ABSTRACT
Current WHO recommendations for monitoring treatment response in adult pulmonary tuberculosis (TB) are sputum smear microscopy and/or culture conversion at the end of the intensive phase of treatment. These methods either have suboptimal accuracy or a long turnaround time. There is a need to identify alternative biomarkers to monitor TB treatment response. We conducted a systematic review of active pulmonary TB treatment monitoring biomarkers. We screened 9,739 articles published between 1 January 2008 and 31 December 2020, of which 77 met the inclusion criteria. When studies quantitatively reported biomarker levels, we meta-analyzed the average fold change in biomarkers from pretreatment to week 8 of treatment. We also performed a meta-analysis pooling the fold change since the previous time point collected. A total of 81 biomarkers were identified from 77 studies. Overall, these studies exhibited extensive heterogeneity with regard to TB treatment monitoring study design and data reporting. Among the biomarkers identified, C-reactive protein (CRP), interleukin-6 (IL-6), interferon gamma-induced protein 10 (IP-10), and tumor necrosis factor alpha (TNF-α) had sufficient data to analyze fold changes. All four biomarker levels decreased during the first 8 weeks of treatment relative to baseline and relative to previous time points collected. Based on limited data available, CRP, IL-6, IP-10, and TNF-α have been identified as biomarkers that should be further explored in the context of TB treatment monitoring. The extensive heterogeneity in TB treatment monitoring study design and reporting is a major barrier to evaluating the performance of novel biomarkers and tools for this use case. Guidance for designing and reporting treatment monitoring studies is urgently needed.
KEYWORDS: tuberculosis, treatment monitoring, biomarkers
INTRODUCTION
In 2018, the global treatment success rate for people with drug-susceptible tuberculosis (TB) was 85% (1). Among the 7.0 million people reported to have received TB treatment in 2018, over 1 million individuals did not receive their treatment. Treatment success drops significantly among people with multidrug-resistant (MDR) TB and people living with HIV, with success rates of 57% and 76%, respectively (1). Continuous monitoring and early identification of people with TB who are at risk of poor treatment outcomes could reduce the number of people who do not complete treatment.
The World Health Organization (WHO) currently recommends sputum smear microscopy or culture conversion at the end of the intensive phase of treatment for monitoring treatment response in adults with pulmonary TB (2). However, these microbiology-based methods have limitations. Both smear microscopy and culture rely on sputum samples, which are not readily available in all populations (e.g., pediatric TB, people living with HIV, extrapulmonary TB) (3–5). Further, both methods are highly operator dependent (6). Smear microscopy is also not able to differentiate viable from nonviable TB, resulting in poor sensitivity and specificity for outcome prediction (7). For TB culture, the limited availability in primary care settings and the delay in time to results constrain its clinical use (8).
There is a clinical and public health need for new treatment monitoring biomarkers and assays that provide quick and accurate predictions of treatment outcomes. To meet the clinical needs for TB treatment monitoring, novel tests that detect biomarkers of interest would ideally be performed on noninvasive samples (e.g., blood, urine) and require limited laboratory expertise and infrastructure. Developments of tests based on host or pathogen biomarkers have previously been summarized in a narrative review article (9). A systematic assessment of these biomarkers is needed to identify those that might represent promising options to optimize treatment monitoring.
In this systematic review, we summarize, for the first time, a set of assays and biomarkers that correlate with TB treatment and, thus, may be of interest for TB treatment monitoring. We provide a summary of the biomarkers and assays identified as well as a more in-depth exploratory evaluation of the longitudinal change in levels of C-reactive protein (CRP), interferon gamma-induced protein 10 (IP-10), interleukin-6 (IL-6), and tumor necrosis factor alpha (TNF-α) during anti-TB treatment.
MATERIALS AND METHODS
We conducted a systematic review of active pulmonary TB treatment monitoring biomarkers and assays that are commercial or have commercial potential. Study selection criteria for this review are illustrated in the PRISMA checklist (Table S1 in the supplemental material).
Search strategy.
We searched six academic databases, including PubMed/MEDLINE, Embase, Web of Science, BIOSIS, Latin American and Caribbean Health Sciences (LILACS), and the Cochrane Database of Systematic Reviews. The full search strategy is presented in Table S2.
Eligibility criteria.
Relevant studies published between 1 January 2008 and 31 December 2020 that were written in English were included. We included randomized clinical trials (RCTs), cohort studies, case-control studies, and cross-sectional studies that investigated the longitudinal change in biomarker levels during anti-TB treatment. We excluded case series, reviews, commentaries/editorials, case reports, mathematical modeling studies, economic analyses, and conference abstracts. We also excluded any study that did not perform reference standard testing at multiple time points throughout treatment and studies with a sample population of less than 10. Studies on children (age less than or equal to 15 years) were excluded given the difficulty of establishing a reference standard in this population. No restrictions were placed on the geographic area or the type of health system setting from where the participants were recruited. We extracted data for three categories of assays and biomarkers identified in consultation with the Foundation for Innovative New Diagnostics (FIND) technology scouting team, including (i) assays that are commercially available in a kit for research purposes, (ii) biomarkers not currently available in a commercial kit but are either under commercial development or have the potential to become commercial (e.g., transcriptomic signatures), and (iii) TB-specific biomarkers or commonly recognized laboratory procedures that are not necessarily commercialized (e.g., 16s rRNA molecular bacterial load assay [MBLA]). We did not include radiological methods or well-established assays such as sputum smear microscopy, culture, and nucleic acid amplification tests (e.g., GeneXpert, Hain). We included studies that used reference standards acceptable for treatment monitoring, which includes sequential measurements of Mycobacterium tuberculosis culture, Xpert MTB/RIF, smear microscopy, and/or clinical outcome. Measurements in comparison to the reference standard were included when at least one time point during treatment follow-up measured the reference standard.
Screening and data extraction.
All publications identified from the search strategy were imported into the reference management database EndNote (version X9), after which duplicate citations were removed. Studies were screened by title and abstract by at least two reviewers (A.J.Z., C.C., N.A.V., and F.L.) before full-text screening. Prior to extraction, two authors (A.J.Z. and C.C.) piloted the data extraction forms independently on a random sample of five papers. An additional reviewer (M.K. and C.M.D.) screened studies for which the inclusion/exclusion criteria were not immediately clear. Two separate Google forms were piloted for data extraction, including (i) summary assessment to extract information relevant to the assay characteristics and study design, and (ii) quantitative assessment to extract biomarker levels (mean/median) and measures of spread (standard deviation, interquartile range) at each follow-up time point. All studies were extracted by at least two reviewers (A.J.Z., C.C., N.A.V., and F.L.). For the quantitative assessment, we only extracted data on biomarkers when quantitative changes in biomarker levels were reported by five or more studies (CRP, IP-10, IL-6, and TNF-α). For the biomarker levels and measures of spread, data were extracted directly from the texts or tables when available. If quantitative data were not available and authors did not respond to the request for data, the data (biomarker level and measure of spread) were extracted directly from available figures (dot plots, box plots, etc.) (10). These data were extracted in duplicate (N.A.V., F.L., and A.J.Z.), and one author (A.J.Z.) reviewed the extracted data and resolved any conflicts. When data were extracted from figures, one author (A.J.Z.) averaged the data across the two extractions.
Assessment of quality and risk of bias.
We evaluated the quality and risk of bias of all included studies for the four domains of the Quality Assessment of Diagnostic Accuracy Score 2 (QUADAS-2), including patient selection, index test, reference standard, and flow and timing. Each domain was evaluated using a set of QUADAS-2 guiding questions (Table S3). Items were scored as “high concern,” “low concern,” or “unclear concern.” The overall risk of bias (Fig. S4) was evaluated as “high risk” for studies with more than one area of high concern, “intermediate risk” for all studies that included one area of high concern, “low risk” for all studies with two or more areas of low concern and no high risk, and “unclear risk” for all studies with three or more areas of unclear concern and no high risk. No commercialized assay for the specific use case of treatment monitoring was identified. Given this, no validated cutoff exists for biomarkers with a quantitative output for monitoring TB response. Thus, cutoffs were not part of the QUADAS-2 assessment for the included studies.
Data analysis.
We investigated how biomarker levels changed over time. When biomarkers had five or more studies that numerically or graphically presented the measures of central tendency and measures of spread at different follow-up time points (10), we evaluated the fold change in biomarker levels relative to the previous time point collected. For this analysis, we did not include studies that used TB-antigen stimulated samples.
We first standardized the data extracted into sample mean and standard deviation values. Specifically, we applied the Box-Cox (BC) method proposed by McGrath et al. to estimate the sample mean and standard deviation from studies that reported the median and first and third quartiles (11, 12). In one study with highly skewed data at some time points (CRP from Ferrian et al.), the BC method produced estimates that were biologically implausible. For this study, we estimated the sample means and standard deviations by maximum likelihood with several candidate models (normal, log-normal, gamma, Weibull) and selected the model with the largest likelihood.
The fold change at each follow-up time was calculated as the difference between the current and previously recorded value divided by the previously recorded value as follows:
Fold changes were plotted separately for each study (Fig. S7). When studies reported the change in biomarker level across different groups of patients (e.g., fast responders, slow responders), we pooled the results to examine the average changes in biomarker level across patients that responded to treatment. Fast responders were generally defined as individuals who experienced culture conversion before 8 to 12 weeks of treatment, while individuals who experienced culture conversion beyond 8 to 12 weeks of treatment were defined as slow responders.
