Abstract
Phototrophic Gemmatimonadetes evolved the ability to use solar energy following horizontal transfer of photosynthesis-related genes from an ancient phototrophic proteobacterium. The electron cryo-microscopy structure of the Gemmatimonas phototrophica photosystem at 2.4 Å reveals a unique, double-ring complex. Two unique membrane-extrinsic polypeptides, RC-S and RC-U, hold the central type 2 reaction center (RC) within an inner 16-subunit light-harvesting 1 (LH1) ring, which is encircled by an outer 24-subunit antenna ring (LHh) that adds light-gathering capacity. Femtosecond kinetics reveal the flow of energy within the RC-dLH complex, from the outer LHh ring to LH1 and then to the RC. This structural and functional study shows that G. phototrophica has independently evolved its own compact, robust, and highly effective architecture for harvesting and trapping solar energy.
Cryo EM reveals a unique double-ring light-harvesting complex in the phototrophic bacterium Gemmatimonas phototrophica.
INTRODUCTION
Photosynthesis is an ancient and fundamental biological process (1–3), in which phototrophic organisms transform solar radiation into the metabolic energy that fuels most of the world’s ecosystems. The relatively simple photosynthetic (PS) apparatus of bacteriochlorophyll-based phototrophs consists of a single type of reaction center (RC) surrounded by a ring of light-harvesting 1 (LH1) subunits, sometimes embellished with smaller rings of LH2 complexes (4). There are seven phyla that contain chlorophototrophic bacteria, and remarkably two phyla have been discovered since 2000: the Acidobacteria (5) and, most recently, Gemmatimonadetes (6). Present-day phototrophic Gemmatimonadetes are commonly found in freshwater bacterioplankton (7). The first phototrophic member of this phylum, Gemmatimonas phototrophica, was isolated from a freshwater lake in the Gobi desert. This slowly growing microaerophile received a complete package of photosynthesis-related genes (fig. S1) by a distant horizontal gene transfer from an ancient phototrophic proteobacterium (6). Thus, G. phototrophica provides an important insight into the evolution of photosynthesis, documenting the transfer of photosynthesis from one phylum to another, conferring phototrophy on a nonphototrophic host (6). This intriguing episode in evolution now has relevance to synthetic biology and to attempts to reprogram a heterotrophic bacterium for light-powered metabolism (8). Here, we purified the PS core complex from G. phototrophica and use a combination of single-particle cryo–electron microscopy (cryo-EM) and ultrafast spectroscopy to resolve the structure to 2.4-Å resolution and to show how its unique evolution created a novel and highly stable architecture for absorbing, transferring, and trapping harvested solar energy.
RESULTS AND DISCUSSION
Overall structure of the RC-dLH complex
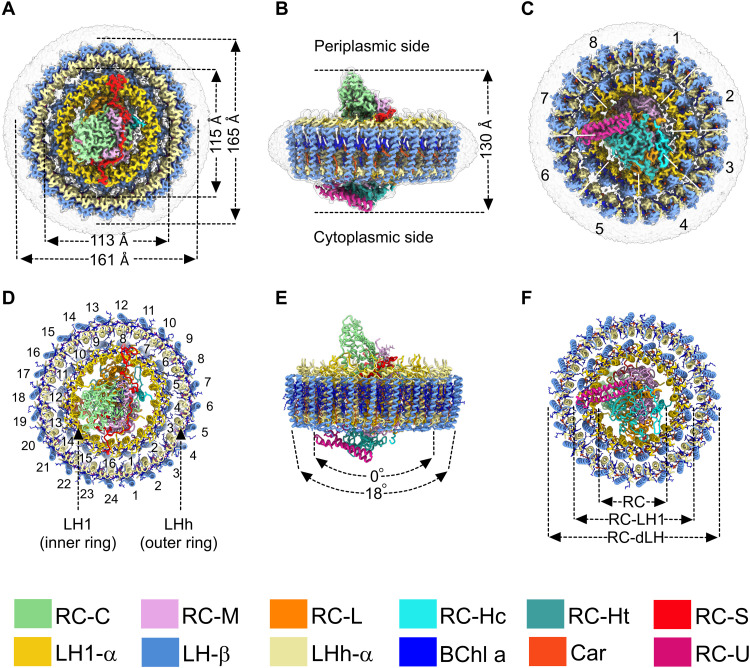
From the raw cryo-EM images of the purified complex, it was apparent that the particles, ~20 nm in size (fig. S2, A and B), were much larger than previously described single-ring RC-LH1 complexes, which typically have diameters of ~16 nm including the detergent belt (9–12). The complex consists of three parts: the central RC, an inner LH1 ring, of the kind found in RC-LH1 complexes (9, 11, 13), and a new type of outer LH ring. The diameter of the outer ring is 161 to 165 Å (Fig. 1, A to C), and the height of the RC-dLH complex measured from the bottom of the Hc subunit to the tip of the C subunit is 130 Å (Fig. 1B). LH1 forms a cylinder, while LHh is slightly conical (Fig. 1E). The total molecular weight calculated for the final model, including all pigments, lipids, detergent, and water molecules, is ~800 kDa (table S1), in agreement with the earlier estimate (14).
Fig. 1. Cryo-EM structure of the RC-dLH complex from G. phototrophica.
(A to C) Views of the color-coded RC-dLH density map. Detergent and other disordered molecules are in gray. (A) Perpendicular view from the periplasmic side of the membrane, with the diameters of the two LH rings indicated. (B) View within the plane of the membrane showing the overall height of the complex. (C) Perpendicular view from the cytoplasmic side. White dashed lines indicate pseudo-C8 symmetry of the double LHh-LH1 ring. (D to F) Ribbon models corresponding to (A) to (C). In (D), the LH1 and LHh subunits are numbered in black. (E) The two arrows indicate the degree of curvature of the outer and inner LH rings. (F) Concentric nature of the complex proceeding outward from the central RC. The color code is shown below the panels. Images were produced using ChimeraX (15).
The overall RC architecture is similar to that found in phototrophic Proteobacteria (9, 11), with transmembrane L and M subunits, and a bound cytochrome c exposed on the periplasmic side of the complex. The H subunit, normally a single entity, is fragmented into two parts: a smaller transmembrane polypeptide (~7.7 kDa), Ht, and a larger polypeptide (~19.9 kDa), Hc, located on the cytoplasmic side of the complex. We identified two open reading frames (ORFs) within the photosynthesis gene cluster encoding Ht and Hc and named them puhA1 and puhA2 (fig. S1). Mass spectrometry analysis confirmed the presence of the Ht and Hc polypeptides (fig. S3). Figure S4 shows aligned sequences of RC-H subunits from several RC-LH1 complexes, highlighting a six-residue region present in Proteobacteria that contain single-subunit RC-H polypeptides. The absence of this possible joining sequence from G. phototrophica could account for the separate Ht and Hc polypeptides. Fits of structural models of the Ht and Hc polypeptides, as well as other RC components, within their respective cryo-EM densities are shown in fig. S5 (A and B). The RC has the same pigment architecture as in purple phototrophic bacteria: two special pair and two accessory bacteriochlorophyll a (BChl a) molecules, two bacteriopheophytin a (BPhe a) molecules, and one carotenoid, spirilloxanthin, in the 15-cis configuration (fig. S5C). Three menaquinone-8 (MQ8) molecules were resolved: one at the QA position, a second at a position analogous to the QP site recently found in some RC-LH1 complexes (9, 11, 16, 17), and another adjacent to the inner face of the LH1 ring, near the QB site, which we designate QF. The locations of these three MQ molecules are shown in the cofactor map of the complex (fig. S6), and their identity as MQ was verified by high-performance liquid chromatography (HPLC) analysis (fig. S7A). Figure S5C shows the fits of MQ8 molecules within the QA, QP, and QF cryo-EM densities.
The RC is surrounded by a double, concentric LH complex built on a repeating modular principle that forms near-circular assemblies from heterodimer units of transmembrane α- and β-polypeptides. The inner ring, denoted LH1, consists of 16 pairs of α/β apoproteins and is encircled by an outer 24-subunit assembly, named LHh (Fig. 1D). Together, LH1 and LHh form the antenna component of the double-ring complex designated as RC-dLH (Fig. 1, D and F). The same pufB gene encodes β-subunits for both LH rings, whereas the α-polypeptides differ and are encoded by pufA1 and pufA2 for the inner and outer rings, respectively (fig. S8). PufA1 clusters with pufA genes from Gammaproteobacteria, whereas the pufA2 gene is only distantly related to pufA genes in Proteobacteria (fig. S9). A pair of BChl a molecules and an all-trans carotenoid, gemmatoxanthin (18) (figs. S5C and S7B), are noncovalently bound between the α- and β-polypeptides in each LH heterodimer. Both the inner and outer LH rings are slightly elliptical (eccentricity = 0.22; Fig. 1A) and have pseudo-C8 symmetry (Fig. 1C). Figure S10 also shows that our three-dimensional (3D) reconstructions, with resolutions between 2.4 and 2.5 Å (fig. S11B), generate a mixture of four different structural conformations of the RC-dLH complex. Figure S11C shows the maps of the four structures colored by local resolution, showing that there could be more mobility of the LH complexes near the cytoplasmic face of the membrane. The relationship between these structural models is depicted in fig. S10, where we superimposed models 1a and 2a, with 2a rotated by 0°, 7.5°, and 22.5°. When the central RC-LH1 components in models 1a and 2a are aligned, the outer LHh rings are displaced (fig. S10C-a, 0° rotation of model_2a). A 7.5° rotation of model_2a with respect to 1a brings the outer rings into alignment (fig. S10C-b), but now both the inner LH1 ring and the RC fall out of alignment. Figure S10C-c shows the consequence of an overall 22.5° rotation of model_2a, which brings the LHh and LH1 rings into alignment, but which leaves misaligned RCs. This analysis shows that 1a/2a and 1b/2b are two different complexes. We can also see another major difference, namely, that the 1b/2b pair contains an additional subunit on the cytoplasmic side denoted as RC-U. The other new subunit, RC-S, is present in all four classes of complex (fig. S10A). Last, Fig. 1 displays model_1b, which contains all of the subunits.
