ABSTRACT
In the major human pathogen Klebsiella pneumoniae, MgrB inactivation by disruptive insertion sequence (IS) elements and mutations leading to early termination are known to play an important role in polymyxin resistance. In this study, we examined a collection of invasive blaKPC-2-producing K. pneumoniae isolates belonging to the high-risk clone sequence type 258 (ST258) displaying high rates of resistance to many antimicrobials, including polymyxins. We identified a deleterious substitution (W20S) in MgrB and confirmed by genetic complementation analysis that this variant was inactive, leading to increased polymyxin B and colistin MICs.
IMPORTANCE Carbapenem-resistant Gram-negative bacteria are designated critical pathogens by the World Health Organization. Polymyxins (i.e., polymyxin B and colistin) are last-resort antibiotics and particularly useful against these multidrug-resistant bacteria. In Klebsiella pneumoniae, the inactivation of MgrB, a negative regulator of PhoPQ, was shown to be the major pathway leading to colistin resistance. While gene disruption by insertion sequence (IS) elements and mutations leading to early termination (stop codons) are frequent, deleterious mutations are not observed frequently and have not been characterized. Here, we identified a deleterious substitution (W20S) in MgrB among a collection of bloodstream infection, blaKPC-2-producing K. pneumoniae sequence type 258 (ST258) isolates, displaying high rates of resistance to polymyxins and associated with a high mortality rate. The dissemination of such a MgrB-W20S mutation leading to polymyxin resistance within the ST258 high-risk clone background is problematic and thus warrants particular attention.
KEYWORDS: colistin, Enterobacterales, KPC, multidrug resistance
OBSERVATION
Polymyxins are last-resort antibiotics, particularly against carbapenem-resistant Gram-negative bacteria. This family of polycationic antimicrobial peptides includes polymyxin B and polymyxin E (i.e., colistin). In Klebsiella pneumoniae, it is well established that the most frequent mechanism of polymyxin resistance is the inactivation of chromosomally encoded mgrB. The small (47 amino acids) membrane protein MgrB is a negative regulator of the two-component system PhoPQ that controls lipopolysaccharide (LPS) modifications. MgrB prevents PhoPQ hyperactivation by directly interacting in the membrane with the PhoQ sensor kinase, while unfunctional or the absence of MgrB leads to enhanced PhoPQ activity and downstream addition of 4-amino-4-deoxy-l-arabinose (l-Ara4N) on lipid A decreasing the LPS negative charge (1). MgrB inactivation has been shown to arise by (i) gene interruption by an insertion sequence (IS) element, (ii) nucleotide deletion/insertion leading to frameshift and premature stop codons, or (iii) nucleotide nonsense substitution creating a premature stop codon (2, 3). Here, we identified for the first time a single amino acid substitution (W20S) responsible for MgrB inactivation increasing polymyxin MICs in carbapenemase blaKPC-2-producing K. pneumoniae sequence type 258 (ST258) invasive isolates.
We recently described the clinical and epidemiological features associated with a cohort of 165 polyclonal blaKPC-2-producing K. pneumoniae bloodstream infections in a Brazilian tertiary hospital between 2014 and 2016 (4). Among the 42 blaKPC-2 isolates belonging to the sequence type 258 (ST258), we observed unexpectedly high rates of resistance to colistin (MIC50, 8 μg/mL; MIC90, 128μg/mL; 80% resistant). ST258 blaKPC-2 isolates carried multiple resistance genes, including rmtB aminoglycoside 16S-methylase, severely limiting therapeutic options (see Table S1 in the supplemental material) (5). The clinical burden of these 42 ST258 K. pneumoniae bloodstream infection cases is described in Table S1 (Hospital Sao Paulo/Universidade Federal de São Paulo Ethics Committee for Clinical Research protocol 1.814.158). Overall (all-cause) 30-day mortality was 59.5%. The low number of polymyxin susceptible isolates precluded a deeper analysis of the impact of polymyxin resistance on the outcome.
