Abstract
The 22 mitochondrial and ∼45 cytosolic tRNAs in human cells contain several dozen different post-transcriptional modified nucleotides such that each carries a unique constellation that complements its function. Many tRNA modifications are linked to altered gene expression, and deficiencies due to mutations in tRNA modification enzymes (TMEs) are responsible for numerous diseases. Easily accessible methods to detect tRNA hypomodifications can facilitate progress in advancing such molecular studies. Our laboratory developed a northern blot method that can quantify relative levels of base modifications on multiple specific tRNAs ∼10 yr ago, which has been used to characterize four different TME deficiencies and is likely further extendable. The assay method depends on differential annealing efficiency of a DNA-oligo probe to the modified versus unmodified tRNA. The signal of this probe is then normalized by a second probe elsewhere on the same tRNA. This positive hybridization in the absence of modification (PHAM) assay has proven useful for i6A37, t6A37, m3C32, and m2,2G26 in multiple laboratories. Yet, over the years we have observed idiosyncratic inconsistency and variability in the assay. Here we document these for some tRNAs and probes and illustrate principles and practices for improved reliability and uniformity in performance. We provide an overview of the method and illustrate benefits of the improved conditions. This is followed by data that demonstrate quantitative validation of PHAM using a TME deletion control, and that nearby modifications can falsely alter the calculated apparent modification efficiency. Finally, we include a calculator tool for matching probe and hybridization conditions.
Keywords: TRIT1, anticodon loop modification, base methylation, i6A37, t6A37
INTRODUCTION
Among all RNA species, the tRNAs are the most extensive post-transcriptionally modified both in density (% modified nucleotides) and diversity (different modifications), collectively carrying >100 modified nucleotides (nt) (Vare et al. 2017; Krutyholowa et al. 2019; Suzuki 2021). Human cytoplasmic (cy) tRNAs carry as many as 17 modified nucleotides, with an average of 13 per tRNA (Pan 2018). Each of the ∼45 major cy-tRNA species has unique identity with a distinct combination of modified nucleotides. Each of the 22-human mitochondrial (mt) tRNAs carry three to nine modifications (Suzuki et al. 2020). Some mt-tRNA modifications are not found on cy-tRNAs and vice versa (Suzuki et al. 2020). Inborn errors of tRNA hypomodification lead to numerous pathological conditions of varying severity (Chujo and Tomizawa 2021; Suzuki 2021). Sequence-specific detection of the presence or absence of modifications on specific tRNAs can be helpful in assessing the extent of suspected or actual TME dysfunction (Abbott et al. 2014; Yarham et al. 2014; Bednarova et al. 2017). This paper reviews a method that can do so for a subset of common modifications. Because it is applicable to modifications that share a certain physiochemical characteristic, we will first provide a general review of this aspect of the method to aid understanding and its practical use.
tRNA modifications comprise a wide range of chemical complexity. A relevant example is the diversity at position 37, 3′ to the anticodon. Several tRNAs carry t6A37, while others are modified with either N6-isopentenyladenosine-37 (i6A37), a methyl addition on N1 of G (m1G37), or after deamination of adenosine to inosine to form m1I37, others remain unmodified, and tRNAsPhe carry wybutosine (yW37) whose synthesis requires sequential enzymatic activities (Lorenz et al. 2017; Boccaletto et al. 2018).
The anticodon loop (ACL) which spans positions 32–38 is most variably modified due to positions 34 and 37 (Han and Phizicky 2018; Han et al. 2018), with wobble base 34 being most diverse. Modifications to 34 and 37 are important for balance of accuracy, flexibility, and efficiency of translation by controlling the thermodynamic and spatial limits of codon–anticodon pairing in the ribosome (Vare et al. 2017). Modifications to nucleotide 34 are thought to promote correct pairing to codon position 3, especially important for wobble decoding (Agris et al. 2018). Conversely, modifications to 37 interfere with Watson:Crick (W:C) bonding and are thought to maintain the reading frame by preventing pairing with an upstream base (Vare et al. 2017). These modifications are relevant to the assay reviewed here, referred to as positive hybridization in the absence of modification (PHAM), and can be listed as i6A37, t6A37, m1G37, m1I37, and yW37, as well as hypermodified forms, for example, mS2i6A37 resulting from methylthiolation of i6A37 in mammalian but not yeast mitochondria, and in bacteria (Wei et al. 2015; Lamichhane et al. 2016). The PHAM method has also been applied to N2,N2-dimethylguanosine-26 (m2,2G26) and N3-methylcytosine (m3C) at position 32 in tRNAsSer, Thr and Arg (and other locations), which also have this characteristic.
The anticodon stem–loop (ASL) directs “modification circuits” in which a modification at one position is a prerequisite for a sequential modification at one or more positions elsewhere in the ACL. Some of the known circuits are disrupted by disease-causing mutations in TMEs (Guy and Phizicky 2014; Guy et al. 2015; Li et al. 2020; for review, see Barraud and Tisné 2019; and in Sokolowski et al. 2017). The PHAM assay has been used to monitor three ACL circuit modifications, i6A37, t6A37, and m3C32, and to document heritable errors of metabolism due to mutations to the TMEs involved in their synthesis (Yarham et al. 2014; Edvardson et al. 2017; Lentini et al. 2020; Lentini et al. 2021). It was also used for another translationally active ASL modification, m2,2G26, whose deficiency also causes a human disease (Dewe et al. 2017).
Notable gel-based assays can distinguish between the presence and absence of a particular modified nucleotide in a tRNA-specific manner. The modified nucleotide, queuosine (Q), has been targeted by multiple approaches (Igloi and Kössel 1985; Zaborske et al. 2014; Wang et al. 2018; Matuszek and Pan 2019). Acryloylamino-phenylboronic (APB) acid is used to exploit a chemical reactivity highly limited to Q (Zaborske et al. 2014). APB interacts with Q in tRNA, slowing its mobility in APB-gels (in addition to above, also see Kessler et al. 2018; Cirzi and Tuorto 2021); in species in which Q is glycosylated, an adaptation can be incorporated (Zhang et al. 2020). A unique tRNA-specific approach was developed for northern blot detection of yW37 found on a single tRNA, the isoacceptor for Phe (Nostramo and Hopper 2020). This exploits the specific reactivity of yW with aniline followed by hydrolysis with mild acid. Upon specific probing, tRNAPhe that contains yW37 exhibits a distinctive cleavage pattern, whereas tRNAPhe lacking yW37 remains full length (Nostramo and Hopper 2020).
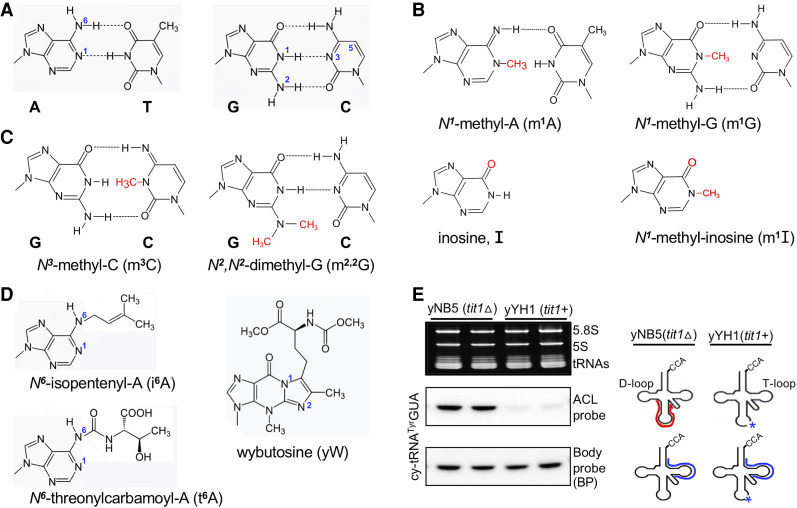
Annealing of cellular RNA to DNA-oligo arrays was found sensitive to certain modifications that interfere with base-pairing, as discussed above for PHAM, and these were verified using TME deletion strains (Hiley et al. 2005). Methylation of atoms normally involved in hydrogen bond formation that enable W:C base-pairing interferes with this, such as m1A, m1G, m3C and m2,2G (Fig. 1A–C). These methylated bases can be discerned by some reverse transcriptase (RT)-mediated cDNA-based tRNA-seq methods (Ryvkin et al. 2013; Arimbasseri et al. 2015, 2016; Cozen et al. 2015; Zheng et al. 2015; Clark et al. 2016; Gogakos et al. 2017; Hrabeta-Robinson et al. 2017; Guo et al. 2020; Behrens et al. 2021; for review, see Motorin and Marchand 2021). In cDNA-seq methodologies, the RTs have trouble “identifying” bases that are methylated/modified on atoms involved in W:C hydrogen bond formation and a “mis”-matched dNTP is “mis”-incorporated, sometimes with a particular frequency or “signature” specific to a particular RT (e.g., Supplemental Figures 2, 3 in Arimbasseri et al. 2015; Gogakos et al. 2017; Behrens et al. 2021). Mismatches in cDNA hydro-tRNA-seq data at positions in specific tRNAs known to be modified with m2,2G26 and m3C32 were validated by their appearance as the correct sequence in S. pombe strains deleted of the TME genes for trm1+ and trm140+, respectively, encoding the respective methyltransferases (Arimbasseri et al. 2015, 2016). Likewise, m1G9, m2,2G26, and yW37-related mismatches in cDNA mim-tRNA-seq data from S. cerevisiae were validated by the trm10Δ, trm1Δ, and trm7Δ strains, respectively (Behrens et al. 2021). Pretreatment of RNA samples with demethylases convert mismatches in cDNA-seq data to the correct nucleotides (Cozen et al. 2015; Zheng et al. 2015; Clark et al. 2016; Behrens et al. 2021). Thanks to tRNA modification databases, not only can mismatches in cDNA data be “called” as modifications but they can also be used to substantially improve tRNA alignment to reference genomes (Behrens et al. 2021). In this way, cDNA–tRNA-seq has been advanced to reliably quantify relative tRNA abundances (Behrens et al. 2021). In any case, conventional methods should validate or determine modifications at unexpected mismatch positions in cDNA-based sequences (Helm and Motorin 2017).