Stabilizing RC-LH1 interactions
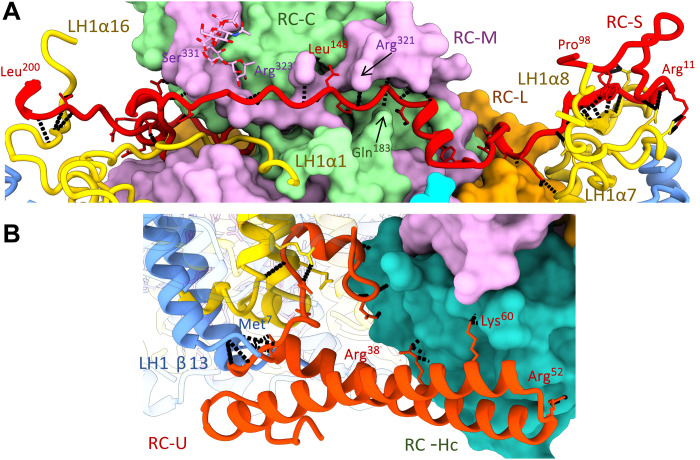
A unique arrangement of stabilizing, membrane-extrinsic polypeptides distinguishes the RC-LH1 inner subcomplex of G. phototrophica from all known RC-LH1 complexes from purple photosynthetic bacteria. RC-S (string like) lies across the periplasmic surface of the inner RC-LH1 complex, spanning the diameter of the LH1 ring (Figs. 1, A and D, and 2A). N-terminal processing verified by mass spectrometry (fig. S3) removes 60% of RC-S, and the remainder of this elongated polypeptide (8 kDa) binds to the surface regions of the RC and LH1 complexes. There are five hydrogen bonds (H-bonds) between the N-terminal region of RC-S and the periplasmic, C-terminal region of LH1-α8 on one side of the LH1 ring and four H-bonds between the RC-S C terminus with LH1-α16 on the other side. The central region of RC-S interacts with the RC-M, RC-C, and RC-L subunits through a series of H-bonds (Fig. 2A); the overall effect is to bind together the LH1 and RC components of the RC-dLH complex. On the cytoplasmic side, RC-U (U clamp–like) links the inner LH1 and the RC-Hc, through H-bonds with the N-terminal regions of LH1-α/β (13), and with RC-Hc, located at the cytoplasmic side of the complex (Fig. 2B). These surface interactions impart additional stability to the inner RC-LH1 complex, relative to other such complexes.
Fig. 2. Protein-protein interactions between the LH1 and RC complexes.
(A) The RC-S polypeptide occupies a deep groove spanning the periplasmic surface of the RC complex and involves residues from RC-C, RC-M, and RC-L. Its N- and C-terminal ends engage the C termini of LH1 α7 and α16 chains, respectively. Intermolecule H-bonds involving RC-S are shown in black. (B) RC-U forms a short coiled-coil spanning between the cytoplasmic surface of RC-Hc and LH1 subunit 13, with a largely hydrophobic N-terminal protrusion buried deep in the transmembrane space.
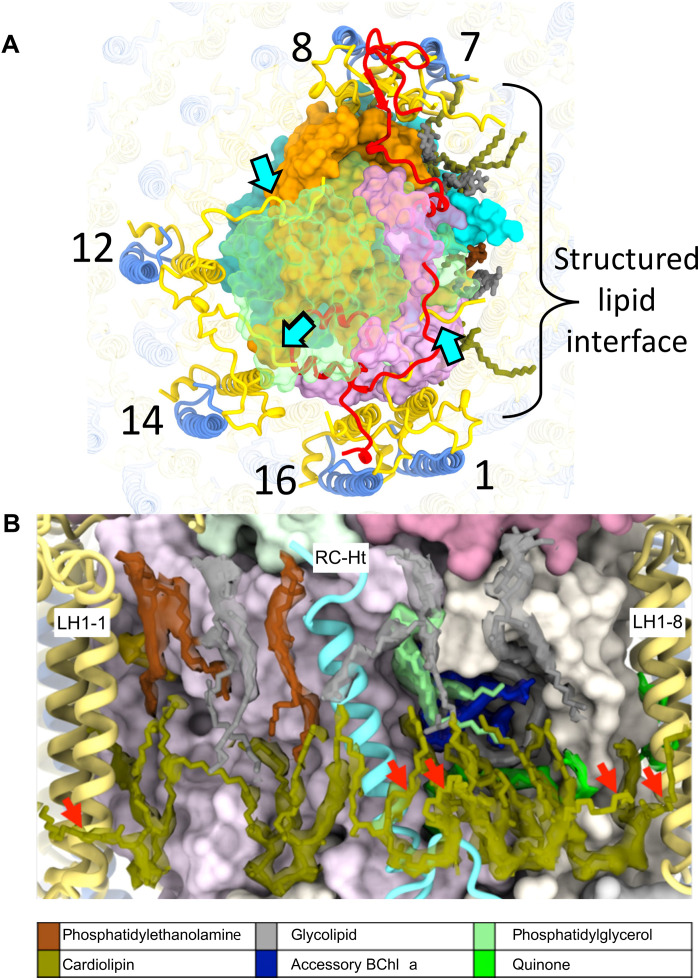
The complex is further stabilized by interactions on the periplasmic side between LH1-α1, 12, and 14 C termini, whose sequences were almost fully resolved, and the RC-M and RC-C subunits (Fig. 3A). In addition to these surface interactions between LH1 and RC, three close contact points between the RC and LH1α are evident, namely, (i) transmembrane helix 1 (TM1) from RC-L (L34-54) to LH1-α8, (ii) RC-Ht to LH1-α5, and (iii) TM1 from RC-M (M54-79) to LH1α1/α16 (Fig. 3A). We also identified a tightly packed region of structured lipids surrounding RC-Ht, within which six cardiolipins interdigitate between RC and LH1 chains, further stabilizing the LH1-RC interaction (Fig. 3B).
Fig. 3. Stabilization of RC-LH1 complex.
(A) View of the periplasmic face of the RC-dLH complex, showing interactions (cyan arrows) between the C termini of LH1-α1, 12, and 14 (yellow) and the RC-M and RC-C subunits (RC-C subunit is shown in translucent green). The region surrounding RC-Ht (covering the QA and spirilloxanthin sites) is dominated by a tightly packed lipid interface. On the cytoplasmic side, six cardiolipins (olive) interdigitate between RC and LH1 chains. The periplasmic interface consists of three uncharged glyceroglycolipids (gray), two phosphatidylethanolamines (brown), and a single phosphatidylglycerol (pale green). (B) View of the structured lipid interface in (A) with a 6-σ density map around the lipids. Outward-projecting cardiolipin tails are marked with red arrows.
Protein-pigment interactions
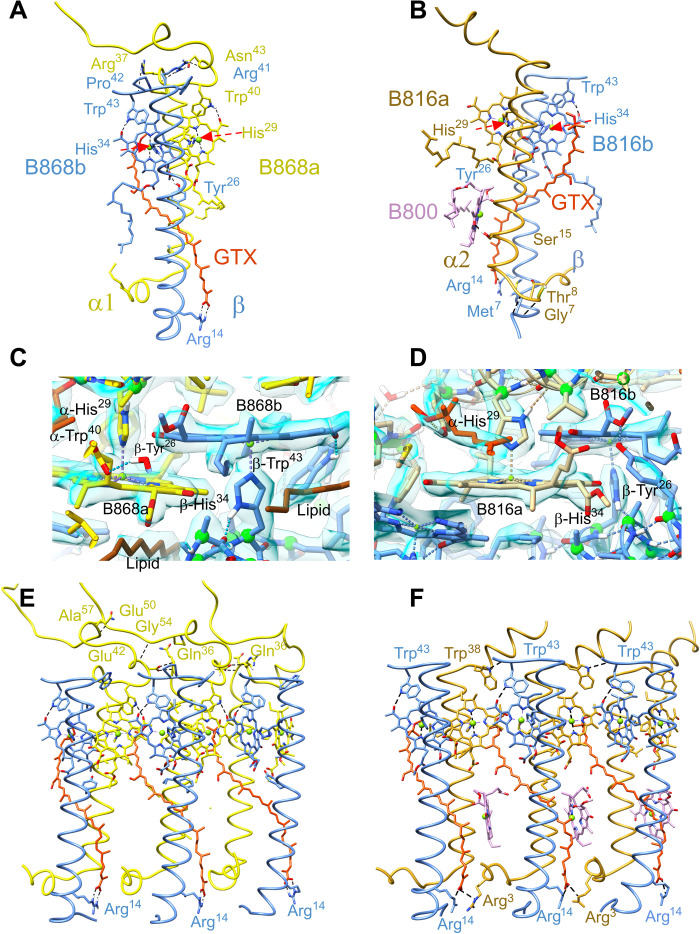
The central Mg2+ ions of two LH1 BChl a molecules, B868α and B868β, are coordinated by α-His29 and β-His34, respectively (Fig. 4, A and C). H-bonds between α-Trp40 and the α-BChl C31 keto group (2.2 Å) and between β-Trp43 and the β-BChl a C31 keto group (2.0 Å) fix the orientation of these BChls, with additional stabilization imparted by another H-bond between β-Tyr26 and the C133 ester group of β-BChl a (2.1 Å). The carboxylic acid on the end of the carotenoid is stabilized by two strong H-bonds with β-Arg14 (1.9 and 1.8 Å). Four interprotein H-bonds between α-Arg37, α-Asn43 and β-Pro42, β-Arg41 on the periplasmic surface stabilize the subunit further.