To characterize how biomarker levels change with respect to treatment, we performed two meta-analyses, (i) a meta-analysis of the fold change in biomarker levels between baseline and 8 weeks of treatment for studies that reported biomarker levels at 8 weeks (the end of the intensive phase of treatment), and (ii) a meta-analysis pooling fold change since previous time point using a random intercept model. Both analyses used the metafor package for R (version 4.0.6) at the study level (13). For estimated fold change of each biomarker, 95% confidence intervals were also calculated to assess the statistical significance for each biomarker. As is common in longitudinal meta-analyses, the primary studies did not report data on the correlation between the effect estimates at the different follow-up times. For each biomarker, we constructed approximate covariance matrices of the study-specific effect estimates by assuming that the correlation between all pairs of mean biomarker values in a given study was the same value ρ (14). We used the correlation parameter, ρ = 0.5, in the primary analysis and used ρ values of 0, 0.25, and 0.75 in sensitivity analyses (Table S8). We also included a sensitivity analysis for the random intercept CRP model, including and excluding Ferrian et al (Table S9). The list of studies included in the week 8 meta-analysis and the fold change meta-analysis can be found in Table S6. The code for all analyses is publicly available on GitHub (https://github.com/stmcg/tmsr).
RESULTS
Search results.
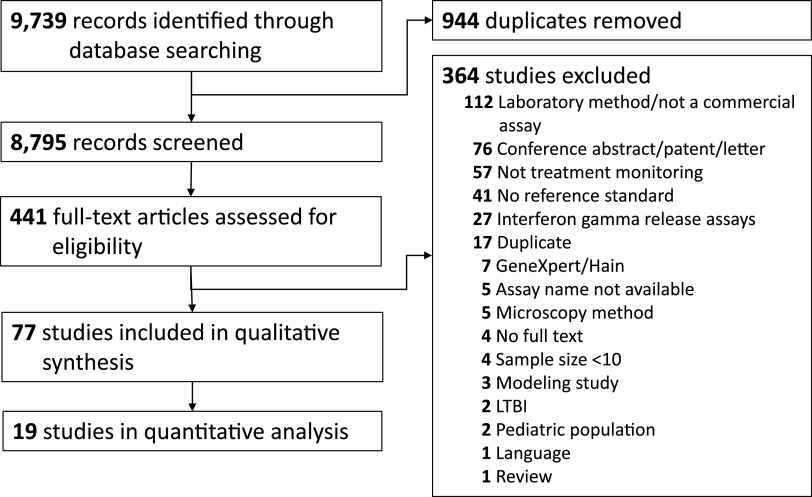
After removing duplicate records, 8,795 publications were screened (title and abstract). Among these, 441 were identified for full-text review, of which 77 were included in the review for the summary (qualitative) assessment (15–91). Nineteen of the records were included in the detailed (quantitative) assessment, including the meta-analyses for the biomarkers CRP, IP-10, IL-6, and TNF-α.
Out of the 441 records that underwent full-text screening, 112 were excluded because the assays did not align with any of the three predefined assays of interest (with regard to commercialization and relevance to TB treatment monitoring). The majority of these were in-house laboratory methods (Fig. 1). Fifty-seven of the studies excluded were not treatment monitoring studies, while 41 did not utilize reference standards during follow-up and/or by the end of therapy. Studies that examined well-established diagnostics, such as interferon gamma release assays (IGRAs; n = 27), GeneXpert/Hain (n = 7), and microscopy methods (n = 5), were excluded, as prior systematic reviews have already characterized the treatment monitoring capabilities of these assays (7, 92–94).
FIG 1.
PRISMA flow diagram for literature search and paper selection.
Characteristics of included studies.
General study demographics and characteristics are summarized in Table 1. Most of the studies were limited to the discovery phase and were conducted in single-center studies in a single country (96%). All but two examined patients from medium-high TB burden countries (94%). Participants with drug resistance (including multidrug resistance) at baseline were included in 26% of studies. More than half of the studies did not indicate whether participants had a history of prior TB, and 88% of studies did not indicate whether participants had previously received the bacillus Calmette-Guérin vaccine. Finally, about one-quarter of studies included people living with HIV.
TABLE 1.
Study characteristics of the 77 studies included in the qualitative synthesis
| Study characteristicc | Value (no. [%]) (n = 77) |
|---|---|
| TB burden of country of enrollmenta | |
| Low (<10 cases per 100,000 population per yr) | 2 (2.60) |
| Middle (11 to 40 cases per 100,000 population per yr) | 8 (10.39) |
| High (>40 cases per 100,000 population per yr and/or WHO list of 30 highest burden countries) | 64 (83.12) |
| Multisiteb | 3 (3.90) |
| Persons with drug resistance at baseline included | |
| Yes | 20 (25.97) |
| No | 20 (25.97) |
| Unclear/not reported | 37 (48.05) |
| Persons with a history of prior TB included | |
| Yes | 17 (22.08) |
| No | 21 (27.27) |
| Unclear/not reported | 39 (50.65) |
| Persons with BCG vaccination included | |
| Yes | 9 (11.69) |
| Unclear/Not reported | 68 (88.31) |
| Persons living with HIV included | |
| Yes | 19 (24.68) |
| No | 45 (58.44) |
| Unclear/not reported | 13 (16.88) |
Based on 2019 data.
Three studies recruited participants from different countries with different TB burden status.
TB, tuberculosis; WHO, World Health Organization; BCG, Bacillus Calmette-Guérin.
Quality and risk of bias assessment (QUADAS-2).
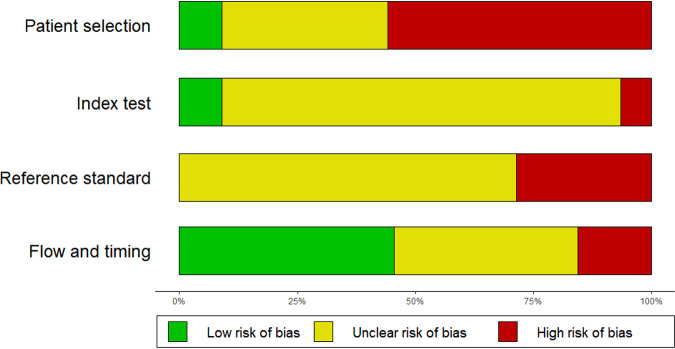
When considering the four main categories of the QUADAS-2 quality and risk of bias assessment tool, “patient selection,” “index test,” “reference standard,” and “flow and timing,” only three studies (4%) had an overall low risk of bias (Fig. S4 in the supplemental material). The QUADAS-2 assessments are summarized in Fig. 2. Specifically, the risk of bias for patient selection was high for studies that used a case-control study design. Many studies excluded smear-negative participants, which also introduced bias in the patient selection strategy. Most studies did not report whether the reference standard was blinded while interpreting the results of the index. Regarding treatment monitoring reference standards, all studies that used culture as a reference standard received an “unclear risk of bias” since the accuracy of culture for this use case is not 100%. Studies that used smear microscopy received a “high risk of bias.” Finally, the flow and timing of the study were generally “low risk of bias,” as the majority of samples were either frozen or processed immediately. For some studies, the loss to follow-up throughout the treatment monitoring period resulted in a high risk of bias.
FIG 2.
Summary of the QUADAS-2 risk of bias assessment.
Summary assessment of treatment monitoring biomarkers.
Across all studies, 81 different biomarkers were identified (Table S5). Forty-nine biomarkers were evaluated in just one study. Most of the biomarkers were host-response markers, with the exception of lipoarabinomannan (LAM) in urine, sputum, and plasma (30, 34, 48, 86), 16s rRNA in sputum (39, 41, 42, 75, 87), 85B mRNA in sputum (19, 60, 64, 81), and IS6110 insertion element in sputum (60). Among the host-response biomarkers, most biomarkers were cytokine proteins measured in blood, both proinflammatory (e.g., IL-1, IL-6, and TNF-α) and anti-inflammatory (e.g., IL-4 and IL-10), which were most commonly analyzed using plasma or serum samples on commercially available research enzyme-linked immunosorbent assay (ELISA) kits. Several chemokines were also investigated, including interferon-inducible T cell alpha chemoattractant (I-TAC), monokine induced by interferon gamma (MIG), and IP-10 (also known as CXCL11, CXCL9, and CXCL10, respectively) (26, 27, 56). Of all biomarkers, IP-10 was the most frequently analyzed biomarker for treatment monitoring, with 11 studies investigating longitudinal changes in marker level (23, 27, 29, 33, 36, 43, 45, 47, 51, 59, 72, 90). Several blood-based transcriptomic and gene expression signatures were examined as treatment monitoring markers, including the parsimonious 3-gene Sweeney3 signature (35, 85), the RISK6 signature (69), and the RISK11 signature (28). Two studies explored changes in breath-based markers such as fractional exhaled nitric oxide (FeNO) and volatile organic compounds (VOCs) (71, 88), while another study by Lee et al. characterized changes in cough frequency throughout treatment (55). Table 2 summarizes the biomarkers that were included in the detailed quantitative assessment, as well as the transcriptomic signatures.
TABLE 2.
