Abstract
Aberrant activation of the RAS family of guanosine triphosphatases (GTPases) is prevalent in lung adenocarcinoma, with somatic mutation of KRAS occurring in ~30% of tumors. We previously identified somatic mutations and amplifications of the gene encoding RAS family GTPase RIT1 in lung adenocarcinomas. To explore the biological pathways regulated by RIT1 and how they relate to the oncogenic KRAS network, we performed quantitative proteomic, phosphoproteomic, and transcriptomic profiling of isogenic lung epithelial cells in which we ectopically expressed wild-type or cancer-associated variants of RIT1 and KRAS. We found that both mutant KRAS and mutant RIT1 promoted canonical RAS signaling, and that overexpression of wild-type RIT1 partially phenocopied oncogenic RIT1 and KRAS, including induction of epithelial-to-mesenchymal transition. Our findings suggest that RIT1 protein abundance is a factor in its pathogenic function. Therefore, chromosomal amplification of wild-type RIT1 in lung and other cancers may be tumorigenic.
Introduction
The RAS-family GTPase-encoding gene RIT1 was found to harbor somatic mutations in lung cancer (1) and myeloid leukemias (2). Disrupted regulation of RAS family genes and RAS effector pathways is a predominant feature of many human cancers. In particular, somatic mutation of the KRAS proto-oncogene is prevalent in lung adenocarcinoma, where KRAS is mutated in up to 30% of tumors. Cancer-associated KRAS variants include G12V and Q61H, which alter the normal regulation of KRAS GTPase activity by disrupting GTP hydrolysis or reducing the RAS protein’s ability to physically associate with GTPase-activating proteins (GAPs) (3, 4). This uncontrolled increase in GTP-bound KRAS results in heightened downstream cellular signaling through the canonical RAS effector pathways RAF–MEK and PI3K–AKT, as well as others. Following the discovery of cancer-associated RAS mutations in the 1980s (5, 6), thousands of studies have delineated the critical pathways involved in RAS-mediated cellular transformation, metastasis, and metabolism.
Notably, in addition to somatic mutations in cancer, germline RIT1 mutations are found in families with Noonan syndrome, a developmental “RAS”-opathy involving altered craniofacial morphology and cardiac abnormalities (7), that is caused by germline mutations in KRAS itself or other RAS-pathway genes such as SOS1, SOS2, LZTR1, and SHOC2 (https://omim.org/). In cancer and Noonan syndrome, RIT1 mutations are found in tumors or individuals that lack canonical KRAS mutations, suggesting that activated RIT1 may impart the same phenotypes conferred by activation of RAS.
Although prior studies have characterized the role of RIT1 in neural development (8) and we and others have described the role of mutant RIT1 in cellular transformation (1, 9, 10), knowledge of the function of cancer- and Noonan-associated RIT1 variants is relatively limited. Unlike in KRAS, mutations in RIT1 are rarely observed near the critical glycine residues involved in GTP hydrolysis, which are Gly12/13 in KRAS and Gly30/31 in RIT1. Instead, RIT1 mutations occur most frequently near the switch II domain, the region also altered by Gln61 mutations in KRAS (Fig. 1A). Gln61 and Gly12/13 mutations in RAS reduce association with regulatory GAPs or alter intrinsic hydrolysis rate (11). It is possible these differences in mutational spectrum reflect functional differences in regulation of RIT1 and KRAS GTPase activity, and no GAPs that regulate RIT1 have been identified to date.
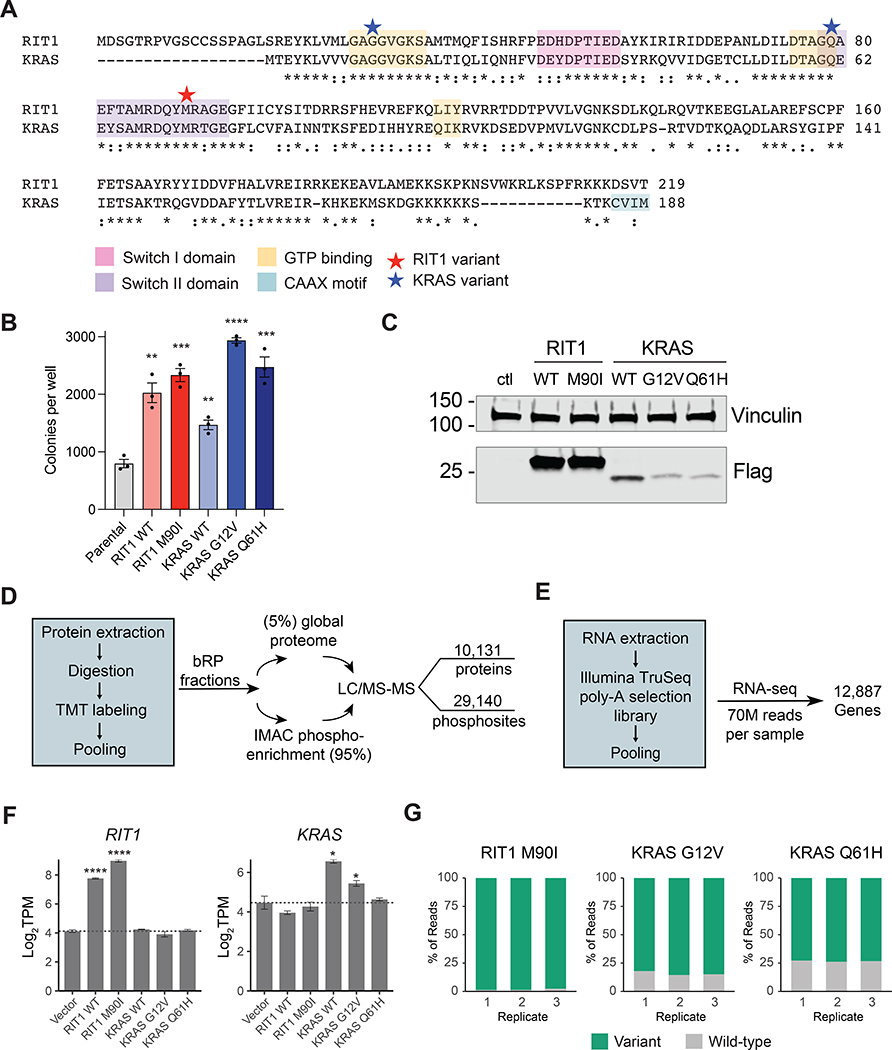
Figure 1. Multi-omic profiling of KRAS- and RIT1-mutant human lung epithelial cells.
(A) Protein alignment of KRAS-4B (Uniprot #P01116-2) and RIT1 Isoform 1 (Uniprot #Q92963-1) generated by ClustalW2 (51). Stars indicate the position of the RIT1M90I, KRASG12V, and KRASQ61 variants used in this study. Asterisks indicate fully conserved residues; colons indicate strongly conserved residues; periods indicate weakly conserved residues. (B) Soft agar colony formation assay of isogenic AALE human lung epithelial cells. Data shown is of N = 3 wells, representative of 3 independent experiments. ** P < 0.01, *** P < 0.001, and **** P < 0.0001 by two-tailed t-test against the parental cell data. (C) Western blot confirmation of ectopic expression of FLAG-tagged RIT1 and KRAS in isogenic AALE cells using an anti-FLAG antibody. Vinculin was used as a loading control. The data shown is representative of N > 3 independent experiments. (D) Workflow for the LC-MS/MS–based generation of proteome and phosphoproteome profiles. bRP, basic reverse phase chromatography. IMAC, immobilized metal affinity chromatography. (E) Workflow for the RNA-seq analysis. (F) mRNA quantification in transcripts per million (TPM) showing mean ± SD of RIT1 (left) or KRAS (right) in isogenic AALE cells, N = 3 RNA-seq libraries per cell line. * P < 0.05 and **** P < 0.0001 by two-tailed Student’s t-test compared to vector controls. (G) RNA-seq quantification of variant allele expression. Data shown is the percentage of reads at the M90I, G12V, or Q61H mutation site for the variant allele or wild-type allele. N = 3 RNA-seq libraries for each isogenic AALE cell line.
