Abstract
A single-tube real-time reverse transcription-PCR (RT-PCR) assay for enterovirus detection in cerebrospinal fluid (CSF) was developed based on a fluorogenic probe and primers directed to highly conserved sequences in the 5′ untranslated region of the enterovirus genome. Quantitative detection of enterovirus genome was demonstrated in a linear range spanning at least 5 logs. Endpoint titration experiments revealed that the in-tube detection limit of the assay was 11.8 enterovirus genome equivalents (95% detection rate) corresponding in our current extraction protocol to 592 enterovirus genome equivalents per ml of CSF. Twenty CSF specimens not suspected of viral meningitis were all found to be negative, and no cross-reactivity with herpes simplex virus type 1 and type 2, varicella-zoster virus, rhinovirus type 53, and influenza viruses A and B was observed. Nineteen CSF specimens from 70 patients suspected of viral meningitis were determined to be positive by PCR (27.1%), whereas only 17 were found to be positive by viral culture (24.3%). The sensitivity of the assay was 100% and the specificity was 96.2% compared to viral culture. Data from the real-time RT-PCR assay were available within 4 h. Our data suggest that the novel real-time RT-PCR assay may offer a reliable but significantly faster alternative to viral culture. Owing to the elimination of postamplification detection steps, its conduct required considerably less hands-on time and was associated with a substantially reduced carryover risk compared to previously described PCR-based enterovirus detection assays.
Enterovirus infections account for a substantial number of aseptic meningitis and encephalitis patients requiring hospitalization in the summer and fall (21). Viral culture is the “gold standard” used to diagnose enteroviral infections in cerebrospinal fluid (CSF) specimens, but it takes several days to be conclusive. A rapid diagnostic test may therefore have a strong impact on the diagnosis and clinical management of viral meningitis (16). During the last decade, different reverse transcription-PCR (RT-PCR) assays were developed as a more convenient alternative to viral culture (1, 2, 15, 18, 22, 24). Various panenterovirus primer sets directed to highly conserved sequences in the 5′ untranslated region (5′UTR) of the enterovirus genome have been designed (16). Many of these assays require gel electrophoresis to detect the amplified product and are in a (semi-)nested format, which is associated with considerable hands-on time, poses a serious hazard for amplification product carryover, and limits the number of specimens that can be processed simultaneously. Specimen throughput is increased in commercial assays based on reverse hybridization to detect the amplification product in microplate format, but the cross-contamination risk, as well as the laborious and time-consuming postamplification detection steps, remains (19).
In recent years, TaqMan technology combined the 5′ nuclease activity of Taq DNA polymerase and Förster resonance energy transfer to detect and quantify amplification product in a closed tube format (3, 7). This real-time quantitative PCR method has proven to be very sensitive and renders post-PCR detection steps obsolete, thereby reducing hands-on and total assay time and also the risk of amplification product carryover. The technology has recently been applied to a variety of target sequences, including the detection and/or quantification of viral genomes in clinical samples (4–6, 8, 10, 14, 20, 23). In the present study, a single-tube fluorogenic 5′ nuclease RT-PCR assay for enterovirus detection is described. After determination of the analytical sensitivity and specificity of the assay, CSF specimens from patients clinically suspected of having viral meningitis were tested and compared with the results from the corresponding viral cultures.
(Data from this study were presented in part at the 11th European Congress of Clinical Microbiology and Infectious Diseases, Istanbul, Turkey, April 1 to 4, 2001.)
MATERIALS AND METHODS
Primers and probe.
Primers (Gibco-BRL, Merelbeke, Belgium) and probe (Eurogentec, Seraing, Belgium) were designed using Primer Express 1.0 software (Applied Biosystems, Foster City, Calif.) and were directed to the conserved sequences in the 5′UTR of the enterovirus genome (Table 1). The reporter (FAM) and quencher dyes (TAMRA) were attached to the 5′ end and 3′ end of the probe, respectively.
TABLE 1.
