Abstract
We report sporadic cases of a severe gastroenteritis associated with Vibrio cholerae serogroup O141. Like O1 and O139 serogroup strains of V. cholerae isolated from cholera cases, the O141 clinical isolates carry DNA sequences that hybridize to cholera toxin (CT) gene probes. The CT genes of O1 and O139 strains are carried by a filamentous bacteriophage (termed CTX phage) which is known to use toxin-coregulated pili (TCP) as its receptor. In an effort to understand the mechanism of emergence of toxigenic O141 V. cholerae, we probed a collection of O141 clinical and environmental isolates for genes involved in TCP production, toxigenicity, virulence regulation, and other phylogenetic markers. The collection included strains isolated between 1964 and 1995 from diverse geographical locations, including eight countries and five U.S. states. Information collected about the clinical and environmental sources of these isolates suggests that they had no epidemiological association. All clinical O141 isolates hybridized to probes specific for genes encoding CT (ctx), zonula occludens toxin (zot), repetitive sequence 1 (RS1), RTX toxin (rtxA), the major subunit of TCP (tcpA), and the essential regulatory gene that controls expression of both CT and TCP (toxR). In contrast, all but one of the nonclinical O141 isolates were negative for ctx, zot, RS1, and tcpA, although these strains were positive for rtxA and toxR. The one toxigenic environmental O141 isolate was also positive for tcpA. Ribotyping and CT typing showed that the O141 clinical isolates were indistinguishable or closely related, while a toxigenic water isolate from Louisiana showed a distantly related ribotype. Nonclinical O141 isolates displayed a variety of unrelated ribotypes. These data support a model for emergence of toxigenic O141 that involves acquisition of the CTX phage sometime after these strains had acquired the pathogenicity island encoding TCP. The clonal nature of toxigenic O141 strains isolated from diverse geographical locations suggests that the emergence is a rare event but that once it occurs, toxigenic O141 strains are capable of regional and perhaps even global dissemination. This study stresses the importance of monitoring V. cholerae non-O1, non-O139 serogroup strains for their virulence gene content as a means of assessing their epidemic potential.
The two most important virulence factors of Vibrio cholerae are cholera toxin (CT), a potent enterotoxin, and the toxin-coregulated pilus antigen (TCP), an essential intestinal colonization factor (16). These virulence factors are encoded by accessory genetic elements which are nearly always present in clinical isolates of V. cholerae but are frequently absent in strains isolated from environmental sources, such as water or shellfish (6, 16, 17). The ctxAB genes are located on the CTX genetic element, which is composed of a 4.5-kb central core region flanked by one or more copies of a repetitive sequence (RS1 or RS2) (34). The core and RS2, a portion of the CTX element, are now known to correspond to the genome of a filamentous bacteriophage (designated CTX phage) (44). An essential CTX phage assembly gene called zot has also been reported to encode a biological activity called zonula occludens toxin (2, 44). The CTX phage uses TCP as its receptor for infecting V. cholerae cells (44). Once infection has occurred, CTX phage DNA can either integrate into the chromosome via a specific attachment site (attRS), forming stable lysogens, or replicate extrachromosomally as a plasmid (17, 44). The expression of both ctx and tcp genes is regulated coordinately by ToxR and other transcriptional regulatory genes in a complicated network that continues to be intensively studied (16).
TCP is encoded by chromosomal DNA that is present in pathogenic strains and absent in most nonpathogenic strains of V. cholerae (25). This element has been referred to as the TCP pathogenicity island (37) as well as the V. cholerae pathogenicity island (VPI) (23) because it encodes several other virulence genes (25). A recent report also suggested that the TCP pathogenicity island may correspond to the genome of another filamentous bacteriophage, termed VPI phage (24). The horizontal acquisition of the pathogenicity island encoding TCP has been proposed as a likely prerequisite for the acquisition of CTX phage and thus for the emergence of new toxigenic strains of V. cholerae with human colonization properties (44).
Support for this model for emergence of toxigenic V. cholerae comes from the characterization of environmental and clinical non-O1, non-O139 strains. Non-O1 and non-O139 strains that are positive for CT but negative for TCP are exceedingly rare (17). These observations have led to the proposal that possession of tcp genes may be characteristic of the O1 serotypes (17). V. cholerae O139 strains are TCP positive only because they emerged as O-antigen recombinants of a CT- and TCP-positive El Tor O1 strain (17). However, Echeverria et al. (15) demonstrated that V. cholerae serogroup O44, O49, and O8 strains isolated from flies in northeastern Thailand in 1981 were positive for both CT and TCP genes. Additionally, in a comparison of ribotypes and serogroups of clinical V. cholerae non-O1, non-O139 isolates, Dalsgaard et al. (9) noticed that strains of the O141 serogroup were frequently CT positive, although the presence of other virulence genes such as those encoding TCP was not assessed.
Although V. cholerae non-O1, non-O139 strains rarely contain CT and TCP genes, they have been associated with sporadic cases of gastroenteritis, including cholera-like diarrhea, mainly in tropical areas (3, 8, 40). As the principal reservoir for V. cholerae is the aquatic environment, non-O1, non-O139 strains have been isolated from surface waters in most parts of the world, including North America. They are commonly isolated from shellfish, and most cases of V. cholerae non-O1, non-O139 gastroenteritis acquired in the United States are associated with eating raw or undercooked oysters (30). However, the bacterial factors responsible for the apparent pathogenicity of these CT-negative strains have not been elucidated. Recently, the genomic sequence of V. cholerae has revealed the presence of a toxin gene cluster related to the family of RTX toxins commonly produced by several different pathogenic gram-negative bacteria (5). Lin et al. (26) demonstrated that these genes encode a product that is responsible for a cytotoxic activity observed when mammalian cells are exposed to V. cholerae cells. Furthermore, the RTX element was present in several environmental isolates of V. cholerae, including non-O1 and O1, CT-negative strains. These results suggest that this toxin could be responsible for pathogenic properties of non-O1 and non-O139 strains.
