Abstract
The two subunits of the eukaryotic ribosome are produced through quasi-independent pathways involving the hierarchical actions of numerous trans-acting biogenesis factors and the incorporation of ribosomal proteins. The factors work together to shape the nascent subunits through a series of intermediate states into their functional architectures. One of the earliest intermediates of the small subunit (SSU or 40S) is the SSU processome which is subsequently transformed into the pre-40S intermediate. This transformation is, in part, facilitated by the binding of the methyltransferase Bud23. How Bud23 is released from the resultant pre-40S is not known. The ribosomal proteins Rps0, Rps2, and Rps21, termed the Rps0-cluster proteins, and several biogenesis factors bind the pre-40S around the time that Bud23 is released, suggesting that one or more of these factors could induce Bud23 release. Here, we systematically examined the requirement of these factors for the release of Bud23 from pre-40S particles. We found that the Rps0-cluster proteins are needed but not sufficient for Bud23 release. The atypical kinase/ATPase Rio2 shares a binding site with Bud23 and is thought to be recruited to pre-40S after the Rps0-cluster proteins. Depletion of Rio2 prevented the release of Bud23 from the pre-40S. More importantly, the addition of recombinant Rio2 to pre-40S particles affinity-purified from Rio2-depleted cells was sufficient for Bud23 release in vitro. The ability of Rio2 to displace Bud23 was independent of nucleotide hydrolysis. We propose a novel role for Rio2 in which its binding to the pre-40S actively displaces Bud23 from the pre-40S.
Keywords: BUD23, RIO2, pre-40S, ribosome biogenesis
INTRODUCTION
In eukaryotes, ribosome biogenesis is a metabolically expensive and essential task that entails the transcription, processing, and folding of the ribosomal RNA (rRNA), incorporation of at least 79 ribosomal proteins (RPs), and the concerted actions of more than 200 trans-acting biogenesis factors (Warner 1999; Woolford and Baserga 2013). In the yeast Saccharomyces cerevisiae, ribosome biogenesis begins in the nucleolus with the transcription of a polycistronic precursor rRNA (pre-rRNA) encoding the 18S rRNA of the small subunit (SSU or 40S) and the 5.8S and 25S rRNAs of the large subunit (LSU or 60S), each flanked by external and internal transcribed spacer regions (ETS and ITS, respectively). Approximately 70 biogenesis factors and RPs bind cotranscriptionally to the pre-rRNA to form an early, metastable 40S biogenesis intermediate, the SSU processome or 90S preribosome (henceforth “processome”) (Dragon et al. 2002; Grandi et al. 2002; Osheim et al. 2004; Pérez-Fernández et al. 2007; Barandun et al. 2017; Cheng et al. 2017; Sun et al. 2017). The processome ultimately transitions, through a series of disassembly intermediates, into an early pre-40S intermediate (Cheng et al. 2020; Du et al. 2020; Lau et al. 2021). This transition involves the nucleolytic removal of the 5′-ETS and cleavage within ITS1, the shedding of most processome factors, and the binding of pre-40S factors. Together, these events drive the compaction of its architecture that produces the pre-40S (Chaker-Margot 2018; Black and Johnson 2021) and is thought to release the nascent 40S into the nucleoplasm (Erdmann et al. 2021; Tartakoff et al. 2021). The remaining maturation of the nascent 40S involves its nuclear export into the cytoplasm, the exchange and release of the pre-40S factors, the incorporation of additional RPs, and the generation of the mature 3′-end of the rRNA.
Among the factors that enter the 40S biogenesis pathway during the transition of the processome into the pre-40S is Bud23. Bud23 is a highly conserved methyltransferase that modifies guanosine 1575 (G1575) of the 18S rRNA in yeast (White et al. 2008), and the corresponding base G1639 in humans (Zorbas et al. 2015), that is located within the P-site of the 40S. Bud23 forms a conserved heterodimeric complex with the methyltransferase adaptor Trm112 (Figaro et al. 2012; Sardana and Johnson 2012; Létoquart et al. 2014). In humans, haploinsufficiency for the chromosomal region encoding human Bud23, WBSCR22, is linked to a rare developmental disorder known as Williams–Beuren Syndrome (Doll and Grzeschik 2001; Merla et al. 2002; Õunap et al. 2013; Létoquart et al. 2014; Haag et al. 2015). Despite being nonessential in yeast, the deletion of BUD23 (bud23Δ) causes a strong growth defect caused by a substantial loss of 40S production (White et al. 2008). The enzymatic activity of Bud23 is fully dispensable in both yeast and humans suggesting that the presence of Bud23 confers its primary role (White et al. 2008; Lin et al. 2012; Létoquart et al. 2014; Zorbas et al. 2015). The defects of bud23Δ in yeast can be partially overcome by extragenic mutations in several processome factors, indicating that Bud23 plays an active role in the processome-to-pre-40S transition (Sardana et al. 2013, 2014; Zhu et al. 2016; Black et al. 2020). During this transition, the central pseudoknot (CPK) forms. The CPK is a universally conserved structural feature formed by long-range contacts between the stem–loop of helix 1 at the 5′-end of the 18S rRNA and nucleotides A1137–U1144 in S. cerevisiae to form helix 2 (Brink et al. 1993; Poot et al. 1998). The CPK, along with helices 28, 35, 36, and 37, comprise the “neck” of the subunit (Supplemental Fig. S1A), the point about which the head region of the subunit rotates relative to its body during translation (Korostelev et al. 2008). CPK formation is a crucial step in 40S biogenesis and proper folding of the CPK appears to be a highly coordinated event requiring the timely release of the U3 snoRNA from the partially disassembled processome. U3 release is catalyzed by the helicase Dhr1 (Sardana et al. 2015), and we recently proposed that the binding of Bud23 to the partially disassembled processome, the Dis-C complex, coordinates the enzymatic activities of Dhr1 and the GTPase Bms1 to promote CPK folding (Fig. 1; Black et al. 2020; Black and Johnson 2021). Rps2 (uS5) is thought to bind to the folded CPK in the subsequent pre-40S intermediates (Supplemental Fig. S1B; Ameismeier et al. 2018). The notion that Bud23 promotes CPK formation suggests that Bud23 function is a prerequisite for the recruitment of Rps2, but this has not been experimentally demonstrated. Furthermore, Bud23 remains in complex with the pre-40S, but how it is released is unknown.
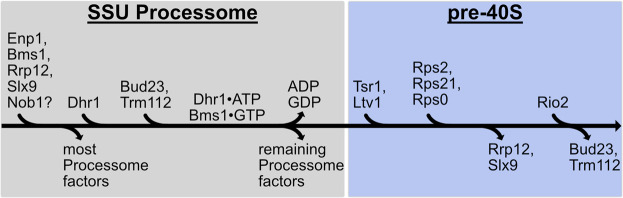
FIGURE 1.
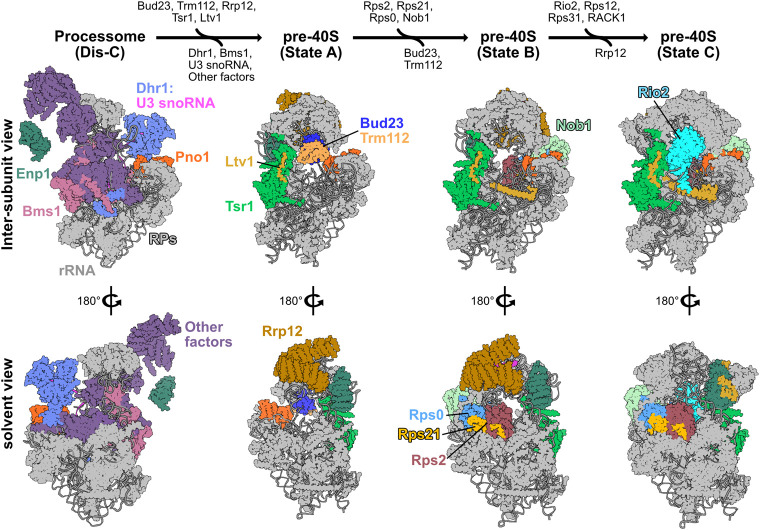
The proposed pathway for binding and release of Bud23 from 40S precursors. A pathway based on structures from yeast and human 40S biogenesis intermediates is shown to visualize the stage-specific association of Bud23 with the pre-40S intermediates. Each intermediate is shown from the intersubunit and the solvent views. The final metastable intermediate of processome disassembly, Dis-C (PDB 6ZQG), contains multiple Processome factors, including the U3 snoRNA, Dhr1, Bms1, Enp1, and Pno1 (Cheng et al. 2020). The binding of Bud23, and its partner Trm112, to Dis-C promotes its transformation into the pre-40S (Black et al. 2020). This transition comes with the release of most Processome factors and yields the earliest pre-40S intermediate, State A (PDB 6G4W) (Ameismeier et al. 2018). During this transition, Tsr1 and Ltv1 also are observed on the pre-40S. Rrp12, which is resolved in earlier Processome intermediates (Singh et al. 2021) but is not in Dis-C, is also seen in State A. Between State A to State B (PDB 6G4S), Nob1, more of Ltv1, and the Rps0-cluster proteins become resolved, and Bud23 and Trm112 are no longer observed. Rrp12 and the rRNA near it also undergo structural remodeling during this transition. Between State B and State C (PDB 6G18), Rio2 and several RPs become resolved while Rrp12 appears to be released. Molecular visualization was generated in UCSF ChimeraX v0.93 (Goddard et al. 2018).
Multiple cryogenic electron microscopy (cryo-EM) studies on pre-40S particles from both yeast and humans provide a framework for understanding the approximate timing of when factors associate with preribosomes (Heuer et al. 2017; Ameismeier et al. 2018, 2020; Scaiola et al. 2018; Mitterer et al. 2019; Shayan et al. 2020; Plassart et al. 2021; Rai et al. 2021). Notably, Bud23 and Trm112 are present in the earliest human pre-40S intermediate, State A (Ameismeier et al. 2018), which also contains the large HEAT-repeat protein Rrp12, the pre-40S-specific factor Tsr1, and the Processome/pre-40S factors Enp1 and Pno1 (Dim1) (Fig. 1). Here, Bud23 binds directly to the rRNA encompassing its target guanosine. In State B, Bud23 and Trm112 were not observed suggesting that they may be released during the transition between these two intermediates. Meanwhile, the endonuclease Nob1, and the Rps0-cluster proteins containing Rps0 (uS2), Rps21 (eS21), and Rps2 appear in State B. The transition from States A to B also comes with structural rearrangements of Rrp12 and the helices 35–37 of the rRNA that contribute to the “neck” of the subunit (Supplemental Fig. S1C). In State C, the atypical kinase/ATPase Rio2 is observed on the pre-40S bound to the P-site where Bud23 was previously located. This order of events is consistent with a recent report from yeast that found that Rio2 is recruited after the Rps0-cluster proteins (Linnemann et al. 2019). The transition between States B and C also coincides with the loss of Rrp12 from the pre-40S as well as the incorporation of several additional RPs and rRNA remodeling (Fig. 1; Supplemental Fig. S1C), suggesting that Rrp12 may be released during this transition. The absence of Bud23 and Trm112 from State B and the simultaneous appearance of several factors and RPs lead us to hypothesize that the binding of one or more of these proteins promotes the release of Bud23 and Trm112.
Here, we explore the timing of Bud23 release from the nascent 40S. We found that Bud23 promotes the binding of the Rps0-cluster proteins to the 40S precursor, consistent with its role in generating the CPK. Conversely, we found that the Rps0-cluster proteins are needed for the subsequent release of Bud23 and Trm112. However, in contrast to the proposed pathway (Fig. 1), we found that Bud23 coexists with the Rps0-cluster proteins on pre-40S particles, indicating that the recruitment of the Rps0-cluster proteins is not sufficient to release Bud23. As Rio2 and Bud23 share overlapping binding sites (Fig. 1), we explored the possibility that Rio2 displaces Bud23 from the pre-40S. Indeed, we showed that Rio2 can induce the release of Bud23. Together, our data supports a model in which the concerted binding of the Rps0-cluster proteins and Rio2 serve to release Bud23 and Trm112 from the pre-40S.