Whereas the regulation of enzymatic activity is thought to be the predominant determinant of RAS oncogenic function, both protein abundance and GTP binding may be key to RIT1 function. Biochemical assays have established that oncogenic RIT1 variants do enhance GTP-bound levels of RIT1 to varying degrees (12, 13). However, the molecular consequences of RIT1 switch II domain mutations may additionally be linked to the loss of RIT1’s physical interaction with LZTR1, a ubiquitin-conjugating enzyme responsible for stimulating degradation of RIT1 (12). Cancer- and Noonan-associated RIT1 variants lose the ability to interact with LZTR1 and consequently are highly overexpressed, resulting in increased signaling activity through the RAF–MEK pathway (12). Because germline loss-of-function mutations in LZTR1 result in increased abundance of wild-type RIT1 protein and also confer Noonan syndrome, overexpression of RIT1 in the absence of mutation may be sufficient to drive altered RIT1 signaling.
The pathogenic role of increased RIT1 expression in LZTR1-mutant Noonan syndrome raises the possibility that wild-type RIT1 overexpression in cancer may also play a pathogenic role. Indeed, 7–14% of lung adenocarcinomas (14, 15) harbor amplification of the RIT1 gene on 1p21.3 which is typically associated with overexpression of RIT1 mRNA. Amplification of RIT1 was found in an independent lung adenocarcinoma genomic study of samples from the Fudan University Shanghai Cancer Center (16). Focal RIT1 amplications have also been noted in uterine carcinosarcoma (17), and RIT1 amplification and/or overexpression may play a role in hepatocellular carcinoma and endometrial cancer (18, 19).
Based on RIT1’s homology to RAS proteins, candidate effectors of RIT1 function have been identified (12, 20–22) but no unbiased mapping of downstream RIT1-regulated pathways in human cells has been performed to date. Here, we sought to broadly describe the changes in the cellular proteome, phosphoproteome, and transcriptome induced by overexpressed wild-type RIT1 and by expression of RIT1M90I, a cancer- and Noonan-associated variant, and to compare these changes to those induced by expression of wild-type and oncogenic KRAS. Because RIT1 mutations occur frequently in lung cancer, we focused this study on analysis of RIT1 and KRAS signaling in a human lung epithelial cell line.
Results
Multi-omic profiling of RIT1- and RAS-transformed human lung epithelial cells
Although the cellular signaling regulated by KRAS has been extensively characterized, little is known about how RIT1 or its oncogenic variants perturb cellular signaling. To address this question in the context of lung cancer, we ectopically expressed either wild-type or variant forms of KRAS and RIT1 in AALE cells, which are immortalized, non-transformed human lung epithelial cells (23). To confirm the functional activity of each expressed variant, we quantified the anchorage-independent growth in soft agar of each isogenic cell line (Fig. 1B). Expression of wild-type or oncogenic variants of KRAS and RIT1 promoted growth in soft agar, confirming the biological activity of each expressed variant (Fig. 1, B and C).
To comprehensively identify the cellular signaling events associated with this phenotypic transformation, we profiled each isogenic cell line using both RNA-seq and deep proteome and phosphoproteome profiling by liquid chromatography tandem mass spectrometry (LC-MS/MS) Fig. 1, D and E). For LC-MS/MS, creation of two overlapping 10-plex tandem mass tag (TMT) pools enabled the relative quantification of the proteome and the phosphoproteome across replicates of the five isogenic cell lines compared to a vector control line (RIT1WT, RIT1M90I, KRASWT, KRASG12V, KRASQ61H; Fig. 1D and fig. S1, A to C). In total, 10,131 proteins were identified, 9002 of which were detected and quantified in every sample; additionally, 29,140 phosphopeptides were detected, 12,325 of which were identified in every sample, enabling comparative analysis across cell lines (tables S1 and S2 and Data Files S1 and S2).
For transcriptome analysis, RNA sequencing was performed in triplicate for each isogenic cell line to a median read-depth per replicate of 70.1 million reads (Fig. 1E, tables S3 and S4 and fig. S1D). 12,887 gene transcripts were quantified, including 7,749 whose corresponding protein was detected in the LC-MS/MS profiling. We note that no compensatory feedback regulation of RIT1 to KRAS expression or vice versa was observed (Fig. 1F). Despite relatively low protein expression of KRAS variants in the AALE lines (Fig. 1C), the majority of KRAS transcripts in each isogenic cell line corresponded to G12V or Q61H variants, respectively, with 84.1% of reads harboring the G12V mutation in KRASG12V cells, and 73.3% of reads harboring the Q61H mutation in KRASQ61H cells (Fig. 1G).
Next, we interrogated the phosphoproteome regulated by KRAS and RIT1. Protein phosphorylation is a reversible and dynamic mechanism of intracellular signaling that enables rapid intracellular transduction of signals controlling cell proliferation, survival, and metabolism. Although RIT1 and KRAS act as GTPase switches, they both additionally stimulate activation of cellular protein kinases, such as BRAF, resulting in substantial reorganization of the phosphoproteome. Because the stoichiometric ratio of phosphorylated to non-phosphorylated proteins is often believed to play an important role in cell signaling, we normalized phosphosite abundance to the total protein abundance for each phosphoprotein (table S2). Recognizing that in some cases absolute abundance of a phospho-peptide may be relevant, non-normalized phosphosite data is also provided (table S5).
Global similarity between oncogenic RIT1 and KRAS signaling networks
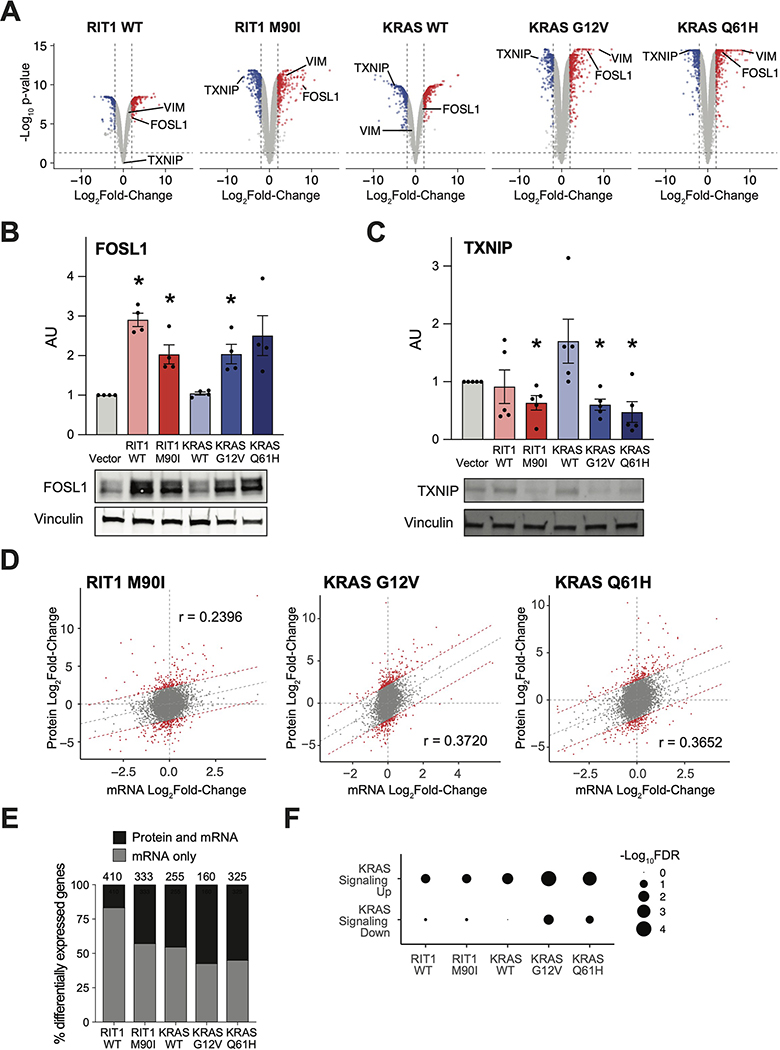
With our LC-MS/MS proteome profiles, we identified differentially abundant proteins in each condition compared to the vector control cells using a two-tailed moderated t-test (Fig. 2A). One of the top proteins with increased abundance in RIT1WT, RIT1M90I, KRASG12V, and KRASQ61H cells was the transcription factor FOSL1 (Fig. 2B), also known as FRA1, a basic leucine zipper transcription factor in the FOS family (24). Activation of RAS is known to promote transcriptional upregulation and protein stabilization of FOSL1 (25, 26). Western blot of independently-derived AALE isogenic lines confirmed greater abundance of FOSL1 in RIT1WT, RIT1M90I, and KRASG12V cells compared to vector expressing cells (Fig. 2B). Consistent with FOSL1 upregulation downstream of the MAPK pathway, analysis of our previous L1000 mRNA profiling dataset revealed common regulation of FOSL1 by RIT1, KRAS, and other RAS pathway oncogenes ARAF, BRAF, EGFR, and NRAS, but not other oncogenes such as MYC, MDM2, or NFE2L2 (fig. S2A).
