ABSTRACT
Patients with occult hepatitis B infection (OBI) have undetectable hepatitis B surface antigen (HBsAg) by conventional assays but detectable hepatitis B virus (HBV) DNA in blood/liver. We evaluated the key performance characteristics of a sensitive HBsAg assay (Architect HBsAg Next qualitative assay, referred to as NEXT) with respect to HBsAg detection. Assay precision, sample carryover, and seroconversion sensitivity of NEXT were evaluated. HBsAg was measured by NEXT in 1,138 individuals, including 1,038 patients who attended liver clinics in a tertiary hospital (200 HBV DNA-positive blood donors whose HBsAg was undetectable by conventional assays, 38 patients receiving immunosuppressive therapy, and 800 chronic hepatitis B patients with HBsAg seroclearance) and 100 HBsAg-negative subjects recruited from a community project. The within-run and within-laboratory coefficients of variation were <6% for the positive sample pools. In 9 seroconversion panels tested, NEXT allowed an earlier HBsAg detection than conventional assays. NEXT detected HBsAg in 10/200 (5%) HBsAg-negative blood donors, 1/20 (5%) and 0/18 HBsAg-negative patients with and without HBV reactivation, respectively, and 59/800 (7.3%) patients with HBsAg seroclearance. HBsAg was detectable by NEXT in 27.8%, 8.2%, 6.9%, 3.8%, and 1.9% samples at <3, 3 to 5, >5 to 8, >8 to 11, and >11 years after HBsAg seroclearance, respectively. Seven out of 100 HBsAg-negative community-identified subjects were tested positive by NEXT. Compared with conventional HBsAg assays, NEXT demonstrated a higher sensitivity and conferred an increment of 5 to 7% detection rate in patients with OBI, thereby helping in identifying HBV carriers and prevention of OBI-associated HBV transmission and reactivation.
KEYWORDS: hepatitis B virus, diagnostic test, hepatitis B surface antigens, blood donors, laboratories
INTRODUCTION
According to the latest report published by the World Health Organization (WHO), there were 1.5 million new cases of hepatitis B virus (HBV) infection in 2019, causing 820,000 hepatitis B-related deaths (1). Diagnosis of both acute and chronic hepatitis B (CHB) infection is based on the detection of hepatitis B surface antigen (HBsAg) in blood. The majority of the conventional HBsAg assays have a lower detection limit around 0.05 IU/mL (ranging from 0.01 to 0.095 IU/mL) (2). Advances in the development of nucleic acid testing (NAT) reveal cases in which patients with undetectable HBsAg may still harbor HBV DNA. This is an important consideration in the setting of early detection of acute hepatitis B infection during the window-phase period and also in the setting of occult hepatitis B infection (OBI).
OBI is defined as the presence of HBV DNA in blood and/or liver in subjects who have undetectable HBsAg by conventional HBsAg assays (3). Although OBI may be considered a quiescent form of CHB, it has several important clinical implications. First, patients with OBI are still at risk of developing liver-related complications or hepatocellular carcinoma (HCC), albeit that the estimated risk is lower than CHB patients (4). Second, OBI patients can potentially transmit HBV via blood transfusion or organ transplant (5). In the setting of liver transplant, the risk of HBV transmission by donors who had past exposure to HBV to recipients is as high as 22 to 100% (6, 7). Third, and more importantly, OBI is a major cause of HBV reactivation, a potentially fatal disease (8). This problem is especially prominent in patients undergoing immunosuppressive therapy such as anti-CD20 monoclonal antibody rituximab (9–11) and hematopoietic stem cell transplantation (12). In recent years, there has been a great advance in the development of various forms of immunosuppressive agents, such as B cell-depleting agents like rituximab and ofatumumab, corticosteroids, and tumor necrosis factor alpha inhibitors or other cytokine inhibitors (11–15). The rapidly expanding use of immunosuppressive agents increases the risk of HBV reactivation in patients with undiscovered OBI. The reported HBV reactivation rate was 41.5% at 2 years for patients with rituximab therapy (11) and 3 to 43% for patients receiving hematopoietic stem cell transplantation (12, 15, 16).
The diagnosis of OBI mainly relies on the detectability of serum and liver HBV DNA in individuals with undetectable HBsAg. However, HBV DNA is often too low to be detected in serum of patients with OBI, and detection of liver HBV DNA is hindered by the requirement of liver biopsy specimen (3). To meet the need for the accurate diagnosis of OBI and HBV infection, an improvement in the sensitivity of both HBsAg and HBV DNA detection assays is eagerly awaited. Sensitive detection of HBV DNA and/or HBsAg would help to identify individuals who are carriers of HBV and thereby identify those at risk of liver-related complications and HBV reactivation, as well as minimizing the risk of HBV transmission.
Recent advance in immunoassay technology enables a more sensitive detection of HBsAg. Newer generations of more sensitive HBsAg assays have been described (17–20). One such assay is the Architect HBsAg NEXT assay (referred to as NEXT; Abbott Laboratories, Abbott Park, IL), the prototype of which has been reported to have a lower limit of detection of 0.0052 IU/mL (21, 22), which is numerically 2 to 18 times more sensitive than conventional assays.
NEXT has been previously demonstrated to be able to detect HBsAg in samples with HBsAg mutants and commercially available panels from patients with vaccine breakthrough and late and early acute hepatitis (23, 24). In the present study, we aimed to evaluate the key performance characteristics of HBsAg NEXT and compare it with other conventional HBsAg assays. We also evaluated the performance of NEXT in the detection of HBsAg in cohorts of patients with OBI.
MATERIALS AND METHODS
Patients.
