Abstract
Large segmental osseous defects heal poorly. Recombinant, human bone morphogenetic protein-2 (rhBMP-2) is used clinically to promote bone healing, but it is applied at very high doses that cause adverse side effects and raise costs while providing only incremental benefit. We describe a previously unexplored, alternative approach to bone regeneration using chemically modified messenger RNA (cmRNA). An optimized cmRNA encoding BMP-2 was delivered to critical-sized femoral osteotomies in rats. The cmRNA remained orthotopically localized and generated BMP locally for several days. Defects healed at doses ≥25 μg of BMP-2 cmRNA. By 4 weeks, all animals treated with 50 μg of BMP-2 cmRNA had bridged bone defects without forming the massive callus seen with rhBMP-2. Moreover, such defects recovered normal mechanical strength quicker and initiated bone remodeling faster. cmRNA regenerated bone via endochondral ossification, whereas rhBMP-2 drove intramembranous osteogenesis; cmRNA provides an innovative, safe, and highly translatable technology for bone healing.
cmRNA encoding BMP-2 can heal large, osseous segmental defects in rats in a superior manner to recombinant BMP-2.
INTRODUCTION
Large osseous segmental defects do not heal well and remain a major clinical problem that can lead to amputation. Regenerative medicine offers new possibilities for restoring lost bone and producing a regenerate that is indistinguishable from its native counterpart. The identification and cloning of morphogens that guide the behavior of osteoprogenitor cells provide opportunities for biologically based tissue regeneration.
Recombinant, human bone morphogenetic protein-2 (rhBMP-2) and rhBMP-7 have been approved for use in the United States and Europe, but the clinical efficacy of these molecules has been disappointing and adverse side effects have been associated with their use (1). The latter are exacerbated, and the financial cost increased, by the very high amounts of recombinant protein that need to be applied to overcome the inadequacies of the collagen sponge used as a scaffold for protein delivery. Several approaches to improving protein delivery, including the use of smart scaffolds (2), have been investigated. However, none of these have advanced to clinical trials. Furthermore, rhBMP-7 has been withdrawn from the market and is no longer available for clinical use. Alongside, restrictions have been imposed in the clinical use of rhBMP-2, and this product is not available in many countries.
Local gene delivery provides a promising, alternative approach to delivering osteogenic proteins at therapeutic levels within an osseous lesion for an extended period of time (3). Successful intralesional delivery and expression of complementary DNA (cDNA) encoding an osteogenic product such as a BMP will result in the sustained in situ production of authentic BMP by the endogenous biological machinery of the cell. Substantial preclinical progress has been made (3, 4), and the results confirm that low amounts of BMP-2 synthesized endogenously within the osseous lesion for a short period of time by genetically modified cells efficiently heal bone (3). Despite these encouraging results, traditional gene therapy has yet to generate a clinical product. The reasons for this are several including the disadvantages of current genetic delivery strategies, cost, safety concerns, translational barriers, and the regulatory complexity of advancing gene therapies for non-Mendelian, non–life-threatening conditions.
Taking the above into consideration, messenger RNA (mRNA), the intermediary between a gene and its encoded protein, may offer the best of both worlds and confer numerous advantages. After delivery into a cell, mRNA will almost immediately begin to be translated into its cognate protein, unlike the case with traditional gene delivery of DNA, which needs first to translocate to the nucleus of the cell before its expression can begin. The protein encoded by mRNA is expressed from the introduced message for a finite period of time, after which the mRNA is degraded by innate physiological processes of the cell, leaving no residue. There is therefore no risk of insertional mutagenesis or other genetic damage, unlike the case with DNA therapeutics. Moreover, synthetic mRNAs encoding specific therapeutic proteins are relatively easy and straightforward to produce and can be readily delivered into the cell with inexpensive, nonviral vectors. Together, these advantages endow mRNA therapeutics with several important advantages over their DNA equivalents. They are expected to be safer and easier to translate to a large patient population, as demonstrated by the mRNA vaccines in use against coronavirus disease 2019 (COVID-19).
The application of mRNA to regenerative medicine has been hampered by its instability, cytotoxicity, and inflammatory properties (5). However, these limitations can be overcome by transcript engineering. Modifications that have been investigated to affect the stability and biocompatibility of mRNA include changes in the 5′ and 3′ untranslated regions (UTRs), open reading frame alterations, poly(A) tail length, and the use of modified nucleosides (6–10). The latter markedly reduce immunogenicity (11, 12). Recent advances in mRNA technology, led by these structural modifications and by the use of lipid vectors for delivery, have kindled interest in the use of mRNA in regenerative medicine as a safe and effective means of tissue restoration (13).