Figure 2. Quantitative proteomic and transcriptomic profiling identifies similarity of oncogenic RIT1 and KRAS signaling.
(A) Volcano plots of global proteome data from isogenic AALE cells showing the Log2Fold-Change in protein abundance in each cell line compared to vector control cells. The y-axis displays the negative Log10(P-value) calculated from a one sample moderated t-test with multiple hypothesis correction by the Benjamini-Hochberg method. Data shown are generated from 2 to 4 technical LC-MS/MS replicates per cell line generated from N=2 biological replicates per cell line. (B and C) Western blotting of FOSL1 (B) and TXNIP (C) protein abundance in RIT1-variant and KRAS-variant AALE cells. Quantification of N = 4 (B) and 5 (C) independent experiments alongside a representative blot for each. * P < 0.05, compared to vector condition, by two-tailed t-test. (D) Global proteome-transcriptome correlation analysis for RIT1-variant and KRAS-variant AALE cell lines. Gray dashed line indicates linear regression between Log2Fold-Change of mRNA to respective Log2Fold-Change of protein. Red denotes genes outside the 95% prediction interval. r = Pearson correlation coefficient. Data shown are generated from 2 to 4 technical LC-MS/MS replicates per cell line generated from N=2 biological replicates per cell line. N = 3 RNA-seq replicates for each isogenic cell line. (E) In RIT1- or KRAS-expressing AALE cell lines compared to vector control AALE cells, the proportion of genes that are differentially expressed at the mRNA level (log2Fold-Change > 1, FDR < 0.05) which are also differentially abundant at the protein level (log2Fold-Change > 1, adjusted P-value < 0.05). Number of total differentially expressed mRNA transcripts is shown above the bar for each cell line. N is as described in (D). (F) Gene set enrichment analysis of differentially expressed gene transcripts between KRAS or RIT1 AALE cell lines and vector controls using GOseq (56). mSigDB hallmark gene sets specific to KRAS signaling are shown. Circle size = −Log10FDR of enrichment significance determined by GOseq. Analysis performed with N = 3 RNA-seq replicates per isogenic cell line.
Among the top proteins with decreased abundance in RIT1M90I, KRASG12V, and KRASQ61H cells was TXNIP, an inhibitor of thioredoxin involved in both redox regulation and glucose metabolism (27, 28). Prior studies indicate that HRASG12V suppresses TXNIP transcription and protein translation (29, 30). TXNIP abundance was also decreased in Western blot analysis of RIT1M90I, KRASG12V, and KRASQ61H cells compared to vector control (Fig. 2C). These validation data demonstrate the utility of LC-MS/MS to describe protein expression changes and additionally suggest the mechanism of RAS-mediated modulation of FOSL1 and TXNIP is shared with RIT1M90I.
Previous studies of steady-state cell lines show that approximately 40% of relative protein abundance is explained by mRNA expression levels (31, 32). Similarly, in the stable AALE cell lines overexpressing KRAS variants or RIT1M90I, we see a moderate correlation between protein abundance and gene transcript levels (Pearson r = 0.2 to 0.4, Fig. 2D and fig. S2B). Correspondingly, many genes which are differentially regulated at the mRNA level are also dysregulated at the protein level (Fig. 2E).
As expected, both KRAS variants and, to a lesser degree, wild-type and variant RIT1 induced KRAS transcriptional signatures (Fig. 2F). To determine whether oncogenic RIT1 and KRAS also induced similar phosphoproteomic changes, we performed Kinase-Substrate Enrichment Analysis (KSEA) (33), which uses kinase-substrate pairings from PhosphoSitePlus (34) and NetworKIN (35) to identify differential phosphorylation of kinase-substrate families (table S6). This analysis identified ribosomal S6 kinase (RPS6KA1), MAPKAPK2, AKT1, and protein kinase C (PRKCA) as top kinases driving phosphorylation changes in RIT1M90I and KRASG12V/KRASQ61H cells (fig. S3, A to G, and table S5). Substrates of Aurora kinase B (fig. S3A to C, and H) and CDK1 and PAK1 were suppressed in RIT1- and KRAS-variant cells (fig. S3, B and C). Although the total phosphorylation of each substrate reflects the balance between kinases and phosphatases in the cell, these data suggest that RIT1M90I, like oncogenic KRAS, can activate canonical RAS effector pathways.
Wild-type RIT1 overexpression phenocopies RIT1 mutational activation
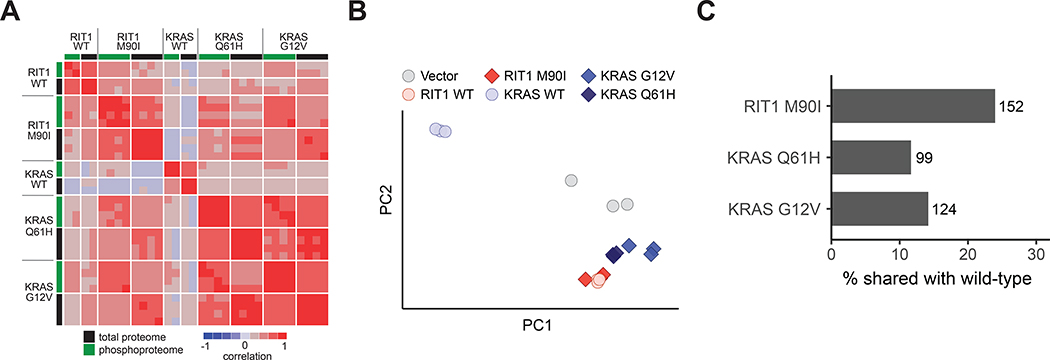
Calculation of a correlation matrix from the proteome and phosphoproteome data showed high correlations of all profiles with the exception of the wild-type KRAS cells. KRASG12V and KRASQ61H proteomes and phosphoproteomes were highly correlated with each other (proteome r = 0.85 and phosphoproteome r = 0.79; Fig. 3A), and with RIT1WT and RIT1M90I cells (r = 0.70–0.80 and 0.72–0.75 for proteome and phosphoproteome, respectively; Fig. 3A). In contrast, wild-type KRAS replicates were the most divergent of all profiles, showing limited correlation to either the KRAS variant or RIT1 profiles.
Figure 3. Wild-type RIT1 overexpression mimics oncogenic RIT1 activation.
(A) Correlation heatmap showing pairwise correlations of each proteome and phosphoproteome replicate to every other replicate. To enable correlation of proteome with phosphoproteome, phosphosites were collapsed to the protein level by taking the median of all phosphosites for each protein. Rows are Spearman correlation; columns are Pearson correlation. Proteome and phosphoproteome data were generated from 2 to 4 technical LC-MS/MS replicates per cell line generated from N=2 biological replicates per cell line. (B) Principal component analysis of RNA-seq data from AALE cells. Circles denote individual control vector, RIT1WT, or KRASWT transcriptomes, as labeled; diamonds denote RIT1-variant or KRAS-variant transcriptomes, as labeled. PC1 explains 96.2% of variance, PC2 explains 2.16% of variance. N = 3 RNA-seq libraries per cell line. (C) Bar plot showing percentage of differentially abundant proteins (|Log2Fold-Change| > 2) in wild-type AALE cells that are also differentially abundant in variant AALE cells. Numbers next to bars indicate number of proteins represented; N as described in (A).
A relatively recent study found that RIT1 mutations, including M90I, may function by relieving negative regulation of RIT1 by a LZTR1-dependent proteasomal degradation mechanism (12). Accordingly, overexpression of wild-type RIT1 should largely phenocopy expression of RIT1M90I. Consistent with this idea, RIT1WT cells more closely resembled both RIT1M90I (proteome r = 0.49) and KRAS variant cells (proteome r = 0.36–0.48) than KRASWT cells (proteome r = 0.19; Fig. 3A). These data highlight a critical divergence between KRAS and RIT1: expression of wild-type KRAS is not capable of activating downstream oncogenic RAS pathways, whereas expression of wild-type RIT1 in part resembles activation of RIT1 or KRAS by mutation. We confirmed this observation in a principal component analysis of the transcriptome data, which further revealed a high degree of similarity between RIT1WT and RIT1M90I-regulated gene expression (Fig. 3B and table S4). Further, of the differentially abundant proteins in RIT1WT cells, 25% were differentialy abundant in RIT1M90I cells. In contrast, 7–10% of proteins with altered abundance levels in KRASWT cells had differentially altered abundance levels in KRASmut cells (Fig. 3C). In the phosphoproteome, unsupervised hierarchical clustering of the phospho-signatures identified the RIT1M90I phosphoproteome as most similar to KRASG12V and KRASQ61H phospho-signatures (fig. S3I), suggesting that RIT1M90I induces similar cellular phosphorylation changes as KRAS variants. Additionally, the phosphoproteome signatures of RIT1WT cells clustered more closely than wild-type KRAS to the RIT1M90I, KRASG12V, and KRASQ61H phosphoproteome signatures (fig. S3I).
