final sample from the donor chamber was taken in order to calculate the mass balance of the compound. The samples (100 μL) were transferred to a 96-well plate containing 100 μL of methanol and warfarin as IS and was sealed until analysis by LC−MS/MS.
Liquid Chromatography Coupled to Triple Quadrupole Mass Spectrometry (LC–MS/MS)
The test compounds were optimized on a Waters Acquity UPLC XEVO TQ-S micro system (Waters Corp.) operating in multiple reaction monitoring (MRM) mode with positive or negative electrospray ionization. Compounds were optimized by using the QuanOptimize software (Waters Corp.). The MS conditions listed in Table 2 were used.
Table 2. MS Conditions.
| transition m/z | dwell time (s) | cone voltage | collision energy |
|---|---|---|---|
| 324.4 > 128.04 | 0.028 | 10 | 60 |
| 324.4 > 171.03 | 0.028 | 10 | 50 |
For chromatographic separation, a C18 BEH 1.7 μm column was used with a general gradient of 5–1000% of mobile phase B over a total running time of 2 min. Mobile phase A consisted of 0.1% formic acid in purified water and mobile phase B of 0.1% formic acid in 100% acetonitrile. The flow rate was set to 0.5 mL/min, and 5 μL of the sample was injected.
Acknowledgments
The computations were enabled by resources provided by the Swedish National Infrastructure for Computing (SNIC) at NSC and UPPMAX, partially funded by the Swedish Research Council through grant agreement no. 2018-05973. The study was facilitated by the Protein Science Facility (Emila Strandback) at Karolinska Institutet and the Compound Center (Åsa Slevin) at the Chemical Biology Consortium Sweden (CBCS) at SciLifeLab. A.L. and J.C. thank Enamine Ltd. for sharing the REAL space database and support in supplying chemicals. The authors thank the staff of the BioMAX beamline at the MAX IV laboratory for the excellent support with diffraction data collection and operation of the equipment. The authors thank the Lund Protein Production Platform (Lund University) for providing access to crystallization equipment and Cytiva (Uppsala, Sweden) for providing a Biacore S200 instrument. The authors thank the Uppsala University Drug Optimization and Pharmaceutical Profiling Platform (UDOPP) for determination of in vitro pharmacokinetic properties. This study made use of the NMR Uppsala infrastructure, which is funded by the Department of Chemistry–BMC and the Disciplinary Domain of Medicine and Pharmacy. L.W.K.M. thanks the Uppsala Antibiotic Centre. L.W.K.M. and A.C. thank the Carl Tryggers Stiftelse för Vetenskaplig Forskning. The authors thank Petra Lukacik, Frank von Delft, and Martin Walsh (Diamond Light Source, UK) for sharing of protein and protocols. Peter Sjö at the Drugs for Neglected Diseases initiative (DNDi, Switzerland) is acknowledged for insightful discussions and for providing reference samples.
Supporting Information Available
The Supporting Information is available free of charge at https://pubs.acs.org/doi/10.1021/jacs.1c08402.
Crystallographic data collection and refinement statistics, physicochemical properties of compounds, counter screen data, dose–response curves, SPR sensorgrams, biological evaluation of key compounds, results from docking to Mpro mutants, selectivity of compound 19, experimental details and characterization (NMR) of synthesized compounds (PDF)
Chemical structures, SMILES, chemical identifiers, inhibition assay data, binding assay data of compounds (XLSX)
Author Contributions
‡ H.G., E.A., D.D.V., and D.A. contributed equally. A.L. and J.C. wrote the manuscript with input from the other authors. All authors have given approval to the final version of the manuscript.
This study was supported by grants from the SciLifeLab National COVID-19 Research Program, financed by the Knut and Alice Wallenberg Foundation (2020.0182). The COVID-19 research program funded the projects NEVERMORE COVID and FRAGMENT2DRUG, which were supported by the Drug Discovery and Development platform at SciLifeLab. J.C. received funding from the European Research Council (ERC) under the European Union’s Horizon 2020 research and innovation program (Grant Agreement 715052) and the Swedish Research Council (2021-03464). The research conducted at MAXIV was supported by the Swedish Research Council (2018-07152 and 2018-06454), the Swedish Governmental Agency for Innovation Systems (2018-04969), and Formas (2019-02496). Part of this research work was performed using the “Caps-It” research infrastructure (project ZW13-02) that was financially supported by the Hercules Foundation and Rega Foundation, KU Leuven. This project has received funding from the Covid-19-Fund KU Leuven/UZ Leuven and the COVID-19 call of FWO (G0G4820N).
