Abstract
Non-human primates play a key role in the preclinical validation of pluripotent stem cell-based cell replacement therapies. Pluripotent stem cells used as advanced therapy medical products boost the possibility to regenerate tissues and organs affected by degenerative diseases. Therefore, the methods to derive human induced pluripotent stem cell and embryonic stem cell lines following clinical standards have quickly developed in the last 15 years. For the preclinical validation of cell replacement therapies in non-human primates, it is necessary to generate non-human primate pluripotent stem cell with a homologous quality to their human counterparts. However, pluripotent stem cell technologies have developed at a slower pace in non-human primates in comparison with human cell systems. In recent years, however, relevant progress has also been made with non-human primate pluripotent stem cells. This review provides a systematic overview of the progress and remaining challenges for the generation of non-human primate induced pluripotent stem cells/embryonic stem cells for the preclinical testing and validation of cell replacement therapies. We focus on the critical domains of (1) reprogramming and embryonic stem cell line derivation, (2) cell line maintenance and characterization and, (3) application of non-human primate pluripotent stem cells in the context of selected preclinical studies to treat cardiovascular and neurodegenerative disorders performed in non-human primates.
Key Words: embryonic stem cells, induced pluripotent stem cells, non-human primates, pluripotent stem cells, preclinical, regeneration, reprogramming, translational research
Introduction
Degenerative diseases are characterized by the loss of functional cells in a tissue or an organ. This heterogeneous group of diseases accounts for a high and still increasing proportion of morbidity and mortality worldwide (Skovronsky et al., 2006; Roth et al., 2020). Therapies to treat these diseases are limited, and organ transplantation is often the only option. However, organ transplantation has severe limitations due to donor organ shortage. One novel approach to circumvent this problem is to treat degenerative diseases by replacing the lost or non-functional cells with healthy cells to restore organ functionality. One promising alternative for the generation of tissue-specific cell types is to use pluripotent stem cells (PSC) as a source of functional cells for the patients.
PSC can be differentiated into all cell types of the adult body. Additionally, they are easily expandable, making it possible to generate therapeutically sufficient numbers of differentiated cells in a relatively short time. Further, PSC can be genetically manipulated in order to reduce immunogenicity issues (Shi et al., 2017; Wang et al., 2020). Embryonic stem cells (ESC) are pluripotent cells that are derived from the preimplantation embryo. However, the clinical application of ESC is limited due to ethical controversies associated with their derivation. Furthermore, the use of ESC essentially represents an immunologically non-preferred allograft transplant situation. In contrast, induced pluripotent stem cells (iPSC), derived from somatic cells by reprogramming, circumvent both problems, the ethical issue and the use of allogeneic cells. Takashi and Yamanaka (2006, 2007) discovered a method to induce pluripotency in adult cells and generated the first iPSC lines. Since their discovery, therapies using iPSC as advanced therapy medical products (ATMP) to regenerate tissues and organs have quickly developed and are currently in an advanced state close to translation to the clinics (Nagoshi et al., 2020; Yamanaka, 2020). However, some important points need to be addressed before the final translation of these technologies. There are still open questions regarding the minimal effective dose, route of administration, medium/long-term rejection risks, long-term efficiency, or the feasibility for the implementation of an efficient infrastructure for a broad administration of these therapies (Shi et al., 2017; Liu et al., 2020; Wang et al., 2020; Tao et al., 2021). This needs to be individually evaluated for each disease, organ, therapeutic strategy, and possibly also for the different sexes and ages. Therefore, it is necessary to perform highly predictive studies using preclinical animal models for safe clinical translation. Non-human primates (NHP) are excellent models to test cell replacement therapies due to their close phylogenetic relationship with humans, which is reflected by high genetic, metabolic, and physiological similarities (Johnsen et al., 2012; Cox et al., 2017; Cardoso-Moreira et al., 2020; Figure 1).
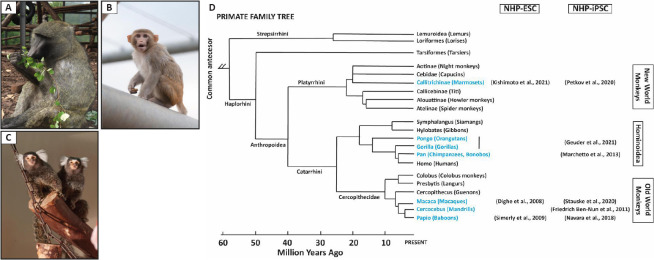
Figure 1.
Non-human primate models for the preclinical testing of cell replacement therapies.
(A) Baboon, (B) rhesus macaque, and (C) marmoset (taken by Karin Tilch and Christian Kiel with the permission from German Primate Center (DPZ)). (D) Family tree of primates. Adapted from Petkov and Jarvis (2012), in blue relevant non-human primate species in stem cell research. Selected references are indicated. NHP-ESC: Non-human primate embryonic stem cells; NHP-iPSC: non-human primate induced pluripotent stem cells.
NHP models were traditionally established in infection biology and have a long history in biomedical research. Selected Old World (Cercopithecidae) and New World monkey (OWM/NWM) species have been established as regenerative medicine models over the last years. In particular, OWM have been key to test ATMP (Johnsen et al., 2012; Kobayashi et al., 2012; Morizane et al., 2013; Shiba et al., 2016; Liu et al., 2018; Ishigaki et al., 2021; Figure 1). In contrast, NWM have not been used extensively since these animal models were not so established and, they present for some organs, e.g., the heart, key physiological differences with humans (Mattisona and Vaughan, 2017). However, NWM models, e.g., the marmoset, have become increasingly important over the last years (Figure 1). NWM present significant practical advantages compared to OWM, including litter size, early sexual maturity, lower costs, and easier handling (Kobayashi et al., 2012; Wu et al., 2012).
NHP are a link between basic research and clinical application (Yang et al., 2018). Notwithstanding, meaningful preclinical testing of PSC-based regenerative therapies in NHP requires the generation of NHP-PSC lines of homologous quality to their human counterparts. However, these technologies have developed at a slower pace for NHP compared to human. This review analyzes the state of the art of NHP-PSC generation, maintenance, and characterization. Additionally, we highlight selected achieved milestones and remaining challenges in the field.
Search Strategy
The literature search for this review was performed using PubMed. We used the search terms “non-human primates”, “NHP” and “primates” in combination with the keywords: “iPSC”, “ESC” “translational research”, “biomedical research”, “regeneration”, “reprogramming” and “cell replacement therapies”. Additionally, the bibliography sections of selected publications found in the first screen were systematically analyzed in order to find additional relevant publications. This review was written from a historical perspective and includes citations from 1995 to 2021.
Non-Human Primate Pluripotent Stem Cells
The preclinical testing of PSC cell replacement therapies requires both xenogenic (Kobayashi et al., 2012; Liu et al., 2018; Wang et al., 2020) and allogenic/autologous (Shiba et al., 2016) studies (Daadi et al., 2014). It is crucial to consider both approaches in parallel since they address different research questions. In xenogenic studies using NHP, usually human cells are transplanted into NHP. These studies focus on the validation of the ATMP itself. This approach is crucial to test the functionality and survival of the cells that will, later on, be used in clinics. In allogenic studies, species-specific PSC are transplanted into the animal model. These studies focus mainly on evaluating the safety, e.g., immune rejection or side effects of the allogenic ATMP, in the context of an organism (Wu et al., 2012). Both approaches work synergistically and are required to generate comprehensive preclinical data.
In order to perform allogeneic/autologous cell replacement studies in NHP, it is necessary to generate PSC lines from NHP species with biomedical relevance. Therefore, a valuable repertoire of PSC lines from the translationally most relevant NHP species has been developed (Figure 1; Additional Tables 1 and 2).
Additional Table 1.
Selected publications of non-human primate embryonic stem cell (NHP-ESC) derivation
| Reference | Source of cells | Derivation/maintenance (Feeders/feeder-free) | Species |
|---|---|---|---|
| Thomson et al., 1995 | Blastocyst (NF) | Feeders/F eeders | Rhesus monkey |
| Thomson et al., 1996 | Blastocyst (NF) | Feeders/F eeders | Marmoset |
| Suemori et al., 2001 | Blastocyst (IVF and ICSI) | Feeders/F eeders | Cynomolgus monkey |
| Cibelli et al., 2002 | Egg (Parthenogenetic) | Feeders/F eeders | Cynomolgus monkey |
| Vrana et al., 2003 | Egg (Parthenogenetic) | Feeders/F eeders | Cynomolgus monkey |
| Sasaki et al., 2005 | Blastocyst (NF) | Feeders/F eeders | Marmoset |
| Mitalipov et al., 2006 | Blastocyst (ICSI) | Feeders/F eeders | Rhesus monkey |
| Suemori and Nakatsuji, 2006 | Blastocyst (IVF and ICSI) | Feeders/F eeders | Cynomolgus monkey |
| Navara et al., 2007 | Blastocyst (ICSI) | Feeders/F eeders | Rhesus monkey |
| Byrne et al., 2007 | Blastocyst (SCNT) | Feeders/F eeders | Rhesus monkey |
| Dighe et al., 2008 | Egg (Parthenogenetic) | Feeders/F eeders | Rhesus monkey |
| Muller et al., 2009 | Blastocyst (NF) | Feeders/F eeders | Marmoset |
| Simerly et al., 2009 | Blastocyst (ICSI) | Feeders/F eeders | Baboon |
| Debowski et al., 2016 | Morula (NF) | Feeders/F eeders | Marmoset |
| Kishimoto et al., 2021 | Blastocyst (NF and IVF) | Feeder-free/Feeder-free (Feeder conditioned medium) | Marmoset |
In "source of cells", it is stated from where the cells were obtained to derive the different NHP-ESC, including oocyte (egg) for the parthenogenic cells lines and blastocyst/morula for the lines derived from fertilized embryos. Additionally, in the lines derived from embryos, how this was obtained by natural fertilization (NF), in vitro fertilization (IVF), intracytoplasmic sperm injection (ICSI), or somatic cell nuclear transfer (SCNT). Additionally, the table specifies how the different cells lines were derived and maintained, focusing on the usage or not of feeder cells to support their expansion and culture (Feeders/Feeder-free). For each one of the studies included in the table, it is also specified the NHP species from which the cell lines were derived, including marmoset (Callithrix jacchus), rhesus monkey (Macaca mulatta), cynomolgus monkey (Macaca fascicularis), marmoset (Callithrix jacchus) and baboon (Papio anubis).
Additional Table 2.
