Abstract
Tracking severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) variants through whole genome sequencing (WGS) is vital for effective infection control and prevention (IPC) measures, but can be time-consuming and resource-heavy. We describe an in-house validation of an allele-specific polymerase chain reaction (ASP) variant assay to detect variants of concern (VOC). ASP sensitivity for detecting Delta, Alpha and Beta was 99.45 %, 100 %, and 66.67 %, respectively, compared with WGS. Specificity was 100 % in detecting all three VOC. ASP generated results 1.3 days faster compared with WGS. These findings suggest using variant assays such as ASP may enhance epidemiological surveillance and IPC measures.
Keywords: Allele-specific PCR, Variant assay, Variants of concerns, Whole genome sequencing, SARS-CoV-2
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) continues to pose a global threat despite recent advances in diagnostics, therapeutics and the development of effective vaccines, mainly due to the emergence of new variants of concern (VOC).(Twohig et al., 2021; “Variants: distribution of case data, 17 September 2021 - GOV.UK,” n.d.) At the time of this study Delta (B.1.617.2 lineage, VOC-21-APR-02) was the predominant variant in the United Kingdom, due to its high transmissibility, and had been associated with increased hospital admissions compared with Alpha (B.1.1.7; VOC-20DEC-01). (Twohig et al., 2021; “Variants: distribution of case data, 17 September 2021 - GOV.UK,” n.d.)
Whole genome sequencing (WGS) of SARS-CoV-2 has been essential in detecting and monitoring these variants, and has been utilised to guide infection prevention and control (IPC) practices in real time, particularly in nosocomial settings.(LW et al., 2020) However, there are limitations to WGS. In Scotland there are only few reference laboratories that have sequencing capabilities, which are performed in partnership with the COVID-19 Genomics UK consortium.(“COVID-19 Genomics UK Consortium,” n.d.) Depending on the sequencing method, WGS can be quite resource-heavy and time-consuming, and the turnaround time reported in the literature varies from hours to weeks.(Baker et al., 2021; Funk et al., 2021; LW, M. et al., 2020) Another challenge is poor quality genomic data from samples with low viral loads, which are generally those with reverse-transcriptase polymerase chain reaction (RT-PCR) CT values >32, depending on the specific RT-PCR assay used and the sequencing method utilised.(Baker et al., 2021; Lam et al., 2021; LW et al., 2020) These limitations led us to investigate alternate methods for identifying variants in the general population.
This study investigated the accuracy of allele-specific PCR (ASP) for detection of SARS-CoV-2 variants and evaluated how it could be used in conjunction with WGS to monitor SARS-CoV-2 infection in the Viral Genotyping Reference Laboratory based in the Royal Infirmary of Edinburgh, NHS Lothian which provides sequencing services for the east of Scotland and the Scottish Islands.
WGS and ASP was performed on total nucleic acid extracted by Biomerieux Easymag from SARS-CoV-2 positive nasopharyngeal samples collected from May–July 2021. The assays used to identify these positive samples were Cepheid’sGeneXpert®, Seegene NIMBUS, Thermo Fisher Applied Biosystems™ 7500, GenMark ePlex® and Alinity m Abbott.
SARS-CoV-2 samples with CT values ≤30 were sequenced using the Artic LoCost V3 Nanopore method. (https://artic.network/resources/ncov/ncov-amplicon-v3.pdf). For ASP the customizable Thermo Fisher Applied Biosystems™ TaqMan™ SARS-CoV-2 Mutation Assay was used, which contains sequence-specific forward and reverse primers to amplify the target sequence region. The three mutation targets chosen were E484 K, L452R and P681R, based on the circulating variant epidemiology at the time. (https://www.thermofisher.com/uk/en/home/clinical/clinical-genomics/pathogen-detection-solutions/real-time-pcr-research-solutions-sars-cov-2/mutation-panel.html) In combination, detection of these mutations allows for identification of SARS-CoV-2 VOC/variants under investigation (VUI), see Table 1 .
Table 1.