Assays and study characteristics for treatment monitoring biomarkers of interesta
| Biomarker | Author (yr) | Reference no. | Assay name (manufacturer) | County | Follow-up times | Sample(s) (state) | Reference(s) used |
|---|---|---|---|---|---|---|---|
| C-reactive protein (CRP) | Almeida (2009) | 15 | Roche CRPLX kit on the Roche modular analyzer (Roche) | Brazil | 0, W1, W3, W5, W8 | Plasma (frozen) | Solid culture (NR), clinical outcome, smear microscopy (NR) |
| Djoba Siawaya (2008) | 78 | CRP ELISA kit (Bender MedSystems) | South Africa | 0, W1, W5, W13, W26 | Serum (frozen) | Chest X-ray, liquid culture (BACTEC) | |
| Ferrian (2017) | 33 | CRP on a multiplex (ProcartaPlex human kits; eBioscience) read on a luminometer (Bio-Plex 200; BioRad) | South Africa | 0, M2, M4, M6 | Plasma (frozen) | Smear microscopy (NR), liquid culture (MGIT 960), line probe assay (Hain) | |
| Francisco (2017) | 35 | Quantikine ELISA (R&D Systems Inc., Minneapolis, MN, USA) | China | 0, M4, M6, M7 | Whole blood (NR) | Clinical outcome | |
| Jayakumar (2015) | 45 | ichroma CRP point-of-care reader (BodiTech Med Inc.) | Uganda | 0, W8, W20 | Serum (frozen) | Solid culture (NR), liquid culture (MGIT 960) | |
| Khalil (2020) | 49 | NycoCard CRP reader II (Abbott) | Egypt | M1, M2, M3 | Whole blood (NR), plasma (NR), serum (NR) | Solid culture (LJ), clinical outcome, smear microscopy (ZN), GeneXpert (MTB/RIF) | |
| Mendy (2016) | 61 | CRP ELISA kit (Immunology Consultants Laboratory) | Gambia | 0, M2, M6 | Plasma (frozen) | Liquid culture (MGIT), smear microscopy (ZN), chest X-ray | |
| Mesquita (2016) | 62 | CRP ELISA kit (Ebioscience) | Brazil | 0, D60 | Serum (frozen) | Culture (NR), chest X-ray | |
| Miranda (2017) | 63 | CRP ELISA kit (Ebioscience) | Brazil | 0, D30, D60 | Serum (frozen) | Solid culture (LJ) | |
| Moraes (2014) | 65 | CRP BNII nephelometer (Dade Behring) | Brazil | 0, D30, D60 | Serum (fresh) | Solid culture (LJ), clinical outcome, smear microscopy (ZN) | |
| Sigal (2017) | 80 | CRP V-Plex human vascular injury panel 2 (Meso Scale Diagnostics) | North America, Spain, South Africa, Uganda | 0, W8, W12 | Serum (frozen) | Solid culture (LJ), liquid culture (MGIT 960), clinical outcome, chest X-ray | |
| IL-6 | Chowdhury (2014) | 25 | IL-6 ELISA kit (RayBiotech) | India | 0, M2, M4, M6 | Serum (NR) | Smear microscopy (ZN), chest X-ray |
| Djoba Siawaya (2009) | 29 | IL-6 Lincoplex human cytokine 29-plex assays (Millipore) | South Africa | 0, W1, W5, W13, W26 | Plasma (frozen) | Liquid culture (BACTEC 460T), smear microscopy (NR) | |
| Feng (2020) | 32 | IL-6 Quantikine ELISA kit (R&D Systems) | Taiwan | 0, W8 | Peripheral blood mononuclear cells (frozen) | Culture (NR), smear microscopy (NR) | |
| Luo (2018) | 57 | IL-6 ELISA kits (Siemens Healthcare Diagnostics Products Ltd., Llanberis, Gwynedd, UK) | China | 0, M2 | Serum (fresh) | Smear microscopy (FM) | |
| Mvungi (2019) | 66 | IL-6 multiplex assay (human premixed multianalyte kit; catalog no. LXSAHM) on the Luminex 200 system | Tanzania | 0, M2 | Plasma (frozen) prepared using QuantiFERON-TB Gold Plus | Clinical outcome | |
| Riou (2012) | 72 | IL-6 multiplex bead array (bulletin no. 10014905; Bio-Rad) on a luminometer (Luminex) | South Africa | 0, W2, W4, W8, W12, W26, W52, W78 | Plasma (frozen) | Solid culture (LJ), liquid culture (MGIT), smear microscopy (FM) | |
| Sigal (2017) | 80 | IL-6 V-Plex human proinflammatory panel 1 (Meso Scale Diagnostics) | North America, Spain, South Africa, Uganda | 0, W8, W12 | Serum (frozen) | Solid culture (LJ), liquid culture (MGIT 960), clinical outcome, chest X-ray | |
| IP-10 | Chen (2011) | 23 | IP-10 ELISA (R&D Systems) | Taiwan | 0, M2, M6 | Serum (NR) | Solid culture (LJ), liquid culture (MGIT), smear microscopy (ZN), chest X-ray |
| Chung (2016) | 27 | IP-10 ELISA (R&D systems, Minneapolis, MN) | South Korea | 0, M2 | Serum (frozen) | Culture (NR), smear microscopy (NR), clinical outcome, chest X-ray | |
| Djoba Siawaya (2009) | 29 | IP-10 Lincoplex human cytokine 29-plex assays (Millipore) | South Africa | 0, W1, W5, W13, W26 | Plasma (frozen) | Liquid culture (BACTEC 460T), smear microscopy (NR) | |
| Ferrian (2017) | 33 | IP-10 on a multiplex (ProcartaPlex human kits; eBioscience) read on a luminometer (Bio-Plex 200; BioRad) | South Africa | 0, M2, M4, M6 | Plasma (frozen) | Smear microscopy (NR), liquid culture (MGIT 960) | |
| Francisco (2017) | 35 | ELISA kit (RayBiotech, Inc.) | China | 0, M4, M6, M7 | Whole blood (NR) | Clinical outcome | |
| Garcia-Basteiro (2017) | 36 | IP-10 ELISA kit (Becton Dickinson and Company) | Mozambique | 0, D7, D60 | Serum (frozen) | Smear microscopy (ZN), GeneXpert (MTB/RIF), liquid culture (MGIT 960) | |
| Hong (2014) | 43 | IP-10 ELISA kit (R&D Systems) | South Korea | 0/within W2, after M6-M9 | Serum (NR) | Culture (NR), chest X-ray, CT scan | |
| Jayakumar (2015) | 45 | IP-10 ELISA kit (R&D Systems) | Uganda | 0, W8, W20 | Serum (frozen) | Solid culture (NR), liquid culture (MGIT 960) | |
| Kabeer (2011) | 47 | IP-10 ELISA kit (R&D Systems) in response to QFT-IT and RD1 | India | 0, M6 | Plasma (fresh) | Solid culture (LJ), liquid culture (BacT) | |
| Kim (2018) | 51 | IP-10 ELISA kit (R&D Systems, Minneapolis, MN, USA) | South Korea | 0, M6, M12 | Urine (NR), Serum (NR) | Culture (NR), chest X-ray | |
| Matsushita (2015) | 59 | IP-10 27-plex assay on the Bio-Plex suspension array system (Bio-Rad) | Vietnam | 0, M2, M7 | Plasma (frozen) | Smear microscopy (NR), chest X-ray | |
| Riou (2012) | 72 | IP-10 multiplex bead array (bulletin no. 10014905; Bio-Rad) on a luminometer (Luminex) | South Africa | 0, W2, W4, W8, W12, W26, W52, W78 | Plasma (frozen) | Solid culture (LJ), liquid culture (MGIT), smear microscopy (FM) | |
| Zhu (2015) | 90 | IP-10 ELISA kit (eBioscience) | China | 0, W2-8 | Plasma (frozen) | Smear microscopy (NR) | |
| TNF-α | Chowdhury (2014) | 25 | TNF-α high-sensitivity human ELISA set (ImmunoTools) | India | 0, M2, M4, M6 | Serum (NR) | Smear microscopy (ZN), chest X-ray |
| Djoba Siawaya (2009) | 29 | TNF-α Lincoplex human cytokine 29-plex assays (Millipore) | South Africa | 0, W1, W5, W13, W26 | Plasma (frozen) | Liquid culture (BACTEC 460T), smear microscopy (NR) | |
| Luo (2018) | 57 | TNF-α ELISA kit (Siemens Healthcare Diagnostics) | China | 0, M2 | Serum (fresh) | Smear microscopy (FM) | |
| Mvungi (2019) | 66 | TNF-α multiplex assay (Human premixed multianalyte kit; catalog no. LXSAHM) on the Luminex 200 system | Tanzania | 0, M2 | Plasma (frozen) using QTF-TB Gold Plus | Clinical outcome | |
| Nie (2020) | 67 | TNF-α ELISA kit (BioLegend) | China | 0, M1-2, M6 | Serum (frozen) | Culture (NR), smear microscopy (NR), chest computed tomography | |
| Riou (2012) | 72 | TNF-α multiplex bead array (bulletin no. 10014905; Bio-Rad) on a luminometer (Luminex) | South Africa | 0, W2, W4, W8, W12, W26, W52, W78 | Plasma (frozen) | Solid culture (LJ), liquid culture (MGIT), smear microscopy (FM) | |
| Zhu (2015) | 90 | TNF-α ELISA kit (eBioscience) | China | 0, W2-8 | Plasma (frozen) | Smear microscopy (NR) | |
| Transcriptomic/gene signatures | Bloom (2012) | 21 | 664-Transcript signature, 320-transcript signature | South Africa | 0, W2, M2, M6, M12 | Whole blood (frozen) | Clinical outcome, chest X-ray |
| Darboe (2019) | 28 | RISK11 signature | South Africa | 0, M2, M6, M8, M14 | Whole blood (NR) | Culture (NR) | |
| Francisco (2017) | 35 | 3-gene signature (GBP5, DUSP3, and KLF2) | China | 0, M4, M6, M7 | Whole blood (NR) | Clinical outcome | |
| Gebremicael (2019) | 38 | 105 genes expression profiling by dual-color reverse-transcription multiplex ligation-dependent probe amplification (dcRT-MLPA) platform | Ethiopia | 0, M6, M18 | Whole blood (NR) | Smear microscopy (ZN) | |
| Penn-Nicholson (2020) | 69 | RISK6 signature | South Africa | 0, M2, treatment completion, 6 to 8 mo posttreatment | Whole blood (frozen) | Culture (NR), smear microscopy (NR), GeneXpert (MTB/RIF) | |
| Sivakumaran (2020) | 82 | 198-gene set profiled using dc-RT MLPA platform | India | 0, M1, M2, M6 | Whole blood (frozen) | Liquid culture (MGIT), smear microscopy (FM), clinical outcome | |
| Warsinske (2018) | 85 | 3-gene signature (GBP5, DUSP3, and KLF2) | South Africa | 0, W1, W4, W24 | Whole blood (NR) | Liquid culture (MGIT), PET-CT |
D, day; W, week; M, month; NR, not reported; MGIT, mycobacteria growth indicator tube; LJ, Löwenstein-Jensen; ZN, Ziehl-Neelsen; FM, fluorescent microscopy; PET-CT, positron emission tomography-computed tomography; QFT-IT, QuantiFERON-TB Gold In-Tube test; dcRT-MLPA, dual-color reverse-transcription multiplex ligation-dependent probe amplification.
