Abstract
Characterization of RNA–protein interaction is fundamental for understanding the metabolism and function of RNA. UV crosslinking has been widely used to map the targets of RNA-binding proteins, but is limited by low efficiency, requirement for zero-distance contact, and biases for single-stranded RNA structure and certain residues of RNA and protein. Here, we report the development of an RNA–protein crosslinker (AMT–NHS) composed of a psoralen derivative and an N-hydroxysuccinimide ester group, which react with RNA bases and primary amines of protein, respectively. We show that AMT–NHS can penetrate into living yeast cells and crosslink Cbf5 to H/ACA snoRNAs with high specificity. The crosslinker induced different crosslinking patterns than UV and targeted both single- and double-stranded regions of RNA. The crosslinker provides a new tool to capture diverse RNA–protein interactions in cells.
Keywords: RNA-binding protein, chemical crosslinker, CLIP, psoralen, H/ACA snoRNP
INTRODUCTION
Interactions between RNA and protein are important at every stage of the life cycle of mRNA, including transcription, processing, modification, translation, localization, and decay. Many noncoding RNAs exert their functions in the form of RNA–protein complex (RNP). Experimental characterization of RNA–protein interactions has greatly accelerated the study of RNA biology.
Over the past few decades, methods have been developed to map the targets and binding sites of RNA-binding proteins (RBPs) at the transcriptome level (Ule et al. 2018; Wheeler et al. 2018). In RNA immunoprecipitation (RIP), an RBP is purified by binding to its antibody and the associated RNAs are subsequently profiled, for example, by high-throughput sequencing. As RIP needs to be carried out under mild conditions to preserve noncovalent RNA–protein interactions, indirectly or nonspecifically bound RNAs are frequently detected. In the crosslinking and immunoprecipitation (CLIP) technique (Ule et al. 2003), RNA and protein are covalently linked, which allows stringent conditions to be used in RNP purification. Crosslinked RNPs can be resolved in denaturing SDS-PAGE and transferred to nitrocellulose that binds only protein, but not free RNA. The RNP of interest can be selectively cut out for RNA extraction. These measures greatly improve the specificity of analysis.
Radiation with UV light at 254-nm wavelength can induce covalent bonds between RNA bases and proteins and has been widely used in CLIP experiments (Ule et al. 2003; Licatalosi et al. 2008). UV is applicable to unmodified cells or tissues and highly selective by only crosslinking amino acid residues and RNA bases that directly contact. However, UV crosslinking has a low efficiency of 1%–3% and biases for certain amino acid residues, pyrimidines, and single-stranded RNA. Double-stranded RNA (dsRNA) is poorly crosslinked by UV because base pairs are often shielded from protein contact. Only RNA and protein with favorable interacting configuration are amenable to UV crosslinking. Variants of CLIP have been developed to optimize the steps of UV crosslinking, RNP purification and sequencing library construction (Granneman et al. 2009; Hafner et al. 2010; Konig et al. 2010; Van Nostrand et al. 2016; van Nues et al. 2017; Gu et al. 2018), but do not change the inherent biases of UV crosslinking.
Alternatively, RNA and protein can be crosslinked by chemical agents. Formaldehyde has been used to crosslink dsRNA and protein (Kim and Kim 2019), but its application is limited by its promiscuous activity to crosslink protein–protein and protein–DNA. There is still a need to develop a chemical crosslinker to capture diverse types of protein–RNA interactions in vivo with high efficiency and specificity. In this study, we reported the synthesis and characterization of a novel RNA–protein crosslinker that can permeate living yeast cells and crosslink protein with diverse RNA structures.
RESULTS
Synthesis of AMT–NHS
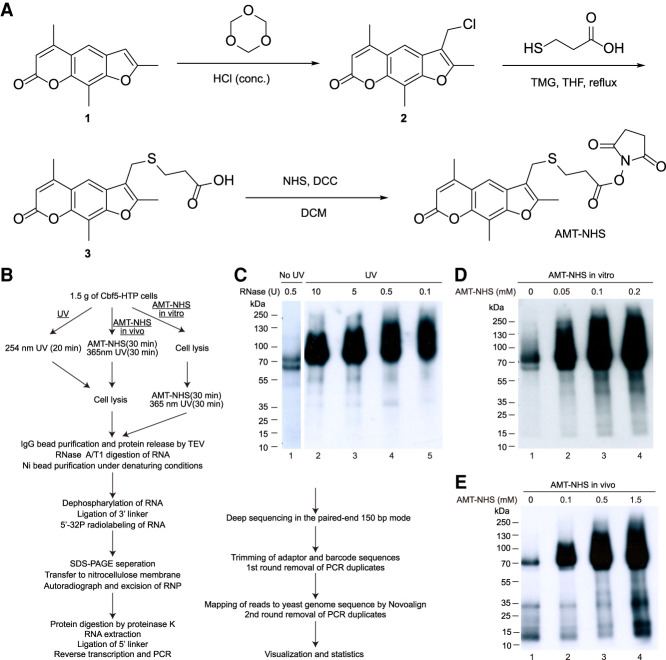
To develop a specific RNA–protein chemical crosslinker, a psoralen-derivative 4′-aminomethyltrioxsalen (AMT) was linked to an N-hydroxysuccinimide ester group (NHS) (Fig. 1A). Psoralen is a three-ring aromatic compound which can insert into RNA duplex and undergo cycloaddition with pyrimidines upon activation by 365-nm UV, forming monoadducts or diadducts (Cimino et al. 1985). The NHS group reacts with the ε-amine group in lysine residue and the amino terminus of polypeptide.
FIGURE 1.
RNA–protein crosslinking activity of AMT–NHS. (A) Synthesis of AMT–NHS. (B) Flowchart of crosslinking, RNP purification, sequencing library construction, and data processing. (C–E) Autoradiogram of crosslinked Cbf5–RNA complex resolved in SDS-PAGE. RNA and protein were crosslinked by 254-nm UV light (C) or by AMT–NHS in vitro (D) or in vivo (E). RNA was digested by increasing amounts of RNase A/T1 in UV crosslinking and by 0.5 units of RNase A/T1 in all AMT–NHS crosslinking.
AMT–NHS was synthesized in three steps (Fig. 1A). Briefly, trioxsalen was chloromethylated at its 3′-carbon with formaldehyde and hydrogen chloride. A 3-mercaptopropanoic acid linker was added through the substitution of arylmethyl chloride. The terminal carboxyl group was further activated by N-hydroxysuccinimide to afford AMT–NHS. The compounds produced at each step were verified by hydrogen nuclear magnetic resonance (1H-NMR).