FIGURE 1.
Schematic structures of modifications that interfere with W:C base-pairing. (A) The schematics show canonical Watson:Crick (W:C) base-pairing through hydrogen bonding between Adenine (A):Thiamine (T) and Guanine (G):Cytosine (C). The N1 and N6 atoms of adenosine, N1 and N2 of guanosine, as well as N3 and C5 of cytosine, are indicated. (B) Schematic structures of N1-methyl-A (m1A), N1-methyl-G (m1G), showing hydrogen bond disruption between A:T, G:C, respectively (compare with panel A). Inosine (I) and N1-methylinosine (m1I) are also shown. (C) N3-methylcytosine (m3C) and N2,N2-dimethylguanosine (m2,2G) showing hydrogen bond disruption between G:C (compare panel A). (D) The bulky, isopentenyl- and threonylcarbamoyl- groups attached to N6 of adenosine-37 and another position-37 modification, wybutosine are shown. (E) Illustration of PHAM assay results after separating total RNA from S. pombe yNB5 (tit1Δ) and yYH1 (tit1+) on a 10% TBE-Urea gel. Top left panel is the ethidium bromide-stained gel prior to transfer. Middle and lower panels show the ACL probe and corresponding Body probe results for to the same tRNA species (indicated to the left). On the right is the ACL probe oligo schematically indicated in red; it can bind efficiently only in the absence of the modification (asterisk), as in the Tit1-deleted strain. The body probe oligo is schematically indicated in blue.
Not only base methylations, but a limited number of other modifications including to the ribose, can also be detected by cDNA-seq approaches (Clark et al. 2016; Guo et al. 2020), as well as inosine, which is misread as guanosine due to loss of hydrogen bonding by deamination of adenosine (Arimbasseri et al. 2015). Still, the number of modifications discernable by mismatches is limited, even though this can be increased by examination of other patterns in the cDNA data such as positional stops (Clark et al. 2016).
Certain base methylations to atoms involved in hydrogen bonding on the W:C face, for example, m6A and m2G, do not lead to mismatches in cDNA tRNA-seq data; methylation of the primary amines at N6 of A and N2 of G leave one of the hydrogens available for hydrogen bond formation. Even large modifications to N6A such as i6A37 and t6A37 were not associated with mismatches in the cDNA tRNA-seq data (Arimbasseri et al. 2015; Clark et al. 2016) nor did they block RT unless hypermodified (Behrens et al. 2021). Yet these have been very good candidates for PHAM (below).
RESULTS
General description of the PHAM method and its application
High-throughput tRNA-seq methods can deliver information on all cellular tRNAs and some modifications but requires significant investment in bioinformatics and other resources. The PHAM blot method requires less investment and setup. It exploits the high sensitivity of DNA-oligo probe annealing to a tRNA sequence due to impaired base-pairing caused by modification of the nucleotide under study. A single PHAM blot can be used to survey multiple specific tRNAs and can detect and quantify some modifications that cDNA-seq methods cannot.
As illustrated in Figure 1A and D, the N6 isopentenyl group on adenine (i6A37) would limit W:C base-pairing. As shown previously and illustrated in Figure 1E, presence of i6A37 is discernable by the PHAM assay (Lamichhane et al. 2011, 2013a,b, 2016; Yarham et al. 2014; Khalique et al. 2020), and is also applicable to t6A37 (Rojas-Benitez et al. 2015; Edvardson et al. 2017; Bacusmo et al. 2018; Beenstock et al. 2020). The m3C32 and m2,2G26 which block the possibility for hydrogen bond formation for W:C base-pairing (Fig. 1C) are also discernable by PHAM (Arimbasseri et al. 2015; Bacusmo et al. 2018; Lentini et al. 2020, 2021). Schematics for m1A and m1G which also block hydrogen bond formation are shown along with I and m1I (Fig. 1B).
tRNA isopentenyltransferase-1 (Tit1) is the TME responsible for i6A37 formation. Figure 1E illustrates PHAM for cy-tRNATyrGUA; the left shows RNA from two fission yeast strains, Schizosaccharomyces pombe yNB5 (tit1Δ, deleted of the tit1+ gene) and yYH1 (tit1+). The right side illustrates that the ACL probe efficiently anneals to A37-unmodified tRNATyrGUA from tit1Δ cells but not to i6A37-modified tRNATyrGUA from tit1+ cells. After initial probing, the blot was stripped of ACL probe, rescanned to ensure that the signal was efficiently removed, and reprobed with a “body probe” (BP) to the T stem–loop region of the cy-tRNATyrGUA showing more or less equal loading (Fig. 1E, lower left panel).
We want to highlight that for i6A37, t6A37, m2,2G26, and m3C32, PHAM was used to document tRNA hypomodification as a functional manifestation of newly described TME deficiencies in human disease syndromes due to mutations in TRIT1 (Yarham et al. 2014), OSGEP (Edvardson et al. 2017), TRMT1 (Dewe et al. 2017), and DALRD3(Lentini et al. 2020), respectively. In addition, a pathogenic mutation at the 38 position of mt-tRNASerUCN was shown to result in tRNA-i6A hypomodification at position 37 (Yarham et al. 2014), consistent with the A36–A37–A38 recognition sequence of IPTases (Motorin et al. 1997).
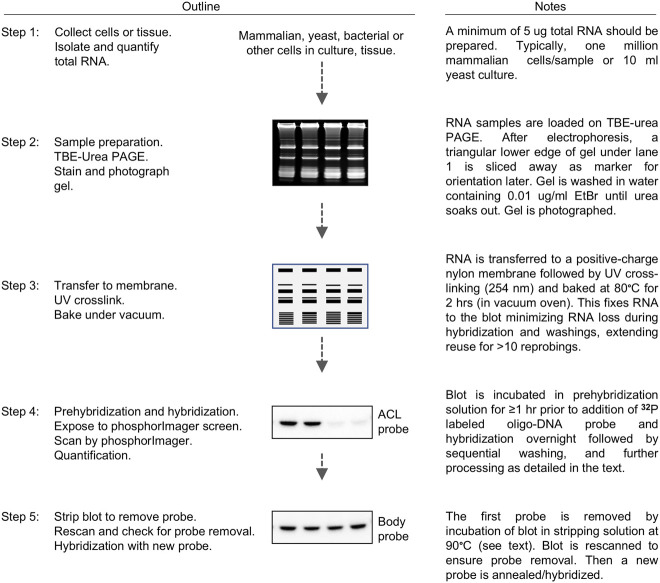
A five-step outline with graphics and notes comprises Figure 2. The major steps are: (i) RNA preparation/purification; (ii) polyacrylamide gel electrophoresis-northern blotting; (iii) probing with a modification-detection DNA-oligo followed by washing and exposure; (iv) blot stripping and probing by a calibration DNA-oligo, washing and exposure; and (v) quantification of hybridization band signals. Steps 1 and 2 each take one day or more, whereas the other steps take longer depending on signal strength and exposure times needed for quantifiable results.
FIGURE 2.
Schematic outline representation of the PHAM method assay. See text for detailed protocol. A five-step protocol is outlined with brief descriptions (see text for details).
Calculation of apparent modification efficiency (AME) %
The following basic equation is used to convert quantified data into AME % = [1 − (ACL (test sample)/BP (test sample))/(ACL (background)/BP (background))] × 100, where ACL = quantity of counts from the ACL probe, and BP = quantity of counts from the BP (control) probe. See below and figures for specific cases.
The PHAM method described with real data to illustrate key points
Of the three points we want to convey in the next sections, the first is the most important, (i) Different tRNAs exhibit different response profiles to ACL probe washing at temperatures above the incubation temperature (Ti). This led to a protocol in which wash temperature must be empirically determined for each ACL probe the first time it is used/tested for optimal performance and quantification. The second and third points are (ii) in general, mt-tRNAs exhibit greater sensitivity to increasing ACL probe wash temperatures than cy-tRNAs, and (iii) nearby modifications lead to false elevation of the apparent modification efficiency (AME) of the target nucleotide.
Other cautionary notes are worthy of mention. The PHAM assay is based on differential annealing efficiency of a DNA-oligo to two or more sample tRNAs that vary in their levels of modification at a particular position of interest. Another DNA-oligo probe that anneals to a different region of the same tRNA serves as a control for normalization/calibration. One should be cautious as discussed above if the target nucleotide is part of a modification circuit, because of the possibility that another modification in the region complementary to the probe would alter annealing. Thus, one should consider reviewing the known circuits (Barraud and Tisné 2019) and perhaps a specific appropriate control experiment.
For probe design, the target nucleotide is generally opposite a central position in the oligo sequence. Supplemental Table S1 is a spreadsheet that can calculate the hybridization Ti for the DNA-oligo ACL probe and body probe (BP) for the salt conditions of the hybridization solution (2× SSC). In the next section, we compare fission yeast S. pombe strains that differ in the presence and absence of the tit1+ gene responsible for i6A37 formation.
Different tRNAs exhibit different profiles to ACL probe washing at temperatures above Ti
Because each tRNA has a unique nucleotide sequence and modification profile in an organism, the DNA probe annealing can be different for each and may also vary among species for similar tRNAs. The i6A37 AME for different tRNAs were analyzed by PHAM at multiple increments of wash temperatures above the Ti of the ACL probe. Annealing of an ACL probe is diminished by modification-induced loss of hydrogen bond formation and possibly steric hindrance in the presence of i6A37. This results in lower probe signal on the northern blot for i6A37-modified tRNA relative to unmodified tRNA. The goal below was to determine if wash temperature altered differential ACL:BP signal ratio and therefore the calculated AME %, and importantly if it would be different for different tRNAs.