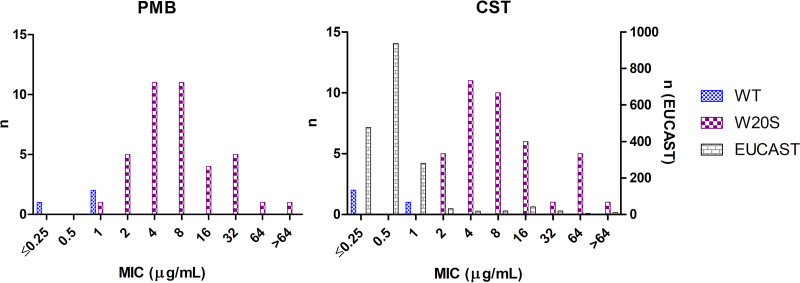
All ST258 isolate genomes (previously sequenced using an Illumina MiSeq instrument [5]) were analyzed for the presence of polymyxin resistance determinants. The mcr resistance genes were not detected in any of the isolates, and none of two-component-systems, namely, phoPQ and crrAB, carried mutations. The pmrB mutation R256G was present in all ST258s from our collection, including susceptible isolates. In mgrB, we identified a single nucleotide substitution, namely, 59G>C, leading to amino acid change W20S that was present in 93% (39/42) of the ST258 isolates and carried by the strains displaying the highest MICs (MIC distributions are shown in Fig. 1). Strains carrying MgrB W20S variants showed statistically higher polymyxin resistance levels than the EUCAST epidemiological cutoff (ECOFF) MIC panel distribution (6) (Wilcoxon-Mann-Whitney one-sided test, exact P = 2.2e-16; using the R Stats Package, version 4.0.3 [7]). We hypothesized that this mutation resulted in a nonfunctional MgrB leading to an increase in polymyxin MICs.
FIG 1.
Polymyxin B and colistin MIC distribution of K. pneumoniae ST258 isolates exhibiting wild-type (WT) mgrB (blue) and a W20S mgrB mutation (violet) analyzed in this study. The number of isolates was displayed on the left axis. The gray bars show the colistin MIC distribution from the EUCAST ECOFF (n = 1841) database (6) with the number of isolates displayed on the right axis.
To confirm its role in polymyxin resistance, we performed complementation experiments in the following three ST258 K. pneumoniae isolates carrying the mgrBW20S mutation: two KPC-2-producers (P27 and HSP12) from the collection and one additional KPC-negative (P52) ST258 strain that was isolated from the same hospital. For genetic complementation, an apramycin resistance gene was amplified from pIJ773 (8) using the primers Xce-apra-F (5′-CCACATGTATCCGTCGACCTGCAGTTCG-3′) and apra-Xce-R (5′-CCACATGTGTGTAGGCTGGAGCTGCTTCG-3′). The resulting product was digested using the FastDigest restriction enzyme XceI (Invitrogen, Thermo Fisher Scientific Inc.) and cloned into pBluescript SK+ (Stratagene Inc.) to build pskA (9). The wild-type (WT) and mutant mgrB were amplified with its own promoter from the strains MGH78578 (mgrB-WT) and P52 (mgrB-W20S), respectively, using the primers Eco-mgrB-ext-F (5′-GGAATTCCTTAAGAAGGCCGTGCTATCC-3′) and mgrB-BamHI-ext-R (5′-CGGGATCCCGAAGGCGTTCATTCTACCACC-3′) adapted from Cannatelli et al. (10). After restriction with BamHI and EcoRI, the PCR products were cloned in pskA and their sequences were verified by Sanger sequencing (Fasteris, Geneva, Switzerland). Polymyxin B and colistin MICs were determined in triplicate by broth microdilution following EUCAST guidelines, using Escherichia coli ATCC 25922 as a quality control. To prevent spontaneous plasmid loss, MICs of all strains carrying pskA plasmid had to be determined in the presence of 25 μg/mL apramycin. Strains carrying the empty pskA plasmid were used as a control, and presence of the plasmid did not significantly influence the MICs (Table 1). The expression of mgrB-WT on the pskA plasmid was able to restore susceptibility to both polymyxin B and colistin in the three different strains harboring the chromosomal W20S mutation with more than 64-fold MIC reduction (Table 1). On the other hand, when the complementation was performed with mgrB-W20S, the strains remained resistant to polymyxins confirming that this MgrB variant was not functional.