Selected publications on the reprogramming of non-human primates (NHP) cells into induced pluripotent stem cells (iPSC)
| Reference | Somatic cell source (isolated from)* | Reprogramming | Reprogramming ^ Maintenance (Feeders/Feeder-free) | Directed differentiation*** | Species**** | |
|---|---|---|---|---|---|---|
|
| ||||||
| Method | Factors used** | |||||
| Liu et al., 2008 | Fibroblasts (Adult ear skin) | Retrovirus | OSKC / Macaque | Feeders/Feeders | Rhesus monkey | |
| Wu etal., 2010 | Fibroblasts (Newborn skin) | Retrovirus | OSKC/Human | Feeders/Feeder-free | Marmoset | |
| Tomioka et al., 2010 | Liver cells (Fetal) | Retrovirus | OSKCNL/Human | Feeders/Feeders | Neuron-like cells | Marmoset |
| Chan etal., 2010 | Fibroblasts (Fetal skin) | Lentivirus | OSK/Macaque | Feeders/Feeders | Neuron-like cells | Rhesus monkey |
| Zhong et al., 2011 | Fibroblasts (Oral mucosa) | Retrovirus | OSKC/Human | Feeders/Feeder-free | Neuron-, cardiomyocyte-, hepatocyte- and hematopoietic- cell-like cells | Pig-tailed macaque |
| Friedrich Ben-Nun et al., 2011 | Fibroblasts (Adult skin) | Retrovirus | OSKC/Human | Feeders/Feeders | Drill | |
| Okahara-Narita et al., 2012 | Fibroblasts (Adult and fetal skin) | Retrovirus | OSKC/Human | Feeders/Feeders | Cynomolgus monkey | |
| Wunderlich et al., 2012 | Fibroblasts (Adult skin) | Simian immunodeficiency virus (SIV)-based vector | OSKC/Human | Feeders/Feeders | Cardiomyocyte-like cells | Cynomolgus monkey |
| Wiedemann et al., 2012 | Mesenchymal stem cells (Adult/ Bone marrow) | Excisable lentiviral vector | OSKC/Human | Feeder-free/Feeders | Neuron-like cells, megakaryocytes, adipocytes, chondrocytes, and osteogenic cells | Marmoset |
| Shimozawa et al., 2013 | Skin and liver cells (Newborn and fetal) | Retrovirus | OSKC / Macaque | Feeders/Feeders | Cynomolgus monkey | |
| Marchetto etal., 2013 | Fibroblasts (Adult skin) | Retrovirus | OSKC/Human | Feeders/Feeder-free | Chimpanzee and bonobo | |
| Morizane et al., 2013 | Fibroblasts (Adult oral mucosa) and peripheral blood mononuclear cells (Adult blood) | Retrovirus / Episomal vectors | OSKL plus L-MYC/ Human | Feeders/Feeders | DA neuron-like cells | Cynomolgus monkey |
| Navara etal., 2013 | Fibroblasts (Fetal skin) | Retrovirus | OSKC/Human | Feeders/Feeders | Baboon | |
| Emborg et al., 2013 | Fibroblasts (Adult skin and subcutaneous tissue) | Retrovirus | OSKC/Human | Feeders/Feeders | DA neuron-like cells | Rhesus monkey |
| Fang et al., 2014 | Fibroblasts (Adult ear skin) | Retrovirus | OK/Human | Feeders/Feeders | Rhesus monkey | |
| Wunderlich et al., 2014 | Endothelial cells or Fibroblasts | Lentivirus | OSKC/OSNL / Human | Feeders/Feeders | Cynomolgus monkey, Gorilla, and Bonobo | |
| Debowski et al., 2015 | Fibroblasts (Newborn skin) | PiggyBac | OSKCNL / Marmoset | Feeders/Feeders | Marmoset | |
| Wang et al., 2015 | Mesenchymal stem cells (Adult/ Bone marrow) | Retrovirus | OSKC/Human | Feeders/Feeders | DA neuron-like cells | Cynomolgus monkey |
| Coppiello et al., 2017 | Fibroblasts (Adult skin) | Sendai virus | OSKC/Human | Feeders/Feeders | Cynomolgus monkey | |
| Vermilyea et al., 2017 | Fibroblasts (Adult skin and subcutaneous tissue) | Episomal vectors | OSKCNL/Human | Feeder-free/Feeder-free | DA neuron-like cells | Marmoset |
| Navara etal., 2018 | Peripheral blood mononuclear cells (Adult blood) | Sendai virus | OSKC/Human | Feeders/Feeders (Feeder conditioned medium) | Baboon | |
| Yang etal., 2018 | Fibroblasts (Adult skin) | Episomal vectors | OSKL plus L-MYC /Human | Feeders/Feeders | DA neuron-like cells | Marmoset |
| Watanabe etal., 2019 | Liver cells (Newborn liver) and fibroblasts (Fetal, newborn and adult skin) | mRNAs | OSKMNL/Human | Feeders/Feeder-free (Feeder conditioned medium) | Marmoset and cynomolgus monkey | |
| Nakajima etal., 2019 | Fibroblasts (Fetal skin) | mRNAs | OSKCNL/Human | Feeder-free / Feeders | Marmoset | |
| Rodriguez-Polo et al., 2019 | Fibroblasts (Adult skin) | PiggyBac | OSKCNL / Marmoset | Feeders/Feeder-free | Baboon | |
| Aron Badin etal., 2019 | Peripheral blood mononuclear cells (Adult blood) | Sendai virus | OSKC/Human | Feeders/Feeders | Striatal neuron precursors | Cynomolgus monkey |
| Stauske et al., 2020 | Fibroblasts (Adult and newborn skin) | Episomal vectors | OSKL plus L-MYC / Human | Feeder-free/Feeder-free | Cardiomyocyte-like cells | Rhesus monkey and baboon |
| Petkov et al., 2020 | Fibroblasts (Fetal skin) | VEE-mRNAs | OSKC /Human | Feeder-free/Feeder-free | Neuron- and cardiomyocyte-like cells | Marmoset |
| Geuder et al., 2021 | Urinary cells (Urine) | Episomal vectors / Sendai virus | OSKC /Human | Feeder-free/Feeder-free | Neural stem cells | Gorilla and orangutan |
| Ishigaki et al., 2021 | Fibroblasts (Adult skin) Peripheral blood T cells (Adult blood) | Lentivirus vectors / Sendai virus | OSKC /Human | Feeders/Feeder-free | Mesenchymal Stem Cells | Rhesus monkey |
| Yoshimatsu et al., 2021 | Fibroblasts (Fetal and adult skin) | Episomal vectors | OSKL plus L-MYC GLIS1, and KDM4D /Human | Feeders/Feeders | Primordial germ cell-like cells | Marmoset |
| Rodriguez-Polo et al., 2021 | Fibroblasts (Adult skin) | PiggyBac | OSKCNL / Marmoset | Feeders/Feeder-free | Rhesus monkey | |
(*) Somatic cell source includes the information about the somatic cells used for reprogramming, including the tissue they were extracted from and the developmental stage of the NHP. (**) Pluripotency factors used for reprogramming. OCT4 (O), SOX2 (S), KLF4 (K), and c-MYC (C), NANOG (N) and LIN28 (L). Some studies include additional non-coding factors to increase the efficiency ofreprogramming, e.g., miRNAs, which have not been included in the table. (***) Directed differentiation of the generated NHP-iPSC towards a specific cell type. DA neuron-like
Non-human primate embryonic stem cells
Human ESC have limited applicability for cell replacement therapies. Therefore, NHP-iPSC are preferentially used for the validation of cell replacement therapies in NHP. However, ESC are the developmental gold standard for PSC and share high similarities with iPSC. Moreover, the first NHP-ESC lines were available years before the first iPSC lines were generated. Hence many of the NHP-PSC methods have been developed first with NHP-ESC and were then translated to iPSC. Therefore, to get the full picture of the progress in NHP-iPSC biotechnology, it is crucial to consider NHP-ESC studies.
The first NHP-ESC lines were derived from rhesus macaque (Macaca mulatta) and marmoset (Callithrix jacchus) in 1995/1996 (Thomson et al., 1995, 1996). Since then, a few groups have extended this pioneering work by deriving additional ESC lines from different NHP species, including cynomolgus monkey (Suemori et al., 2001), rhesus macaque (Navara et al., 2007), baboon (Simerly et al., 2009), and marmoset (Sasaki et al., 2005; Müller et al., 2009; Debowski et al., 2015; Additional Table 1). ESC lines have been derived from both naturally fertilized and artificially fertilized embryos using in vitro fertilization (IVF). Furthermore, NHP-ESC have been derived from cloned embryos after somatic cell nuclear transfer (SCNT) (Suemori et al., 2001; Navara et al., 2007; Simerly et al., 2009; Kishimoto et al., 2021; Additional Table 1). ESC lines are usually derived from the early embryo; however, it is also possible to generate ESC from non-fertilized oocytes. The resultant ESC lines are parthenogenetic and hold great promise for biomedical research since they are usually homozygous for the major histocompatibility complex (MHC). MHC homozygosity allows a better immune matching in comparison with normal ESC lines. Towards transplantation, these cells promise to reduce the adaptive immune response and, consequently, reduce the need for immunosuppression. Over the last years, NHP parthenogenic lines have been reported. However, only a few lines are available, and all of them from OWM (Cibelli et al., 2002; Vrana et al., 2003; Dighe et al., 2008).
During ESC line derivation, one key aspect is the transition of the small and only transiently existing population of pluripotent cells of the inner cell mass (embryoblast) of the blastocyst to permanently self-renewing ESC which are “frozen” in their pluripotent developmental state. The conversion is usually achieved by plating destructed embryos on mouse embryonic fibroblasts (MEFs) feeder cells. Using MEFs entails disadvantages discussed below; however, one of the major disadvantages is that the cell culture medium is not chemically defined. Recently, Kishimoto et al. (2021) report, to our knowledge, the first NHP-ESC lines derived in feeder-free conditions. In this study, naturally fertilized and in vitro fertilization embryos were plated in either iMatrix- or MEF-coated culture dishes. No significant differences were found in the rates of ESC line derivation between naturally fertilized and in vitro fertilization-derived embryos. However, the establishment of novel ESC lines was more efficient in feeder-free conditions, demonstrating the robustness of the culture conditions (Kishimoto et al., 2021).
In conclusion, several NHP-ESC lines are available to date. These cell lines have greatly contributed to establishing the conditions to maintain NHP-PSC in vitro. Additionally, NHP-ESC can contribute to many other research fields, including transgenesis, regulation of pluripotency, and developmental and evolutionary studies.
Non-human primate induced pluripotent stem cells
Technologies to generate iPSC have greatly evolved since the first reports 15 years ago. There are several ongoing clinical trials using PSC to regenerate different tissues and organs. This impressively fast translation from experimental studies to clinical application has been possible due to the refinement in iPSC generation and maintenance methodologies. One of the groundbreaking improvements was the invention of chemically defined culture media and adherence substrates. This allowed the generation and culture of human iPSC and their application in compliance with good manufacturing practices required for the transition of these cells from the experimental laboratory to the clinics. NHP-iPSC have closely followed the progress made with human and mouse iPSC. Nonetheless, there are still domains in iPSC technology that have been challenging to be applied to NHP-iPSC successfully. Therefore, it is crucial to comparatively analyze the state of the art of human and NHP-iPSC generation and culture.
Factors for reprogramming non-human primate cells
Nuclear reprogramming of somatic cells into iPSC was first achieved by the overexpression of the reprogramming factors OCT4, SOX2, KLF4, and c-MYC (OSKM). These factors trigger chromatin structure modifications and subsequent transcriptomic adaptations necessary for the transition of differentiated cells to a pluripotent state. OSKM have the ability to bind pluripotency-related recognition sites in the genome to initiate the autoregulatory circuit that activates endogenous pluripotency genes (Teshigawara et al., 2017). Most reprogramming methods available were originally designed for human cells, and are hence based on human reprogramming factors. As mentioned above, it is crucial that the exogenous reprogramming factors bind to genomic DNA for successful iPSC generation. This could have been a problem for the generation of NHP-iPSC using xenogenic (human) OSKM. However, NHP share high genetic similarities with humans in comparison with mice. This is also reflected in the OSKM genes (Table 1).
Table 1.
Pluripotency factor identity on the protein level of selected animal models versus human
| Mouse | Marmoset | Macaque | Baboon | |
|---|---|---|---|---|
| OCT4 | 90.54% | 97.50% | 99.39% | 99.39% |
| SOX2 | 98.42% | 98.10% | 100.00% | 99.68% |
| KLF4 | 96.55% | 98.10% | 99.71% | 99.71% |
| C-MYC | 96.00% | 96.30% | 98.00% | 99.77% |
Values are given in percentage of similarity on the protein level. Mouse (Mus musculus), Marmoset (Callithrix jacchus), macaque (Macaca mulatta), and baboon (Papio anubis).