The authors declare the following competing financial interest(s): A.L., D.D.V., and J.C. have submitted a patent application on the compounds.
Notes
Crystal structures have been deposited in the PDB with accession codes: 7B2U, 7AU4, 7B2J, 7B5Z, 7B77, 7BIJ, 7NEO, 7O46, 7QBB, and 7NBT.
Supplementary Material
References
- WHO. https://www.who.int/emergencies/diseases/novel-coronavirus-2019/situation-reports (accessed 2021-11-15).
- Li Y.; Tenchov R.; Smoot J.; Liu C.; Watkins S.; Zhou Q. A Comprehensive Review of the Global Efforts on COVID-19 Vaccine Development. ACS Cent. Sci. 2021, 7, 512–533. 10.1021/acscentsci.1c00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siemieniuk R. A.; Bartoszko J. J.; Ge L.; Zeraatkar D.; Izcovich A.; Kum E.; Pardo-Hernandez H.; Qasim A.; Martinez J. P. D.; Rochwerg B.; Lamontagne F.; Han M. A.; Liu Q.; Agarwal A.; Agoritsas T.; Chu D. K.; Couban R.; Cusano E.; Darzi A.; Devji T.; Fang B.; Fang C.; Flottorp S. A.; Foroutan F.; Ghadimi M.; Heels-Ansdell D.; Honarmand K.; Hou L.; Hou X.; Ibrahim Q.; Khamis A.; Lam B.; Loeb M.; Marcucci M.; McLeod S. L.; Motaghi S.; Murthy S.; Mustafa R. A.; Neary J. D.; Rada G.; Riaz I. B.; Sadeghirad B.; Sekercioglu N.; Sheng L.; Sreekanta A.; Switzer C.; Tendal B.; Thabane L.; Tomlinson G.; Turner T.; Vandvik P. O.; Vernooij R. W.; Viteri-García A.; Wang Y.; Yao L.; Ye Z.; Guyatt G. H.; Brignardello-Petersen R. Drug Treatments for Covid-19: Living Systematic Review and Network Meta-Analysis. BMJ. 2020, 370, m2980. 10.1136/bmj.m2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan H.; Peto R.; Henao-Restrepo A.-M.; Preziosi M.-P.; Sathiyamoorthy V.; Abdool Karim Q.; Alejandria M. M.; Hernández García C.; Kieny M.-P.; Malekzadeh R.; Murthy S.; Reddy K. S.; Roses Periago M.; Abi Hanna P.; Ader F.; Al-Bader A. M.; Alhasawi A.; Allum E.; Alotaibi A.; Alvarez-Moreno C. A.; Appadoo S.; Asiri A.; Aukrust P.; Barratt-Due A.; Bellani S.; Branca M.; Cappel-Porter H. B. C.; Cerrato N.; Chow T. S.; Como N.; Eustace J.; García P. J.; Godbole S.; Gotuzzo E.; Griskevicius L.; Hamra R.; Hassan M.; Hassany M.; Hutton D.; Irmansyah I.; Jancoriene L.; Kirwan J.; Kumar S.; Lennon P.; Lopardo G.; Lydon P.; Magrini N.; Maguire T.; Manevska S.; Manuel O.; McGinty S.; Medina M. T.; Mesa Rubio M. L.; Miranda-Montoya M. C.; Nel J.; Nunes E. P.; Perola M.; Portolés A.; Rasmin M. R.; Raza A.; Rees H.; Reges P. P. S.; Rogers C. A.; Salami K.; Salvadori M. I.; Sinani N.; Sterne J. A. C.; Stevanovikj M.; Tacconelli E.; Tikkinen K. A. O.; Trelle S.; Zaid H.; Røttingen J.-A.; Swaminathan S. Repurposed Antiviral Drugs for Covid-19 - Interim WHO Solidarity Trial Results. N. Engl. J. Med. 2021, 384, 497–511. 10.1056/NEJMoa2023184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannalire R.; Cerchia C.; Beccari A. R.; Di Leva F. S.; Summa V. Targeting SARS-CoV-2 Proteases and Polymerase for COVID-19 Treatment: State of the Art and Future Opportunities. J. Med. Chem. 2020, 10.1021/acs.jmedchem.0c01140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. The Design of Drugs for HIV and HCV. Nat. Rev. Drug Discovery 2007, 6, 1001–1018. 10.1038/nrd2424. [DOI] [PubMed] [Google Scholar]