TABLE 1.
Polymyxin B and colistin MIC of complemented P27, P52, and HSP12 strains
| Strain | MIC (μg/mL) of: |
Interpretation | |
|---|---|---|---|
| Polymyxin B | Colistin | ||
| P27 | 16 | 16 | R |
| P27 + pskA-emptya | 16 | 16 | R |
| P27 + pskA-mgrB-WTa | ≤0.25 | ≤0.25 | S |
| P27 + pskA-mgrB-W20Sa | 16 | 8 | R |
| P52 | 16 | 16 | R |
| P52 + pskA-emptya | 8 | 16 | R |
| P52 + pskA-mgrB-WTa | ≤0.25 | ≤0.25 | S |
| P52 + pskA-mgrB-W20Sa | 8 | 16 | R |
| HSP12 | 32 | 32 | R |
| HSP12 + pskA-emptya | 16 | 32 | R |
| HSP12 + pskA-mgrB-WTa | ≤0.25 | ≤0.25 | S |
| HSP12 + pskA-mgrB-W20Sa | 8 | 16 | R |
MIC in the presence of apramycin 25 μg/mL to circumvent plasmid loss.
This description of a deleterious mutation in MgrB at position W20 is in agreement with a recent biochemical functional analysis of MgrB in Escherichia coli showing that W20 is a key residue for a MgrB/PhoQ interaction (11). Its role in polymyxin resistance was not characterized because in E. coli mgrB plays a minor role compared with the acquisition of mcr (12). Two K. pneumoniae isolates carrying a mgrB W20R mutation have been reported previously, but this mutation was not further investigated (3, 13). Based on our results, we can speculate that the W20R substitution is also a loss-of-function mutation influencing polymyxin MICs. The proposed EUCAST ECOFFs for colistin (2 μg/mL) did not fully discriminate isolates possessing a W20S mgrB mutation from the wild-type population; five (5/39) isolates were still classified as susceptible to colistin by the current breakpoint. It is possible that other factors interfered with MgrB/PhoPQ polymyxin resistance pathways in these isolates.
To investigate any fitness cost linked with a W20S mutation, growth curves were performed in triplicates after dilution (1/100) of the log-phase culture (at optical density [OD], 1) in 1 mL of fresh Mueller-Hinton broth (MHB), distributed in a 24-well plate under continuous shaking (180 rpm) at 37°C in the plate reader Infinite 200Pro (Tecan Trading AG, Switzerland). Doubling times were calculated for the three MgrB WT isolates and three randomly chosen W20S isolates. The W20S mutation did not impair the fitness of the tested strains since the generation times of the three strains with a W20S mutation (HSP12, 23.27 min; P27, 22.67 min; P52, 22.84 min) were similar to those of the strains with WT mgrB (HSP87, 23.87 min; P15, 23.71 min; P39, 22.02 min). Growth curves are shown in Fig. S1 in the supplemental material. The presence of a compensatory mutation explaining this absence of fitness cost could not be ruled out in our experiment. This observation is in line with previous publications showing that MgrB inactivation does not impose a fitness cost both in mutant isogenic strains and in the context of a clinical outbreak (14–16).