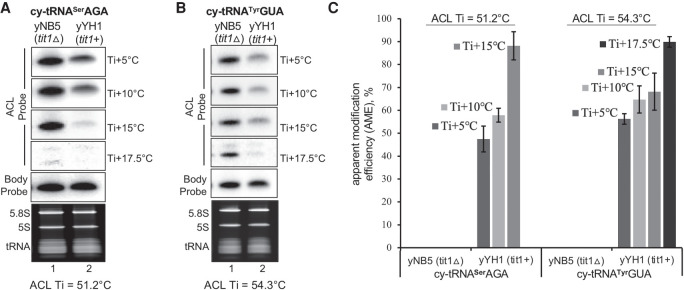
Two cy-tRNA substrates, tRNASerAGA and tRNATyrGUA were examined at increasing wash temperatures above the Ti of the ACL probes, Ti +5°C, +10°C, +15°C, and +17.5°C. The ACL Ti for tRNASerAGA = 51.2°C, and for tRNATyrGUA = 54.3°C. These tRNAs carry i6A37 in yYH1 (tit1+) cells but lack it in yNB5 (tit1Δ). Both probes anneal better to tRNAs from tit1Δ than from tit1+ cells (Fig. 3A,B).
FIGURE 3.
Different tRNAs exhibit different ACL probe wash profiles at temperatures above Ti. (A,B) PHAM northern blot assay of total RNA from S. pombe yYH1 (tit1+) and yNB5 (tit1Δ). The figures show signals from the i6A37-sensitive anticodon loop probe (ACL) probes specific to S. pombe cy-tRNASerAGA (A), and cy-tRNATyrGUA (B) after washing at temperatures indicated to the right of the panels, where the Ti for each tRNA is provided below the panels. The T stem–loop body probe (BP) results are also shown. (C) Graphic display of quantification of AME based on duplicate experiments for both tRNAs in A and B at different wash temperatures according to the equation: AME % = [1 − (ACL tit1+/BP tit1+)/(ACL tit1Δ/BP tit1Δ)] × 100. The graph shows the mean ± standard deviation (error bars), N = 2 biological replicates.
As is routinely observed, probe annealing and signal strength decreases as wash temperature increases, as expected (data not shown). Importantly, however, we observed that the differences in changes for the two tRNAs differed as the wash temperature increased, and with different optima (Fig. 3A,B). The AME in Figure 3C, based on duplicate experiments also reflects the differential patterns. The tRNASerAGA showed a steeper incline than tRNATyrGUA to the optimal temperature (Fig. 3C). More important, wash temperatures of Ti +15°C or higher led to higher AMEs in tit1+ cells (Fig. 3C). This latter point demonstrated a most relevant benefit in the case of mt-tRNAs (see below and Discussion).
The data suggest that quantification of intermediate levels of modification can be assessed when positive controls are at ∼90% and negative controls (0%–10%), the latter based on TME deletion or persistent high efficiency knockdown, because tRNA half-lives can be ∼60 h (Abelson et al. 1974). Synthetic unmodified RNA could suffice as a null control although its probe annealing efficacy may differ from cellular tRNA that lack a modification due to a TME deficiency. It may be useful to note alternative methods for estimating TME deficiency, for example in patient cells compared to control cells. A prior report set the quantifications of BP/ACL for control cells to 100% and compared this to BP/ACL from patient cells (Yarham et al. 2014).
Mitochondrial tRNA–ACL probes may show wide range or acute sensitivity to wash temperatures
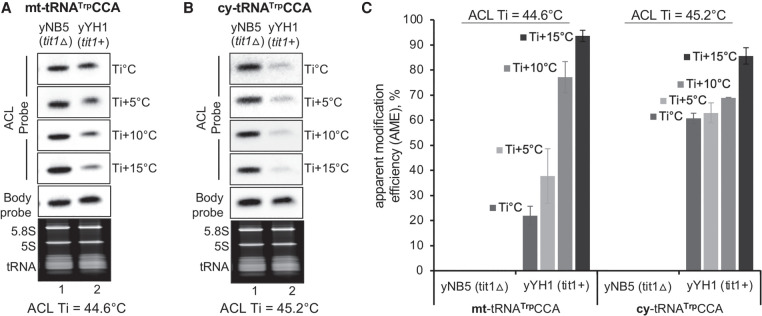
While a larger number of modifications are found on cy-tRNAs than on mt-tRNAs, some are unique to either, and a small subset is found on both (Suzuki et al. 2020). During our studies, we observed that some mt-tRNAs exhibit a steep incline in response to wash temperature which is observed in Figure 4 for S. pombe mt-tRNATrpCCA. We compared S. pombe mt-tRNATrpCCA (ACL Ti = 44.6°C) and cy-tRNATrpCCA (ACL Ti = 44.6°C), both of which carry i6A37 in yYH1, tit1+ and lack it in yNB5, tit1Δ (Fig. 4A,B). The mt-tRNA retained more ACL probe at lower temperature relative to the cy-tRNA, but it washed off as the temperature was increased (Fig. 4A,B). Quantification of data from duplicate experiments led to a greater range AME % for the mt-tRNA at varying wash temperatures as compared to the cy-tRNA (Fig. 4C).
FIGURE 4.
ACL probes to mt-tRNAs can exhibit wide range or acute sensitivity to wash temperatures. PHAM northern blot assay of total RNA from S. pombe yYH1 (tit1+) and yNB5 (tit1Δ). (A) Signals from the i6A37-sensitive anticodon loop (ACL) probe specific to S. pombe mt-tRNATrpCCA. (B) The same blot was examined for ACL hybridization to cy-tRNATrpCCA. The various wash temperatures are indicated to the right of the panels, where the Ti for each tRNA is provided below the panels. T stem–loop body probe (BP) results are also shown. (C) Quantification was done as for Figure 3, on triplicate results, N = 3 biological replicates.
We note that conditions previously reported for the PHAM assay led to calculated AME for S. pombe mt-tRNATrpCCA at only ∼25% using tit1Δ as negative control (0% mod) (Fig. 4A; Lamichhane et al. 2013a,b, 2016). Likewise, human mt-tRNASer (UCN) and mt-tRNATrp was calculated at 50% and 40%, respectively and others lower, while human cy-tRNASerUGA was at ∼95% (Lamichhane et al. 2013b). Notably, AME here went to ∼40% at Ti +5°C for S. pombe mt-tRNATrpCCA but to ∼95% at Ti +15°C (Fig. 4A,C). Thus, Figure 4A–C demonstrate relatively large differences between mt- and cy-tRNAs in response to wash conditions. We have also observed similar disparities for S. cerevisiae mt-tRNATyr and cy-tRNATyrGUA and for human mt-tRNAs Ser (UCN) and Tyr. In summary, the data indicate tRNA-specific sensitivities to wash temperatures for some mt-tRNAs and suggest careful monitoring using positive and negative controls as in Figures 3, 4 to obtain optimal performance. In a later section, we demonstrate PHAM performance to quantify intermediate levels of i6A37 modification.
Nearby modifications may falsely alter the apparent modification efficiency of a target nucleotide
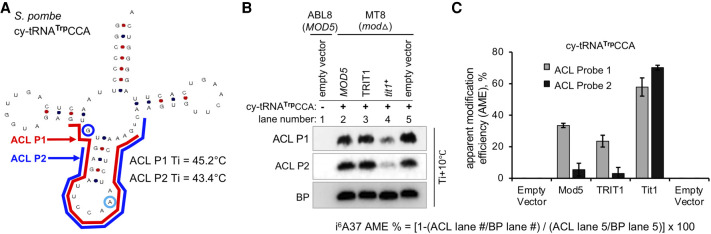
If the target modification-sensitive probe is complementary to a second (nearby) modification that interferes with W:C base-pairing, the second can offset the calculated AME. To examine the effect of a second modification, m2,2G26 on detection and quantification of the target modification of interest, i6A37, we made use of three IPTases with different activities for a tRNA substrate to reveal it to different extents. While S. pombe Tit1 has relative high activity for tRNATrpCCA, S. cerevisiae Mod5 and human TRIT1 are specifically less active for modification of this tRNA, which unlike all other sense-decoding IPTase substrates has pyrimidines in the 34 and 35 positions (Lamichhane et al. 2011; Khalique et al. 2020).
We designed two ACL probes to detect i6A37, one overlapping and the other nonoverlapping the G26 position of cy-tRNATrpCCA expressed from a plasmid in IPTase-deleted (mod5Δ) MT8 cells, also expressing Tit1, Mod5 or TRIT1 (Fig. 5). ACL probe 1 (P1) is complementary to positions 24–46 of tRNATrpCCA, including G26 which is modified to m2,2G26 in this tRNA species (Fig. 5A; Arimbasseri et al. 2015). ACL probe 2 (P2) is complementary to positions 28–50, avoiding G26 (Fig. 5A). The control body probe (BP) was to the T stem–loop region. As expected, the probes detected no tRNA in cells lacking the cy-tRNATrpCCA expression plasmid (Fig. 5B, lane 1). ACL probes P1 and P2 were comparably washed at Ti +10°C. High signal with both ACL probes in MOD5 and TRIT1 cells reflects low i6A37 levels on cy-tRNATrpCCA relative to tit1+ which as expected shows lower ACL signal (Fig. 5B; Khalique et al. 2020). Visual inspection reveals that the ACL signal ratios for MOD5:tit1+ and TRIT1:tit1+ are higher with P2 than with P1 (Fig. 5B, compare lanes 2 & 4, and 3 & 4). The P1 and P2 quantification data, each corrected by the corresponding BP data led to substantial differences in the calculated AMEs (Fig. 5C). This analysis provided evidence that a nearby second modification can interfere with the annealing of an overlapping complimentary probe and falsely alter the calculated AME of the target nucleotide. Use of IPTases with relatively low activity for cy-tRNATrpCCA suggest that such artifactual elevation of AME may be enhanced at low levels of modification of the principal target nucleotide. These data indicate that it is important to design probes to avoid overlap of a second modified nucleotide(s) with base-pair disrupting characteristics.
FIGURE 5.
Nearby modifications may falsely alter the apparent modification efficiency of a target nucleotide. (A) Clover leaf model structure of S. pombe cy-tRNATrpCCA showing locations of the ACL probe-1 (P1) in red, complementary to positions G26–G46, and ACL probe-2 (P2) in blue, complementary to positions G28–C48. The primary ACL modification-detection residue, A37, is indicated within a light blue circle, and the second modification site residue, G26 is indicated within a blue circle. The clover leaf model schematic was produced by tRNAscan-SE within GtRNAdb 2.0 (Chan and Lowe 2015). (B) PHAM assay with total RNA from S. cerevisiae ABL8 (MOD5+) and MT8 (Mod5Δ) cells transformed with S. pombe cy-tRNATrpCCA as one plasmid, together with an expression plasmid for S. cerevisiae Mod5, human TRIT1, S. pombe Tit1 or pREP82X empty vector as indicated above the lanes. Hybridization results with ACL probe-1 (P1, upper panel), ACL probe-2 (P2, middle), and body probe (BP, lower panel) are shown. (C) Quantification of AME was as for Figure 3 using the equation shown in the figure where # would correspond to the lane in B to match panel C.