RIT1 promotes an epithelial-to-mesenchymal transition
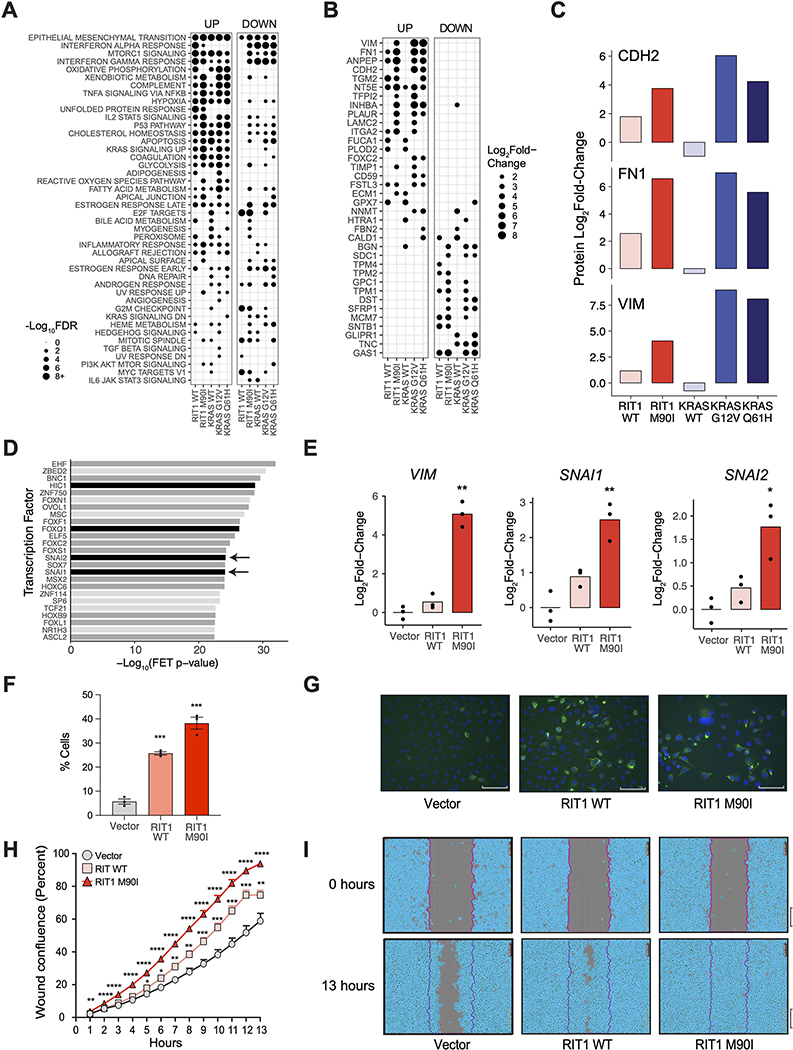
To further investigate the phenotypic changes conferred by these proteome and transcriptome changes, we performed gene set overlap analysis using MSigDB Hallmark Pathway gene sets (36) (Fig. 4A). The epithelial-to-mesenchymal transition (EMT) pathway was the most significant gene set enriched among up-regulated proteins across all KRAS- and RIT1-expressing cells compared to vector control (Fig. 4A). EMT is a cellular transdifferentiation process promoted by cell-extrinsic signaling proteins and orchestrated by activation of transcription factors such as Twist, Snail, and Zeb family transcription factors (37). It has long been observed that oncogenic RAS proteins—including KRAS variants—promote EMT, but little is known about the involvement of RIT1 in this process.
Figure 4. Wild-type RIT1 and RIT1M90I promote an epithelial-to-mesenchymal (EMT) transition phenotype.
(A) Gene set overlap analysis of up-regulated (“Up”; Log2Fold-Change>2) and down-regulated (“down”; Log2Fold-Change<−2) proteins in each condition in AALE cells, using MSigDB Hallmark Pathways (36). Gene sets sorted by descending average −Log10FDR across all conditions. Circle size = −Log10FDR of gene set overlap analysis determined by MSigDB. The analysis was based on data generated from 2 to 4 technical LC-MS/MS replicates per cell line generated from N=2 biological replicates per cell line. (B) Log2Fold-Change in protein abundance of hallmark EMT genes in AALE cells expressing wild-type or variant RIT1 or KRAS relative to abundance in cells expressing the control vector. Proteins are sorted by average Log2Fold-Change across all conditions. N is as described in (A). (C) Log2Fold-Change in protein abundance of N-cadherin (CDH2), fibronectin (FN1), and vimentin (VIM) in AALE cells expressing wild-type or variant RIT1 or KRAS. N is as described in (A). (D) Transcription factor target enrichment analysis of differentially expressed genes in RIT1M90I AALE cells using Enrichr libraries through ChEA3. FET, Fisher’s exact test. Analysis performed on N = 3 RNA-seq libraries compared to 3 vector controls. Arrows indicate proteins of the Snail family. Black denote confirmed EMT genes in dbEMT (49); dark gray denote EMT-associated genes in the literature. (E) Real-time PCR of relative Log2Fold-Change of mRNA expression in perturbed AALE cells of vimentin (VIM), Snail (SNAI1), and Slug (SNAI2). N=3 biological replicates; * P <0.05 and ** P < 0.01 by two-tailed t-test. (F) Immunofluorescence of Vimentin stained cells. N=3 biological replicates; *** P <0.001 by two-tailed t-test. (G) Representative immunofluorescence images of Vimentin stained cells. Scale bar corresponds to 89.1μm. N = 3 replicates per cell line. (H) Wound healing scratch assay. Percent wound confluence at hourly time points are shown with SEM error bars. N=16 replicates per cell line; data shown is representative of N = 3 independent experiments; * P <0.05, ** P <0.01, *** P <0.001, and **** P <0.0001 by two-tailed t-test each time point, corrected for multiple testing. (I) Representative images of wound healing at time of wound making (0 hours) and at end time point (13 hours). Data are representative of 3 independent experiments. Scale bar, 300μm.
Notably, RIT1M90I and, to a lesser degree, wild-type RIT1 were capable of promoting expression changes of key EMT markers, including up-regulation of vimentin, N-cadherin, and fibronectin, and downregulation of keratin-19 (Fig. 4, B and C, and fig. S4A). Although the canonical EMT transcription factors Snail (SNAI1) and Slug (SNAI2) were not detected by proteomic analysis, transcriptomic analysis of RIT1M90I cells showed increased activity of these EMT transcription factors by transcription factor target enrichment analysis (Fig. 4D and fig. S4, B and C). Real-time qPCR confirmed induction of vimentin, Snail, and Slug by RIT1M90I (Fig. 4E). Quantitative immunofluorescence analysis revealed both RIT1WT and RIT1M90I induced vimentin protein expression in AALE cells (Fig. 4, F and G).
To functionally determine if EMT characteristics are present in RIT1-transformed AALE cells, we performed a scratch assay to quantify cellular migration. Consistent with a functional EMT, wild-type RIT1 and RIT1M90I cells showed enhanced migration, closing the scratch wound up to 1.5x and 1.9x faster than vector control cells respectively (Fig. 4, H and I). Therefore, we find that both wild-type RIT1 overexpression and RIT1M90I promote an EMT phenotype in lung epithelial cells involving increased expression of vimentin and increased cellular migration.
Together these data strengthen the notion that oncogenic RIT1 hijacks canonical RAS effector pathways to promote tumorigenesis. In addition, these data demonstrate that wild-type RIT1 overexpression can induce many of these same processes to a weaker degree, supporting a role for protein abundance in regulation of RIT1 activity and RIT1 amplification/overexpression in contributing to malignant phenotypes.