Here, we reported and confirmed by genetic experiments a novel amino acid substitution leading to polymyxin resistance in clinical high-risk K. pneumoniae isolates. This KPC-2-producing ST258 subclone carrying the W20S mutation was able to disseminate locally provoking 39 bloodstream infections over a 3-year period (2014 to 2016) (4). The heavy usage of polymyxin B in this hospital, in both empirical and directed therapies, due to a high rate of carbapenem-resistant isolates, might play a role in the efficient dissemination of this clone. In a previous study, we showed that the polymyxin B resistance rate has increased dramatically in this hospital, raising from 0% to 30.6% between the years 2009 and 2015 among 224 K. pneumoniae isolates recovered from blood cultures (17). The local dissemination of this polymyxin-resistant variant of the major international clone ST258 is thus worrisome, justifying that specific attention is needed to detect this new polymyxin resistance determinant.
Data availability.
Sequences are available under BioProject accession numbers PRJNA510003, PRJNA628956, PRJNA629307, PRJNA628957, PRJNA629309, PRJNA628954, PRJNA628953, and PRJEB41225.
ACKNOWLEDGMENTS
D.O.A. benefited from a Swiss National Science Foundation SNSF Mobility Postdoctoral Fellowship (APM P300PB_171601) and a Geneva University Hospitals Overseas Training Grant. D.O.A. has a scientific senior resident position provided by the faculty of Medicine, University of Geneva and The Geneva University Hospitals, and is the recipient of grants provided by The Sir Julius Thorn Trust Foundation (Switzerland), The Schmidheiny Foundation (Switzerland), and the Geneva University Hospitals Fund. Coordenação de Aperfeiçoamento de Pessoal de Nível Superior provided a grant to W.M.B.S.M. (88881.133245/2016-01). The National Council for Science and Technological Development provided a grant to A.C.G. (process number 312066/2019-8).
A.C.G. recently received research funding and/or consultation fees from Bayer, Eurofarma, Cristalia, Entasis Therapeutics, InfectoPharm, Merck Sharp & Dohme, Pfizer, and Zambon.
Footnotes
Supplemental material is available online only.
Contributor Information
Diego O. Andrey, Email: Diego.Andrey@unige.ch.
Philip N. Rather, Emory University School of Medicine
REFERENCES
- 1.Olaitan AO, Morand S, Rolain JM. 2014. Mechanisms of polymyxin resistance: acquired and intrinsic resistance in bacteria. Front Microbiol 5:643. doi: 10.3389/fmicb.2014.00643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cannatelli A, Giani T, D'Andrea MM, Di Pilato V, Arena F, Conte V, Tryfinopoulou K, Vatopoulos A, Rossolini GM, COLGRIT Study Group. 2014. MgrB inactivation is a common mechanism of colistin resistance in KPC-producing Klebsiella pneumoniae of clinical origin. Antimicrob Agents Chemother 58:5696–5703. doi: 10.1128/AAC.03110-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Macesic N, Nelson B, McConville TH, Giddins MJ, Green DA, Stump S, Gomez-Simmonds A, Annavajhala MK, Uhlemann AC. 2020. Emergence of polymyxin resistance in clinical Klebsiella pneumoniae through diverse genetic adaptations: a genomic, retrospective cohort study. Clin Infect Dis 70:2084–2091. doi: 10.1093/cid/ciz623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.DO A, Pereira Dantas P, Martins WBS, Marques De Carvalho F, Almeida LGP, Sands K, Portal E, Sauser J, Cayo R, Nicolas MF, Vasconcelos ATR, Medeiros EA, Walsh TR, Gales AC. 2020. An emerging clone, Klebsiella pneumoniae carbapenemase 2-producing K. pneumoniae sequence Type 16, associated with high mortality rates in a CC258-endemic setting. Clin Infect Dis 71:e141–e150. doi: 10.1093/cid/ciz1095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Roch M, Sierra R, Sands K, Martins W, Schrenzel J, Walsh TR, Gales AC, Andrey DO. 2021. Vertical and horizontal dissemination of an IncC plasmid harbouring rmtB 16S rRNA methylase gene, conferring resistance to plazomicin, among invasive ST258 and ST16 KPC-producing Klebsiella pneumoniae. J Glob Antimicrob Resist 24:183–189. doi: 10.1016/j.jgar.2020.12.006. [DOI] [PubMed] [Google Scholar]
- 6.European Committee on Antimicrobial Susceptibility Testing. 2021. Antimicrobial wild type distributions of microorganisms. https://mic.eucast.org/.