In the present study, we examined the virulence-associated gene content of clinical and environmental isolates of V. cholerae O141.
MATERIALS AND METHODS
Patient information and bacteriology.
Information about patients and V. cholerae strains isolated in the United States was provided to the Centers for Disease Control and Prevention (CDC), Atlanta, Ga., by physicians through health departments of various states. In addition to the O141 strains isolated in the United States, an additional three clinical and six environmental V. cholerae O141 strains received for serotyping at the National Institute of Infectious Diseases, Tokyo, Japan, were included in the study for comparison purposes. Strain designations and other information are provided in Table 1.
TABLE 1.
Characterization of the V. cholerae 0141 strains studied
| Strain | Source | Place and yr of isolation | Reported clinical diagnosis | Ribotypea | CT genotypea |
|---|---|---|---|---|---|
| 609-84b | Stool | New York, 1984 | Cholera | B | CTk |
| 2454-85b | Stool | Tennessee, 1985 | Diarrhea (V. cholerae syndrome) | A | CTh |
| 2466-85b | Stool | North Carolina, 1985 | Gastroenteritis | A | CTi |
| 2527-87b | Stool | Maryland, 1987 | Diarrhea, nausea | A | CTk |
| 2533-86b | Stool | California, 1986 | None listed | B | CTi |
| 3176-78b | Water | Louisiana, 1978 | E | CTj | |
| F2031b | Stool | Spain, 1994 | None listed | C | CTh |
| 1178-96b | Stool | Taiwan, 1992 | None listed | C | CTl |
| 234-93b | Stool | India, 1993 | None listed | A | CTm |
| 574-94 | Water | Bolivia, 1992 | D | ||
| 930122 | Water | Cambodia, 1964 | F | ||
| CH236 | Shrimp | Germany, 1995 | G | ||
| 827-95 | Water | Brazil, 1995 | H | ||
| 834-95 | Water | Brazil, 1995 | I | ||
| 849-95 | Water | Brazil, 1995 | I |
Ribotype and CT genotype were established using the restriction enzyme BglI.
Strain hybridized with the CT probe.
All strains were identified biochemically as V. cholerae non-O1 based on standard biochemical reactions (39) and negative reactions in the agglutination tests employing polyvalent O1 antisera. The U.S. isolates were sent by the CDC to the National Institute of Infectious Diseases for serotyping. It should be noted that analyses of the files with the original strain information revealed that the two environmental strains reported isolated in the United States by Dalsgaard et al. (9) were identical. Thus, only a single environmental V. cholerae O141 strain, designated 3176-78, was isolated. Each of the V. cholerae non-O1, non-O139 strains listed in Table 1 was examined serologically by the slide agglutination test and designated according to an extended serotyping system which contains more than 193 different O serotypes (42; T. Shimada, personal communication). Preparation of O antisera and slide agglutination were performed as previously described (42). Although hybridization with CT and NAG-ST (heat-stable enterotoxin) probes, serotyping, and ribotyping had been done in a previous study (9), these characterization techniques were repeated in the present study as described below.
Antibiotic susceptibility testing and isolation of plasmid DNA.
Each isolate was tested for susceptibility to 12 antibacterial agents by the disk (Oxoid Ltd., Basingstoke, United Kingdom) diffusion method using Mueller-Hinton agar (Difco, Detroit, Mich.) as described by the National Committee for Clinical Laboratory Standards (31). The following antibiotics were used (micrograms per disk): ampicillin, 30; chloramphenicol, 30; ciprofloxacin, 5; colistin, 10; gentamicin, 10; kanamycin, 30; nalidixic acid, 30; neomycin, 30; streptomycin, 10; sulfamethoxazole, 100; tetracycline, 30; and trimethoprim-sulfamethoxazole, 5.2/240. Isolates were also tested for susceptibility to the vibriostatic agent O/129 (2,4-diamino-6,7-diisopropylpteridine phosphate), 150 μg per disk.
Plasmid extraction was carried out using the method of Kado and Liu (22), modified by incubating the cells at elevated pH (12.54) for 30 min at 56°C during the lysis step. V. cholerae O1 strain 1075/25 carrying a 150-kb plasmid was used as a positive control (43). Electrophoresis and visualization of plasmids were carried out essentially as previously described (33).
DNA probes for detection of virulence-associated genes.
The presence of several virulence-associated genes was determined through hybridization with DNA probes. These genes included ctxA (the A subunit of the CT gene), the NAG-ST gene, tcpA (the major subunit of the TCP gene), RS1 (the repetitive sequence 1 gene), zot (zonula occludens toxin gene), toxR (an essential toxin-regulating gene), attRS1 (specific attachment site 1 gene), CTXp (a CT phage gene), pTLC (a toxin-linked cryptic plasmid gene), and rtx (a RTX toxin gene) (Table 2). Although not specific for V. cholerae O141, the organization of the V. cholerae chromosome, including pTLC, rtx, CTX phage, and the CTX element, is shown in Fig. 1. Colony blots were prepared with nylon (Hybond; Amersham International plc, Aylesbury, United Kingdom) or Whatman filters and processed by standards methods (21). Positive and negative V. cholerae control strains were included on all filters.
TABLE 2.
DNA probes and PCR primers used to study virulence genes in V. cholerae 0141
| Gene | DNA probe and label | Reference |
|---|---|---|
| ctxA | 23-bp oligonucleotide, alkaline phosphatase | 45 |
| NAG-ST | 16-bp oligonucleotide alkaline phosphatase | 32 |
| tcpA | 2-kb HindIII digest of pRT110, 32P | 41 |
| RS1 | 1-kb BglI/EcoRV digest of pJM17, 32P | 29 |
| zot | 2.5-kb XbaI/PstI digest of pJM17, 32P | 19 |
| toxR | 0.9-kb XbaI-SalI of plasmid pToxRII, digoxigenin | 40 |
| attRS1 | 18-bp oligonucleotide alkaline phosphatase | 40 |
| CTXp | pCTX-km, digoxigenin | 44 |
| rtx | pWL80, digoxigenin | 26 |
| pTLC | p9A, digoxigenin | 36 |
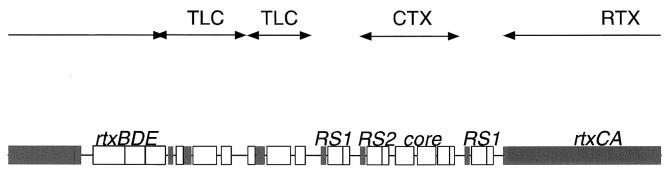
FIG. 1.