This study included a total of 1,138 patients/subjects, including 1,038 patients who had undetectable HBsAg by conventional assays but with clinical or serological evidence of OBI and 100 HBsAg-negative subjects recruited from the community. Based on how they were recruited, the participants were grouped into 4 groups. Group 1 comprised 200 HBsAg-negative, HBV DNA-positive (by NAT) blood donors who were referred from the Hong Kong Red Cross Blood Transfusion Services to the Liver Clinics at the Queen Mary Hospital, Hong Kong, for clinical follow-up between June 2009 and April 2020. Group 2 included 38 HBsAg-negative, antibodies against hepatitis B core protein (anti-HBc)-positive patients who subsequently received either rituximab-containing therapy or allogeneic hematopoietic stem cell transplantation and were followed up in our center from September 2011 to November 2014 (13). Group 3 included 800 chronic hepatitis B patients who have been followed up in the Liver Clinics at the Queen Mary Hospital since 1998 and had documented HBsAg seroclearance, defined as negative serum HBsAg for 2 consecutive follow-ups at least 6 months apart. The 100 HBsAg-negative subjects (group 4) were randomly selected HBsAg-negative participants in a previous territory-wide community surveillance study conducted from 2015 to 2016 (25). All 4 groups of patients/subjects had HBsAg undetectable by conventional assays (mentioned below). Stored samples were retrieved from our samples archive for analysis in the present study. This study was approved by the Institutional Review Board of the University of Hong Kong and the Hospital Authority Hong Kong West Cluster, Hong Kong (UW-19-429).
Laboratory analyses.
The performance of NEXT was assessed using the Architect i2000SR analyzer (Abbott Laboratories) according to the manufacturer’s instructions. During HBsAg detection by NEXT, samples with a signal over cutoff (S/CO) of ≥1 were considered initial reactive. The initial reactive samples were retested in duplicate. If at least one of the duplicate retest results was reactive, the samples were tested with the NEXT confirmatory test to confirm the results.
The conventional HBsAg assays used were the Prism HBsAg assay (referred to as PRISM; Abbott Laboratories; analytical sensitivity 0.011 IU/mL) (2), the Architect HBsAg Qualitative II assay (referred to as Qual II; Abbott Laboratories; analytical sensitivity 0.02 IU/mL) (21), the Elecsys HBsAg II assay (referred to as Elecsys II; Roche Diagnostics, Indianapolis, IN; analytical sensitivity 0.032 IU/mL) (21), and the Murex HBsAg V3 Assay (referred to as MUREX; DiaSorin, Saluggia, Italy; analytical sensitivity 0.05 IU/mL) (2). NAT was performed using the Procleix Ultrio Plus assay on the Tigris platform (referred to as Procleix; Grifols Diagnostic Solutions, Emeryville, CA; lower limit of detection [LLOD] of 3.4 IU/mL for HBV DNA). Quantitative HBV DNA measurement was performed using the Cobas HBV test (Roche Diagnostic GmbH, Mannheim, Germany; LLOD, 10 IU/mL). Anti-HBc was measured by the Elecsys anti-HBc assay (Roche Diagnostics). Anti-HBs were measured by either the Elecsys anti-HBs assay (Roche Diagnostics; LLOD, 2 IU/L) or the Architect anti-HBs assay (Abbott Laboratories; LLOD, 10 IU/L).
Assay precision.
Evaluation of assay precision was performed according to Clinical and Laboratory Standards Institute (CLSI) Evaluation of Precision of Quantitative Measurement Procedures EP5A Guidelines. A precision panel consisted of one negative quality control and one positive quality control (both supplied in the NEXT assay kit) and three clinical specimen pools. The 3 patient specimen pools included a negative specimen pool (S/CO, ∼0.6), a low-positive patient pool (S/CO, ∼1.4), and a moderate-positive specimen pool (S/CO, ∼5). Each panel member was tested in 4 replicates each day for five consecutive days.
Sample carryover.
Sample carryover contamination was assessed using HBsAg-negative samples running subsequently to a sample with high HBsAg titer. Serum obtained from a CHB patient with high HBsAg titer (named high-positive sample; HBsAg at approximately 200,000 IU/mL) and a negative sample pool (HBsAg < 0.05 IU/mL) were used, the HBsAg titers of which were measured by the Elecsys HBsAg Quant II assay (Roche Diagnostics). Two separate runs, separated by 2 h, were conducted. In each run, 5 replicates of each sample were tested in the following order: negative sample pool (named protected negative), high-positive sample, and negative sample pool (named unprotected negative). The mean S/CO readings of the protected and unprotected negatives were compared.
Seroconversion sensitivity.
Seroconversion sensitivity was evaluated using commercially available HBsAg seroconversion panels (Zeptometrix, Buffalo NY). Nine seroconversion panels (Panel ID numbers 6271, 6272, 6275, 6277, 6284, 11000, 11003, 11008, and 11017), consisting of longitudinal plasma samples collected from seroconverting donors were used. Of them, 6272 and 11000 were previously identified as probable vaccine breakthrough infections (26). The performance of NEXT was compared with that of Elecsys II. Additionally, the previously published seroconversion results of Qual II assay were used as a comparison (22, 27). HBV NAT results provided in the package insert of each panel using HBV real-time PCR method (Abbott Laboratories) were used as reference.
Statistical analysis.
Statistical analysis was performed using SPSS version 27 (IBM, Armonk, NY). Continuous samples were compared using either the Student's t test or the Mann-Whitney U test when appropriate.
RESULTS
Evaluation of assay performance of HBsAg NEXT.
(i) Precision.