Fig. 4. Protein-protein and protein-pigment interactions within the two rings of light-harvesting antenna complexes.
(A) Key interactions involved in stabilizing a single α/β subunit of the 16-member LH1 ring. Histidine residues that are involved in coordination with central Mg atom of the BChls are indicated by red arrows. (B) Single α/β subunit of the 24-member LHh ring. (C) Interaction between BChl a molecules and protein in LH1 subunit. The density map is shown in blue. (D) Interaction between BChl a molecules and protein in the LHh subunit. (E) Three consecutive subunits of the LH1 ring. Interfaces between subunits are primarily mediated by the bound pigments, with few specific protein-protein interactions. (F) Three consecutive subunits of the outer LHh ring, showing H-bonds between the C-terminal domains in three LHh α/β units. GTX, gemmatoxanthin. Colors as in Fig. 1 with the exception of B816 and B868 BChls, which are colored by chain, and B800 is colored pink.
The outer LHh ring comprises similar heterodimeric α/β subunits, and the same β-polypeptide is found in the inner and outer rings. The central Mg2+ ion of α- and β-BChls a binds α-His29 and β-His34, respectively (Fig. 4, B and D). Two H-bonds between β-Tyr26 and the C133 ester group of α-BChl a and between β-Trp43 and the C31 keto group of β-BChl a (2.1 Å) establish the orientation of this pigment. The N-terminal domains of the α- and β-polypeptides are linked by H-bonds between α-Gly7–β-Met7 (1.8 Å) and α-Thr8–β-Met7 (2.0 Å) (Fig. 4B). Figure 4B shows that the outer LHh ring binds monomeric B800 BChls that are oriented nearly perpendicular to the plane of the membrane. This positioning is unusual for such pigments when they appear in LH2 complexes (19–21), and it is also notable that there is no clear bond coordinating the central Mg2+ ion of the B800 BChl, which forms only one H-bond between its C31 keto group and α-Ser15. This lack of interactions might account for the relatively weak density for B800 observed at some positions of the LHh ring, reflecting a lower occupancy of this site or inconsistent orientation of the pigment.
Three LH1 α-β heterodimers are shown in Fig. 4E; they are interlocked by partially overlapping bacteriochlorin rings of the BChls and further stabilized by a chain of C-terminal intersubunit H-bonds, in which the C-terminal region of a central α(0) has four H-bonds with α(+1) and three H-bonds with the α(−1) polypeptide. Further stability is imparted by hydrophobic interactions between the all-trans carotenoid gemmatoxanthin inserted between each pair of LH1 α/β-polypeptides and the N and C termini of the preceding and following α chains, respectively. In addition, the maps reveal 16 tightly bound phospholipids with unusually well-resolved tails modeled as 1-palmitoleyl-2-myristoyl-sn-phosphatidylethanolamine; these insert between successive α/β subunits such that each lipid tail lies across the non–histidine-complexed face of a BChl a ring, while the head groups insert between the LH1 and LHh rings (fig. S6).
As with the LH1 ring, the outer LHh complex is stabilized by a chain of C-terminal intersubunit H-bonds (3.1 Å) between α(0)-Trp38 and β(+1)-Trp43 and from the carotenoid carboxyl group to α-Arg3 on a neighboring subunit (2.9 Å) (Fig. 4F). We also note that the distances of the LH1-β helices to the LHh-α helices are much shorter than the helical distances within LH1 rings (see fig. S10D), indicating a significant protein-protein interaction.
Quinol export from the RC-dLH complex
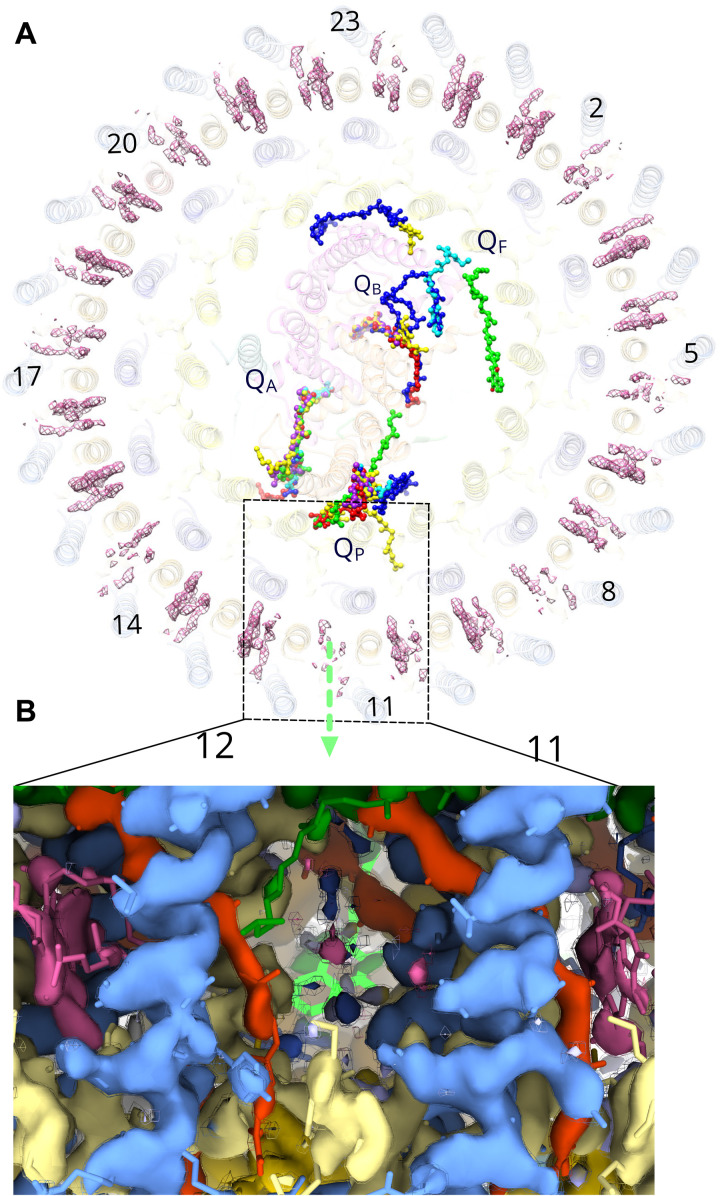
Purple bacteria use a variety of strategies for ensuring quinone traffic between the RC QB site and the cytochrome bc1 complex (9, 11, 22–24), which necessarily involve traversing a single LH1 ring. The α and β helices in the inner LH1 ring of the G. phototrophica RC-dLH complex are arranged similarly to RC-LH1 complexes found in some purple bacteria where the RC is completely surrounded by a single LH1 ring (11, 12, 22, 25), and in these cases, “breathing” motions and small gaps between polypeptides could allow quinone traffic (11, 16, 22). The outer LHh ring potentially represents an additional obstacle to quinone diffusion, although we note that the α-α distance in the outer LHh ring (16.4 Å) is larger than in LH1 (14.8 Å). To compare the quinone arrangements in G. phototrophica and some other purple bacteria, we superimposed the top views of the respective PS complexes (Fig. 5A). The overlay shows that QA is conserved and tightly bound, but quinones at the QB site are much more dispersed, particularly in the case of G. phototrophica. In this case, the distance of the nearest resolved quinone from the QB site is such that we named it as QF (“free” quinone). All known structures have a conserved QP quinone, adjacent to a possible exit point in the inner ring (Fig. 5A), and which coincides with a notably weak B800 density in the outer LHh ring (Fig. 5, A and B) that could arise from either low occupancy of this site or local disorder associated with quinone diffusion at this point in the ring. Weak densities for B800 at other points in the outer ring (Fig. 5A) could signify the location of other Q-channels.
Fig. 5. The QP quinol export site in the RC-dLH complex.
(A) The RC-dLH complex viewed from the cytoplasmic side of the membrane, with all details partly obscured, and showing the densities for B800 BChls (pink) in the outer LHh ring. Structurally resolved quinones for the G. phototrophica complex are shown (green), with those for the RC-LH1 complexes from Rhodopseudomonas palustris W-minus (16) (blue), R. palustris W-plus (16) (cyan), T. tepidum (11) (yellow), Thiorhodovibrio stain 970 (17) (purple), and Blastochloris viridis (9) (red). The green arrow indicates a likely path for quinol export, at a position of weak B800 density. The positions of LHh subunit are numbered. (B) Two LHh α/β units 11 and 12 viewed in the plane of the membrane showing the proposed position for quinol export that corresponds to a low or zero occupancy B800 site; LHh-α (pale yellow), LHh-β (cornflower blue), gemmatoxanthin (orange red), B800 (pink), B816 (dark green), and QP (bright green). The cryo-EM map is shown at two contour levels (opaque surface = 7.0σ; wireframe = 7.5σ).