- 7.R Core Team. 2020. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. [Google Scholar]
- 8.Gust B, Challis GL, Fowler K, Kieser T, Chater KF. 2003. PCR-targeted Streptomyces gene replacement identifies a protein domain needed for biosynthesis of the sesquiterpene soil odor geosmin. Proc Natl Acad Sci USA 100:1541–1546. doi: 10.1073/pnas.0337542100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lenzi MH, Martins W, Roch M, Ramos PL, Sands K, Cayo R, Walsh TR, Andrey DO, Gales AC. 2021. A new mutation in mgrB mediating polymyxin resistance in Klebsiella variicola. Int J Antimicrob Agents 58:106424. doi: 10.1016/j.ijantimicag.2021.106424. [DOI] [PubMed] [Google Scholar]
- 10.Cannatelli A, D'Andrea MM, Giani T, Di Pilato V, Arena F, Ambretti S, Gaibani P, Rossolini GM. 2013. In vivo emergence of colistin resistance in Klebsiella pneumoniae producing KPC-type carbapenemases mediated by insertional inactivation of the PhoQ/PhoP mgrB regulator. Antimicrob Agents Chemother 57:5521–5526. doi: 10.1128/AAC.01480-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yadavalli SS, Goh T, Carey JN, Malengo G, Vellappan S, Nickels BE, Sourjik V, Goulian M, Yuan J. 2020. Functional determinants of a small protein controlling a broadly conserved bacterial sensor kinase. J Bacteriol 202:e00305-20. doi: 10.1128/JB.00305-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Poirel L, Jayol A, Nordmann P. 2017. Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin Microbiol Rev 30:557–596. doi: 10.1128/CMR.00064-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nordmann P, Jayol A, Poirel L. 2016. Rapid detection of polymyxin resistance in Enterobacteriaceae. Emerg Infect Dis 22:1038–1043. doi: 10.3201/eid2206.151840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cannatelli A, Santos-Lopez A, Giani T, Gonzalez-Zorn B, Rossolini GM. 2015. Polymyxin resistance caused by mgrB inactivation is not associated with significant biological cost in Klebsiella pneumoniae. Antimicrob Agents Chemother 59:2898–2900. doi: 10.1128/AAC.04998-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Otter JA, Doumith M, Davies F, Mookerjee S, Dyakova E, Gilchrist M, Brannigan ET, Bamford K, Galletly T, Donaldson H, Aanensen DM, Ellington MJ, Hill R, Turton JF, Hopkins KL, Woodford N, Holmes A. 2017. Emergence and clonal spread of colistin resistance due to multiple mutational mechanisms in carbapenemase-producing Klebsiella pneumoniae in London. Sci Rep 7:12711. doi: 10.1038/s41598-017-12637-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kidd TJ, Mills G, Sa-Pessoa J, Dumigan A, Frank CG, Insua JL, Ingram R, Hobley L, Bengoechea JA. 2017. A Klebsiella pneumoniae antibiotic resistance mechanism that subdues host defences and promotes virulence. EMBO Mol Med 9:430–447. doi: 10.15252/emmm.201607336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Braun G, Cayo R, Matos AP, de Mello Fonseca J, Gales AC. 2018. Temporal evolution of polymyxin B-resistant Klebsiella pneumoniae clones recovered from blood cultures in a teaching hospital during a 7-year period. Int J Antimicrob Agents 51:522–527. doi: 10.1016/j.ijantimicag.2017.08.031. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material. Download SPECTRUM01766-21_Supp_1_seq4.pdf, PDF file, 0.8 MB (793.1KB, pdf)
Data Availability Statement
Sequences are available under BioProject accession numbers PRJNA510003, PRJNA628956, PRJNA629307, PRJNA628957, PRJNA629309, PRJNA628954, PRJNA628953, and PRJEB41225.