Two quality controls (one positive and one negative) and three patient pool samples (two positive and one negative) were analyzed (Table 1). Twenty tests (4 replicates per day for 5 days) were run for each sample. For both the negative quality control and negative patient pool (patient pool 1), all runs gave nonreactive results (S/CO < 1). The S/CO for the negative quality control ranged from 0.21 to 0.28 (mean, 0.25), and that for the negative patient pool ranged from 0.52 to 0.74 (mean, 0.62), with a within-run and within-laboratory percent coefficient of variation (%CV) ranging from 5.31% to 10.69% (Table 1).
TABLE 1.
Precision analysis of HBsAg NEXTa
| Sample | Mean (S/CO) | Within-run precision |
Within-laboratory precision |
||
|---|---|---|---|---|---|
| SD (S/CO) | %CV | SD (S/CO) | %CV | ||
| Negative control | 0.25 | 0.013 | 5.31 | 0.020 | 8.16% |
| Positive control | 3.07 | 0.089 | 2.90 | 0.106 | 3.46 |
| Patient pool 1 | 0.62 | 0.060 | 9.72 | 0.066 | 10.69 |
| Patient pool 2 | 1.41 | 0.063 | 4.46 | 0.070 | 4.95 |
| Patient pool 3 | 4.52 | 0.099 | 2.19 | 0.257 | 5.69 |
Mean signal over cutoff (S/CO) value of each sample was generated from testing in 4 replicates each day for five consecutive days. Patient pool 1 was the HBsAg-negative specimen pool, and patient pools 2 and 3 were positive specimen pools.
All results for the positive quality control and positive patient pools were reactive (S/CO > 1). The within-run %CV was 2.90% for the positive quality control (mean, S/CO 3.07) and 4.46% and 2.19% for the two positive patient pools (mean S/CO, 1.41 and 4.52), respectively. The within-laboratory %CVs for the positive quality control and the two positive patient pools were 3.46%, 4.95%, and 5.69%, respectively (Table 1). Overall, the within-run and within-laboratory %CVs for the positive controls and samples were less than 6%.
(ii) Sample carryover analysis.
In both runs with consecutive 5 replicates each of the samples, the difference in the mean S/CO between the protected negative (the negative sample prior to the high-positive sample) and the unprotected negative (the negative sample following the high-positive sample) was 0.01 (approximately 5%). There was no significant difference between the S/CO in the unprotected and protected negative samples (both P > 0.05; Table 2).
TABLE 2.
Sample carryover test results of HBsAg NEXTa
| Run | Mean S/CO of: |
S/CO difference between unprotected and protected negative samples | P valueb (unprotected versus protected) | ||
|---|---|---|---|---|---|
| Protected negative | High positive sample | Unprotected negative | |||
| 1 | 0.20 | 946.68 | 0.21 | 0.01 | 0.21 |
| 2 | 0.19 | 954.15 | 0.18 | −0.01 | 0.54 |
Data were generated from two runs, each with 5 replicates of each sample. Protected negative and unprotected negative samples were run preceding and following the high-positive sample, respectively. S/CO, signal over cutoff.
P value was determined by t test. A P value of <0.05 was considered statistically significant.
(iii) Seroconversion sensitivity analysis.
Out of 115 specimens from the 9 seroconversion panels, 69 specimens (60%) were confirmed positive by NEXT, whereas 40 (35%) and 44 (38%) specimens had detectable HBsAg by the Elecsys II and Qual II, respectively (Fig. 1 and Table 3). In all of the 9 seroconversion panels tested, the number of days to the first confirmed result was less for the NEXT (mean, 38.8 days) than those of Elecsys II (mean, 54.2 days) and Qual II (mean,: 52.0 days), but higher than that of the HBV NAT (mean, 29.6 days). On average, NEXT reduced the early seroconversion window period by 15.4 days compared with Elecsys II and by 13.2 days compared with Qual II, corresponding to an increased seroconversion sensitivity of 22 to 25%.
FIG 1.
Schematic illustration of the seroconversion sensitivity of NEXT and the comparator assays. Individual seroconversion panel members are represented by the vertical arrows in each panel. Numbers preceding the horizontal arrow bars indicate the number of days to the first repeatedly reactive and confirmed result. Asterisk represents the last panel member, 6275-7, who was not included in analysis due to insufficient sample volume.
TABLE 3.
Seroconversion data of HBsAg NEXT and comparator assays
| Panel ID | No. of panel members | Days to the first detection (no. of panel members detected) in: |
|||
|---|---|---|---|---|---|
| NEXT | Qual IIb | Elecsys II | HBV NATc | ||
| 6271 | 5 | 0 (5) | 7 (3) | 7 (3) | 0 (5) |
| 6272 | 25 | 46 (15) | 94 (7) | 94 (7) | 0 (25) |
| 6275 | 6a | 2 (5) | 7 (4) | 22 (2) | 0 (6) |
| 6277 | 11 | 26 (8) | 33 (6) | 33 (6) | 21 (9) |
| 6284 | 19 | 46 (8) | 50 (7) | 50 (7) | 36 (11) |
| 11000 | 9 | 0 (9) | 26 (3) | 26 (3) | 0 (9) |
| 11003 | 8 | 133 (5) | 142 (3) | 142 (3) | 133 (5) |
| 11008 | 18 | 62 (7) | 69 (5) | 72 (4) | 51 (10) |
| 11017 | 14 | 34 (7) | 40 (6) | 42 (5) | 25 (9) |
| Mean days | NA | 38.8 | 52 | 54.2 | 29.6 |
| Sum of panel members (% of total no.) | 115 | 69 (60) | 44 (38) | 40 (35) | 89 (77) |
The last panel member, 6275-7, was not included in analysis due to insufficient sample volume.
Based on the data published previously (22).
Nucleic acid test based on the data provided in the package insert, determined by the HBV real-time PCR assay (Abbott Laboratories).