RESULTS
Bud23 promotes the recruitment of the Rps0-cluster proteins to 40S precursors
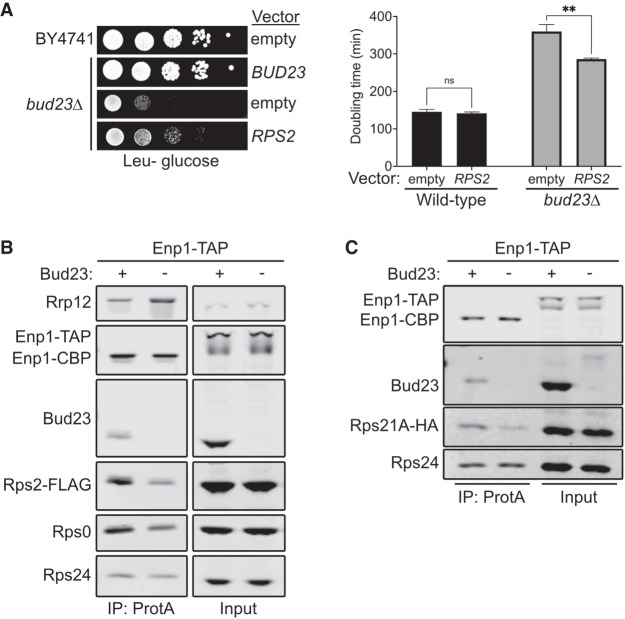
While characterizing the spontaneous extragenic suppressors of the growth defect caused by bud23Δ (Black et al. 2020), we also screened for high-copy suppressors. To this end, we transformed bud23Δ cells with a 2µ genomic plasmid library (Connelly and Hieter 1996) and screened for colonies with improved growth over background. We isolated two vectors that contained overlapping genomic regions of chromosome VII encoding the full coding sequences for the genes RPS2, NAB2, and GPG1 (Supplemental Fig. S2A,B). RPS2 encodes the small ribosomal subunit protein Rps2, making it the likely candidate gene responsible for suppression. To verify that the suppression phenotype was due to ectopic RPS2, we independently cloned RPS2 into centromeric and 2µ vectors and tested it for suppression of bud23Δ. Indeed, ectopic expression of RPS2 partially restored growth in the bud23Δ cells (Fig. 2A; Supplemental Fig. S2A). Notably, there was no obvious difference in suppression by RPS2 expressed from a low-copy centromeric vector or a high-copy 2µ vector (Supplemental Fig. S2A). Our previous analyses of extragenic suppressors of bud23Δ showed that these suppressors partially alleviate the 40S biogenesis defect of bud23Δ cells (Sardana et al. 2013; Black et al. 2020). To assess whether the partial suppression of bud23Δ due to ectopic RPS2 also improved 40S production, we analyzed ribosome subunit levels by separating extracts on sucrose density gradients. Compared to wild-type cells, there was a loss of free 40S, a strong increase of free 60S and a reduction of actively translating polysomes in bud23Δ mutant cells (Supplemental Fig. S2C). Ectopic expression of RPS2 led to a modest decrease in the amount of free 60S indicative of a partial restoration of 40S biogenesis in bud23Δ cells. Tsr4 was recently identified as an essential, dedicated chaperone that cotranslationally binds Rps2 to facilitate its expression (Black et al. 2019; Rössler et al. 2019; Landry-Voyer et al. 2020). We wondered if suppression by ectopic RPS2 could be enhanced by coexpression of TSR4. Indeed, we saw that bud23Δ cells harboring both CEN-borne RPS2 and 2µ-borne TSR4 grew better than cells expressing RPS2 alone (Supplemental Fig. S2D). This result supports the conclusion that increased levels of Rps2 suppress the growth defect of bud23Δ cells.
FIGURE 2.
Bud23 promotes the recruitment of the Rps0-cluster proteins to 40S precursors. (A, left) Ectopic expression of RPS2 suppressed the growth defect of bud23Δ as shown by 10-fold serial dilutions of wild-type (BY4741) or bud23Δ (AJY2676) cells transformed with an empty vector (pRS415) or a vector encoding BUD23 (pAJ2154) or RPS2 (pAJ2960) spotted on SD Leu- media and grown for 2 d at 30°C. (Right) Ectopic expression of RPS2 suppressed the growth defect of bud23Δ, as shown by the quantification of doubling time in minutes of the same strains used in A. See Materials and Methods for details. Data are shown as mean doubling time ± standard deviation of biological duplicates. Significance was calculated by two-way analysis of variance (ANOVA) with Sidak correction for multiple comparisons using GraphPad Prism 9 for Mac iOS (www.graphpad.com) (adjusted P = 0.9155 [ns]; P = 0.0035 [**]). (B,C) Bud23-depletion decreased the association of Rps0, Rps2, and Rps21-HA as shown by western blotting of specific factors associated with affinity-purified particles using Enp1-TAP as bait. Strains AJY4748 (Bud23+) and AJY4750 (Bud23−) that were transformed with pAJ5124 for B and AJY4749 (Bud23+) and AJY4751 (Bud23−) for C were each cultured in SD Ura- or YPD to early exponential phase, then treated with 0.5 mM auxin for 10 min prior to collection.
Bud23 promotes the transition from the Processome to the pre-40S (Black et al. 2020), which occurs upstream of Rps2 binding. Thus, the simplest interpretation of the genetic interaction between RPS2 and BUD23 is that the absence of Bud23 reduces Rps2 recruitment, and the increased expression of Rps2 partially compensates for its reduced recruitment. To address whether Bud23 influences the recruitment of Rps2, we affinity purified pre-40S from Bud23-depleted cells and monitored the levels of Rps2. We used a strain containing a previously characterized system in which an auxin-inducible degron (AID) is fused to the carboxyl terminus of Bud23 (Bud23–AID) to enable a controlled and rapid depletion of Bud23 (Black et al. 2020). To monitor the levels of Rps2, we used a CRISPR–Cas9 method (Akhmetov et al. 2018) to integrate a carboxy-terminal FLAG epitope in the genomic locus of RPS2 (Rps2–FLAG). We then purified particles via Enp1 containing a carboxy-terminal tandem affinity purification tag (Enp1–TAP) from either wild-type or Bud23-depleted cells. Affinity-purified complexes were enzymatically eluted with TEV protease and subsequently sedimented through sucrose cushions to separate preribosomal particles from any possible extraribosomal bait and other copurifying proteins. The pellet fraction was then separated by SDS-PAGE and subjected to western blotting for specific factors. This analysis revealed that Bud23-depletion reduced the amount of Rps2–FLAG on SSU precursors relative to those purified from wild-type cells (Fig. 2B). Therefore, Bud23 is needed for the efficient recruitment of Rps2.
Rps2 appears to enter the pre-40S concurrently with Rps0 and Rps21 (Fig. 1; Ameismeier et al. 2018) as part of a heterotrimeric cluster of RPs known as the Rps0-cluster proteins (Linnemann et al. 2019). We wondered if Rps0 and Rps21 would also be reduced in pre-40S particles from Bud23-depleted cells. Indeed, blotting for endogenous Rps0 revealed that its association with nascent pre-40S particles is reduced in the absence of Bud23 (Fig. 2B). To monitor Rps21, we carboxy-terminally tagged the genomic locus of Rps21A with HA (Rps21A–HA). We saw reduced levels of Rps21A–HA on SSU precursors purified in the absence of Bud23 (Fig. 2C). The reduction of the Rps0-cluster proteins appears specific, as Rps24 levels remained relatively constant. Furthermore, the abundance of the Processome/pre-40S factor Rrp12 increased in response to Bud23-depletion (Fig. 2B), indicating that loss of the Rps0-cluster proteins was not due to a general loss of 40S precursors, but rather a reduced ability to transition the Processome into the pre-40S as our previous analyses of such Bud23-depleted particles showed (Black et al. 2020). Because Bud23-depletion reduced the recruitment of the Rps0-cluster proteins, we also tested if the ectopic expression of RPS21A and RPS0B could suppress the growth defect of bud23Δ cells, however they did not (Supplemental Fig. S3). We also asked if ectopic coexpression of all three Rps0-cluster proteins could enhance the suppression effect of ectopic RPS2; however, there was no substantial enhancement of suppression compared to cells carrying RPS2 alone. Thus, despite Bud23 promoting the recruitment of the three Rps0-cluster proteins (Fig. 2B,C), the suppression of bud23Δ is specific to RPS2, perhaps due to the direct interaction between Rps2 and the CPK. We recently proposed that Bud23 binding enhances the rate of productive CPK folding upon Dhr1 displacement of U3 snoRNA (Black and Johnson 2021), and we speculate that the overexpression of RPS2 partially restores productive CPK folding in the absence of Bud23,. The notion that Rps2 binding can influence CPK structure is supported by structural studies of bacterial SSU assembly in which S5, the ortholog of Rps2, is thought to stabilize the CPK (Yang et al. 2014) and the observation that an S5 mutant induces CPK misfolding (Roy-Chaudhuri et al. 2010). Together, these data further support the notion that Bud23 promotes CPK formation.
The Rps0-cluster proteins promote the release of Bud23 from the pre-40S
Based on recent cryo-EM structures of human pre-40S (Ameismeier et al. 2018), the release of Bud23 appears coincident with the appearance of the Rps0-cluster proteins and Nob1 (Fig. 1). We hypothesized that the binding of one or more of these proteins promotes the release of Bud23 from the pre-40S. To test this hypothesis, we asked whether Bud23 would accumulate on 40S precursors affinity-purified from cells depleted of Rps2. We also depleted the Processome helicase Dhr1 or Rps3 for comparison because loss of Dhr1 arrests particles prior to the association of either Bud23 or Rps2 (Sun et al. 2017) and Rps3 is incorporated after Rps2 incorporation and Bud23 release (Ameismeier et al. 2018). In these experiments, the genes encoding each of these factors were placed under the control of a glucose-repressible GAL1 promoter (PGAL1). Cells were treated with glucose for five and a half hours to repress DHR1 and 2 h to repress RPS2 and RPS3. These time points were chosen because they were previously shown to sufficiently deplete the target gene to cause 40S biogenesis defects (Ferreira-Cerca et al. 2005; Sardana et al. 2015; Linnemann et al. 2019). We then affinity purified SSU precursors via carboxy-terminally FLAG-TEV-Protein A tagged Enp1 (Enp1–FTP), as it associates with 40S precursors that span the time of association of Bud23 and Rps2 (Fig. 1; Schäfer et al. 2003; Chaker-Margot et al. 2015; Zhang et al. 2016; Ameismeier et al. 2018).
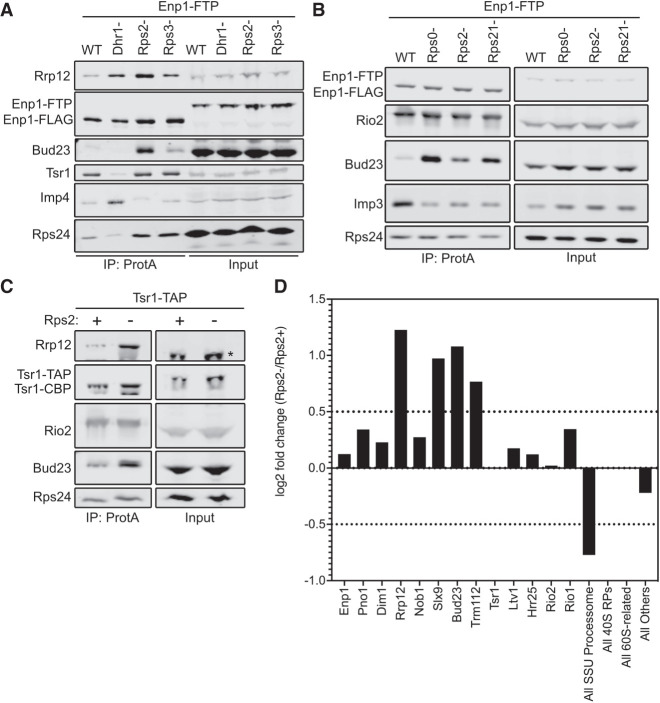
Following affinity purification, we used western blotting to monitor the presence of Bud23, the Processome/pre-40S factor Rrp12, the pre-40S-specific factor Tsr1, the Processome factor Imp4, and the RP Rps24. Low levels of Bud23 copurified with the Enp1–FTP particles isolated from wild-type cells (Fig. 3A). Depletion of Dhr1 caused both Imp4 and Rrp12 to increase while Tsr1 levels decreased, indicating the expected arrest at the Processome stage. Conversely, Rps2- and Rps3-depletion did not cause Imp4 levels to increase nor did they reduce Tsr1 levels, indicating an arrest at the pre-40S stage rather than at the Processome stage. Rps2-depletion but not Rps3 depletion caused a marked accumulation of Rrp12, consistent with a recent report (Linnemann et al. 2019). Notably, Rps2-depletion but not Rps3 depletion caused a strong accumulation of Bud23 (Fig. 3A). Because Rps0 and Rps21 appear to enter the pre-40S with Rps2 (Fig. 1; Ameismeier et al. 2018), we asked if their depletion would also cause Bud23 to accumulate on pre-40S particles. Indeed, the depletion of Rps21 and Rps0 each caused Bud23 to accumulate on pre-40S particles purified via Enp1–FTP (Fig. 3B). Interestingly, the depletion of each RP led to differing extents of Bud23 accumulation. Rps0-depletion showed the strongest accumulation of Bud23 while the depletion of Rps21 and Rps2 showed less accumulation of Bud23. While we cannot formally rule out that this differential accumulation of Bud23 is not due to potential differences in the rates of depletion for each RP, we note that the depletion of each RP caused an equivalent accumulation of Rrp12 (Supplemental Fig. S4). This suggests that the Rps0-cluster proteins are each needed for Bud23 release, but their depletion has differing effects on the extent of Bud23 release from the pre-40S.