Absorption tuning and energy transfer
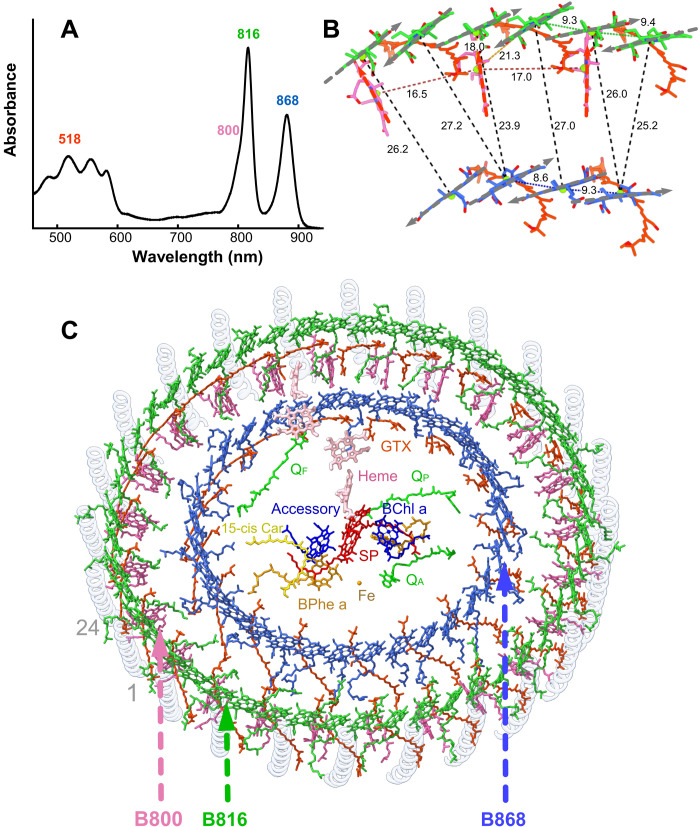
Purple phototrophic bacteria usually have relatively small LH2 antenna rings that absorb light and pass excitation energy on to LH1 and then to the central RC where charge separation occurs (26). Although G. phototrophica lacks LH2, the extra LHh ring plays a similar role, adding 800- and 816-nm absorption bands to the 868-nm absorption of the inner LH1 ring (Fig. 6A) and creating a spectral gradient to funnel energy toward the RC. The B800 lies 18.5 Å from the nearest outer ring B816 BChl, a distance comparable with those found for LH2 complexes (19–21), and the structure allows fast energy transfer (ET) to inner ring B868 BChls, over a distance of 24 to 26 Å.
Fig. 6. Structural and kinetic basis for the ET network in RC-dLH.
(A) The 77K absorption spectrum of the RC-dLH complex, colored according to the approximate absorption ranges of carotenoids (orange) and the B800 (pink), B816 (green), and B868 (blue) BChls. (B) Relative positions of BChl a molecules in the inner and outer rings for a unit of pseudo-C8 symmetry. Gray arrows indicate QY dipole moments of the BChls. The proteins have been removed for clarity. (C) Outer B800-B816 and inner B868 rings of BChls, and gemmatoxanthin (GTX, red orange), enclosing the central RC pigments. Other colors as in Figs. 1 and 2.
Differing extents of H-bonding in the inner and outer LH rings create the B868 and B816 absorption bands, respectively; α-Trp40 and β-Trp43 of LH1 H-bond to C31 keto groups of the respective α- and β-bound BChl a pigments. This effect is reinforced by the spacing of BChl pigments; the larger intrasubunit (9.4 Å) and intersubunit (9.3 Å) Mg-Mg distances for BChl a in the outer LHh ring tend to blue-shift absorption, relative to the 9.3- and 8.6-Å Mg-Mg distances for the LH1 ring (Fig. 6B). The 77K absorption spectrum of the RC-dLH complex (Fig. 6A) shows the contributions from the individual pigment pools in Fig. 6 (B and C). The relative orientations of BChl a molecules in the LHh and LH1 rings contribute to efficient ET; the QX and QY dipole moments for outer ring BChls are almost perfectly aligned (~3° deviation) with those in the inner ring (Fig. 6B); this, together with the close, ~2.5-nm separation between outer and inner ring BChls (Fig. 6B), ensures high-efficiency inter-ring ET.
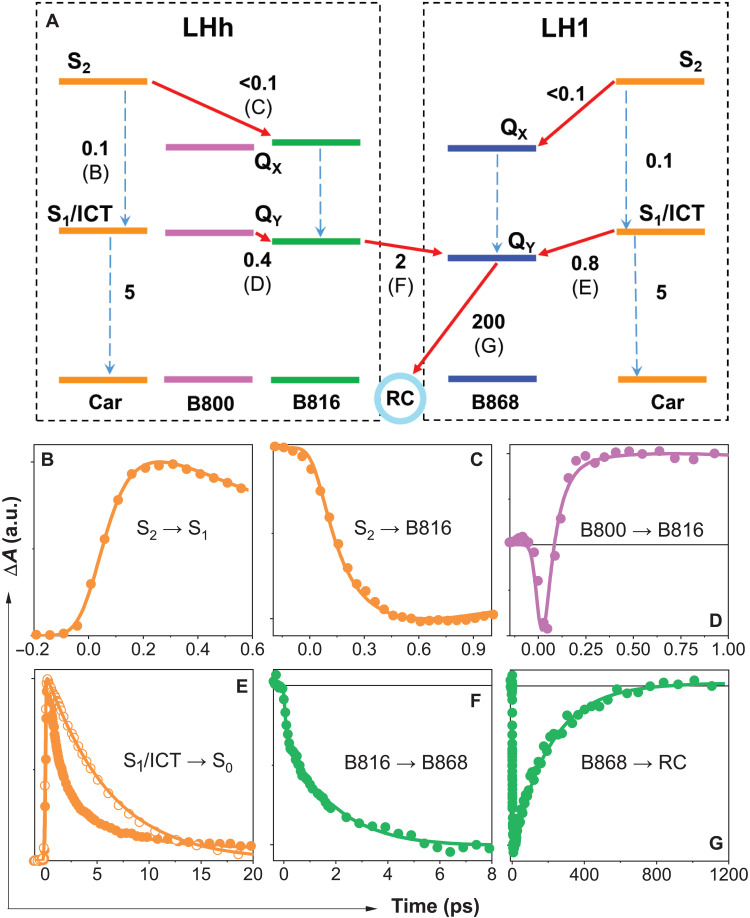
Femtosecond transient absorption spectroscopy was used to identify individual ET routes within the network of antenna pigments shown in Fig. 6C. The ET scheme in Fig. 7A summarizes ET reactions in the outer LHh and inner LH1 antenna rings. Excitation of carotenoids in the complex at 540 nm reveals ET pathways between carotenoids (gemmatoxanthin) and BChls, and a comparison of absorption and fluorescence excitation spectra shows that the overall process of ET from carotenoids to BChls is ~80% efficient (fig. S12). The initially excited carotenoid S2 state is quickly depopulated on a sub–100 fs time scale (Fig. 7B). The sub–300 fs rise of the B816 signal after carotenoid excitation (Fig. 7C) shows that the pathway to BChl QY proceeds via the S2 state. The S2 pathway is active also in B868 as evidenced by a strong B868 nm signal at 0.3 ps after carotenoid excitation (fig. S13B).
Fig. 7. ET kinetics in the RC-dLH1 complex.
(A) The two boxes (black dashed lines) map the ET events in the outer LHh and inner LH1 antenna rings. Solid red arrows represent ET channels; dashed arrows correspond to internal conversion processes. A time constant (in picoseconds) is associated with each process. The letters B to G associated with ET/internal conversion processes in the scheme correspond to the kinetics shown in (B) to (G), which monitor the dynamics of these processes. Color of each kinetic corresponds to excitation of carotenoids (orange), B800 (purple), and B816 (green). The probing wavelengths are within the bands corresponding to the gemmatoxanthin S1/ICT state (620 nm; B and E), B816 bleaching (820 nm; C), B816 excited-state absorption (805 nm; D), and B868 bleaching (880 nm; F and G). The kinetics shown as open symbols in (E) corresponds to the S1/ICT lifetime of gemmatoxanthin in methanol. Full transient absorption spectra showing the spectral bands and a detailed description are shown in fig. S13. a.u., arbitrary units.
A fraction of the excited carotenoid S2 population not transferred via the S2 pathway relaxes to the lowest carotenoid excited state. The carotenoid features an intramolecular charge transfer (ICT) state (fig. S13A); thus, the lowest energy state is a coupled S1/ICT state (27). S1/ICT is also an energy donor, as evidenced by the S1/ICT lifetime that is shorter in RC-dLH1 than for the carotenoid in MeOH (Fig. 7E), showing that the lower-energy BChls can act as energy acceptors because the shortening of the S1/ICT lifetime is caused by opening ET channel to BChl. The expected S1/ICT energy of gemmatoxanthin, which based on the S1/ICT lifetime should be comparable to spheroidenone, is too low to enable transfer to B816 BChls; thus, the S1/ICT pathway is likely active solely in the inner LH1 ring. The involvement of B800 in carotenoid-BChl ET cannot be a priori ruled out, but the transient absorption spectra shown in fig. S13B do not identify any signal associated with the B800 band after carotenoid excitation. Thus, carotenoid-B800 ET must be negligible, if present at all.