RNA–protein crosslinking activity of AMT–NHS in yeast
We chose Cbf5, a major RNA-binding protein in the box H/ACA snoRNP of Saccharomyces cerevisiae, to test the RNA–protein crosslinking activity of AMT–NHS. An H/ACA snoRNP is composed of a distinct H/ACA snoRNA and four core proteins: Cbf5, Nop10, Nhp2, and Gar1 (Kiss et al. 2010; Yu and Meier 2014). Most H/ACA snoRNPs catalyze site-specific conversion of uridine to pseudouridine in rRNA and snRNA with specificity determined by H/ACA snoRNA. All H/ACA snoRNAs share a consensus secondary structure of hairpin-hinge-hairpin-tail and contain the conserved H (ANANNA) and ACA motifs in the hinge and 3′ tail regions, respectively. Each hairpin can harbor a large internal loop, termed the pseudouridylation pocket, which binds a substrate by forming two short duplexes with the sequences flanking the target uridine to be modified. Each hairpin is associated with one set of four core proteins. Cbf5 extensively binds the lower stem of the hairpin and the H or ACA motif with its PUA domain and loosely contacts the empty pseudouridylation pocket with its catalytic domain (Li and Ye 2006). As the interaction between Cbf5 and H/ACA snoRNA has been structurally characterized and H/ACA snoRNAs contain both single- and double-stranded RNA structures, Cbf5 would be ideal to test the efficiency, specificity, and structural preference of AMT–NHS mediated crosslinking.
We adopted the procedure of UV crosslinking and cDNA analysis (CRAC) to tag and purify Cbf5 and prepare sequencing libraries (Fig. 1B; Granneman et al. 2009; Lin et al. 2013). The genomic CBF5 gene was carboxy-terminally fused to a His6-TEV cleavage site-Protein A (HTP) tag. After crosslinking with AMT–NHS or 254-nm UV light as comparison, Cbf5-HTP was bound to IgG-coated beads and released with TEV protease digestion. Following partial digestion of crosslinked RNA, Cbf5-His6 was further purified with Ni beads under denaturing conditions. The crosslinked RNA on beads was dephosphorated, linked to the 3′ adaptor, and 5′-32P radiolabeled. Cbf5-His6 was eluted, resolved in SDS-PAGE, and blotted to nitrocellulose membranes. Proteins crosslinked to radiolabeled RNAs can be visualized by an autoradiograph (Fig. 1C–E). The Cbf5 proteins bound with RNA fragments of various sizes appeared as up-shifted smeared bands. In the case of UV crosslinking, the size of RNP was gradually reduced when the crosslinked RNA was digested by increasing amounts of nucleases, as expected (Fig. 1C). Following excision of RNP bands and digestion of proteins, the associated RNA was extracted, ligated to the 5′ adaptor, and reverse transcribed into cDNA. The cDNA libraries were amplified with indexed primers and sequenced by Illumina HiSeq X10 in the paired-end 150 bp mode. After the adaptor and barcode sequences were trimmed and PCR duplicates cleaned, the reads were aligned to the yeast genomic sequence.
AMT–NHS was added to lysed cells to test its ability to crosslink RNA and protein (called in vitro treatment) or directly to living cells to additionally assess its ability to enter yeast cells (called in vivo treatment). Crosslinking of AMT–NHS to RNA was induced by radiation of 365-nm light for 30 min according to the previous application of AMT (Lu et al. 2016). Visualization of radiolabeled RNPs showed that AMT–NHS induced RNA crosslinking of Cbf5 in both lysed and living cells in a dose-dependent manner (Fig. 1D,E). Background levels of Cbf5 were radiolabeled even without crosslinking treatments (lanes 1 in Fig. 1D,E). This could be due to phosphorylation by contaminating protein kinases (Tawk et al. 2017) or RNA-crosslinking by environmental UV light (see below).
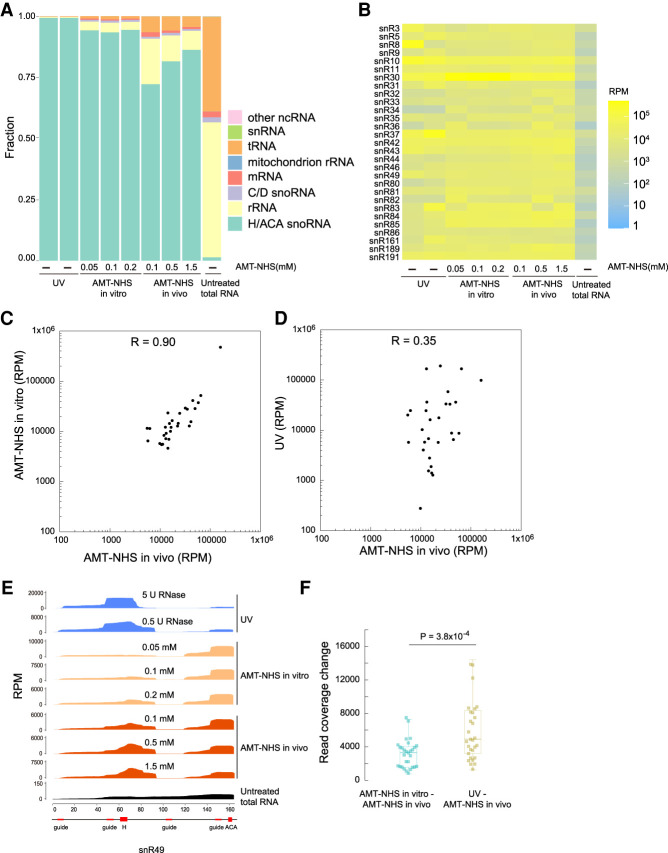
UV crosslinking of Cbf5 was extremely specific as the reads were exclusively (99.3%) contributed by a total of 29 H/ACA snoRNAs in yeast (Fig. 2A). In AMT–NHS in vitro crosslinking, 94% of reads came from H/ACA snoRNAs for all three tested concentrations of crosslinker. This suggested that AMT–NHS crosslinking was slightly less specific than UV crosslinking. The specificity did not improve with increasing concentrations of crosslinker, indicating that the minimal concentration used (0.05 mM) was sufficient for crosslinking in vitro. For AMT–NHS in vivo crosslinking, the percentage of H/ACA snoRNA reads increased from 72% to 86% when the concentration of crosslinker rose from 0.1 to 1.5 mM. Higher concentrations of crosslinker allowed more crosslinker molecules to enter yeast cells, hence improved the yield of crosslinked H/ACA snoRNA and the specificity of crosslinking. These data indicate that AMT–NHS can enter yeast cells and crosslink Cbf5 to H/ACA snoRNAs.