Genomic organization of the pTLC (TLC) element, the CTX prophage, and the RTX gene cluster on the V. cholerae chromosome. The pTLC element is 842 bp upstream of the CTX element whereas the RTX gene cluster is 693 bp downstream of the CTX element. Open and filled boxes represent ORFs that are oriented left to right and right to left, respectively. The pTLC element is tandemly duplicated on the chromosome. In El Tor strains, the first copy contains an insertion sequence that shares sequence similarity to IS911. The CTX prophage is also commonly tandemly duplicated in toxigenic strains. In classical strains, the RTX gene cluster is disrupted by a deletion that removes part of rtxA and rtxB and all of rtxC.
Ribotyping and CT genotyping.
The ribotypes of the V. cholerae O141 isolates were determined previously (9) and established by using BglI to digest chromosomal DNA (35). Ribotyping was performed by the procedure described by Dalsgaard et al. (7) with digoxigenin-labeled 16S and 23S rRNA probes. In our repeated ribotyping of the O141 strains, V. cholerae O1 strains O70, 1083/30, and 2722/33 (11) and O139 strains ADC 1125/9 and NIH 178 (12) were included for comparison purposes. Restriction fragment length polymorphism analysis of DNA sequences associated with CT genes (CT genotyping) was performed by hybridization of nylon membranes containing BglI-digested DNA with a digoxigenin-labeled oligonucleotide CT probe (45). A 1-kb DNA molecular size standard (GIBCO BRL, Gaithersburg, Md.) was used as a size marker in ribotyping, and λ/HindIII was used for CT genotyping. CT genotypes were determined for each of the nine strains that hybridized with the CT probe. V. cholerae O1 strains 9868, isolated in Guinea-Bissau in 1996, and 1724/34, isolated in Thailand in 1991 (11), were included in CT genotyping for comparison purposes. BglI was selected because it does not have any recognition sequence within the ctxA gene but it has a single cleavage site located upstream and adjacent to the ctxA gene (29). Thus, the number of bands comprising each CT genotype pattern represents the number of copies of the ctxA harbored by each strain.
RESULTS
Patient information.
Limited epidemiologic and clinical information accompanied the isolates sent to the CDC by the health departments of various states (Table 1). All U.S. patients were adults, ranging in age from 34 to 72 years old; three of the five patients were men. The reported clinical diagnoses are shown in Table 1. No food or travel histories were reported, with the exception of the patient from California, who ate shellfish in Atlanta, Ga., before his illness began. Four of the patients survived their illness; the outcome of the remaining case from Maryland was not reported. Clinical details were reported only for the patient from New York (diagnosis, cholera). This previously healthy 55-year-old man presented to a New York City hospital with complaints of malaise and diarrhea. He was passing approximately 8 to 12 liters of rice water stool per day and had indications of acute renal failure, including anuria and significant metabolic acidosis. The patient responded well to hospitalization, rehydration, and tetracycline treatment, with a gradual decrease in diarrhea over a 1-week period. Information about patients and clinical manifestations of the cases reported in Spain, Taiwan, and India was not available (Table 1).
Biochemical reactions, antibiotic susceptibility patterns, and plasmid analysis.
The V. cholerae non-O1 strains produced yellow colonies on thiosulfate-citrate-bile salt-sucrose agar and grew in NaCl concentrations of 0, 3, 6, and 7% but not 8%. All were oxidase and indole positive, fermented glucose, produced lysine and ornithine decarboxylases but not arginine dehydroxylase, and were ONPG (o-nitrophenyl-β-d-galactopyranoside) positive. Only the three water isolates from Brazil utilized cellobiose. All isolates were sensitive in susceptibility testing to the vibriostatic agent O/129. The growth shown by the O141 strains at relatively high salinities of 6 and 7% NaCl corroborates previous findings with V. cholerae non-O1, non-O139 (10). Thus, strains showed biochemical reactions typical of those of V. cholerae (31). Using the extended serotyping scheme, the V. cholerae non-O1 strains agglutinated in O141 antisera in repeated testing (42).
Each of the strains tested showed resistance to colistin but were sensitive to most of the remaining antibacterial agents tested, including tetracycline, trimethoprim, chloramphenicol, and the quinolones. V. cholerae non-O1 strains are normally resistant to colistin. Strains 234-93 and 1178-96 showed resistance to ampicillin. Plasmid analysis revealed that none of the clinical isolates contained plasmids. Among the environmental isolates, strain CH236, isolated from shrimp imported into Germany, contained plasmids of 7.2 and 23 kb, and strain 574-94, isolated from water in Bolivia, contained a single plasmid of 5.7 kb.
Virulence-associated genes.
The clinical and environmental V. cholerae O141 were further characterized to provide information about the presence of several genes which either encode virulence factors or are genetically linked to virulence genes (i.e., virulence-associated genes) (Table 2). In the colony hybridization studies, each of the clinical O141 strains and one strain isolated from a Louisiana water sample contained sequences that hybridized to the ctxA, RS1, and zot probes (Table 3). Since all three of the probes hybridize to sequences located on CTX phage, these data strongly suggest that all CT-positive O141 strains contain a copy of the CTX prophage inserted in their chromosomes. This was corroborated by hybridization of these strains with the CTXp probe. Inserted upstream of CTX phage in El Tor O1 strains are two tandem copies of a 4.7-kbp cryptic plasmid termed pTLC (36). Evidence suggests that this plasmid may stimulate the replication of CTX phage in V. cholerae O1 El Tor strains (W. Lin and J. Mekalanos, unpublished results). All O141 strains were probed with a pTLC probe, but all were negative (Table 3). This result strongly suggests that O141 strains are not derived from toxigenic El Tor O1 strains, a conclusion which is also supported by the ribotyping data (see below).