FIGURE 3.
The Rps0-cluster proteins promote the release of Bud23 from the pre-40S. (A) Rps2-depletion caused Bud23 to accumulate on preribosomal particles as shown by western blotting for specific factors that copurified with Enp1–FTP-associated particles in the presence (WT) or absence of Dhr1 (Dhr1-), Rps2 (Rps2-), or Rps3 (Rps3-). Strains AJY4724 (WT), AJY4283 (Rps2-), and AJY4725 (Rps3-) were cultured in YPGal media until early exponential phase, then cultured for 2 h following the addition of 2% glucose to deplete Rps2 and Rps3. Strain AJY4285 (Dhr1-) was cultured for 2.5 h in YPGal, then cultured for 6 h following the addition of 2% glucose to deplete Dhr1. (B) The depletion Rps0, Rps2, and Rps21 each caused Bud23 to accumulate on preribosomal particles, as shown by western blotting for specific factors that copurified with Enp1–FTP-associated particles in the presence (WT) or absence of Rps0 (Rps0-), Rps2 (Rps2-), or Rps21 (Rps21-). Strains AJY4745 (WT), AJY4705 (Rps0-), AJY4706 (Rps2-), and AJY4707 (Rps21-) were cultured in YPGal to early exponential phase, then treated with 2% glucose for 2 h to repress the RP gene transcription. Equal amounts of the affinity-purified particles were separated by SDS-PAGE and subjected to western blotting for the indicated factors. (C) Bud23 accumulated in Rps2-depleted particles as shown by western blotting for specific factors that copurify with Tsr1–TAP-associated pre-40S particles. The asterisks (*) denotes Tsr1–TAP signal in the Input that cross-reacted with the Rrp12 antibody due to the presence of the Protein A tag. Strains AJY4754 (Rps2+) and AJY4755 (Rps2−) were cultured in the same manner as the WT and Rps2− cells in A. (D) Proteomic composition of Tsr1-associated preribosomal particles from C as measured by semiquantitative mass spectrometry is shown as the log2 fold change between Rps2− and Rps2+ samples. These values were calculated as described in Materials and Methods. A factor displaying a log2 fold change value greater than 0.5 (dashed line) is considered to have increased in abundance. Individual factors are ordered by the approximate timing of their known association with 40S precursors. The supporting data are provided in Supplemental File 1.
To support the above findings, we repeated the affinity-purifications using an alternative bait, Tsr1. Tsr1 is thought to specifically associate with pre-40S particles and enter the nascent SSU shortly after the Processome has transitioned into the pre-40S and remains associated with the particle after Bud23 is thought to be released (Fig. 1). To this end, we affinity purified pre-40S particles via Tsr1–TAP from wild-type or PGAL1–RPS2 cells that were treated with glucose for 2 h. Like the particles isolated via Enp1–FTP, western blot analysis of the Tsr1–TAP particles showed a strong accumulation of both Bud23 and Rrp12 in response to Rps2-depletion (Fig. 3C). To gain broader insight into the effect that Rps2-depletion has on the protein composition of the affinity-purified particles, we also performed semi-quantitative mass spectrometry on these particles. Total spectral counts of each protein identified by mass spectrometry were normalized to the molecular weight of each protein (Supplemental File 1). These values were subsequently normalized to the corresponding value of Tsr1 to quantify the approximate stoichiometry of each protein as done previously (Black et al. 2018, 2020; Sun et al. 2017; An et al. 2018). From these values, the log2 fold change of Rps2-depleted particles to the wild-type particles was calculated for each factor to identify how Rps2-depletion affects their presence on the pre-40S. This analysis revealed that the levels of most pre-40S factors remain relatively unchanged (log2 fold change <0.5) while Rrp12, Slx9, Bud23, and its binding partner Trm112 increased in response to Rps2-depletion (log2 fold change >0.5) (Fig. 3D). The accumulation of Rrp12 and Slx9 is consistent with a recent report that showed these proteins accumulate on Rio2-purified pre-40S particles in response to Rps2-depletion (Linnemann et al. 2019). Notably, Bud23 and Trm112 accumulated on ∼20% of particles in the absence of Rps2 (Supplemental File 1), suggesting that they are present in substoichiometric amounts on the Rps2-depleted particles. Meanwhile, other accumulating factors such as Rrp12 and Slx9 accumulate to nearly 70% stoichiometry and suggests that there is a heterogenous population of Bud23-containing and Bud23-absent particles in the affinity-purified particles. As noted above, Rps0- and Rps21-depletion had greater accumulation of Bud23 than Rps2 did (Fig. 3B; Supplemental Fig. S4), suggesting that the Rps0- and Rps21-depleted particles are more homogenous than the Rps2-depleted sample. Nevertheless, these results suggest that the binding of the Rps0-cluster proteins is needed for the release of Bud23 (and Trm112) from the pre-40S and are consistent with predictions based on the structures of human pre-40S intermediates (Fig. 1; Ameismeier et al. 2018).
Depletion of Tsr4 also causes Bud23 to accumulate on pre-40S
To further support the idea that Rps2 is needed for Bud23 release, we asked if Bud23 accumulates on pre-40S particles in cells depleted of the dedicated chaperone for Rps2, Tsr4 (Black et al. 2019; Rössler et al. 2019; Landry-Voyer et al. 2020). The transcriptional repression of RP genes quickly leads to measurable defects in ribosome biogenesis because of the rapid depletion of available free RPs during the assembly of ribosomes. On the other hand, the depletion of biogenesis factors typically requires considerably longer times to impact ribosome biogenesis, dependent on the half-life of the assembly factor. Consequently, we again turned to the AID system to induce rapid degradation of Tsr4 in an auxin-dependent manner (Nishimura et al. 2009). We were unable to generate a genomically integrated TSR4–AID strain using an existing AID system that expresses the Oryza sativa E3 ligase, OsTir1, from a strong constitutive promoter (Nishimura et al. 2009). We considered the possibility that the expression of OsTir1 even in the absence of auxin was sufficient to target Tsr4 for degradation, as has been reported for some essential proteins (Mendoza-Ochoa et al. 2019). To circumvent this, we engineered a system in which the OsTIR1 gene was under the control of the galactose-inducible promoter (Supplemental Fig. S5A) and controlled by a β-estradiol-inducible GAL4 DNA binding domain, akin to a recently published system (Mendoza-Ochoa et al. 2019). Using this system, we successfully generated a TSR4–AID strain that was inviable on media containing auxin and β-estradiol but fully viable on media lacking them (Supplemental Fig. S5B). Time-course analysis of Tsr4–AID (which contains the HA epitope) following the addition of auxin and β-estradiol showed that the protein is almost fully depleted 2 h post-treatment (Supplemental Fig. S5C). Purification of Enp1–FTP particles from cells in which Tsr4–AID was depleted for 2 h revealed that both Rrp12 and Bud23 accumulated in the absence of Tsr4 (Supplemental Fig. S5D). Because Tsr4 promotes Rps2 expression (Black et al. 2019; Rössler et al. 2019; Landry-Voyer et al. 2020), this result further supports the notion that Rps2 is needed for the release of Bud23 from the pre-40S.
Nob1 and Ltv1 are not needed for Bud23 release
In addition to the Rps0-cluster proteins, Nob1 and an additional segment of Ltv1 also become resolved by cryo-EM on the pre-40S particle as Bud23 and Trm112 appear to be released (Fig. 1; Ameismeier et al. 2018), raising the possibility that these two factors may also play a role in release of Bud23. To test this idea, we generated LTV1–AID and NOB1–AID strains in the ENP1–FTP background. Both strains grew as well as wild-type cells on media lacking auxin and β-estradiol but showed growth defects in their presence (Supplemental Fig. S6A). Ltv1–AID was almost fully depleted after 2 h of treatment with both auxin and β-estradiol, while Nob1–AID was nearly undetectable after 1.5 h (Supplemental Fig. S6B). However, neither the depletion of Ltv1 nor Nob1 caused Bud23 to accumulate on SSU precursors while Tsr4-depletion did (Supplemental Fig. S6C,D). These results support the idea that the Rps0-cluster proteins have a proximal role in the release of Bud23.
The Rps0-cluster proteins copurify with Bud23
The above results implicate the Rps0-cluster proteins in the release of Bud23. This is consistent with the structures of human pre-40S intermediates that suggest Bud23 leaves upon the binding of these RPs (Ameismeier et al. 2018). However, it was unknown whether the presence of these RPs directly induces the release of Bud23 or if their association leads to a particle competent for Bud23 release by a downstream event. To this end, we immunoprecipitated carboxy-terminally tagged Bud23 (Bud23–GFP) and assayed for the presence of the Rps0-cluster proteins by western blotting. We also immunoprecipitated Utp9–GFP and Rio2–GFP in parallel with Bud23–GFP as controls that bracket the time in which Bud23 associates with pre-40S particles. Utp9 is a Processome-specific factor (Dragon et al. 2002) that associates with the Processome before the loading of the Rps0-cluster proteins, while Rio2 is exclusively a pre-40S factor known to bind to particles after the assembly of the Rps0-cluster proteins (Heuer et al. 2017; Scaiola et al. 2018). To detect Rps2, we carboxy-terminally tagged it with a FLAG epitope (Rps2–FLAG) and expressed it ectopically in a strain disrupted for genomic RPS2. Rps2–FLAG was fully functional (Supplemental Fig. S7A) and coprecipitated with Bud23–GFP and Rio2–GFP but not with Utp9–GFP, while the Processome/pre-40S factor Pno1 copurified with all three baits (Fig. 4A). In an independent experiment, we asked whether Rps0 and Rps21 could coprecipitate with the same GFP-tagged baits. Rps21 and Rps0 are each encoded by paralogous genes—RPS21A/RPS21B and RPS0A/RPS0B, respectively. To monitor Rps21, we ectopically expressed functional Rps21A–HA (Supplemental Fig. S7B) in cells disrupted for both RPS21A and RPS21B, while endogenous Rps0 was detected with an anti-Rps0 antibody. Like Rps2–FLAG, Rps21A–HA, and Rps0 copurified with Bud23–GFP and Rio2–GFP but not with Utp9–GFP, while the Processome-specific factor Imp4 copurified with only Utp9–GFP (Fig. 4B). When taken together, these data indicate that Bud23 and the Rps0-cluster proteins can coexist on pre-40S particles in contrast to what was predicted by the structures of human pre-40S intermediates (Fig. 1; Ameismeier et al. 2018).
FIGURE 4.
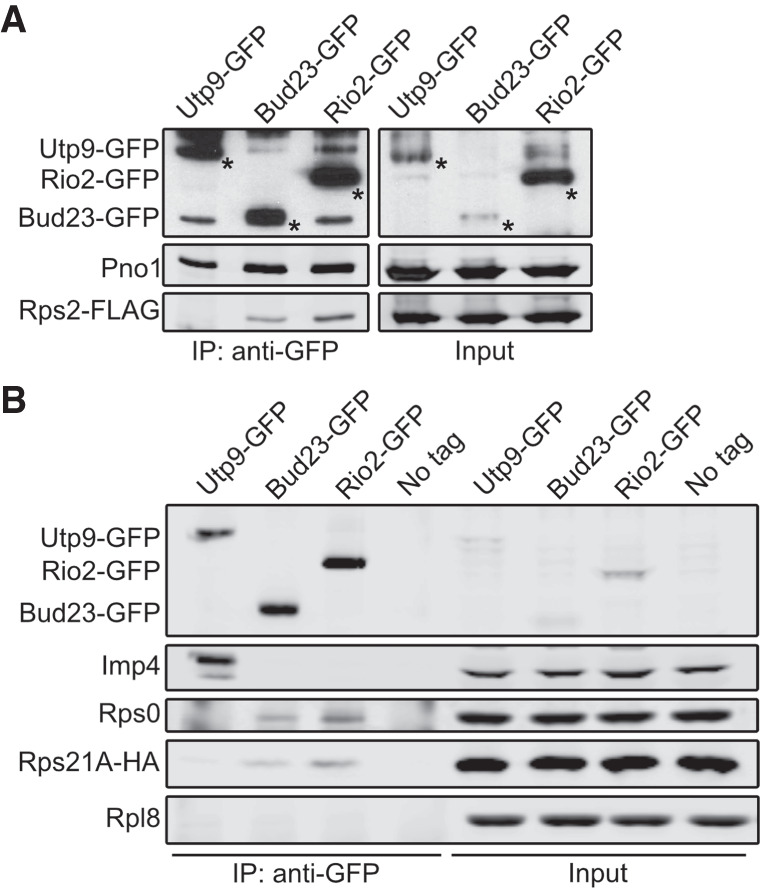
Bud23 coexists on 40S precursors with the Rps0-cluster proteins. Bud23 and Rio2 associate with pre-40S particles containing the Rps0-cluster proteins as shown by the coimmunoprecipitation of Rps2–FLAG, Rps0, or Rps21-HA with the indicated GFP-tagged baits. (A) Rps2–FLAG coimmunoprecipitated with Bud23–GFP (AJY4722) and Rio2-GFP (AJY4723) but not the Processome factor Utp9-GFP (AJY4721). Asterisks (*) denote the GFP-tagged baits. (B) Rps0 and Rps21A-HA coimmunoprecipitated with Bud23–GFP (AJY4767) and Rio2-GFP (AJY4768) but not Utp9-GFP (AJY4766) nor were they present in a negative control that used cells with no bait (No Tag; AJY4732). The Processome/pre-40S factor Pno1 is shown in A, and the Processome factor Imp4 is shown in B as positive controls for the copurification of preribosomal particles. Rpl8 is shown in B as a specificity control for contamination of mature ribosomes.