Therefore, most studies reprogrammed NHP cells using human factors (Additional Table 2). However, some studies used species-specific factors (Shimozawa et al., 2013; Debowski et al., 2015). One of the first reports of macaque iPSC by Shinozawa et al. (2013) infected fetal liver cells and skin fibroblasts with retroviral vectors containing macaque OSKM cDNAs (Additional Table 2). In another study in 2015, we used marmoset OSKM in combination with LIN28 (L) and NANOG (N) to reprogram marmoset fibroblasts using the piggyBac system (Debowski et al., 2015). Later, the same transposon system containing marmoset cDNA sequences was used to reprogram baboon and macaque fibroblasts (Rodriguez-Polo et al., 2019, 2021a). These studies demonstrated that primate reprogramming factors can be used to reprogram other primate species successfully. Both studies used integrative (retroviral) or reversibly integrative (piggyBac) reprogramming approaches. Transplantation of NHP-iPSC that constitutively express xenogeneic genes can evoke unintended immune responses and increase the risk of teratoma formation (Aoi, 2016; Sekine et al., 2020). Hence, according to the current standards, iPSC with exogenous expression of reprogramming factors are not candidates for preclinical testing and clinical application. Furthermore, cross-species combination of cells to be reprogrammed and reprogramming factors may lead to lower reprogramming efficiency. However, studies comparing reprogramming efficiency using human versus NHP exogenous factors to reprogram NHP cells are still missing.
Reprogramming methods
Since the pioneering studies performed by Yamanaka using retroviral vectors to deliver OSKM, many groups have developed novel reprogramming strategies (Schlaeger et al., 2015). The aim is to generate iPSC with high efficiency and that meet the requirements for safe clinical application. Reprogramming methods can be divided into two main groups, integrative and non-integrative, depending on whether the expression vectors integrate into the host cell genome or not. Most reprogramming strategies have been successfully tested in one or more NHP species, generally a few years after their validation in human cells.
Integrative reprogramming approaches: Retroviral vectors were initially used to ectopically express reprogramming factors in NHP cells according to the experiments performed with mouse and human cells (Takahashi and Yamanaka, 2006; Takahashi et al., 2007) as they facilitate robust expression of the reprogramming factors. Only a few years after the first human iPSC lines were reported, the first NHP-iPSC were correspondingly generated from the macaque (Liu et al., 2008; Chan et al., 2010; Morizane et al., 2013; Shimozawa et al., 2013), marmoset (Tomioka et al., 2010), baboon (Navara et al., 2013), as well as chimpanzees and bonobos (Marchetto et al., 2013; Gallego Romero et al., 2015; Additional Table 2). These early NHP-iPSC, together with the already available NHP-ESC lines, established the bases of NHP-PSC maintenance and early standards for characterization. One interesting viral reprogramming approach, to our knowledge the only one designed to be NHP specific, was followed by Wunderlich and collaborators. In 2012 they reported cynomolgus macaque iPSC generated by a viral vector derived from simian immunodeficiency virus aiming to increase the low infection efficiency using human-derived lentiviral vectors in NHP cells (Wunderlich et al., 2012).
Notwithstanding, virus-based reprogramming is associated with the modification of gDNA entailing risks of insertional mutagenesis. Additionally, the constitutive expression of the reprogramming factors can lead to tumorogenesis and failure of differentiation of the iPSC (Sekine et al., 2020). These problems propelled the development of novel reprogramming approaches. New methods like non-integrating viral vectors and reversibly integrating systems, e.g., the piggyBac transposon, allowed the generation of transgene-free iPSC (Woltjen et al., 2011; Wiedemann et al., 2012; Zou et al., 2012). In NHP, the piggyBac system has been employed to generate marmoset (Debowski et al., 2015), baboon (Rodriguez-Polo et al., 2019), and macaque iPSC (Rodriguez-Polo et al., 2021a).
Non-integrative reprogramming approaches: Non-DNA-integrative methods work less efficiently in comparison with integrative methods (Schlaeger et al., 2015). However, these systems allow reprogramming without modification of the genome. Non-integrative DNA methods like episomal vectors provide the advantage of easy amplification of the reprogramming tools and allow generating transgene-free iPSC. Stauske et al. (2020) used episomal vectors (Okita et al., 2011) based on the EBNA sequence to reprogram skin fibroblasts from macaque and baboon (Figure 2A-C). This system has also been successfully used to reprogram marmoset iPSC (Vermilyea et al., 2017; Additional Table 2). The plasmid sequences contain Epstein-Barr virus-derived sequences (EBNA) that allow the episomes to replicate in coordination with the cellular genomic DNA. However, EBNA expression gets progressively silenced during the first passages of the iPSC after reprogramming, leading to the progressive loss of the episomes. DNA-based reprogramming methods, however, even the non-integrating ones, entail the risk of unintended genomic integration. Therefore, RNA-based methods were developed to circumvent the risk of intended or unintended genomic alterations by DNA-based approaches. The use of RNA to reprogram somatic cells discards the possibility of genomic integration. Sendai virus-based vectors can be used to deliver self-replicating reprogramming factor mRNA (Fusaki et al., 2009; Nishimura et al., 2011). Several groups have reported novel, transgene-free NHP-iPSC generated using this system, including baboon (Navara et al., 2018), gorilla, orangutan (Geuder et al., 2021), and macaque (Coppiello et al., 2017). Watanabe et al. (2019) also used an RNA method to reprogram cynomolgus monkey and marmoset cells. They performed serial transfections of mRNA (Watanabe et al., 2019) and followed a workflow developed for human cells, combining reprogramming factor mRNAs (OSKMLN) with ESC-specific miRNAs (302a-d, 367) and vaccinia virus-derived interferon response suppressor mRNAs (E3, K3, and B18R). Petkov et al. (2020, 2021) used a similar reprogramming approach, but in order to avoid the need for serial transfections, self-replicating mRNA vectors based on the Venezuelan equine encephalitis virus (VEE-mRNAs) were used to reprogram marmoset fetal fibroblasts. This method allowed the generation of marmoset iPSC and neural stem cells with a single RNA transfection (Petkov et al., 2020; Petkov and Behr, 2021; Figures 1 and 2D). Alternative to DNA and RNA methods, protein-based reprogramming has been reported for human iPSC generation (Kim et al., 2009; Zhang et al., 2012; Seo et al., 2017). However, this method has low efficiency and, to our knowledge, has not been successfully applied to reprogram NHP cells.
Figure 2.
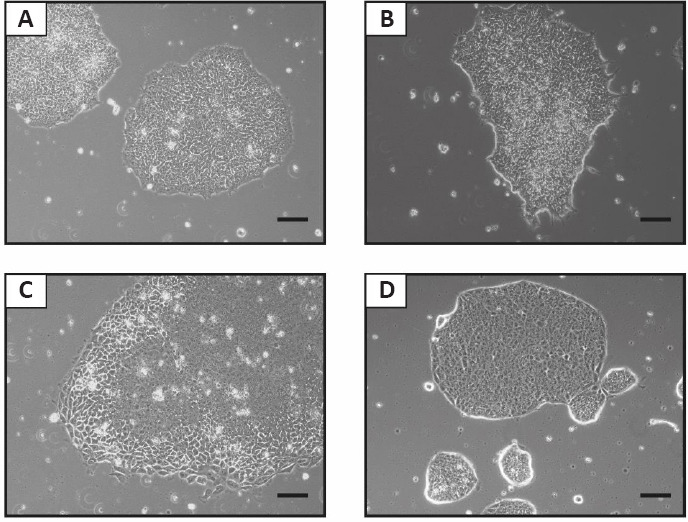
Representative bright field pictures of undifferentiated human and non-human primate transgene-free induced pluripotent stem cells (iPSC) colonies in feeder-free conditions.
(A) Human, (B) baboon, and (C) rhesus macaque iPSC were generated using episomes (Stauske et al., 2020; Rodríguez-Polo et al., 2021b). (D) Marmoset iPSC were generated using VEE-mRNAs (Petkov et al., 2020). The iPSC are in a primed state. Scale bars: 100 µm. Unpublished data.
Although hard data on the relatively low efficiency of NHP-iPSC generation are scarce (Stauske et al. 2020), it is generally a common experience that NHP-iPSC generation is significantly less efficient than human or mouse iPSC generation. The reasons for that are not known to us. However, several options have been considered to improve the efficiency of NHP cell reprogramming based on protocols established for human cell reprogramming. This includes the use of epigenetic modifiers, e.g., valproic acid or sodium butyrate (Wu et al., 2010; Stauske et al., 2020; Rodríguez-Polo et al., 2021b). Additionally, inhibition of P53 (chemical compounds, dominant-negative forms, or shpRNA) (Yang et al., 2018; Watanabe et al., 2019; Stauske et al., 2020) was used. Finally, small molecules modulating ROCK (e.g., DDD00033325), TGF-β/Activin/NODAL (e.g., SB431542), MEK (e.g., PD0325901), or GSK3 (e.g., CHIR99021) pathways have been used to increase NHP reprogramming efficiency (Wu et al., 2010; Watanabe et al., 2019; Petkov et al., 2020).
Considering all progress made over the last years in the generation of NHP-iPSC it is possible to say that reprogramming methods for human cells can be translated directly to NHP (Additional Table 2). The state-of-the-art reprogramming methods include, from our point of view, episomes, Sendai virus, RNA vectors, and mRNAs (Figure 3 and Additional Table 2). However, differences in reprogramming efficiency between human and NHP cells have been pointed out by a few studies (Friedrich Ben-Nun et al., 2011; Schlaeger et al., 2015; Stauske et al., 2020). This can theoretically be due, as mentioned above, to the difference between human exogenous and NHP endogenous pluripotency factors. However, the expression of marmoset exogenous factors in marmoset fibroblasts also did not result in high reprogramming efficiency (Debowski et al., 2015). Alternatively, most of the culture media, small molecules, inhibitors, and other supplements used during reprogramming, are designed and optimized for human cells. Additive effects of small differences in the performance of the growth factors and supplements in human cells and cells from other species may be the main reason for the lower reprogramming efficiency seen in many non-human cells. Lastly, unknown endogenous differences between human and NHP cells may account for the observed differences in the respective reprogramming efficiencies.
Figure 3.
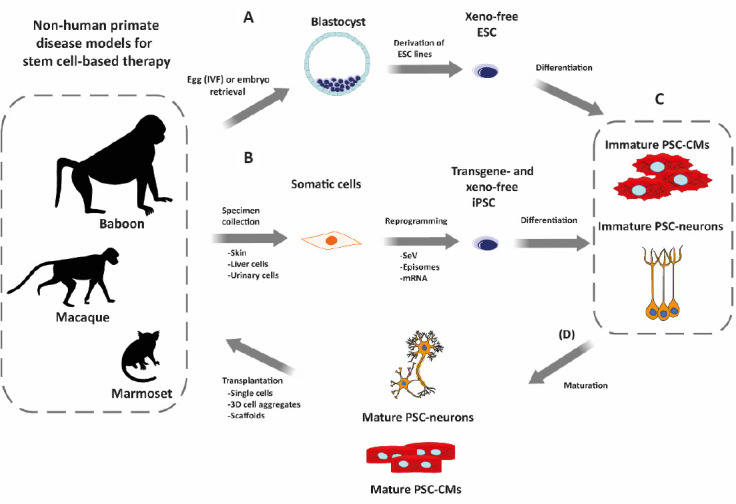
Overview of the workflow for testing pluripotent stem cells (PSC)-based regeneration therapies in non-human primates (NHP).
(A) Embryonic stem cell (ESC) are derived from naturally or artificially fertilized embryos. (B) Alternatively, induced PSC (iPSC) are generated from NHP somatic cells obtained by biopsy (invasive) or non-invasively from, e.g., urinary tract cells present in urine. Non-integrative reprogramming methods (RNA-based like Sendai virus (SeV) or mRNA; DNA-based like episomes) are used to deliver the reprogramming factors. During ESC derivation and iPSC reprogramming, cells are maintained under xeno-free conditions. (C) Upon NHP-PSC generation and characterization, the PSC can be differentiated into tissue-specific cell types, e.g., cardiomyocytes (PSC-CMs) or neurons (PSC-neurons). PSC-derived cells, however, present mostly immature, fetal-like phenotypes. (D) Before transplantation, different approaches can be employed to mature the differentiated cells. The differentiated cell types can be used directly for transplantation or after aggregation to tissues in 3D molds/scaffolds. PSC-derived cells are then transplanted back into the NHP model of choice following allogeneic or autologous approaches. CM: Cardiomyocytes.