TABLE 3.
Presence of virulence-associated genes among V. cholerae 0141 strains isolated from patients or environmental samples
| Strain | Presence of virulence-associated genea
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| ctxA | RS1 | zot | toxR | attRS1 | tcpA | CTXp | rtx | pTLC | NAG-ST | |
| 609-84 | + | + | + | + | + | + | + | + | − | − |
| 2454-85 | + | + | + | + | + | + | + | + | − | − |
| 2466-85 | + | + | + | + | + | + | + | + | − | − |
| 2527-87 | + | + | + | + | + | + | + | + | − | − |
| 2533-86 | + | + | + | + | + | + | + | + | − | − |
| 3176-78 | + | + | + | + | + | + | + | + | − | − |
| F2031 | + | + | + | + | + | + | + | + | − | − |
| 1178-96 | + | + | + | + | + | + | + | + | − | − |
| 234-93 | + | + | + | + | + | + | + | + | − | − |
| 574-94 | − | − | − | + | + | − | − | + | − | − |
| 930122 | − | − | − | + | + | − | − | + | − | − |
| CH236 | − | − | − | + | + | − | − | + | − | + |
| 827-95 | − | − | − | + | + | − | − | + | − | − |
| 834-95 | − | − | − | + | + | − | − | + | − | − |
| 849-95 | − | − | − | + | + | − | − | + | − | − |
The presence of virulence-associated genes was detected with DNA probes (see Materials and Methods for details).
All O141 strains tested hybridized with the toxR and attRS1 gene probes, suggesting that environmental isolates have the potential to integrate the CTX prophage and to regulate CT expression (Table 3). However, all CT-negative environmental O141 strains lacked sequences that hybridized to tcpA, suggesting that they are unlikely to serve as efficient recipients of CTX phage (17, 44). Furthermore, all O141 strains tested, including these TCP-negative environmental strains, were positive for the rtx probe, suggesting that RTX toxin might be a virulence factor produced by these environmental strains.
One environmental strain hybridized to the NAG-ST probe, a gene which is known to exist on an integron-like genetic element in other strains of V. cholerae (27).
Ribotyping and CT genotyping.
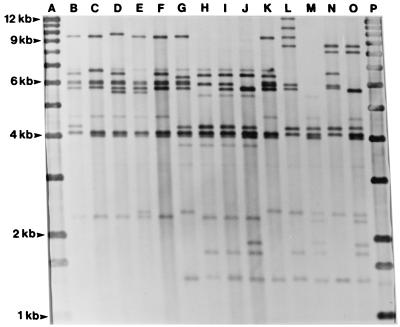
Examples of the different V. cholerae O141 ribotypes are shown in Fig. 2 together with ribotypes of the O1 and O139 strains. The clinical U.S. V. cholerae O141 strains 2454-85, 2466-85, and 2527-87 and the clinical reference strain 234-93 from India showed an indistinguishable ribotype A (Fig. 2, lane I). The two remaining clinical U.S. strains, 609-84 and 2533-83, showed an indistinguishable ribotype B, which was very closely related to type A, as the two types differed by a single fragment only. The two ribotypes shown by the clinical U.S. strains were also closely related to the indistinguishable ribotype C demonstrated by the two clinical strains from Spain and Taiwan (Fig. 2 and Table 1). Each of the clinical O141 strains showed ribotypes which differed from the types shown by O1 and O139 strains by two or more fragments (Fig. 2).
FIG. 2.
Examples of BglI ribotypes of V. cholerae O141 and serotype O1 and O139 reference strains. Lanes (strain designation, O serogroup, and ribotype): B, O79, O1, unnamed ribotype; C, 1083/30, O1, unnamed type; D, 2722/33, O1, unnamed type; E, ADC 1125/9, O139, unnamed type; F, NIH 178, O139, unnamed type; G, 3176-78, O141, ribotype E; H, 609-84, O141, type B; I, 2527-86, O141, type A; J, F2031, O141, type C; K, 574-94, O141, type D; L, 930122, O141, type F; M, CH236, O141, type G; N, 827-95, O141, type H; O, 834-95, O141, type I. Lanes A and P are 1-kb molecular size standards.
None of the environmental O141 isolates showed ribotypes identical to types demonstrated by clinical strains. With the exception of strain 574-94, the CT-negative O141 strains showed ribotypes which differed from the ribotypes observed among the clinical O141 strains by several fragments. The CT-positive strain 3176-78, recovered from a water sample in the United States, showed ribotype E, which demonstrated a 9.5-kb fragment not shown by any of the CT-positive clinical O141 strains, whereas ribotypes of both O1 and O139 reference strains demonstrated the 9.5-kb fragment. Although they had a common 9.5-kb fragment, the ribotypes of strain 3176-78, O1 strains, and O139 strains differed by several fragments (Fig. 2)
Southern blot hybridization with the CT probe of BglI-digested genomic DNAs of the O141 strains showed six different genotypes consisting of two or three fragments of between 6.8 and 25 kb (results not shown). Each of the strains tested showed a common 7.1-kb fragment which was also found in the two V. cholerae O1 reference strains. Little correspondence was found between ribotype and CT genotype, as strains with an identical ribotype most often showed different CT genotypes (Table 1).