Detailed quantitative assessment of treatment monitoring biomarkers.
For biomarkers where there were five or more studies that numerically or graphically presented the measures of central tendency and measures of spread at different follow-up time points, we further characterized the week 8 fold change and fold changes with respect to previously reported time points. Results of the meta-analysis found that CRP, IP-10, IL-6, and TNF-α (Table 3) decreased by week 8 of treatment compared to baseline (week 0). CRP experienced the greatest week 8 fold change of −76.1% (95% confidence interval [CI], −89.4% to −62.9%) while TNF-α had the smallest fold change of −10.3 (95% CI, −24.7% to −4.2%). Both IL-6 and IP-10 experienced fold changes of −24.7% (95% CI, −50.7% to 1.3%) and −38.2% (95% CI, −61.3% to −15.0%), respectively, though the confidence interval for IL-6 crossed the null.
TABLE 3.
Pooled week 8 fold change and fold change since previously recorded time point of CRP, IL-6, IP-10, and TNF-α among people with TB on therapyd
| Biomarker | Data for baseline to week 8 |
Data since previously recorded time point |
||||
|---|---|---|---|---|---|---|
| No. of studiesa | No. of participants | Avg fold change (% [95% CI]) | No. of Studies | No. of participantsb | Avg fold change (% [95% CI]) | |
| CRP | 5 | 275 | −76.1 (−89.4 to −62.9) | 7 | 447c | −53.9 (−70.2 to −37.5) |
| IL-6 | 4 | 522 | −24.7 (−50.7 to 1.3) | 5 | 558 | −31.0 (−59.5 to −2.5) |
| IP-10 | 4 | 154 | −38.2 (−61.3 to −15.0) | 9 | 430 | −36.2 (−49.0 to −23.4) |
| TNF-α | 4 | 497 | −10.3 (−24.7 to −4.2) | 6 | 517c | −17.7 (−31.3 to −4.0) |
Only includes studies that collected data at week 8 (Table S6 in the supplemental material).
At enrollment.
Number of participants in Zhu et al. (90) was not specified.
CRP, C-reactive protein; IL-6, interleukin-6; IP-10, interferon gamma-induced protein 10; TNF-α, tumor necrosis factor alpha; CI, confidence interval.
We further investigated the fold change of these four biomarkers with respect to the previously recorded time point (Fig. S7). The results of our meta-analysis found that there was a statistically significant decrease in levels of all four biomarkers with respect to the previous recorded value. Results of this meta-analysis complement the findings of the week 8 meta-analysis. CRP had the largest average change in biomarker level of −53.9% (95% CI, −70.2% to −37.5%) relative to the previously recorded time point. TNF-α had the smallest average fold change of −17.7% (95% CI, −31.3% to −4.0%) relative to previously recorded time points. Both IL-6 and IP-10 levels experienced a similar average changes of −31.0% (95% CI, −59.5% to −2.5%) and −36.2% (95% CI, −49.0% to −23.5%). For all biomarkers, confidence intervals were narrow, and the same conclusions were obtained in the sensitivity analyses where we varied the assumed correlation value (Table S8).
DISCUSSION
In this systematic review, we examined the current landscape of assays used to evaluate changes in biomarker levels with respect to TB treatment. Host inflammation markers, including CRP, IP-10, IL-6, and TNF-α, were some of the most commonly evaluated biomarkers for TB treatment response. CRP and IP-10 have been particularly well characterized as a biomarker for TB screening and diagnosis in other studies (95, 96). However, we identified 81 different biomarkers that were evaluated in the context of TB treatment monitoring. Thus, while these four biomarkers appear to be promising given our exploratory quantitative analyses and should be further investigated, research into other, more novel biomarkers for TB treatment monitoring remains important.
From our quantitative analyses, we observed that, for the average fold change between baseline and week 8 of treatment, CRP, IP-10, and TNF-α had a statistically significant decrease. This analysis informs us of the average magnitude of the decrease in biomarker level between baseline and the end of the intensive phase of treatment. The results of our meta-analysis found that, on average, all four biomarkers decreased with respect to previously recorded time points. Out of the four biomarkers analyzed, CRP had the largest absolute week 8 fold change value of −76.1% (95% CI, −89.4% to −62.9%) and fold change relative to previous recorded time points of −53.9% (95% CI, −70.2% to −37.5%). This early response during the intensive phase of TB treatment and continued fold change throughout treatment may help with clinical decision-making by identifying people who respond favorably to treatment, though further analyses are needed to characterize how this fold change differs between people who respond to treatment and people who do not respond to treatment or are lost to follow-up. In addition, further investigations are required, as most of the included studies recruited a narrow patient spectrum, making the generalizability of the results a challenge.
Since these host inflammation markers are usually obtained from blood, serum, and/or plasma samples, they provide an advantage over traditional sputum-based methods such as microscopy and culture. However, as the detected changes were small, obtaining accurate readings in a timely, near-patient manner will be difficult. Nevertheless, the changes were statistically significant, which may suggest they may have potential to support clinical decision-making for TB treatment monitoring.
Among the host noninflammatory biomarkers, blood-based transcriptomic and gene expression signatures have gained significant momentum for TB diagnostics and treatment monitoring. The ability to detect the up- or downregulation of specific genes may allow for simpler and earlier identification of people who respond both favorably and unfavorably to treatment. As these signatures become increasingly parsimonious, their potential for commercialization into assays that run on standard PCR machines increases. Cepheid (USA) recently developed a prototype cartridge assay that runs on the GeneXpert platform for the Sweeney3 (3-gene signature) called the Xpert MTB Host Response or Xpert-MTB-HR-Prototype. A recent study performed a preliminary investigation on the performance of the Xpert-MTB-HR-Prototype as a treatment monitoring tool among 31 patients with pulmonary TB and found that the signature correlated with treatment progression (97). So far, each of the transcriptomic signatures identified in this review has only been evaluated in a limited number of cohorts, preventing us from meta-analyzing the fold change of these markers throughout treatment. Additional well-conducted studies are needed to quantitatively evaluate the performance of these signatures for treatment monitoring. Promising gene signatures that should be evaluated further in the context of TB treatment monitoring include Sweeney3, RISK6, and RISK11 (28, 35, 69, 85).
There are several limitations associated with this study. First, because not all studies reported the exact biomarker levels for CRP, IP-10, IL-6, and TNF-α, some of the data had to be extracted from figures, which may have introduced measurement error in the quantitative analyses. We attempted to mitigate this bias by extracting the estimates in duplicate. Further, a recent study by Mierden et al. found that the error from empirical evaluation of data from figures is often inconsequential and that “data extraction from graphs is a good method to harvest data if it is not provided in the text or tables” (10). Second, most, if not all, biomarkers were evaluated using different assays in each study. For example, across the 10 studies that evaluated CRP, 9 different assays were used, including 3 different ELISA kits (61–63, 78), 2 multiplex kits (33, 80), 1 nephelometer (65), 2 assays on point-of-care modules (one by Abbott, the other by BodiTech Med) (45, 49), and 1 assay on the Roche modular analyzer (15). This heterogeneity and consequent variability in assay performances could not be accounted for in the analyses. Third, we compared studies with different patient characteristics (e.g., different HIV status levels, fast versus slow responders, different proportion of drug-resistant or multidrug-resistant TB, etc.). Because the majority of studies did not disaggregate biomarker-level data by patient characteristic or treatment regimen, we were unable to perform subgroup analyses comparing how the fold change in biomarkers differed across populations. Fourth, the results of the meta-analysis for the fold change relative to previously recorded time points is entirely dependent on the data collected in the included studies. Given the high risk of bias and extensive heterogeneity across the studies, the quantitative fold change results are exploratory and limited in interpretation outside the context of our systematic review. Nevertheless, these preliminary data may help inform future studies to investigate these biomarkers in a more rigorous and standardized manner for TB treatment monitoring. Finally, this study does not explain the biological reason for the change in marker levels over time, which is essential for understanding the treatment monitoring potential of the biomarker. Further evaluations are needed to understand whether such changes in biomarker levels directly inform us that the treatment is effective. Additionally, studies are needed to characterize how biomarkers would respond to partially effective and ineffective regimens.