Quantitative validation of PHAM using a yeast TME deletion control
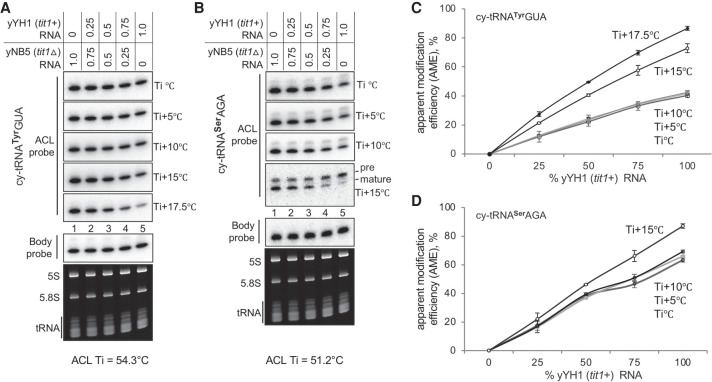
We did a mixing experiment, of S. pombe yYH1 (tit+) and yNB5 (tit1Δ RNAs, to examine quantitative performance of the PHAM northern blot method. We quantified and calculated i6A37 AME % for cy-tRNAs SerAGA and TyrGUA in duplicate, of mixes of total RNAs purified from tit+ and tit1Δ, containing 0%, 25%, 50%, 75%, and 100% of each as indicated above the lanes of Figure 6A and B. The blots were hybridized, washed, stripped, processed, and quantified according to the PHAM protocol method. Figure 6A and B shows one blot processed for both tRNAs. Figure 6A shows that as the fraction of modified tRNA increased (tit+, from left to right), annealing of the ACL probe to tRNATyrGUA decreased as the wash temperature increased, which resulted in higher AME approximating 90% with all tit+ RNA (Fig. 6C). The tRNASerAGA also showed decreased ACL probe annealing with increasing tit+ tRNA as wash temperature increased (Fig. 6B, mature), also resulting in increasing AME (Fig. 6D). This experiment demonstrated the quantitative nature of the PHAM method.
FIGURE 6.
Quantitative validation of the PHAM method using a yeast TME deletion control. (A,B) PHAM assay results for cy-tRNATyrGUA and cy-tRNASerAGA on the same mixture of total RNAs purified from the yYH1 (tit1+) and yNB5 (tit1Δ) strains as indicated above the lanes. The various wash temperatures are indicated to the right of the panels, where the Ti for each tRNA is provided below the panels. For B, we noted that as wash temperature increased the ratio of an upper band to the main, mature tRNA band increased. As S. pombe tRNASerAGA genes do not contain introns, the upper band may reflect one or more nuclear unmodified pre-tRNASerAGA isodecoders with a 3′ trailer. (C,D) Quantitation of AMEs was calculated from duplicate experiments at each different wash temperature using the following equation: AME % = [1 − (ACL tit1+/BP tit1+)/(ACL tit1Δ/BP tit1Δ)] × 100. N = 2, error bars represent the SD. For C and D, linear regression using the trendline function of Microsoft Excel revealed R2 values of >0.985 for the lines representing Ti +17.5°C and Ti +15°C (not shown).
For tRNASerAGA, the substantial reduction of ACL signal after the highest wash temperature revealed a reproducible appearance of an upper band, which unlike the lower band, did not decrease as the fraction of tit+ RNA increased (Fig. 6B, ACL probe, bottom panel, left to right, and data not shown). Inspection analysis of duplicate blots suggest the following plausible explanation. The upper band reflects a precursor-tRNASerAGA that has not acquired i6A37 in tit+ nor tit1Δ cells prior to its processing to the mature size tRNA. Accordingly, the unmodified upper band would exhibit efficient ACL probe annealing relative to mature tRNA-i6A37. As S. pombe tRNASerAGA genes do not contain introns, the upper band may reflect a nuclear pre-tRNASerAGA isodecoder with a unique 3′-trailer, for example, tRNA-Ser-AGA 1-3 (Chan and Lowe 2015).
DISCUSSION
PHAM is a relatively accessible and versatile method that has been used in several laboratories to examine i6A37, t6A37, m2,2G26, and m3C32 levels on numerous tRNAs in 14 publications. Yet, during the use of the method in our laboratory over the past several years, variability was observed that we suspect is common due to the nature of hybridization technology. It is typical for PHAM blot data to compare signals from the ACL (or other target region) probe and the control probe, by visual inspection, or accompanied by quantification and expressed as a ratio or modification index, sometimes with inexplicable variability for similar modification-sensitive probes for different tRNAs. Here we report that our attempts to understand and minimize the variability led to an overall improved method that more reliably produces optimal results. Most notably, we found that despite guidance by a formula to estimate DNA-oligo melting temperature (Tm) and hybridization incubation (Ti) (Leonard Davis 1986), when used on the heavily modified structured cellular tRNAs, each probe-tRNA pair should be tested empirically, subjected to multiple increasing wash temperature increments above the calculated Ti.
That the PHAM method is versatile refers to the multiple times a blot can be used to obtain data for several different tRNAs/modifications. In addition to the empiric conditions that must be determined for each tRNA-probe pair the first time characterized, other limitations apply. PHAM would not be expected to locate positions of previously unknown modifications. Also, although northern blotting normally uses agarose gels for large RNAs, PHAM uses polyacrylamide-urea gels which yield high resolution separation. Finally, PHAM alone does not identify a modification nor is a standard for absolute quantification; other gold standard methods are available (see Helm and Motorin 2017).
Improved PHAM conditions revealed new findings and clarified previous results
Although it might be suspected that a second modified base that is complementary to an ACL probe would interfere with annealing, it was not previously documented. Our data suggest that the commonly modified m2,2G26 found on many tRNAs (see Arimbasseri et al. 2015) can lead to substantial false increases in calculated AMEs when the target ACL nucleotide of interest is modified at relatively low levels. This was important to demonstrate because it suggests scrutiny when comparing samples, for example, when a TME is partially inactivated to different extents by different mutations. Thus, Figure 5 illustrated and emphasized the importance of careful probe design.
Another advance revealed by data in this report is that apparently similar tRNAs can respond very differently to near identical washings. A comparison case was S. pombe cy-tRNATrpCCA and mt-tRNATrpCCA whose sequences at 73 and 72 nt are also of the same clover-leaf architecture. Although these tRNAs have ACLs of identical primary sequence and the respective ACL probes differ by ≤1°C in Ti, their wash profiles differed significantly (Fig. 4).
The disparity for mt-tRNAs was important because it helped clarify previous unresolved results. Attempts to quantify i6A37 modification of S. pombe and human mt-tRNAs by PHAM led to calculated levels of ∼25% and ∼50% washing at Ti for mt-tRNAs, whereas cy-tRNAs showed substantially higher levels (Lamichhane et al. 2013a,b, 2016), consistent with Figure 4. More specifically, human mt-tRNASer (UCN), and mt-tRNATrp were estimated at 50% and 40%, whereas mt-tRNACys was ≤10% by PHAM, the latter much lower than noted at 99% for mt-tRNACys determined by gold standard methods (Suzuki et al. 2020). Although we have not systematically reexamined/quantified human mt-tRNAs, it was reassuring to find that the calculated AME for S. pombe mt-tRNATrp was increased to 95% after Ti +15°C (Fig. 4C). We suspect that the disparity observed between responses of mt-and cy-tRNAs to ACL probes of similar length and Ti, reflect overall general differences in flexibility/rigidity of their anticodon stem–loop structures and consequent ability to stably pair with DNA-oligo (Vare et al. 2017).
A third advance is that the PHAM assay was subjected to a calibration analysis of its ability to estimate AME for tRNA fractional i6A37 content by examining RNAs that contained different relative amounts of modified and unmodified tRNAs from yeast TME replete and deleted strains (Fig. 6). This provides confidence in calculations of AME including when the content of tRNA from the modified source was as low as 25% (Fig. 6). This suggests that it may be possible to monitor significant loss of other modifications, for example, m1A at specific mRNA sites using PHAM, although this remains to be determined.
Advancing molecular studies of TMEs and effects on substrate-tRNA modifications
Deficiency in a TME can lead to differential effects on distinct tRNA substrates. The PHAM assay can be considered as an alternative to LC-MS/MS and other methods (see Introduction) to detect i6A37, m2,2G26, m3C32, and t6A37. It has been used to document tRNA hypomodification resulting from newly described TME deficiencies resulting in human disease syndromes due to mutations in TRIT1 (Yarham et al. 2014), TRMT1 (Dewe et al. 2017), DALRD3(Lentini et al. 2020), and OSGEP (KAE1/TCS3) (Edvardson et al. 2017), respectively.
OSGEP is one of five subunits of the human KEOPS (kinase, endopeptidase, and other proteins of small size) enzyme complex that catalyzes t6A37 synthesis on the multitude of cy-tRNAs that decode ANN codons (Edvardson et al. 2017). Not too long after the initial report of neurodegeneration and evidence of kidney pathophysiology associated with kae1-mutations and hypomodification of t6A37, it was reported that mutations in genes encoding all four of the other KEOPS subunits lead to Galloway-Mowat syndrome (Braun et al. 2017; Arrondel et al. 2019). Yeast KEOPS is comprised of four subunits (apparently lacking a Gon7 homolog) all of which contribute to substrate tRNA binding, and mutations in each of which cause t6A37 hypomodification of multiple tRNAs as monitored by PHAM (Beenstock et al. 2020).
A pathogenic mutation in a mt-tRNA sequence can lead to hypomodification at another position, for example if it alters a TME substrate recognition site, as was shown using PHAM for the A38G mutation in mt-tRNASer (UCN) resulting in tRNA-i6A37 hypomodification (Yarham et al. 2014). It seems reasonable to expect that the PHAM method can be extended to molecular studies and other pathogenic mutations, including those involved in tRNA modifications that disrupt base-pairing, such as m1A, m1G, and yW.