Discussion
Herein, we described quantitative proteomic, phosphoproteomic, and transcriptomic datasets that provide a systems-level view of the RIT1-regulated signaling network in lung epithelial cells. These datasets were generated from isogenic human lung epithelial cells to provide a physiological view of the consequences of RIT1 activation in the same cellular compartment that is involved in lung adenocarcinoma, a tumor type with prevalent mutations in KRAS and RIT1. Broadly, we find that proteome and transcriptome signatures from RIT1M90I-expressing cells partially phenocopy those from cells with overexpression of wild-type RIT1. This finding lends further support to the notion that oncogenic RIT1 variants function at least in part through increasing RIT1 abundance (12). This is in contrast to KRAS, where overexpression of wild-type KRAS induces signatures less related to that of oncogenic variants KRASG12V and KRASQ61H. The opposing functions of wild-type and variant KRAS is consistent with recent evidence suggesting that KRAS functions as a dimer and that wild-type KRAS directly inhibits the function of oncogenic KRAS variants via physical dimerization (38).
This divergence in the function of wild-type RIT1 and KRAS hints at fundamental differences in molecular regulation of each wild-type GTPase. The ability of RIT1 to promote downstream RAF/MEK/ERK signaling when aberrantly expressed suggests that RIT1 may not be subject to the same tight regulation by GTPase-activating proteins (GAPs) that normally keep RAS in an inactive state. Furthermore, these data point to the importance of studying wild-type RIT1 amplification and overexpression in cancers. RIT1, on chromosome 1q, is frequently amplified in uterine carcinosarcoma, liver hepatocellular cancer, cholangiocarcinoma, breast cancer, lung adenocarcinoma, and ovarian cancer (fig. S5A) (39). RIT1 mRNA expression is increased in amplified cases, regardless of tissue type (fig. S5B), raising the possibility that RIT1 overexpression could play a role in tumorigenesis in these cancers.
Although this resource should enable identification of differences between RIT1 and KRAS networks, we observe marked similarity between the transcriptional, proteomic and phopshoproteomic changes induced by RIT1M90I, KRASG12V, and KRASQ61H. Oncogenic RIT1 and KRAS share the ability to activate canonical RAS effector pathways such as those involved in regulation of FOSL1 and TXNIP abundance. These cellular signaling changes are accompanied by phenotypic changes to the lung epithelial cells including acquisition of anchorage-independent growth capability as well as induction of an EMT phenotype with enhanced cellular migration and increased mRNA or protein abundance of EMT markers vimentin, Snail, and Slug.
Together, these results demonstrate the power of quantitative proteomics and transcriptomics to provide global views of oncogene signaling and provide a resource for understanding the altered cell signaling potentially underlying Noonan syndrome and cancer.
Materials and Methods
Isogenic cell line generation
Plasmid constructs were cloned using Gateway Technology (Invitrogen/ThermoFisher) using pLX301 destination vector (Broad Institute) and pDONR223-RIT1 donor vectors previously described(1). Lentivirus was generated by transfection of HEK293T cells with packaging and envelope vectors using standard protocols. AALE cells were a kind gift from Jesse Boehm (Broad Institute). Isogenic cells were generated by transduction of lentivirus generated from pLX317-Renilla luciferase or pLX301-RIT1WT, pLX301-RIT1M90I, pLX301-KRASWT, pLX301-KRASG12V, or pLX301-KRASQ61H and selection with puromycin. Stable pools of cells were maintained in small airway growth medium (Lonza).
Soft agar assay
1×105 cells were suspended in 1 ml of 0.33% select agar in small airway growth medium (Lonza) and plated on a bottom layer of 0.5% select agar in the same media in six-well dishes. Each cell line was analyzed in triplicate. Colonies were photographed after 3–5 weeks and quantified using Fiji software.
Transcriptome profiling
Three replicates per cell line were harvested at ~90% confluence (n = 18 total culture dishes). Cells were lysed and total RNA was extracted using Direct-zol RNA Miniprep plus (Zymo Research). Libraries were constructed using the non-strand-specific poly-A selection Illumina TruSeq kit for 50bp paired-end reads. Libraries were pooled and sequenced on the Illumina NovaSeq platform (Fred Hutch Genomics Core). Reads were aligned to the human reference genome build hg19/GRCh37 using STAR v.2.5.3a (40). Alignments were annotated for duplicates and read groups then reordered and indexed using Picard Tools v.1.114 (41). Read statistics for each RNA-seq sample were calculated using RSeQC (42). Quantification of gene transcripts was performed by the featureCounts program within the Subread package (43), using hg19 gene annotations from UCSC. Gene level counts per million reads (CPM) and reads per kilobase of transcript mapped reads (RPKM) values were calculated with edgeR v.3.22.3 (44), and converted into transcripts per million reads (TPM) values with an in-house script. In total, 12,887 genes were identified with average logCPM at least 0.1 across all samples. Differential expression analyses comparing KRAS or RIT1 perturbed cell lines against vector control lines were performed using the R package edgeR (44).
High performance liquid chromatography tandem mass spectrometry (LC-MS/MS)
Cells were washed in ice-cold PBS, scraped into PBS, pelleted, and snap frozen in liquid nitrogen. The experimental workflow for sample processing, TMT-labeling, peptide enrichment, and LC-MS/MS were largely as previously described (45). Briefly, pellets were lysed in 200 μl of chilled urea lysis buffer (8 M urea, 75 mM NaCl, 50 mM Tris (pH 8.0), 1 mM EDTA, 2 μg/ml aprotinin, 10 μg/ml leupeptin, 1 mM PMSF, 1:100 (vol/vol) Phosphatase Inhibitor Cocktail 2, 1:100 Phosphatase Inhibitor Cocktail 3, 10 mM NaF, and 20 μM PUGNAc) for each ~50 mg portion of wet-weight tissue. Lysates were reduced with 5mM DTT, alkylated with 10 mM IAM, and digestion performed in solution with 1 mAU LysC per 50 μg of total protein and trypsin at an enzyme/substrate ratio of 1:49. Reactions were quenched with FA and brought to pH = 3 with FA. Peptides were desalted on 200 mg tC18 SepPak cartridges and dried by vacuum centrifugation. 340 μg of peptides were labeled with 10-plex Tandem Mass Tag reagents (TMT10, Fisher Scientific), according to manufacturer’s instructions. To enable quantification of peptides across all 12 samples, the samples were labeled in sets of 10 across two different TMT10 pools in a crossover design with 8 of 12 samples analyzed in both TMT10 pools. A 50/50 mix of both AALE vector control lysates was used as an internal reference in both TMT10 runs (fig. S1B).
Each TMT10-plex was desalted in a 200 mg tC18 SepPak cartridge and fractionated using offline HPLC. 5% of each fraction was collected into an HPLC vial for proteome analysis by LC-MS/MS. The remaining 95% was processed for phospho-peptide enrichment via immobilized metal affinity chromatography (IMAC). IMAC enrichment was performed using Ni-NTA Superflow Agarose beads incubated with peptides solubilized in a final concentration of 80% MeCN/0.1% TFA. Phospho-enriched peptides were desalted and collected into an HPLC vial for analysis by LC-MS/MS.
Online fractionation was performed using a nanoflow Proxeon EASY-nLC 1200 UHPLC system (Thermo Fisher Scientific) and separated peptides were analyzed on a benchtop Orbitrap Q Exactive Plus mass spectrometer (Thermo Fisher Scientific) equipped with a nanoflow ionization source (James A. Hill Instrument Services). In-house packed columns (20 cm × 75 μm diameter C18 silica picofrit capillary column; 1.9 μm ReprosIl-Pur C18-AQ beads, Dr. Maisch GmbH, r119.aq; Picofrit 10 μm tip opening, New Objective, PF360-75-10-N-5). Mobile phase flow rate was 200 nL/min, comprised of 3 % acetonitrile/0.1 % formic acid (solvent A) and 90 % acetonitrile /0.1 % formic acid (solvent B). The 110 min LC-MS/MS method consisted of a 10 min column-equilibration procedure; a 20 min sample-loading procedure; and the following gradient profile: (min: % B) 0:2; 2:6; 85:30; 94:60; 95:90; 100:90; 101:50; 110:50 (the last two steps at 500 nL/min flow rate). Data-dependent acquisition was performed using Xcalibur QExactive v2.4 software in positive ion mode at a spray voltage of 2.00 kV. MS1 Spectra were measured with a resolution of 70,000, an AGC target of 3e6 and a mass range from 300 to 1800 m/z. Up to 12 MS/MS spectra per duty cycle were triggered at a resolution of 35,000, an AGC target of 5e4, an isolation window of 0.7 m/z, a maximum ion time of 120 msec, and normalized collision energy of 30. Peptides that triggered MS/MS scans were dynamically excluded from further MS/MS scans for 20 sec. Charge state screening was enabled to reject precursor charge states that were unassigned, 1, or >6. Peptide match was set to preferred for monoisotopic precursor mass assignment.