Variants and associated mutations.
| Lineage | VOC | Word Health Organization | E484 | L452 | P681 |
|---|---|---|---|---|---|
| B.1.1.7 | VOC- 20DEC-01 | Alpha | E | L | H |
| B.1.351 | VOC- 20DEC-02 | Beta | K | L | P |
| B.1.617.2 | VOC- 21APR-02 | Delta | E | R | R |
The runs were performed on the ABI 7500 instrument and data analysed using Thermo Fisher QuantStudio Design and Analysis Software v2.5. A total of three sample controls were included in every run: a wild-type strain positive control, a mutant strain positive control, and a negative control. The wild-type and mutant controls were either obtained commercially or were clinical isolates which had been previously characterised using WGS. For the run to be valid all three had to give the expected result as determined by the Thermo Fisher QuantStudio Design and Analysis Software v2.5 using a 95 % confidence limit. The limit of detection was comparable with diagnostic SARS-CoV-2 assays with limit of detection of approximately 0/25 copies/μL of a quantified SARS-CoV-2 viral lysate in all three targets (see Table 2 ).
Table 2.
Limit of Detection of the three targets determined using a quantified SARS-CoV-2 viral lysate.
| Copies/μl | E484K |
P681R |
L452R |
|||
|---|---|---|---|---|---|---|
| Replicate 1 (CT value) |
Replicate 2 (CT value) |
Replicate 1 (CT value) |
Replicate 2 (CT value) |
Replicate 1 (CT value) |
Replicate 2 (CT value) |
|
| 250 | 30.85 | 29.98 | 30.53 | 30.72 | 30.31 | 30.7 |
| 25 | 34.14 | 32.65 | 33.25 | 33.45 | 34.19 | 34.58 |
| 2.5 | 38.25 | 37.23 | 37.49 | 37.14 | 38 | 38.41 |
| 0.25 | 41.05 | 40.2 | 40.86 | – | 41.09 | – |
| 0.025 | – | – | – | – | – | – |
Turnaround times (TAT) for both ASP and WGS were calculated as the number of days between the date when samples were booked into the laboratory to the date the results were authorized. CT value comparisons were only performed on NHS Lothian samples as we did not have access to the CT values of samples from outwith Lothian and had requested that samples sent to us had CT values ≤30. CT analysis was also based on a single CT target value (E target) for each sample, therefore samples that were detected using RT-PCR assays that do not test for E target were not included in the analysis.
All statistical analyses were carried out using GraphPad Prism 9.2.0. 95 % Confidence Intervals (CI) were computed using the hybrid Wilson/Brown method. CT value comparisons were analysed using the Mann-Whitney test. P < 0.05 was considered statistically significant.
We first looked at the sensitivity and specificity of ASP in detecting VOC compared with WGS, the gold standard approach. Of a total of 969 samples taken over the time period, 653 were included in this analysis, after excluding samples with no sequencing data. The main VOCs identified through WGS were Delta (n = 545), Alpha (n = 102) and Beta (n = 3), with 3 samples having lineages not associated with known VOCs. There were no discordant results between ASP and WGS; however, there were 4 samples that ASP was unable to identify. WGS identified 3 of these samples as Delta, and 1 as Beta.
The sensitivity of ASP in detecting Delta was 99.45 % (95 % CI 98.39–99.85), Alpha, 100 % (95 % CI 96.37–100) and Beta, 66.67 % (95 % CI 11.85–98.29) respectively compared to WGS. The specificity was 100 % in detecting all three VOC (95 % CI for Delta: 96.57–100; Alpha: 99.31–100; and Beta: 99.41–100).