Rio2 is needed for the release of Bud23 from the pre-40S
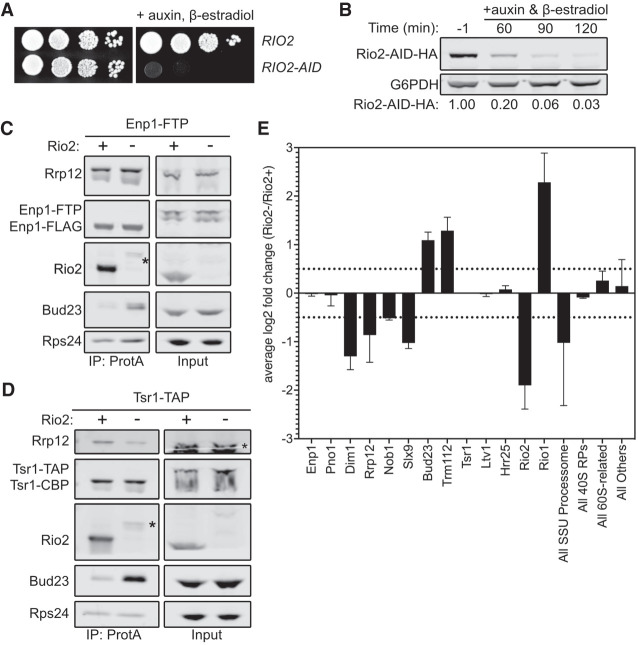
The ability of the Rps0-cluster proteins and Bud23 to coexist on the pre-40S particle (Fig. 4) suggests that a step downstream from the loading of the Rps0-cluster proteins releases Bud23. The Rps0-cluster proteins were recently reported to promote the recruitment of the ATPase Rio2 to the pre-40S (Linnemann et al. 2019). Notably, Rio2 and Bud23 share an overlapping binding site within the head of the pre-40S particle (Fig. 1; Ameismeier et al. 2018). Therefore, a simple model is one in which the Rps0-cluster proteins recruit Rio2 to the pre-40S particle to actively displace Bud23. Alternatively, it is possible that the Rps0-cluster proteins promote Rio2 binding by inducing the release of Bud23. However, we did not observe any Rps0-, Rps2-, or Rps21-dependent loss of Rio2 in the affinity-purified Enp1–FTP and Tsr1–TAP (Fig. 3B–D). The reason for this discrepancy between our results and those of the previous study (Linnemann et al. 2019) is not clear, but it may be attributable to the different stringencies of buffers used for affinity purification or different lengths of time of RP-depletion (Linnemann et al. 2019). Nevertheless, because Rio2 and Bud23 share a binding site (Fig. 1), we decided to explore the possibility that Rio2 is needed for Bud23 release. RIO2 is an essential gene, and the depletion of native Rio2 is slow after transcriptional repression (Vanrobays et al. 2003). To generate a more acute depletion, we generated RIO2–AID strains containing ENP1–FTP or TSR1–TAP to test if Rio2 promotes Bud23 release. As anticipated, cells harboring RIO2–AID were unable to grow on media containing β-estradiol and auxin but grew like wild-type on media lacking them (Fig. 5A). As with the other AID-tagged proteins, Rio2–AID was nearly undetectable after 2 h of treatment with β-estradiol and auxin (Fig. 5B). Affinity purification of SSU precursors using either Enp1–FTP and Tsr1–TAP as baits from wild-type cells or cells in which Rio2–AID was depleted for 2 h showed that, like the Rps0-cluster proteins, Bud23 increased on particles depleted of Rio2 (Fig. 5C,D). These data implicate Rio2 in the release of Bud23.
FIGURE 5.
Rio2 is needed for Bud23 release from pre-40S particles. (A) The growth phenotypes of the wild-type (AJY4724) or RIO2-AID (AJY4733) strains as shown by 10-fold serial dilutions of cells on YPD media with or without 0.5 mM auxin and 1 µM β-estradiol. (B) Western blot of time-course of the depletion of Rio2-AID-HA using equivalent amounts of total protein from AJY4733 cells cultured to exponential phase, then collected prior to or after the addition of 0.5 mM auxin and 1 µM β-estradiol for the indicated time points. G6PDH was used as the loading control. The ratio Rio2-AID-HA signal to G6PDH signal for each time point relative to that of the untreated time point (−1) is shown below. (C,D) Bud23 accumulated in Rio2-depleted particles as shown by western blotting for specific factors on pre-40S particles that copurify with Enp1–FTP (C) and Tsr1–TAP (D). Asterisks (*) denote residual Rio2-AID-HA signal that is detected by the anti-Rio2 antibody. For C, strains AJY4724 (Rio2+) and AJY4733 (Rio2−) were cultured in YPD to early exponential phase, then treated with 0.5 mM auxin and 1 µM β-estradiol to deplete Rio2-AID-HA. For D, strains AJY4754 (Rio2+) and AJY4757 (Rio2−) were similarly cultured in YPD. (E) Proteomic composition of Tsr1-associated preribosomal particles as measured by semiquantitative mass spectrometry is shown as the average log2 fold change (Rio2−/Rio2+ samples) for two independent biological replicates. Error bars denote the standard deviation. Individual factors are ordered by the timing of their approximate known association with 40S precursors. The log2 fold change values were calculated as described in the Materials and Methods section. The supporting data are provided in Supplemental File 2.
To further interrogate the composition of the Rio2-depleted particles, we performed semiquantitative mass spectrometry analysis on pre-40S particles purified via Tsr1–TAP. As done with the Rps2-depleted particles (Fig. 3D), we compared the log2 fold change values of Rio2-depleted versus wild-type particles to determine the effect that Rio2-depletion has on pre-40S particles. This revealed that both Bud23 and its binding partner Trm112 increased in abundance upon Rio2-depletion in biological duplicate experiments (Fig. 5E; Supplemental File 2). The Rio2-related ATPase Rio1 also accumulated in the Rio2-depleted sample while Dim1, Rrp12, and Slx9 all strongly decreased in response to Rio2-depletion. This reduction of Rrp12 is consistent with western blots of the Tsr1–TAP particles that showed Rrp12 decreases in abundance (Fig. 5D). Furthermore, the levels of Rrp12 did not change in a Rio2-dependent manner in the Enp1–FTP samples indicating that Rio2-depletion does not induce Rrp12 accumulation (Fig. 5C). This is notable as the depletion of the Rps0-cluster proteins or Tsr4 consistently showed a concurrent increase of both Bud23 and Rrp12 (Fig. 3A,C,D; Supplemental Figs. S4, S5D). Thus, Rio2-depletion appears to uncouple Bud23 accumulation from Rrp12 accumulation and indicates that the depletion of the Rps0-cluster proteins and Rio2 have different effects on pre-40S composition. These differences became more apparent when the mass spectrometry data for the Rps2- and Rio2-depleted pre-40S particles were directly compared (Supplemental Fig. S8). This comparison showed that Bud23 and Trm112 are the only proteins that increase in response to the loss of both Rps2 and Rio2. In contrast, the depletion of Rps2 and Rio2 had differing effects on Rrp12 and Slx9 which accumulated upon loss of Rps2 but were reduced upon loss of Rio2. Notably, Bud23 and Trm112 accumulated on ∼20% of the Rio2-depleted particles (Supplemental File 2), again indicating that it is present in substoichiometric amounts and suggesting that there is a heterogenous population of particles that accumulate in the absence of Rio2. These data further support the notion that Rio2 is needed for the release of Bud23 and Trm112 and indicate that the release of Rrp12 and Slx9 is independent of Bud23 release.
The binding of Rio2 displaces Bud23 from the pre-40S
The above data indicate that Rio2 is needed for the release of Bud23. Because these two proteins share binding sites (Fig. 1), we hypothesized that Rio2 actively displaces Bud23 from the pre-40S. To test this hypothesis, we developed an in vitro release assay in which we asked whether the addition of recombinant Rio2 (Rio2-HA) could release Bud23 from purified particles. To this end, we affinity-purified pre-40S particles from Rio2-depleted cells using Tsr1–TAP as bait. Following enzymatic elution, the particles were incubated with buffer or excess recombinant Rio2. The reactions were then fractionated on a sucrose cushion by ultracentrifugation to separate extraribosomal proteins from pre-40S-bound proteins and analyzed by SDS-PAGE and western blotting. In the reaction lacking Rio2, most of the Bud23 signal cosedimented with pre-40S particles (Fig. 6A; Supplemental Fig. S9A; lanes 1,2). Strikingly, the majority of Bud23 signal shifted to the free protein fraction upon the addition of recombinant Rio2 (Fig. 6A; Supplemental Fig. S9A; compare lanes 1/2 to 3/4). This change was concurrent with the cosedimentation of Rio2 with the pre-40S particles indicating that the presence of Rio2 can directly induce Bud23 release. Importantly, the binding of recombinant Rio2 appeared to be stoichiometric with pre-40S as evident by the comparable intensities of Coomassie staining with respect to proteins of similar molecular weights. Rio2 is an atypical kinase whose ATPase activity promotes autophosphorylation that induces its own release (Ferreira-Cerca et al. 2012), however it is not clear if its enzymatic activity is needed for Bud23 release. To this end we also compared the effects of the addition of recombinant Rio2 without any nucleotide (Fig. 6A; Supplemental Fig. S9A; compare lanes 3,4) to Rio2 preincubated with either excess ATP (Fig. 6A; Supplemental Fig. S9A; compare lanes 5,6) or the nonhydrolysable analog AMP–PNP (Fig. 6A; Supplemental Fig. S9A; compare lanes 7,8). This analysis revealed that recombinant Rio2 binds the purified pre-40S particles and induced Bud23 release independent of its nucleotide-binding status. Interestingly, we noticed that the addition of recombinant Rio2 preincubated with ATP, but not with AMP–PNP or without nucleotide, promoted the phosphorylation and release of Ltv1 (Fig. 6A; compare lanes 5/6 to 3/4 and 7/8). This is consistent with recent reports that found Rio2 is needed for the Hrr25-driven phosphorylation and release of Ltv1 (Mitterer et al. 2019; Huang et al. 2020). Together, these in vitro data strongly suggest that the binding of Rio2, and not its enzymatic activity, displaces Bud23 from the pre-40S.
FIGURE 6.