Maintenance of non-human primates-pluripotent stem cells
In order to use human ESC and iPSC in clinical settings, it is necessary to keep them undifferentiated in culture in compliance with good manufacturing practices rules. These standards, well defined for human PSC (Baghbaderani et al., 2015; Jo et al., 2020), also need to be applied (as much as possible) to NHP-PSC to ensure the safety of the animals and the translatability of the studies. Human and NHP-PSC have been traditionally maintained using feeder cells and medium containing serum (Liu et al., 2008; Tomioka et al., 2010) (Additional Table 2). The use of feeder-cells, usually of murine origin, increases the risk of immune reaction and precludes chemically definded the culture medium (Baghbaderani et al., 2015). Additionally, serum adds another source of xenogenic molecules to the PSC. The complete removal of cell culture components of xenogenic origin and standardizing the culture conditions was one milestone progressively achieved for human iPSC (Sun et al., 2009).
In contrast to the reprogramming methods, the translatability of state-of-the-art human PSC culture conditions has been challenging for NHP. During iPSC reprogramming, most studies were still dependent on MEFs, with only a few reports of reprogramming using feeder-free conditions (Vermilyea et al., 2017; Petkov et al., 2020; Stauske et al., 2020; Geuder et al., 2021; Additional Table 2). However, there are several reports of adaptation of putative NHP-iPSC after successfully reprogramming to feeder-free conditions (Zhong et al., 2011; Marchetto et al., 2013; Vermilyea et al., 2017) (Additional Tables 2 and 3; Selected publications using chemically defined pluripotent stem cell (PSC) medium in feeder-free conditions). Additionally, feeder-free culture conditions for NHP-PSC are apparently more complex than for human cells (Petkov et al., 2020; Stauske et al., 2020; Kishimoto et al., 2021) (Additional Table 3). Usually, it is necessary to supplement the culture medium with different sets of supplements and inhibitors to keep NHP-PSC undifferentiated (Shimada et al., 2012; Navara et al., 2018). This becomes more evident in studies focusing on NWM reprogramming. Until a few years ago, maintaining marmoset PSC without feeders was not successful (Petkov et al., 2020; Kishimoto et al., 2021) (Additional Table 2). In contrast, human PSC could be easily maintained using commercial media, e.g., StemMACS™ iPS-Brew XF, Essential 8TM Medium, or NutriStem® hPSC XF Medium. This points towards underestimated differences between human and NHP pluripotency. This fact had a clear impact on the translatability/replicability of the studies since the PSC maintenance conditions affect their identity. Therefore, it is necessary to continue refining maintenance protocols considering both human and NHP cell lines in parallel. Additionally, basic research in NHP pluripotency is still necessary to pinpoint possible differences between species.
Additional Table 3.
Selected publications using chemically defined pluripotent stem cell (PSC) medium in feeder-free conditions
| Reference | Cell type | Species | Coating | Medium | Supplements |
|---|---|---|---|---|---|
| Marchetto et al., 2013 | iPSC | Chimpanzee and bonobo | Matrigel (BD) | TeSR1 (StemCell Technologies) | |
| Vermilyea et al., 2017 | iPSC | Marmoset | Factor-reduced Matrigel (BD) | Essential 8™ Medium (Life Technologies) | 100ng/mL Nodal (R&D Systems), GlutaMAX (Life Technologies), chemically defined lipid concentrate (Life Technologies), and 1.94 ^g/mL reduced glutathione (Sigma-Aldrich) |
| Rodriguez-Polo et al., 2019 | iPSC | Baboon | Geltrex LDEV-Free Reduced Growth Factor (Thermo Fisher Scientific) | Essential 8™ Medium (Life Technologies) | |
| Stauske et al., 2020 | iPSC | Rhesus monkey and baboon | Geltrex LDEV-Free Reduced Growth Factor (Thermo Fisher Scientific) | StemMACS™ iPS-Brew (Miltenyi Biotec) | 1 ^MIWR1 (Sigma-Aldrich) and 0.5 ^MCHIR99021 (Merck) |
| Petkov et al., 2020 | iPSC | Marmoset | Geltrex LDEV-Free Reduced Growth Factor (Thermo Fisher Scientific) | StemMACS™ iPS-Brew (Miltenyi Biotec) | 3 ^MIWR1 (Sigma-Aldrich), 0.5 ^MCHIR99021 (Merck), 0.3 ^M CGP77675 (Selleckchem), 10 ng/mL human recombinant leukemia inhibitory factor (hrLIF) (PeproTech), and 7 ^M Forskolin (Selleckchem) |
| Geuder et al., 2021 | iPSC | Gorilla and orangutan | Geltrex LDEV-Free Reduced Growth Factor (Thermo Fisher Scientific) | StemFit iPSC/ESC Culture Media (Ajinomoto) | 100 ng/ml recombinant human basic FGF (Peprotech) |
| Ishigaki et al., 2021 | iPSC | Rhesus monkey | Cellartis DEF-CS 500 COAT-1 (Takara Bio) | Cellartis® DEF-CS™ 500 Culture System (Takara Bio) |
Publications using commercial medium in feeders or conditioned medium in feeder-free conditions have been excluded from this table. Columns left to right: reference of the study, type of cells, species, the coating used, medium, and supplements.
Standards for non-human primate pluripotent stem cell characterization
After the establishment of novel PSC lines, it is crucial to determine the developmental state, chromosomal integrity, and differentiation potential of the generated cell lines in order to reduce heterogeneity between studies. Although the standards for the characterization of human PSC are well defined (Baghbaderani et al., 2015; Jo et al., 2020), no standards in NHP-PSC characterization have been set so far (Yang et al., 2018). NHP-PSC are currently characterized following the human PSC workflow. Accordingly, human PSC morphology, structure, proliferation, differentiation potential, and transcriptomic profile, among others, are taken as references to characterize novel NHP-PSC lines. However, for every laboratory working with NHP- and human PSC, there are differences between human and NHP-PSC (Hong et al., 2016). In some cases, these differences are or seem to be subtle, e.g., cell/colony morphology. In other cases the differences are more important, e.g., non-reactivity to a selected differentiation protocol. In the future, using an expanded panel of NHP-PSC lines, it is crucial to perform more studies focusing on dissecting primate-specific pluripotency traits and signaling pathways involved in differentiation. For PSC characterization, it is essential to exclude a pathogenic contamination of the novel PSC lines. This seems intuitive; however, together with the common tests performed routinely in cell culture, it is necessary to include specific tests for NHP. In the case of PSC from some OWM species, it is necessary to exclude the presence of Herpes-B-Virus (McHV-1), Simian immunodeficiency virus, Simian-T-lymphotropic virus or Respiratory Syncytial Virus. Additionally, the presence of NHP-specific pathogen strains has to be evaluated, e.g., mycoplasma. Defined panels of PCR/RT-PCR-based tests need to be developed for efficient detection of contaminations.
Additionally, the identity and developmental potency of the PSC lines need to be assessed. Clonal variability between lines is a problem hindering the progress in stem cell research (Aoi, 2016). Individual lines can be biased towards differentiation into a specific lineage or unable to meet pluripotency criteria (Lee et al., 2017). This has been reported for NHP-PSC, particularly with marmoset iPSC that tend to differentiate towards the neural lineage (Shimada et al., 2012; Torrez et al., 2012; Qiu et al., 2015; Vermilyea et al., 2017). Aoi (2016) compiled three in vivo assays to assess pluripotency in mouse pluripotent stem cells: (1) teratoma assay, (2) chimera formation, and (3) tetraploid complementation. Chimera formation is considered to be the gold standard to probe the pluripotent state of putative PSC. However, the generation of chimeric NHP using NHP-PSC turns out to be challenging despite the progress made in the last years (Chen et al., 2015; Roodgar et al., 2019; Aksoy et al., 2021). Moreover, the generation of chimeric or cloned (tetraploid complementation) NHP only to validate PSC lines seems neither feasible nor ethical. Teratoma formation can be used alternatively. However, studies have suggested that the results from teratoma assay provide limited quantitative information on the quality of the cell lines (Müller et al., 2010; Yang et al., 2018). Genomic assays could be used in parallel to obtain quantitative data on the quality and functional potency of novel NHP-PSC lines. Yang et al. (2018) addressed the standardization of the generation and characterization of NHP-PSC lines. They used a genomic assay, Scorecard, to characterize novel marmoset iPSC (Bock et al., 2011; Müller et al., 2011). Genomic assays, e.g., PluriTest (Müller et al., 2011) or Scorecard (Bock et al., 2011; Tsankov et al., 2015), measure the molecular signatures of pluripotency. Additionally, Scorecard allows the evaluation of the ability of the cells to differentiate into the three germ layers, also described as functional pluripotency (Yang et al., 2018).
Finally, it is necessary to perform routine identity controls of the NHP-PSC lines. Molecular signatures generated by Scorecard can be used for this (Tsankov et al., 2015). Additionally, other methods, e.g., DNA fingerprinting and PCR-based short tandem repeats, allow the identity testing of the cell lines (Yang et al., 2018). However, some of these methods have limitations when PSC lines are derived from NHP from colonies with high levels of familial relationship.
Non-Human Primate Pluripotent Stem Cell for the Preclinical Testing of Cell Replacement Therapies
The potential of PSC as ATMP has revolutionized regenerative medicine. Several approaches to (re)generate different types of tissues and organs using PSC are in a late stage of preclinical development. All progress culminated, in 2017, in the first transplantation of PSC-derived cells into a patient to treat macula degeneration (Mandai et al., 2017). NHP played a key role in the development of these therapies and their translation to the clinics (Johnsen et al., 2012; Hong et al., 2016). In particular, NHP have special relevance in studies focusing on the regeneration of the nervous and cardiovascular systems due to the physiological similarities of NHP with humans (Johnsen et al., 2012).
NHP offer unique opportunities to develop and validate pluripotent stem cell-based interventions (Daadi et al., 2014). They have a longer life span and bigger body size compared to other animal models, e.g., mice. Thus, it is possible to perform long-term longitudinal studies to evaluate the efficacy and safety of ATMP upon transplantation. Furthermore, NHP anatomy allows the use of imaging techniques, surgical procedures, and equipment used in clinics with little or no adaptation (Johnsen et al., 2012; Daadi et al., 2014; Cox et al., 2017). Finally, NHP-PSC resemble their human counterparts more closely than PSC for other animal models, e.g., in comparison with their leukemia inhibitory factor dependent murine analogs (Liu et al., 2008; Gallego Romero et al., 2015; Hong et al., 2016). Even though only a few studies have compared human- and NHP-PSC, the few data available in gene expression point toward high similarities in cell identity, behavior, and execution of pluripotency (Gallego Romero et al., 2015; Hong et al., 2016; Cardoso-Moreira et al., 2020). Nonetheless, research with NHP entails some challenges not found with other animal models. These include high costs, complex approval processes, and some research tools are not fully developed for this species, e.g., genome annotation (Cox et al., 2017). Therefore, it is necessary to carefully plan and conduct preclinical studies in NHP to provide the translatability of the studies.
Immunogenicity of primate pluripotent stem cell derivatives
One main challenge for the successful translation of PSC technologies into clinics is the allogenic immune rejection of PSC-derivatives by the recipient. PSC-derived ATMP that are not perfectly immunologically MHC (major histocompatibility complex) -matched with the recipients cause a T cell-mediated immune response and ultimately rejection (Liu et al., 2017; Tao et al., 2021). Even though immune suppressants prevent immune rejection, a chronic regimen can greatly impact patients’ health and burden healthcare systems (Liu et al., 2017). The discovery of iPSC came with the promise to circumvent this problem with the possibility of autologous transplantation. However, this approach seems challenging or even non-realistic to put into practice due to the high costs and long time needed to generate, characterize and validate novel iPSC lines. Additionally, some studies indicate that perfectly matched iPSC-derived products can also be immunogenic (Liu et al., 2017; Aron Badin et al., 2019).