Protein-peptide identification, phosphosite localization, and quantification
MS data was interpreted using the Spectrum Mill software package v6.0 pre-release (Agilent Technologies). MS/MS spectra were merged if they were acquired within +/− 45 sec of each other with the same precursor m/z. Also, MS/MS spectra that did not have a sequence tag length > 0 (meaning, minimum of two masses separated by the in chain mass of an amino acid) or did not have a precursor MH+ in the range of 750–6000 were excluded from searching. MS/MS spectra searches were performed against a concatenated UniProt human reference proteome sequence database containing 58,929 human proteins including isoforms (obtained 10/17/2014) and 150 additional common laboratory contaminants. ESI-QEXACTIVE-HCD-3 scoring parameters were used for both whole proteome and phosphoproteome datasets. Spectra were allowed +/− 20 ppm mass tolerance for precursor as well as product ions, 30% minimum matched peak intensity, and “trypsin allow P” was set as enzyme specificity with up to 4 missed cleavages allowed. Carbamidomethylation at cysteine was set as fixed modification together with TMT10 isobaric labels at lysine residues (N-termini would be considered regardless if it was TMT labelled). Acetylation of protein N-termini andoxidized methionine were set as variable modifications with a precursor MH+ shift range of −18 to 64 Da for the proteome searches. For the phosphoproteome searches the precursor MH+ shift range was set to 0 to 272 Da and variable modifications of phosphorylation of serine, threonine, and tyrosine. Identities interpreted for individual spectra were automatically designated as confidently assigned using the Spectrum Mill autovalidation module to use target-decoy based false discovery rate (FDR) estimates to apply score threshold criteria. For the whole proteome datasets, thresholding was done at the spectral (< 1.2%) and protein levels (< 0.1%). For the phosphoproteome datasets, thresholding was done at the spectral (< 1.2%) and phosphosite levels (< 1.0%).
Replicates across TMT-plexes were highly correlated (fig. S1C) with median Pearson r = 0.87 and 0.69 for proteome and phosphoproteome, respectively. Technical replicates and biological replicates were merged to generate final total proteome and phospho-proteome profiles for each isogenic cell line (tables S1 and S2). Replicate-level profiles are also supplied as JavaScript Object Notation (.json) files that can be visualized and analyzed using the Morpheus Matrix Visualization and Analysis Software at https://software.broadinstitute.org/morpheus (Data Files S1 and S2). Differential protein and phospho-site signatures were generated by computing the mean log2(fold change) of the abundance of each site in each sample compared to the vector control cells. Statistical significance of differentially abundant proteins and phosphosites was determined by performing a one sample moderated t-test with multiple hypothesis correction (tables S1 and S2).
Integrative Proteome and Transcriptome Analysis
Correlation of changes in protein expression and changes in RNA expression was modeled with a linear model. 95% prediction intervals were calculated to highlight genes with weak concordance between protein and RNA expression. Differentially expressed transcripts were determined by filtering edgeR results (described above) to transcripts with |log2Fold-Change| > 1 and FDR < 0.05. Differentially abundant proteins were determined by filtering to proteins with |log2Fold-Change | > 1 and adjusted P-value < 0.05.
Gene Set Enrichment Analysis
Analysis of enrichment of KRAS signaling in differential RNA expression profiles was performed in R with GOseq (46). KRAS signaling gene sets were taken from MSigDB hallmark gene sets (36, 47).
Transcription factor target enrichment analysis
Analysis of over-representation of transcription factor targets was performed with ChIP-X Enrichment Analysis 3 (ChEA3) by submitting lists of differentially expressed genes (|log2Fold-Change| > 1 and FDR < 0.05). ChEA3 performs Fisher’s Exact Tests to compare the input gene set to transcription factor target gene sets in six different libraries (48). Analysis of the Enrichr Queries library was selected as the focus of the present study. Transcription factors resulting from this analysis were manually annotated as one of three groups of EMT association. These groups were: (i) confirmed EMT genes defined by dbEMT(49), (ii) genes shown to be associated with EMT in at least one study in literature, and (iii) genes unrelated to EMT.
Antibodies and immunoblotting
Antibodies against FOSL1 (D80B4), TXNIP (D5F3E), and Vimentin (D21H3) were purchased from Cell Signaling Technology. Vinculin (V9264) was purchased from Sigma Aldrich. Secondary antibodies StarBright Blue 700 goat anti-rabbit IgG, StarBright Blue 520 goat anti-rabbit IgG and StarBright Blue 520 Goat anti-Mouse IgG (12005867) were purchased from Bio-Rad. Antibody against RIT1 (#53720) was purchased from Abcam. Cell lysates were prepared in RTK lysis buffer with protease (11836153001, Roche) and phosphatase (04906837001, Roche) inhibitors added and quantified by the BCA assay (Thermo Scientific). Samples were then boiled in Laemmli buffer (1610747, Bio-Rad) and 50 μg of protein was loaded onto 4–15% Mini-Protean TGX (4561084, Bio-Rad) gels. Protein gels were run and transferred to PVDF membranes (1704274, Bio-Rad) according to manufacturer’s instructions. Proteins were detected by specific primary antibody and secondary antibody then visualized using the ChemiDoc MP Imaging System (Bio-Rad) or Odyssey Imager (Li-Cor).
KSEA analysis
Kinase-substrate enrichment analysis (KSEA) (50) was performed using the KSEA App (33) (https://casecpb.shinyapps.io/ksea/) using kinase-substrate mappings from PhosphoSitePlus(34) and a P value threshold of < 0.05. A minimum of five detected phospho-site substrates were needed for kinases to be included in the analysis. The full list of kinase scores and number of substrates are shown in table S6. 36 kinases had sufficient substrate sites detected to be included in the analysis. Kinase-substrate mappings are shown in table S6.
Scratch assay
Cells were seeded in 96-well ImageLock plates (Sartorius) at a concentration of 46,000 cells/well. The cells were then incubated overnight in SAGM media without EGF (Lonza). After incubation, with the cells forming a monolayer, a scratch wound is made using the 96-well WoundMaker tool (Sartorius). Plate was imaged every hour for 13 hours and analyzed using the IncuCyte ZOOM v2016A (Sartorius). Further analysis was performed in GraphPad Prism v9 using multiple comparison two-tailed t-tests and reporting q values.
qRT-PCR
Extracted RNA for each biological replicate of the 6 cell lines were used to generate cDNA by reverse transcription with the SuperScript IV First-Strand Synthesis kit (Invitrogen). Quantitative RT-PCR reactions were set up in technical triplicates using the Taqman Gene Expression Master Mix (Thermo Fisher) and a TaqMan Gene Expression Assay for a reference gene, 18S (Hs99999901_s1), as well as for each gene of interest: VIM (Hs00958111_m1), SNAI1 (Hs00195591_m1), and SNAI2 (Hs00161904_m1). PCR reactions were performed and gene expression quantified using the CFX384 Real-Time System (Bio-Rad). Relative gene expression was calculated by the Livak method using an in-house script.
Immunofluorescence
For immunofluorescence detection of Vimentin, 2.5 × 105 AALE stable cells (passage 5) were plated in triplicate on 4-well Nunc Lab-Tek II CC2 chamber slides and allowed to adhere overnight at 37°C and 5% CO2. The next day, media was aspirated and cells were washed once with PBS, then fixed with 4% paraformaldehyde in PBS for 10 min at room temperature. Cells were then washed twice with ice cold PBS and permeabilized with 0.25% Triton-X-100 for 10 minutes followed by three washes with PBS. Cells were blocked with 10% normal goat serum (Thermo Fisher Scientific) in PBS with 0.1% Tween 20 (PBST) followed by overnight incubation at 4 °C with vimentin rabbit antibody (D21H3 XP® monoclonal antibody, Cell Signaling #5741, 1:1000 dilution) in PBST. The next day, cells were washed with PBST three times for 5 min then incubated with Alexa Fluor® 488 preadsorbed Goat anti-rabbit IgG antibody (Abcam #ab150117 at 1:1000 dilution) for 1 hour at room temperature in the dark. Cells were then washed three times with PBST and slides mounted using Vectashield antifade mounting medium with 1.5μg/mL DAPI (Vector Laboratories). The slides were sealed with clear nail polish and imaged on a Nikon Eclipse E800 microscope with 20X/0.75 NA PlanApo objective using a DAPI filter set (excitation wavelength: 330–380 nm; emission wavelength: 435–485) and fixed 300 ms exposure using the FITC filter set (excitation wavelength: 460–500 nm; emission wavelength: 510–560 nm) using a AxioCam HRm camera and AxioVision 40 software v4.8.20. To determine the percent of positive cells, images were analyzed using TissueQuest software v7.0.1.139 (TissueGnostics, Vienna, Austria). Images were segmented using nuclear DAPI fluorescence and then a ring mask around nuclei was applied to the vimentin image. The mean fluorescence intensity within each masked area was quantified and the percentage of cells above a threshold intensity were summarized by the software. Data were analyzed using a two-tailed t-test in GraphPad Prism v9, with P < 0.05 considered significant.