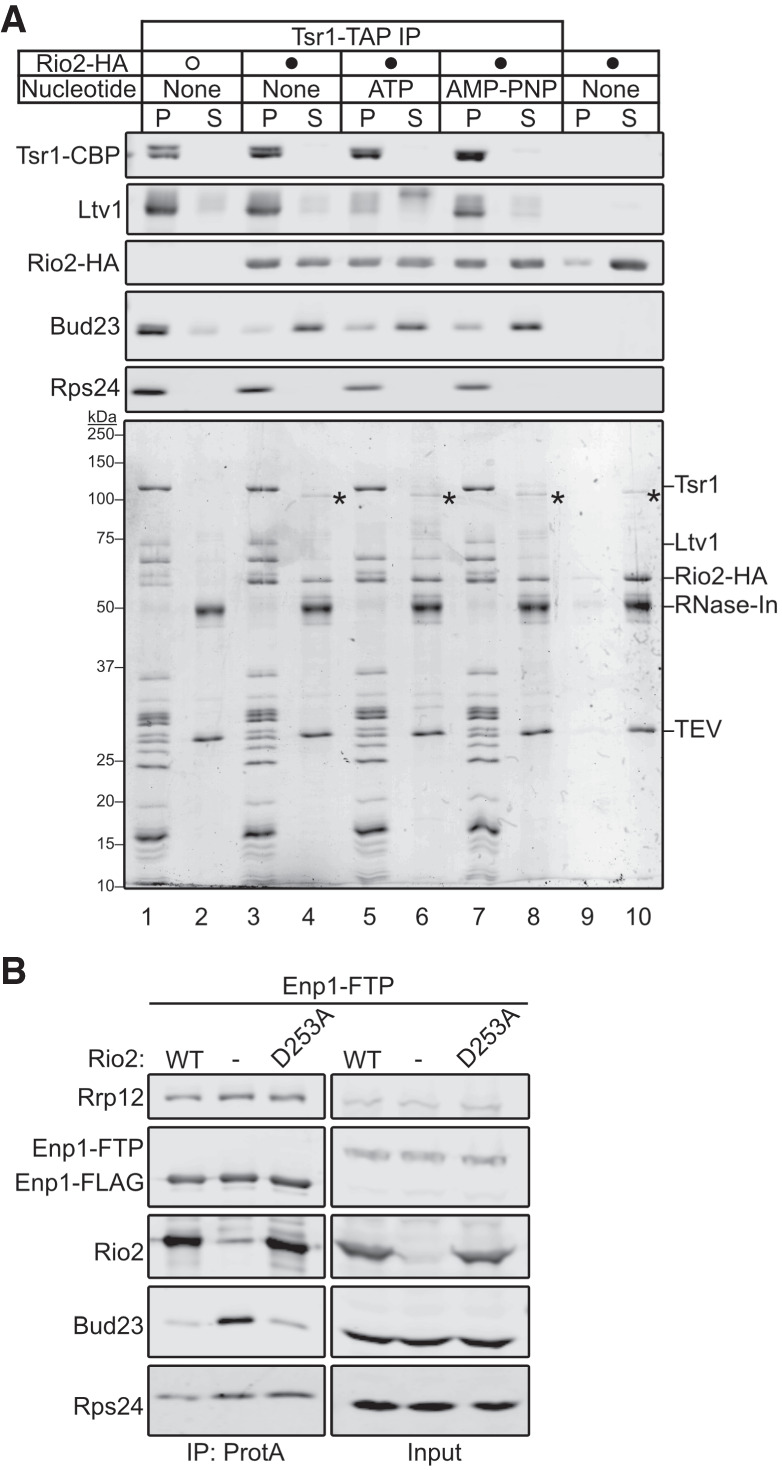
The binding of Rio2 displaces Bud23 from pre-40S particles. (A) The addition of recombinant Rio2 (Rio2-HA) to affinity-purified pre-40S particles depleted of Rio2 induces the release of Bud23 as shown by western blotting for specific factors on pre-40S particles that copurified with Tsr1–TAP. The Coomassie-stained gel is shown for reference. The absence or presence of Rio2-HA is indicated by a hollow or filled circle, respectively. The reactions were incubated for 10 min at 20°C and performed in the absence or presence of 0.5 mM ATP and AMP–PNP. The reactions were subsequently overlayed on sucrose cushions and subjected to ultracentrifugation. The pellet (P) and supernatant (S) fractions, respectively, containing preribosome-bound and extraribosomal proteins, were collected, TCA precipitated, and separated on SDS-PAGE gels. See the Materials and Methods section for a full description of experimental details. In the Coomassie stained gel, asterisks (*) denote residual uncleaved recombinant MBP-Rio2-HA that remain in the supernatant fraction. (B) Bud23 does not accumulate on preribosomes containing Rio2–D253A as shown by western blotting for specific factors on pre-40S particles that copurify with Enp1–FTP. Strain AJY4733 was transformed with an empty vector (pAJ5103) or vectors encoding RIO2 (pAJ4657) or rio2–D253A (pAJ4698). Cells were cultured in SD Leu- media to early exponential phase, then cultured for 2 h in the presence of 0.5 mM auxin and 1 µM β-estradiol.
To test the notion that the binding of Rio2 promotes Bud23 release in vivo, we purified Enp1–FTP particles from Rio2-depleted cells harboring an empty vector or a vector expressing either wild-type Rio2 or the catalytically inactive Rio2 mutant, Rio2–D253A. D253 of Rio2 binds to the magnesium ion that coordinates the gamma phosphate of ATP and becomes transiently phosphorylated during ATP hydrolysis (Ferreira-Cerca et al. 2012). Importantly, Rio2–D253A is a mutant that retains its ability to bind pre-40S particles (Ferreira-Cerca et al. 2012). This experiment showed that Bud23 accumulated in the pre-40S particles lacking Rio2 but not in those containing Rio2–D253A (Fig. 6B) further suggesting that the binding of Rio2 to the pre-40S induces the release of Bud23. We also asked whether Rio2 and Bud23 could coexist on particles. For this, we affinity-purified Rio2–TAP particles and probed for the copurification of Bud23. This approach showed that Rio2–TAP did not copurify Bud23 (Supplemental Fig. S10A). Moreover, we found that Rps2-depletion did not enable Bud23 to coprecipitate with Rio2–TAP, suggesting that the loss of Rps2 does not stabilize Bud23 on Rio2–TAP particles. Notably, we also did not see Rio2–TAP decrease in its association with pre-40S particles upon Rps2-depletion as was reported (Linnemann et al. 2019) but consistent with our previous results showing that Rps2-depletion did not affect Rio2 recruitment (Fig. 3B–D). We also performed the inverse experiment and saw that neither amino-terminally tagged Protein A-TEV-His Bud23 (PTH-Bud23) nor carboxy-terminally tagged Bud23–FTP copurified Rio2 (Supplemental Fig. S10B). Thus, Bud23 and Rio2 binding to pre-40S particles appear to be mutually exclusive. Taken together, these in vivo and in vitro data support the conclusion that the binding of Rio2 displaces Bud23 from the pre-40S.
DISCUSSION
Bud23 is a conserved methyltransferase that modifies G1575 of 18S rRNA in yeast (G1539 in humans) during 40S biogenesis (White et al. 2008; Figaro et al. 2012; Létoquart et al. 2014; Zorbas et al. 2015). We previously proposed that Bud23 binds to a partially disassembled Processome where it coordinates the activities of the GTPase Bms1 and the RNA helicase Dhr1 to ensure the productive folding of the central pseudoknot (CPK) as the nascent particle transitions into the pre-40S (Black et al. 2020; Black and Johnson 2021). Consistent with the role of Bud23 in CPK formation, we found here that the overexpression of Rps2, which binds to the CPK, partially bypassed the defects caused by bud23Δ (Fig. 2A; Supplemental Fig. S2). We also showed that the binding of the Rps0-cluster proteins (Fig. 3) and Rio2 (Figs. 5, 6) promote the release of Bud23 from pre-40S, an event that could only be inferred from the structures of human intermediates (Fig. 1; Ameismeier et al. 2018). Based on our results, we propose a revised timeline for when Bud23, and by extension Trm112, associate with and are released from preribosomes (Fig. 7). Bud23 and Trm112 are recruited to a partially disassembled Processome during the transition to a pre-40S. Subsequently, the Rps0-cluster proteins, Rps0, Rps2, and Rps21 are recruited followed by the release of Rrp12 and Slx9. Bud23 and Trm112 are then displaced by Rio2, whose binding site in the P-site overlaps that of Bud23. The ability of Rio2 to release Bud23 was independent of its nucleotide-binding status (Fig. 6) indicating that it is the binding of Rio2, rather than its ATPase activity, that drives Bud23 release. Surprisingly, despite the finding that Rio2 binding displaces Bud23 from the pre-40S (Figs. 5, 6), we repeatedly found that Rio2 was present in the Rps0-cluster-depleted particles in which Bud23 accumulated (Fig. 3B–D). However, Bud23 and Rio2 did not copurify one another (Supplemental Fig. S10). We suggest these results can be explained by particle heterogeneity (Supplemental Files 1, 2). Bud23 appears to be a limiting factor and is not essential in 40S assembly. Its accumulation on particles may lead to stalling of those particles while additional particles continue to be assembled in the absence of Bud23, allowing Rio2 to join these new particles lacking Bud23. Nevertheless, we speculate that Bud23 release is controlled by a mechanism that requires the binding of both the Rps0-cluster proteins and Rio2.
FIGURE 7.
A proposed pathway for events surrounding the binding and release of Bud23 from 40S precursors. A revised pathway for the incorporation and release of factors is shown. Late assembling Processome factors, including Enp1, Rrp12, Slx9, Bms1, Dhr1, and possibly Nob1 complete the Processome. The transition of the Processome to pre-40S requires the enzymatic activities of Dhr1 and Bms1, which work in concert with the recruitment of Bud23–Trm112. During this transformation, the remaining Processome factors are released and Tsr1 and Ltv1 are recruited. Next, the Rps0-cluster proteins incorporate into the pre-40S and coexist with Bud23 and Trm112 on the pre-40S. Continued maturation of the pre-40S particle allows the release of Rrp12 and Slx9. The binding of Rio2 to the nascent P-site of the pre-40S actively displaces Bud23 and Trm112, and the nascent 40S continues down its biogenesis pathway.
We found that the binding of Rps0-cluster proteins is necessary for the release of Bud23 (Fig. 3; Supplemental Figs. S4, S5D). The structures of human pre-40S intermediates suggest that the binding of the Rps0-cluster proteins is mutually exclusive with Bud23 (Fig. 1; Ameismeier et al. 2018). However, we provide evidence that Bud23 coexists with the Rps0-cluster proteins on pre-40S particles in yeast (Fig. 4), indicating that Bud23 is released downstream from their incorporation. We also found that Rrp12 release was independent of Bud23 release (Supplemental Fig. S8), counter to our prevailing understanding of the pathway from these structural analyses (Fig. 1; Ameismeier et al. 2018). These differences in the order of events surrounding Bud23 release may be explained by differences in the assembly pathways between yeast and human as is thought for other steps in the 40S pathway (Nieto et al. 2020; Ameismeier et al. 2020; Plassart et al. 2021). Alternatively, the differences might be attributable to the limitations of the techniques used or the use of mutants to arrest intermediates versus isolation of the most stable intermediates from wild-type cells. It is important to note that resolving a protein on a preribosomal intermediate by cryo-EM requires low conformational heterogeneity of the factor whereas western blotting and mass spectrometry can detect proteins despite structural heterogeneity. Additional work is needed to determine if our proposed pathway for Bud23 release (Fig. 7) is conserved from yeast to humans.
As Rio2 and Bud23 have overlapping binding sites at the P-site (Supplemental Fig. S1C), it is straightforward to rationalize how Rio2 releases Bud23; however, it is not obvious how the Rps0-cluster proteins promote its release. The structures of human pre-40S intermediates show that significant architectural rearrangements occur during the transition from States A to B (Ameismeier et al. 2018) that enable maturation of the neck region, composed of the CPK and the helices 28 and 35–37. In State A, h37 is positioned about 79 Å away from its mature position where Rrp12 embraces it (Supplemental Fig. S1C). The observation that h37, but not h35 and h36, was resolved in this state is consistent with a study of early pre-40S intermediates from yeast showing that these helices are highly flexible (Hector et al. 2014). In State B, h37 has been released from Rrp12 and has moved into its mature position, and h35, h36, and the Rps0-cluster proteins become resolved (Supplemental Fig. S1C). Here, h35 and h36 interact with Rps0, while h36 also interacts with the minor groove of the CPK. Furthermore, Rps2 binding to both h36 and the CPK completes the maturation of the neck. The interactions between these helices and Rps0-cluster proteins may stabilize these RNA rearrangements, as studies of bacterial SSU intermediates show a correlation between the flexibility of h35–37 and reduced levels of S2 and S5, the bacterial counterparts of Rps0 and Rps2, respectively (Clatterbuck Soper et al. 2013; Sashital et al. 2014; Sharma et al. 2018). However, it is unclear whether the repositioning of h35–37 is actively induced by the binding of Rps0 and Rps2 or if the RPs bind after h35–37 remodeling has occurred. Nevertheless, we speculate that the release of Bud23 depends on the repositioning of h35–37, functionally connecting the release of Bud23 to the maturation of the neck of the SSU. Interestingly, we saw that the depletion of each of the Rps0-cluster proteins had a differential effect on the extent of Bud23 accumulation (Fig. 3B; Supplemental Fig. S4), suggesting that these proteins may impact the pre-40S differently. Consistent with this notion, RPS2—but not RPS0B nor RPS21A—suppressed the growth defect of bud23Δ (Supplemental Fig. S3). Therefore, it may be that these three proteins may not load simultaneously as originally suggested by the structural observations (Ameismeier et al. 2018). Future investigation into how the Rps0-cluster proteins associate with the pre-40S may shed light on how they influence the rearrangements of h35–37 and the mechanism by which they promote the release of Bud23.
MATERIALS AND METHODS
Strains, plasmids, and growth media
The S. cerevisiae strains used in this study and their sources are listed in Table 1. Detailed descriptions of how each strain was constructed are provided in Supplemental Methods 1. All yeast were cultured at 30°C in either YPD (2% peptone, 1% yeast extract, 2% dextrose), YPGal (2% peptone, 1% yeast extract, 1% galactose), or synthetic dropout (SD) medium containing 2% dextrose unless otherwise noted. Solid media contained 2% agar. When appropriate, media were supplemented with 150 to 250 µg/mL G418 or 100 µg/mL nourseothricin. The plasmids used in this study are listed in Table 2.