Primer and probe sequences used in the enterovirus real-time RT-PCR assay
| Primer or probe | Sequence | Tma (°C) | Length (no. of nucleotides) |
|---|---|---|---|
| Forward primer | 5′-CCCTGAATGCGGCTAATCC-3′ | 59.1 | 19 |
| Reverse primer | 5′-ATTGTCACCATAAGCAGCCA-3′ | 55.3 | 20 |
| Probe | 5′-AACCGACTACTTTGGGTGTCCGTGTTTC-3′ | 67.5 | 28 |
Calculated by using Primer Express software version 1.0.
Enterovirus RNA standard.
The enterovirus RNA standard (Armored RNA from Ambion RNA Diagnostics, Austin, Tex.) comprises 196 bp of the 5′UTR from the poliovirus 1 genome (GenBank accession number POLIOS1), including the conserved sequences to which primers and probe are directed. The RNA sequence is protected from degradation because it has been packaged into an RNA phage sequence (13). The standard stock solution contained 4,600 enterovirus genome equivalents (Geq) per μl (C. Walker-Peach, unpublished data) and was stored at 4°C. It was diluted in nuclease-free water prior to use.
RNA extraction.
RNA was extracted from 140 μl of CSF or viral culture supernatant by using the QIAamp viral RNA extraction kit (Qiagen, Hilden, Germany) according to the manufacturer's instructions. RNA was recovered in 70 μl of nuclease-free water and either used immediately or stored at −80°C until further analysis. Enterovirus RNA standard was released from its phage coat either by incubation at 75°C for 3 min or by extraction as described above.
Real-time RT-PCR.
Assay conditions were optimized in an ABI Prism 7700 SDS (Applied Biosystems, Foster City, Calif.). Then, 10 μl of RNA extract was added to the reaction mix, resulting in a final concentration of 300 nM forward primer, 900 nM reverse primer, 150 nM dual-labeled fluorogenic probe, 1× TaqMan universal master mix, and 1× RT mix (One-Step RT-PCR TaqMan reagent kit from Applied Biosystems, Foster City, Calif.). RNA extracts were substituted by nuclease-free water in negative control reactions. Samples were assayed in duplicate reactions in a total volume of 25 μl. RT was performed at 48°C for 30 min and immediately followed by activation of AmpliTaq Gold DNA polymerase at 95°C for 10 min. A total of 40 cycles was performed, consisting of a denaturation step at 95°C for 15 s and a combined annealing-extension step at 60°C for 1 min. Fluorescence data were collected during each annealing-extension step and analyzed by using ABI Prism SDS software version 1.6.3. Reaction tubes were discarded unopened after completion of the run.
Dynamic range and limit of detection.
To test the dynamic range of the assay, the RNA extract of viral culture supernatant from echovirus 30 cultured as described below was diluted from 10 to 107 times. Each dilution and a negative control sample containing nuclease-free water were tested in duplicate. To determine the limit of detection the enterovirus RNA standard (Armored RNA) was serially diluted in a pool of negative CSF specimens ranging from 31,250 to 2 Geq/ml and extracted as described above. Eight replicates of each extract were tested.
Specimens for serotype reactivity and specificity testing.
Serotype reactivity was tested on freeze-dried quality control samples of viral culture supernatants from coxsackieviruses A16 and B6 and echoviruses 6 and 30 from the College of American Pathologists Surveys Program and from coxsackieviruses A9 and B5, echovirus 11, and enterovirus 71 from the European Union Quality Control Concerted Action. Likewise, specificity was tested on freeze-dried quality control samples of viral culture supernatants from herpesvirus type 1 and type 2, varicella-zoster virus, and influenza virus A or B, also obtained from the College of American Pathologists Surveys Program. Samples were reconstituted by using the volume of sterile water as stated by the package insert and kept frozen at −80°C until analysis. Viral culture supernatant of rhinovirus 53 (American Type Culture Collection, Manassas, Va.) and the reconstituted quality control samples were extracted as described above and tested in duplicate with the enterovirus real-time RT-PCR assay. Organism-specific PCR-based assays confirmed the presence of the viral genomes in the extracts used for specificity testing (data not shown).
Comparison of real-time RT-PCR with viral culture.