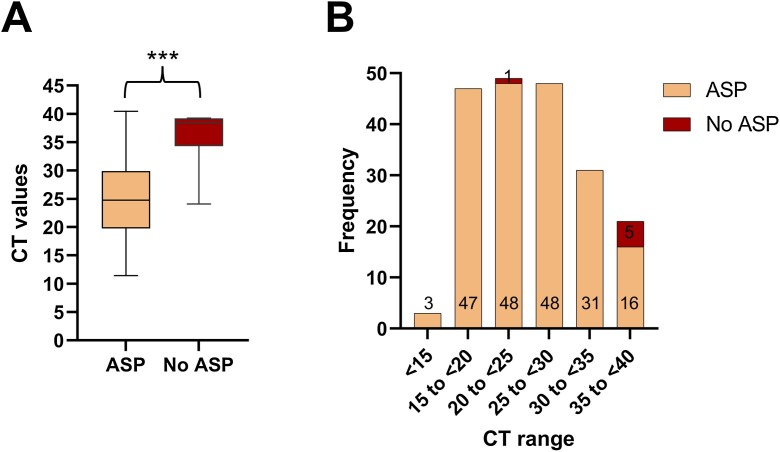
When validating WGS in Edinburgh we found that it did not generate good coverage throughout the genome in samples with CT > 30 (data not shown), thus we did not routinely sequence these samples. ASP provided VOC/VUI results in 121 samples where sequencing was not attempted due to high CT values. Samples that failed to generate ASP data had a statistically significant higher median CT value, compared with samples that had ASP data (38.43, n = 6 versus 24.76, n = 194; p=<0.001, see Fig. 1 A). However, there didn’t appear to be a threshold CT value that did not provide ASP data, as we were still able to generate an ASP profile in 16 samples where CT > 35, see Fig. 1B. We were also able to obtain ASP data on samples where WGS failed to generate results due to either low genome coverage (n = 49) or run failure (n = 48).
Fig. 1.
A: Box plot demonstrating median CT values for samples with ASP data and for samples with no ASP data, *** p=<0.001.B: CT ranges for samples that have ASP data and samples with no ASP data.
TAT from receiving samples to releasing variant information was 1.3 days faster for ASP than WGS (5 days versus 6.3 days respectively).
Comparison of results generated from WGS and ASP demonstrated that ASP is >99 % accurate at detecting Delta and Alpha. The sensitivity of ASP in detecting Beta was reduced as we only had 3 samples available for testing within the study timeframe. There were 4 samples that ASP was unable to identify. Of these samples, 2 had relatively low CT values and 2 were from outwith Lothian, and so had CT ≤ 30. The samples were in different ASP runs and the ASP runs themselves did not fail. This suggests technical errors with these specific samples were likely responsible for the discrepancy, rather than any inherent inability of ASP to identify the samples. Due to service pressures at the time, we did not attempt to repeat ASP in these samples.
Several RT- PCR assays detecting specific variants have been developed, however this is still relatively new and only a few studies demonstrating its validity are available. One study done in the Netherlands demonstrated concordant results between a variant assay (SARS-CoV-2 Lightmix® kit) and WGS, although the VOC epidemiology was different to the present study, with Alpha being the prevalent variant. Furthermore, only 56 samples were validated against WGS.(Ong et al., 2021) Another study in France compared the performance of two variant assays, the Thermo Fisher® TaqPath™ COVID-19 CE-IVD RT-PCR kit (TaqPath) and ID solutions® ID™ SARS- CoV-2/UK/SA Variant Triplex RT-PCR (ID triplex). Both assays were similarly effective to each other in detecting Alpha, with the ID triplex being able to detect Beta/ Gamma lineage as well. However only a subset of samples that were discordant between WGS or ASP or with Beta/Gamma were sequenced, so sensitivity and specificity could not be evaluated.(Migueres et al., 2021) In our study, the majority of samples had both ASP and WGS performed, allowing us to evaluate the sensitivity and specificity of our variant assay to WGS.