FIGURE 2.
Different crosslinking patterns by AMT–NHS and UV. (A) RNA class distribution of reads. Two UV crosslinking samples (lanes 1 and 2) were digested by 5 and 0.5 U of RNase A/T1. The last sample was the sequencing data of total RNA from non-crosslinked cells. (B) Heatmaps of H/ACA snoRNAs drawn on read per million (RPM). (C,D) Scatter plots of H/ACA snoRNA read counts from AMT–NHS in vivo crosslinking versus AMT–NHS in vitro crosslinking (C) or UV crosslinking (D). Read counts are the averages of two or three data sets with similar treatment. (E) Read coverage of snR49. The sequence motifs of snR49 are shown beneath the x-axis. (F) Box plot showing read coverage change between two types of crosslinking. Read coverage was normalized to an area of 10,000 for each H/ACA snoRNA and averaged from two or three data sets with similar treatment. Absolute changes of read count at each position of snoRNA were summed. P-value was from two-tailed t-test.
AMT–NHS and UV induce different crosslinking patterns
The sequencing data showed that AMT–NHS and UV crosslinked Cbf5 to H/ACA snoRNAs in different patterns (Fig. 2B). The abundance of 29 H/ACA snoRNAs was strongly correlated (Pearson's correlation coefficient R = 0.9) between AMT–NHS in vitro and in vivo crosslinking (Fig. 2C), suggesting that the crosslinker reacted similarly in cell lysates and living cells. In contrast, the abundance of snoRNAs was weakly correlated (R = 0.35) between AMT–NHS and UV crosslinking (Fig. 2D). Moreover, the read coverage profiles of snoRNAs were significantly more dissimilar between AMT–NHS and UV crosslinking than between AMT–NHS in vivo and in vitro crosslinking (Fig. 2E,F). These data suggest that Cbf5 and H/ACA snoRNA were crosslinked differently by AMT–NHS and UV.
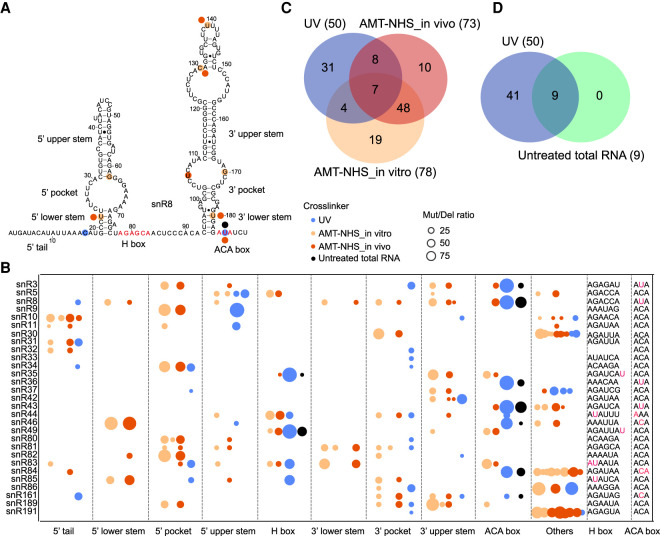
Crosslinked RNA reads often contain mutations and deletions (mut/del) that are indicative of the actual sites of crosslinking. Crosslinked RNAs contain at least one covalently linked amino acid residue after protein digestion. The amino acid adduct would interfere with reverse transcriptase, causing mutations or deletions in reads or blocking cDNA synthesis. Any site with ≥20 reads and a ≥5% rate of mut/del was considered as a crosslinking site and analyzed for the location in secondary structures of H/ACA snoRNAs (Fig. 3A,B), the overlap between different crosslinking treatments (Fig. 3C,D), the enrichment of nearby short sequence motifs (Fig. 4A), and the structural contexts (Fig. 4B,C).
FIGURE 3.
AMT–NHS and UV crosslink Cbf5 to different sites on H/ACA snoRNAs. (A) Crosslinking sites on snR8. The secondary structures and sequence motifs are labeled. Crosslinking sites are shown as circles colored by crosslinking approach. (B) Distribution of crosslinking sites on H/ACA snoRNA secondary structures. Others refer to large insertions present in several snoRNAs. Each crosslinking site is denoted by a circle whose radius is proportional to mut/del rates. The sequences around the H and ACA motifs are shown and the mut/del sites of UV crosslinking are colored magenta. (C) Venn plot of crosslinking sites in UV, AMT–NHS in vivo and AMT–NHS in vitro crosslinked H/ACA snoRNAs. (D) Venn plot of crosslinking sites in H/ACA snoRNAs from UV crosslinked samples and an untreated sample.
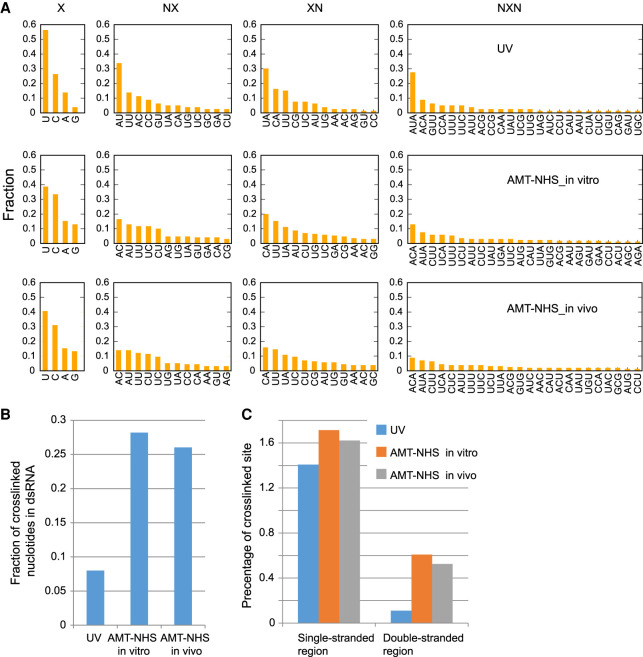
FIGURE 4.
AMT–NHS and UV have different sequence and structure preferences in crosslinking of Cbf5 and H/ACA snoRNAs. (A) Histogram of 1-mer, 2-mer (NX and XN), and 3-mer (NXN) motifs around mut/del sites (X). (B) Fraction of crosslinked nucleotides in double-stranded regions. (C) Percentage of crosslinked nucleotides in double- and single-stranded regions. Yeast H/ACA snoRNAs have 3621 and 3268 nt in double- and single-stranded regions, respectively.