MATERIALS AND METHODS
Protocol: total RNA isolation
Use a method applicable to the organism of choice. Importantly, be aware that some RNA purification kits, methods and protocols are mistakenly misnamed useful for “Total RNA,” which refers to inclusion of rRNA versus purification of poly(A) and are oblivious to–and designed to–exclude tRNAs and smaller species by precipitation with LiCl or sizing columns. A broad-use method for extraction and purification from tissues or cells is one based on guanidinium thiocyanate-acidic phenol (MacDonald et al. 1987; Maraia 1991), which is similar to TRIzol/TriPure (Invitrogen/Sigma)-based methods that use, for example, ammonium acetate followed by alcohol precipitation to not exclude tRNAs (see below).
Total RNA isolation from yeast
Here we used the acidic hot phenol method to isolate total RNA from S. cerevisiae and S. pombe. Transfer an overnight yeast culture to a 50 mL conical flask containing 10 mL of media at OD600 ∼ 0.1 and grow to mid log phase (OD600 ∼ 0.55) at 30°C for S. cerevisiae, 32°C for S. pombe.
Transfer the culture to a 50 mL centrifuge tube and spin at 3000 rpm for 3 min at 4°C. Discard the supernatant and resuspend pellet in 1 mL ice cold ddH2O. Transfer the culture to a 1.5 mL microcentrifuge tube, and pellet by centrifugation at 8000 rpm for 1 min at 4°C. Discard the supernatant. Resuspend the cell pellet in 400 µL TES solution (10 mM Tris-HCl pH 7.5, 10 mM EDTA, 0.5% SDS) followed by addition of 400 µL acid phenol (Sigma P4682). Close the cap tightly. Vortex vigorously for 30 sec and incubate in a shaker for 1 h at 65°C; alternatively, frequent brief vortexing can also be done.
Incubate on ice for 5 min and spin down at 12K rpm (benchtop centrifuge, e.g., 12,000–16,000g) for 5 min at 4°C. Carefully transfer the aqueous phase (top) to a clean, RNase-free 1.5 mL microcentrifuge tube (use extreme care to not approach, penetrate, disturb, or aspirate any of the white interphase material). Add 400 µL acid phenol (Sigma P4682) to the separated aqueous phase, vortex vigorously, incubate on ice for 5 min and centrifuge at 12K rpm for 5 min at 4°C. Again, transfer the aqueous phase to a clean, RNase-free 1.5 mL microcentrifuge tube, add 400 µL chloroform, vortex vigorously and repeat the centrifugation step. Transfer the aqueous phase to a clean, RNase-free 1.5 mL microcentrifuge tube, add 40 µL of 3M sodium acetate pH 5.3, mix well and add 1-mL ice cold 100% ethanol, mix again and incubate for ≥1 h, or overnight at −20°C. Centrifuge at 12K rpm for 5 min at 4°C. Visualize the pellet and wash it with 1 mL of ice cold 70% ethanol, vortex and centrifuge briefly at max speed. Visualize the pellet, sharply invert the tube to decant the supernatant onto paper towel. Air-dry the pellet for 10 min at room temperature. Be careful to not over-dry, and do not use a vacuum dryer; if the pellet is too dry the RNA will not dissolve well.
Total RNA isolation from mammalian cells (see total RNA isolation above)
Standard TRIzol/TriPure (Invitrogen/Sigma)-based methods can be used. This protocol is for HEK293 cells cultured in media of choice. Combining two wells of a six-well plate will yield more than enough RNA to perform the PHAM assay. Harvest cells at 70%–80% confluency. Wash twice with 2 mL PBS per well followed by addition of 500 µL TriPure per well. Transfer all solution, 1 mL, to a 1.5 mL centrifuge tube. Add 200 µL chloroform (Sigma 2432) and shake vigorously for 15 sec, incubate at room temperature for 2 min. Centrifuge at 12,000g for 15 min at 4°C.
Carefully transfer the (top) aqueous phase only to an RNase-free 1.5 mL tube (use extreme care to not approach, penetrate, disturb, or aspirate any of the white interphase material). Add 450 µL isopropanol to the isolated aqueous phase. Mix thoroughly by inversion several times, with intermittent vortexing. Incubate at room temperature for 5 min and centrifuge at 12,000g, 4°C for 10 min. Visualize the pellet, then wash it at least three times with 1 mL 75% ethanol, vortexing each time shortly and centrifuging for 2 min at max speed between each wash. Visualize the pellet, and sharply invert the tube to decant the supernatant onto paper towel. Air-dry pellet for 10 min at room temperature. Be careful to not over-dry; do not use a vacuum dryer (if the pellet is too dry the RNA will not dissolve well).
At this point, it is important to minimize the time that purified RNA will be in aqueous solution susceptible to hydrolysis/degradation. Resuspend the pellet in 30 µL RNase-free ddH2O. Keep at room temperature for 5 min, vortex once or twice very shortly and spin down; make sure the pellet is completely dissolved. Remove an aliquot to be used to quantify the RNA concentration and immediately put the tube with the remainder of the RNA into a bucket of dry ice to rapid-freeze, then transfer to −80°C.
Protocol: northern blot prep, part 1; urea-denaturing polyacrylamide gel electrophoresis of RNA
We use precast 10% TBE-Urea gels (Thermo Fisher, EC68752BOX) run in XCell SureLock Mini-Cell (EI0001, Thermo Fisher); however, other precast or homemade TBE-Urea gels and running systems can be used. Slowly and carefully remove the comb from the gel, noting any misshaped wells due to difficult comb removal. Consider use of a soft, thin pipette tip with 1× TBE to expunge loose polyacrylamide from the bottom of the wells and to straighten walls of the wells. Pre-run in 1× TBE running buffer at 180V for 30 min. During the pre-run, the 8M urea denaturant will diffuse from the gel into the wells; it is not necessary to remove this denaturant; one can carefully load the sample under it in the lowest part of the well. The sample will remain under the urea layer (do not puncture or misshape the well as it will misshape the bands).
RNA sample preparation, loading buffer, and gel loading
Add an equal volume of prewarmed 2× RNA RNase-free loading dye (NEB B0363S, 95% formamide, 0.02% SDS, 0.02% bromophenol blue, 0.01% Xylene cyanole, 1 mM EDTA) to 5 or 10 µg of each total RNA sample and heat denature at 70°C for 3 min. Load directly from the heat block to prevent renaturation by placing at the bottom of the well.
Load the gel by slow pipetting samples at the bottom of the wells. Run at 180V until the xylene cyanole (XC, the upper dye) reaches near but not run-off the bottom of the gel (∼1.5 h). In 10% TBE-Urea PAGE, XC has mobility corresponding to a 55-nt single stranded DNA.
Gel dismantling, RNA staining with ethidium bromide and photo documentation
Put on clean gloves and wash in ddH20. Whatever contacts the gel surface from this point onward may transfer to the blot membrane may affect the hybridization results. Be prepared to note and keep track of the position of lane 1, so mark it immediately after dismantling the gel from the electrophoresis apparatus. With a clean rinsed spatula/knife, slice off a substantial triangular lower gel segment under lane 1 as an important orientation marker to which a similar triangle will be cut from the nylon blot membrane of matching dimensions. Shake the gel in a substantial volume of ddH2O containing ≤0.01 µg/mL EtBr for 5 min, followed by 3 × 10 min washes with ddH2O. Take a high quality full-gel picture on a UV illuminator using minimal exposure time and intensity to avoid UV damage to the RNA. Keep the gel wet until ready to transfer.
Protocol: northern prep part 2; transfer RNA to membrane, mark-up, UV-crosslink, and vacuum dry
The key to obtaining clean results is preparation of a clean blot, free of unwanted micro-debris and other 32P-ATP-interacting material that may bond to the membrane as indelible sources of background noise for every probing. Good blots crosslinked and baked can be useful for more than 10 tRNA ACL-BP probe cycles.
Transfer RNA from the gel using a semidry or wet transfer method to a positive charge nylon membrane (Gene Screen Plus, PerkinElmer) that has been pre-cut fit with a triangle to match that under lane 1 of the gel. Rinse the gel and set it on the transfer stage using plenty of liquid to prevent it from getting stretched and deformed and to ease away wrinkles. Rinse the gel surface before placing a clean rinsed membrane on it. We find it useful to stream transfer the buffer from a pipette over the gel surface to clear away micro-debris, including tiny polyacrylamide particles, before putting the membrane on it. Ensure that the membrane and gel are correctly situated/oriented according to the proper function of the apparatus. Add a pre-wet filter paper and remove air bubbles by rolling a plastic pipette over the assembly. Add some buffer to make sure the conduction, and thus the transfer, will be optimal. For semidry blotting, allow the liquid between membrane and gel to absorb away.
After the transfer is complete, dissemble the blot, keeping track of which side of the blot was in contact with the gel/RNA. Place the blot on a clean damp Whatman 3MM paper with its RNA surface facing up, and write on it with a sharp point marker under lane 1. Clean both sides of the membrane surface, including of any polyacrylamide specs or micro-debris that may have attached during transfer, prior to UV crosslinking. Quickly rinse the membrane briefly in 2× SSC, allow it to drip damp, then UV crosslink at 254 nm wavelength with the RNA side facing the UV source (in a Stratalinker UV-1800, Stratagene) on auto-crosslink mode. The membrane should be damp, not dry, during crosslinking. It is important at this moment to note and document (mark it) which side of the membrane has the RNA, because it will have significant effects on the efficiencies of (i) UV crosslinking, (ii) hybridization, (iii) exposure to phosphorimager and/or X-ray film, thereby affecting the resolution of the bands observed, reproducibility of hybridization and quantification of one tRNA to the next, and therefore the overall success of the experiment. If you have a handheld UV lamp, you should verify that one side has RNA and not the other (it should be apparent); this may also reveal irregularities (bubbles) in the transfer. After letting the membrane dry, label it on the side that contains the transferred RNA, along the bottom edge with your initials, the date and other unique identifying information, making an indelible impression with a marker or ballpoint pen. Next, thoroughly dry the membrane, preferably by baking at 80°C for 2 h under vacuum.