There is variation in the literature when using the ARTIC LoCost V3 Nanopore method, with some studies generating results with CT > 38 but others only up to CT 32–33.(Charre et al., 2020; Lam et al., 2021) When this protocol was validated in our lab, we found that we did not get good quality genomic coverage in samples with CT > 30. ASP was able to provide information when WGS could not be obtained such as in high CT samples. This is in contrast with the study in Netherlands where the variant assay provided inconclusive data in samples with CT > 32.(Ong et al., 2021)
Two significant advantages of ASP are the faster TAT and reduced resource requirement when compared with WGS. Together, these factors help to provide rapid vital epidemiological data to inform IPC in real time. ASP could also be utilised in most diagnostic laboratories which do not have sequencing capabilities. (Kami et al., 2021; Ong et al., 2021)
In this study, the mean TAT of 5 days for ASP is longer than the theoretical best TAT. Indeed, it is possible to obtain results within 2–6 h. (Kami et al., 2021; Ong et al., 2021) Our TATs included the time period that elapsed between samples being received and extracted, as well as the time between ASP being completed and the results being authorized. Therefore, the reported TATs could be explained by bottlenecks occurring at either of these stages. Factors such as receiving a high volume and batching of samples, staffing levels, access to equipment and the fact that the service was only available 5 days a week could have contributed to these delays in the real-world setting that this study was based in.
The most significant limitation is that ASP alone would not be able to identify new variants, such as the now dominant Omicron. New variants can only be identified through sequencing, and ASP would need to be revalidated so that the most relevant targets are utilised depending on contemporaneous circulating variants. Using only 3 targets does limit the ability to monitor the prevalence of known variants, as not all can be detected using these markers. However, increasing the number of targets would further increase the number of PCR reactions required, which could become a bottleneck for testing. The three targets evaluated in this study were chosen to balance lab throughput and distinction between known variants circulating at the time. ASP is to be used in conjunction with, rather than instead of WGS to monitor the prevalence of new dominant variants and shifts in prevalence between known variants.
A further limitation of this study is that we only included CT analysis for samples that were detected as SARS-CoV-2 positive by platforms in NHS Lothian. It is also based on a single CT target value (E target) for each sample. Finally, in samples where sequencing was not attempted or failed, the accuracy of ASP data couldn’t be compared with WGS. However, they are likely to be accurate given the high sensitivity and specificity of ASP.
This study demonstrates that variant assays such as ASP can be utilised in combination with WGS to provide rapid and accurate monitoring and identification of VOC/VUIs, guiding the ongoing response to the pandemic and enhancing epidemiological surveillance and infection control measures.
Author Declaration
We the undersigned declare that this manuscript is original, has not been published before and is not currently being considered for publication elsewhere.
We wish to confirm that there are no known conflicts of interest associated with this publication and there has been no significant financial support for this work that could have influenced its outcome.
We confirm that the manuscript has been read and approved by all named authors and that there are no other persons who satisfied the criteria for authorship but are not listed. We further confirm that the order of authors listed in the manuscript has been approved by all of us.
We confirm that we have given due consideration to the protection of intellectual property associated with this work and that there are no impediments to publication, including the timing of publication, with respect to intellectual property. In so doing we confirm that we have followed the regulations of our institutions concerning intellectual property.
We understand that the Corresponding Author is the sole contact for the Editorial process (including Editorial Manager and direct communications with the office). He/she is responsible for communicating with the other authors about progress, submissions of revisions and final approval of proofs. We confirm that we have provided a current, correct email address which is accessible by the Corresponding Author.
Author Statement
Sanjita Brito-Mutunayagam: Formal analysis, Investigation, Data Curation, Writing-Original Draft, Visualization
Daniel Maloney: Software, Methodology, Resources, Data Curation, Writing-Review and Editing,
Gina McAllister: Writing-Review and Editing, Data Curation, Resources, Methodology, Software, Validation, Investigation
Rebecca Dewar: Methodology, Validation, Resources, Data Curation
Martin McHugh: Conceptualization, Supervision, Writing-Review and Editing, Data Curation, Resources, Methodology, Software, Validation, Investigation
Kate Templeton: Conceptualization, Writing-Review and Editing, Supervision.
Declaration of Competing Interest
None
Data availability
Data will be made available on request.
The data that has been used is confidential.