TABLE 1.
Strains used in this study
TABLE 2.
Plasmids used in this study
Screen for high-copy suppressors of bud23Δ
The bud23Δ strain, AJY2161, was transformed with a 2µ URA3 plasmid-based genomic library (Connelly and Hieter 1996). Transformed cells were plated on SD Ura- agar media and grown at 30°C and plasmids were isolated from colonies that grew faster than the background. The overwhelming majority of these plasmids contained BUD23 (data not shown), but two independently isolated plasmids contained genomic regions other than BUD23 (pAJ1652 and pAJ1653).
Quantitative growth assay
To measure the doubling times of cells, strains BY4741 and AJY2676 were each transformed with pRS415 and pAJ2960. Two independent cultures of each genetic background were first grown to saturation in SD Leu- media at 30°C, then diluted to an OD600nm of 0.03 in fresh SD Leu- media and grown in 200 µL micro-cultures in a 48-well plate with continuous shaking at 30°C on a BioTek Synergy HTX microplate reader. OD600nm was measured every 10 min for 48 h. The slopes of the growth curves were calculated as described (www.github.com/decarpen/growth-curves), and doubling times were calculated as Ln(2)/slope (Toussaint and Conconi 2006).
Affinity purification of preribosomes
For the Bud23-AID depletion experiments, cells were grown in 500 mL YPD or SD Ura- media until early exponential phase, then treated with 0.5 mM auxin for 10 min. For the Rps0-, Rps2-, and Rps21-depletion experiments cells were grown in 500 mL or 1 L of YPGal media until early exponential phase, then cultured for 2 h following the addition of 2% glucose. For the Tsr4–AID, Rio2–AID, Ltv1–AID, and Nob1–AID depletion experiments, cells were grown in 500 mL or 1 L YPD or SD Leu- until early exponential phase, then treated with 0.5 mM auxin and 1 µM β-estradiol for 2 h to deplete Tsr4–AID, Rio2–AID, and Ltv1–AID and 1.5 h to deplete Nob1–AID. Cells were harvested by centrifugation at 4°C, frozen in liquid nitrogen, and stored at −80°C. All subsequent steps were performed on ice or at 4°C.
For the affinity purification of preribosomes using various Protein A-tagged biogenesis factors as baits, cell pellets were thawed and washed with Lysis Buffer (50 mM Tris-HCl pH 7.5/7.6 (25°C), 100 mM KCl, 5 mM MgCl2, 5 mM β-mercaptoethanol (βME), 1 mM PMSF and benzamidine, and 1 µM leupeptin and pepstatin) supplemented with EDTA-free Pierce Protease Inhibitor Mini Tablet cocktail (Thermo Scientific). Cells were then resuspended in 1–2 volumes of Lysis Buffer. Extracts were generated by glass bead lysis, separated from the glass beads by centrifugation through a 10 mL column (Pierce) at 1000g for 2 min, and then clarified by centrifugation at 18,000g for 10 min. Clarified extracts were normalized according to absorbance at 260 nm (A260nm) and supplemented with 0.03% TritonX-100. Normalized extracts were incubated for 1–1.5 h with 3.75 mg of Dynabeads (Invitrogen) coupled with rabbit IgG (Sigma), prepared as previously described (Oeffinger et al. 2007). After binding, the beads were washed thrice with Wash Buffer (Lysis Buffer supplemented with 0.03% TritonX-100). The beads were then resuspended in Wash Buffer supplemented with TEV protease and Murine RNase Inhibitor (New England Biolabs), and the bait-associated complexes were eluted for 1–1.5 h. The resultant eluates were overlaid onto sucrose cushions (15% sucrose, 50 mM Tris-HCl pH 7.6 (25°C), 100 mM KCl, 5 mM MgCl2), then centrifuged at 70,000 rpm for 15 min in a Beckman Coulter TLA100 rotor to isolate factors associated with preribosomal particles. The resultant pellets were collected, and the associated proteins were precipitated with 15% trichloroacetic acid (TCA) to remove the sucrose, then resuspended in 1× Laemmli buffer. Equivalent volumes or approximately equivalent amounts of isolated proteins were either separated on 6%–18% SDS-PAGE gels for western blotting (see below) and/or prepared for mass spectrometric analysis (see below).
Immunoprecipitation of GFP-tagged factors
Strains were cultured to mid-exponential phase in 500 mL YPD. Cells were harvested as in the above section. All subsequent steps were performed on ice or at 4°C. For immunoprecipitation, cell pellets were thawed and washed with Lysis Buffer (50 mM Tris-HCl pH 7.6 (25°C), 100 mM KCl, 5 mM MgCl2, 5 mM βME, 1 mM PMSF and benzamidine, and 1 µM leupeptin and pepstatin) supplemented with EDTA-free Pierce Protease Inhibitor Mini Tablet cocktail (Thermo Scientific). Cells were then resuspended in approximately 1.5 volumes and broken by glass bead lysis, and extracts were clarified by centrifugation at 18,000g for 10–15 min. Extracts were normalized according to A260nm in a final volume of 500 µL and supplemented with 0.03% TritonX-100. An amount of 2 µL of rabbit anti-GFP (M.P. Rout) was added to each normalized extract and rotated for 60–80 min. Each sample was subsequently rotated with 30 µL of Protein G-coupled Dynabeads (Invitrogen) for 30 min. The beads were then washed thrice with Lysis Buffer supplemented with 0.03% TritonX-100. Beads were resuspended in 1× Laemlli buffer and proteins were eluted by heating at 99°C for 3 min. Proteins were then separated on 6%–18% SDS-PAGE gels and specific factors were detected by western blotting analysis (see below).
Western blotting analysis
Primary rabbit antibodies used in this study were anti-calmodulin binding peptide (CBP; Millipore), anti-Glucose-6-phosphate dehydrogenase (G6PDH; Sigma ImmunoChemicals), anti-Rps24 (our laboratory), anti-Bud23 (C. Wang), anti-Rrp12 (M. Dosil), anti-Rps0 (L. S. Valášek), anti-Rpl8 (K.-Y. Lo), anti-Tsr1, anti-Rio2, anti-Pno1, and anti-Ltv1 (K. Karbstein). Other primary antibodies used in this study were guinea pig anti-Imp3 and anti-Imp4 (S. Baserga), rat anti-FLAG (Agilent), mouse anti-GFP (Invitrogen) and anti-HA (Biolegend), and goat anti-GFP-HRP (Rockland). Secondary antibodies were goat anti-mouse antibody-IRDye 800CW, anti-rabbit antibody-IRDye 680RD, anti-guinea pig antibody-IRDye 800CW, and anti-rat antibody-IRDye 800CW; all secondary antibodies were from Li-Cor Biosciences. All blots except the anti-GFP blot in Figure 5A were imaged with an Odyssey CLx infrared imaging system (Li-Cor Biosciences) using Image Studio (Li-Cor Biosciences). The anti-GFP blot in Figure 6A was imaged using SuperSignal West Pico PLUS Chemiluminescent Substrate (Thermo Scientific) and imaged on film.
Mass spectrometry and analysis
Proteins were electrophoresed ∼5 mm into NuPAGE Novex 4%–12% Bis-Tris gels followed by in-gel Trypsin digestion and preparation for mass spectrometry as previously described (Black et al. 2018). Protein identification was done by LC-MS/MS on a Thermo Orbitrap Fusion with a 2-h run time (the University of Texas at Austin Center for Biomedical Research Support Biological Mass Spectrometry Facility [RRID:SCR_021728]). The resultant data were processed in Scaffold v5.0.0 (Proteome Software, Inc.). A protein threshold of 99% minimum with two peptides minimum and peptide threshold of 1% false discovery rate were applied for total spectral counts. The data were exported, and bespoke Python 2.7 scripts were used to calculate a peptides per molecular weight (PPMW) factor for each protein as previously described (Black et al. 2018). The PPMW for each protein was normalized to that of the bait, Tsr1, to generate the relative stoichiometry against bait factor (RSAB). The log2 fold change (log2FC) for each protein was then manually calculated by comparing the RSAB for each protein in the Rio2- or Rps2-depleted sample to that of the wild-type sample in Microsoft Excel. For the log2FC calculation, all zero RSAB values were replaced with an RSAB value of 0.001. Graphs and heatmaps were generated in GraphPad Prism 9 for Mac iOS (www.graphpad.com). Supplemental Files 1 and 2 contain relevant spectral counts and processed data from the mass spectrometry experiments.
Sucrose density gradient analysis
Strain AJY2676 was transformed with plasmids pAJ2154, pAJ2960, and pRS315 and grown in 200 mL of SD Leu- media until mid-exponential phase. Cycloheximide (CHX) was added to a final concentration of 150 µg/mL, and the cultures were shaken for 10 min at 30°C. The cells were then poured over ice, collected by centrifugation, frozen in liquid nitrogen, and stored at −80°C. Sucrose density gradients were performed as previously described (Black et al. 2020), except the Lysis Buffer consisted of 50 mM Tris-HCl pH 7.6 (25°C), 100 mM KCl, 5 mM MgCl2, 150 µg/ml CHX, 5 mM βME, 1 mM PMSF and benzamidine, and 1µM leupeptin and pepstatin.
Expression and purification of recombinant Rio2-HA
Yeast MBP-10xHis-TEV-Rio2-HA was purified from BL21-CodonPlus (DE3)-RIL Escherichia coli cells (Stratagene) transformed with pAJ4720. An amount of 1 L of bacterial culture was grown at 37°C to an OD600nm of 0.5 and induced with 1 mM IPTG for 3 h at 30°C. Cells were harvested and washed with Lysis Buffer (40 mM Tris pH 8.0, 500 mM NaCl, 10% glycerol). Cells were frozen in liquid nitrogen and stored at −80°C. All subsequent steps were carried out on ice or at 4°C. The cell pellet was resuspended in 40 mL of Lysis Buffer supplemented with 10 mM imidazole, and protease inhibitors (1 mM PMSF and benzamidine, and 1 µM leupeptin and pepstatin), and disrupted by sonication. Lysate was clarified by centrifugation at 20,000g for 20 min. The clarified lysate was supplemented with 5 mM βME, then bound to a 1 mL Ni-NTA column (HisTrap HP, GE Healthcare). The column was first washed with 20 mL Lysis Buffer supplemented with 10 mM imidazole, protease inhibitors, and 5 mM βME and then with 10 mL Lysis Buffer supplemented with 25 mM imidazole, protease inhibitors, and 5 mM βME. Bound protein was eluted from the column with Lysis Buffer supplemented with 250 mM imidazole, protease inhibitors, and 5 mM βME. Fractions containing MBP-10xHis-TEV-Rio2-HA were pooled, supplemented with TEV protease, and dialyzed overnight in Lysis Buffer supplemented with 10 mM imidazole. The dialyzed protein was rotated with 100 µL Ni-NTA slurry (Invitrogen) for 1 h to separate Rio2-HA from the MBP-10xHis tag and uncleaved protein. The resultant flow-through containing Rio2-HA was recovered, aliquoted, and stored at −80°C.
In vitro Bud23 release assays
Two liters of strain AJY4757, expressing Tsr1–TAP, were grown in YPD to early exponential phase, then treated with 0.5 mM auxin and 1 µM β-estradiol for 2 h. Cells were collected by centrifugation, split evenly four ways, frozen in liquid nitrogen, and stored at −80°C. All subsequent steps were performed on ice or at 4°C, unless otherwise noted. A cell pellet was thawed and washed with 2 mL of Lysis Buffer (50 mM Tris-HCl pH 7.6 [25°C], 100 mM KCl, 5 mM MgCl2, 5 mM β-mercaptoethanol [βME], 1 mM PMSF and benzamidine, and 1 µM leupeptin and pepstatin) supplemented with EDTA-free Pierce Protease Inhibitor Mini Tablet cocktail (Thermo Scientific). Cells were resuspended in 1.5 volumes of Lysis Buffer and broken by glass bead lysis. The extract was clarified by centrifugation at 20,000g for 10 min. Clarified extract (500 µL) was supplemented with 0.03% TritonX-100 and incubated with 7.5 mg of Dynabeads (Invitrogen) coupled to rabbit IgG (Sigma), prepared as previously described (Oeffinger et al. 2007), for 1 h. Beads were washed thrice with Wash Buffer (Lysis Buffer supplemented with 0.03% TritonX-100) and resuspended in 250 µL Elution Buffer (Wash Buffer supplemented with homemade TEV and Murine RNase Inhibitor [New England Biolabs]). Preribosomal complexes were eluted from the beads for 70 min. The concentration of pre-40S complexes was estimated by comparison with a titration of highly purified 40S subunits by SDS-PAGE and Coomassie staining.