Protocol: blot prehybridization and hybridization; setup
Except for the probe in the latter, prehybridization and hybridization buffers are the same: 2× SSC +1× Denhardt's solution +0.5% SDS +100 µg/mL yeast total RNA (Invitrogen AM7118) from Torula, related to S. cerevisiae. Note: do not use this RNA for analysis of S. cerevisiae RNA, it will decrease detection; use sheared Salmon sperm DNA (Invitrogen AM9680).
We use dry ovens of two types for probe hybridization, with rotating glass cylindrical sealed tubes (Techne Hybrigene), or with a flat rocking platform that holds a container of choice. The advantage of the rocking platform is the blot(s) can be more easily placed into and removed from the container than from the cylinders. This also helps avoid air bubbles and other irregularities/difficulties associated with cylinders such as placement of blots. Also, membranes tend not to “free float” in the cylinders but rather stick to the glass, sometimes trapping air bubbles. It is therefore very important to apply the membrane with the RNA side surface facing the solution. If other options are unavailable, a water bath can be used for hybridization; it is best if the blot is in a watertight pouch using a plastic bag sealer and submerged with shaking at the Ti °C.
Rehydrate the blot with 2× SSC buffer and transfer it into the hybridization tube/container with the RNA side facing the solution. Add 14 mL prehybridization buffer and incubate at the DNA-oligo probe-specified Ti for ≥1–2 h with optimal access of all parts of the blot to the solution.
Blot hybridization
There is no need to discard the prehybridization buffer. The 32P radioactive labeled DNA-oligo probe (below) that has been heat denatured for 10 min at 65°C in 1 mL of hybridization buffer is added to the blot-incubation 14 mL solution and further incubated for 8–16 h at Ti °C. When labeled to high specific activity (below), DNA-oligo probes are used at 2 million cpm/mL of hybridization solution. For 15 mL hybridization, incubate a 30 million cpm 32P-DNA probe in 0.5–1 mL hybridization solution for 5 min at 80°C just before adding this to the preheated hybridization solution already on the blot. Add the 32P-DNA probe solution to the hybridization liquid while mixing rather than directly onto the membrane.
Hybridization uses a large excess of probe and time, allowing equilibrium annealing important for reproducible results and quantification. Caution: Alteration of the recommended conditions, including reduced probe amounts (below) and/or use to quantify high abundance, for example, rRNAs, can fail due to probe exhaustion/consumption and nonequilibrium conditions. For quantification of high abundance rRNA, addition of a large excess of unlabeled DNA-oligo identical to the labeled probe is suggested.
Initial washing at the Ti
The next day, preheat 2× SSC, 0.1% SDS and hold it at the Ti. Carefully decant all the 32P hybridization solution to an appropriately labeled holding container behind a plexiglass shield for possible reuse. Completely drain all remaining liquid from the hybridization vessel to a radiation waste container. Using a flat head, blunt end Millipore filter forceps, transfer the blot to a clean tray with the RNA side up. It is useful to monitor washing of the probe; examine the blot with a Geiger counter, noting approximate cpm on the 0.1×, 1×, or 10× meter, and the distance of the recorder above the membrane and in the vicinity of expected bands. Be sure that there is no contaminating source of radioactivity nearby, including under the tray. Add 150 mL of room temperature wash buffer (2× SSC, 0.1% SDS). Rinse the hybridization vessel with wash buffer and decant in the radioactive waste. Add wash buffer to the incubation vessel and place it back in the hybridization oven to shake/rotate to clean the vessel further and keep it at Ti. Decant the wash buffer on the blot in the radioactive waste and wash the blot two more times for 5 min at room temperature with 150 mL wash buffer, and perform a final wash of the blot with 20 mL of the preheated wash buffer in the hybridization vessel at Ti for 30 min. Repeat examination of the blot, RNA side up with the Geiger counter, and note the cpm, which should have decreased substantially.
Seal the blot in plastic, wrinkle-free, and with minimal liquid and air bubbles, so the RNA side is visible and will be in close/direct contact with a phosphorimager screen or X-ray film. If necessary, place filler cardboard behind the blot to press it closely against the screen for high resolution bands.
It is wise to obtain multiple exposures for long and short times. When not in use, blots in sealed/wrapped plastic should be put in a storage folder for protection from light, UV, and scratches/damage to their surface, in the refrigerator, preferably in a hard notebook, for reuse (subsequent probing).
Rewashing at temperatures above Ti
It is recommended that blots be rewashed at multiple increments of 5°C and/or 2.5°C above the Ti with autoradiography after each such wash is complete, until the majority of the signal is removed from the sample that represents the target modification-containing sample. Wet the blot with wash buffer at room temperature, then discard. Pour 20 mL prewarmed wash buffer and incubate for 30 min at the new wash temperature. In accordance with calculations for DNA-oligo melting (Leonard Davis 1986), it should be expected that exposure times be lengthened after increases in wash temperature in order to achieve sufficient signal above background useful for quantitative purposes. Use the Geiger counter method to follow the progress of each subsequent wash.
Stripping blots for reuse
A general principle of strategy for the order of which probes to use before others is to first examine tRNAs with probes expected to give the weakest signals, that is, ACL probes followed by sequential stringent washing against multiple different tRNAs, less abundant before more abundant, before hybridizing with body probes. This is because stronger signals due to more abundant RNA are generally more refractory to stripping. The most abundant RNAs including loading controls will last as the most enduring, as the blot is sequentially stripped of its RNA and “wears out” due to repeated reuse.
Protect the blot from unnecessarily harsh stripping. Use the Geiger counter as a guide. Most conditions will require stripping of a previously annealed probe, especially if it will block analysis of the second probe. If the previous probe is to a different RNA and is no longer visible, it does not necessarily need to be stripped away. However, a preexposure of the blot before a reprobing should always be done to ensure that no bands will be present that may confound the new probing results. Also, if a blot with efficiently annealed probe inadvertently dries, it may become “fixed” to the tRNA and block subsequent reprobings.
To strip, pour preheated stripping buffer (0.1× SSC + 0.1% SDS), at 85°C–90°C, onto the blot with the RNA side up, and shake with incubation for 10 min. Confirm removal of the 32P labeled DNA probe using a phosphorimager for at least several hours. Save the image, quantitate the signals, and calculate the signal per pixel (“counts”) per minute of exposure time, which may be important for later analysis.
Protocol: DNA-oligo probe 5′-end labeling and purification
In an Eppendorf tube, mix: 10 µL desired DNA-oligo (1 pmol/µL) (more than 10 pmol deceases specific activity of the probe), 3.3 µL 10× PNK buffer (final reaction volume is 33 µL), 1 of T4 PNK (NEB), 5 µL 32P-γATP (allow time to defrost, PerkinElmer, BLU002A001MC, 3000 Ci/mmol, at 3.3 µM); add H2O to final volume of no more than 33 µL (final ATP concentration should be ≥1 µM). Mix by pipetting up and down. Incubate at 37°C for 1–1.5 h. Stop the reaction by adding 4 µL of 0.5 M EDTA (pH 8.0).
Carefully apply the total mixture onto the center of a freshly packed, that is, centrifuged Sephadex MicroSpin G-25 column (GE Healthcare) that is in a 1.5 mL Eppendorf tube. Spin for 2 min at 750g in a microcentrifuge. The flow-through into the Eppendorf has the labeled 32P-probe; the column retains the unincorporated 32P-γATP. Carefully measure and record with a Geiger counter the relative amounts of 32P-γATP in each: the Eppendorf and the resin/column. Record the volume of the probe liquid fraction recovered in microliters for future reference and calculation of SA (specific activity).
If the flow-through in the Eppendorf tube has ≥70% of the total 32P-cpm, the separation of the labeled DNA-oligo from free 32P-γATP was likely inefficient, that is, it contains free 32P-γATP. Too much free 32P-γATP can be a source of dirty hybridizations and high blot background that cannot be washed away, obscuring tRNA bands and making quantification difficult or impossible. Therefore, repeat the purification by applying the flow-through material onto one or two freshly prepared Sephadex MicroSpin G-25 columns (overloading can lead to poor column performance).
Determine specific activity (SA) of 32P-labeled probe
Using appropriate scintillation fluid, measure the cpm of the probe in the final flow-through using a scintillation counter. To do this, also make duplicate 1/10th and 1/50th dilutions of the probe, measure and convert to cpm/µL probe. This is used to calculate SA.
Calculate SA based on the starting amount of oligo, that is, assume 100% recovery, expressed as cpm/µg probe: SA = cpm/µg probe for purposes below. Optimally, SA can be >109 cpm/ug oligo DNA of ∼24 nt, or106 cpm/pmole.
Guidelines for DNA-oligo probe design
Modification-sensitive probes are designed according to similar principles; a near central position in the DNA-oligo targets the modified nucleotide in the tRNA. DNA-oligo lengths are typically 20–25 nt depending on G + C-content, which determines Tm (melting temperature), based on the formula: Tm = [16.6 log(M) + 0.41(Pgc) + 81.5 − (675/L) − 0.65] (Leonard Davis 1986). Here, M = molar concentration of Na+, to a maximum of 0.5 (2× SSC = 0.39 M Na+; saline-sodium citrate buffer = 0.3 M NaCl, 30 mM Na3C6H5O7), Pgc = %G + C content, and L = nucleotide length of the DNA-oligo (Leonard Davis 1986). The Ti is 15°C lower than the Tm.
As discussed above, avoid covering a nearby nucleotide that is part of a modification circuit and/or whose modification alters base-pairing. Use similar guidance to design the body probe (BP) used as a control. Distribution, frequencies and other information about tRNA modifications can be found at http://genesilico.pl/trnamodviz (Machnicka et al. 2014).
An Excel file in the Supplemental Material (Supplemental Table S1) will automatically calculate the Tm, Ti, and other information after the 5′-to-3′ sequence of the DNA-oligo probe (complementary to the tRNA) is entered into the appropriate space in column C. The file can also serve as an inventory of probe sequences.