UV crosslinking predominantly occurred at uridine (56%) and cytidine (26%) and at single-stranded regions (92%) (Fig. 4A,B), which is consistent with its known preferences. The H and ACA motifs were hot spots for UV crosslinking. The crosslinking sites in the H and HACA motifs showed much higher mut/del rates (average 62.2% for 13 sites) than those of the other sites (average 12.5% for 37 sites), indicating a high degree of crosslinking at the conserved protein-binding sites. Moreover, crosslinking occurred in seven of eight cases where a uridine occupies the second position of the ACA or H motif (Fig. 3B). The uridine is stacked on an isoleucine in the PUA domain of Cbf5 (Li and Ye 2006) and this particular configuration appears to render the uridine highly susceptible for UV crosslinking. This preference also caused “AUA” as the most frequent 3-mer motif near UV crosslinking sites (Fig. 4A).
Surprisingly, nine mut/del sites could be detected in the H and ACA motifs of H/ACA snoRNAs from total RNA of an untreated sample (Fig. 3B). These mut/del sites completely overlapped with the UV crosslinking sites (Fig. 3D), suggesting that they resulted from crosslinking by environmental UV light. In support of this suggestion, background levels of radiolabeled Cbf5 were detected even without UV or AMT–NHS treatment (lane 1 in Fig. 1D–E).
AMT–NHS in vitro and in vivo crosslinking shared 71% and 75% of crosslinking sites with each other, but shared only 14.1% and 20.5%, or 12.8% and 13.7% if excluding those background UV crosslinking sites present in the untreated sample, of crosslinking sites with UV crosslinking (Fig. 3C). The comparison indicated that AMT–NHS and UV select crosslinking sites in different ways. The crosslinking sites of AMT–NHS were distributed all over the structures of H/ACA snoRNAs (Fig. 3A,B) and located in both double-stranded (∼27%) and single-stranded (∼73%) regions (Fig. 4B). Single-stranded regions were crosslinked by UV and AMT–NHS with similar frequency, but double-stranded regions were five times more likely crosslinked by AMT–NHS than UV (Fig. 4C). Mutations and deletions most frequently occurred at uridine (∼40%) and cytidine (∼32%) in AMT–NHS crosslinking (Fig. 4A). Psoralen is thought to preferentially crosslink staggered uridines located on the two strands of RNA duplex. AU and UA were among the top dinucleotide motifs, but not significantly enriched over other motifs. These data suggest that AMT–NHS preferentially crosslinks uridine and cytidine located in diverse structural contexts.
DISCUSSION
We have synthesized AMT–NHS and demonstrated that it was able to enter yeast cells and efficiently crosslink Cbf5 with H/ACA snoRNAs. This study provided a proof-of-concept that AMT–NHS can be used to map RNA–protein interactions in vivo. In the case of Cbf5, the efficiency and specificity of AMT–NHS crosslinking appears to be less than that of UV crosslinking, but is still practically decent. It is noteworthy that UV crosslinking of Cbf5 and snoRNA was remarkably efficient and even induced by background UV light.
We found that AMT–NHS crosslinks H/ACA snoRNA on sites that are distinct from those induced by UV, which would subsequently affect the abundance and distribution of reads. As a medium-sized chemical compound (MW = 443.47 g/mol), AMT–NHS should crosslink RNA bases and amine groups that are <10.5 Å apart (distance between the center of the psoralen ring and the ester oxygen). Compared with UV crosslinking that requires zero-distance interaction between RNA and protein, the chemical crosslinker is capable of capturing interactions involving more distantly spaced sites and more diverse RNA structures, especially dsRNAs. These unique properties should allow AMT–NHS to capture interactions that are otherwise hard to detect by UV crosslinking. The full potential of AMT–NHS should be further assessed on RBPs that bind dsRNAs and are crosslinked poorly with UV.
AMT–NHS has several disadvantages as compared to UV. As a chemical compound, AMT–NHS only reacts with sites on the surface of RNPs, but the penetrable UV is able to crosslink buried sites. AMT–NHS needs to be applied to a concentrated solution of cells and requires nearly an hour to enter yeast cells and react with RNA and protein. The long incubation time would disturb gene expression patterns and alter transient RNA–protein interactions. In contrast, UV can be directly applied to actively growing cells and induce crosslink in seconds, allowing detection of transient and dynamic interactions (Hahn et al. 2012; Turowski et al. 2014; van Nues et al. 2017). Like UV, AMT–NHS also has biases to crosslink pyrimidines. At the protein side, AMT–NHS reacts specifically with lysine residues which are often, but not always, present at RNA-binding interfaces.
Compared to highly specific UV crosslinking of Cbf5, AMT–NHS crosslinking produced approximately 6% of reads that were not from H/ACA snoRNAs at in vitro saturating conditions and even larger percentages of nonspecific reads when the concentration of crosslinker was limiting at in vivo conditions. The long arm of AMT–NHS may allow it to access and crosslink nonspecific RNAs. To further assess the potential of AMT–NHS, we crosslinked another yeast protein Rnt1 by UV and AMT–NHS (Supplemental Fig. S1). Rnt1 is an RNase III endonuclease that cleaves double-stranded regions on pre-rRNA, pre-snoRNA, pre-snRNA, and pre-mRNA (Elela et al. 1996; Chanfreau et al. 1998; Kufel et al. 1999; Gagnon et al. 2015; Abou Elela and Ji 2019). We found that Rnt1 could be crosslinked to RNA by UV and AMT–NHS (Supplemental Fig. S1A,B), but mostly in a nonspecific manner. Most reads were from tRNAs and mature rRNAs that are not targets of Rnt1 (Supplemental Fig. S1C). Of the reads from known substrates of Rnt1, only a small fraction were derived from the Rnt1-binding sites (Supplemental Fig. S1D). Rnt1 appears to make very transient interactions with substrates and cannot be efficiently crosslinked to substrates by either UV or AMT–NHS. The data of Rnt1 showed that AMT–NHS can induce significant nonspecific crosslinking of protein with abundant tRNAs and rRNAs when specific binding is difficult to capture.