- Vuong W.; Khan M. B.; Fischer C.; Arutyunova E.; Lamer T.; Shields J.; Saffran H. A.; McKay R. T.; van Belkum M. J.; Joyce M. A.; Young H. S.; Tyrrell D. L.; Vederas J. C.; Lemieux M. J. Feline Coronavirus Drug Inhibits the Main Protease of SARS-CoV-2 and Blocks Virus Replication. Nat. Commun. 2020, 11, 4282. 10.1038/s41467-020-18096-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs J.; Grum-Tokars V.; Zhou Y.; Turlington M.; Adrian Saldanha S.; Chase P.; Eggler A.; Dawson E. S.; Baez-Santos Y. M.; Tomar S.; Mielech A. M.; Baker S. C.; Lindsley C. W.; Hodder P.; Mesecar A.; Stauffer S. R. Discovery, Synthesis, And Structure-Based Optimization of a Series of N-(tert-Butyl)-2-(N-Arylamido)-2-(pyridine-3-yl) Acetamides (ML188) as Potent Noncovalent Small Molecule Inhibitors of the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 3CL Protease. J. Med. Chem. 2013, 56, 534–546. 10.1021/jm301580n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y.; Liang C.; Xin L.; Ren X.; Tian L.; Ju X.; Li H.; Wang Y.; Zhao Q.; Liu H.; Cao W.; Xie X.; Zhang D.; Wang Y.; Jian Y. The Development of Coronavirus 3C-Like Protease (3CL) Inhibitors from 2010 to 2020. Eur. J. Med. Chem. 2020, 206, 112711. 10.1016/j.ejmech.2020.112711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faucher F.; Bennett J. M.; Bogyo M.; Lovell S. Strategies for Tuning the Selectivity of Chemical Probes That Target Serine Hydrolases. Cell. Chem. Biol. 2020, 27, 937–952. 10.1016/j.chembiol.2020.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yao J.-F.; Yang H.; Zhao Y.-Z.; Xue M. Metabolism of Peptide Drugs and Strategies to Improve Their Metabolic Stability. Curr. Drug Metab. 2018, 19, 892–901. 10.2174/1389200219666180628171531. [DOI] [PubMed] [Google Scholar]
- Lenci E.; Trabocchi A. Peptidomimetic Toolbox for Drug Discovery. Chem. Soc. Rev. 2020, 49, 3262–3277. 10.1039/D0CS00102C. [DOI] [PubMed] [Google Scholar]
- Grygorenko O. O.; Radchenko D. S.; Dziuba I.; Chuprina A.; Gubina K. E.; Moroz Y. S. Generating Multibillion Chemical Space of Readily Accessible Screening Compounds. iScience 2020, 23, 101681. 10.1016/j.isci.2020.101681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lynch M. L.; Snell E. H.; Bowman S. E. J. Structural Biology in the Time of COVID-19: Perspectives on Methods and Milestones. IUCrJ. 2021, 8, 335–341. 10.1107/S2052252521003948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irwin J. J.; Shoichet B. K. Docking Screens for Novel Ligands Conferring New Biology. J. Med. Chem. 2016, 59, 4103–4120. 10.1021/acs.jmedchem.5b02008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Graaf C.; Kooistra A. J.; Vischer H. F.; Katritch V.; Kuijer M.; Shiroishi M.; Iwata S.; Shimamura T.; Stevens R. C.; de Esch I. J. P.; Leurs R. Crystal Structure-Based Virtual Screening for Fragment-like Ligands of the Human Histamine H1 Receptor. J. Med. Chem. 2011, 54, 8195–8206. 10.1021/jm2011589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lyu J.; Wang S.; Balius T. E.; Singh I.; Levit A.; Moroz Y. S.; O’Meara M. J.; Che T.; Algaa E.; Tolmachova K.; Tolmachev A. A.; Shoichet B. K.; Roth B. L.; Irwin J. J. Ultra-Large Library Docking for Discovering New Chemotypes. Nature 2019, 566, 224–229. 10.1038/s41586-019-0917-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein R. M.; Kang H. J.; McCorvy J. D.; Glatfelter G. C.; Jones A. J.; Che T.; Slocum S.; Huang X.-P.; Savych O.; Moroz Y. S.; Stauch B.; Johansson L. C.; Cherezov V.; Kenakin T.; Irwin J. J.; Shoichet B. K.; Roth B. L.; Dubocovich M. L. Virtual Discovery of Melatonin Receptor Ligands to Modulate Circadian Rhythms. Nature 2020, 579, 609–614. 10.1038/s41586-020-2027-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgulla C.; Padmanabha Das K. M.; Leigh K. E.; Cespugli M.; Fischer P. D.; Wang Z.