Here, we have evaluated the ability of an optimized chemically modified mRNA (cmRNA) encoding BMP-2 (12) to heal a critical-sized segmental defect in the rat femur. The data demonstrate the complete healing of the damaged tissue with almost no escape of the cmRNA from the lesion into the circulation or major organs. De novo formed bone showed mechanical properties comparable to those of the native bone while lacking the massive callus formed by rhBMP-2 in this model. Unlike the latter, BMP-2 cmRNA induced bone healing via the native endochondral route. The work summarized here demonstrates the efficacy and safety of an affordable transcript therapy for expedited bone healing.
RESULTS
Local, intraosseous expression of cmRNA without escape from the site of application
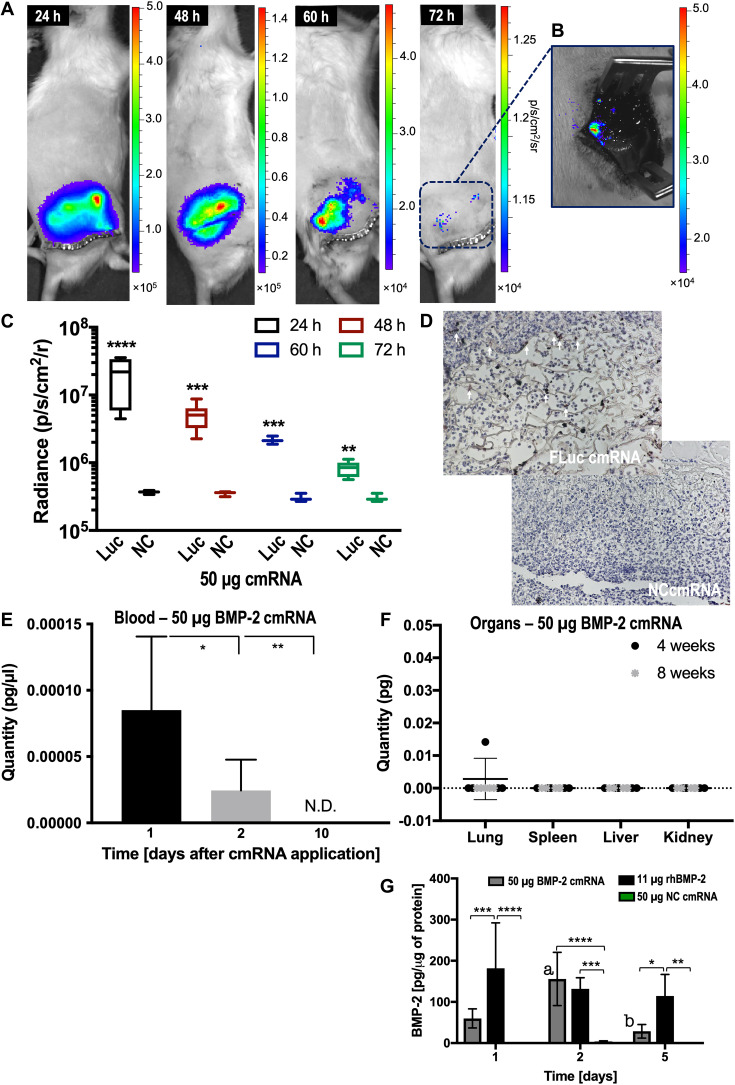
Figure S1 summarizes the study design. We used a standardized rat, mid-diaphyseal, femoral, 5-mm defect model stabilized by a bridging plate (fig. S1B). If left untreated, this defect does not heal. Intralesional implantation of 50 μg of cmRNA encoding firefly luciferase (FLuc) on a collagen sponge resulted in orthotopically localized expression of FLuc at a declining rate for at least 3 days (Fig. 1, A to D). No luciferase activity was detected when a truncated, noncoding (NC) cmRNA was administered (Fig. 1, C and D). No expression of luciferase was detected at other sites in the body by in vivo imaging (Fig. 1A).
Fig. 1. Single application of FLuc cmRNA induces robust and localized luciferase activity in the bone but is absent from the circulation and essential organs.