Therefore, it is necessary to continue assessing the immune reaction risk of allogeneic versus autologous transplantation of PSC-derivatives. Moreover, it is required to do it in different organs and following different transplantation strategies to assess the risk of a specific PSC treatment. With this aim, NHP together with NHP-PSC offer advantages not found in other animal models since their immune system more faithfully resembles the human immune system (Cox et al., 2017; Navara et al., 2018). Several studies have focused on the analysis of the immune reaction to the transplantation of PSC-derivatives in NHP. However, many studies reach contradictory conclusions (Liu et al., 2017; Aron Badin et al., 2019; Chen and Niu, 2019; Tao et al., 2021). These problems may be addressed by extending the duration of the studies and better study designs, particularly the inclusion of appropriate control groups.
Non-human primate-pluripotent stem cell differentiation
Currently, there are robust protocols available to differentiate human PSC into a broad panel of cell types. These protocols were fine-tuned over the past years based on the progressive gain of knowledge in development (Aoi, 2016). Several of these workflows have been successfully validated in NHP-iPSC (Maria et al., 2014; Wang et al., 2015; Shiba et al., 2016; Tiburcy et al., 2017; Additional Table 2). However, it is important to continue investigating the (potential) differences between human and NHP PSC-derived cells, e.g., cardiomyocyte- and neuron-like cells (Maria et al., 2014; Pavlovic et al., 2018; Zhao et al., 2018). One example is the evaluation of NHP versus human iPSC-derived cardiomyocytes for the treatment of myocardial infarction. In 2018, Zhao et al. performed a study directly comparing the potential of iPSC-cardiomyocytes from both species for left ventricular remodeling. They found that both human and NHP iPSC-cardiomyocytes provide similar cardioprotection in the rat infarction model (Zhao et al., 2018). Another study performed by Marchetto and collaborators analyzed differential migration patterns between human, chimpanzee, and bonobo iPSC-derived neural progenitor cells, in vitro and in vivo. Differences in transcriptomic profiles and migration patterns between human and NHP neural progenitor cells were found. This and other studies point toward the need for more comparative studies to highlight potential differences between human and NHP PSC-derived cells (Marchetto et al., 2019). The maturation state of the transplanted PSC-derived cells and their long-term stability are also often overlooked (Tiburcy et al., 2017; Guo and Pu, 2020; Silva et al., 2020). Nowadays, many efforts have been put into obtaining more mature cells derived from PSC, including neurons and cardiomyocytes. Such maturation protocols, mainly developed for human cells, still need to be validated with NHP-PSC.
Selected aspects of the regeneration of the cardiovascular system
Non-human primate cardiovascular system share many similarities with humans (Johnsen et al., 2012). Additionally, NHP can naturally develop many human cardiovascular diseases (Cox et al., 2017). Furthermore, OWM present a similar progression of age-related comorbidities to humans, e.g., diabetes (Cox et al., 2017). There are also protocols to surgically induce other cardiac conditions, like myocardial infarction (Shi et al., 2013; Liu et al., 2018). Therefore, NHP- cardiovascular disease models in combination with NHP-PSC (or their derivatives) represent an excellent system for preclinical testing of novel therapies currently under development.
PSC-derived cardiac-specific cell types can be transplanted to a diseased heart to restore its functionality (Shiba et al., 2016; Liu et al., 2018). PSC-derived cardiomyocytes can be transplanted alone or together with other heart cells, e.g., fibroblasts or endothelial cells. Additionally, therapeutic cells can be directly applied to the heart or as 3D engineered tissue aggregates (Funakoshi et al., 2016; Tiburcy et al., 2017) (Figure 3). The considerable diversity of the approaches to treat degenerative cardiac diseases reflects the need for preclinical models to evaluate which approach provides optimal results under which condition. Additionally, it is possible to generate iPSC-derived endothelial cell progenitors to explore the potential of these cells to regenerate vasculature or enhance the fidelity and survival of the ATMP. Towards this goal, Shi et al. (2013) differentiated baboon ESC into endothelial precursors and transferred the generated angioblasts into a decellularized arterial segment ex vivo. Fourteen days after inoculation, survival and maturation of the precursor cells into CD31 positive endothelial cells was demonstrated (Shi et al., 2013; Cox et al., 2017). Another study performed by Shiba et al. (2016) focused on evaluating NHP-PSC derived cardiomyocytes to restore heart function in a NHP infarction model. Cynomolgus macaque iPSC-cardiomyocytes were injected into macaque hearts after a surgically induced infarction (Shiba et al., 2016). Twelve weeks after injection, the monkeys were euthanized, and the remuscularization of the myocardium by the transplanted cells was evaluated. Partial contribution to remuscularization and vascularization in the scar tissue was found. Furthermore, the contractile function of the heart improved. However, transient tachycardia was also encountered (Shiba et al., 2016).
Selected aspects of the regeneration of the central nervous system
Due to the similarities between the human and the NHP brain, NHP are also crucial for testing PSC-based regeneration therapies of the central nervous system. In these studies, PSC-derived neurons, or neuronal progenitors, are transplanted into a specific brain area or the spinal cord to restore impaired functionality (Figure 3). These therapies appear especially attractive to treat degenerative diseases like Parkinson’s or Huntington’s disease that have a high incidence in some populations (Chen and Niu, 2019).
Several NHP models of degenerative brain diseases have been established in the last years (Chen and Niu, 2019). In 2013, Emborg et al. (2013) differentiated macaque iPSC into neurons and glia and performed autologous transplantation into a drug-induced NHP model of Parkinson’s disease. They analyzed the graft six months after transplantation, confirming the survival of the transplanted cells in the primate brain (Emborg et al., 2013). However, few graft-derived tyrosine hydroxylase positive neurons survived after six months, and no improvement in the motor function was observed after transplantation. Following this study, Wang and collaborators used a different protocol to generate dopaminergic neurons to transplant in a homologous Parkinson’s disease NHP model. In this study, they described a higher survival rate of the cells in the grafts and a behavioral improvement of the treated monkeys in comparison with the controls (Wang et al., 2015). In a recent study, Tao et al. transplanted allogeneic or autologous DA neural progenitors in a Parkinson’s NHP disease model (Tao et al., 2021). Importantly, this macaque study covered a 2-year observation period with no immunosuppression in order to mimic a potential future clinical scenario. NHP with autologous transplants showed substantial mitigation of the disease symptoms compared to the non immunologically matched group. Histological analysis confirmed these observations, showing higher graft survival in the autologous compared to the allogeneic group. Additionally, the allogenic group showed higher immune responses (T cells, leukocytes, microglia, and astrocytes) in and around grafts in comparison with the allogeneic group (Tao et al., 2021).
The relevance of NHP models for preclinical testing of PSC-based regeneration for Parkinson’s and other degenerative diseases of the nervous system has been demonstrated (Emborg et al., 2013; Chen and Niu, 2019; Tao et al., 2021). However, it is necessary to improve knowledge about the progression of degenerative diseases in NHP while at the same time iPSC derivation and differentiation protocols are further refined (Chen and Niu, 2019).
Summary and Future Directions
This review provides a systematic overview of the progress made in NHP-PSC generation, maintenance and preclinical application. NHP have demonstrated to be crucial for the development of novel treatments, including PSC-based ATMP. Even though some cell replacement therapies are close to clinical application, it is crucial to continue developing and testing state-of-the-art (“clinical-grade”) NHP-PSC lines. Further refinement of reprogramming and maintenance methods and standardization of the characterization deserves the highest priority in order to reproducibly generate high-quality PSC lines to be used in preclinical transplantation studies in NHP. Furthermore, the immunological properties of the NHP-PSC may be modulated by gene editing so that rejection of cells upon transplantation is minimized or even abolished (Yamanaka, 2020). This will also enhance the analysis of the structural and functional integration of the transplanted cells after extended observation periods. In general, we believe that it is important to freely share cell lines and protocols between research groups. This may also include the development of public NHP-PSC repositories.
We are living in exciting times of stem cell research. Over the last years, new stem cell technologies have arisen, promising a revolution in regenerative medicine. Translational NHP studies are the key to realizing this revolution in the treatment of degenerative diseases.
Additional files:
Additional file 1: Open peer review report 1 (86.1KB, pdf) .
Additional Table 1: Selected publications of non-human primate embryonic stem cell (NHP-ESC) derivation.
Additional Table 2: Selected publications on the reprogramming of non-human primates (NHP) cells into induced pluripotent stem cells (iPSC)
Additional Table 3: Selected publications using chemically defined pluripotent stem cell (PSC) medium in feeder-free conditions.
Acknowledgments:
We thank Angelina Berenson, Nicole Umland, Ulrike Gödecke, Simone Kalinowski, for excellent technical assistance. We also thank Irene Rodríguez Polo for the help illustrating the figures. We are grateful to Kerstin Zaft for administrative support.
Footnotes
Conflicts of interest: The authors declare no conflicts of interest.
Availability of data and materials: All data generated or analyzed during this study are included in this published article and its supplementary information files.
Open peer reviewer: Tomo Šarić, Uniklinik Koln, Germany.
Funding: This work was partially supported by the German Centre for Cardiovascular Research (DZHK) and the German Primate Center – Leibniz Institute for Primate Research, which is financed by the Bundesrepublik Deutschland and the Bundesländer (Federal states) (Grant number 81Z0300201 to RB).