Panels 6272 and 11000 were previously identified as probable vaccine breakthrough infections (26). NEXT could detect HBsAg starting on day 46 for panel 6272 and day 0 (first blood draw) for panel 11000, which is 48 days and 26 days, respectively, earlier than both Elecsys II and Qual II. Both panels 6272 and 11000 have detectable anti-HBs starting from day 0, suggesting that NEXT is able to detect HBsAg in the presence of anti-HBs significantly earlier than Elecsys II and Qual II. Seroconversion data of panel 11000 are listed in Table 4.
TABLE 4.
Seroconversion data of Panel 11000 by HBsAg NEXT and comparator assaysa
| Panel member | Days from first blood samplingb | HBV DNA copies/mL* | S/CO or COI of: |
||
|---|---|---|---|---|---|
| NEXT | Qual II | Elecsys II | |||
| 11000-01 | 0 | 1,210 | 5.75 | 0.23 | 0.458 |
| 11000-02 | 3 | 650 | 3.52 | 0.24 | 0.47 |
| 11000-03 | 12 | 2,772 | 14.15 | 0.38 | 0.53 |
| 11000-04 | 14 | 3,739 | 5.47 | 0.5 | 0.652 |
| 11000-05 | 19 | 4,817 | 7.83 | 0.72 | 0.826 |
| 11000-06 | 21 | 6,064 | 14.53 | 0.94 | 0.958 |
| 11000-07 | 26 | 13,892 | 19.73 | 2.54 | 1.08 |
| 11000-08 | 29 | 12,964 | 30.33 | 4.19 | 2.76 |
| 11000-09 | 33 | 36,539 | 97.15 | 20.13 | 9.5 |
Signal over cutoff (S/CO) or cutoff index (COI) >1 is considered reactive. Repeatedly reactive and confirmed results are highlighted in gray.
Based on the data provided in the package insert of the panel determined by the HBV real-time PCR assay (Abbott Laboratories).
Detection of HBsAg in patient groups.
(i) Group 1: HBsAg-negative, NAT-positive blood donors.
Group 1 included 200 blood donors who were diagnosed to have OBI based on the undetectable HBsAg results as determined by Prism but detectable HBV DNA results by NAT (determined by Procleix; LLOD, 3.4 IU/mL), followed by confirmation of positive HBV DNA by an in-house PCR assay (28). They were referred from the Hong Kong Red Cross Blood Transfusion Services to the Liver Clinics at the Queen Mary Hospital, Hong Kong, for clinical follow-up. Stored samples from 200 such patients were retrieved and tested with NEXT. Of the 200 samples, 10 (5%) were repeatedly positive, with a mean S/CO of 3.78 (standard deviation, 1.17). These NEXT-reactive samples were cross-checked with either MUREX or Elecsys II, which showed negative HBsAg detection. Anti-HBs data were available for 140 patients, of whom 68 (49%) had undetectable anti-HBs (<10 IU/L). Nine out of the 10 patients with positive HBsAg by NEXT had available anti-HBs data, and all 9 had anti-HBs <10 IU/L.
(ii) Group 2: HBsAg-negative patients with HBV reactivation.
Group 2 included 38 patients with hematological malignancies who received either rituximab-containing immunosuppressive therapy or allogeneic hematopoietic stem cell transplantation (11–13). All 38 patients had undetectable HBsAg (by either Qual II or Elecsys II) and detectable anti-HBc before starting therapy. In the present study, we retrieved stored samples from these 38 patients. At baseline, all 38 patients (20 with HBV reactivation and 18 without HBV reactivation) had HBsAg undetectable by NEXT. Median anti-HBs levels at baseline tended to be higher in the patients without HBV reactivation (56.5 IU/L) than those with HBV reactivation (28 IU/L; P = 0.07). At the end of follow-up, HBsAg was also undetectable by NEXT in the 18 patients without HBV reactivation. We attempted to detect HBsAg in the patients with HBV reactivation prior to the occurrence of HBV reactivation. Of the 20 serum samples collected at 4 to 20 weeks before HBV reactivation, one (5%) had detectable HBsAg by NEXT. The sample which yielded positive HBsAg by NEXT was collected at 4 weeks before HBV reactivation from a patient receiving rituximab-containing therapy.
(iii) Group 3: patients with spontaneous HBsAg seroclearance.