We examined ET in the LHh outer ring BChls, using 785-nm excitation for the B800 band and 820 nm for the B816 absorption band. Following direct excitation of B800 at 785 nm, we monitored the excited state absorption signal of B816 (Fig. 7D); the rise contains a 0.4-ps component that is present exclusively after 785-nm excitation and we assign this time constant to B800-B816 ET. The presence of ET between B800 and B816 rings in the LHh outer ring is also justified by time evolution of transient absorption spectra shown in fig. S13. The ET dynamics measured after excitation at 785 and 820 nm provide strong evidence for a set of B800 BChls that are distinct from B816, and which absorb on the blue side of the B816 band. These B800 pigments, by analogy with similar monomeric pigments in LH2 complexes (19–21), are those identified structurally in the LHh outer ring (Figs. 4 to 6). The time evolution of transient absorption spectra shows that the B800 BChls are preferentially excited at 785 nm and transfer energy to the BChls absorbing at 820 nm. The fact that no such dynamics are observed after 820-nm excitation proves that there are two groups of BChls: one absorbing at ~800 nm, and the other at ~820 nm.
We monitored energy flow from the outer to the inner BChl ring by exciting LHh B816 and recording the rise of the LH1 B868 BChl a signal (Fig. 7F). The 2-ps time constant of inter-ring B816-B868 energy flow is faster than the 3- to 5-ps ET between LH2 and LH1 complexes in purple bacteria (28). Last, B868 decay (Fig. 7G) monitors the final slow ET step from B868 to the RC.
Thus, the concentric, double-ring RC-dLH complex not only represents a unique, compact, and exceptionally stable complex for efficient harvesting, transfer, and trapping solar energy but also exemplifies some interesting evolutionary innovations. The β-polypeptide serves for both LH rings, so adding a second LH ring requires only one additional gene, pufA2. This represents an economical strategy for increasing the light harvesting capacity of a single RC-LH1 complex. Because no pufA2 homologs were found in Proteobacteria (fig. S9), it seems that its evolution occurred only after Gemmatimonadetes received their photosynthesis genes via horizontal gene transfer.
The new membrane-extrinsic linker proteins that cross-link the complex, and the extensive LH bonding and lipid networks revealed by this RC-dLH structure, likely impart great stability. Although the assembly of this complex requires a large investment in energy, this could be offset by its extraordinary stability and the robustness of the RC-dLH complex likely represents an evolutionary advantage.
MATERIALS AND METHODS
Cultivation
G. phototrophica AP64T cells were streaked on agar plates containing per liter: 1 g of yeast extract, 0.2 g of pyruvate, 0.2 g of glucose, 1 g of K2HPO4 (pH 7.7), 1.5 g of Difco agar, and 1 ml of SL8 trace metal solution (0.2 mg of CoCl2.6H2O, 5.2 mg of Na2.EDTA, 20 μg of NiCl2.6H2O, 20 μg of CuCl2.2H2O, 1.8 mg of SrCl2.6H2O, 70 μg of ZnCl2, and 62 μg of H3BO3 per ml). The inoculated plates were incubated under microaerophilic conditions (10% O2 + 90% N2) at 28 ± 1°C in the dark for 2 weeks to maximize the expression of the photosystem complexes. The grown cells were then scraped from the agar plates and stored at −20°C until needed.
Identification of genes encoding the subunits of the PS complex in G. phototrophica
All the genes encoding the subunits of the PS complexes were identified in its photosynthesis gene cluster (fig. S1) located at the main chromosome of G. phototrophica (GenBank BioProject: PRJNA213561). While the pufL, pufM, and pufC genes encoding L, M, and C subunits of the RC were automatically recognized by the National Center for Biotechnology Information (NCBI) annotation pipeline, the puhA gene encoding the H subunit was missing. After the manual inspection of the genome, we have identified two ORFs, 204 and 546 bp long, located upstream of the lhaA gene. The first ORF was assigned as a hypothetical protein (locus tag = GEMMAAP_06435), and the longer one was not automatically recognized. We assigned these ORFs as puhA1 and puhA2. puhA1 gene encodes a smaller [molecular weight (MW) = 7.7 kDa] transmembrane polypeptide denoted as Ht. The larger one (MW = 19.9 kDa) encodes the cytoplasmic subunit Hc. The deduced amino acid sequences matched the observed electron densities (fig. S5A).
Purification of PS complexes
Harvested G. phototrophica cells (1 to 2 g wet weight) were resuspended in 20 mM tris-Cl and 50 mM NaCl (pH 8.0) and homogenized thoroughly with a few grains of deoxyribonuclease and a few milligrams of MgCl2. The cells were broken by passage three times through an Emulsiflex-CS cell disrupter, and unbroken cells were removed by low-speed centrifugation (10 min, 12,000g, 4°C). The membranes were pelleted by ultracentrifugation (120 min, 180,000g, 4°C) and gently resuspended in 20 mM tris-Cl (pH 8.0) and adjusted to an optical density = ~10 cm−1 at the QX band (~580 nm). The resuspended membranes were then solubilized with 2% n-dodecyl β-d-maltoside (β-DDM) and 0.2% Triton X-100 for 60-min stirring at room temperature in the dark and then centrifuged to remove any nonsolubilized materials. The solubilized sample was carefully layered on top of a stepwise sucrose gradient and run overnight (208,000g, 16 hours, 4°C). The gradient consisted of sucrose dissolved in 20 mM tris-Cl and 0.02% b-DDM (pH 8.0; TD buffer) and layered in 0.2 M steps from 0.4 to 1.2 M. The RC-dLH band was carefully removed the following day and loaded onto a sintered-glass gravity column containing ToyoPearl 650S anion-exchange resin (Tosoh). The complex was eluted with increasing concentrations of NaCl in TD buffer and assayed spectrophotometrically for purity.
The best fractions were pooled and concentrated before gel filtration. Gel filtration was performed with TD buffer using a Knauer Azura system equipped with ultraviolet-visible-near-infrared (UV-VIS-NIR) diode array detector and on a Superdex S-300 (1 m XK-16 column, GE Healthcare) column at a flow rate of 0.5 ml min−1 at 10°C (fig. S14A). The collected fractions with the best A816/A260 absorption ratio were pooled and concentrated as required. The final RC-dLH complex pool typically had an A816/A260 = ~1.4 to 1.3. Its purity was analyzed using SDS–polyacrylamide gel electrophoresis (fig. S14B). Separated bands on the gel were assigned by mass spectrometry analysis.
Pigment and quinone analyses
Respiratory quinones were analyzed by HPLC as described previously (29). Natural menaquinones extracted from Micrococcus luteus were used as control standards. The analysis revealed the presence of two respiratory quinones, MQ8 with a smaller quantity (15%) of MQ9 (fig. S7A). Pigments were analyzed by HPLC with photodiode array UV-VIS-NIR detection as described before (29). The extracted complex contains phytol and geranylgeranyl esterified BChl a molecules and a linear carotenoid gemmatoxanthin with two structural isomers (see fig. S7B).
Femtosecond spectroscopy
Femtosecond spectroscopy was carried out using an amplified femtosecond laser system (Spectra Physics) producing ~100-fs pulses centered at 800 nm with a 1-kHz repetition rate. The output was divided into excitation and probe beams by a beam splitter. The excitation pulses were generated by an optical parametric amplifier (TOPAS Prime, Light Conversion, Lithuania). The probe pulses were generated by sending the 800-nm beam to a homebuilt optical parametric amplifier to produce a 1300-nm output that was focused to a 3-mm CaF2 plate, generating a white light continuum covering the 450- to 1100-nm range. The white-light beam was collimated by an off-axis parabolic mirror and split by a broadband, 50/50 beam splitter to reference and probe beams. The probe beam was focused by a 250-mm spherical mirror to the sample where it overlapped with the excitation beam. Probe and reference beams were directed to the entrance slit of a grating spectrograph and detected by a double charge-coupled device array (Pascher Instruments, Sweden). The transient absorption spectra were measured in two spectral windows, 480 to 760 nm and 730 to 1000 nm. The resulting two datasets were stitched and chirp-corrected by a software routine implemented in CarpetView fitting software (Light Conversion, Lithuania). The pump polarization was always set at magic angle (54.7°) relative to the probe polarization. For visible pumping, the photon density was less than 1014 photons cm−2 pulse−1.
Proteomic analysis of the purified RC-dLH complex
Two samples of PS complex containing 15 μg of protein were solubilized in 25 μl of 2% (w/v) SDS, 40 mM tris base, and 60 mM dithiothreitol at 60°C for 5 min. Proteins were then extracted by precipitation using a 2D clean up kit (Cytiva) according to the manufacturer’s protocol. After centrifugation (15,700g for 10 min), the protein pellets were separately dissolved in (i) 10 μl of 8 M urea and 100 mM tris-Cl (pH 8.5) and (ii) 10 μl of 0.2% (v/v) formic acid, both containing 5 mM tris(2-carboxyethyl)phosphine-HCl. The proteins were incubated at 37°C for 30 min to reduce disulfide bonds followed by cooling to room temperature and S-sulfenylation by the addition of 1 μl of 100 mM methyl methanethiosulfonate in isopropanol. After 10 min, proteolysis of the samples was performed by the addition of 0.6 μg of (i) endoproteinase Lys-C/trypsin mixture (Promega) and (ii) pepsin (Promega) with incubation at 37°C. After 2 hours, (i) was diluted by the addition of 75 μl of 50 mM tris (pH 8.5) and 10 mM CaCl2 and both digestions continued for 16 hours. The resultant peptides were desalted using C18 spin columns (Thermo Fisher Scientific) and dried by vacuum centrifugation.