DISCUSSION
In this study, we report sporadic cases of gastroenteritis associated with V. cholerae serogroup O141. We sought to further define the virulence-associated gene content of the O141 strains. Specifically, we hoped to address (i) whether clinical O141 strains possessed the same critical virulence genes as O1 and O139 strains, (ii) whether environmental O141 strains were similar to or distinct from these clinical isolates in terms of their virulence gene content, and (iii) whether ribotyping could be used to establish the relatedness of O141 strains to each other as well as other V. cholerae serogroups. Of particular importance for understanding the emergence of toxigenic O141 strains was their TCP status, given the role of this pilus colonization factor as the receptor for the CTX phage (44). We found that all clinical O141 isolates contained TCP genes and at least one copy of the CTX prophage, based on their reactivity to tcpA, ctx, zot, and RS1 probes. In contrast, all but one of the environmental O141 isolates were negative for ctx, zot, RS1, and tcpA. The single environmental O141 isolate from Louisiana that was positive for tcpA was also ctx positive. Ribotyping confirmed that all CT- and TCP-positive O141 strains were closely related regardless of their geographical origin, while all environmental O141 strains were diverse and largely unrelated to each other or to O1, O139, or O141 clinical isolates. In general, these data support a model for emergence of toxigenic O141 that involves first the acquisition of the pathogenicity island encoding TCP followed by the acquisition of the CTX phage (37, 44). Our data suggest that once toxigenic O141 emerged, these strains spread regionally as clones rather than emerging independently in diverse geographic locations from resident nontoxigenic O141 environmental strains.
Our conclusions appear to be consistent with data from recent studies addressing similar questions. For example, Faruque et al. (17) found that only O1 and O139 strains and none of 132 non-O1, non-O139 strains tested carried TCP genes. Similarly, Sharma et al. (40) reported that of 18 CT-negative strains of non-O1, non-O139 V. cholerae isolated from several outbreaks in India in 1996, none were positive for TCP genes. Accordingly, we think that a TCP-positive, CTX-negative O141 strain was the most likely precursor of the toxigenic O141 strains characterized here. Unfortunately, we could not identify an environmental O141 isolate that was TCP positive or that even displayed a ribotype indistinguishable from or closely related to those of the toxigenic O141 strains. However, clonally related, TCP-positive, CT-negative V. cholerae O1 strains have been documented as environmental isolates from the Gulf Coast of the United States, and these strains are clearly closely related to toxigenic strains isolated from water and clinical samples from the region prior to 1991 (36). Echeverria et al. (14) also reported that non-O1, non-O139 strains that are TCP positive and CT negative can be isolated from environmental sources. Thus, TCP-positive V. cholerae strains, regardless of serogroup, remain potential precursors of epidemic strains because of their dual capacity to serve as efficient recipients of CTX phage (particularly in vivo) and to colonize the human intestine by TCP-dependent mechanisms.
Nonetheless, it is possible that toxigenic O141 strains emerged by acquisition of the CTX phage first, followed by the TCP island. A few TCP-negative, CT-positive strains were identified by Faruque et al. (17), but these strains can be explained by either loss of the TCP island after CTX phage acquisition or acquisition of the CTX phage by a TCP-independent mechanism. TCP-independent acquisition of CTX phage has been reported by Boyd and Waldor (4) and by Faruque et al. (18). However, these mechanisms involved generalized transduction by another lytic phage and a far less efficient undefined mechanism, respectively.
How V. cholerae strains acquire the pathogenicity island that encodes TCP remains controversial. A recent report by Karaolis et al. (24) concluded that the pathogenicity island corresponded to the genome of a filamentous bacteriophage termed VPI phage which putatively could move between El Tor O1 strains and at least one recipient strain of the O10 serogroup. However, bioinformatic analysis of the open reading frames (ORFs) present in the island has failed to uncover genes that have significant homology to phage assembly genes (20). Nonetheless, the G+C content of the island together with other DNA composition and codon usage analyses supports the conclusion that this island has an origin other than V. cholerae (20, 23). Thus, it is likely that the TCP genes have been recently acquired by V. cholerae O1, but we cannot say how this has occurred or whether this mechanism informs us further about the likely evolutionary steps that lead to the emergence of toxigenic TCP-positive O141 strains. Previous studies have proposed that when and if horizontal transfer of TCP and the CTX genetic elements occur, it may be linked to concomitant changes in the somatic antigen (40). However, our findings, together with the report by Echeverria et al. (15) of non-O1, non-O139 environmental strains containing tcpA and the CTX element, suggest that horizontal transfer of the O antigen does not necessarily occur before or after the acquisition of the TCP and CT genes.
In this study we used ribotyping as a phylogenetic tool for accessing the baseline similarity between the O141 strains in our collection. Ribotyping using BglI demonstrated indistinguishable or closely related ribotypes among the O141 strains isolated from stool specimens. The loss or gain of a BglI restriction site may result in the loss of a fragment and the creation of two new fragments; thus, the minor differences in fragment patterns shown by the clinical strains suggest that they originated from the same clone. That genetic events responsible for changes in ribotypes and pulsed-field gel electrophoresis types are occurring over time was demonstrated by Dalsgaard et al. (13), who found several closely related V. cholerae O1 ribotypes among strains isolated in Lima, Peru, during a 5-year period following the introduction of V. cholerae O1 into Peru in 1991. The close relationship of toxigenic O141 strains to each other is further supported by the fact that all these strains lack sequences that hybridize to the cryptic plasmid pTLC. All toxigenic O1 and O139 strains tested to date possess this integrated plasmid (36). Thus, together with our ribotyping results, the absence of pTLC in toxigenic O141 strains strongly suggests that these strains did not emerge as an O-antigen recombinant of an O1 or O139 strain.