References
- 1.Aksoy I, Rognard C, Moulin A, Marcy G, Masfaraud E, Wianny F, Cortay V, Bellemin-Ménard A, Doerflinger N, Dirheimer M, Mayère C, Bourillot PY, Lynch C, Raineteau O, Joly T, Dehay C, Serrano M, Afanassieff M, Savatier P. Apoptosis G1 phase stall and premature differentiation account for low chimeric competence of human and rhesus monkey naive pluripotent stem cells. Stem Cell Reports. 2021;16:56–74. doi: 10.1016/j.stemcr.2020.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aoi T. 10th anniversary of iPS cells: The challenges that lie ahead. J Biochem Adv Access. 2016;160:121–129. doi: 10.1093/jb/mvw044. [DOI] [PubMed] [Google Scholar]
- 3.Aron Badin R, Bugi A, Williams S, Vadori M, Michael M, Jan C, Nassi A, Lecourtois S, Blancher A, Cozzi E, Hantraye P, Perrier AL. MHC matching fails to prevent long-term rejection of iPSC-derived neurons in non-human primates. Nat Commun. 2019;10:4357. doi: 10.1038/s41467-019-12324-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baghbaderani BA, Tian X, Neo BH, Burkall A, Dimezzo T, Sierra G, Zeng X, Warren K, Kovarcik DP, Fellner T, Rao MS. CGMP-manufactured human induced pluripotent stem cells are available for pre-clinical and clinical applications. Stem Cell Reports. 2015;5:647–659. doi: 10.1016/j.stemcr.2015.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bock C, Kiskinis E, Verstappen G, Gu H, Boulting G, Smith ZD, Ziller M, Croft GF, Amoroso MW, Oakley DH, Gnirke A, Eggan K, Meissner A. Reference maps of human es and ips cell variation enable high-throughput characterization of pluripotent cell lines. Cell. 2011;144:439–452. doi: 10.1016/j.cell.2010.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrne JA, Pedersen DA, Clepper LL, Nelson M, Sanger WG, Gokhale S, Wolf DP, Mitalipov SM. Producing primate embryonic stem cells by somatic cell nuclear transfer. Nature. 2007;450:497–502. doi: 10.1038/nature06357. [DOI] [PubMed] [Google Scholar]
- 7.Cardoso-Moreira M, Sarropoulos I, Velten B, Mort M, Cooper DN, Huber W, Kaessmann H. Developmental gene expression differences between humans and mammalian models. Cell Rep. 2020;33:108308. doi: 10.1016/j.celrep.2020.108308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chan AWS, Cheng PH, Neumann A, Yang JJ. Reprogramming huntington monkey skin cells into pluripotent stem cells. Cell Reprogram. 2010;12:509–517. doi: 10.1089/cell.2010.0019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, Niu Y, Li Y, Ai Z, Kang Y, Shi H, Xiang Z, Yang Z, Tan T, Si W, Li W, Xia X, Zhou Q, Ji W, Li T. Generation of cynomolgus monkey chimeric fetuses using embryonic stem cells. Cell Stem Cell. 2015;17:116–124. doi: 10.1016/j.stem.2015.06.004. [DOI] [PubMed] [Google Scholar]
- 10.Chen ZZ, Niu YY. Stem cell therapy for Parkinson’s disease using non-human primate models. Zool Res. 2019;40:349–357. doi: 10.24272/j.issn.2095-8137.2019.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cibelli JB, Grant KA, Chapman KB, Cunniff K, Worst T, Green HL, Walker SJ, Gutin PH, Vilner L, Tabar V, Dominko T, Kane J, Wettstein PJ, Lanza RP, Studer L, Vrana KE, West MD. Parthenogenetic stem cells in nonhuman primates. Science. 2002;295:819. doi: 10.1126/science.1065637. [DOI] [PubMed] [Google Scholar]
- 12.Coppiello G, Abizanda G, Aguado N, Iglesias E, Arellano-Viera E, Rodriguez-Madoz JR, Carvajal-Vergara X, Prosper F, Aranguren XL. Generation of Macaca fascicularis iPS cell line ATCi-MF1 from adult skin fibroblasts using non-integrative Sendai viruses. Stem Cell Res. 2017;21:1–4. doi: 10.1016/j.scr.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 13.Cox LA, Olivier M, Spradling-Reeves K, Karere GM, Comuzzie AG, VandeBerg JL. Nonhuman primates and translational research-Cardiovascular disease. ILAR J. 2017;58:235–250. doi: 10.1093/ilar/ilx025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Daadi MM, Barberi T, Shi Q, Lanford RE. Nonhuman primate models in translational regenerative medicine. Stem Cells Dev. 2014;23:83–87. doi: 10.1089/scd.2014.0374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Debowski K, Drummer C, Lentes J, Cors M, Dressel R, Lingne T, Salinas-riester G, Fuchs S, Sasaki E, Behr R. The transcriptomes of novel marmoset monkey embryonic stem cell lines reflect distinct genomic features. Sci Rep. 2016;6:29122. doi: 10.1038/srep29122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Debowski K, Warthemann R, Lentes J, Salinas-Riester G, Dressel R, Langenstroth D, Gromoll J, Sasaki E, Behr R. Non-viral generation of marmoset monkey iPS cells by a six-factor-in-one-vector approach. PLoS One. 2015;10:e0118424. doi: 10.1371/journal.pone.0118424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dighe V, Clepper L, Pedersen D, Byrne J, Ferguson B, Gokhale S, Penedo MCT, Wolf D, Mitalipov S. Heterozygous embryonic stem cell lines derived from nonhuman primate parthenotes. Stem Cells. 2008;26:756–766. doi: 10.1634/stemcells.2007-0869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Emborg ME, Liu Y, Xi J, Zhang X, Yin Y, Lu J, Joers V, Swanson C, Holden JE, Zhang SC. Induced pluripotent stem cell-derived neural cells survive and mature in the nonhuman primate brain. Cell Rep. 2013;3:646–650. doi: 10.1016/j.celrep.2013.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fang R, Liu K, Zhao Y, Li H, Zhu D, Du Y, Xiang C, Li X, Liu H, Miao Z, Zhang X, Shi Y, Yang W, Xu J, Deng H. Generation of naive induced pluripotent stem cells from rhesus monkey fibroblasts. Cell Stem Cell. 2014;15:488–497. doi: 10.1016/j.stem.2014.09.004. [DOI] [PubMed] [Google Scholar]
- 20.Friedrich Ben-Nun I, Montague SC, Houck ML, Tran HT, Garitaonandia I, Leonardo TR, Wang YC, Charter SJ, Laurent LC, Ryder OA, Loring JF. Induced pluripotent stem cells from highly endangered species. Nat Methods. 2011;8:829–831. doi: 10.1038/nmeth.1706. [DOI] [PubMed] [Google Scholar]
- 21.Funakoshi S, Miki K, Takaki T, Okubo C, Hatani T, Chonabayashi K, Nishikawa M, Takei I, Oishi A, Narita M, Hoshijima M, Kimura T, Yamanaka S, Yoshida Y. Enhanced engraftment proliferation and therapeutic potential in heart using optimized human iPSC-derived cardiomyocytes. Sci Rep. 2016;6:19111. doi: 10.1038/srep19111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fusaki N, Ban H, Nishiyama A, Saeki K, Hasegawa M. Efficient induction of transgene-free human pluripotent stem cells using a vector based on Sendai virus an RNA virus that does not integrate into the host genome. Proc Japan Acad Ser B Phys Biol Sci. 2009;85:348–362. doi: 10.2183/pjab.85.348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gallego Romero I, Pavlovic BJ, Hernando-Herraez I, Zhou X, Ward MC, Banovich NE, Kagan CL, Burnett JE, Huang CH, Mitrano A, Chavarria CI, Friedrich Ben-Nun I, Li Y, Sabatini K, Leonardo TR, Parast M, Marques-Bonet T, Laurent LC, Loring JF, Gilad Y. A panel of induced pluripotent stem cells from chimpanzees: a resource for comparative functional genomics. Elife. 2015;4:e07103. doi: 10.7554/eLife.07103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Geuder J, Wange LE, Janjic A, Radmer J, Janssen P, Bagnoli JW, Müller S, Kaul A, Ohnuki M, Enard W. A non-invasive method to generate induced pluripotent stem cells from primate urine. Sci Rep. 2021;11:3516. doi: 10.1038/s41598-021-82883-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo Y, Pu WT. Cardiomyocyte maturation: new phase in development. Circ Res. 2020:1086–1106. doi: 10.1161/CIRCRESAHA.119.315862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong SG, Lin Y, Dunbar CE, Zou J. The role of nonhuman primate animal models in the clinical development of pluripotent stem cell therapies. Mol Ther. 2016;24:1165–1169. doi: 10.1038/mt.2016.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishigaki H, Pham VL, Terai J, Sasamura T, Nguyen CT, Ishida H, Okahara J, Kaneko S, Shiina T, Nakayama M, Itoh Y, Ogasawara K. No tumorigenicity of allogeneic induced pluripotent stem cells in major histocompatibility complex-matched cynomolgus macaques. Cell Transplant. 2021 doi: 10.1177/0963689721992066. doi: 10.1177/0963689721992066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jo HY, Han HW, Jung I, Ju JH, Park SJ, Moon S, Geum D, Kim H, Park HJ, Kim S, Stacey GN, Koo SK, Park MH, Kim JH. Development of genetic quality tests for good manufacturing practice-compliant induced pluripotent stem cells and their derivatives. Sci Rep. 2020;10:3939. doi: 10.1038/s41598-020-60466-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Johnsen DO, Johnson DK, Whitney RA. Nonhuman Primates in Biomedical Research. In: Abee CR, Mansfield K, Tardif S, Morris T, editors. 2nd ed. Amsterdam: Elsevier; 2012. [Google Scholar]
- 30.Kim D, Kim CH, Moon J Il, Chung YG, Chang MY, Han BS, Ko S, Yang E, Cha KY, Lanza R, Kim KS. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kishimoto K, Shimada A, Shinohara H, Takahashi T, Yamada Y, Higuchi Y, Yoneda N, Suemizu H, Kawai K, Kurotaki Y, Hanazawa K, Takashima Y, Sasaki E. Establishment of novel common marmoset embryonic stem cell lines under various conditions. Stem Cell Res. 2021;53:102252. doi: 10.1016/j.scr.2021.102252. [DOI] [PubMed] [Google Scholar]
- 32.Kobayashi Y, Okada Y, Itakura G, Iwai H, Nishimura S, Yasuda A, Nori S, Hikishima K, Konomi T, Fujiyoshi K, Tsuji O, Toyama Y, Yamanaka S, Nakamura M, Okano H. Pre-evaluated safe human iPSC-derived neural stem cells promote functional recovery after spinal cord injury in common marmoset without tumorigenicity. PLoS One. 2012;7:e52787. doi: 10.1371/journal.pone.0052787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee JH, Laronde S, Collins TJ, Shapovalova Z, Tanasijevic B, McNicol JD, Fiebig-Comyn A, Benoit YD, Lee JB, Mitchell RR, Bhatia M. Lineage-specific differentiation is influenced by state of human pluripotency. Cell Rep. 2017;19:20–35. doi: 10.1016/j.celrep.2017.03.036. [DOI] [PubMed] [Google Scholar]
- 34.Liu G, David BT, Trawczynski M, Fessler RG. Advances in pluripotent stem cells: history mechanisms technologies and applications. Stem Cell Rev Rep. 2020;16:3–32. doi: 10.1007/s12015-019-09935-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu H, Zhu F, Yong J, Zhang P, Hou P, Li H, Jiang W, Cai J, Liu M, Cui K, Qu X, Xiang T, Lu D, Chi X, Gao G, Ji W, Ding M, Deng H. Generation of induced pluripotent stem cells from adult rhesus monkey fibroblasts. Cell Stem Cell. 2008;3:587–590. doi: 10.1016/j.stem.2008.10.014. [DOI] [PubMed] [Google Scholar]
- 36.Liu X, Li W, Fu X, Xu Y. The immunogenicity and immune tolerance of pluripotent stem cell derivatives. Front Immunol. 2017;8:6–11. doi: 10.3389/fimmu.2017.00645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu YW, Chen B, Yang X, Fugate JA, Kalucki FA, Futakuchi-Tsuchida A, Couture L, Vogel KW, Astley CA, Baldessari A, Ogle J, Don CW, Steinberg ZL, Seslar SP, Tuck SA, Tsuchida H, Naumova AV, Dupras SK, Lyu MS, Lee J, et al. Human embryonic stem cell–derived cardiomyocytes restore function in infarcted hearts of non-human primates. Nat Biotechnol. 2018;36:597–605. doi: 10.1038/nbt.4162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mandai M, Watanabe A, Kurimoto Y, Hirami Y, Morinaga C, Daimon T, Fujihara M, Akimaru H, Sakai N, Shibata Y, Terada M, Nomiya Y, Tanishima S, Nakamura M, Kamao H, Sugita S, Onishi A, Ito T, Fujita K, Kawamata S, et al. Autologous induced stem-cellderived retinal cells for macular degeneration. N Engl J Med. 2017;376:1038–1046. doi: 10.1056/NEJMoa1608368. [DOI] [PubMed] [Google Scholar]