-F.; Tesseyre G.; Pandita S.; Shnapir A.; Calderaio A.; Gechev M.; Rose A.; Lewis N.; Hutcheson C.; Yaffe E.; Luxenburg R.; Herce H. D.; Durmaz V.; Halazonetis T. D.; Fackeldey K.; Patten J. J.; Chuprina A.; Dziuba I.; Plekhova A.; Moroz Y.; Radchenko D.; Tarkhanova O.; Yavnyuk I.; Gruber C.; Yust R.; Payne D.; Näär A. M.; Namchuk M. N.; Davey R. A.; Wagner G.; Kinney J.; Arthanari H. A Multi-Pronged Approach Targeting SARS-CoV-2 Proteins Using Ultra-Large Virtual Screening. iScience 2021, 24, 102021. 10.1016/j.isci.2020.102021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ton A.-T.; Gentile F.; Hsing M.; Ban F.; Cherkasov A. Rapid Identification of Potential Inhibitors of SARS-CoV-2 Main Protease by Deep Docking of 1.3 Billion Compounds. Mol. Inform. 2020, 39, e2000028 10.1002/minf.202000028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clyde A.; Galanie S.; Kneller D. W.; Ma H.; Babuji Y.; Blaiszik B.; Brace A.; Brettin T.; Chard K.; Chard R.; Coates L.; Foster I.; Hauner D.; Kertesz V.; Kumar N.; Lee H.; Li Z.; Merzky A.; Schmidt J. G.; Tan L.; Titov M.; Trifan A.; Turilli M.; Van Dam H.; Chennubhotla S. C.; Jha S.; Kovalevsky A.; Ramanathan A.; Head M. S.; Stevens R. High Throughput Virtual Screening and Validation of a SARS-CoV-2 Main Protease Non-Covalent Inhibitor. BioRxiv 2021, 10.1101/2021.03.27.437323. [DOI] [PubMed] [Google Scholar]
- Schuller M.; Correy G. J.; Gahbauer S.; Fearon D.; Wu T.; Díaz R. E.; Young I. D.; Carvalho Martins L.; Smith D. H.; Schulze-Gahmen U.; Owens T. W.; Deshpande I.; Merz G. E.; Thwin A. C.; Biel J. T.; Peters J. K.; Moritz M.; Herrera N.; Kratochvil H. T.; Aimon A.; Bennett J. M.; Brandao Neto J.; Cohen A. E.; Dias A.; Douangamath A.; Dunnett L.; Fedorov O.; Ferla M. P.; Fuchs M. R.; Gorrie-Stone T. J.; Holton J. M.; Johnson M. G.; Krojer T.; Meigs G.; Powell A. J.; Rack J. G. M.; Rangel V. L.; Russi S.; Skyner R. E.; Smith C. A.; Soares A. S.; Wierman J. L.; Zhu K.; O’Brien P.; Jura N.; Ashworth A.; Irwin J. J.; Thompson M. C.; Gestwicki J. E.; von Delft F.; Shoichet B. K.; Fraser J. S.; Ahel I. Fragment Binding to the Nsp3 Macrodomain of SARS-CoV-2 Identified through Crystallographic Screening and Computational Docking. Sci. Adv. 2021, 7, eabf8711. 10.1126/sciadv.abf8711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douangamath A.; Fearon D.; Gehrtz P.; Krojer T.; Lukacik P.; Owen C. D.; Resnick E.; Strain-Damerell C.; Aimon A.; Ábrányi-Balogh P.; Brandão-Neto J.; Carbery A.; Davison G.; Dias A.; Downes T. D.; Dunnett L.; Fairhead M.; Firth J. D.; Jones S. P.; Keeley A.; Keserü G. M.; Klein H. F.; Martin M. P.; Noble M. E. M.; O’Brien P.; Powell A.; Reddi R. N.; Skyner R.; Snee M.; Waring M. J.; Wild C.; London N.; von Delft F.; Walsh M. A. Crystallographic and Electrophilic Fragment Screening of the SARS-CoV-2 Main Protease. Nat. Commun. 2020, 11, 5047. 10.1038/s41467-020-18709-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen D. R.; Allerton C. M. N.; Anderson A. S.; Aschenbrenner L.; Avery M.; Berritt S.; Boras B.; Cardin R. D.; Carlo A.; Coffman K. J.; Dantonio A.; Di L.; Eng H.; Ferre R.; Gajiwala K. S.; Gibson S. A.; Greasley S. E.; Hurst B. L.; Kadar E. P.; Kalgutkar A. S.; Lee J. C.; Lee J.; Liu W.; Mason S. W.; Noell S.; Novak J. J.; Obach R. S.; Ogilvie K.; Patel N. C.; Pettersson M.; Rai D. K.; Reese M. R.; Sammons M. F.; Sathish J. G.; Singh R. S. P.; Steppan C. M.; Stewart A. E.; Tuttle J. B.; Updyke L.; Verhoest P. R.; Wei L.; Yang Q.; Zhu Y. An Oral SARS-CoV-2 Mpro Inhibitor Clinical Candidate for the Treatment of COVID-19. Science 2021, eabl4784. 10.1126/science.abl4784. [DOI] [PubMed] [Google Scholar]
- Mesecar A. D.www.rcsb.org/structure/6W63. (accessed 2020-04-15).