It is important to highlight that the overall quality of studies evaluated was poor, suggesting an overall high risk of bias with respect to the reference standard, index test, and patient selection of QUADAS-2 domains. What is most concerning, however, is the extensive heterogeneity in the study design and data reporting strategies across TB treatment monitoring studies. This heterogeneity limited our ability to properly evaluate the performance of the biomarkers and assays. Lack of uniform follow-up time points and reporting strategies, inconsistent definitions of treatment success versus treatment failure, and variability in the type and timing of reference standards were some of the key issues that complicated the evaluation of biomarkers and assays. Treatment monitoring of active pulmonary TB is an essential part of TB care, and yet, there is very little guidance on best practices for researchers on how to design studies and evaluate the accuracy and characteristics of treatment monitoring biomarkers and assays, and even less guidance for clinicians to use these different biomarkers to inform TB treatment progression among patients. Our systematic review and meta-analysis highlights that while TB treatment monitoring is an active area of research, additional work is needed to formulate appropriate study guidelines, gain clear consensus regarding stakeholder needs through WHO-endorsed TB target product profiles (TPPs), and inform clinical decision-making (98).
ACKNOWLEDGMENTS
We thank Genevieve Gore (McGill University) for helping us with the systematic search strategy.
C.C., C.M.D., and M.K. designed and conceptualized the study. A.J.Z., F.L., N.A.V., and C.C. screened all studies and performed the primary data extraction. M.R., C.M.D., and M.K. validated the data. A.J.Z., S.M., A.B., and M.K. performed the formal analyses. A.J.Z. wrote the original draft of the manuscript. F.L., N.A.V., C.C., S.M., A.B., E.M., M.R., C.M.D., and M.K. edited and reviewed the final version of the manuscript.
We do not have any conflicts of interest to declare.
S.M. acknowledges support from the National Science Foundation Graduate Research Fellowship Program under grant no. DGE1745303, National Library Of Medicine of the National Institutes of Health under award no. T32LM012411, and Fonds de recherche du Québec-Nature et technologies B1X research scholarship.
Footnotes
Supplemental material is available online only.
Contributor Information
Claudia M. Denkinger, Email: Claudia.Denkinger@Uni-Heidelberg.de.
Mikashmi Kohli, Email: Mikashmi.Kohli@finddx.org.
Alexander J. McAdam, Boston Children's Hospital
REFERENCES
- 1.Vinet L, Zhedanov A. 2011. A “missing” family of classical orthogonal polynomials. J Phys A Math Theor 44:085201. 10.1088/1751-8113/44/8/085201. [DOI] [Google Scholar]
- 2.World Health Organization. 2010. Treatment of tuberculosis guidelines. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 3.Marais BJ, Schaaf HS. 2010. Childhood tuberculosis: an emerging and previously neglected problem. Infect Dis Clin North Am 24:727–749. 10.1016/j.idc.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 4.Purohit M, Mustafa T. 2015. Laboratory diagnosis of extra-pulmonary tuberculosis (EPTB) in resource-constrained setting: state of the art, challenges and the need. J Clin Diagn Res 9:EE01–EE06. 10.7860/JCDR/2015/12422.5792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Peter JG, Theron G, Singh N, Singh A, Dheda K. 2014. Sputum induction to aid diagnosis of smear-negative or sputum-scarce tuberculosis in adults in HIV-endemic settings. Eur Respir J 43:185–194. 10.1183/09031936.00198012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nahid P, Kim PS, Evans CA, Alland D, Barer M, Diefenbach J, Ellner J, Hafner R, Hamilton CD, Iademarco MF, Ireton G, Kimerling ME, Lienhardt C, MacKenzie WR, Murray M, Perkins MD, Posey JE, Roberts T, Sizemore C, Stevens WS, Via L, Williams SD, Yew WW, Swindells S. 2012. Clinical research and development of tuberculosis diagnostics: moving from silos to synergy. J Infect Dis 205:S159–S168. 10.1093/infdis/jis194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horne DJ, Royce SE, Gooze L, Narita M, Hopewell PC, Nahid P, Steingart KR. 2010. Sputum monitoring during tuberculosis treatment for predicting outcome: systematic review and meta-analysis. Lancet Infectious Diseases 10:387–394. 10.1016/S1473-3099(10)70071-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nema V. 2012. Tuberculosis diagnostics: challenges and opportunities. Lung India 29:259–266. 10.4103/0970-2113.99112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Goletti D, Lindestam Arlehamn CS, Scriba TJ, Anthony R, Cirillo DM, Alonzi T, Denkinger CM, Cobelens F. 2018. Can we predict tuberculosis cure? What tools are available? Eur Respir J 52:1801089. 10.1183/13993003.01089-2018. [DOI] [PubMed] [Google Scholar]
- 10.Van der Mierden S, Spineli LM, Talbot SR, Yiannakou C, Zentrich E, Weegh N, Struve B, Brügge TFZ, Bleich A, Leenaars CHC. 2021. Extracting data from graphs: a case-study on animal research with implications for meta-analyses. Res Synth Methods 12:701–710. 10.1002/jrsm.1481. [DOI] [PubMed] [Google Scholar]
- 11.McGrath S, Zhao XFei, Steele R, Thombs BD, Benedetti A, Levis B, Riehm KE, Saadat N, Levis AW, Azar M, Rice DB, Sun Y, Krishnan A, He C, Wu Y, Bhandari PM, Neupane D, Imran M, Boruff J, Cuijpers P, Gilbody S, Ioannidis JPA, Kloda LA, McMillan D, Patten SB, Shrier I, Ziegelstein RC, Akena DH, Arroll B, Ayalon L, Baradaran HR, Baron M, Beraldi A, Bombardier CH, Butterworth P, Carter G, Chagas MH, Chan JCN, Cholera R, Chowdhary N, Clover K, Conwell Y, de Man-van Ginkel JM, Delgadillo J, Fann JR, Fischer FH, Fischler B, Fung D, Gelaye B, Goodyear-Smith F, et al. 2020. Estimating the sample mean and standard deviation from commonly reported quantiles in meta-analysis. Stat Methods Med Res 29:2520–2537. 10.1177/0962280219889080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McGrath S, Zhao XF, Steele R, Benedetti A. 2020. estmeansd: estimating the sample mean and standard deviation from commonly reported quantiles in meta-analysis R package version 0.2.1, vol 29. https://cran.r-project.org/package=estmeansd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Viechtbauer W. 2010. Conducting meta-analyses in R with the metafor package. J Stat Softw 36:1–48. 10.18637/jss.v036.i03. [DOI] [Google Scholar]
- 14.Cooper H, Hedges LV, Valentine JC. 2019. The handbook of research syntheses and meta-analysis. Russell Sage Foundation, New York, NY. [Google Scholar]
- 15.Almeida MLD, Barbieri MA, Gurgel RQ, Abdurrahman ST, Baba UA, Hart CA, Shenkin A, Silva AM, de Souza L, Cuevas LE. 2009. α1-acid glycoprotein and α1-antitrypsin as early markers of treatment response in patients receiving the intensive phase of tuberculosis therapy. Trans R Soc Trop Med Hyg 103:575–580. 10.1016/j.trstmh.2008.11.024. [DOI] [PubMed] [Google Scholar]
- 16.Altunoglu E, Erdenen F, Gelisgen R, Kar O, Korkmaz G, Muderrisoglu C, Tabak O, Uzun H. 2014. Serum adenosine deaminase activity and neopterin levels during therapy in patients with pulmonary tuberculosis and community-acquired pneumonia. Istanbul Med J 15:78–82. 10.5152/imj.2014.24855. [DOI] [Google Scholar]
- 17.Andrade BB, Pavan Kumar N, Mayer-Barber KD, Barber DL, Sridhar R, Rekha VVB, Jawahar MS, Nutman TB, Sher A, Babu S. 2013. Plasma heme oxygenase-1 levels distinguish latent or successfully treated human tuberculosis from active disease. PLoS One 8:e62618. 10.1371/journal.pone.0062618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Anusiem CA, Okonkwo PO. 2017. The impact of treatment on the serum concentration of interleukin-1 beta in pulmonary tuberculosis. Am J Ther 24:e329–e332. 10.1097/MJT.0000000000000360. [DOI] [PubMed] [Google Scholar]
- 19.Atahan E, Saribas S, Demirci M, Babalık A, Akkus S, Balıkcı A, Satana D, Ziver T, Dinc HO, Keskin M, Ozbey D, Kocak BT, Gareayaghi N, Kirmusaoglu S, Tokman HB, Kocazeybek B. 2020. Evaluating the effectiveness of anti-tuberculosis treatment by detecting Mycobacterium tuberculosis 85B messenger RNA expression in sputum. J Infect Public Health 13:1490–1494. 10.1016/j.jiph.2020.05.016. [DOI] [PubMed] [Google Scholar]