(A) Bioluminescence images at 24, 48, 60, and 72 hours after administration (n = 6). Scale denotes light intensity as protons s−1 cm−2 sr−1. (B) Bioluminescence image of an exposed bony defect 72 hours after administration. (C) Quantification of light intensity over time. “Luc” indicates FLuc cmRNA administration, while “NC” shows NC cmRNA used as control. Six samples were plotted and analyzed by multiple t test corrected by Holm-Sidak for comparison. (D) Luciferase stained by IHC 72 hours after administration of FLuc cmRNA or NC cmRNA control. White arrows indicate luciferase staining. (E) BMP-2 cmRNA concentration in blood 1, 2, and 10 days after administration. Scale indicates picograms of BMP-2 cmRNA per microliter of blood. N.D., not determined. (F) BMP-2 cmRNA content of lung (right, lower lobe), spleen (entire), liver (right, caudate lobe), and kidney (right, entire) 4 and 8 weeks after administration. In (E) and (F), eight samples were used. One-way analysis of variance (ANOVA), corrected by Tukey for multiple comparison, was used for statistical analysis. (G) Levels of BMP-2 produced in vivo within the femoral defect 1, 2, and 5 days after BMP-2 cmRNA administration. Animals treated with rhBMP-2 and NC cmRNA were also analyzed for comparison. Six samples were analyzed per group using ELISA. Two-way ANOVA, corrected by Tukey for multiple comparison, was used for statistical analysis. Statistical significance is shown by *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001. Also, statistical significance revealed by the letter “a” indicates P = 0.0085 for increased levels of BMP-2 at day 2 compared to day 1 after BMP-2 cmRNA administration. Similarly, the letter “b” denotes P = 0.0005 for decreased levels of BMP-2 at day 5 when compared to day 2 after BMP-2 cmRNA administration.
In agreement with the imaging data, only femtogram amounts of BMP-2 cmRNA [blood (≥0.08 fg/μl)] were detected in the circulation at days 1 and 2 after administration of the 50-μg dose, and cmRNA was undetectable by day 10 (Fig. 1E). No BMP-2 cmRNA was detected in lung, spleen, liver, or kidney at 4 and 8 weeks after administration of the 50-μg dose (Fig. 1F), apart from one outlier with a femtogram signal in its lung.
Administration of 50 μg of BMP-2 cmRNA resulted in significant in vivo BMP-2 production (Fig. 1G), whereas no BMP-2 production was detected in the NC cmRNA control group. In vivo BMP-2 levels showed a different temporal profile after delivery as rhBMP-2. For the cmRNA group, production peaked at day 2 and then decayed to very low levels by day 5 after administration. In the rhBMP-2 group where 11 μg of protein, the clinically equivalent dose, was implanted at the time of surgery, in vivo protein levels were highest at day 1 with much slower subsequent decay. The total amount of BMP-2 generated by cmRNA was considerably lower than the amount present after administration of rhBMP-2.
Dose-dependent bridging of critical-sized defects by BMP-2 cmRNA
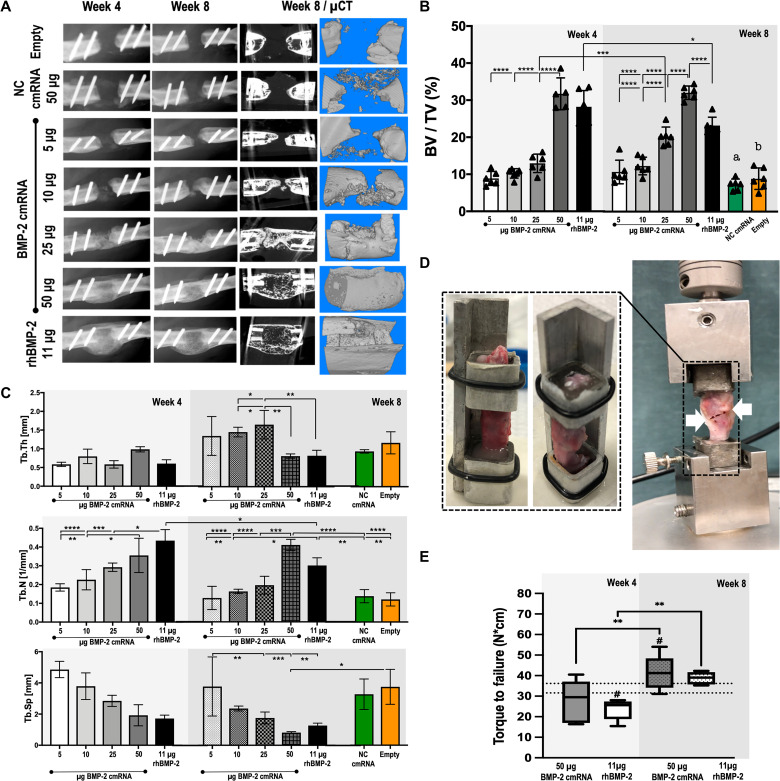
The in situ production of BMP-2 healed bone defects at doses ≥25 μg of BMP-2 cmRNA (Fig. 2A). A clear dose dependency was already evident in the x-ray images and micro–computed tomography (μCT) reconstruction at 4 weeks after administration. As early as 4 weeks after treatment, all animals treated with 50 μg of BMP-2 cmRNA had bridged bone defects indicating complete healing (Fig. 2A). While lower doses of 5 to 10 μg of BMP-2 cmRNA seemed to stimulate de novo tissue formation, this was insufficient to bridge the defect. At a dose of 25 μg of cmRNA, there was a 50% union rate, while 50 μg of BMP-2 cmRNA succeeded in completely bridging all bone defects. The massive, ectopic callus formed in response to rhBMP-2 was not observed during healing with cmRNA (Fig. 2A).