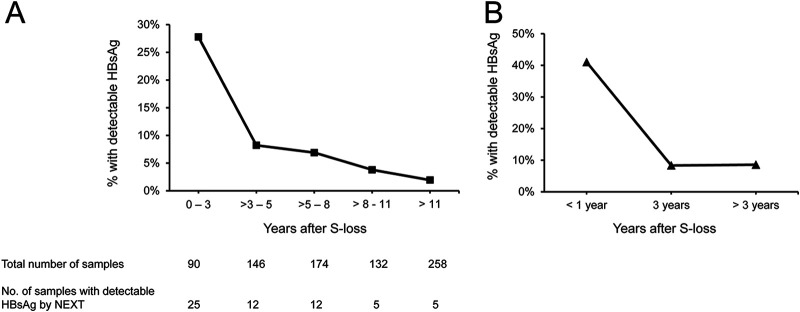
Group 3 included 800 CHB patients who have been followed up at our center and had documented spontaneous HBsAg seroclearance. Samples collected 0.5 to 29.7 years (median, 7.8 years; interquartile range [IQR], 4.6 to 13.4 years) after documented HBsAg seroclearance were subjected to HBsAg measurement by NEXT. Of them, 90 (11%), 146 (18%), 174 (22%) 132 (17%), and 258 (32%) were collected at 0 to 3 years, >3 to 5 years, >5 to 8 years, >8 to 11, and >11 years after HBsAg seroclearance, respectively. All 800 samples had HBsAg undetectable by conventional Qual II or MUREX assays. Anti-HBs data were available for 787 (98%) patients, of whom 445 (57%) had anti-HBs of ≥10 IU/L. The median anti-HBs level was 15 IU/L (IQR, 10 to 91 IU/L). Of these 800 samples, 59 (7.3%) had HBsAg detectable by NEXT (S/CO range, 1.04 to 21.7). Patients with detectable HBsAg by NEXT had lower median anti-HBs levels (10 IU/L) than those with undetectable HBsAg by NEXT (median, 17 IU/L; P < 0.001). There was a decreasing trend of HBsAg detectability rate through time: HBsAg was detectable in 25/90 (27.8%), 12/146 (8.2%), 12/174 (6.9%), 5/132 (3.8%), and 5/258 (1.9%) samples collected at 0 to 3 years, >3 to 5 years, >5 to 8 years, >8 to 11, and >11 years after HBsAg seroclearance, respectively (Fig. 2A). The decreasing trend of HBsAg detectability was also observed in patients with available serial samples. Of the 800 patients, 39 patients had serial samples collected at 0 to 1 year, 3 years, and/or >3 years after HBsAg seroclearance. In these 39 patients, HBsAg was detectable by NEXT in 16 (41%) samples collected within 1 year after seroclearance, 3 (8%) samples collected at year 3, and 3 (8%) samples collected at >3 to 15 years after seroclearance (Fig. 2B).
FIG 2.
Detection of HBsAg by HBsAg NEXT in patients with HBsAg seroclearance. (A) HBsAg detectable rate in the 800 samples collected at various time point after HBsAg seroclearance. (B) Longitudinal samples collected from 39 patients.
(iv) Group 4: healthy individuals from a community project.
Previously, we conducted a territory-wide community study in which viral hepatitis serology testing was provided to participants of health seminars in all 18 municipal districts in Hong Kong (25). In that study, 10,256 participants were recruited, among whom 9,453 (92.2%) were tested HBsAg negative by Elecsys II. In the present study, we retrieved 100 samples from HBsAg-negative subjects in this cohort and tested with NEXT. These 100 samples were confirmed by the Elecsys II to be HBsAg negative. Of these 100 subjects, 29 (29%) were anti-HBc positive. Using NEXT, we found that HBsAg was detectable in 7 (7%) subjects, all of whom were anti-HBc positive, whereas only 22 of the 93 NEXT-tested HBsAg-negative subjects (23.7%) were anti-HBc positive (P < 0.0001). HBsAg detectability by NEXT was also associated with a lower anti-HBs titer (<2.0 IU/L in HBsAg-positive subjects versus 39.8 IU/L in HBsAg-negative subjects; P = 0.013).
Summary.
The detection rate of HBsAg by NEXT in the 4 groups of patients is summarized in Table 5. Excluding the 18 non-OBI patients (in group 2) who did not have HBV reactivation, the results of the 1,120 individuals were tabulated. Among the 1,020 OBI patients (groups 1 to 3) tested, 70 (6.9%) yielded positive HBsAg detection by NEXT. Even for the apparently noninfected healthy individuals with negative HBsAg in our locality (group 4), 7% had detectable HBsAg by NEXT. The increment of HBsAg detection rate was roughly 6 to 7% compared with conventional assays.
TABLE 5.
Summary of HBsAg detection by NEXT in the 4 patient cohorts
| Patient cohort | No. of samples tested | Incremental detection by NEXT |
|
|---|---|---|---|
| No. | % | ||
| Group 1, HBsAg-negative, NAT-yield blood donors | 200 | 10 | 5 |
| Group 2, HBsAg-negative patients with HBV reactivationa | 20 | 1 | 5 |
| Group 3, patients with spontaneous HBsAg seroclearance | 800 | 59 | 7.3 |
| Group 4, HBsAg-negative individuals from a community project | 100 | 7 | 7 |
The 18 HBsAg-negative patients who did not have HBV reactivation upon receiving immunosuppressive therapy were not included in this table.
DISCUSSION
In this study, we evaluated the performance of a newly developed sensitive HBsAg assay (Architect HBsAg Next qualitative assay). This assay has been demonstrated to have a consistent sensitivity across major HBV genotypes A to H and common mutants in previous studies (21). Assay precision was satisfactory, with a within-run and within-laboratory %CV of <6% for positive samples and controls. A relatively higher %CV was observed for the negative control and patient pool (8.16% and 10.69%, respectively), likely due to the very low signals produced by the negative samples associated with assay noise. Of note, all the negative samples produced S/CO readings below 1 (standard deviation < 0.1), which validated the performance claim by the manufacturer that the S/CO standard deviation should be ≤0.1 for all negative samples. There was no contamination carryover, even when negative samples were run subsequently to samples of high titer. Using commercially available seroconversion panels, we found that NEXT was superior to the two comparator assays (Elecsys II and Qual II). In line with previous findings (23, 27), NEXT could detect HBsAg on average of 14 days earlier than Elecsys II and Qual II, the two comparator conventional assays used, and the seroconversion sensitivity was increased by 22 to 25%. This finding also implies that NEXT has a potential to diagnose acute hepatitis B during the window phase of acute hepatitis B infection where HBsAg may not be detectable by the conventional assays.
Although OBI represents a hidden menace to the society, patients with OBI are often undiagnosed, as their apparent HBsAg is negative, whereas their HBV DNA is usually not tested, mostly due to the cost burden associated with the extra HBV DNA tests. Using a high-sensitivity HBsAg immunoassay such as NEXT, which is offered at a similar cost to other chemiluminescent-based HBsAg assays and as a less expensive solution than PCR-based HBV DNA tests, could be provided to address this issue.