CSF specimens with clinical suspicion of viral meningitis were received during the summer and fall of 2000. From a total of 70 specimens, 58 were obtained from pediatric patients (age 14 years or younger). All specimens were immediately stored at 4°C and processed within 24 h of receipt. Specimens were extracted as described above and immediately analyzed in duplicate by real-time RT-PCR, as well as negative control reactions and positive control reactions containing 72 enterovirus Geq.
Viral culture was performed on human embryonic lung fibroblast cells (MRC-5) and human embryonic rhabdomyosarcoma cells (RD) (both from BioWhittaker Europe, Verviers, Belgium) grown in Eagle minimal essential medium (EMEM; BioWhittaker Europe) supplemented with 10% (MRC-5) or 8% (RD) fetal calf serum (Biochrom, Berlin, Germany). After inoculation by allowing 0.5 ml of CSF specimen to adsorb to the cells for 1 h, the cultures were maintained in EMEM supplemented with 2% fetal calf serum at 36°C in 5% CO2. The cells were examined daily for the presence of viral cytopathogenic effect (CPE). The cultures were passed (cells were lysed and lysate was used for inoculation of a new monolayer) after 7 days and again examined daily for another 7 days. Cultures showing CPE suggestive for enterovirus were passed once more in the same cell line for confirmation.
Statistical analysis.
A detailed probit analysis was performed using Systat statistical software version 10. Probability plot curve-fitting parameters were determined by using Sigmaplot version 10 graphical software.
RESULTS
Dynamic range and limit of detection.
The 10-fold dilution series of viral culture supernatant from echovirus 30 demonstrated that the dynamic range of the assay spans at least 5 logs. The lowest dilution factor of 10 corresponded to a threshold cycle (Ct) of 16.2, while the highest dilution factor of 106 corresponded to a Ct of 34.1. Eight replicates from an endpoint dilution series of the enterovirus RNA standard in a pool of negative CSF were tested with the new assay. A positive signal was detected in all eight replicates down to 1,250 enterovirus Geq/ml of CSF. At 250 Geq/ml, six of eight replicates were positive (75%) and at 50 Geq/mL two of eight were positive (25%), whereas at 10 Geq/ml one of eight replicates still yielded a positive signal. Probit analysis of these numbers indicated that the limit of detection (95% detection rate) was 11.8 Geq per reaction, corresponding to 592 Geq/ml of CSF in our current extraction protocol. The 50% detection rate was 2.4 Geq/reaction, corresponding to 120 Geq/ml of CSF. From the standard curve of the enterovirus RNA standard it could be concluded that the presence of a single enterovirus Geq in the reaction theoretically corresponded to a Ct of 37.3.
Extraction efficiency.
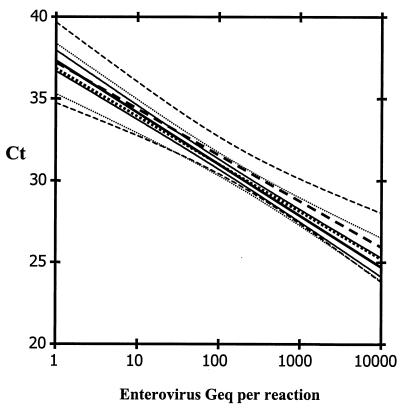
Enterovirus RNA standard released from the phage coat proteins and diluted in nuclease-free water was compared with the enterovirus RNA standard diluted prior to extraction, either in nuclease-free water or in a pool of negative CSF specimens. No significant differences between slopes or y intercepts of the respective standard curves were observed (Fig. 1). Hence, the extraction efficiency was equal at all dilutions and no evidence for PCR inhibition was observed.
FIG. 1.
Standard curves with 95% confidence limits from a dilution series of the stabilized enterovirus RNA standard (solid lines), the stabilized enterovirus RNA standard extracted from water (dotted lines), or from a pool of negative CSF specimens (dashed lines). RNA extracts were assayed in duplicate.
Serotype reactivity and specificity.