Quantification of tRNA-detected bands by DNA-oligo probes for calculation of AME
Scan blots using a phosphorimager (GE-Typhoon FLA 9500, Storm, FujiFilm or similar equipment with a large dynamic range). Quantify bands using Multi Gauge V3.0 (FujiFilm) or other appropriate (ImageQuant) software.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Division of Intramural Research, Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, Maryland, USA. R.M., S.M., and A.K. would like to thank M. Bayfield for discussions, and to acknowledge the NICHD Scientific Director's Award.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.078929.121.
Freely available online through the RNA Open Access option.
MEET THE FIRST AUTHOR
Abdul Khalique.

Meet the First Author(s) is a new editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. Abdul Khalique is the first author of this paper, “A versatile tRNA modification-sensitive northern blot method with enhanced performance.” Abdul is a visiting postdoctoral fellow in the section of Molecular and Cellular Biology at the Eunice Kennedy Shriver National Institute of Child Health and Human Development, NIH, studying the molecular genetics of tRNA modification and processing in humans and yeast.
What are the major results described in your paper and how do they impact this branch of the field?
Three major points are described in the paper. (i) Different tRNAs exhibit different response profiles to anticodon loop (ACL) probe washing at temperatures above the incubation temperature (Ti). This leads to a protocol in which the wash temperature must be empirically determined for each ACL probe the first time it is used/tested for optimal performance and quantification. (ii) In general, mitochondrial tRNAs exhibit greater sensitivity to increasing ACL probe wash temperatures than cytoplasmic tRNAs, and (iii) nearby modifications lead to false elevation of the apparent modification efficiency (AME) of the target nucleotide. The PHAM method has been used in many laboratories to examine and quantify the t6A37, i6A37, m2,2G26, and m3C32 modification levels on multiple tRNAs. Improved sensitivity would help to accurately quantify the unmodified versus modified tRNA and better understand the gene expression profile.
What led you to study RNA or this aspect of RNA science?
RNA is the central part of the central dogma that plays a very important role in gene expression regulation. In humans, ≤2% of genes are coded into protein; however, many genes are transcribed into RNA but function as regulatory molecules such as noncoding small RNAs. The RNA field is largely unexplored due to its complex nature and difficulty in handling. More specifically, tRNAs are highly modified, but the specific functions of this complex modification mostly remain unknown. This led me to study the complex nature of the tRNA and its role in gene expression and develop a more accessible and easy method to study tRNA modification.
During the course of these experiments, were there any surprising results or particular difficulties that altered your thinking and subsequent focus?
Yes. While I was optimizing the PHAM method with sequential washing of the blots at temperatures above the incubation temperature (Ti), I observed that the ACL probe that is targeted to i6A37 and complementary to a second nearby modification, for example, m2,2G26, alters the calculated apparent modification efficiency (AME); and that led to a substantial false increase in AME. So, it is important to design such probes to avoid overlap of a second modified nucleotide(s) with base-pair disrupting characteristics.
What are some of the landmark moments that provoked your interest in science or your development as a scientist?
My father is a schoolteacher. When I was young, around 10 years old, I used to go to his school on special occasions and roam around the chemistry or biology labs. I remember one day some of the senior graduate students were doing experiments in the laboratory, where they were mixing two liquids in flasks to produce a pink color; now I can recall it may have been an acid-base titration. It was very fascinating and impressive, inducing my interest toward science.
If you were able to give one piece of advice to your younger self, what would that be?
There should be balance in professional and personal life. Choose what you like the most and what you wanted to become if you have the opportunities, but be aware that sometimes opportunities are not exactly the same as you would have liked, so be patient, focus and do the hard work, and one day you will get there.
Are there specific individuals or groups who have influenced your philosophy or approach to science?
Yes, two persons have influenced me in my approach to science. One of them is my mentor when I was doing graduate work and the second one is my post-doc PI.
What are your subsequent near- or long-term career plans?
My long-term career plan is to become an independent research scientist at a research institute or university.
REFERENCES
- Abbott JA, Francklyn CS, Robey-Bond SM. 2014. Transfer RNA and human disease. Front Genet 5: 158. 10.3389/fgene.2014.00158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abelson H, Johnson L, Penman S, Green H. 1974. Changes in RNA in relation to growth of the fibroblast: II. The lifetime of mRNA, rRNA, and tRNA in resting and growing cells. Cell 1: 161–165. 10.1016/0092-8674(74)90107-X [DOI] [PubMed] [Google Scholar]
- Agris PF, Eruysal ER, Narendran A, Väre VYP, Vangaveti S, Ranganathan SV. 2018. Celebrating wobble decoding: half a century and still much is new. RNA Biol 15: 537–553. 10.1080/15476286.2017.1356562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimbasseri AG, Blewett NH, Iben JR, Lamichhane TN, Cherkasova V, Hafner M, Maraia RJ. 2015. RNA polymerase III output is functionally linked to tRNA dimethyl-G26 modification. PLoS Genet 11: e1005671. 10.1371/journal.pgen.1005671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arimbasseri AG, Iben J, Wei FY, Rijal K, Tomizawa K, Hafner M, Maraia RJ. 2016. Evolving specificity of tRNA 3-methyl-cytidine-32 (m3C32) modification: a subset of tRNAsSer requires N6-isopentenylation of A37. RNA 22: 1400–1410. 10.1261/rna.056259.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arrondel C, Missoury S, Snoek R, Patat J, Menara G, Collinet B, Liger D, Durand D, Gribouval O, Boyer O, et al. 2019. Defects in t6A tRNA modification due to GON7 and YRDC mutations lead to Galloway-Mowat syndrome. Nat Commun 10: 3967. 10.1038/s41467-019-11951-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bacusmo JM, Orsini SS, Hu J, DeMott M, Thiaville PC, Elfarash A, Paulines MJ, Rojas-Benitez D, Meineke B, Deutsch C, et al. 2018. The t6A modification acts as a positive determinant for the anticodon nuclease PrrC, and is distinctively nonessential in Streptococcus mutans. RNA Biol 15: 508–517. 10.1080/15476286.2017.1353861 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barraud P, Tisné C. 2019. To be or not to be modified: miscellaneous aspects influencing nucleotide modifications in tRNAs. IUBMB Life 71: 1126–1140. 10.1002/iub.2041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bednarova A, Hanna M, Durham I, VanCleave T, England A, Chaudhuri A, Krishnan N. 2017. Lost in translation: defects in transfer RNA modifications and neurological disorders. Front Mol Neurosci 10: 135. 10.3389/fnmol.2017.00135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenstock J, Ona SM, Porat J, Orlicky S, Wan LCK, Ceccarelli DF, Maisonneuve P, Szilard RK, Yin Z, Setiaputra D, et al. 2020. A substrate binding model for the KEOPS tRNA modifying complex. Nat Commun 11: 6233. 10.1038/s41467-020-19990-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens A, Rodschinka G, Nedialkova DD. 2021. High-resolution quantitative profiling of tRNA abundance and modification status in eukaryotes by mim-tRNAseq. Mol Cell 81: 1802–1815.e1807. 10.1016/j.molcel.2021.01.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boccaletto P, Machnicka MA, Purta E, Piatkowski P, Baginski B, Wirecki TK, de Crecy-Lagard V, Ross R, Limbach PA, Kotter A, et al. 2018. MODOMICS: a database of RNA modification pathways. 2017 update. Nucleic Acids Res 46: D303–D307. 10.1093/nar/gkx1030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun DA, Rao J, Mollet G, Schapiro D, Daugeron MC, Tan W, Gribouval O, Boyer O, Revy P, Jobst-Schwan T, et al. 2017. Mutations in KEOPS-complex genes cause nephrotic syndrome with primary microcephaly. Nat Genet 49: 1529–1538. 10.1038/ng.3933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM. 2015. GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res 44: D184–D189. 10.1093/nar/gkv1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chujo T, Tomizawa K. 2021. Human transfer RNA modopathies: diseases caused by aberrations in transfer RNA modifications. FEBS J 288: 7096–7122. 10.1111/febs.15736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cirzi C, Tuorto F. 2021. Analysis of queuosine tRNA modification using APB northern blot assay. Methods Mol Biol 2298: 217–230. 10.1007/978-1-0716-1374-0_14 [DOI] [PubMed] [Google Scholar]