References
- Baker D.J., Aydin A., Le-Viet T., Kay G.L., Rudder S., de Oliveira Martins L., Tedim A.P., Kolyva A., Diaz M., Alikhan N.F., Meadows L., Bell A., Gutierrez A.V., Trotter A.J., Thomson N.M., Gilroy R., Griffith L., Adriaenssens E.M., Stanley R., Charles I.G., Elumogo N., Wain J., Prakash R., Meader E., Mather A.E., Webber M.A., Dervisevic S., Page A.J., O’Grady J. CoronaHiT: high-throughput sequencing of SARS-CoV-2 genomes. Genome Med. 2021;13 doi: 10.1186/s13073-021-00839-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charre C., Ginevra C., Sabatier M., Regue H., Destras G., Brun S., Burfin G., Scholtes C., Morfin F., Valette M., Lina B., Bal A., Josset L. Evaluation of NGS-based approaches for SARS-CoV-2 whole genome characterisation. Virus Evol. 2020;6:75. doi: 10.1093/VE/VEAA075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Funk T., Pharris A., Spiteri G., Bundle N., Melidou A., Carr M., Gonzalez G., Garcia-Leon A., Crispie F., O’Connor L., Murphy N., Mossong J., Vergison A., Wienecke-Baldacchino A.K., Abdelrahman T., Riccardo F., Stefanelli P., Martino A.Di, Bella A., Presti A.Lo, Casaca P., Moreno J., Borges V., Isidro J., Ferreira R., Gomes J.P., Dotsenko L., Suija H., Epstein J., Sadikova O., Sepp H., Ikonen N., Savolainen-Kopra C., Blomqvist S., Möttönen T., Helve O., Gomes-Dias J., Adlhoch C., groups, on behalf of C. study Characteristics of SARS-CoV-2 variants of concern B.1.1.7, B.1.351 or P.1: data from seven EU/EEA countries, weeks 38/2020 to 10/2021. Eurosurveillance. 2021;26 doi: 10.2807/1560-7917.ES.2021.26.16.2100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kami W., Kinjo T., Arakaki W., Oki H., Motooka D., Nakamura S., Fujita J. Rapid and simultaneous identification of three mutations by the NovaplexTM SARS-CoV-2 variants I assay kit. J. Clin. Virol. 2021;141 doi: 10.1016/j.jcv.2021.104877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam C., Gray K., Gall M., Sadsad R., Arnott A., Johnson-Mackinnon J., Fong W., Basile K., Kok J., Dwyer D.E., Sintchenko V., Rockett R.J. Sars-CoV-2 genome sequencing methods differ in their ability to detect variants from low viral load samples. J. Clin. Microbiol. 2021 doi: 10.1128/jcm.01046-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LW, M, WL, H, B, W, CJ, H, M, H, AS, J, MD, C, et al. Rapid implementation of SARS-CoV-2 sequencing to investigate cases of health-care associated COVID-19: a prospective genomic surveillance study. Lancet Infect. Dis. 2020;20:1263–1272. doi: 10.1016/S1473-3099(20)30562-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Migueres M., Lhomme S., Trémeaux P., Dimeglio C., Ranger N., Latour J., Dubois M., Nicot F., Miedouge M., Mansuy J.M., Izopet J. Evaluation of two RT-PCR screening assays for identifying SARS-CoV-2 variants. J. Clin. Virol. 2021;143 doi: 10.1016/J.JCV.2021.104969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ong D.S.Y., Koeleman J.G.M., Vaessen N., Breijer S., Paltansing S., de Man P. Rapid screening method for the detection of SARS-CoV-2 variants of concern. J. Clin. Virol. 2021;141 doi: 10.1016/j.jcv.2021.104903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Twohig K.A., Nyberg T., Zaidi A., Thelwall S., Sinnathamby M.A., Aliabadi S., et al. Hospital admission and emergency care attendance risk for SARS-CoV-2 delta (B.1.617.2) compared with alpha (B.1.1.7) variants of concern: a cohort study. Lancet Infect. Dis. 2021;0 doi: 10.1016/S1473-3099(21)00475-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.
The data that has been used is confidential.