It is difficult to explain how V. cholerae O141 strains isolated from stool specimens of different geographical and chronological origins apparently belong to the same clone. Although the patient data are limited, there is nothing to suggest that the patients in the United States, Spain, and India were epidemiologically related. The toxigenic O141 strains could be disseminated by cargo chips, e.g., contaminated ballast, bilge, and/or sewage, a mode of transmission which has been described for toxigenic O1 strains from the Latin American epidemic into the United States (28). It is uncertain if the toxigenic O141 strain 3176-78, isolated from a water sample in Louisiana in 1978, could have been the source strain from which the toxigenic U.S. clinical strains emerged. It is possible that the differences in ribotype patterns between strain 3176-78 and the clinical strains may have evolved during the ≥6-year time span between their isolation. However, it is most interesting that an O141 strain containing the CTX phage and TCP was isolated from a U.S. Gulf Coast water sample in 1978. The U.S. Gulf Coast continues to constitute a permissive environmental site for persistence of a unique clone of V. cholerae that includes both TCP+ nontoxigenic and TCP+ toxigenic strains of V. cholerae O1 (36). It is tempting to speculate that TCP and perhaps CT might contribute to the fitness of V. cholerae in some aquatic environments. If so, toxigenic O141 strains might persist and expand globally in incidence once virulent strains have emerged. It will be interesting to determine more precisely the intimate relationships in which V. cholerae persists within the aquatic environment and how these influence the fitness of pathogenic versus nonpathogenic strains or other interactions important for emergence, such as interaction between the bacterium and its converting phages (16, 44).
All serogroup O141 strains tested possessed the gene encoding the regulatory protein ToxR, which controls the coordinate expression of genes associated with pathogenicity in toxigenic V. cholerae and the 17-bp attRS1 target sequence, in which the CTX phage integrates into the chromosome of V. cholerae (16). A study of V. cholerae non-O1, non-O139 strains associated with an upsurge in the incidence of cholera-like diarrhea in Calcutta, India, reported similar findings with all strains containing toxR and attRS1 genes (40) and proposed that such non-O1, non-O139 strains could be proto-cholera agents. Although this may be true, the relative risk of these strains becoming positive for both TCP and CT genes seems lower than that of strains that are already positive for TCP and thus can acquire CTX phage by simple transduction. With the few exceptions noted above, most V. cholerae strains that carry CT genes also carry TCP genes (17). Two explanations have been proposed to explain why TCP-positive and CT-negative strains are rarely found (17). One reason may be that such strains do not cause full-blown cholera and hence are not adequately enriched through the explosive replication within the human host that fully virulent strains enjoy. Alternatively, most TCP-positive strains are rapidly converted to toxigenic strains by infection with CTX phage either within the host intestine or in the aquatic environment (15). Given that we were unable to identify any O141 strains that were TCP positive but CT negative, our results do not differentiate between these two explanations. More curious is why toxigenic O141 strains have not yet caused a serious cholera epidemic. Our genetic analysis of these strains suggest that they have this potential especially in areas of the world where immunity to the O1 and O139 serogroups provides a selective edge for a heterologous serogroup such as O141.
V. cholerae non-O1, non-O139 serotypes are increasingly isolated from patients with diarrhea. Dalsgaard et al. (8) found that non-O1, non-O139 strains were isolated at rates similar to or higher than those of serotype O1 strains in a study conducted in Thailand from 1993 to 1995. Such single cases of diarrhea seem most often to be associated with a wide range of serotypes (8, 40). However, outbreaks of diarrhea have been associated with certain serotypes, e.g., O10 and O12 strains in 1994 in Peru (6), O6 and O14 strains among Khmers in a refugee camp in Thailand (1), and O10 strains in India (38). The non-O1, non-O139 strains from both single and outbreak cases very rarely contain the tcp and the CTX genetic elements, and although a number of studies have proposed several potential virulence factors among non-O1, non-O139 strains, the factor(s) responsible for diarrhea and its mode of action remain to be identified. With the recent finding by Lin et al. (26) that the RTX genes encode a product that has cytotoxic activity for mammalian cells, it was proposed that the RTX toxin may play an important role in the virulence of CTX-negative strains. All O141 strains tested in this study possessed the RTX genes, including several CTX-negative strains isolated from water samples. Preliminary results from ongoing studies in our laboratories corroborate the findings that a very high proportion of both environmental and clinical non-O1, non-O139 strain contain RTX genes (unpublished results). Thus, it appears that the cytotoxic activity showed by RTX-positive strains may be a widely distributed virulence factor among V. cholerae non-O1, non-O139 strains.
ACKNOWLEDGMENTS
We are grateful for the technical assistance provided by Anne-Mette Petersen at the Royal Veterinary and Agricultural University in Denmark. We also thank the state health departments for providing information about patients and V. cholerae strains.
Anders Dalsgaard was supported by the Danish Council for Development Research, Danida grant 90928. The laboratory of John J. Mekalanos was supported by grant AI-18045 from the National Institute of Allergy and Infectious Diseases.