We applied NEXT to detect HBsAg in 4 groups of individuals. In the 200 HBsAg-negative blood donors with confirmed OBI by HBV DNA test (group 1), we demonstrated that if NEXT were used instead of conventional assays, 5% more patients harboring HBV could be identified. With the enhanced sensitivity of NEXT, it can be envisaged that the assay can be used together with NAT to detect OBI and reduce the risk of HBV transmission by OBI in a blood donation setting. We would like to point out that we did not investigate any samples from HBsAg-negative, NAT-negative blood donors. This is because performing such a study with retrospectively archived samples would raise an ethical issue, as the blood products have already been transfused to recipients, and a call-back arrangement would be required if any transfused blood products were found to be HBsAg positive. We suggest a prospective study be conducted in the future in collaboration with blood transfusion facilities so that the NEXT assay can be used as a complementary screening test before blood transfusion.
In group 2, all 38 patients had undetectable HBsAg at the start of immunosuppressive therapy. During the course of 2 years’ follow-up, none of the patients without HBV reactivation had detectable HBsAg by NEXT, while 1 out of 20 patients with HBV reactivation had detectable HBsAg at 4 weeks before the emergence of detectable HBV DNA. Although only 20 patients with HBV reactivation were included, we showed that the use of a sensitive HBsAg assay (NEXT), together with HBV DNA assays. can allow an earlier detection of HBV reactivation in prevention of this potentially fatal condition.
In patients with confirmed HBsAg loss by conventional HBsAg assays (group 3), 59 (7.3%) still harbored HBsAg (detectable by NEXT). This suggested that the standard of detection of HBsAg can be elevated, in which patients who were HBsAg negative may still harbor a low titer of circulating HBsAg. The positive detection rate of HBsAg by NEXT was higher at the time of documented HBsAg seroclearance and decreased with time afterward. It is notable that HBsAg was still detectable in some patients even at >15 years after HBsAg seroclearance. It is possible that, in patients with HBsAg seroclearance who subsequently develop HCC, there exists a low titer of HBsAg which can only be detectable when a sensitive HBsAg assay is used. While expression of detectable HBsAg after HBsAg seroclearance had been reported previously (18, 29), the implementation of a standardized and reliable sensitive HBsAg assay is much awaited. Whether this small amount of HBsAg is the main culprit of HCC development in patients with HBsAg seroclearance remains to be studied.
We also tested whether HBsAg was detectable by NEXT in the general population of HBsAg-negative subjects. Among the 100 apparently healthy participants whose HBsAg was negative, 7 had HBsAg detectable by NEXT. All these 7 patients had detectable anti-HBc. Among the 29 HBsAg-negative, anti-HBc positive subjects, 7 (24%) had detectable HBsAg by NEXT. The present finding suggests that approximately one-quarter of the HBsAg-negative, anti-HBc-positive individuals in the general public in areas of endemicity such as Hong Kong may still have a very low titer of HBsAg. Thus, the use of sensitive HBsAg assays such as NEXT can identify more “HBsAg carriers” in the general public, especially in those who were anti-HBc positive. It will help in risk stratification of patients and in the prevention of HBV transmission.
Taken together, the present study showed that the use of NEXT can provide help in the identification of HBsAg carriers who would have been missed if conventional assays were used. In general, detection of HBsAg by NEXT conferred an increment of a 6 to 7% HBsAg detection rate when compared with conventional assays (Table 5). This is especially helpful in areas of endemicity such as Asia and Africa, in which, for each population of 1 million, 60,000 to 70,000 more HBsAg-positive cases can be identified. The use of this sensitive HBsAg assay can definitely improve the prevention of HBV transmission and HBV reactivation, as well as allow better policy implementation regarding the prevention HBV-related complications.
ACKNOWLEDGMENTS
We thank John Yuen, Kate Wang, and staff of Abbott Laboratories; the Red Cross Blood Transfusion Service; and the Hong Kong Liver Foundation for their assistance.
D.K.-H.W. received speaker’s fees from Abbott Laboratories and travel support from Gilead Sciences and Chong Lap. C.C. is an employee of Abbott Laboratories, Singapore. W.-K.S. received speaker’s fees from AstraZeneca and Mylan, is an advisory board member of CSL Behring, is an advisory board member and received speaker’s fees from AbbVie, and is an advisory board member and received speaker’s fees and researching funding from Gilead Sciences. M.-F.Y. serves as advisor/consultant for AbbVie, Assembly Biosciences, Aligos Therapeutics, Arbutus Biopharma, Bristol Myer Squibb, Clear B Therapeutics, Dicerna Pharmaceuticals, Finch Therapeutics, GlaxoSmithKline, Gilead Sciences, Immunocore, Janssen, Merck Sharp and Dohme, Hoffmann-La Roche, Springbank Pharmaceuticals, and Vir Biotechnology and receives grant/research support from Assembly Biosciences, Aligos Therapeutics, Arrowhead Pharmaceuticals, Bristol Myer Squibb, Fujirebio Incorporation, Gilead Sciences, Immunocore, Merck Sharp and Dohme, Hoffmann-La Roche, Springbank Pharmaceuticals, and Sysmex Corporation. The remaining authors have no conflict of interest to declare.
This study is supported by Abbott Laboratories.
Contributor Information
Wai-Kay Seto, Email: wkseto@hku.hk.
Man-Fung Yuen, Email: mfyuen@hku.hk.