Supplementary Material
Acknowledgements:
We thank Drs. Athea Vichas and Jon Cooper (Fred Hutchinson Cancer Research Center) for advice, discussion, and critical reading of the manuscript. We thank Dr. D.R. Mani (Broad Institute) for guidance on statistical analysis, and Jesse Boehm (Broad Institute) for the AALE cells. We thank the Fred Hutch Cellular Imaging Shared Resource for assistance with microscopy and image analysis. We thank the Fred Hutch Genomics Shared Resource for performing RNA sequencing and assisting with analysis.
Funding:
This research was funded in part through the National Cancer Institute (NCI) K99/R00 CA197762 and R37 CA25205 to AHB, NIH/NCI Cancer Center Support Grant P30 CA015704, NCI Clinical Proteomic Tumor Analysis Consortium grants NIH/NCI U24-CA210986 and NIH/NCI U01 CA214125 to SAC. AL was supported in part by NSF IGERT DGE-1258485. KH was supported in part by PHS NRSA T32GM007270 from NIGMS.
Footnotes
Competing interests: S.A.C. is a member of the scientific advisory boards of Kymera, PTM BioLabs, and Seer.
Data and materials availability:
pLX301 lentiviral expression vectors for expression of RIT1 and KRAS are available from A.H.B. under a material agreement with Fred Hutchinson Cancer Research Center. The RNA-seq data have been deposited in the NCBI Gene Expression Omnibus database with accession number GSE146479. The original mass spectra and the protein sequence database used for searches have been deposited in the public proteomics repository MassIVE (https://massive.ucsd.edu) and are accessible at ftp://MSV000085225@massive.ucsd.edu with username: MSV000085225 and password: oncogenic. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.
References and Notes
- 1.Berger AH, Imielinski M, Duke F, Wala J, Kaplan N, Shi G-X, Andres DA, Meyerson M, Oncogenic RIT1 mutations in lung adenocarcinoma. Oncogene. 33, 4418–4423 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gómez-Seguí I, Makishima H, Jerez A, Yoshida K, Przychodzen B, Miyano S, Shiraishi Y, Husseinzadeh HD, Guinta K, Clemente M, Hosono N, McDevitt MA, Moliterno AR, Sekeres MA, Ogawa S, Maciejewski JP, Novel recurrent mutations in the RAS-like GTP-binding gene RIT1 in myeloid malignancies. Leukemia. 27, 1943–1946 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Trahey M, McCormick F, A cytoplasmic protein stimulates normal N-ras p21 GTPase, but does not affect oncogenic mutants. Science. 238, 542–545 (1987). [DOI] [PubMed] [Google Scholar]
- 4.Pai Emil F., Kabsch Wolfgang, Krengel Ute, Holmes Kenneth C., John Jacob & Wittinghofer Alfred, Structure of the guanine-nucleotide-binding domain of the Ha-ras oncogene p21 in the triphosphate conformation. Nature. 2, 1105–1111 (1989). [DOI] [PubMed] [Google Scholar]
- 5.Nakano H, Yamamoto F, Neville C, Evans D, Mizuno T, Perucho M, Isolation of transforming sequences of two human lung carcinomas: structural and functional analysis of the activated c-K-ras oncogenes. Proc. Natl. Acad. Sci. U. S. A. 81, 71–75 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rodenhuis S, van de Wetering ML, Mooi WJ, Evers SG, van Zandwijk N, Bos JL, Mutational activation of the K-ras oncogene. A possible pathogenetic factor in adenocarcinoma of the lung. N. Engl. J. Med. 317, 929–935 (1987). [DOI] [PubMed] [Google Scholar]
- 7.Aoki Y, Niihori T, Banjo T, Okamoto N, Mizuno S, Kurosawa K, Ogata T, Takada F, Yano M, Ando T, Hoshika T, Barnett C, Ohashi H, Kawame H, Hasegawa T, Okutani T, Nagashima T, Hasegawa S, Funayama R, Nagashima T, Nakayama K, Inoue S-I, Watanabe Y, Ogura T, Matsubara Y, Gain-of-Function Mutations in RIT1 Cause Noonan Syndrome, a RAS/MAPK Pathway Syndrome. Am. J. Hum. Genet. 93, 173–180 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cai W, Carlson SW, Brelsfoard JM, Mannon CE, Moncman CL, Saatman KE, Andres DA, Rit GTPase signaling promotes immature hippocampal neuronal survival. J. Neurosci. 32, 9887–9897 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rusyn EV, Reynolds ER, Shao H, Grana TM, Chan TO, Andres DA, Cox AD, Rit, a non-lipid-modi/-ed Ras-related protein, transforms NIH3T3 cells without activating the ERK, JNK, p38 MAPK or PI3K/Akt pathways. Oncogene. 19, 4685–4694 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Sakabe K, Teramoto H, Zohar M, Behbahani B, Miyazaki H, Chikumi H, Gutkind JS, Potent transforming activity of the small GTP-binding protein Rit in NIH 3T3 cells: evidence for a role of a p38gamma-dependent signaling pathway. FEBS Lett. 511, 15–20 (2002). [DOI] [PubMed] [Google Scholar]
- 11.Smith MJ, Neel BG, Ikura M, NMR-based functional profiling of RASopathies and oncogenic RAS mutations. Proc. Natl. Acad. Sci. U. S. A. 110, 4574–4579 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Castel P, Cheng A, Cuevas-Navarro A, Everman DB, Papageorge AG, Simanshu DK, Tankka A, Galeas J, Urisman A, McCormick F, RIT1 oncoproteins escape LZTR1-mediated proteolysis. Science. 363, 1226–1230 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fang Z, Marshall CB, Yin JC, Mazhab-Jafari MT, Gasmi-Seabrook GMC, Smith MJ, Nishikawa T, Xu Y, Neel BG, Ikura M, Biochemical Classification of Disease-associated Mutants of RAS-like Protein Expressed in Many Tissues (RIT1). J. Biol. Chem. 291, 15641–15652 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.TCGA, Comprehensive molecular profiling of lung adenocarcinoma. Nature. 511, 543–550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campbell JD, Alexandrov A, Kim J, Wala J, Berger AH, Pedamallu CS, Shukla SA, Guo G, Brooks AN, Murray BA, Imielinski M, Hu X, Ling S, Akbani R, Rosenberg M, Cibulskis C, Ramachandran A, Collisson EA, Kwiatkowski DJ, Lawrence MS, Weinstein JN, Verhaak RGW, Wu CJ, Hammerman PS, Cherniack AD, Getz G, Cancer Genome Atlas Research Network, Artyomov MN, Schreiber R, Govindan R, Meyerson M, Distinct patterns of somatic genome alterations in lung adenocarcinomas and squamous cell carcinomas. Nat. Genet. 48, 607–616 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen H, Carrot-Zhang J, Zhao Y, Hu H, Freeman SS, Yu S, Ha G, Taylor AM, Berger AC, Westlake L, Zheng Y, Zhang J, Ramachandran A, Zheng Q, Pan Y, Zheng D, Zheng S, Cheng C, Kuang M, Zhou X, Zhang Y, Li H, Ye T, Ma Y, Gao Z, Tao X, Han H, Shang J, Yu Y, Bao D, Huang Y, Li X, Zhang Y, Xiang J, Sun Y, Li Y, Cherniack AD, Campbell JD, Shi L, Meyerson M, Genomic and immune profiling of pre-invasive lung adenocarcinoma. Nat. Commun. 10, 5472 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cherniack AD, Shen H, Walter V, Stewart C, Murray BA, Bowlby R, Hu X, Ling S, Soslow RA, Broaddus RR, Zuna RE, Robertson G, Laird PW, Kucherlapati R, Mills GB, Cancer Genome Atlas Research Network, Weinstein JN, Zhang J, Akbani R, Levine DA, Integrated Molecular Characterization of Uterine Carcinosarcoma. Cancer Cell. 31, 411–423 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song Z, Liu T, Chen J, Ge C, Zhao F, Zhu M, Chen T, Cui Y, Tian H, Yao M, Li J, Li H, HIF-1α-induced RIT1 promotes liver cancer growth and metastasis and its deficiency increases sensitivity to sorafenib. Cancer Lett. 460, 96–107 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Xu F, Sun S, Yan S, Guo H, Dai M, Teng Y, Elevated expression of RIT1 correlates with poor prognosis in endometrial cancer. Int. J. Clin. Exp. Pathol. 8, 10315–10324 (2015). [PMC free article] [PubMed] [Google Scholar]
- 20.Shao H, Andres DA, A novel RalGEF-like protein, RGL3, as a candidate effector for rit and Ras. J. Biol. Chem. 275, 26914–26924 (2000). [DOI] [PubMed] [Google Scholar]