Fig. 2. Bone healing with BMP-2 cmRNA is dose-dependent; doses >25 μg are necessary to heal critically sized bone defects reliably in rats.
(A) X-ray images and μCT reconstruction images of the defect after BMP-2 cmRNA administration. Representative x-ray images are shown for weeks 4 and 8 after administration, while μCT images are shown at 8 weeks. (B) Bone volume–to–tissue volume ratios (BV/TV), shown as percentages. (C) Trabecular thickness (Tb.Th; mm), trabecular number (Tb.N; 1/mm), and trabecular separation (Tb.Sp; mm). Values shown in (B) and (C) were derived by μCT 4 and 8 weeks after BMP-2 cmRNA administration at different doses. NC cmRNA and empty defects were used as controls. Six samples were plotted per group. Data were analyzed using two-way ANOVA corrected by Holm-Sidak for multiple comparisons. Letters a and b [in (B)] indicate significantly lower (P < 0.0001) BV/TV ratio for NC cmRNA (a) and empty defect (b) when compared to 25 μg of BMP-2 cmRNA, 50 μg of BMP-2 cmRNA, or 11 μg of rhBMP-2 at 8 weeks after treatment. (D) Custom-made sample holder and machine for torsional testing used in this study. White arrows indicate the breaking point within the newly formed tissue for a BMP-2 cmRNA–treated specimen. (E) Results of the biomechanics testing are plotted as torque to failure (N*cm). Samples (n = 8) of animals treated with either 50 μg of BMP-2 cmRNA or 11 μg of rhBMP-2 were analyzed 4 and 8 weeks after administration. Normal femora (n = 8) were also evaluated; the range of failure torque values for normal bone is plotted as double, discontinuous line in the graph. Data were analyzed using one-way ANOVA corrected by Tukey for multiple comparisons; *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001; # indicates comparison to normal bone (P < 0.03).
The dose-dependent induction of bone by cmRNA was confirmed by μCT (Fig. 2B). The 50-μg BMP-2 cmRNA group showed the highest bone volume (BV) values as early as 4 weeks when compared to doses of cmRNA <50 μg (P < 0.0001), comparable to the rhBMP-2 control (P > 0.05). By 8 weeks after treatment, the 50-μg cmRNA group showed significantly higher BV–to–total volume (BV/TV) ratios than all other treatment groups, including the recombinant protein counterpart (P < 0.0001; Fig. 2B). It was interesting to observe that the BV/TV ratios of the rhBMP-2 treatment peaked at 4 weeks, followed by a significant decrease at 8 weeks (P = 0.01). This was not the case for either the 25- or 50-μg BMP-2 cmRNA groups, where BV/TV values increased or remained unaltered, respectively.
Quantitative bone parameters further indicated the superior quality of the 50-μg BMP-2 cmRNA treatment. A significantly higher number of bone trabeculae that featured less separation were confirmed in this group when compared to all other cmRNA groups (Fig. 2C). Moreover, the newly formed trabeculae were thinner for the 50-μg BMP-2 cmRNA and rhBMP-2 groups. Values of trabecular thickness (Tb.Th), trabecular number (Tb.N), and trabecular separation (Tb.Sp) were similar for both the 50-μg BMP-2 cmRNA and rhBMP-2 groups (P > 0.05; Fig. 2C). Of note, Tb.N significantly decreased at 8 weeks for the rhBMP-2 group (P = 0.018; Fig. 2C), while it increased in the 50-μg BMP-2 cmRNA group. For rhBMP-2, Tb.N decreased, while Tb.Th maintained approximately constant over time. The decrease in Tb.N while maintaining constant thickness and separation speaks to an accelerated bone loss, which might explain the lower BV/TV value of rhBMP-2 from 4 to 8 weeks. We did not observe this loss of bone in the BMP-2 cmRNA group.