- 39.Marchetto MC, Hrvoj-Mihic B, Kerman BE, Yu DX, Vadodaria KC, Linker SB, Narvaiza I, Santos R, Denli AM, Mendes AP, Oefner R, Cook J, McHenry L, Grasmick JM, Heard K, Fredlender C, Randolph-Moore L, Kshirsagar R, Xenitopoulos R, Chou G, et al. Species-specific maturation profiles of human, chimpanzee and bonobo neural cells. Elife. 2019;8:e37527. doi: 10.7554/eLife.37527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marchetto MCN, Narvaiza I, Denli AM, Benner C, Lazzarini TA, Nathanson JL, Paquola ACM, Desai KN, Herai RH, Weitzman MD, Yeo GW, Muotri AR, Gage FH. Differential L1 regulation in pluripotent stem cells of humans and apes. Nature. 2013;503:525–529. doi: 10.1038/nature12686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maria S, Helle B, Tristan L, Gaynor S, Michele M, Teresia O, Oliver C, Roger S, Ole I. Improved cell therapy protocol for Parkinson’s disease based on differentiation efficiency and safety of hESC-hiPSC and non-human primate iPSC-derived DA neurons. Stem Cells. 2014;31:1548–1562. doi: 10.1002/stem.1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mattisona JA, Vaughan KL. An overview of nonhuman primates in aging research. Exp Gerontol. 2017;94:41–45. doi: 10.1016/j.exger.2016.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mitalipov S, Kuo H-C, Byrne J, Clepper L, Meisner L, Johnson J, Zeier R, Wolf D. Isolation and characterization of novel rhesus monkey embryonic stem cell lines. Stem Cells. 2006;24:2177–2186. doi: 10.1634/stemcells.2006-0125. [DOI] [PubMed] [Google Scholar]
- 44.Morizane A, Doi D, Kikuchi T, Okita K, Hotta A, Kawasaki T, Hayashi T, Onoe H, Shiina T, Yamanaka S, Takahashi J. Direct comparison of autologous and allogeneic transplantation of IPSC-derived neural cells in the brain of a nonhuman primate. Stem Cell Reports. 2013;1:283–292. doi: 10.1016/j.stemcr.2013.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Müller FJ, Goldmann J, Löser P, Loring JF. A call to standardize teratoma assays used to define human pluripotent cell lines. Cell Stem Cell. 2010;6:412–414. doi: 10.1016/j.stem.2010.04.009. [DOI] [PubMed] [Google Scholar]
- 46.Müller FJ, Schuldt BM, Williams R, Mason D, Altun G, Papapetrou EP, Danner S, Goldmann JE, Herbst A, Schmidt NO, Aldenhoff JB, Laurent LC, Loring JF. A bioinformatic assay for pluripotency in human cells. Nat Methods. 2011;8:315–317. doi: 10.1038/nmeth.1580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Müller T, Fleischmann G, Eildermann K, Mätz-Rensing K, Horn PA, Sasaki E, Behr R. A novel embryonic stem cell line derived from the common marmoset monkey (Callithrix jacchus) exhibiting germ cell-like characteristics. Hum Reprod. 2009;24:1359–1372. doi: 10.1093/humrep/dep012. [DOI] [PubMed] [Google Scholar]
- 48.Nagoshi N, Okano H, Nakamura M. Regenerative therapy for spinal cord injury using iPSC technology. Inflamm Regen. 2020;40:40. doi: 10.1186/s41232-020-00149-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Nakajima M, Yoshimatsu S, Sato T, Nakamura M, Okahara J, Sasaki E, Shiozawa S, Okano H. Establishment of induced pluripotent stem cells from common marmoset fibroblasts by RNA-based reprogramming. Biochem Biophys Res Commun. 2019;515:593–599. doi: 10.1016/j.bbrc.2019.05.175. [DOI] [PubMed] [Google Scholar]
- 50.Navara CS, Chaudhari S, McCarrey JR. Optimization of culture conditions for the derivation and propagation of baboon (Papio anubis) induced pluripotent stem cells. PLoS One. 2018;13:e0193195. doi: 10.1371/journal.pone.0193195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Navara CS, Hornecker J, Grow D, Chaudhari S, Hornsby PJ, Ichida JK, Eggan K, McCarrey JR. Derivation of induced pluripotent stem cells from the baboon: a nonhuman primate model for preclinical testing of stem cell therapies. Cell Reprogram. 2013;15:495–502. doi: 10.1089/cell.2012.0093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Navara CS, Mich-Basso JD, Redinger CJ, Ben-Yehudah A, Jacoby E, Kovkarova-Naumovski E, Sukhwani M, Orwig K, Kaminski N, Castro CA, Simerly CR, Schatten G. Pedigreed primate embryonic stem cells express homogeneous familial gene profiles. Stem Cells. 2007;25:2695–2704. doi: 10.1634/stemcells.2007-0286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nishimura K, Sano M, Ohtaka M, Furuta B, Umemura Y, Nakajima Y, Ikehara Y, Kobayashi T, Segawa H, Takayasu S, Sato H, Motomura K, Uchida E, Kanayasu-Toyoda T, Asashima M, Nakauchi H, Yamaguchi T, Nakanishia M. Development of defective and persistent Sendai virus vector: a unique gene delivery/expression system ideal for cell reprogramming. J Biol Chem. 2011;286:4760–4771. doi: 10.1074/jbc.M110.183780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okahara-Narita J, Umeda R, Nakamura S, Mori T, Noce T, Torii R. Induction of pluripotent stem cells from fetal and adult cynomolgus monkey fibroblasts using four human transcription factors. Primates. 2012;53:205–213. doi: 10.1007/s10329-011-0283-1. [DOI] [PubMed] [Google Scholar]
- 55.Okita K, Matsumura Y, Sato Y, Okada A, Morizane A, Okamoto S, Hong H, Nakagawa M, Tanabe K, Tezuka KI, Shibata T, Kunisada T, Takahashi M, Takahashi J, Saji H, Yamanaka S. A more efficient method to generate integration-free human iPS cells. Nat Methods. 2011;8:409–412. doi: 10.1038/nmeth.1591. [DOI] [PubMed] [Google Scholar]
- 56.Pavlovic BJ, Blake LE, Roux J, Chavarria C, Gilad Y. A comparative assessment of human and chimpanzee iPSC-derived cardiomyocytes with primary heart tissues. Sci Rep. 2018;8:15312. doi: 10.1038/s41598-018-33478-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petkov CI, Jarvis ED. Birds primates and spoken language origins: Behavioral phenotypes and neurobiological substrates. Front Evol Neurosci. 2012;4:12. doi: 10.3389/fnevo.2012.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Petkov S, Dressel R, Rodriguez-Polo I, Behr R. Controlling the switch from neurogenesis to pluripotency during marmoset monkey somatic cell reprogramming with self-replicating mRNAs and small molecules. Cells. 2020;9:2422. doi: 10.3390/cells9112422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Petkov SG, Behr R. Generation of marmoset monkey iPSCs with self-replicating VEEmRNAs in feeder-free conditions. Methods Mol Biol. 2021 doi: 10.1007/7651_2021_381. doi: 10.1007/7651_2021_381. [DOI] [PubMed] [Google Scholar]
- 60.Qiu Z, Mishra A, Li M, Farnsworth SL, Guerra B, Lanford RE, Hornsby PJ. Marmoset induced pluripotent stem cells: Robust neural differentiation following pretreatment with dimethyl sulfoxide. Stem Cell Res. 2015;15:141–150. doi: 10.1016/j.scr.2015.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rodriguez-Polo I, Mißbach S, Petkov S, Mattern F, Maierhofer A, Grządzielewska I, Tereshchenko Y, Urrutia-Cabrera D, Haaf T, Dressel R, Bartels I, Behr R. A piggyBac based platform for genome editing and clonal rhesus macaque iPSC line derivation. Sci Rep. 2021a;11:15439. doi: 10.1038/s41598-021-94419-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rodriguez-Polo I, Stauske M, Becker A, Bartels I, Dressel R, Behr R. Baboon induced pluripotent stem cell generation by piggyBac transposition of reprogramming factors. Primate Biol. 2019;6:75–86. doi: 10.5194/pb-6-75-2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Rodríguez-Polo I, Stauske M, Behr R. Generation and cultivation of transgene-free macaque and baboon iPSCs under chemically defined conditions. Methods Mol Biol. 2021b doi: 10.1007/7651_2021_380. doi: 10.1007/7651_2021_380. [DOI] [PubMed] [Google Scholar]
- 64.Roodgar M, Suchy FP, Bajpai V, Viches-Moure JG, Bhadury J, Oikonomopoulos A, Wu JC, Mankowski JL, Loh KM, Nakauchi H, VandeVoort CA, Snyder MP. Cross-species blastocyst chimerism between nonhuman primates using iPSCs Morteza. bioRxiv. 2019 doi:10.1101/635250. [Google Scholar]
- 65.Roth GA, Mensah GA, Johnson CO, Addolorato G, Ammirati E, Baddour LM, Barengo NC, Beaton AZ, Benjamin EJ, Benziger CP, Bonny A, Brauer M, Brodmann M, Cahill TJ, Carapetis J, Catapano AL, Chugh SS, Cooper LT, Coresh J, Criqui M, et al. Global Burden of Cardiovascular Diseases and Risk Factors, 1990–2019: Update From the GBD 2019 Study. J Am Coll Cardiol. 2020;76:2982–3021. doi: 10.1016/j.jacc.2020.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Sasaki E, Hanazawa K, Kurita R, Akatsuka A, Yoshizaki T, Ishii H, Tanioka Y, Ohnishi Y, Suemizu H, Sugawara A, Tamaoki N, Izawa K, Nakazaki Y, Hamada H, Suemori H, Asano S, Nakatsuji N, Okano H, Tani K. Establishment of novel embryonic stem cell lines derived from the common marmoset (Callithrix jacchus) Stem Cells. 2005;23:1304–1313. doi: 10.1634/stemcells.2004-0366. [DOI] [PubMed] [Google Scholar]
- 67.Schlaeger TM, Daheron L, Brickler TR, Entwisle S, Chan K, Cianci A, DeVine A, Ettenger A, Fitzgerald K, Godfrey M, Gupta D, McPherson J, Malwadkar P, Gupta M, Bell B, Doi A, Jung N, Li X, Lynes MS, Brookes E, et al. A comparison of non-integrating reprogramming methods. Nat Biotechnol. 2015;33:58–63. doi: 10.1038/nbt.3070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Sekine K, Tsuzuki S, Yasui R, Kobayashi T, Ikeda K, Hamada Y, Kanai E, Camp JG, Treutlein B, Ueno Y, Okamoto S, Taniguchi H. Robust detection of undifferentiated iPSC among differentiated cells. Sci Rep. 2020;10:10293. doi: 10.1038/s41598-020-66845-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Seo BJ, Hong YJ, Do JT. Cellular reprogramming using protein and cell-penetrating peptides. Int J Mol Sci. 2017;18:552. doi: 10.3390/ijms18030552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shi Q, Hodara V, Simerly CR, Schatten GP, Vandeberg JL. Ex vivo reconstitution of arterial endothelium by embryonic stem cell-derived endothelial progenitor cells in baboons. Stem Cells Dev. 2013;22:631–642. doi: 10.1089/scd.2012.0313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Shi Y, Inoue H, Wu JC, Yamanaka S. Induced pluripotent stem cell technology: a decade of progress. Nat Rev Drug Discov. 2017;16:115–130. doi: 10.1038/nrd.2016.245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Shiba Y, Gomibuchi T, Seto T, Wada Y, Ichimura H, Tanaka Y, Ogasawara T, Okada K, Shiba N, Sakamoto K, Ido D, Shiina T, Ohkura M, Nakai J, Uno N, Kazuki Y, Oshimura M, Minami I, Ikeda U. Allogeneic transplantation of iPS cell-derived cardiomyocytes regenerates primate hearts. Nature. 2016;538:388–391. doi: 10.1038/nature19815. [DOI] [PubMed] [Google Scholar]