Psoralen preferentially reacts with pyrimidines in double helical regions of RNA, but is also able to react with pyrimidines in single-stranded regions with much less efficiency (Cimino et al. 1985). In the case of helical regions, psoralen first reacts with a pyrimidine in one strand to form a monoadduct, and then reacts with an adjacent pyrimidine in the opposite strand to generate a diadduct. We expect that the diadduct that crosslinks two RNA strands would severely affect the processivity of reverse transcriptase, yielding prematurely terminated cDNA that cannot be amplified. Our method likely mainly detects monoadducts formed on double- and single-stranded regions. This would cause reduction of crosslinking signals from helical regions and account for the observation that AMT–NHS crosslinked nucleotides are more enriched in single-stranded regions than double-stranded regions (Fig. 4B,C). The iCLIP approach is probably more suitable for library preparation of AMT–NHS crosslinked RNA since it includes a step of cDNA circulation and is capable of recovering prematurely terminated cDNA (Konig et al. 2010).
MATERIALS AND METHODS
Synthesis of AMT–NHS
Step 1: Synthesis of 3-(chloromethyl)-2,5,9-trimethyl-7H-furo[3,2-g]chromen-7-one (compound 2)
A mixture of trioxane (217 mg, 2.4 mmol) and concentrated HCl (20 mL) was vigorously stirred when trioxsalen (compound 1, 500 mg, 2.2 mmol) was added portion-wise. The reaction was stirred overnight and then diluted with 100 mL water. Subsequently, the mixture was extracted with dichloromethane, washed three times with water, and dried with sodium sulfate. The organic solvent was concentrated to obtain compound 2 as a white solid (420 mg, 70%): 1H NMR (CDCl3, 500 MHz) δ 7.60 (s, 1H), 6.26 (t, J = 9.2 Hz, 1H), 4.75 (s, 2H), 2.62–2.54 (m, 3H), 2.54–2.45 (m, 6H).
Step 2: Synthesis of 3-(((2,5,9-trimethyl-7-oxo-7H-furo[3,2-g]chromen-3-yl)methyl)thio) propanoic acid (compound 3)
To a solution of compound 2 (100 mg, 0.36 mmol) and 3-mercaptopropanoic acid (38 µL, 0.44 mmol) in THF (4 mL), dimethylbiguanide (100 µL, 0.79 mmol) was added at room temperature. The reaction was heated to reflux for 24 h and then the solvent was evaporated to dryness. The mixture was dissolved in dichloromethane and washed with water and brine. Afterward, the solvent was dried by sodium sulfate and evaporated to obtain a crude product. Further purification by column chromatography afforded compound 3 as a white solid (75 mg, 60%):1H NMR (d-DMSO, 500 MHz) δ 7.77 (s, 1H), 6.31 (s, 1H), 3.92 (s, 2H), 2.60 (t, J = 6.0 Hz, 2H), 2.53 (t, J = 7.0 Hz, 2H), 2.46 (m, 6H), 2.43 (s, 3H).
Step 3: Synthesis of 2,5-dioxopyrrolidin-1-yl 3-(((2,5,9-trimethyl-7-oxo-7H-furo[3,2-g] chromen-3-yl)methyl)thio)propanoate (AMT-NHS)
Compound 3 (20 mg, 0.055 mmol) was dissolved in dichloromethane (3 mL), followed by the addition of N-hydroxysuccinimide (7 mg, 0.061 mmol) and dicyclohexylcarbodiimide (12.5 mg, 0.061 mmol) at room temperature. After stirring for 2 h, the mixture was extracted with dichloromethane and the organic phase was washed with water and brine. Solvent was evaporated under vacuum to give the crude product, which was purified by column chromatography to afford AMT-NHS as a white solid (15 mg, 60%):1H NMR (CDCl3, 500 MHz) δ 7.64 (s, 1H), 6.26 (s, 1H), 3.89 (s, 2H), 2.90 (t, J = 7.0 Hz, 2H), 2.85 (s, 4H), 2.82 (t, J = 6.5 Hz, 2H), 2.59 (s, 3H), 2.53 (s, 3H), 2.50 (s, 3H). AMT–NHS was dissolved in DMSO in a concentration of 30 mM.
Yeast plasmid and strains
Yeast experiments were conducted in accordance with standard protocols. All strains were derived from BY4741 (Mat a, leu2Δ0, Met15Δ0, ura3Δ0). A cassette containing the HTP tag was PCR amplified from plasmid pBS1539 (Granneman et al. 2009) and inserted at the end of CBF5 and RNT1 genes by homologous recombination. Positive clones were selected with Synthetic Complete medium lacking Ura.
RNA–protein crosslinking
Yeast cells were grown in YPD media (1% yeast extract, 2% peptone, and 2% glucose) to A600 = 0.8–1.0, collected and washed once with phosphate buffered saline (PBS). Each crosslinking experiment used 1.5 g of yeast cells.
For UV crosslinking, the cells were dissolved in 1.5 mL of PBS and dispersed in a plastic Petri dish. The cells were placed on ice and irradiated at 254-nm wavelength for a total dose of 1.6 J with a UVP CL-1000 UV CrossLinker. The cells were collected by centrifugation and resuspended in 1.5 mL of lysis buffer (20 mM HEPES-Na [pH 7.5], 1.5 mM MgCl2, 150 mM NaCl, 0.1% Nonidet P-40, and 5 mM β-mercaptoethanol) supplemented with Complete EDTA-free Protease Inhibitor Cocktail (Roche). The cell suspension was frozen by dropping into liquid nitrogen and broken with steel balls in a Tissuelyser-24 (JingXin) or stored at −80°C.
For AMT–NHS in vivo crosslinking, the cells were resuspended in 1.5 mL of PBS containing 0.1, 0.5, or 1.5 mM of AMT–NHS or DMSO as control. The samples were incubated at 30°C in the dark with low-speed rotation for 30 min. The cells were washed by PBS to remove crosslinker, resuspended in 1.5 mL of PBS, and transferred to a Petri dish. The cells were placed on ice, irradiated at 365-nm wavelength for 30 min with a UVP CL-1000 UV CrossLinker and agitated every 10 min. The cells were then lysed as described above.
For AMT–NHS in vitro crosslinking, the cells were first lysed as described above. The supernatant was collected by centrifugation at 30,000g for 30 min and added with 0.05, 0.1, or 0.2 mM of AMT–NHS or DMSO. The samples were incubated at 30°C in the dark with low-speed rotation for 30 min. The crosslinker was neutralized by the addition of 20 mM Tris-HCl (pH 7.5). The samples were irradiated at 365-nm wavelength as described above.