Elitza S. Theel, Mayo Clinic
REFERENCES
- 1.World Health Organization. 2021. Global progress report on HIV, viral hepatitis and sexually transmitted infections, 2021. Accountability for the global health sector strategies 2016–2021: actions for impact. World Health Organization, Geneva, Switzerland. [Google Scholar]
- 2.Servant-Delmas A, Mercier-Darty M, Ly TD, Wind F, Alloui C, Sureau C, Laperche S. 2012. Variable capacity of 13 hepatitis B virus surface antigen assays for the detection of HBsAg mutants in blood samples. J Clin Virol 53:338–345. 10.1016/j.jcv.2012.01.003. [DOI] [PubMed] [Google Scholar]
- 3.Raimondo G, Locarnini S, Pollicino T, Levrero M, Zoulim F, Lok AS, Taormina Workshop on Occult HBV Infection Faculty Members. 2019. Update of the statements on biology and clinical impact of occult hepatitis B virus infection. J Hepatol 71:397–408. 10.1016/j.jhep.2019.03.034. [DOI] [PubMed] [Google Scholar]
- 4.Yuen MF, Wong DK, Sablon E, Tse E, Ng IO, Yuan HJ, Siu CW, Sander TJ, Bourne EJ, Hall JG, Condreay LD, Lai CL. 2004. HBsAg seroclearance in chronic hepatitis B in the Chinese: virological, histological, and clinical aspects. Hepatology 39:1694–1701. 10.1002/hep.20240. [DOI] [PubMed] [Google Scholar]
- 5.Yuen MF, Wong DK, Lee CK, Tanaka Y, Allain JP, Fung J, Leung J, Lin CK, Sugiyama M, Sugauchi F, Mizokami M, Lai CL. 2011. Transmissibility of hepatitis B virus (HBV) infection through blood transfusion from blood donors with occult HBV infection. Clin Infect Dis 52:624–632. 10.1093/cid/ciq247. [DOI] [PubMed] [Google Scholar]
- 6.Dickson RC, Everhart JE, Lake JR, Wei Y, Seaberg EC, Wiesner RH, Zetterman RK, Pruett TL, Ishitani MB, Hoofnagle JH. 1997. Transmission of hepatitis B by transplantation of livers from donors positive for antibody to hepatitis B core antigen. The National Institute of Diabetes and Digestive and Kidney Diseases Liver Transplantation Database. Gastroenterology 113:1668–1674. 10.1053/gast.1997.v113.pm9352871. [DOI] [PubMed] [Google Scholar]
- 7.Munoz SJ. 2002. Use of hepatitis B core antibody-positive donors for liver transplantation. Liver Transpl 8:S82-7. 10.1053/jlts.2002.35783. [DOI] [PubMed] [Google Scholar]
- 8.Perrillo RP, Gish R, Falck-Ytter YT. 2015. American Gastroenterological Association Institute technical review on prevention and treatment of hepatitis B virus reactivation during immunosuppressive drug therapy. Gastroenterology 148:221–244.e3. 10.1053/j.gastro.2014.10.038. [DOI] [PubMed] [Google Scholar]
- 9.Evens AM, Jovanovic BD, Su YC, Raisch DW, Ganger D, Belknap SM, Dai MS, Chiu BC, Fintel B, Cheng Y, Chuang SS, Lee MY, Chen TY, Lin SF, Kuo CY. 2011. Rituximab-associated hepatitis B virus (HBV) reactivation in lymphoproliferative diseases: meta-analysis and examination of FDA safety reports. Ann Oncol 22:1170–1180. 10.1093/annonc/mdq583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeo W, Chan TC, Leung NW, Lam WY, Mo FK, Chu MT, Chan HL, Hui EP, Lei KI, Mok TS, Chan PK. 2009. Hepatitis B virus reactivation in lymphoma patients with prior resolved hepatitis B undergoing anticancer therapy with or without rituximab. J Clin Oncol 27:605–611. 10.1200/JCO.2008.18.0182. [DOI] [PubMed] [Google Scholar]
- 11.Seto WK, Chan TS, Hwang YY, Wong DK, Fung J, Liu KS, Gill H, Lam YF, Lie AK, Lai CL, Kwong YL, Yuen MF. 2014. Hepatitis B reactivation in patients with previous hepatitis B virus exposure undergoing rituximab-containing chemotherapy for lymphoma: a prospective study. JCO 32:3736–3743. 10.1200/JCO.2014.56.7081. [DOI] [PubMed] [Google Scholar]
- 12.Seto WK, Chan TS, Hwang YY, Wong DK, Fung J, Liu KS, Gill H, Lam YF, Lau EHY, Cheung KS, Lie AKW, Lai CL, Kwong YL, Yuen MF. 2017. Hepatitis B reactivation in occult viral carriers undergoing hematopoietic stem cell transplantation: a prospective study. Hepatology 65:1451–1461. 10.1002/hep.29022. [DOI] [PubMed] [Google Scholar]
- 13.Seto WK, Wong DK, Chan TY, Hwang YY, Fung J, Liu KS, Gill H, Lam YF, Cheung KS, Lie AK, Lai CL, Kwong YL, Yuen MF. 2016. Association of hepatitis B core-related antigen with hepatitis B virus reactivation in occult viral carriers undergoing high-risk immunosuppressive therapy. Am J Gastroenterol 111:1788–1795. 10.1038/ajg.2016.436. [DOI] [PubMed] [Google Scholar]
- 14.Perez-Alvarez R, Diaz-Lagares C, Garcia-Hernandez F, Lopez-Roses L, Brito-Zeron P, Perez-de-Lis M, Retamozo S, Bove A, Bosch X, Sanchez-Tapias JM, Forns X, Ramos-Casals M, Group BS, BIOGEAS Study Group. 2011. Hepatitis B virus (HBV) reactivation in patients receiving tumor necrosis factor (TNF)-targeted therapy: analysis of 257 cases. Medicine (Baltimore, MD) 90:359–371. 10.1097/MD.0b013e3182380a76. [DOI] [PubMed] [Google Scholar]