Enterovirus serotype reactivity was tested to confirm that primers and probe recognize various enterovirus serotypes. Eight different quality control samples containing coxsackieviruses A9 and A16, coxsackieviruses B5 and B6, echoviruses 6, 11, and 30, and enterovirus 71 were processed identically to clinical CSF specimens and were found to be positive when tested by the enterovirus real-time RT-PCR. The specificity of the assay was demonstrated by testing nucleic acid extracts of viral culture supernatant. No positive signals were observed with nucleic acid extracts from herpes simplex virus type 1 and 2, varicella-zoster virus, rhinovirus type 53, and influenza virus A and B viral culture supernatants. In addition, CSF specimens from 20 patients with no clinical suspicion of viral meningitis were all negative.
Comparison of the real-time RT-PCR assay with viral culture.
A total of 70 CSF specimens were tested in parallel by real-time RT-PCR assay and viral culture. Results from the real-time RT-PCR assay were generally available in less than 4 h, including extraction of the CSF specimens. Enterovirus was detected in 17 specimens by viral culture (24.3%), whereas real-time RT-PCR detected enterovirus in 2 additional specimens (27.1%). The Ct value of the 17 culture-positive samples was 35.2 ± 2.4 compared to 35.4 and 35.6 from the 2 culture-negative samples. Hence, the sensitivity of the real-time RT-PCR was 100.0% compared to viral culture. The specificity, positive predictive value, and negative predictive value were 96.2, 89.5, and 100.0%, respectively. For 48 of the 51 patients with a negative enterovirus test, a final diagnosis unrelated to viral meningitis was eventually retained.
DISCUSSION
A sensitive and rapid 5′ nuclease-based RT-PCR assay to detect enterovirus genome in CSF specimens was developed. Detection rates in CSF specimens indicate that the new RT-PCR assay is at least as sensitive as viral culture as performed in our lab. The currently developed assay format is furthermore amenable to reproducible quantification over a wide dynamic range. This may open perspectives for patient follow-up once antiviral therapy is available. A new antiviral agent, pleconaril, interfering with enterovirus replication is currently being tested in phase III clinical trials (11, 17).
Eight different enteroviruses of which the serotypes were known were positive when tested with our assay. Primer-probe combinations directed to the 5′UTR conserved sequences should be able to detect all enterovirus strains (16), except for echoviruses 22 and 23, which have recently been reclassified as parechovirus because of their divergent genome sequence (12). Although the assay did not show cross-reactivity with rhinovirus type 53, cross-reactivity with any other rhinoviruses cannot be completely excluded due to their close homology with enteroviruses. The presence of rhinovirus in CSF is unlikely, but sufficient care should be taken during specimen collection and processing to avoid contaminating the sample with rhinoviruses from other sources.
Enterovirus was detected in 19 of 70 patients clinically suspected of having viral meningitis. This fraction may seem rather low but represents only specimens for which a sufficient volume was available to perform both real-time RT-PCR and viral culture. Additional enterovirus infections were identified in a number of specimens for which no viral culture was performed (data not shown). For most enterovirus-negative patients, a final diagnosis of viral meningitis was excluded. However, no other etiology could be identified for three patients presenting with clinical and laboratory findings indicative of viral meningitis. The symptoms may have been caused by other unrelated viruses or by an enterovirus serotype with a divergent primer or probe binding site. Also, the viral load may have been below the limit of detection of the assay in its current format. Finally, the presence of inhibitory substances may have compromised RT or amplification efficiency.
A lower viral load does not seem to explain why two PCR-positive samples were not confirmed by viral culture. Indeed, the Ct values of those two specimens were not different from the Ct values of the PCR-positive, viral-culture-positive specimens. False positivity due to cross-contamination cannot entirely be excluded but seems unlikely. In addition to the closed tube format of the assay and four separate rooms for specimen handling, RT-PCR master mix preparation, RT-PCR setup, and RT-PCR conduct, stringent precautions were taken to prevent carryover of amplification product, and no other positive specimens were handled during the processing of these two particular specimens. The presence of a positive signal in both duplicate reactions further decreases the likelihood of accidental contamination. Unfortunately, serotype information is lacking to exclude the presence of an enterovirus serotype that is difficult to grow.