- 20.Barry SE, Ellis M, Yang Y, Guan G, Wang X, Britton WJ, Saunders BM. 2018. Identification of a plasma microRNA profile in untreated pulmonary tuberculosis patients that is modulated by anti-mycobacterial therapy. J Infect 77:341–348. 10.1016/j.jinf.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 21.Bloom CI, Graham CM, Berry MPR, Wilkinson KA, Oni T, Rozakeas F, Xu Z, Rossello-Urgell J, Chaussabel D, Banchereau J, Pascual V, Lipman M, Wilkinson RJ, O'Garra A. 2012. Detectable changes in the blood transcriptome are present after two weeks of antituberculosis therapy. PLoS One 7:e46191. 10.1371/journal.pone.0046191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chedid C, Kokhreidze E, Tukvadze N, Banu S, Uddin MKM, Biswas S, Russomando G, Acosta CCD, Arenas R, Ranaivomanana PP, Razafimahatratra C, Herindrainy P, Rakotosamimanana N, Hamze M, Ismail MB, Bayaa R, Berland J-L, Delogu G, Endtz H, Ader F, Goletti D, Hoffmann J. 2020. Association of baseline white blood cell counts with tuberculosis treatment outcome: a prospective multicentered cohort study. Int J Infect Dis 100:199–206. 10.1016/j.ijid.2020.09.017. [DOI] [PubMed] [Google Scholar]
- 23.Chen Y-C, Chin C-H, Liu S-F, Wu C-C, Tsen C-C, Wang Y-H, Chao T-Y, Lie C-H, Chen C-J, Wang C-C, Lin M-C. 2011. Prognostic values of serum IP-10 and IL-17 in patients with pulmonary tuberculosis. Dis Markers 31:101–110. 10.1155/2011/938794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Choi R, Kim K, Kim M-J, Kim S-Y, Kwon OJ, Jeon K, Park HY, Jeong B-H, Shin SJ, Koh W-J, Lee S-Y. 2016. Serum inflammatory profiles in pulmonary tuberculosis and their association with treatment response. J Proteomics 149:23–30. 10.1016/j.jprot.2016.06.016. [DOI] [PubMed] [Google Scholar]
- 25.Chowdhury IH, Ahmed AM, Choudhuri S, Sen A, Hazra A, Pal NK, Bhattacharya B, Bahar B. 2014. Alteration of serum inflammatory cytokines in active pulmonary tuberculosis following anti-tuberculosis drug therapy. Mol Immunol 62:159–168. 10.1016/j.molimm.2014.06.002. [DOI] [PubMed] [Google Scholar]
- 26.Chung W, Lee K, Jung Y, Kim Y, Park J, Sheen S, Lee J, Kang D, Park K. 2015. Serum CXCR3 ligands as biomarkers for the diagnosis and treatment monitoring of tuberculosis. Int J Tuber Lung Dis 19:1476–1484. 10.5588/ijtld.15.0325. [DOI] [PubMed] [Google Scholar]
- 27.Chung WY, Yoon D, Lee KS, Jung YJ, Kim YS, Sheen SS, Park KJ. 2016. The usefulness of serum CXCR3 ligands for evaluating the early treatment response in tuberculosis: a longitudinal cohort study. Medicine (Baltimore) 95:e3575. 10.1097/MD.0000000000003575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Darboe F, Mbandi SK, Naidoo K, Yende-Zuma N, Lewis L, Thompson EG, Duffy FJ, Fisher M, Filander E, van Rooyen M, Bilek N, Mabwe S, McKinnon LR, Chegou N, Loxton A, Walzl G, Tromp G, Padayatchi N, Govender D, Hatherill M, Karim SA, Zak DE, Penn-Nicholson A, Scriba TJ, SATVI Clinical Immunology Team. 2019. Detection of tuberculosis recurrence, diagnosis and treatment response by a blood transcriptomic risk signature in HIV-infected persons on antiretroviral therapy. Front Microbiol 10:1441. 10.3389/fmicb.2019.01441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Djoba Siawaya JF, Beyers N, Van Helden P, Walzl G. 2009. Differential cytokine secretion and early treatment response in patients with pulmonary tuberculosis. Clin Exp Immunol 156:69–77. 10.1111/j.1365-2249.2009.03875.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Drain PK, Gounder L, Grobler A, Sahid F, Bassett IV, Moosa MYS. 2015. Urine lipoarabinomannan to monitor antituberculosis therapy response and predict mortality in an HIV-endemic region: a prospective cohort study. BMJ Open 5:e006833. 10.1136/bmjopen-2014-006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ehtesham NZ, Nasiruddin M, Alvi A, Kumar BK, Ahmed N, Peri S, Murthy KJR, Hasnain SE. 2011. Treatment end point determinants for pulmonary tuberculosis: human resistin as a surrogate biomarker. Tuberculosis 91:293–299. 10.1016/j.tube.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 32.Feng J-Y, Ho L-I, Chuang F-Y, Pan S-W, Chen Y-Y, Tung C-L, Li C-P, Su W-J. 2021. Depression and recovery of IL-17A secretion in mitogen responses in patients with active tuberculosis-a prospective observational study. J Formos Med Assoc 120:1080–1089. 10.1016/j.jfma.2020.09.012. [DOI] [PubMed] [Google Scholar]
- 33.Ferrian S, Manca C, Lubbe S, Conradie F, Ismail N, Kaplan G, Gray CM, Fallows D. 2017. A combination of baseline plasma immune markers can predict therapeutic response in multidrug resistant tuberculosis. PLoS One 12:e0176660. 10.1371/journal.pone.0176660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feruglio SL, Trøseid M, Damås JK, Kvale D, Dyrhol-Riise AM. 2013. Soluble markers of the Toll-like receptor 4 pathway differentiate between active and latent tuberculosis and are associated with treatment responses. PLoS One 8:e69896. 10.1371/journal.pone.0069896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Francisco NM, Fang Y-M, Ding L, Feng S, Yang Y, Wu M, Jacobs M, Ryffel B, Huang X. 2017. Diagnostic accuracy of a selected signature gene set that discriminates active pulmonary tuberculosis and other pulmonary diseases. J Infect 75:499–510. 10.1016/j.jinf.2017.09.012. [DOI] [PubMed] [Google Scholar]
- 36.García-Basteiro AL, Mambuque E, den Hertog A, Saavedra B, Cuamba I, Oliveras L, Blanco S, Bulo H, Brew J, Cuevas LE, Cobelens F, Nhabomba A, Anthony R. 2017. IP-10 kinetics in the first week of therapy are strongly associated with bacteriological confirmation of tuberculosis diagnosis in HIV-infected patients. Sci Rep 7:1–8. 10.1038/s41598-017-13785-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gebremicael G, Alemayehu M, Sileshi M, Geto Z, Gebreegziabxier A, Tefera H, Ashenafi N, Tadese C, Wolde M, Kassa D. 2019. The serum concentration of vitamin B12 as a biomarker of therapeutic response in tuberculosis patients with and without human immunodeficiency virus (HIV) infection. Int J Gen Med 12:353–361. 10.2147/IJGM.S218799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gebremicael G, Kassa D, Alemayehu Y, Gebreegziaxier A, Kassahun Y, van Baarle D, H M Ottenhoff T, M Cliff J, C Haks M. 2019. Gene expression profiles classifying clinical stages of tuberculosis and monitoring treatment responses in Ethiopian HIV-negative and HIV-positive cohorts. PLoS One 14:e0226137. 10.1371/journal.pone.0226137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hai HT, Vinh DN, Thu DDA, Hanh NT, Phu NH, Srinivasan V, Thwaites GE, Tt Thuong N. 2019. Comparison of the Mycobacterium tuberculosis molecular bacterial load assay, microscopy and GeneXpert versus liquid culture for viable bacterial load quantification before and after starting pulmonary tuberculosis treatment. Tuberculosis 119:101864. 10.1016/j.tube.2019.101864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Honeyborne I, Lipman MC, Eckold C, Evangelopoulos D, Gillespie SH, Pym A, McHugh TD. 2015. Effective anti-tuberculosis therapy correlates with plasma small RNA. Eur Respir J 45:1741–1744. 10.1183/09031936.00221214. [DOI] [PubMed] [Google Scholar]
- 41.Honeyborne I, McHugh TD, Phillips PPJ, Bannoo S, Bateson A, Carroll N, Perrin FM, Ronacher K, Wright L, van Helden PD, Walzl G, Gillespie SH. 2011. Molecular bacterial load assay, a culture-free biomarker for rapid and accurate quantification of sputum Mycobacterium tuberculosis bacillary load during treatment. J Clin Microbiol 49:3905–3911. 10.1128/JCM.00547-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Honeyborne I, Mtafya B, Phillips PPJ, Hoelscher M, Ntinginya EN, Kohlenberg A, Rachow A, Rojas-Ponce G, McHugh TD, Heinrich N, Pan African Consortium for the Evaluation of Anti-tuberculosis Antibiotics. 2014. The molecular bacterial load assay replaces solid culture for measuring early bactericidal response to antituberculosis treatment. J Clin Microbiol 52:3064–3067. 10.1128/JCM.01128-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hong JY, Lee HJ, Kim SY, Chung KS, Kim EY, Jung JY, Park MS, Kim YS, Kim SK, Chang J, Cho S-N, Kang YA. 2014. Efficacy of IP-10 as a biomarker for monitoring tuberculosis treatment. J Infect 68:252–258. 10.1016/j.jinf.2013.09.033. [DOI] [PubMed] [Google Scholar]
- 44.Ige O, Edem VF, Arinola OG. 2016. Plasma adenosine deaminase enzyme reduces with treatment of pulmonary tuberculosis in Nigerian patients: indication for diagnosis and treatment monitoring. Niger J Physiol Sci 31:53. [PubMed] [Google Scholar]
- 45.Jayakumar A, Vittinghoff E, Segal MR, MacKenzie WR, Johnson JL, Gitta P, Saukkonen J, Anderson J, Weiner M, Engle M, Yoon C, Kato-Maeda M, Nahid P. 2015. Serum biomarkers of treatment response within a randomized clinical trial for pulmonary tuberculosis. Tuberculosis 95:415–420. 10.1016/j.tube.2015.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jiang T-T, Shi L-Y, Chen J, Wei L-L, Li M, Hu Y-T, Gan L, Liu C-M, Tu H-H, Li Z-B, Yi W-J, Li J-C. 2018. Screening and identification of potential protein biomarkers for evaluating the efficacy of intensive therapy in pulmonary tuberculosis. Biochem Biophys Res Commun 503:2263–2270. 10.1016/j.bbrc.2018.06.147. [DOI] [PubMed] [Google Scholar]