- 21.Shi GX, Andres DA, Rit Contributes to Nerve Growth Factor-Induced Neuronal Differentiation via Activation of B-Raf-Extracellular Signal-Regulated Kinase and p38 Mitogen-Activated Protein Kinase Cascades. Mol. Cell. Biol. 25, 830–846 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Castel P, Holtz-Morris A, Kwon Y, Suter BP, McCormick F, DoMY-Seq: A yeast two-hybrid-based technique for precision mapping of protein-protein interaction motifs. J. Biol. Chem. (2020), doi: 10.1074/jbc.RA120.014284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lundberg AS, Randell SH, Stewart SA, Elenbaas B, Hartwell KA, Brooks MW, Fleming MD, Olsen JC, Miller SW, Weinberg RA, Hahn WC, Immortalization and transformation of primary human airway epithelial cells by gene transfer. Oncogene. 21, 4577–4586 (2002). [DOI] [PubMed] [Google Scholar]
- 24.Young MR, Colburn NH, Fra-1 a target for cancer prevention or intervention. Gene. 379, 1–11 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Mechta F, Lallemand D, Pfarr CM, Yaniv M, Transformation by ras modifies AP1 composition and activity. Oncogene. 14, 837–847 (1997). [DOI] [PubMed] [Google Scholar]
- 26.Casalino L, De Cesare D, Verde P, Accumulation of Fra-1 in ras-transformed cells depends on both transcriptional autoregulation and MEK-dependent posttranslational stabilization. Mol. Cell. Biol. 23, 4401–4415 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoshihara E, Masaki S, Matsuo Y, Chen Z, Tian H, Yodoi J, Thioredoxin/Txnip: redoxisome, as a redox switch for the pathogenesis of diseases. Front. Immunol. 4, 514 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parikh H, Carlsson E, Chutkow WA, Johansson LE, Storgaard H, Poulsen P, Saxena R, Ladd C, Schulze PC, Mazzini MJ, Jensen CB, Krook A, Björnholm M, Tornqvist H, Zierath JR, Ridderstråle M, Altshuler D, Lee RT, Vaag A, Groop LC, Mootha VK, TXNIP regulates peripheral glucose metabolism in humans. PLoS Med. 4, e158 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Elgort MG, O’Shea JM, Jiang Y, Ayer DE, Transcriptional and Translational Downregulation of Thioredoxin Interacting Protein Is Required for Metabolic Reprogramming during G(1). Genes Cancer. 1, 893–907 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ye Z, Ayer DE, Ras Suppresses TXNIP Expression by Restricting Ribosome Translocation. Mol. Cell. Biol. 38 (2018), doi: 10.1128/MCB.00178-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lundberg E, Fagerberg L, Klevebring D, Matic I, Geiger T, Cox J, Algenäs C, Lundeberg J, Mann M, Uhlen M, Defining the transcriptome and proteome in three functionally different human cell lines. Mol. Syst. Biol. 6, 450 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y, Beyer A, Aebersold R, On the Dependency of Cellular Protein Levels on mRNA Abundance. Cell. 165, 535–550 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Wiredja DD, Koyutürk M, Chance MR, The KSEA App: a web-based tool for kinase activity inference from quantitative phosphoproteomics. Bioinformatics (2017), doi: 10.1093/bioinformatics/btx415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hornbeck PV, Kornhauser JM, Tkachev S, Zhang B, Skrzypek E, Murray B, Latham V, Sullivan M, PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res. 40, D261–D270 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linding R, Jensen LJ, Ostheimer GJ, van Vugt MATM, Jørgensen C, Miron IM, Diella F, Colwill K, Taylor L, Elder K, Metalnikov P, Nguyen V, Pasculescu A, Jin J, Park JG, Samson LD, Woodgett JR, Russell RB, Bork P, Yaffe MB, Pawson T, Systematic discovery of in vivo phosphorylation networks. Cell. 129, 1415–1426 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P, The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye X, Weinberg RA, Epithelial-Mesenchymal Plasticity: A Central Regulator of Cancer Progression. Trends Cell Biol. 25, 675–686 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ambrogio C, Köhler J, Zhou Z-W, Wang H, Paranal R, Li J, Capelletti M, Caffarra C, Li S, Lv Q, Gondi S, Hunter JC, Lu J, Chiarle R, Santamaría D, Westover KD, Jänne PA, KRAS Dimerization Impacts MEK Inhibitor Sensitivity and Oncogenic Activity of Mutant KRAS. Cell. 172, 857–868.e15 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Cancer Genome Atlas Research Network, Weinstein JN, Collisson EA, Mills GB, Shaw KRM, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM, The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 45, 1113–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, Gingeras TR, STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Picard Tools - By Broad Institute, (available at http://broadinstitute.github.io/picard).
- 42.Wang L, Wang S, Li W, RSeQC: quality control of RNA-seq experiments. Bioinformatics. 28, 2184–2185 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Liao Y, Smyth GK, Shi W, FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 44.Robinson MD, McCarthy DJ, Smyth GK, edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 26, 139–140 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mertins P, Tang LC, Krug K, Clark DJ, Gritsenko MA, Chen L, Clauser KR, Clauss TR, Shah P, Gillette MA, Petyuk VA, Thomas SN, Mani DR, Mundt F, Moore RJ, Hu Y, Zhao R, Schnaubelt M, Keshishian H, Monroe ME, Zhang Z, Udeshi ND, Mani D, Davies SR, Townsend RR, Chan DW, Smith RD, Zhang H, Liu T, Carr SA, Reproducible workflow for multiplexed deep-scale proteome and phosphoproteome analysis of tumor tissues by liquid chromatography-mass spectrometry. Nat. Protoc. 13, 1632–1661 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Young MD, Wakefield MJ, Smyth GK, Oshlack A, Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 11, R14 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP, Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U. S. A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keenan AB, Torre D, Lachmann A, Leong AK, Wojciechowicz ML, Utti V, Jagodnik KM, Kropiwnicki E, Wang Z, Ma’ayan A, ChEA3: transcription factor enrichment analysis by orthogonal omics integration. Nucleic Acids Res. 47, W212–W224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhao M, Liu Y, Zheng C, Qu H, dbEMT 2.0: An updated database for epithelial-mesenchymal transition genes with experimentally verified information and precalculated regulation information for cancer metastasis. J. Genet. Genomics. 46, 595–597 (2019). [DOI] [PubMed] [Google Scholar]
- 50.Casado P, Rodriguez-Prados J-C, Cosulich SC, Guichard S, Vanhaesebroeck B, Joel S, Cutillas PR, Kinase-substrate enrichment analysis provides insights into the heterogeneity of signaling pathway activation in leukemia cells. Sci. Signal. 6, rs6 (2013). [DOI] [PubMed] [Google Scholar]
- 51.Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG, Clustal W and Clustal X version 2.0. Bioinformatics. 23, 2947–2948 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
pLX301 lentiviral expression vectors for expression of RIT1 and KRAS are available from A.H.B. under a material agreement with Fred Hutchinson Cancer Research Center. The RNA-seq data have been deposited in the NCBI Gene Expression Omnibus database with accession number GSE146479. The original mass spectra and the protein sequence database used for searches have been deposited in the public proteomics repository MassIVE (https://massive.ucsd.edu) and are accessible at ftp://MSV000085225@massive.ucsd.edu with username: MSV000085225 and password: oncogenic. All other data needed to evaluate the conclusions in the paper are present in the paper or the Supplementary Materials.