- Irwin J. J.; Tang K. G.; Young J.; Dandarchuluun C.; Wong B. R.; Khurelbaatar M.; Moroz Y. S.; Mayfield J.; Sayle R. A. ZINC20-A Free Ultralarge-Scale Chemical Database for Ligand Discovery. J. Chem. Inf. Model. 2020, 60, 6065–6073. 10.1021/acs.jcim.0c00675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman R. G.; Carchia M.; Sterling T.; Irwin J. J.; Shoichet B. K. Ligand Pose and Orientational Sampling in Molecular Docking. PLoS One 2013, 8, e75992 10.1371/journal.pone.0075992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischer A.; Smieško M.; Sellner M.; Lill M. A. Decision Making in Structure-Based Drug Discovery: Visual Inspection of Docking Results. J. Med. Chem. 2021, 64, 2489–2500. 10.1021/acs.jmedchem.0c02227. [DOI] [PubMed] [Google Scholar]
- Open Science Discovery of Oral Non-Covalent SARS-CoV-2 Main Protease Inhibitors. ChemRxiv 2021, 10.33774/chemrxiv-2021-585ks-v2. [DOI] [Google Scholar]
- Hopkins A. L.; Keserü G. M.; Leeson P. D.; Rees D. C.; Reynolds C. H. The Role of Ligand Efficiency Metrics in Drug Discovery. Nat. Rev. Drug Discovery 2014, 13, 105–121. 10.1038/nrd4163. [DOI] [PubMed] [Google Scholar]
- Johnson T. W.; Gallego R. A.; Edwards M. P. Lipophilic Efficiency as an Important Metric in Drug Design. J. Med. Chem. 2018, 61, 6401–6420. 10.1021/acs.jmedchem.8b00077. [DOI] [PubMed] [Google Scholar]
- Dahlin J. L.; Nissink J. W. M.; Strasser J. M.; Francis S.; Higgins L.; Zhou H.; Zhang Z.; Walters M. A. PAINS in the Assay: Chemical Mechanisms of Assay Interference and Promiscuous Enzymatic Inhibition Observed during a Sulfhydryl-Scavenging HTS. J. Med. Chem. 2015, 58, 2091–2113. 10.1021/jm5019093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng B. Y.; Shoichet B. K. A Detergent-Based Assay for the Detection of Promiscuous Inhibitors. Nat. Protoc. 2006, 1, 550–553. 10.1038/nprot.2006.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu Y.; McCauley J. GISAID: Global Initiative on Sharing All Influenza Data - from Vision to Reality. Euro Surveill. 2017, 10.2807/1560-7917.ES.2017.22.13.30494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomar S.; Johnston M. L.; John S. E. S.; Osswald H. L.; Nyalapatla P. R.; Paul L. N.; Ghosh A. K.; Denison M. R.; Mesecar A. D. Ligand-Induced Dimerization of Middle East Respiratory Syndrome (MERS) Coronavirus Nsp5 Protease (3CLpro): Implications for Nsp5 Regulation and the Development of Antivirals. J. Biol. Chem. 2015, 290, 19403–19422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han S. H.; Goins C. M.; Arya T.; Shin W.-J.; Maw J.; Hooper A.; Sonawane D. P.; Porter M. R.; Bannister B. E.; Crouch R. D.; Abigail Lindsey A.; Lakatos G.; Martinez S. R.; Alvarado J.; Akers W. S.; Wang N. S.; Jung J. U.; Macdonald J. D.; Stauffer S. R. Structure-Based Optimization of ML300-Derived, Noncovalent Inhibitors Targeting the Severe Acute Respiratory Syndrome Coronavirus 3CL Protease (SARS-CoV-2 3CLpro). J. Med. Chem. 2021, 10.1021/acs.jmedchem.1c00598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warren G. L.; Andrews C. W.; Capelli A.-M.; Clarke B.; LaLonde J.; Lambert M. H.; Lindvall M.; Nevins N.; Semus S. F.; Senger S.; Tedesco G.; Wall I. D.; Woolven J. M.; Peishoff C. E.; Head M. S. A Critical Assessment of Docking Programs and Scoring Functions. J. Med. Chem. 2006, 49, 5912–5931. 10.1021/jm050362n. [DOI] [PubMed] [Google Scholar]