Other closed-tube detection formats have recently been developed. During the conduct of our study, a real-time 5′ nuclease assay to detect and quantify the presence of enterovirus in sludge water was published, with slightly different primer and probe sequences (9). The authors of that study supplemented their reaction mix with considerable amounts of T4 gene protein 32. The protein acts as a helix destabilizer and may improve amplification efficiency by rendering secondary structures in single-stranded nucleic acid strands more accessible to primers, probe, and/or Taq DNA polymerase. A different reaction mix composition may explain the difference in sensitivity in our assay. The absence of extra additives in our assay should increase its robustness and reproducibility and make it less prone to human error. Also, a nucleic acid sequence-based amplification-based molecular beacon assay for enterovirus detection in CSF was recently presented (P. Sillekens, M. van de Wiel-van de Meer, H. Foolen, A. Malhotra, C. Ginocchio, and J. Fox, ESCV Winter Meet., 2001). Although data are collected in real time, the assay has only limited quantitative potential since it relies on endpoint detection.
In summary, this novel single-tube real-time RT-PCR assay for the detection of enterovirus in CSF specimens offers the combination of high sensitivity, speed, reduced hands-on time, and minimal risk on amplification product carryover. However, further tests should be performed to evaluate its performance in a routine diagnostic setting.
ACKNOWLEDGMENT
We thank M. Knaapen for his valuable advice and criticism.
REFERENCES
- 1.Chapman N M, Tracy S, Gauntt C J, Fortmueller U. Molecular detection and identification of enteroviruses using enzymatic amplification and nucleic acid hybridization. J Clin Microbiol. 1990;28:843–850. doi: 10.1128/jcm.28.5.843-850.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Halonen P, Rocha E, Hierholzer J, Holloway B, Hyypia T, Hurskainen P, Pallansch M. Detection of enteroviruses and rhinoviruses in clinical specimens by PCR and liquid-phase hybridization. J Clin Microbiol. 1995;33:648–653. doi: 10.1128/jcm.33.3.648-653.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Holland P M, Abramson R D, Watson R, Gelfand D H. Detection of specific polymerase chain reaction product by utilizing the 5′–3′ exonuclease activity of Thermus aquaticus DNA polymerase. Proc Natl Acad Sci USA. 1991;88:7276–7280. doi: 10.1073/pnas.88.16.7276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kessler H H, Muhlbauer G, Rinner B, Stelzl E, Berger A, Dorr H W, Santner B, Marth E, Rabenau H. Detection of herpes simplex virus DNA by real-time PCR. J Clin Microbiol. 2000;38:2638–2642. doi: 10.1128/jcm.38.7.2638-2642.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kimura H, Kido S, Ozaki T, Tanaka N, Ito Y, Williams R K, Morishima T. Comparison of quantitations of viral load in varicella and zoster. J Clin Microbiol. 2000;38:2447–2449. doi: 10.1128/jcm.38.6.2447-2449.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kleiber J, Walter T, Haberhausen G, Tsang S, Babiel R, Rosenstraus M. Performance characteristics of a quantitative, homogeneous TaqMan RT-PCR test for HCV RNA. J Mol Diagn. 2000;2:158–166. doi: 10.1016/S1525-1578(10)60632-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Livak K J, Flood S J, Marmaro J, Giusti W, Deetz K. Oligonucleotides with fluorescent dyes at opposite ends provide a quenched probe system useful for detecting PCR product and nucleic acid hybridization. PCR Methods Appl. 1995;4:357–362. doi: 10.1101/gr.4.6.357. [DOI] [PubMed] [Google Scholar]
- 8.Martell M, Gomez J, Esteban J I, Sauleda S, Quer J, Cabot B, Esteban R, Guardia J. High-throughput real-time reverse transcription-PCR quantitation of hepatitis C virus RNA. J Clin Microbiol. 1999;37:327–332. doi: 10.1128/jcm.37.2.327-332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Monpoeho S, Dehee A, Mignotte B, Schwartzbrod L, Marechal V, Nicolas J C, Billaudel S, Ferre V. Quantification of enterovirus RNA in sludge samples using single tube real-time RT-PCR. BioTechniques. 2000;29:88–93. doi: 10.2144/00291st03. [DOI] [PubMed] [Google Scholar]