- Clark WC, Evans ME, Dominissini D, Zheng G, Pan T. 2016. tRNA base methylation identification and quantification via high-throughput sequencing. RNA 22: 1771–1784. 10.1261/rna.056531.116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cozen AE, Quartley E, Holmes AD, Hrabeta-Robinson E, Phizicky EM, Lowe TM. 2015. ARM-seq: AlkB-facilitated RNA methylation sequencing reveals a complex landscape of modified tRNA fragments. Nat Methods 12: 879–884. 10.1038/nmeth.3508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewe JM, Fuller BL, Lentini JM, Kellner SM, Fu D. 2017. TRMT1-catalyzed tRNA modifications are required for redox homeostasis to ensure proper cellular proliferation and oxidative stress survival. Mol Cell Biol 37: e00214-17. 10.1128/MCB.00214-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edvardson S, Prunetti L, Arraf A, Haas D, Bacusmo JM, Hu JF, Ta-Shma A, Dedon PC, de Crecy-Lagard V, Elpeleg O. 2017. tRNA N6-adenosine threonylcarbamoyltransferase defect due to KAE1/TCS3 (OSGEP) mutation manifest by neurodegeneration and renal tubulopathy. Eur J Hum Genet 25: 545–551. 10.1038/ejhg.2017.30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogakos T, Brown M, Garzia A, Meyer C, Hafner M, Tuschl T. 2017. Characterizing expression and processing of precursor and mature human tRNAs by hydro-tRNAseq and PAR-CLIP. Cell Rep 20: 1463–1475. 10.1016/j.celrep.2017.07.029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo LT, Adams RL, Wan H, Huston NC, Potapova O, Olson S, Gallardo CM, Graveley BR, Torbett BE, Pyle AM. 2020. Sequencing and structure probing of long RNAs using MarathonRT: a next-generation reverse transcriptase. J Mol Biol 432: 3338–3352. 10.1016/j.jmb.2020.03.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy MP, Phizicky EM. 2014. Conservation of an intricate circuit for crucial modifications of the tRNAPhe anticodon loop in eukaryotes. RNA 21: 61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guy MP, Shaw M, Weiner CL, Hobson L, Stark Z, Rose K, Kalscheuer VM, Gecz J, Phizicky EM. 2015. Defects in tRNA anticodon loop 2′-O-methylation are implicated in nonsyndromic X-linked intellectual disability due to mutations in FTSJ1. Hum Mutat 36: 1176–1187. 10.1002/humu.22897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Phizicky EM. 2018. A rationale for tRNA modification circuits in the anticodon loop. RNA 24: 1277–1284. 10.1261/rna.067736.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han L, Guy MP, Kon Y, Phizicky EM. 2018. Lack of 2′-O-methylation in the tRNA anticodon loop of two phylogenetically distant yeast species activates the general amino acid control pathway. PLoS Genet 14: e1007288. 10.1371/journal.pgen.1007288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helm M, Motorin Y. 2017. Detecting RNA modifications in the epitranscriptome: predict and validate. Nat Rev Genet 18: 275–291. 10.1038/nrg.2016.169 [DOI] [PubMed] [Google Scholar]
- Hiley SL, Jackman J, Babak T, Trochesset M, Morris QD, Phizicky E, Hughes TR. 2005. Detection and discovery of RNA modifications using microarrays. Nucleic Acids Res 33: e2. 10.1093/nar/gni002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hrabeta-Robinson E, Marcus E, Cozen AE, Phizicky EM, Lowe TM. 2017. High-throughput small RNA sequencing enhanced by AlkB-facilitated RNA de-methylation (ARM-Seq). Methods Mol Biol 1562: 231–243. 10.1007/978-1-4939-6807-7_15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igloi GL, Kössel H. 1985. Affinity electrophoresis for monitoring terminal phosphorylation and the presence of queuosine in RNA. Application of polyacrylamide containing a covalently bound boronic acid. Nucleic Acids Res 13: 6881–6898. 10.1093/nar/13.19.6881 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler AC, Kulkarni SS, Paulines MJ, Rubio MAT, Limbach PA, Paris Z, Alfonzo JD. 2018. Retrograde nuclear transport from the cytoplasm is required for tRNATyr maturation in T. brucei. RNA Biol 15: 528–536. 10.1080/15476286.2017.1377878 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalique A, Mattijssen S, Haddad AF, Chaudhry S, Maraia RJ. 2020. Targeting mitochondrial and cytosolic substrates of TRIT1 isopentenyltransferase: specificity determinants and tRNA-i6A37 profiles. PLoS Genet 16: e1008330. 10.1371/journal.pgen.1008330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krutyholowa R, Zakrzewski K, Glatt S. 2019. Charging the code: tRNA modification complexes. Curr Opin Struct Biol 55: 138–146. 10.1016/j.sbi.2019.03.014 [DOI] [PubMed] [Google Scholar]
- Lamichhane TN, Blewett NH, Maraia RJ. 2011. Plasticity and diversity of tRNA anticodon determinants of substrate recognition by eukaryotic A37 isopentenyltransferases. RNA 17: 1846–1857. 10.1261/rna.2628611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane TN, Blewett NH, Cherkasova VA, Crawford AK, Iben JR, Farabaugh PJ, Begley TJ, Maraia RJ. 2013a. Lack of tRNA modification isopentenyl-A37 alters mRNA decoding and causes metabolic deficiencies in fission yeast. Mol Cell Biol 33: 2918–2929. 10.1128/MCB.00278-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane TN, Mattijssen S, Maraia RJ. 2013b. Human cells have a limited set of tRNA anticodon loop substrates of the tRNA isopentenyltransferase TRIT1 tumor suppressor. Mol Cell Biol 33: 4900–4908. 10.1128/MCB.01041-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamichhane TN, Arimbasseri AG, Rijal K, Iben JR, Wei FY, Tomizawa K, Maraia RJ. 2016. Lack of tRNA-i6A modification causes mitochondrial-like metabolic deficiency in S. pombe by limiting activity of cytosolic tRNATyr, not mito-tRNA. RNA 22: 583–596. 10.1261/rna.054064.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentini JM, Alsaif HS, Faqeih E, Alkuraya FS, Fu D. 2020. DALRD3 encodes a protein mutated in epileptic encephalopathy that targets arginine tRNAs for 3-methylcytosine modification. Nat Commun 11: 2510. 10.1038/s41467-020-16321-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lentini J, Bargabos R, Chen C, Fu D. 2021. METTL8 is required for 3-methylcytosine modification in human mitochondrial tRNAs. bioRxiv 10.1101/2021.05.02.442361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonard Davis G. 1986. Basic Methods in Molecular Biology. Elsevier. [Google Scholar]
- Li J, Wang YN, Xu BS, Liu YP, Zhou M, Long T, Li H, Dong H, Nie Y, Chen PR, et al. 2020. Intellectual disability-associated gene ftsj1 is responsible for 2'-O-methylation of specific tRNAs. EMBO Rep 21: e50095. 10.15252/embr.202050095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz C, Lünse CE, Mörl M. 2017. tRNA modifications: impact on structure and thermal adaptation. Biomolecules 7: 35. 10.3390/biom7020035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacDonald RJ, Swift GH, Przybyla AE, Chirgwin JM. 1987. Isolation of RNA using guanidinium salts. In Methods in Enzymology (ed. Berger SL, Kimmel AR), Vol. 152, pp. 219–226. Academic Press, Boston. [DOI] [PubMed] [Google Scholar]
- Machnicka MA, Olchowik A, Grosjean H, Bujnicki JM. 2014. Distribution and frequencies of post-transcriptional modifications in tRNAs. RNA Biol 11: 1619–1629. 10.4161/15476286.2014.992273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia R. 1991. The subset of mouse B1 (Alu-equivalent) sequences expressed as small processed cytoplasmic transcripts. Nucleic Acids Res 19: 5695–5702. 10.1093/nar/19.20.5695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matuszek Z, Pan T. 2019. Quantification of queuosine modification levels in tRNA from human cells using APB gel and northern blot. Bio Protoc 9: e3191. 10.21769/BioProtoc.3191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y, Marchand V. 2021. Analysis of RNA modifications by second- and third-generation deep sequencing: 2020 update. Genes (Basel) 12: 278. 10.3390/genes12020278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motorin Y, Bec G, Tewari R, Grosjean H. 1997. Transfer RNA recognition by the Escherichia coli Δ2-isopentenyl-pyrophosphate:tRNA Δ2-isopentenyl transferase: dependence on the anticodon arm structure. RNA 3: 721–733. [PMC free article] [PubMed] [Google Scholar]
- Nostramo RT, Hopper AK. 2020. A novel assay provides insight into tRNAPhe retrograde nuclear import and re-export in S. cerevisiae. Nucleic Acids Res 48: 11577–11588. 10.1093/nar/gkaa879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan T. 2018. Modifications and functional genomics of human transfer RNA. Cell Res 28: 395–404. 10.1038/s41422-018-0013-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas-Benitez D, Thiaville PC, de Crecy-Lagard V, Glavic A. 2015. The levels of a universally conserved tRNA modification regulate cell growth. J Biol Chem 290: 18699–18707. 10.1074/jbc.M115.665406 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryvkin P, Leung YY, Silverman IM, Childress M, Valladares O, Dragomir I, Gregory BD, Wang LS. 2013. HAMR: high-throughput annotation of modified ribonucleotides. RNA 19: 1684–1692. 10.1261/rna.036806.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sokolowski M, Klassen R, Bruch A, Schaffrath R, Glatt S. 2017. Cooperativity between different tRNA modifications and their modification pathways. Biochim Biophys Acta 1861: 409–419. 10.1016/j.bbagrm.2017.12.003 [DOI] [PubMed] [Google Scholar]
- Suzuki T. 2021. The expanding world of tRNA modifications and their disease relevance. Nat Rev Mol Cell Biol 22: 375–392. 10.1038/s41580-021-00342-0 [DOI] [PubMed] [Google Scholar]
- Suzuki T, Yashiro Y, Kikuchi I, Ishigami Y, Saito H, Matsuzawa I, Okada S, Mito M, Iwasaki S, Ma D, et al. 2020. Complete chemical structures of human mitochondrial tRNAs. Nat Commun 11: 4269. 10.1038/s41467-020-18068-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vare VY, Eruysal ER, Narendran A, Sarachan KL, Agris PF. 2017. Chemical and conformational diversity of modified nucleosides affects tRNA structure and function. Biomolecules 7: 29. 10.3390/biom7010029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Matuszek Z, Huang Y, Parisien M, Dai Q, Clark W, Schwartz MH, Pan T. 2018. Queuosine modification protects cognate tRNAs against ribonuclease cleavage. RNA 24: 1305–1313. 10.1261/rna.067033.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei FY, Zhou B, Suzuki T, Miyata K, Ujihara Y, Horiguchi H, Takahashi N, Xie P, Michiue H, Fujimura A, et al. 2015. Cdk5rap1-mediated 2-methylthio modification of mitochondrial tRNAs governs protein translation and contributes to myopathy in mice and humans. Cell Metab 21: 428–442. 10.1016/j.cmet.2015.01.019 [DOI] [PubMed] [Google Scholar]
- Yarham JW, Lamichhane T, Mattijssen S, Bruni F, McFarland R, Maraia RJ, Taylor RW. 2014. Defective i6A37 modification of mitochondrial and cytosolic tRNAs results from pathogenic mutations in TRIT1 and its substrate tRNA. PLoS Genet 10: e1004424. 10.1371/journal.pgen.1004424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaborske JM, DuMont VL, Wallace EW, Pan T, Aquadro CF, Drummond DA. 2014. A nutrient-driven tRNA modification alters translational fidelity and genome-wide protein coding across an animal genus. PLoS Biol 12: e1002015. 10.1371/journal.pbio.1002015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang W, Xu R, Matuszek Ż, Cai Z, Pan T. 2020. Detection and quantification of glycosylated queuosine modified tRNAs by acid denaturing and APB gels. RNA 26: 1291–1298. 10.1261/rna.075556.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng G, Qin Y, Clark WC, Dai Q, Yi C, He C, Lambowitz AM, Pan T. 2015. Efficient and quantitative high-throughput tRNA sequencing. Nat Methods 12: 835–837. 10.1038/nmeth.3478 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.