For analysis by reversed-phase liquid chromatography coupled to mass spectrometry, the peptides were dissolved in 10 μl of 0.1% (v/v) formic acid and 8% (v/v) acetonitrile and diluted threefold with 0.1% (v/v) formic acid for a 1-μl injection containing 500 ng. Peptides were separated using an RSLCnano system (Thermo Fisher Scientific) with a PepSwift PS-DVB monolithic column (200 μm by 25 cm, 60°C, Thermo Fisher Scientific) and a 60-min gradient from 97% (v/v) solvent A [0.1% (v/v) formic acid] to 35% (v/v) solvent B [0.1% (v/v) formic acid and 80% (v/v) acetonitrile] at 2.5 μl min−1. Mass spectra were acquired on a Q Exactive HF Quadrupole Orbitrap (Thermo Fisher Scientific) using a heated electrospray ionization source operating with the following parameters: spray voltage, 4 kV positive; capillary temperature, 240°C; sheath gas, 5 U; S-lens radio frequency level, 60 U. For full-scan centroid MS acquisition, the following parameters were used: mass/charge ratio (m/z) range, 375 to 1500; resolution, 120,000; automatic gain control target, 1 × 106; maximum fill time, 60 ms. Data-dependent centroid product ion scans (maximum of 10 per cycle) used the following parameters: intensity threshold, 2.5 × 105; dynamic exclusion time, 60 s; resolution, 30,000; automatic gain control target, 1 × 105; maximum fill time, 60 ms; isolation window, 2.0 Th; normalized collision energy, 27.
Protein identification was performed by database searching using Byonic (v2.9.38, Protein Metrics) operating with the default parameters except that methylthiocysteine (+45.9877 Da) was specified as a fixed modification and methionine sulfoxide (+15.19949 Da) as a common variable modification (with a maximum of two per peptide). Cleavage sites were specified as C-terminal to K and R for the endoproteinase Lys-C/trypsin digest and both N- and C-terminal to F, Y, W, and L (semi-specific) for pepsin. The database was the G. phototrophica strain AP64 reference proteome (www.uniprot.org/proteomes/UP000076404) downloaded on 8 March 2021 and edited to re-annotate RC-dLH subunits for consistency with the nomenclature used here.
Cryo-EM sample preparation and data collection
A HexAuFoil grid (30), manufactured in-house at the Medical Research Council Laboratory of Molecular Biology, Cambridge, was used for data collection. The grid was plasma-cleaned under a mixed atmosphere (O2:Ar = 1:9) for 120 s in a plasma chamber (Fischione, 1070). It was vitrified using a manual plunger of the Talmon type (31) in a 4°C cold room and an ethane cryostat (32) held at 93 K. A 2.7-μl volume of protein solution was applied to the foil side of the cleaned grid and manually blotted for 13 s with filter paper (Whatman No. 1) and plunged in liquid ethane at 93 K. The grid was stored in liquid nitrogen before use. Data were collected on a Thermo Fisher Scientific Titan Krios G3i electron cryomicroscope equipped with a Falcon 4 direct electron detector at the Cambridge Pharmaceutical CryoEM Consortium (33). The microscope was operated at 300 kV with a nominal magnification of ×120,000, corresponding to 0.649 Å per pixel at the specimen level, calibrated using the Au (111) lattice reflections. The detector was operated in counting mode at a flux of 1.8 e− pixel−1 s−1. Each 5.8-s exposure was fractionated into 20 frames, resulting in an electron fluence of 1.24 e− Å−2 frame−1. The defocus range was set to −0.8 to −2.4 μm. Automated data acquisition was performed in a special version of EPU 2.8.1 (Thermo Fisher Scientific), adapted to the HexAuFoil grids, with one exposure per hole in aberration-free image shift mode with 6-μm cluster size. In total, 19,865 movies were collected from a single HexAuFoil grid (fig. S2A and table S1).
Cryo-EM data processing
All data processing, except for particle picking, was performed in RELION 3.1 (34). The movement was corrected per micrograph using RELION’s own implementation of motion correction. The contrast transfer function correction was performed in CTFFIND4.1 (35) within RELION. A total of 1,517,482 particles were autopicked in crYOLO (36), after retraining the neural network with manual picks from 10 random micrographs. The picked particles were extracted into 412 × 412 pixel boxes, rescaled to 128 × 128 pixels, and subjected to reference-free 2D classification, from which 1,373,398 particles were selected (fig. S2B) for initial 3D model generation and refinement. Following the initial 3D refinement, we performed 3D classification without alignment with five classes and T = 4. The class that showed clear secondary structure features (606,327 particles) was selected for further refinement, and the particles were re-extracted in 512 × 512 pixel boxes and rescaled to 256 × 256 pixels. Defocus and astigmatism were refined per micrograph, and beam tilt, trefoil, and fourth-order aberrations were refined for the whole dataset. We then repeated 3D refinement and reached 2.8-Å resolution. We used particle polishing in RELION (37), with optimized parameters σv = 0.5 Å/(e−Å−2), σd = 13,000 Å, and σa = 0.9 Å/(e−Å−2), and verified that no correlated particle movement occurred within the holes during the exposures. Then, particles were re-extracted in 616 × 616 pixel boxes and rescaled to 400 × 400 pixels, corresponding to a pixel size of 0.999 Å per pixel. We repeated 3D refinement and classification without alignment, with four classes and T = 8.
We found two classes showing high-resolution features, which are related by a rotation of the outer LHh ring by 360°/(2 × 24) = 7.5° around the pseudo-symmetry axis of the core RC-LH1 complex (fig. S2C). We selected these two classes (150,315 and 198,029 particles, respectively) for further 3D refinement. At this stage, we reestimated the defocus on a per-particle basis and obtained 2.5- and 2.3-Å reconstructions, respectively. We noticed a weak density for two helices on the cytoplasmic side of the RC, bridging it with the LH1 ring, and performed focused 3D classification without alignment with a soft mask around this region and T = 64 to select for the complexes containing these helices (later identified as RC-U) (fig. S2E). In this way, we obtained four classes, corresponding to the two possible arrangements of the LHh ring, each with and without the density for RC-U (fig. S10).
We subjected all thus far discarded RC-dLH classes with some secondary structure (327,203 particles) to another round of 3D refinement, followed by 3D classification without alignment, with four classes and T = 4. We noticed that some classes contained high-resolution features in the LH1 rings but blurred and fragmented density in place of the RC. We performed focused 3D classification without alignment, using a soft mask around the RC, with four classes and T = 32, and found three classes (fig. S2D), corresponding to the two already identified relative orientations between the RC, the LH1 ring, and the LHh ring. We performed another round of classification, focusing on the cytoplasmic helices as above, and thus found more particles that could be merged with the four classes we identified (some of these required ±45° rotation around the pseudo-symmetry axis of the LH1 rings). After we merged all particles for each of the four classes, we repeated contrast transfer function parameter refinement, Bayesian polishing, 3D refinement with reconstructions in SIDESPLITTER (38), and zero-dose extrapolation (30) to obtain four final maps of these different conformations at 2.4- to 2.5-Å resolution (fig. S11B and table S1). The maps were sharpened by a B factor of 30 Å2. Orientation distributions (fig. S11A) were analyzed using cryoEF (39).
Modeling and refinement
Initial model building was performed in a preliminary reconstruction, later revealed to be derived from a superposition of the different states captured in the final 2.4-Å maps. The G. phototrophica core complex consists of three parts: a central RC with a c-type cytochrome attached, surrounded by two concentric LH rings. Density for the LH rings was readily interpretable, but in this initial map, the RC density was extremely challenging—our final reconstructions showed that this was due to the superposition of two different conformations related by a 22.5° rotation. To build an initial model, the crystal structure of the RC from Thermochromatium tepidum (13) [Protein Data Bank (PDB): 5Y5S] was fitted as a rigid body into the density map using Chimera. For the RC-M and RC-L subunits, amino acid sequences and cofactors in the template were replaced with those from G. phototrophica in COOT (40). The RC-H exists as a single chain in T. tepidum but is separated into two chains in G. phototrophica. The chain was split, renumbered, and mutated accordingly.
Initially, the α/β subunits of LH2 from Phaeospirillum molischianum (17) (PDB: 1LGH) were used as starting templates for the inner LH ring chains. One α/β pair (combined with its associated BChl a pair) was rigid body–fitted to optimize the fit of the two BChl molecules to the density map. The G. phototrophica genome encodes three predicted antenna protein sequences, denoted PufA1, PufA2, and PufB. On the basis of fits to the density map, LH1-β was assigned as PufB, while LH1-α was assigned as PufA1. Addition of an all-trans carotenoid completed a single subunit of the inner LH ring. This was propagated to the remaining 15 LH1 α/β sites in the density map by rigid body fitting in Chimera. The outer LHh ring was constructed in a similar manner; LHh-β was revealed to be the same PufB sequence as in the inner ring, while LHh-α was assigned as PufA2.
Starting from the preliminary model described above, except where otherwise stated, all further model building was performed in ISOLDE (41). First, severe clashes were resolved by sequentially settling each individual chain into its density in isolation from the rest of the model. The model was then manually inspected and rebuilt from end to end. During the course of this, we noticed weak density apparently corresponding to two extra transmembrane helices in the space between RC and LH1, prompting us to collect a new dataset and revisit the particle classification; these were later revealed to be artifacts arising from the 22.5° rotation of RC-LH1 versus LHh seen in the final maps.
For each of the newly identified RC-S and RC-U chains, we first built a poly-alanine trace using COOT and then assigned a tentative de novo sequence to the best-resolved regions by adding side chains in ISOLDE with consideration of both fit to density and physical environment. This tentative sequence was then used as the basis for a BLASTp search of the G. phototrophica genome. Each case yielded a single clear candidate sequence, namely, A0A143BK87 (cytoplasmic chain) and A0A143BID3 (periplasmic chain). Modeling of the full sequences into the map yielded sensible results, and the presence of these chains in the complex was confirmed by mass spectrometry.