- 47.Kabeer BSA, Raja A, Raman B, Thangaraj S, Leportier M, Ippolito G, Girardi E, Lagrange PH, Goletti D. 2011. IP-10 response to RD1 antigens might be a useful biomarker for monitoring tuberculosis therapy. BMC Infect Dis 11:135–139. 10.1186/1471-2334-11-135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kawasaki M, Echiverri C, Raymond L, Cadena E, Reside E, Gler MT, Oda T, Ito R, Higashiyama R, Katsuragi K, Liu Y. 2019. Lipoarabinomannan in sputum to detect bacterial load and treatment response in patients with pulmonary tuberculosis: analytic validation and evaluation in two cohorts. PLoS Med 16:e1002780. 10.1371/journal.pmed.1002780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Khalil M, Halim H, Abdelazeem M. 2020. C-reactive protein versus erythrocyte sedimentation rate in monitoring multidrug-resistant tuberculosis. Egypt J Chest Dis Tuberc 69:458. [Google Scholar]
- 50.Kim ES, Park KU, Song JHan, Lim H-J, Cho Y-J, Yoon HIl, Lee JHo, Lee C-T, Park JS. 2013. The clinical significance of CA-125 in pulmonary tuberculosis. Tuberculosis 93:222–226. 10.1016/j.tube.2012.10.014. [DOI] [PubMed] [Google Scholar]
- 51.Kim SY, Kim J, Kim DR, Kang YA, Bong S, Lee J, Kim S, Lee NS, Sim B, Cho S-N, Kim YS, Lee H. 2018. Urine IP-10 as a biomarker of therapeutic response in patients with active pulmonary tuberculosis. BMC Infect Dis 18:1–6. 10.1186/s12879-018-3144-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kumar NP, Velayutham B, Nair D, Babu S. 2017. Angiopoietins as biomarkers of disease severity and bacterial burden in pulmonary tuberculosis. Int J Tuber Lung Dis 21:93–99. 10.5588/ijtld.16.0565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kumar NP, Banurekha VV, Nair D, Babu S. 2017. Diminished plasma levels of common γ-chain cytokines in pulmonary tuberculosis and reversal following treatment. PLoS One 12:e0176495. 10.1371/journal.pone.0176495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar NP, Banurekha VV, Nair D, Babu S. 2016. Circulating angiogenic factors as biomarkers of disease severity and bacterial burden in pulmonary tuberculosis. PLoS One 11:e0146318. 10.1371/journal.pone.0146318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee GO, Comina G, Hernandez-Cordova G, Naik N, Gayoso O, Ticona E, Coronel J, Evans CA, Zimic M, Paz-Soldan VA, Gilman RH, Oberhelman R. 2020. Cough dynamics in adults receiving tuberculosis treatment. PLoS One 15:e0231167. 10.1371/journal.pone.0231167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee MR, Tsai CJ, Wang WJ, Chuang TY, Yang CM, Chang LY, et al. 2015. Plasma biomarkers can predict treatment response in tuberculosis patients: a prospective observational study. Med (United States) 94:e1628. 10.1097/MD.0000000000001628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Luo X, Wu F, Ma J, Xiao H, Cui H. 2018. Immunological recovery in patients with pulmonary tuberculosis after intensive phase treatment. J Int Med Res 46:3539–3551. 10.1177/0300060518773258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mardining Raras TY, Noor Chozin I. 2010. The soluble plasminogen activator receptor as a biomarker on monitoring the therapy progress of pulmonary TB-AFB(+) patients. Tuberc Res Treat 2010:406346. 10.1155/2010/406346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Matsushita I, Hang NTL, Hong LT, Tam DB, Lien LT, Thuong PH, Cuong VC, Hijikata M, Kobayashi N, Sakurada S, Higuchi K, Harada N, Keicho N. 2015. Dynamics of immune parameters during the treatment of active tuberculosis showing negative interferon gamma response at the time of diagnosis. Int J Infect Dis 40:39–44. 10.1016/j.ijid.2015.09.021. [DOI] [PubMed] [Google Scholar]
- 60.Mdivani N, Li H, Akhalaia M, Gegia M, Goginashvili L, Kernodle DS, Khechinashvili G, Tang Y-W. 2009. Monitoring therapeutic efficacy by real-time detection of Mycobacterium tuberculosis mRNA in sputum. Clin Chem 55:1694–1700. 10.1373/clinchem.2009.124396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mendy J, Togun T, Owolabi O, Donkor S, Ota MOC, Sutherland JS. 2016. C-reactive protein, Neopterin and Beta2 microglobulin levels pre and post TB treatment in The Gambia. BMC Infect Dis 16:115. 10.1186/s12879-016-1447-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mesquita EDD, Gil-Santana L, Ramalho D, Tonomura E, Silva EC, Oliveira MM, Andrade BB, Kritski A, for the Rede-TB Study group. 2016. Associations between systemic inflammation, mycobacterial loads in sputum and radiological improvement after treatment initiation in pulmonary TB patients from Brazil: a prospective cohort study. BMC Infect Dis 16:1–12. 10.1186/s12879-016-1736-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Miranda P, Gil-Santana L, Oliveira MG, Mesquita EDD, Silva E, Rauwerdink A, Cobelens F, Oliveira MM, Andrade BB, Kritski A. 2017. Sustained elevated levels of C-reactive protein and ferritin in pulmonary tuberculosis patients remaining culture positive upon treatment initiation. PLoS One 12:e0175278. 10.1371/journal.pone.0175278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Montenegro RA, Guarines KM, Montenegro LML, Lira LAS, Falcão J, Melo FL, Santos FCF, Nascimento ALA, Zuzarte MS, Leite RC, Schindler HC. 2014. Assessment of messenger RNA (mRNA) of Mycobacterium tuberculosis as a marker of cure in patients with pulmonary tuberculosis. J Appl Microbiol 117:266–272. 10.1111/jam.12508. [DOI] [PubMed] [Google Scholar]
- 65.Moraes MLd, Ramalho DMdP, Delogo KN, Miranda PFC, Mesquita EDD, de Melo Guedes de Oliveira HM, Netto AR, Dos Anjos MJ, Kritski AL, de Oliveira MM. 2014. Association of serum levels of iron, copper, and zinc, and inflammatory markers with bacteriological sputum conversion during tuberculosis treatment. Biol Trace Elem Res 160:176–184. 10.1007/s12011-014-0046-0. [DOI] [PubMed] [Google Scholar]
- 66.Mvungi HC, Mbelele PM, Buza JJ, Mpagama SG, Sauli E. 2019. Blood cytokine responses to early secreted protein antigen-6/culture filtrate protein-10 tuberculosis antigens 2 months after antituberculosis treatment among patients with drug-susceptible pulmonary tuberculosis. Int J Mycobacteriol 8:53–59. 10.4103/ijmy.ijmy_30_19. [DOI] [PubMed] [Google Scholar]
- 67.Nie W, Wang J, Jing W, Shi W, Wang Q, Huang X, Cai B, Ge Q, Nie L, Han X, Du Y, Wang J, Guo R, Chu N. 2020. Value of serum cytokine biomarkers TNF-α, IL-4, sIL-2R and IFN-γ for use in monitoring bacterial load and anti-tuberculosis treatment progress. Cytokine X 2:100028. 10.1016/j.cytox.2020.100028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Osawa T, Watanabe M, Morimoto K, Okumura M, Yoshiyama T, Ogata H, Goto H, Kudoh S, Ohta K, Sasaki Y. 2020. Serum procalcitonin levels predict mortality risk in patients with pulmonary tuberculosis: a single-center prospective observational study. J Infect Dis 222:1651–1654. 10.1093/infdis/jiaa275. [DOI] [PubMed] [Google Scholar]
- 69.Penn-Nicholson A, Mbandi SK, Thompson E, Mendelsohn SC, Suliman S, Chegou NN, Malherbe ST, Darboe F, Erasmus M, Hanekom WA, Bilek N, Fisher M, Kaufmann SHE, Winter J, Murphy M, Wood R, Morrow C, Van Rhijn I, Moody B, Murray M, Andrade BB, Sterling TR, Sutherland J, Naidoo K, Padayatchi N, Walzl G, Hatherill M, Zak D, Scriba TJ, The Adolescent Cohort Study team. 2020. RISK6, a 6-gene transcriptomic signature of TB disease risk, diagnosis and treatment response. Sci Rep 10:1–21. 10.1038/s41598-020-65043-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rabna P, Andersen A, Wejse C, Oliveira I, Gomes VF, Haaland MB, Aaby P, Eugen-Olsen J. 2012. Utility of the plasma level of suPAR in monitoring risk of mortality during TB treatment. PLoS One 7:e43933. 10.1371/journal.pone.0043933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ralph AP, Yeo TW, Salome CM, Waramori G, Pontororing GJ, Kenangalem E, et al. 2013. Impaired pulmonary nitric oxide bioavailability in pulmonary tuberculosis: association with disease severity and delayed mycobacterial clearance with treatment. J Infect Dis 208:616–626. 10.1093/infdis/jit248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Riou C, Perez Peixoto B, Roberts L, Ronacher K, Walzl G, Manca C, Rustomjee R, Mthiyane T, Fallows D, Gray CM, Kaplan G. 2012. Effect of standard tuberculosis treatment on plasma cytokine levels in patients with active pulmonary tuberculosis. PLoS One 7:e36886. 10.1371/journal.pone.0036886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rockwood N, Costa DL, Amaral EP, Du Bruyn E, Kubler A, Gil-Santana L, Fukutani KF, Scanga CA, Flynn JL, Jackson SH, Wilkinson KA, Bishai WR, Sher A, Wilkinson RJ, Andrade BB. 2017. Mycobacterium tuberculosis induction of heme oxygenase-1 expression is dependent on oxidative stress and reflects treatment outcomes. Front Immunol 8:542. 