- Zhang C.-H.; Stone E. A.; Deshmukh M.; Ippolito J. A.; Ghahremanpour M. M.; Tirado-Rives J.; Spasov K. A.; Zhang S.; Takeo Y.; Kudalkar S. N.; Liang Z.; Isaacs F.; Lindenbach B.; Miller S. J.; Anderson K. S.; Jorgensen W. L. Potent Noncovalent Inhibitors of the Main Protease of SARS-CoV-2 from Molecular Sculpting of the Drug Perampanel Guided by Free Energy Perturbation Calculations. ACS Cent. Sci. 2021, 7, 467–475. 10.1021/acscentsci.1c00039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura N.; Sacco M. D.; Ma C.; Hu Y.; Townsend J. A.; Meng X.; Zhang F.; Zhang X.; Ba M.; Szeto T.; Kukuljac A.; Marty M. T.; Schultz D.; Cherry S.; Xiang Y.; Chen Y.; Wang J. Expedited Approach toward the Rational Design of Noncovalent SARS-CoV-2 Main Protease Inhibitors. J. Med. Chem. 2021, 10.1021/acs.jmedchem.1c00509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorgulla C.; Boeszoermenyi A.; Wang Z.-F.; Fischer P. D.; Coote P. W.; Padmanabha Das K. M.; Malets Y. S.; Radchenko D. S.; Moroz Y. S.; Scott D. A.; Fackeldey K.; Hoffmann M.; Iavniuk I.; Wagner G.; Arthanari H. An Open-Source Drug Discovery Platform Enables Ultra-Large Virtual Screens. Nature 2020, 580, 663–668. 10.1038/s41586-020-2117-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Word J. M.; Lovell S. C.; Richardson J. S.; Richardson D. C. Asparagine and Glutamine: Using Hydrogen Atom Contacts in the Choice of Side-Chain Amide Orientation. J. Mol. Biol. 1999, 285, 1735–1747. 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- Weiner S. J.; Kollman P. A.; Singh U. C.; Case D. A.; Ghio C.; Alagona G.; Profeta S.; Weiner P. A New Force Field for Molecular Mechanical Simulation of Nucleic Acids and Proteins. J. Am. Chem. Soc. 1984, 106, 765–784. 10.1021/ja00315a051. [DOI] [Google Scholar]
- Bender B. J.; Gahbauer S.; Luttens A.; Lyu J.; Webb C. M.; Stein R. M.; Fink E. A.; Balius T. E.; Carlsson J.; Irwin J. J.; Shoichet B. K. A Practical Guide to Large-Scale Docking. Nat. Protoc. 2021, 16, 4799–4832. 10.1038/s41596-021-00597-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaiteh M.; Zeifman A.; Saarinen M.; Svenningsson P.; Bréa J.; Loza M. I.; Carlsson J. Docking Screens for Dual Inhibitors of Disparate Drug Targets for Parkinson’s Disease. J. Med. Chem. 2018, 61, 5269–5278. 10.1021/acs.jmedchem.8b00204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudling A.; Gustafsson R.; Almlöf I.; Homan E.; Scobie M.; Warpman Berglund U.; Helleday T.; Stenmark P.; Carlsson J. Fragment-Based Discovery and Optimization of Enzyme Inhibitors by Docking of Commercial Chemical Space. J. Med. Chem. 2017, 60, 8160–8169. 10.1021/acs.jmedchem.7b01006. [DOI] [PubMed] [Google Scholar]
- Gallagher K.; Sharp K. Electrostatic Contributions to Heat Capacity Changes of DNA-Ligand Binding. Biophys. J. 1998, 75, 769–776. 10.1016/S0006-3495(98)77566-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysinger M. M.; Shoichet B. K. Rapid Context-Dependent Ligand Desolvation in Molecular Docking. J. Chem. Inf. Model. 2010, 50, 1561–1573. 10.1021/ci100214a. [DOI] [PubMed] [Google Scholar]
- Meng E. C.; Shoichet B. K.; Kuntz I. D. Automated Docking with Grid-Based Energy Evaluation. J. Comput. Chem. 1992, 13, 505–524. 10.1002/jcc.540130412. [DOI] [Google Scholar]