Initial models of the O-linked glycans found at Thr108 of the cytochrome c subunit and Ser331 of RC-M were copied from the glycan found on heparinase II from Pedobacter heparinus (3e80) (42). Because neither site was covered by the mass spectrometry data due to a lack of nearby protease cleavage sites, the identity of each sugar residue was assigned on the basis of the combination of fit to the map and physical interaction with the surroundings. The G. phototrophica glycan appears to share the same α-d-mannose(α1-2-d-glucuronic acid) α1-4-l-rhamnose core structure as P. heparinus, with the exception that the glucuronic acid shows clear evidence of O-acetylation at the 2- and 3-positions. The final sugar residue, linked to O4 of the glucuronic acid, was modeled on the basis of fit to the map as N-acetyl-α-d-glucosamine (GlcNAc). This branch of the O-glycan could not be matched to any glycan substructure in GlyTouCan (43). However, a complete analysis of the glycosyltransferases encoded in the G. phototrophica genome using the CAZy database (44) identified at least one gene encoding potential transferase activity for N-acetyl-α-d-glucosamine—GEMMAAP_08265, 49% sequence identity to α-N-acetylglucosaminyltransferase from Bacillus anthracis str. Sterne. All carbohydrate models were validated with Privateer (45).
With the improved resolution afforded by the new reconstructions, we were able to identify a second ring of 24 relatively weakly bound BChl a molecules near the cytoplasmic side of the LHh ring. For most BChl a molecules in the map, resolution of the flexible tails was insufficient to distinguish between geranylgeranyl or phytyl groups. All BChl molecules have been modeled as BChl a with phytyl tails.
The 40 all-trans carotenoids were identifiable as gemmatoxanthin based on fit to density and salt bridges between their terminal carboxylic acids and the adjacent LH-α (Arg3) and LH-β (Arg14) chains. However, the single 15-cis carotenoid found in RC-M was identified as spirilloxanthin with clear definition of both head groups. For the quinones QA, QP, and QF, MQ8 gave the best fit to density. The minor pool of MQ9 molecules determined by the HPLC analysis may be part of a free quinone pool unresolved by the cryo-EM analysis (fig. S7A).
After rebuilding and refinement of the above partial models, lipids and β-DDM detergent molecules were hand-built into the residual density, beginning with the highest-resolution map and propagating the additions to the other three. Lipid head-group identities were assigned on the basis of the combination of fit to density and interactions with surrounding residues; where the head-group density of a phospholipid was poorly resolved, we assigned it as phosphatidylethanolamine. Last, water molecules were placed in the best-resolved locations with the map contoured at 6σ, keeping only those waters that were stable in ISOLDE simulations in the positions indicated by the map.
Last, after settling each model in ISOLDE at a simulation temperature of 0 K, each model was subjected to global real-space refinement in phenix.refine. To avoid overfitting of backbone and side-chain conformations to traditional validation metrics, Ramachandran and rotamer restraints were disabled as was conformational grid searching; instead, all torsions were restrained to remain close to their input angles. Explicit restraints were added for each BChl a and heme group to enforce the expected distances between their central Mg/Fe ions and coordinating His/Met side-chain atoms. The refinement statistics are summarized in table S1, and the refined models and corresponding density maps were deposited to PDB and Electron Microscopy Data Bank (EMDB).
Acknowledgments
We thank J. Lawrence for help during the project.
Funding: This study was mainly financed by the Czech Science Foundation, EXPRO project 19-28778X PhotoGemm+ (M.Ko.); Czech Science Foundation, EXPRO project 19-28323X (T.P.); Biotechnology and Biological Sciences Research Council (BBSRC) UK, award number BB/M000265/1 (P.Q. and C.N.H.); European Research Council Synergy Award 854126 (C.N.H.); European Regional Development Fund Project “Mechanisms and dynamics of macromolecular complexes: From single molecules to cells” CZ.02.1.01/0.0/0.0/15_003/0000441 (D.K.); Medical Research Council Grant MC_UP_120117 (C.J.R.); Sir John Bradfield Scholarship from the Cambridge Trust (K.N.); Royal Society Olga Kennard Research Fellow, UF160039 (J.A.); and Wellcome Trust grant 209407/Z/17/Z (T.I.C.).
Author contributions: P.Q., T.P., K.S., H.S., C.J.R., C.N.H., and M.Ko. designed and organized the experiments. A.T.G. purified the PS complexes, with the help of R.S. and D.K. K.N., P.C.-H., B.v.K., K.N.G., C.A.S., and P.Q. collected cryo-EM data. K.N., C.J.R., and P.Q. processed cryo-EM data. T.I.C., J.A., and P.Q. performed model building and refinement. P.J.J. collected and analyzed the mass spectrometry data. N., M.Ku., P.H., J.H., and M.Ko. analyzed pigments and respiratory quinones. Y.Z. and M.Ko. identified genes encoding the protein subunits. T.P., I.Š., M.Kl., P.Č., and D.B. performed and analyzed spectroscopic measurements. T.I.C., P.Q., C.N.H., M.Ko., and T.P. prepared figures. P.Q., C.N.H., and M.Ko. wrote the manuscript. All authors proofread and approved the manuscript.
Competing interests: The authors declare that they have no competing interests.
Data and materials availability: All relevant data are provided in the main text, the Supplementary Materials, or publically accessible databases. Mass spectrometry proteomics data and edited G. phototrophica reference proteome database have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository (http://proteomecentral.proteomexchange.org) with the dataset identifier PXD024609. Cryo-EM raw movies are deposited in Electron Microscopy Public Image Archive with the accession code of EMPIAR-10681. Atomic coordinates and the cryo-EM density maps have been deposited in the PDB under accession numbers 7O0U, 7O0V, 7O0W, and 7O0X and the EMDB under accession numbers EMD-12679, EMD-12680, EMD-12681, and EMD-12682. All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials.
Supplementary Materials
This PDF file includes:
Figs. S1 to S14
Table S1
References
Other Supplementary Material for this manuscript includes the following:
Movie S1
REFERENCES AND NOTES
- 1.R. E. Blankenship, Molecular Mechanisms of Photosynthesis (John Wiley & Sons, ed. 2, 2014), pp. 312. [Google Scholar]
- 2.Hohmann-Marriott M. F., Blankenship R. E., Evolution of photosynthesis. Annu. Rev. Plant Biol. 62, 515–548 (2011). [DOI] [PubMed] [Google Scholar]
- 3.Martin W. F., Bryant D. A., Beatty J. T., A physiological perspective on the origin and evolution of photosynthesis. FEMS Microbiol. Rev. 42, 205–232 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sener M., Strumpfer J., Abhishek S., Hunter C. N., Schulten K., Overall energy conversion efficiency of a photosynthetic vesicle. eLife 5, e09541 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bryant D. A., Costas A. M., Maresca J. A., Chew A. G., Klatt C. G., Bateson M. M., Tallon L. J., Hostetler J., Nelson W. C., Heidelberg J. F., Ward D. M., Candidatus Chloracidobacterium thermophilum: An aerobic phototrophic Acidobacterium. Science 317, 523–526 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Zeng Y., Feng F. Y., Medová H., Dean J., Koblížek M., Functional type 2 photosynthetic reaction centers found in the rare bacterial phylum Gemmatimonadetes. Proc. Natl. Acad. Sci. U.S.A. 111, 7795–7800 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mujakić I., Andrei A.-Ş., Shabarova T., Fecskeová L. K., Salcher M. M., Piwosz K., Ghai R., Koblížek M., Common presence of phototrophic Gemmatimonadota in temperate freshwater lakes. mSystems 6, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen G. E., Canniffe D. P., Barnett S. F. H., Hollingshead S., Brindley A. A., Vasilev C., Bryant D. A., Hunter C. N., Complete enzyme set for chlorophyll biosynthesis in Escherichia coli. Sci. Adv. 4, eaaq1407 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qian P., Siebert C. A., Wang P. Y., Canniffe D. P., Hunter C. N., Cryo-EM structure of the Blastochloris viridis LH1–RC complex at 2.9 Å. Nature 556, 203–208 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Xin Y., Shi Y., Niu T., Wang Q., Niu W., Huang X., Ding W., Yang L., Blankenship R. E., Xu X., Sun F., Cryo-EM structure of the RC-LH core complex from an early branching photosynthetic prokaryote. Nat. Commun. 9, 1568 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yu L. J., Suga M., Wang-Otomo Z. Y., Shen J. R., Structure of photosynthetic LH1–RC supercomplex at 1.9 Å resolution. Nature 556, 209–213 (2018). [DOI] [PubMed] [Google Scholar]