REFERENCES
- 1.Bagchi K, Echeverria P, Arthur J D, Sethabutr O, Serichantalergs O, Hoge C W. Epidemic of diarrhea caused by Vibrio cholerae non-O1 that produced heat-stable toxin among Khmers in a camp in Thailand. J Clin Microbiol. 1993;31:1315–1317. doi: 10.1128/jcm.31.5.1315-1317.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baudry B A, Fasano A, Ketley J, Kaper J B. Cloning of a gene (zot) encoding a new toxin produced by Vibrio cholerae. Infect Immun. 1992;60:428–434. doi: 10.1128/iai.60.2.428-434.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bhattacharya M K, Dutta D, Bhattacharya S K, Deb A, Mukhopadhyay A K, Nair G B, Shimada T, Takeda Y, Chowdhury A, Mahalanabis D. Association of a disease approximating cholera caused by Vibrio cholerae of serogroups other than O1 and O139. Epidemiol Infect. 1998;120:1–5. doi: 10.1017/s0950268897008352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boyd E F, Waldor M K. Alternative mechanism of cholera toxin acquisition by Vibrio cholerae: generalized transduction of CTXΦ by bacteriophage CP-T1. Infect Immun. 1999;67:5898–5905. doi: 10.1128/iai.67.11.5898-5905.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Braun V, Schonherr R, Hobbie S. Enterobacterial hemolysins: activation, secretion and pore formation. Trends Microbiol. 1993;1:211–216. doi: 10.1016/0966-842x(93)90134-d. [DOI] [PubMed] [Google Scholar]
- 6.Dalsgaard A, Albert M J, Taylor D N, Shimada T, Meza R, Serichantalergs O, Echeverria P. Characterization of Vibrio cholerae non-O1 serogroups obtained from an outbreak of diarrhea in Lima, Peru. J Clin Microbiol. 1995;33:2715–2722. doi: 10.1128/jcm.33.10.2715-2722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dalsgaard A, Echeverria P, Larsen J L, Siebeling R, Serichantalergs O, Huss H H. Application of ribotyping for differentiating Vibrio cholerae non-O1 isolates from shrimp farms in Thailand. Appl Environ Microbiol. 1995;61:245–251. doi: 10.1128/aem.61.1.245-251.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dalsgaard A, Forslund A, Bodhidatta L, Serichantalergs O, Pitarangsi C, Pang L, Shimada T, Echeverria P. A high proportion of Vibrio cholerae strains isolated from children with diarrhoea in Bangkok, Thailand are multiple antibiotic resistant and belong to heterogenous non-O1, non-O139 O-serotypes. Epidemiol Infect. 1999;122:217–226. doi: 10.1017/s0950268899002137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dalsgaard A, Forslund A, Mortensen H F, Shimada T. Ribotypes of clinical Vibrio cholerae non-O1 non-O139 strains in relation to O-serotypes. Epidemiol Infect. 1998;121:535–545. doi: 10.1017/s0950268898001654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dalsgaard A, Huss H H, Kittikun A H-, Larsen J L. The prevalence of Vibrio cholerae and Salmonella in a major shrimp production area in Thailand. Int J Food Microbiol. 1995;28:101–113. doi: 10.1016/0168-1605(94)00165-3. [DOI] [PubMed] [Google Scholar]
- 11.Dalsgaard A, Serichantalergs O, Forslund A, Pitarangsi C, Echeverria P. Phenotypic and molecular characterization of Vibrio cholerae O1 isolated in Samutsakorn, Thailand before, during and after the emergence of V. cholerae O139. Epidemiol Infect. 1998;121:259–268. doi: 10.1017/s0950268898001125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalsgaard A, Skov M N, Serichantalergs O, Echeverria P. Comparison of pulsed-field gel electrophoresis and ribotyping for subtyping of Vibrio cholerae O139 isolated in Thailand. Epidemiol Infect. 1996;117:51–58. doi: 10.1017/s0950268800001126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dalsgaard A, Skov M N, Serichantalergs O, Echeverria P, Meza R, Taylor D N. Molecular evolution of Vibrio cholerae O1 isolated in Lima, Peru, from 1991 to 1995. J Clin Microbiol. 1997;35:1151–1156. doi: 10.1128/jcm.35.5.1151-1156.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Echeverria P, Harrison B A, Tirapat C, McFarland A. Flies as a source of enteric pathogens in a rural village in Thailand. Appl Environ Microbiol. 1983;46:32–36. doi: 10.1128/aem.46.1.32-36.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Echeverria P, Hoge C W, Bodhidatta L, Serichantalergs O, Dalsgaard A, Eampokalap B, Perrault J, Pazzaglia G, O'Hanley P, English C. Molecular characterization of Vibrio cholerae O139 isolates from Asia. Am J Trop Med Hyg. 1995;52:124–127. doi: 10.4269/ajtmh.1995.52.124. [DOI] [PubMed] [Google Scholar]
- 16.Faruque S M, Albert M J, Mekalanos J J. Epidemiology, genetics, and ecology of toxigenic Vibrio cholerae. Microbiol Mol Biol Rev. 1998;62:1301–1314. doi: 10.1128/mmbr.62.4.1301-1314.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Faruque S M, Asadulghani, Saha M N, Abdul Alim A R M, Albert M J, Nasirul Islam K M, Mekalanos J J. Analysis of clinical and environmental strains of nontoxigenic Vibrio cholerae for susceptibility to CTX phage: molecular basis for origination of new strains with epidemic potential. Infect Immun. 1998;66:5819–5825. doi: 10.1128/iai.66.12.5819-5825.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faruque S M, Rahman M M, Asadulghani K M, Islam N, Mekalanos J J. Lysogenic conversion of environmental Vibrio mimicus strains by CTXPhi. Infect Immun. 1999;67:5723–5729. doi: 10.1128/iai.67.11.5723-5729.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fesano A, Baudry B, Pumplin A W, Wassermann S S, Tall B D, Ketley J M, Kaper J B. Vibrio cholerae produces a second enterotoxin which affects intestinal tight junctions. Proc Natl Acad Sci USA. 1991;88:5242–5246. doi: 10.1073/pnas.88.12.5242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heidelberg J F, Elsen J A, Nelson W C, et al. DNA sequence of both chromosomes of the cholera pathogen Vibrio cholerae. Nature. 2000;406:477–484. doi: 10.1038/35020000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Honda T, Iida T. The pathogenicity of Vibrio parahaemolyticus and the role of the thermostable direct haemolysin and related haemolysins. Rev Med Microbiol. 1993;4:106–113. [Google Scholar]