Purification of crosslinked RNP and construction of sequencing library
The crosslinked RNA was purified and cloned as previously described (Granneman et al. 2009; Lin et al. 2013). The adaptors and primers for sequencing library construction were previously reported (Wu et al. 2021). After crosslinking and cell lysis, the clarified supernatant was incubated with 300 µL of IgG Sepharose beads (GE Healthcare) at 4°C for 2.5 h with rotation. The beads were washed three times with lysis buffer and incubated with TEV protease for 10 h to release proteins. The eluate was digested with RNase A/T1 mixture (Thermo) at 37°C for 10 min. The reaction was adjusted to contain 6 M guanidine HCl, 300 mM NaCl and 10 mM imidazole and incubated with 30 µL of MagneHis Ni-Particles (Promega) at 4°C for 12 h.
The beads were washed with wash buffer (20 mM HEPES-Na [pH 7.5], 6 M Guanidine HCl, 300 mM NaCl, 20 mM imidazole, 0.1% Nonidet P-40, and 5 mM β-mercaptoethanol) and then with PNK buffer (20 mM HEPES-Na [pH 7.5], 10 mM MgCl2, and 5 mM β-mercaptoethanol). The dephosphorization, ligation, and radiolabeling reactions were all conducted in a ThermoMixer (Eppendorf) with intermittent shaking. For RNA dephosphorization, the beads were added with 4 units of FastAP (Thermo) and 50 units of RNasin Plus RNase Inhibitor (Promega) in 50 µL PNK and incubated at 37°C for 30 min. The beads were washed three times with wash buffer and three times with PNK buffer.
The 3′ DNA adaptor [5′-TNNNTAGNNNTGGAATTCTCGGGTGCAAGG (NH2)-3′, barcodes are underlined] was preadenylated with Mth RNA Ligase (NEB) according to the manufacturer's instructions. To ligate the 3′ adaptor, the beads were added with 25 units of T4 RNA ligase 2, truncated KQ (NEB), 100 pmol of preadenylated 3′-adaptor, 50 units of RNase Inhibitor (Promega), and 15% PEG8000 in 50 µL PNK and incubated at 25°C for 2.5 h and then at 22°C for 16 h. The beads were washed three times with wash buffer and three times with PNK buffer.
To radiolabel RNA, the beads were added with 20 units of T4 polynucleotide kinase (NEB), 2 µL of 32P–γATP (PerkinElmer), and 40 units of RNase Inhibitor in 50 µL PNK and incubated at 37°C for 30 min. After brief centrifugation, the reaction was added with 1 µL of 50 mM cold ATP and incubated for an additional 10 min. The beads were washed four times with wash buffer and four times with PNK buffer.
The beads were resuspended in 16 µL of 2× Ni elute buffer (40 mM HEPES-Na [pH 7.5], 100 mM NaCl, 600 mM Imidazole, 0.2% Nonidet P-40, and 10 mM β-mercaptoethanol) and 16 µL of 2× NuPAGE loading buffer (Invitrogen) and heated at 70°C for 10 min. After centrifugation, the dissolved protein was resolved in a 4%–12% Bis-Tris NuPAGE gel (Invitrogen) and blotted onto nitrocellulose membranes (Bio-Rad).
Radiolabeled RNPs were visualized on X-ray films. Bands of interest were excised and chopped into pieces. Sliced membranes were added with 200 µL of digestion solution containing 20 mM HEPES-Na (pH 7.5), 4 mg/mL proteinase K (Promega), 50 mM NaCl, and 10 mM EDTA and incubated at 37°C for 30 min with shaking. The reaction was added with 200 µL of solution containing 20 mM HEPES-Na (pH 7.5), 50 mM NaCl, 10 mM EDTA, and 7 M Urea and incubated at 60°C for 30 min. The solution containing released RNAs was transferred to a new tube, extracted with phenol/chloroform, precipitated with isopropanol, and dissolved in 6 µL of H2O.
The RNA was mixed with 20 pmol of the 5′ RNA linkers (5′-ACACGACGCUCUUCCGAUCUNNNCGNNNU-3′ and 5′-ACACGACGCUCUUCCGAUCUNNNAUNNNC-3′, barcodes are underlined) and heated at 80°C for 90 sec in a PCR machine and then placed on ice. The RNA mixture was added with 2 µL of 10 mM ATP, 40 units of RNase Inhibitor, 2 µL of T4 RNA Ligase 1 (NEB), 6 µL of 50% PEG8000, 2 µL of 10× PNK, and water to a total volume of 20 µL. The ligation reaction was incubated at 25°C for 2.5 h and then at 22°C for 18 h.
RNA was extracted by phenol/chloroform and precipitated with isopropanol. RNA was resuspended in 11 µL of H2O and added with 10 pmol of reverse transcription primer (5′-GCCTTGGCACCCGAGAATTCCA-3′). The RNA mixture was heated at 80°C for 2 min and placed on ice. The mixture was added with 4 µL of 5× First strand buffer, 1 µL of 0.1 M DTT, 1 µL of 10 mM dNTPs, 40 units of RNase Inhibitor and 1 µL of SuperScript III (Invitrogen) to a total volume of 20 µL. The reaction was incubated in a PCR machine at 55°C for 30 min, 50°C for 30 min, and 70°C for 15 min.
The first round PCR reaction contained 3 µL of cDNA solution, 4 µL of 5× HF buffer (NEB), 0.4 µL of 10 mM dNTPs, 1 µL of 10 mM DP5 primer (5′-ACACGACGCTCTTCCGATCT-3′), 1 µL of 10 mM DP7 primer (5′-CCTTGGCACCCGAGAATTCC-3′), and 0.2 µL of Phusion DNA polymerase (NEB) in a total volume of 20 µL. The DNA was denatured at 98°C for 30 sec, amplified for 21 cycles (98°C for 8 sec, 65°C for 20 sec, and 72°C for 15 sec) and extended at 72°C for 5 min. The DNA was resolved in a 10% native polyacrylamide gel and visualized with DNA SYBR Gold (Invitrogen). DNA bands of 80–200 bp were excised, crushed, and soaked out overnight in buffer containing 10 mM Tris-HCl (pH 8.0), 300 mM NaCl, and 1 mM EDTA.
The second round of PCR was conducted with the first round PCR product as template and 10 pmol each of indexed primers P5 (5′-AATGATACGGCGACCACCGAGATCTACACXXXXXACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′) and P7 (5′-CAAGCAGAAGACGGCATACGAGATXXXXXXGTGACTGGAGTTCCTTGGCACCCGAGAATTCCA-3′) with a similar setting as above. The extension time was changed to 20 sec and a total of five cycles were performed. DNA of 170–300 bp was recovered as described above.