- 12.Qian P., Croll T. I., Swainsbury D. J. K., Castro-Hartmann P., Moriarty N. W., Sader K., Hunter C. N., Cryo-EM structure of the Rhodospirillum rubrum RC–LH1 complex at 2.5 Å. Biochem. J. 478, 3253–3263 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qian P., Swainsbury D. J. K., Croll T. I., Salisbury J. H., Martin E. C., Jackson P. J., Hitchcock A., Castro-Hartmann P., Sader K., Hunter C. N., Cryo-EM structure of the Rhodobacter sphaeroides RC–LH1 core monomer complex at 2.5 Å. Biochem. J. 478, 3775–3790 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dachev M., Bína D., Sobotka R., Moravcová L., Gardian Z., Kaftan D., Šlouf V., Fuciman M., Polívka T., Koblížek M., Unique double concentric ring organization of light harvesting complexes in Gemmatimonas phototrophica. PLOS Biol. 15, e2003943 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pettersen E. F., Goddard T. D., Huang C. C., Meng E. C., Couch G. S., Croll T. I., Morris J. H., Ferrin T. E., UCSF ChimeraX: Structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Swainsbury D. J. K., Qian P., Jackson P. J., Faries K. M., Niedzwiedzki D. M., Martin E. C., Farmer D. A., Malone L. A., Thompson R. F., Ranson N. A., Canniffe D. P., Dickman M. J., Holten D., Kirmaier C., Hitchcock A., Hunter C. N., Structures of Rhodopseudomonas palustris RC-LH1 complexes with open or closed quinone channels. Sci. Adv. 7, abe2631 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tani K., Kanno R., Makino Y., Hall M., Takenouchi M., Imanishi M., Yu L., Overmann J., Madigan M. T., Kimura Y., Mizoguchi A., Humbel B. M., Wang-Otomo Z., Cryo-EM structure of a Ca2+-bound photosynthetic LH1-RC complex containing multiple αβ-polypeptides. Nat. Commun. 11, 4955 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nupur M., Kuzma J., Hájek P., Hrouzek A., Gardiner T., Lukeš M., Moos M., Šimek P., Koblížek M., Structure elucidation of the novel carotenoid gemmatoxanthin from the photosynthetic complex of Gemmatimonas phototrophica AP64. Sci. Rep. 11, 15964 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gardiner A. T., Naydenova K., Castro-Hartmann P., Nguyen-Phan T. C., Russo C. J., Sader K., Hunter C. N., Cogdell R. J., Qian P., The 2.4 Å cryo-EM structure of a heptameric light-harvesting 2 complex reveals two carotenoid energy transfer pathways. Sci. Adv. 8, abe4650 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McDermott G., Prince S. M., Freer A. A., Hawthornthwaite-Lawless A. M., Papiz M. Z., Cogdell R. J., Isaacs N. W., Crystal structure of an integral membrane light-harvesting complex from photosynthetic bacteria. Nature 374, 517–521 (1995). [Google Scholar]
- 21.Koepke J., Hu X. C., Muenke C., Schulten K., Michel H., The crystal structure of the light-harvesting complex II (B800-B850) from Rhodospirillum molischanum. Structure 4, 581–597 (1996). [DOI] [PubMed] [Google Scholar]
- 22.Jamieson S. J., Wang P., Qian P., Kirkland J. Y., Conroy M. J., Hunter C. N., Bullough P. A., Projection structure of the photosynthetic reaction center–antenna complex of Rhodospirillum rubrum at 8.5 Å resolution. EMBO J. 21, 3927–3935 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Qian P., Papiz M. Z., Jackson P. J., Brindley A. A., Ng I. W., Olsen J. D., Dickman M. J., Bullough P. A., Hunter C. N., Three-dimensional structure of the Rhodobacter sphaeroides RC-LH1-PufX complex: Dimerization and quinone channels promoted by PufX. Biochemistry 52, 7575–7585 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Roszak A. W., Howard T. D., Southall J., Gardiner A. T., Law C. J., Isaacs N. W., Cogdell R. J., Crystal structure of the RC-LH1 core complex from Rhodopseudomonas palustris. Science 302, 1969–1972 (2003). [DOI] [PubMed] [Google Scholar]
- 25.Qian P., Addlesee H. A., Ruban A. V., Wang P., Bullough P. A., Hunter C. N., A reaction center-light-harvesting 1 complex (RC-LH1) from a Rhodospirillum rubrum mutant with altered esterifying pigments: Characterization by optical spectroscopy and cryo-electron microscopy. J. Biol. Chem. 278, 23678–23685 (2003). [DOI] [PubMed] [Google Scholar]
- 26.Cogdell R. J., Gall A., Köhler J., The architecture and function of the light-harvesting apparatus of purple bacteria: From single molecules to in vivo membranes. Q. Rev. Biophys. 39, 227–324 (2006). [DOI] [PubMed] [Google Scholar]
- 27.Polivka T., Frank H. A., Molecular factors controlling photosynthetic light harvesting by carotenoids. Acc. Chem. Res. 43, 1125–1134 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hess S., Chachisvilis M., Timpmann K., Jones M. R., Fowler G. J., Hunter C. N., Sundström V., Temporally and spectrally resolved subpicosecond energy transfer within the peripheral antenna complex (LH2) and from LH2 to the core antenna complex in photosynthetic purple bacteria. Proc. Natl. Acad. Sci. U.S.A. 92, 12333–12337 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng Y., Nupur, Wu N., Madsen A. M., Chen X., Gardiner A. T., Koblížek M., Gemmatimonas groenlandica sp. nov. is an aerobic anoxygenic phototroph in the phylum Gemmatimonadetes. Front. Microbiol. 11, 606612 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Naydenova K., Jia P., Russo C. J., Electron cryomicroscopy with sub-Angstrom specimen movement. Science , 223–226 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bellare J. R., David H. T., Scriven L. E., Talmon Y., Controlled environment vitrification system: An improved sample preparation technique. J. Electron Microsc. Tech. 10, 87–111 (1988). [DOI] [PubMed] [Google Scholar]
- 32.Russo C. J., Passmore L. A., Ultrastable gold substrates: Properties of a support for high-resolution electron cryomicroscopy of biological specimens. J. Struct. Biol. 193, 33–44 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sader K., Matadeen R., Hartmann P. C., Halsan T., Schlichten C., Industrial cryo-EM facility setup and management. Acta Cryst. D 76, 313–325 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zivanov J., Nakane T., Forsberg B. O., Kimanius D., Hagen W. J. H., Lindahl E., Scheres S. H. W., New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife 7, e42166 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rohou A., Grigorieff N., CTFFIND4: Fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner T., Merino F., Stabrin M., Moriya T., Antoni C., Apelbaum A., Hagel P., Sitsel O., Raisch T., Prumbaum D., Quentin D., Roderer D., Tacke S., Siebolds B., Schubert E., Shaikh T. R., Lill P., Gatsogiannis C., Raunser S., SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun. Biol. 2, 218 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zivanov J., Nakane T., Scheres S. H. W., A Bayesian approach to beam-induced motion correction in cryo-EM single-particle analysis. Iucrj 6, 5–17 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ramlaul K., Palmer C. M., Nakane T., Aylett C. H. S., Mitigating local over-fitting during single particle reconstruction with SIDESPLITTER. J. Struct. Biol. 211, 107545 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Naydenova K., Russo C. J., Measuring the effects of particle orientation to improve the efficiency of electron cryomicroscopy. Nat. Commun. 8, 629 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P., Cowtan K., Coot: Model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Croll T. I., ISOLDE: A physically realistic environment for model building into low-resolution electron-density maps. Acta Crystallogr. D 74, 519–530 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaya D., Zhao W., Garron M., Xiao Z., Cui Q., Zhang Z., Sulea T., Linhardt R. J., Cygler M., Catalytic mechanism of heparinase II investigated by site-directed mutagenesis and the crystal structure with its substrate. J. Biol. Chem. 285, 20051–20061 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tiemeyer M., Aoki K., Paulson J., Cummings R. D., York W. S., Karlsson N. G., Lisacek F., Packer N. H., Campbell M. P., Aoki N. P., Fujita A., Matsubara M., Shinmachi D., Tsuchiya S., Yamada I., Pierce M., Ranzinger R., Narimatsu H., Aoki-Kinoshita K. F., GlyTouCan: An accessible glycan structure repository. Glycobiology 27, 915–919 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lombard V., Ramulu H. G., Drula E., Coutinho P. M., Henrissat B., The Carbohydrate-active enzymes database (CAZy) in 2013. Nucleic Acids Res. 42, D490–D495 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Agirre J., Iglesias-Fernández J., Rovira C., Davies G. J., Wilson K. S., Cowtan K. D., Privateer: Software for the conformational validation of carbohydrate structures. Nat. Struct. Mol. Boil. 22, 833–834 (2015). [DOI] [PubMed] [Google Scholar]
- 46.Edgar R. C., MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Price M. N., Dehal P. S., Arkin A. P., FastTree 2–approximately maximum-likelihood trees for large alignments. PLOS ONE 5, e9490 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kucukelbir A., Sigworth F. J., Tagare H. D., Quantifying the local resolution of cryo-EM density maps. Nat. Methods 11, 63–65 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zigmantas D., Hiller R. G., Sharples F. P., Frank H. A., Sundströma V., Polívka T., Effect of a conjugated carbonyl group on the photophysical properties of carotenoids. Phys. Chem. Chem. Phys. 6, 3009–3016 (2004). [Google Scholar]
- 51.West R. G., Fuciman M., Staleva-Musto H., Šebelík V., Bína D., Durchan M., Kuznetsova V., Polívka T., Equilibration dependence of fucoxanthin S1 and ICT signatures on polarity, proticity, and temperature by multipulse femtosecond absorption spectroscopy. J. Phys. Chem. B 122, 7264–7276 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Scholes G. D., Fleming G. R., On the mechanism of light harvesting in photosynthetic purple bacteria: B800 to B850 energy transfer. J. Phys. Chem. B 104, 1854–1868 (2000). [Google Scholar]
- 53.Dobáková M., Sobotka R., Tichý M., Komenda J., Psb28 protein is involved in the biogenesis of the photosystem II inner antenna CP47 (PsbB) in the cyanobacterium Synechocystis sp. PCC 6803. Plant Physiol. 149, 1076–1086 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S14
Table S1
References
Movie S1