- 22.Kado C I, Liu S T. Rapid procedure for detection and isolation of large and small plasmids. J Bacteriol. 1981;145:1365–1373. doi: 10.1128/jb.145.3.1365-1373.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karaolis D K R, Johnson J A, Bailey C C, Boedeker E C, Kaper J B, Reeves P R. A Vibrio cholerae pathogenicity island associated with epidemic and pandemic strains. Proc Natl Acad Sci USA. 1998;95:3134–3149. doi: 10.1073/pnas.95.6.3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karaolis D K R, Somara S, Maneval D R J, Johnson J A, Kaper J B. A bacteriophage encoding a pathogenicity island, a type-IV pilus and a phage receptor in cholera bacteria. Nature. 1999;399:375–379. doi: 10.1038/20715. [DOI] [PubMed] [Google Scholar]
- 25.Kovach M E, Shaffer M D, Peterson K M. A putative integrase gene defines the distal end of a large cluster of ToxR-regulated colonization genes in Vibrio cholerae. Microbiology. 1996;142:2165–2174. doi: 10.1099/13500872-142-8-2165. [DOI] [PubMed] [Google Scholar]
- 26.Lin W, Fullner K, Clayton R, Sexton J, Rogers M, Calia K, Calderwood S, Fraser C, Mekalanos J. Proceedings of the U.S./Japan Meeting on Cholera and Enteric Diseases. 1998. Identification of a Vibrio cholerae RTX toxin gene cluster that is tightly linked to the cholera toxin prophage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazel D, Dychinco B, Webb V A, Davies J. A distinctive class of integron in the Vibrio cholerae genome. Science. 1998;280:605–608. doi: 10.1126/science.280.5363.605. [DOI] [PubMed] [Google Scholar]
- 28.McCarthy S A, Khambaty F M. International dissemination of epidemic Vibrio cholerae by cargo ship ballast and other nonpotable waters. Appl Environ Microbiol. 1994;60:2597–2601. doi: 10.1128/aem.60.7.2597-2601.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mekalanos J J. Duplication and amplification of toxin genes in Vibrio cholerae. Cell. 1983;35:253–263. doi: 10.1016/0092-8674(83)90228-3. [DOI] [PubMed] [Google Scholar]
- 30.Morris J G J, Wilson R, Davis B R, Wachsmuth I K, Riddle C F, Wathen H G, Pollard R A, Blade P A. Non-O group 1 Vibrio cholerae gastroenteritis in the United States. Ann Intern Med. 1981;94:656–658. doi: 10.7326/0003-4819-94-5-656. [DOI] [PubMed] [Google Scholar]
- 31.National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing. Approved standard M100–S8. Vol. 18. Wayne, Pa: National Committee for Clinical Laboratory Standards; 1998. [Google Scholar]
- 32.Ogawa A, Kato J, Watanabe H, Nair B G, Takeda T. Cloning and nucleotide sequence of a heat-stable enterotoxin gene from Vibrio cholerae non-O1 isolated from a patient with traveler's diarrhea. Infect Immun. 1990;58:3325–3329. doi: 10.1128/iai.58.10.3325-3329.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olsen J E, Larsen J L. Restriction fragment length polymorphism of the Vibrio anguillarum serovar O1 virulence plasmid. Appl Environ Microbiol. 1990;56:3130–3132. doi: 10.1128/aem.56.10.3130-3132.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pearson G D N, Woods A, Chaing S, Mekalanos J J. CTX genetic element encodes a site-specific recombination system and an intestinal colonization factor. Proc Natl Acad Sci USA. 1993;90:3750–3754. doi: 10.1073/pnas.90.8.3750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Popovic T, Bopp C A, Olsvik Ö, Wachsmuth K. Epidemiologic application of a standardized ribotype scheme for Vibrio cholerae O1. J Clin Microbiol. 1993;31:2474–2482. doi: 10.1128/jcm.31.9.2474-2482.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rubin E J, Lin W, Mekalanos J J, Waldor M K. Replication and integration of a Vibrio cholerae cryptic plasmid linked to the CTX prophage. Mol Microbiol. 1998;28:1247–1254. doi: 10.1046/j.1365-2958.1998.00889.x. [DOI] [PubMed] [Google Scholar]
- 37.Rubin E J, Waldor M K, Mekalanos J J. Mobile genetic elements and the evolution of new epidemic strains of V. cholerae. In: Krause R M, editor. Emerging infections. New York, N.Y: Academic Press; 1998. p. 9.x-y. [Google Scholar]
- 38.Rudra S, Mahajan R, Kathur M, Kathuria K, Talwar V. Cluster of cases of clinical cholera due to Vibrio cholerae O10 in east Delhi. Indian J Med Res. 1996;103:71–73. [PubMed] [Google Scholar]
- 39.Sakazaki R. Bacteriology of vibrio and related organisms. In: Barua D, Greenough B R, editors. Cholera. New York, N.Y: Plenum Medical Book Company; 1992. pp. 37–55. [Google Scholar]
- 40.Sharma C, Thungapathra M, Ghosh A, et al. Molecular analysis of non-O1, non-O139 Vibrio cholerae associated with an unusual upsurge in the incidence of cholera-like disease in Calcutta, India. J Clin Microbiol. 1998;36:756–763. doi: 10.1128/jcm.36.3.756-763.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Shaw C E, Taylor R K. Vibrio cholerae O395 tcpA pilin gene sequence and comparison of predicted protein structural features to those of type 4 pilin. Infect Immun. 1990;58:3042–3049. doi: 10.1128/iai.58.9.3042-3049.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shimada T, Arakawa E, Itoh K, Okitsu T, Matsushima A, Asai Y, Yamai S, Nakazato T, Nair G B, Albert M J, Takeda Y. Extended serotyping scheme for Vibrio cholerae. Curr Microbiol. 1994;28:175–178. [Google Scholar]
- 43.Tabtieng R, Wattanasri S, Echeverria P, Seriwatana J, Bodhidatta L, Chatkaeomorakot A, Rowe B. An epidemic of V. cholerae El Tor Inaba resistant to several antibiotics with a conjugative group C plasmid coding for type II dihydrofolate reductase in Thailand. Am J Trop Med Hyg. 1989;41:680–686. doi: 10.4269/ajtmh.1989.41.680. [DOI] [PubMed] [Google Scholar]
- 44.Waldor M K, Mekalanos J J. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272:1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 45.Wright A C, Guo Y, Johnson J A, Nataro J P, Morris J G., Jr Development and testing of a non-radioactive DNA oligonucleotide probe that is specific for Vibrio cholerae cholera toxin. J Clin Microbiol. 1992;30:2302–2306. doi: 10.1128/jcm.30.9.2302-2306.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]