The total RNA from untreated cells was hydrolyzed at mild alkaline conditions and converted into sequencing libraries according to the RiboMeth-seq procedure as previously described (Wu et al. 2021).
Data processing and analysis
The libraries were sequenced on an Illumina HiSeq X10 machine using the paired-end 150 bp mode by Annoroad. Reads were demultiplexed according to the index sequences in primers P5 and P7. Adaptor sequences were trimmed by cutadapt (v1.18) with a minimal length of 33 nt (including 19 nt of barcodes) for reads (Kechin et al. 2017). PCR duplicates that share the same barcode and RNA sequence were reduced with a home-written Python script. Barcodes at both ends of reads were trimmed and recorded in the name of reads. Reads were aligned to the genome sequence of Saccharomyces cerevisiae (GCF_000146045.2_R64_genomic.fna) with NovoAlign (V3.09.00, http://novocraft.com). The reads that shared the same barcodes, the same start genomic coordinates and the same “CIGAR” words, which encode information of deletion and insertion but no mutation, were reduced in the second round removal of PCR duplicates. These PCR duplicates acquired mutation during PCR amplification and would escape the first round of PCR duplicate removal. The aligned reads were stored as sam or bam files and visualized on IGV (2.7.22.7.2) (Robinson et al. 2011). Statistics of reads and mutation/deletion were analyzed by bedtools, samtools, and home-written scripts (Quinlan and Hall 2010).
For mutation/deletion and crosslinking site analysis, data sets from similar treatments (UV, AMT–NHS in vivo and AMT–NHS in vitro) were combined. The terminal residues of reads were sometimes mutated for unknown reasons, and these terminal mutations were not counted. Crosslinking sites must have at least 20 reads and a mut/del rate equal or greater than 5%. Multiple continuous mut/del sites, which likely resulted from a single crosslinking event, are represented by one middle crosslinking site. Secondary structures of H/ACA snoRNAs were predicted by the RNAcentral database (RNAcentral Consortium 2021).
DATA DEPOSITION
The sequencing data have been deposited into the National Genomics Data Center (bigd.big.ac.cn) under GSA accession code CRA004708.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank Hongjie Zhang for help in radioactivity experiments, Songlin Wu for help in data analysis, and Xiuling Gao for technical assistance. The study was supported by National Key R&D Program of China (2017YFA0504600), National Natural Science Foundation of China (91540201, 31970259, 31430024, 31325007, 91940306, and 31971200), and Strategic Priority Research Program of Chinese Academy of Sciences (XDB37010201).
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078896.121.
MEET THE FIRST AUTHORS
Xuzhen Guo.

Yan Han.

Meet the First Author(s) is a new editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. Yan Han and Xuzhen Guo, the co-first authors of the paper “Development of an RNA–protein crosslinker to capture protein interactions with diverse RNA structures in cells,” were graduate students at the Institute of Biophysics, Chinese Academy of Sciences in the laboratories of Drs. Keqiong Ye and Jiangyun Wang, respectively. Yan focused on the development of RNA–protein crosslinking agents and analysis of ribosome assembly factors by RNA–protein crosslinking and structural biology methods. Xuzhen was in a chemical biology laboratory and interested in synthesizing new compounds to capture RNA–protein, RNA–RNA, and protein–protein interactions.
What are the major results described in your paper and how do they impact this branch of the field?
In this study, we developed a new compound called AMT–NHS to crosslink RNA and protein. AMT–NHS is composed of a psoralen derivative, which can intercalate in RNA helices and react with pyrimidines upon photo-activation, and an N-hydroxysuccinimide ester group, which mainly links lysine residues of protein. We tested AMT–NHS to crosslink H/ACA snoRNPs in yeast and found that it can enter living yeast cells and efficiently crosslink the Cbf5 protein with H/ACA snoRNAs. Compared with UV, AMT–NHS crosslinked RNAs in a different pattern and targeted more sites on double-stranded RNAs. We expected that the crosslinker would become a new tool to detect RNA–protein interactions in cells, particularly those involving double-stranded RNAs.
What led you to study RNA or this aspect of RNA science?
YH: I have been analyzing the RNA binding sites of ribosome assembly factors using classic UV-crosslinking methods and learned several limitations of UV. When our collaborators synthesized a new RNA–protein crosslinker that was expected to better target dsRNA than UV, I readily tested its crosslinking activity in yeast.
XG: I became interested in RNA research when I was involved in developing an enzymatic method for site-specific labeling of RNA. After learning the lack of an efficient RNA–protein crosslinker targeting dsRNA, I embarked to synthesize AMT–NHS.
During the course of these experiments, were there any surprising results or particular difficulties that altered your thinking and subsequent focus?
YH: I initially did not observe any crosslinked Cbf5–RNA complex in SDS-PAGE. I was frustrated after many failed attempts. I then carefully monitored Cbf5 after every experimental step and found that the protein disappeared after phosphatase treatment. I suspected that the phosphatase was contaminated with proteases that degraded my protein. After changing to another brand of phosphatase, strong signals of crosslinked Cbf5 miraculously emerged. From this experience I learned how to troubleshoot when something went wrong.
If you were able to give one piece of advice to your younger self, what would that be?
YH: I would tell myself not to be afraid of failures. When an experiment does not work, we should go back and check every detail.
XG: Rome was not built in a day. Stay calm and patient; you will find appropriate solutions eventually.
What are your subsequent near- or long-term career plans?
YH: I am interested in RNA drug development and want to tackle diseases in my future research.
XG: I hope to set up my own research group in the future and develop novel chemical agents for biomedical research, disease diagnosis, and clinical treatment.
What were the strongest aspects of your collaboration as co-first authors?
The study depended on complementary expertise of the two co-first authors. Yan was experienced in biochemistry and yeast genetics, whereas Xuzhen was well-trained in chemical synthesis.