10.3389/fimmu.2017.00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Ronacher K, Chegou NN, Kleynhans L, Djoba Siawaya JF, Du Plessis N, Loxton AG, Maasdorp E, Tromp G, Kidd M, Stanley K, Kriel M, Menezes A, Gutschmidt A, van der Spuy GD, Warren RM, Dietze R, Okwera A, Thiel B, Belisle JT, Cliff JM, Boom WH, Johnson JL, van Helden PD, Dockrell HM, Walzl G. 2019. Distinct serum biosignatures are associated with different tuberculosis treatment outcomes. Tuberculosis (Edinb) 118:101859. 10.1016/j.tube.2019.101859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sabiiti W, Azam K, Farmer ECW, Kuchaka D, Mtafya B, Bowness R, Oravcova K, Honeyborne I, Evangelopoulos D, McHugh TD, Khosa C, Rachow A, Heinrich N, Kampira E, Davies G, Bhatt N, Ntinginya EN, Viegas S, Jani I, Kamdolozi M, Mdolo A, Khonga M, Boeree MJ, Phillips PPJ, Sloan D, Hoelscher M, Kibiki G, Gillespie SH. 2020. Tuberculosis bacillary load, an early marker of disease severity: the utility of tuberculosis molecular bacterial load assay. Thorax 75:606–608. 10.1136/thoraxjnl-2019-214238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sahin F, Yildiz P. 2012. Serum CA-125: biomarker of pulmonary tuberculosis activity and evaluation of response to treatment. Clin Invest Med 35:E223–E228. [PubMed] [Google Scholar]
- 77.Said AF, Mohamed BI, El-Sharkawy E, Al-Sherif M. 2013. Role of cancer antigen 125 in active pulmonary tuberculosis. Egypt J Chest Dis Tuberc 62:419–424. 10.1016/j.ejcdt.2013.07.016. [DOI] [Google Scholar]
- 78.Djoba Siawaya JF, Bapela NB, Ronacher K, Veenstra H, Kidd M, Gie R, Beyers N, van Helden P, Walzl G. 2008. Immune parameters as markers of tuberculosis extent of disease and early prediction of anti-tuberculosis chemotherapy response. J Infect 56:340–347. 10.1016/j.jinf.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 79.Siawaya JFD, Bapela NB, Ronacher K, Beyers N, Van Helden P, Walzl G. 2008. Differential expression of interleukin-4 (IL-4) and IL-4δ2 mRNA, but not transforming growth factor beta (TGF-β), TGF-βRII, Foxp3, gamma interferon, T-bet, or GATA-3 mRNA, in patients with fast and slow responses to antituberculosis treatment. Clin Vaccine Immunol 15:1165–1170. 10.1128/CVI.00084-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sigal GB, Segal MR, Mathew A, Jarlsberg L, Wang M, Barbero S, Small N, Haynesworth K, Davis JL, Weiner M, Whitworth WC, Jacobs J, Schorey J, Lewinsohn DM, Nahid P. 2017. Biomarkers of tuberculosis severity and treatment effect: a directed screen of 70 host markers in a randomized clinical trial. EBioMedicine 25:112–121. 10.1016/j.ebiom.2017.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Singh UB, Rana T, Kaushik A, Porwal C, Makkar N. 2012. Day zero quantitative mRNA analysis as a prognostic marker in pulmonary tuberculosis category II patients on treatment. Clin Microbiol Infect 18:E473–81. 10.1111/j.1469-0691.2012.04004.x. [DOI] [PubMed] [Google Scholar]
- 82.Sivakumaran D, Jenum S, Vaz M, Selvam S, Ottenhoff THM, Haks MC, Malherbe ST, Doherty TM, Ritz C, Grewal HMS. 2020. Combining host-derived biomarkers with patient characteristics improves signature performance in predicting tuberculosis treatment outcomes. Commun Biol 3:359. 10.1038/s42003-020-1087-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ugarte-Gil CA, Elkington P, Gilman RH, Coronel J, Tezera LB, Bernabe-Ortiz A, Gotuzzo E, Friedland JS, Moore DAJ. 2013. Induced sputum MMP-1, -3 & -8 concentrations during treatment of tuberculosis. PLoS One 8:e61333. 10.1371/journal.pone.0061333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Wang X, Tang J, Wang R, Chen C, Tan S, Yu F, Tao Y, Li Y. 2016. Sputum endothelin-1 level is associated with active pulmonary tuberculosis and effectiveness of anti-tuberculosis chemotherapy. Exp Ther Med 11:1104–1108. 10.3892/etm.2016.2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Warsinske HC, Rao AM, Moreira FMF, Santos PCP, Liu AB, Scott M, Malherbe ST, Ronacher K, Walzl G, Winter J, Sweeney TE, Croda J, Andrews JR, Khatri P. 2018. Assessment of validity of a blood-based 3-gene signature score for progression and diagnosis of tuberculosis, disease severity, and treatment response. JAMA Netw Open 1:e183779. 10.1001/jamanetworkopen.2018.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wood R, Racow K, Bekker L-G, Middelkoop K, Vogt M, Kreiswirth BN, Lawn SD. 2012. Lipoarabinomannan in urine during tuberculosis treatment: association with host and pathogen factors and mycobacteriuria. BMC Infect Dis 12:47. 10.1186/1471-2334-12-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yan L, Xiao H, Zhang Q. 2018. Using simultaneous amplification and testing method for evaluating the treatment outcome of pulmonary tuberculosis. BMC Infect Dis 18:512. 10.1186/s12879-018-3424-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Zetola NM, Modongo C, Matsiri O, Tamuhla T, Mbongwe B, Matlhagela K, Sepako E, Catini A, Sirugo G, Martinelli E, Paolesse R, Di Natale C. 2017. Diagnosis of pulmonary tuberculosis and assessment of treatment response through analyses of volatile compound patterns in exhaled breath samples. J Infect 74:367–376. 10.1016/j.jinf.2016.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Zhao G, Luo X, Han X, Liu Z. 2020. Combining bioinformatics and biological detection to identify novel biomarkers for diagnosis and prognosis of pulmonary tuberculosis. Saudi Med J 41:351–360. 10.15537/smj.2020.4.24989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zhu Y, Jia H, Chen J, Cui G, Gao H, Wei Y, Lu C, Wang L, Uede T, Diao H. 2015. Decreased osteopontin expression as a reliable prognostic indicator of improvement in pulmonary tuberculosis: impact of the level of interferon-γ-inducible protein 10. Cell Physiol Biochem 37:1983–1996. 10.1159/000438559. [DOI] [PubMed] [Google Scholar]
- 91.Alzubaidi LKA, Alyasiri NS, Mehdi LY. 2019. Study the level of interlukin-2 during and after treatment in sputum of Iraqi patients with pulmonary tuberculosis. Ind Jour of Publ Health Rese Develop 10:746–751. 10.5958/0976-5506.2019.02524.5. [DOI] [Google Scholar]
- 92.Clifford V, He Y, Zufferey C, Connell T, Curtis N. 2015. Interferon gamma release assays for monitoring the response to treatment for tuberculosis: a systematic review. Tuberculosis (Edinb) 95:639–650. 10.1016/j.tube.2015.07.002. [DOI] [PubMed] [Google Scholar]
- 93.Shenai S, Ronacher K, Malherbe S, Stanley K, Kriel M, Winter J, Peppard T, Barry CE, Wang J, Dodd LE, Via LE, Barry CE III, Walzl G, Alland D. 2016. Bacterial loads measured by the Xpert MTB/RIF assay as markers of culture conversion and bacteriological cure in pulmonary TB. PLoS One 11:e0160062. 10.1371/journal.pone.0160062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mbelele PM, Mohamed SY, Sauli E, Mpolya EA, Mfinanga SG, Addo KK, Heysell SK, Mpagama SG. 2018. Meta-narrative review of molecular methods for diagnosis and monitoring of multidrug-resistant tuberculosis treatment in adults. Int J Mycobacteriol 7:299–309. 10.4103/ijmy.ijmy_135_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ruhwald M, Aabye MG, Ravn P. 2012. IP-10 release assays in the diagnosis of tuberculosis infection: current status and future directions. Expert Rev Mol Diagn 12:175–187. 10.1586/erm.11.97. [DOI] [PubMed] [Google Scholar]
- 96.Yoon C, Chaisson LH, Patel SM, Allen IE, Drain PK, Wilson D, Cattamanchi A. 2017. Diagnostic accuracy of C-reactive protein for active pulmonary tuberculosis: a meta-analysis. Int J Tuber Lung Dis 21:1013–1019. 10.5588/ijtld.17.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Zimmer AJ, Schumacher SG, Södersten E, Mantsoki A, Wyss R, Persing DH, Banderby S, Strömqvist Meuzelaar L, Prieto J, Gnanashanmugam D, Khatri P, Ongarello S, Ruhwald M, Denkinger CM. 2021. A novel blood-based assay for treatment monitoring of tuberculosis. BMC Res Notes 14:247. 10.1186/s13104-021-05663-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Denkinger CM, Schumacher SG, Gilpin C, Korobitsyn A, Wells WA, Pai M, Leeflang M, Steingart KR, Bulterys M, Schünemann H, Glaziou P, Weyer K. 2019. Guidance for the evaluation of tuberculosis diagnostics that meet the World Health Organization (WHO) target product profiles: an introduction to WHO process and study design principles. J Infect Dis 220:S91–S98. 10.1093/infdis/jiz097. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1, Text S2, Table S3, Fig. S4, Tables S5 and S6, Fig. S7, and Tables S8 to S10. Download jcm.01859-21-s0001.pdf, PDF file, 0.8 MB (768.4KB, pdf)