To assay in vitro release of Bud23, Tsr1–TAP eluate was mixed with a solution with or without Rio2-HA that was preincubated on ice for 10 min with either 1 mM ATP or AMP–PNP or no nucleotide. The final concentrations were ∼40 nM pre-40S, 100 nM Rio2-HA, and 500 nM nucleotide in a final volume of 100 µL. The reactions were then incubated at 20°C for 10 min, then placed on ice for 5 min. An amount of 95µL of each reaction was then overlayed onto a 50 µL sucrose cushion (15% sucrose, 50 mM Tris-HCl pH 7.6 (25°C), 100 mM KCl, 5 mM MgCl2), then centrifuged at 70,000 rpm for 15 min in a Beckman Coulter TLA100 rotor to separate pre-40S-bound factors from extraribosomal factors. The top 105 µL were collected as the supernatant fraction and remaining 40 µL were collected as the pellet fraction. The fractions were precipitated with 15% TCA, washed with 100% acetone, dried, and resuspended 1× Laemmli buffer. Half of each sample was separated on 6%–18% SDS-PAGE gels for western blotting analysis (see above), and a fourth of each sample was separated for Coomassie staining. Two independent replicates were performed using different pellets from the same cell culture.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Yelland and R. Lin for commenting on the manuscript, and we further thank J. Yelland for his assistance with cloning pAJ4392 and pAJ4686. We also thank P. Milkereit for the generous gift of the yeast strains harboring conditional RPS genes, especially strains ToY256, ToY286, ToY327, and Y801; D. Lycan for the yeast strain LY193; P. Hieter for the yeast genomic 2µ URA3 plasmid library; and M. Dosil, L.S. Valášek, S. Baserga, K. Karbstein, M. P. Rout, K.-Y. Lo, and C. Wang for sharing the antibodies listed above. This work was supported by the National Institutes of Health grants GM127127 and GM108823 to A.W.J. and a fellowship from the University of Texas at Austin (www.utexas.edu) Graduate School to J.J.B.
Author contributions: J.J.B. and A.W.J. designed the study. A.W.J. screened for ectopic suppressors of bud23Δ, and J.J.B. designed and performed all other experiments and analyses. J.J.B. and A.W.J. interpreted the results. J.J.B. wrote the first draft of the manuscript, and A.W.J. edited it.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.079025.121.
MEET THE FIRST AUTHOR
Joshua J. Black.

Meet the First Author(s) is a new editorial feature within RNA, in which the first author(s) of research-based papers in each issue have the opportunity to introduce themselves and their work to readers of RNA and the RNA research community. Joshua Black is the first author of this paper, “Release of the ribosome biogenesis factor Bud23 from small subunit precursors in yeast.” Joshua obtained his PhD from the University of Texas at Austin, working in Arlen Johnson's laboratory on how the small subunit of the eukaryotic ribosome is assembled, specifically focusing on several biogenesis factors in yeast that promote the transition of an early preribosomal intermediate, called the SSU processome, into the next intermediate, known as the pre-40S.
What are the major results described in your paper and how do they impact this branch of the field?
I recently proposed that the binding of the methyltransferase Bud23 to the SSU processome promotes structural rearrangements within the nascent particle to help generate the pre-40S, but how Bud23 is released from the resultant pre-40S remained an outstanding question. Notably, several ribosomal proteins and assembly factors were thought to bind around the time that Bud23 is released. From this, I hypothesized that one or more of these factors promotes Bud23 release, which led me to systematically test whether the depletion of such factors could trap Bud23 on pre-40S particles. I found that three ribosomal proteins—Rps0, Rps21, and Rps2—and the ATPase/kinase Rio2 contribute to Bud23 release. Most notably, Bud23 and Rio2 share binding sites at the P-site of the nascent 40S, and my data indicate that the binding of Rio2 actively displaces Bud23 from the pre-40S and ascribes a novel function to the long-studied Rio2. I also speculate that the binding of Rps0, Rps21, and Rps2 induce Bud23 release by promoting rRNA rearrangements; however, this will require further investigation. I think this study nicely demonstrates how the coming and going of the biogenesis factors promote the forward progression of ribosome assembly.
What led you to study RNA or this aspect of RNA science?
I think what sparked my interest in ribosome biogenesis was an appreciation that ribosomes are molecular machines that are gradually built in an assembly line–like pathway. The recent publication of numerous preribosomal structures has really brought this to life, allowing one to envision how biogenesis factors successively promote the formation of the mature ribosomal architecture. Most excitingly, these structures offer the context for understanding the plethora of existing genetic and biochemical information as well as the potential for generating new hypotheses like in my current paper. Together, this information has allowed the field to develop elegant molecular models to explain the ribosome assembly.
What are some of the landmark moments that provoked your interest in science or your development as a scientist?
My path to a science career has not been linear. Prior to college, my science classes felt like a series of static facts that one simply absorbs by rote memorization. However, early in college, I happened to read some of the books written by the late astronomer/science communicator Carl Sagan. His elegant and often poetic writing about the philosophy of science made me realize that science is an extremely active process that is used to understand how the universe works. Sagan's writing kindled my interest in science, and I quickly set chemistry as my college major as this major's degree plan allowed the most flexibility to explore the various scientific disciplines. From here, I found my biology courses to be the most exciting, which led me to some undergraduate research experiences in molecular biology. These experiences sparked my interest in biological research, leading me apply to molecular biology PhD programs. From here, I took advantage of the rotations to find the right mentor and field of study. I think what I would like to convey is that one's trajectory does not have to be fully planned if one has the privilege to freely explore their opportunities.
What are your subsequent near- or long-term career plans?
For the near term, I just wrapped up my PhD training and accepted a postdoctoral fellow position at Johns Hopkins Medical Institute in Baltimore, Maryland, USA. There, I plan to explore the intersection of translation and cellular signaling in the laboratory of Dr. Rachel Green. As for the long term, I remain open to explore opportunities—be they in academia, a governmental institution, or the private sector.
REFERENCES
- Akhmetov A, Laurent JM, Gollihar J, Gardner EC, Garge RK, Ellington AD, Kachroo AH, Marcotte EM. 2018. Single-step precision genome editing in yeast using CRISPR-Cas9. Bio Protoc 8: e2765. 10.21769/BioProtoc.2765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ameismeier M, Cheng J, Berninghausen O, Beckmann R. 2018. Visualizing late states of human 40S ribosomal subunit maturation. Nature 558: 249–253. 10.1038/s41586-018-0193-0 [DOI] [PubMed] [Google Scholar]
- Ameismeier M, Zemp I, van den Heuvel J, Thoms M, Berninghausen O, Kutay U, Beckmann R. 2020. Structural basis for the final steps of human 40S ribosome maturation. Nature 587: 683–687. 10.1038/s41586-020-2929-x [DOI] [PubMed] [Google Scholar]
- An W, Du Y, Ye K. 2018. Structural and functional analysis of Utp24, an endonuclease for processing 18S ribosomal RNA. PLoS One 13: e0195723. 10.1371/journal.pone.0195723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barandun J, Chaker-Margot M, Hunziker M, Molloy KR, Chait BT, Klinge S. 2017. The complete structure of the small-subunit processome. Nat Struct Mol Biol 24: 944–953. 10.1038/nsmb.3472 [DOI] [PubMed] [Google Scholar]
- Black JJ, Johnson AW. 2021. Genetics animates structure: leveraging genetic interactions to study the dynamics of ribosome biogenesis. Curr Genet 67: 729–738. 10.1007/s00294-021-01187-y [DOI] [PubMed] [Google Scholar]
- Black JJ, Wang Z, Goering LM, Johnson AW. 2018. Utp14 interaction with the small subunit processome. RNA 24: 1214–1228. 10.1261/rna.066373.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JJ, Musalgaonkar S, Johnson AW. 2019. Tsr4 is a cytoplasmic chaperone for the ribosomal protein Rps2 in Saccharomyces cerevisiae. Mol Cell Biol 39: 1–20. 10.1128/MCB.00094-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black JJ, Sardana R, Elmir EW, Johnson AW. 2020. Bud23 promotes the final disassembly of the small subunit processome in Saccharomyces cerevisiae. PLoS Genet 16: e1009215. 10.1371/journal.pgen.1009215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brink MF, Verbeet MP, de Boer HA. 1993. Formation of the central pseudoknot in 16S rRNA is essential for initiation of translation. EMBO J 12: 3987–3996. 10.1002/j.1460-2075.1993.tb06076.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaker-Margot M. 2018. Assembly of the small ribosomal subunit in yeast: mechanism and regulation. RNA 24: 881–891. 10.1261/rna.066985.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaker-Margot M, Hunziker M, Barandun J, Dill BD, Klinge S. 2015. Stage-specific assembly events of the 6-MDa small-subunit processome initiate eukaryotic ribosome biogenesis. Nat Struct Mol Biol 22: 920–923. 10.1038/nsmb.3111 [DOI] [PubMed] [Google Scholar]
- Cheng J, Kellner N, Berninghausen O, Hurt E, Beckmann R. 2017. 3.2-Å-resolution structure of the 90S preribosome before A1 pre-rRNA cleavage. Nat Struct Mol Biol 24: 954–964. 10.1038/nsmb.3476 [DOI] [PubMed] [Google Scholar]
- Cheng J, Lau B, La Venuta G, Ameismeier M, Berninghausen O, Hurt E, Beckmann R. 2020. 90S pre-ribosome transformation into the primordial 40S subunit. Science 369: 1470–1476. 10.1126/science.abb4119 [DOI] [PubMed] [Google Scholar]
- Clatterbuck Soper SF, Dator RP, Limbach PA, Woodson SA. 2013. In vivo X-ray footprinting of pre-30S ribosomes reveals chaperone-dependent remodeling of late assembly intermediates. Mol Cell 52: 506–516. 10.1016/j.molcel.2013.09.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connelly C, Hieter P. 1996. Budding yeast SKP1 encodes an evolutionarily conserved kinetochore protein required for cell cycle progression. Cell 86: 275–285. 10.1016/S0092-8674(00)80099-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll A, Grzeschik KH. 2001. Characterization of two novel genes, WBSCR20 and WBSCR22, deleted in Williams-Beuren syndrome. Cytogenet Cell Genet 95: 20–27. 10.1159/000057012 [DOI] [PubMed] [Google Scholar]
- Dragon F, Gallagher JEG, Compagnone-Post PA, Mitchell BM, Porwancher KA, Wehner KA, Wormsley S, Settlage RE, Shabanowitz J, Osheim Y, et al. 2002. A large nucleolar U3 ribonucleoprotein required for 18S ribosomal RNA biogenesis. Nature 417: 967–970. 10.1038/nature00769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Du Y, An W, Zhu X, Sun Q, Qi J, Ye K. 2020. Cryo-EM structure of 90S small ribosomal subunit precursors in transition states. Science 369: 1477–1481. 10.1126/science.aba9690 [DOI] [PubMed] [Google Scholar]
- Erdmann PS, Hou Z, Klumpe S, Khavnekar S, Beck F, Wilfling F, Plitzko JM, Baumeister W. 2021. In situ cryo-electron tomography reveals gradient organization of ribosome biogenesis in intact nucleoli. Nat Commun 12: 5364. 10.1038/s41467-021-25413-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira-Cerca S, Pöll G, Gleizes P-E, Tschochner H, Milkereit P. 2005. Roles of eukaryotic ribosomal proteins in maturation and transport of pre-18S rRNA and ribosome function. Mol Cell 20: 263–275. 10.1016/j.molcel.2005.09.005 [DOI] [PubMed] [Google Scholar]
- Ferreira-Cerca S, Sagar V, Schäfer T, Diop M, Wesseling A-M, Lu H, Chai E, Hurt E, LaRonde-LeBlanc N. 2012. ATPase-dependent role of the atypical kinase Rio2 on the evolving pre-40S ribosomal subunit. Nat Struct Mol Biol 19: 1316–1323. 10.1038/nsmb.2403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figaro S, Wacheul L, Schillewaert S, Graille M, Huvelle E, Mongeard R, Zorbas C, Lafontaine DLJ, Heurgue-Hamard V. 2012. Trm112 is required for Bud23-mediated methylation of the 18S rRNA at position G1575. Mol Cell Biol 32: 2254–2267. 10.1128/MCB.06623-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaemmaghami S, Huh W-K, Bower K, Howson RW, Belle A, Dephoure N, O'Shea EK, Weissman JS. 2003. Global analysis of protein expression in yeast. Nature 425: 737–741. 10.1038/nature02046 [DOI] [PubMed] [Google Scholar]