REFERENCES
- Abou Elela S, Ji X. 2019. Structure and function of Rnt1p: an alternative to RNAi for targeted RNA degradation. Wiley Interdiscip Rev RNA 10: e1521. 10.1002/wrna.1521 [DOI] [PubMed] [Google Scholar]
- Chanfreau G, Legrain P, Jacquier A. 1998. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J Mol Biol 284: 975–988. 10.1006/jmbi.1998.2237 [DOI] [PubMed] [Google Scholar]
- Cimino GD, Gamper HB, Isaacs ST, Hearst JE. 1985. Psoralens as photoactive probes of nucleic acid structure and function: organic chemistry, photochemistry, and biochemistry. Annu Rev Biochem 54: 1151–1193. 10.1146/annurev.bi.54.070185.005443 [DOI] [PubMed] [Google Scholar]
- Elela SA, Igel H, Ares M Jr. 1996. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell 85: 115–124. 10.1016/S0092-8674(00)81087-9 [DOI] [PubMed] [Google Scholar]
- Gagnon J, Lavoie M, Catala M, Malenfant F, Elela SA. 2015. Transcriptome wide annotation of eukaryotic RNase III reactivity and degradation signals. PLoS Genet 11: e1005000. 10.1371/journal.pgen.1005000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Granneman S, Kudla G, Petfalski E, Tollervey D. 2009. Identification of protein binding sites on U3 snoRNA and pre-rRNA by UV cross-linking and high-throughput analysis of cDNAs. Proc Natl Acad Sci 106: 9613–9618. 10.1073/pnas.0901997106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu J, Wang M, Yang Y, Qiu D, Zhang Y, Ma J, Zhou Y, Hannon GJ, Yu Y. 2018. GoldCLIP: Gel-omitted Ligation-dependent CLIP. Genomics Proteomics Bioinformatics 16: 136–143. 10.1016/j.gpb.2018.04.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141: 129–141. 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn D, Kudla G, Tollervey D, Beggs JD. 2012. Brr2p-mediated conformational rearrangements in the spliceosome during activation and substrate repositioning. Genes Dev 26: 2408–2421. 10.1101/gad.199307.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kechin A, Boyarskikh U, Kel A, Filipenko M. 2017. cutPrimers: a new tool for accurate cutting of primers from reads of targeted next generation sequencing. J Comput Biol 24: 1138–1143. 10.1089/cmb.2017.0096 [DOI] [PubMed] [Google Scholar]
- Kim B, Kim VN. 2019. fCLIP-seq for transcriptomic footprinting of dsRNA-binding proteins: lessons from DROSHA. Methods 152: 3–11. 10.1016/j.ymeth.2018.06.004 [DOI] [PubMed] [Google Scholar]
- Kiss T, Fayet-Lebaron E, Jady BE. 2010. Box H/ACA small ribonucleoproteins. Mol Cell 37: 597–606. 10.1016/j.molcel.2010.01.032 [DOI] [PubMed] [Google Scholar]
- Konig J, Zarnack K, Rot G, Curk T, Kayikci M, Zupan B, Turner DJ, Luscombe NM, Ule J. 2010. iCLIP reveals the function of hnRNP particles in splicing at individual nucleotide resolution. Nat Struct Mol Biol 17: 909–915. 10.1038/nsmb.1838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kufel J, Dichtl B, Tollervey D. 1999. Yeast Rnt1p is required for cleavage of the pre-ribosomal RNA in the 3′ ETS but not the 5′ ETS. RNA 5: 909–917. 10.1017/S135583829999026X [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Ye K. 2006. Crystal structure of an H/ACA box ribonucleoprotein particle. Nature 443: 302–307. 10.1038/nature05151 [DOI] [PubMed] [Google Scholar]
- Licatalosi DD, Mele A, Fak JJ, Ule J, Kayikci M, Chi SW, Clark TA, Schweitzer AC, Blume JE, Wang X, et al. 2008. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature 456: 464–469. 10.1038/nature07488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Lu J, Feng Y, Sun M, Ye K. 2013. An RNA-binding complex involved in ribosome biogenesis contains a protein with homology to tRNA CCA-adding enzyme. PLoS Biol 11: e1001669. 10.1371/journal.pbio.1001669 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Z, Zhang QC, Lee B, Flynn RA, Smith MA, Robinson JT, Davidovich C, Gooding AR, Goodrich KJ, Mattick JS, et al. 2016. RNA duplex map in living cells reveals higher-order transcriptome structure. Cell 165: 1267–1279. 10.1016/j.cell.2016.04.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. 2010. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26: 841–842. 10.1093/bioinformatics/btq033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- RNAcentral Consortium. 2021. RNAcentral 2021: secondary structure integration, improved sequence search and new member databases. Nucleic Acids Res 49: D212–D220. 10.1093/nar/gkaa921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson JT, Thorvaldsdottir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. 2011. Integrative genomics viewer. Nat Biotechnol 29: 24–26. 10.1038/nbt.1754 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tawk C, Sharan M, Eulalio A, Vogel J. 2017. A systematic analysis of the RNA-targeting potential of secreted bacterial effector proteins. Sci Rep 7: 9328. 10.1038/s41598-017-09527-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turowski TW, Lebaron S, Zhang E, Peil L, Dudnakova T, Petfalski E, Granneman S, Rappsilber J, Tollervey D. 2014. Rio1 mediates ATP-dependent final maturation of 40S ribosomal subunits. Nucleic Acids Res 42: 12189–12199. 10.1093/nar/gku878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ule J, Jensen KB, Ruggiu M, Mele A, Ule A, Darnell RB. 2003. CLIP identifies Nova-regulated RNA networks in the brain. Science 302: 1212–1215. 10.1126/science.1090095 [DOI] [PubMed] [Google Scholar]
- Ule J, Hwang HW, Darnell RB. 2018. The future of cross-linking and immunoprecipitation (CLIP). Cold Spring Harb Perspect Biol 10: a032243. 10.1101/cshperspect.a032243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Nostrand EL, Pratt GA, Shishkin AA, Gelboin-Burkhart C, Fang MY, Sundararaman B, Blue SM, Nguyen TB, Surka C, Elkins K, et al. 2016. Robust transcriptome-wide discovery of RNA-binding protein binding sites with enhanced CLIP (eCLIP). Nat Methods 13: 508–514. 10.1038/nmeth.3810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Nues R, Schweikert G, de Leau E, Selega A, Langford A, Franklin R, Iosub I, Wadsworth P, Sanguinetti G, Granneman S. 2017. Kinetic CRAC uncovers a role for Nab3 in determining gene expression profiles during stress. Nat Commun 8: 12. 10.1038/s41467-017-00025-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheeler EC, Van Nostrand EL, Yeo GW. 2018. Advances and challenges in the detection of transcriptome-wide protein-RNA interactions. Wiley Interdiscip Rev RNA 9: e1436. 10.1002/wrna.1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S, Wang Y, Wang J, Li X, Li J, Ye K. 2021. Profiling of RNA ribose methylation in Arabidopsis thaliana. Nucleic Acids Res 49: 4104–4119. 10.1093/nar/gkab196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu YT, Meier UT. 2014. RNA-guided isomerization of uridine to pseudouridine-pseudouridylation. RNA Biol 11: 1483–1494. 10.4161/15476286.2014.972855 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.