- Mysinger M. M.; Carchia M.; Irwin J. J.; Shoichet B. K. Directory of Useful Decoys, Enhanced (DUD-E): Better Ligands and Decoys for Better Benchmarking. J. Med. Chem. 2012, 55, 6582–6594. 10.1021/jm300687e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein R. M.; Yang Y.; Balius T. E.; O’Meara M. J.; Lyu J.; Young J.; Tang K.; Shoichet B. K.; Irwin J. J. Property-Unmatched Decoys in Docking Benchmarks. J. Chem. Inf. Model. 2021, 61, 699–714. 10.1021/acs.jcim.0c00598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sterling T.; Irwin J. J. ZINC 15 – Ligand Discovery for Everyone. J. Chem. Inf. Model. 2015, 55, 2324–2337. 10.1021/acs.jcim.5b00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baell J. B.; Holloway G. A. New Substructure Filters for Removal of Pan Assay Interference Compounds (PAINS) from Screening Libraries and for Their Exclusion in Bioassays. J. Med. Chem. 2010, 53, 2719–2740. 10.1021/jm901137j. [DOI] [PubMed] [Google Scholar]
- Landrum G.; Tosco P.; Kelley B.; sriniker; gedeck; Schneider N.; Vianello R.; Ric; Dalke A.; Cole B.; Savelyev A.; Swain M.; Turk S.; Dan N.; Vaucher A.; Kawashima E.; Wójcikowski M.; Probst D.; Godin G.; Cosgrove D.; Pahl A.; Berenger F.; Varjo J. L.; O’Boyle N.; Fuller P.; Jensen J. H.; Sforna G.; Gavid D. Rdkit/rdkit: 2019_09_03 (Q3 2019) Release. Zenodo 2020, 10.5281/zenodo.3603542. [DOI] [Google Scholar]
- Schomburg K.; Ehrlich H.-C.; Stierand K.; Rarey M. From Structure Diagrams to Visual Chemical Patterns. J. Chem. Inf. Model. 2010, 50, 1529–1535. 10.1021/ci100209a. [DOI] [PubMed] [Google Scholar]
- Webb B.; Sali A. Comparative Protein Structure Modeling Using MODELLER. Curr. Protoc. Protein Sci. 2016, 86, 2.9.1–2.9.37. [DOI] [PubMed] [Google Scholar]
- Xue X.; Yang H.; Shen W.; Zhao Q.; Li J.; Yang K.; Chen C.; Jin Y.; Bartlam M.; Rao Z. Production of Authentic SARS-CoV Mpro with Enhanced Activity: Application as a Novel Tag-Cleavage Endopeptidase for Protein Overproduction. J. Mol. Biol. 2007, 366, 965–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akaberi D.; Krambrich J.; Ling J.; Luni C.; Hedenstierna G.; Järhult J. D.; Lennerstrand J.; Lundkvist Å. Mitigation of the Replication of SARS-CoV-2 by Nitric Oxide in Vitro. Redox Biol. 2020, 37, 101734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursby T.; Åhnberg K.; Appio R.; Aurelius O.; Barczyk A.; Bartalesi A.; Bjelčić M.; Bolmsten F.; Cerenius Y.; Doak R. B.; Eguiraun M.; Eriksson T.; Friel R. J.; Gorgisyan I.; Gross A.; Haghighat V.; Hennies F.; Jagudin E.; Norsk Jensen B.; Jeppsson T.; Kloos M.; Lidon-Simon J.; de Lima G. M. A.; Lizatovic R.; Lundin M.; Milan-Otero A.; Milas M.; Nan J.; Nardella A.; Rosborg A.; Shilova A.; Shoeman R. L.; Siewert F.; Sondhauss P.; Talibov V. O.; Tarawneh H.; Thånell J.; Thunnissen M.; Unge J.; Ward C.; Gonzalez A.; Mueller U. BioMAX - the First Macromolecular Crystallography Beamline at MAX IV Laboratory. J. Synchrotron Radiat. 2020, 27, 1415–1429. 10.1107/S1600577520008723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winn; Ballard C. C.; Cowtan K. D.; Dodson E. J.; Emsley P.; Evans P. R.; Keegan R. M.; Krissinel E. B.; Leslie A. G. W.; McCoy A.; McNicholas S. J.; Murshudov G. N.; Pannu N. S.; Potterton E. A.; Powell H. R.; Read R. J.; Vagin A.; Wilson K. S. Overview of the CCP4 Suite and Current Developments. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 235–242. 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCoy A. J.; Grosse-Kunstleve R. W.; Adams P. D.; Winn M. D.; Storoni L. C.; Read R. J. Phaser Crystallographic Software. J. Appl. Crystallogr. 2007, 40, 658–674. 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabsch W. XDS.. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 125–132. 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long F.; Nicholls R. A.; Emsley P.; Graeulis S.; Merkys A.; Vaitkus A.; Murshudov G. N. AceDRG: A Stereochemical Description Generator for Ligands. Acta Crystallogr. D Struct. Biol. 2017, 73, 112–122. 10.1107/S2059798317000067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P.; Lohkamp B.; Scott W. G.; Cowtan K. Features and Development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murshudov G. N.; Skubák P.; Lebedev A. A.; Pannu N. S.; Steiner R. A.; Nicholls R. A.; Winn M. D.; Long F.; Vagin A. A. REFMAC5 for the Refinement of Macromolecular Crystal Structures. Acta Crystallogr. D Biol. Crystallogr. 2011, 67, 355–367. 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams C. J.; Headd J. J.; Moriarty N. W.; Prisant M. G.; Videau L. L.; Deis L. N.; Verma V.; Keedy D. A.; Hintze B. J.; Chen V. B.; Jain S.; Lewis S. M.; Arendall W. B. 3rd; Snoeyink J.; Adams P. D.; Lovell S. C.; Richardson J. S.; Richardson D. C. MolProbity: More and Better Reference Data for Improved All-Atom Structure Validation. Protein Sci. 2018, 27, 293–315. 10.1002/pro.3330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boudewijns R.; Thibaut H. J.; Kaptein S. J. F.; Li R.; Vergote V.; Seldeslachts L.; Van Weyenbergh J.; De Keyzer C.; Bervoets L.; Sharma S.; Liesenborghs L.; Ma J.; Jansen S.; Van Looveren D.; Vercruysse T.; Wang X.; Jochmans D.; Martens E.; Roose K.; De Vlieger D.; Schepens B.; Van Buyten T.; Jacobs S.; Liu Y.; Martí-Carreras J.; Vanmechelen B.; Wawina-Bokalanga T.; Delang L.; Rocha-Pereira J.; Coelmont L.; Chiu W.; Leyssen P.; Heylen E.; Schols D.; Wang L.; Close L.; Matthijnssens J.; Van Ranst M.; Compernolle V.; Schramm G.; Van Laere K.; Saelens X.; Callewaert N.; Opdenakker G.; Maes P.; Weynand B.; Cawthorne C.; Vande Velde G.; Wang Z.; Neyts J.; Dallmeier K. STAT2 Signaling Restricts Viral Dissemination but Drives Severe Pneumonia in SARS-CoV-2 Infected Hamsters. Nat. Commun. 2020, 11, 5838. 10.1038/s41467-020-19684-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nissen K.; Hagbom M.; Krambrich J.; Akaberi D.; Sharma S.; Ling J.; Hoffman T.; Svensson L.; Bondeson K.; Salaneck E. Presymptomatic Viral Shedding and Infective Ability of SARS-CoV-2; a Case Report. Heliyon 2021, 7, e06328 10.1016/j.heliyon.2021.e06328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corman V. M.; Landt O.; Kaiser M.; Molenkamp R.; Meijer A.; Chu D. K. W.; Bleicker T.; Brünink S.; Schneider J.; Schmidt M. L.; Mulders D. G.; Haagmans B. L.; van der Veer B.; van den Brink S.; Wijsman L.; Goderski G.; Romette J.-L.; Ellis J.; Zambon M.; Peiris M.; Goossens H.; Reusken C.; Koopmans M. P. G.; Drosten C. Detection of 2019 Novel Coronavirus (2019-nCoV) by Real-Time RT-PCR. Euro Surveill. 2020, 10.2807/1560-7917.ES.2020.25.3.2000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.