- Goddard TD, Huang CC, Meng EC, Pettersen EF, Couch GS, Morris JH, Ferrin TE. 2018. UCSF ChimeraX: meeting modern challenges in visualization and analysis. Protein Sci 27: 14–25. 10.1002/pro.3235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Rybin V, Bassler J, Petfalski E, Strauss D, Marzioch M, Schäfer T, Kuster B, Tschochner H, Tollervey D, et al. 2002. 90S pre-ribosomes include the 35S pre-rRNA, the U3 snoRNP, and 40S subunit processing factors but predominantly lack 60S synthesis factors. Mol Cell 10: 105–115. 10.1016/S1097-2765(02)00579-8 [DOI] [PubMed] [Google Scholar]
- Haag S, Kretschmer J, Bohnsack MT. 2015. WBSCR22/Merm1 is required for late nuclear pre-ribosomal RNA processing and mediates N7-methylation of G1639 in human 18S rRNA. RNA 21: 180–187. 10.1261/rna.047910.114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hector RD, Burlacu E, Aitken S, Le Bihan T, Tuijtel M, Zaplatina A, Cook AG, Granneman S. 2014. Snapshots of pre-rRNA structural flexibility reveal eukaryotic 40S assembly dynamics at nucleotide resolution. Nucleic Acids Res 42: 12138–12154. 10.1093/nar/gku815 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heuer A, Thomson E, Schmidt C, Berninghausen O, Becker T, Hurt E, Beckmann R. 2017. Cryo-EM structure of a late pre-40S ribosomal subunit from Saccharomyces cerevisiae. Elife 6: e30189. 10.7554/eLife.30189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Ghalei H, Karbstein K. 2020. Quality control of 40S ribosome head assembly ensures scanning competence. J Cell Biol 219: e202004161. 10.1083/jcb.202004161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korostelev A, Ermolenko DN, Noller HF. 2008. Structural dynamics of the ribosome. Curr Opin Chem Biol 12: 674–683. 10.1016/j.cbpa.2008.08.037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landry-Voyer A-M, Bergeron D, Yague-Sanz C, Baker B, Bachand F. 2020. PDCD2 functions as an evolutionarily conserved chaperone dedicated for the 40S ribosomal protein uS5 (RPS2). Nucleic Acids Res 48: 12900–12916. 10.1093/nar/gkaa1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau B, Cheng J, Flemming D, La Venuta G, Berninghausen O, Beckmann R, Hurt E. 2021. Structure of the maturing 90S pre-ribosome in association with the RNA exosome. Mol Cell 81: 293–303.e4. 10.1016/j.molcel.2020.11.009 [DOI] [PubMed] [Google Scholar]
- Létoquart J, Huvelle E, Wacheul L, Bourgeois G, Zorbas C, Graille M, Heurgué-Hamard V, Lafontaine DLJ. 2014. Structural and functional studies of Bud23-Trm112 reveal 18S rRNA N7-G1575 methylation occurs on late 40S precursor ribosomes. Proc Natl Acad Sci 111: E5518–E5526. 10.1073/pnas.1413089111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J-L, Yu H-C, Chao J-L, Wang C, Cheng M-Y. 2012. New phenotypes generated by the G57R mutation of BUD23 in Saccharomyces cerevisiae. Yeast 29: 537–546. 10.1002/yea.2934 [DOI] [PubMed] [Google Scholar]
- Linnemann J, Pöll G, Jakob S, Ferreira-Cerca S, Griesenbeck J, Tschochner H, Milkereit P. 2019. Impact of two neighbouring ribosomal protein clusters on biogenesis factor binding and assembly of yeast late small ribosomal subunit precursors. PLoS One 14: e0203415. 10.1371/journal.pone.0203415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza-Ochoa GI, Barrass JD, Terlouw BR, Maudlin IE, de Lucas S, Sani E, Aslanzadeh V, Reid JAE, Beggs JD. 2019. A fast and tuneable auxin-inducible degron for depletion of target proteins in budding yeast. Yeast 36: 75–81. 10.1002/yea.3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merla G, Ucla C, Guipponi M, Reymond A. 2002. Identification of additional transcripts in the Williams-Beuren syndrome critical region. Hum Genet 110: 429–438. 10.1007/s00439-002-0710-x [DOI] [PubMed] [Google Scholar]
- Mitterer V, Shayan R, Ferreira-Cerca S, Murat G, Enne T, Rinaldi D, Weigl S, Omanic H, Gleizes P-E, Kressler D, et al. 2019. Conformational proofreading of distant 40S ribosomal subunit maturation events by a long-range communication mechanism. Nat Commun 10: 2754. 10.1038/s41467-019-10678-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto B, Gaspar SG, Moriggi G, Pestov DG, Bustelo XR, Dosil M. 2020. Identification of distinct maturation steps involved in human 40S ribosomal subunit biosynthesis. Nat Commun 11: 156. 10.1038/s41467-019-13990-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura K, Fukagawa T, Takisawa H, Kakimoto T, Kanemaki M. 2009. An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat Methods 6: 917–922. 10.1038/nmeth.1401 [DOI] [PubMed] [Google Scholar]
- Oeffinger M, Wei KE, Rogers R, DeGrasse JA, Chait BT, Aitchison JD, Rout MP. 2007. Comprehensive analysis of diverse ribonucleoprotein complexes. Nat Methods 4: 951–956. 10.1038/nmeth1101 [DOI] [PubMed] [Google Scholar]
- Osheim YN, French SL, Keck KM, Champion EA, Spasov K, Dragon F, Baserga SJ, Beyer AL. 2004. Pre-18S ribosomal RNA is structurally compacted into the SSU processome prior to being cleaved from nascent transcripts in Saccharomyces cerevisiae. Mol Cell 16: 943–954. 10.1016/j.molcel.2004.11.031 [DOI] [PubMed] [Google Scholar]
- Õunap K, Käsper L, Kurg A, Kurg R. 2013. The human WBSCR22 protein is involved in the biogenesis of the 40S ribosomal subunits in mammalian cells. PLoS One 8: e75686. 10.1371/journal.pone.0075686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pérez-Fernández J, Román A, De Las Rivas J, Bustelo XR, Dosil M. 2007. The 90S preribosome is a multimodular structure that is assembled through a hierarchical mechanism. Mol Cell Biol 27: 5414–5429. 10.1128/MCB.00380-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plassart L, Shayan R, Montellese C, Rinaldi D, Larburu N, Pichereaux C, Froment C, Lebaron S, O'Donohue M-F, Kutay U, et al. 2021. The final step of 40S ribosomal subunit maturation is controlled by a dual key lock. Elife 10: e61254. 10.7554/eLife.61254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poot RA, van den Worm SH, Pleij CWA, van Duin J. 1998. Base complementarity in helix 2 of the central pseudoknot in 16S rRNA is essential for ribosome functioning. Nucleic Acids Res 26: 549–553. 10.1093/nar/26.2.549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rai J, Parker MD, Huang H, Choy S, Ghalei H, Johnson MC, Karbstein K, Stroupe ME. 2021. An open interface in the pre-80S ribosome coordinated by ribosome assembly factors Tsr1 and Dim1 enables temporal regulation of Fap7. RNA 27: 221–233. 10.1261/rna.077610.120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rössler I, Embacher J, Pillet B, Murat G, Liesinger L, Hafner J, Unterluggauer JJ, Birner-Gruenberger R, Kressler D, Pertschy B. 2019. Tsr4 and Nap1, two novel members of the ribosomal protein chaperOME. Nucleic Acids Res 47: 6984–7002. 10.1093/nar/gkz317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy-Chaudhuri B, Kirthi N, Culver GM. 2010. Appropriate maturation and folding of 16S rRNA during 30S subunit biogenesis are critical for translational fidelity. Proc Natl Acad Sci 107: 4567–4572. 10.1073/pnas.0912305107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardana R, Johnson AW. 2012. The methyltransferase adaptor protein Trm112 is involved in biogenesis of both ribosomal subunits. Mol Biol Cell 23: 4313–4322. 10.1091/mbc.e12-05-0370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardana R, White JP, Johnson AW. 2013. The rRNA methyltransferase Bud23 shows functional interaction with components of the SSU processome and RNase MRP. RNA 19: 828–840. 10.1261/rna.037671.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardana R, Zhu J, Gill M, Johnson AW. 2014. Physical and functional interaction between the methyltransferase Bud23 and the essential DEAH-box RNA helicase Ecm16. Mol Cell Biol 34: 2208–2220. 10.1128/MCB.01656-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardana R, Liu X, Granneman S, Zhu J, Gill M, Papoulas O, Marcotte EM, Tollervey D, Correll CC, Johnson AW. 2015. The DEAH-box helicase Dhr1 dissociates U3 from the pre-rRNA to promote formation of the central pseudoknot. PLoS Biol 13: e1002083. 10.1371/journal.pbio.1002083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sashital DG, Greeman CA, Lyumkis D, Potter CS, Carragher B, Williamson JR. 2014. A combined quantitative mass spectrometry and electron microscopy analysis of ribosomal 30S subunit assembly in E. coli. Elife 3: e04491. 10.7554/eLife.04491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scaiola A, Peña C, Weisser M, Böhringer D, Leibundgut M, Klingauf-Nerurkar P, Gerhardy S, Panse VG, Ban N. 2018. Structure of a eukaryotic cytoplasmic pre-40S ribosomal subunit. EMBO J 37: e98499. 10.15252/embj.201798499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer T, Strauss D, Petfalski E, Tollervey D, Hurt E. 2003. The path from nucleolar 90S to cytoplasmic 40S pre-ribosomes. EMBO J 22: 1370–1380. 10.1093/emboj/cdg121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma IM, Rappé MC, Addepalli B, Grabow WW, Zhuang Z, Abeysirigunawardena SC, Limbach PA, Jaeger L, Woodson SA. 2018. A metastable rRNA junction essential for bacterial 30S biogenesis. Nucleic Acids Res 46: 5182–5194. 10.1093/nar/gky120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shayan R, Rinaldi D, Larburu N, Plassart L, Balor S, Bouyssié D, Lebaron S, Marcoux J, Gleizes P-E, Plisson-Chastang C. 2020. Good vibrations: structural remodeling of maturing yeast pre-40S ribosomal particles followed by cryo-electron microscopy. Molecules 25: 1125. 10.3390/molecules25051125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. 1989. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics 122: 19–27. 10.1093/genetics/122.1.19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Vanden Broeck A, Miller L, Chaker-Margot M, Klinge S. 2021. Nucleolar maturation of the human small subunit processome. Science 373: eabj5338. 10.1126/science.abj5338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Q, Zhu X, Qi J, An W, Lan P, Tan D, Chen R, Wang B, Zheng S, Zhang C, et al. 2017. Molecular architecture of the 90S small subunit pre-ribosome. Elife 6: e22086. 10.7554/eLife.22086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartakoff AM, Chen L, Raghavachari S, Gitiforooz D, Dhinakaran A, Ni C-L, Pasadyn C, Mahabeleshwar GH, Pasadyn V, Woolford JL. 2021. The nucleolus as a polarized coaxial cable in which the rDNA axis is surrounded by dynamic subunit-specific phases. Curr Biol 31: 2507–2519.e4. 10.1016/j.cub.2021.03.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toussaint M, Conconi A. 2006. High-throughput and sensitive assay to measure yeast cell growth: a bench protocol for testing genotoxic agents. Nat Protoc 1: 1922–1928. 10.1038/nprot.2006.304 [DOI] [PubMed] [Google Scholar]
- Vanrobays E, Gelugne J-P, Gleizes P-E, Caizergues-Ferrer M. 2003. Late cytoplasmic maturation of the small ribosomal subunit requires RIO proteins in Saccharomyces cerevisiae. Mol Cell Biol 23: 2083–2095. 10.1128/MCB.23.6.2083-2095.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warner JR. 1999. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci 24: 437–440. 10.1016/S0968-0004(99)01460-7 [DOI] [PubMed] [Google Scholar]
- White J, Li Z, Sardana R, Bujnicki JM, Marcotte EM, Johnson AW. 2008. Bud23 methylates G1575 of 18S rRNA and is required for efficient nuclear export of pre-40S subunits. Mol Cell Biol 28: 3151–3161. 10.1128/MCB.01674-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolford JL, Baserga SJ. 2013. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195: 643–681. 10.1534/genetics.113.153197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Guo Q, Goto S, Chen Y, Li N, Yan K, Zhang Y, Muto A, Deng H, Himeno H, et al. 2014. Structural insights into the assembly of the 30S ribosomal subunit in vivo: functional role of S5 and location of the 17S rRNA precursor sequence. Protein Cell 5: 394–407. 10.1007/s13238-014-0044-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Wu C, Cai G, Chen S, Ye K. 2016. Stepwise and dynamic assembly of the earliest precursors of small ribosomal subunits in yeast. Genes Dev 30: 718–732. 10.1101/gad.274688.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu J, Liu X, Anjos M, Correll CC, Johnson AW. 2016. Utp14 recruits and activates the RNA helicase Dhr1 To undock U3 snoRNA from the preribosome. Mol Cell Biol 36: 965–978. 10.1128/MCB.00773-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zorbas C, Nicolas E, Wacheul L, Huvelle E, Heurgué-Hamard V, Lafontaine DLJ. 2015. The human 18S rRNA base methyltransferases DIMT1L and WBSCR22-TRMT112 but not rRNA modification are required for ribosome biogenesis. Mol Biol Cell 26: 2080–2095. 10.1091/mbc.E15-02-0073 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.