- 10.Morris T, Robertson B, Gallagher M. Rapid reverse transcription-PCR detection of hepatitis C virus RNA in serum by using the TaqMan fluorogenic detection system. J Clin Microbiol. 1996;34:2933–2936. doi: 10.1128/jcm.34.12.2933-2936.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nowak-Wegrzyn A, Phipatanakul W, Winkelstein J A, Forman M S, Lederman H M. Successful treatment of enterovirus infection with the use of Pleconaril in 2 infants with severe combined immunodeficiency. Clin Infect Dis. 2001;32:E13–E14. doi: 10.1086/317523. [DOI] [PubMed] [Google Scholar]
- 12.Oberste M S, Maher K, Pallansch M A. Complete sequence of echovirus 23 and its relationship to echovirus 22 and other human enteroviruses. Virus Res. 1998;56:217–223. doi: 10.1016/s0168-1702(98)00080-x. [DOI] [PubMed] [Google Scholar]
- 13.Pasloske B L, Walkerpeach C R, Obermoeller R D, Winkler M, DuBois D B. Armored RNA technology for production of ribonuclease-resistant viral RNA controls and standards. J Clin Microbiol. 1998;36:3590–3594. doi: 10.1128/jcm.36.12.3590-3594.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pevenstein S R, Williams R K, McChesney D, Mont E K, Smialek J E, Straus S E. Quantitation of latent varicella-zoster virus and herpes simplex virus genomes in human trigeminal ganglia. J Virol. 1999;73:10514–10518. doi: 10.1128/jvi.73.12.10514-10518.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Read S J, Kurtz J B. Laboratory diagnosis of common viral infections of the central nervous system by using a single multiplex PCR screening assay. J Clin Microbiol. 1999;37:1352–1355. doi: 10.1128/jcm.37.5.1352-1355.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Romero J R. Reverse-transcription polymerase chain reaction detection of the enteroviruses. Arch Pathol Lab Med. 1999;123:1161–1169. doi: 10.5858/1999-123-1161-RTPCRD. [DOI] [PubMed] [Google Scholar]
- 17.Romero J R. Pleconaril: a novel antipicornaviral drug. Expert Opin Investig Drugs. 2001;10:369–379. doi: 10.1517/13543784.10.2.369. [DOI] [PubMed] [Google Scholar]
- 18.Rotbart H A. Enzymatic RNA amplification of the enteroviruses. J Clin Microbiol. 1990;28:438–442. doi: 10.1128/jcm.28.3.438-442.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rotbart H A, Sawyer M H, Fast S, Lewinski C, Murphy N, Keyser E F, Spadoro J, Kao S Y, Loeffelholz M. Diagnosis of enteroviral meningitis by using PCR with a colorimetric microwell detection assay. J Clin Microbiol. 1994;32:2590–2592. doi: 10.1128/jcm.32.10.2590-2592.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ryncarz A J, Goddard J, Wald A, Huang M L, Roizman B, Corey L. Development of a high-throughput quantitative assay for detecting herpes simplex virus DNA in clinical samples. J Clin Microbiol. 1999;37:1941–1947. doi: 10.1128/jcm.37.6.1941-1947.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sawyer M H. Enterovirus infections: diagnosis and treatment. Pediatr Infect Dis J. 1999;18:1033–1039. doi: 10.1097/00006454-199912000-00002. [DOI] [PubMed] [Google Scholar]
- 22.Schwab K J, De Leon R, Sobsey M D. Concentration and purification of beef extract mock eluates from water samples for the detection of enteroviruses, hepatitis A virus, and Norwalk virus by reverse transcription-PCR. Appl Environ Microbiol. 1995;61:531–537. doi: 10.1128/aem.61.2.531-537.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schweiger B, Zadow I, Heckler R, Timm H, Pauli G. Application of a fluorogenic PCR assay for typing and subtyping of influenza viruses in respiratory samples. J Clin Microbiol. 2000;38:1552–1558. doi: 10.1128/jcm.38.4.1552-1558.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zoll G J, Melchers W J, Kopecka H, Jambroes G, van der Poel H J, Galama J M. General primer-mediated polymerase chain reaction for detection of enteroviruses: application for diagnostic routine and persistent infections. J Clin Microbiol. 1992;30:160–165. doi: 10.1128/jcm.30.1.160-165.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]