- 15.Vigano M, Vener C, Lampertico P, Annaloro C, Pichoud C, Zoulim F, Facchetti F, Poli F, Scalamogna M, Deliliers GL, Colombo M. 2011. Risk of hepatitis B surface antigen seroreversion after allogeneic hematopoietic SCT. Bone Marrow Transplant 46:125–131. 10.1038/bmt.2010.70. [DOI] [PubMed] [Google Scholar]
- 16.Hammond SP, Borchelt AM, Ukomadu C, Ho VT, Baden LR, Marty FM. 2009. Hepatitis B virus reactivation following allogeneic hematopoietic stem cell transplantation. Biol Blood Marrow Transplant 15:1049–1059. 10.1016/j.bbmt.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Matsubara N, Kusano O, Sugamata Y, Itoh T, Mizuii M, Tanaka J, Yoshizawa H. 2009. A novel hepatitis B virus surface antigen immunoassay as sensitive as hepatitis B virus nucleic acid testing in detecting early infection. Transfusion 49:585–595. 10.1111/j.1537-2995.2008.02026.x. [DOI] [PubMed] [Google Scholar]
- 18.Shinkai N, Matsuura K, Sugauchi F, Watanabe T, Murakami S, Iio E, Ogawa S, Nojiri S, Joh T, Tanaka Y. 2013. Application of a newly developed high-sensitivity HBsAg chemiluminescent enzyme immunoassay for hepatitis B patients with HBsAg seroclearance. J Clin Microbiol 51:3484–3491. 10.1128/JCM.00726-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Takeda K, Maruki M, Yamagaito T, Muramatsu M, Sakai Y, Tobimatsu H, Kobayashi H, Mizuno Y, Hamaguchi Y. 2013. Highly sensitive detection of hepatitis B virus surface antigen by use of a semiautomated immune complex transfer chemiluminescence enzyme immunoassay. J Clin Microbiol 51:2238–2244. 10.1128/JCM.00324-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pronier C, Candotti D, Boizeau L, Bomo J, Laperche S, Thibault V. 2020. The contribution of more sensitive hepatitis B surface antigen assays to detecting and monitoring hepatitis B infection. J Clin Virol 129:104507. 10.1016/j.jcv.2020.104507. [DOI] [PubMed] [Google Scholar]
- 21.Lou S, Taylor R, Pearce S, Kuhns M, Leary T. 2018. An ultra-sensitive Abbott ARCHITECT((R)) assay for the detection of hepatitis B virus surface antigen (HBsAg). J Clin Virol 105:18–25. 10.1016/j.jcv.2018.05.009. [DOI] [PubMed] [Google Scholar]
- 22.Sickinger E, Braun HB, Meyer T, Schmid K, Daghfal D, Oer M, Schultess J. 2020. Performance characteristics of the high sensitivity Alinity i & ARCHITECT HBsAg Next qualitative/confirmatory assays. Diagn Microbiol Infect Dis 97:115033. 10.1016/j.diagmicrobio.2020.115033. [DOI] [PubMed] [Google Scholar]
- 23.Kuhns MC, McNamara AL, Holzmayer V, Cloherty GA. 2019. Molecular and serological characterization of hepatitis B vaccine breakthrough infections in serial samples from two plasma donors. Virol J 16:43. 10.1186/s12985-019-1154-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhns MC, Holzmayer V, McNamara AL, Sickinger E, Schultess J, Cloherty GA. 2019. Improved detection of early acute, late acute, and occult hepatitis B infections by an increased sensitivity HBsAg assay. J Clin Virol 118:41–45. 10.1016/j.jcv.2019.08.001. [DOI] [PubMed] [Google Scholar]
- 25.Liu KSH, Seto WK, Lau EHY, Wong DK, Lam YF, Cheung KS, Mak LY, Ko KL, To WP, Law MWK, Wu JT, Lai CL, Yuen MF. 2019. A territorywide prevalence study on blood-borne and enteric viral hepatitis in Hong Kong. J Infect Dis 219:1924–1933. 10.1093/infdis/jiz038. [DOI] [PubMed] [Google Scholar]
- 26.Keating SM, Heitman JD, Wu S, Deng X, Stramer SL, Kuhns MC, Mullen C, Norris PJ, Busch MP. 2014. Cytokine and chemokine responses in the acute phase of hepatitis B virus replication in naive and previously vaccinated blood and plasma donors. J Infect Dis 209:845–854. 10.1093/infdis/jit563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sickinger E, Braun H, Schultess J, Oer M. 2019. Performance evaluation of a new next-generation high-sensitive architect HBsAg assay. Clinica Chimica Acta 493:S555. 10.1016/j.cca.2019.03.1166. [DOI] [Google Scholar]
- 28.Tsoi WC, Lelie N, Lin CK. 2013. Enhanced detection of hepatitis B virus in Hong Kong blood donors after introduction of a more sensitive transcription-mediated amplification assay. Transfusion 53:2477–2488. 10.1111/trf.12165. [DOI] [PubMed] [Google Scholar]
- 29.Seto WK, Tanaka Y, Wong DK, Lai CL, Shinkai N, Yuen JC, Tong T, Fung J, Hung IF, Yuen MF. 2012. Evidence of serologic activity in chronic hepatitis B after surface antigen (HBsAg) seroclearance documented by conventional HBsAg assay. Hepatol Int 7:98–105. 10.1007/s12072-012-9354-7. [DOI] [PMC free article] [PubMed] [Google Scholar]