- 73.Shimada H, Okada Y, Ibata K, Ebise H, Ota S ichi, Tomioka I, Nomura T, Maeda T, Kohda K, Yuzaki M, Sasaki E, Nakamura M, Okano H. Efficient derivation of multipotent neural stem/progenitor cells from non-human primate embryonic stem cells. PLoS One. 2012;7:e49469. doi: 10.1371/journal.pone.0049469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shimozawa N, Ono R, Shimada M, Shibata H, Takahashi I, Inada H, Takada T, Nosaka T, Yasutomi Y. Cynomolgus monkey induced pluripotent stem cells established by using exogenous genes derived from the same monkey species. Differentiation. 2013;85:131–139. doi: 10.1016/j.diff.2013.02.004. [DOI] [PubMed] [Google Scholar]
- 75.Silva TP, Bekman EP, Fernandes TG, Vaz SH, Rodrigues CAV, Diogo MM, Cabral JMS, Carmo-Fonseca M. Maturation of human pluripotent stem cell-derived cerebellar neurons in the absence of co-culture. Front Bioeng Biotechnol. 2020;8:70. doi: 10.3389/fbioe.2020.00070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Simerly CR, Navara CS, Castro CA, Turpin JC, Redinger CJ, Mich-Basso JD, Jacoby ES, Grund KJ, McFarland DA, Oliver SL, Ben-Yehudah A, Carlisle DL, Frost P, Penedo C, Hewitson L, Schatten G. Establishment and characterization of baboon embryonic stem cell lines: An Old World Primate model for regeneration and transplantation research. Stem Cell Res. 2009;2:178–187. doi: 10.1016/j.scr.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Skovronsky DM, Lee VMY, Trojanowski JQ. Neurodegenerative diseases: new concepts of pathogenesis and their therapeutic implications. Annu Rev Pathol. 2006;1:151–170. doi: 10.1146/annurev.pathol.1.110304.100113. [DOI] [PubMed] [Google Scholar]
- 78.Stauske M, Rodriguez Polo I, Haas W, Knorr DY, Borchert T, Streckfuss-Bömeke K, Dressel R, Bartels I, Tiburcy M, Zimmermann WH, Behr R. Non-human primate iPSC generation cultivation and cardiac differentiation under chemically defined conditions. Cells. 2020;9:1349. doi: 10.3390/cells9061349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Suemori H, Nakatsuji N. Generation and characterization of monkey embryonic stem cells. Methods Mol Biol. 2006;329:81–89. doi: 10.1385/1-59745-037-5:81. [DOI] [PubMed] [Google Scholar]
- 80.Suemori H, Tada T, Torii R, Hosoi Y, Kobayashi K, Imahie H, Kondo Y, Iritani A, Nakatsuji N. Establishment of embryonic stem cell lines from cynomolgus monkey blastocysts produced by IVF or ICSI. Dev Dyn. 2001;222:273–279. doi: 10.1002/dvdy.1191. [DOI] [PubMed] [Google Scholar]
- 81.Sun N, Panetta NJ, Gupta DM, Wilson KD, Lee A, Jia F, Hu S, Cherry AM, Robbins RC, Longaker MT, Wu JC. Feeder-free derivation of induced pluripotent stem cells from adult human adipose stem cells. Proc Natl Acad Sci U S A. 2009;106:15720–15725. doi: 10.1073/pnas.0908450106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 83.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 84.Tao Y, Vermilyea SC, Zammit M, Lu J, Olsen M, Metzger JM, Yao L, Chen Y, Phillips S, Holden JE, Bondarenko V, Block WF, Barnhart TE, Schultz-Darken N, Brunner K, Simmons H, Christian BT, Emborg ME, Zhang SC. Autologous transplant therapy alleviates motor and depressive behaviors in parkinsonian monkeys. Nat Med. 2021;27:632–639. doi: 10.1038/s41591-021-01257-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Teshigawara R, Cho J, Kameda M, Tada T. Mechanism of human somatic reprogramming to iPS cell. Lab Investig. 2017;97:1152–1157. doi: 10.1038/labinvest.2017.56. [DOI] [PubMed] [Google Scholar]
- 86.Thomson JA, Kalishman J, Golos TG, Durning M, Harris CP, Becker RA, Hearn JP. Isolation of a primate embryonic stem cell line. Proc Natl Acad Sci U S A. 1995;92:7844–7848. doi: 10.1073/pnas.92.17.7844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Thomson JA, Kalishman J, Golos TG, Durning M, Harris CP, Hearn JP. Pluripotent cell lines derived from common marmoset (Callithrix jacchus) blastocysts. Biol Reprod. 1996;55:254–259. doi: 10.1095/biolreprod55.2.254. [DOI] [PubMed] [Google Scholar]
- 88.Tiburcy M, Hudson JE, Balfanz P, Schlick S, Meyer T, Chang Liao ML, Levent E, Raad F, Zeidler S, Wingender E, Riegler J, Wang M, Gold JD, Kehat I, Wettwer E, Ravens U, Dierickx P, van Laake LW, Goumans MJ, Khadjeh S, et al. Defined engineered human myocardium with advanced maturation for applications in heart failure modeling and repair. Circulation. 2017;135:1832–1847. doi: 10.1161/CIRCULATIONAHA.116.024145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Tomioka I, Maeda T, Shimada H, Kawai K, Okada Y, Igarashi H, Oiwa R, Iwasaki T, Aoki M, Kimura T, Shiozawa S, Shinohara H, Suemizu H, Sasaki E, Okano H. Generating induced pluripotent stem cells from common marmoset (Callithrix jacchus) fetal liver cells using defined factors including Lin28. Genes to Cells. 2010;15:959–969. doi: 10.1111/j.1365-2443.2010.01437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Torrez LB, Perez Y, Yang J, Zur Nieden NI, Klassen H, Liew CG. Derivation of neural progenitors and retinal pigment epithelium from common marmoset and human pluripotent stem cells. Stem Cells Int. 2012;2012:417865. doi: 10.1155/2012/417865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tsankov AM, Akopian V, Pop R, Chetty S, Gifford CA, Daheron L, Tsankova NM, Meissner A. A qPCR ScoreCard quantifies the differentiation potential of human pluripotent stem cells. Nat Biotechnol. 2015;33:1182–1192. doi: 10.1038/nbt.3387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Vermilyea SC, Guthrie S, Meyer M, Smuga-Otto K, Braun K, Howden S, Thomson JA, Zhang SC, Emborg ME, Golos TG. Induced pluripotent stem cell-derived dopaminergic neurons from adult common marmoset fibroblasts. Stem Cells Dev. 2017;26:1225–1235. doi: 10.1089/scd.2017.0069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Vrana KE, Hipp JD, Goss AM, McCool BA, Riddle DR, Walker SJ, Wettstein PJ, Studer LP, Tabar V, Cunniff K, Chapman K, Vilner L, West MD, Grant KA, Cibelli JB. Nonhuman primate parthenogenetic stem cells. Proc Natl Acad Sci U S A. 2003;100:11911–11916. doi: 10.1073/pnas.2034195100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Wang S, Zou C, Fu L, Wang B, An J, Song G, Wu J, Tang X, Li M, Zhang J, Yue F, Zheng C, Chan P, Zhang YA, Chen Z. Autologous iPSC-derived dopamine neuron transplantation in a nonhuman primate Parkinson’s disease model. Cell Discov. 2015;1:15012. doi: 10.1038/celldisc.2015.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wang X, Lu M, Tian X, Ren Y, Li Y, Xiang M, Chen S. Diminished expression of major histocompatibility complex facilitates the use of human induced pluripotent stem cells in monkey. Stem Cell Res Ther. 2020;11:334. doi: 10.1186/s13287-020-01847-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Watanabe T, Yamazaki S, Yoneda N, Shinohara H, Tomioka I, Higuchi Y, Yagoto M, Ema M, Suemizu H, Kawai K, Sasaki E. Highly efficient induction of primate iPS cells by combining RNA transfection and chemical compounds. Genes to Cells. 2019;24:473–484. doi: 10.1111/gtc.12702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Wiedemann A, Hemmer K, Bernemann I, Göhring G, Pogozhykh O, Figueiredo C, Glage S, Schambach A, Schwamborn JC, Blasczyk R, Müller T. Induced pluripotent stem cells generated from adult bone marrow–derived cells of the nonhuman primate (callithrix jacchus) using a novel quad-cistronic and excisable lentiviral vector. Cell Reprogram. 2012;14:485–496. doi: 10.1089/cell.2012.0036. [DOI] [PubMed] [Google Scholar]
- 98.Woltjen K, Hämäläinen R, Mark Kibschull, Mileikovsky M, Nagy A. Transgene-free production of pluripotent stem cells using piggyBac transposons. Methods Mol Biol. 2011;767:87–103. doi: 10.1007/978-1-61779-201-4_7. [DOI] [PubMed] [Google Scholar]
- 99.Wu Y, Mishra A, Qiu Z, Farnsworth S, Tardif SD, Hornsby PJ. Nonhuman primate induced pluripotent stem cells in regenerative medicine. Stem Cells Int. 2012;2012:767195. doi: 10.1155/2012/767195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Wu Y, Zhang Y, Mishra A, Tardif SD, Hornsby PJ. Generation of induced pluripotent stem cells from newborn marmoset skin fibroblasts. Stem Cell Res. 2010;4:180–188. doi: 10.1016/j.scr.2010.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Wunderlich S, Haase A, Merkert S, Beier J, Schwanke K, Schambach A, Glage S, Göhring G, Curnow EC, Martin U. Induction of pluripotent stem cells from a cynomolgus monkey using a polycistronic simian immunodeficiency virus-based vector differentiation toward functional cardiomyocytes and generation of stably expressing reporter lines. Cell Reprogram. 2012;14:471–484. doi: 10.1089/cell.2012.0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wunderlich S, Kircher M, Vieth B, Haase A, Merkert S, Beier J, Göhring G, Glage S, Schambach A, Curnow EC, Pääbo S, Martin U, Enard W. Primate iPS cells as tools for evolutionary analyses. Stem Cell Res. 2014;12:622–629. doi: 10.1016/j.scr.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 103.Yamanaka S. Pluripotent stem cell-based cell therapy—promise and challenges. Cell Stem Cell. 2020;27:523–531. doi: 10.1016/j.stem.2020.09.014. [DOI] [PubMed] [Google Scholar]
- 104.Yang G, Hong H, Torres A, Malloy KE, Choudhury GR, Kim J, Daadi MM. Standards for deriving nonhuman primate-induced pluripotent stem cells neural stem cells and dopaminergic lineage. Int J Mol Sci. 2018;19:2788. doi: 10.3390/ijms19092788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Yoshimatsu S, Nakajima M, Iguchi A, Sanosaka T, Sato T, Nakamura M, Nakajima R, Arai E, Ishikawa M, Imaizumi K, Watanabe H, Okahara J, Noce T, Takeda Y, Sasaki E, Behr R, Edamura K, Shiozawa S, Okano H. Non-viral induction of transgene-free iPSCs from somatic fibroblasts of multiple mammalian species. Stem Cell Reports. 2021;16:754–770. doi: 10.1016/j.stemcr.2021.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Zhang H, Ma Y, Gu J, Liao B, Li J, Wong J, Jin Y. Reprogramming of somatic cells via TAT-mediated protein transduction of recombinant factors. Biomaterials. 2012;33:5047–5055. doi: 10.1016/j.biomaterials.2012.03.061. [DOI] [PubMed] [Google Scholar]
- 107.Zhao X, Chen H, Xiao D, Yang H, Itzhaki I, Qin X, Chour T, Aguirre A, Lehmann K, Kim Y, Shukla P, Holmström A, Zhang JZ, Zhuge Y, Ndoye BC, Zhao M, Neofytou E, Zimmermann WH, Jain M, Wu JC. Comparison of non-human primate versus human induced pluripotent stem cell-derived cardiomyocytes for treatment of myocardial infarction. Stem Cell Reports. 2018;10:422–435. doi: 10.1016/j.stemcr.2018.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Zhong B, Trobridge GD, Zhang X, Watts KL, Ramakrishnan A, Wohlfahrt M, Adair JE, Kiem HP. Efficient generation of nonhuman primate induced pluripotent stem cells. Stem Cells Dev. 2011;20:795–807. doi: 10.1089/scd.2010.0343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Zou XY, Yang HY, Yu Z, Tan XB, Yan X, Huang GTJ. Establishment of transgene-free induced pluripotent stem cells reprogrammed from human stem cells of apical papilla for neural differentiation. Stem Cell Res Ther. 2012;3:43. doi: 10.1186/scrt134. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.