Mechanical properties of defects healed by BMP-2 cmRNA and rhBMP-2
Because of the small sizes of the rat bone specimens, a custom-made sample holder and test system was used (Fig. 2D and movies S1 and S2). Healed defects showed significantly increased resistance to failure over time (Fig. 2E). This was observed for both the 50-μg BMP-2 cmRNA and rhBMP-2 groups (P < 0.003; Fig. 2E). No statistically significant difference was found for the failure torque (N*cm) at 4 or 8 weeks between these two groups. However, the BMP-2 cmRNA group had recovered the failure torque values of native bone at 4 weeks (P = 0.19), whereas values for the rhBMP-2 group remained significantly lower than normal (P = 0.02; Fig. 2E). At 8 weeks, only the BMP-2 cmRNA–healed specimens showed significantly higher failure torque values than normal bone (P = 0.03; Fig. 2E). This suggests that recovery of mechanical strength may be quicker in response to cmRNA.
Expression of transcripts associated with osteogenesis following application of BMP-2 cmRNA and rhBMP-2
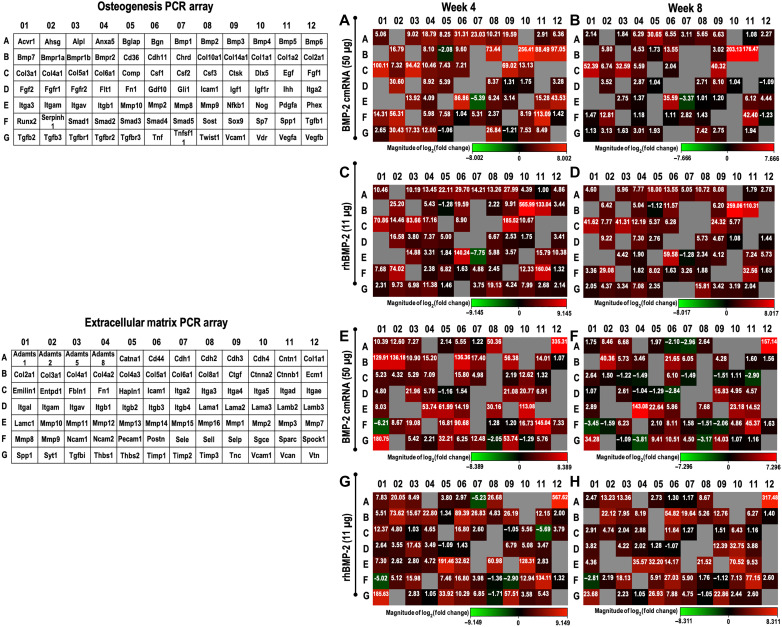
Polymerase chain reaction (PCR) array technology was used to investigate the expression of genes associated with osteogenesis and matrix formation in response to BMP-2 cmRNA and rhBMP-2. The results are presented in Fig. 3, with the top 10 up-regulated genes for each treatment and time point highlighted in table S1. In addition, gene expression data, organized by biological role of the analyzed gene, are summarized in tables S2 to S7.
Fig. 3. Expression of relevant osteogenic and extracellular matrix–related genes in newly formed tissue.
The analyzed set of genes using each array is tabulated (left) using a layout that matches the heat map representation of the results in (A) to (H). Osteogenic (A to D) and extracellular matrix (E to H) PCR array results shown as heat maps for weeks 4 and 8 after treatment with either 50 μg of BMP-2 cmRNA or 11 μg of rhBMP-2. Heat maps provide a visualization of the fold changes in expression between the selected groups (i.e., treatment versus normal bone) for every gene in the array in the context of the array layout. Inserted values provide the fold change. Gray boxes indicate a gene whose average threshold cycle is either not determined or greater than the defined cutoff (35). Following the array manufacturer’s recommendation, this was interpreted as a gene whose expression was undetectable.
Both osteogenic agents stimulated an up-regulation of transcripts encoding key components of the extracellular matrix of bone, particularly collagens, osteonectin, and osteopontin. Osteogenic transcription factors such as Runx2 and Sp7, but also Twist, were mildly up-regulated at 4 weeks, but not at 8 weeks, which might be explained by the late time points used for PCR evaluations (table S2). Certain growth factors and their receptors were also up-regulated in both treatment groups (table S3), especially members of the transforming growth factor–β (TGF-β) superfamily. Up-regulation of Tgfb3 and receptors Tgfbr1 and Fgfr1 was observed 4 weeks after BMP-2 cmRNA treatment. Transcripts encoding enzymes involved in bone remodeling such as matrix metalloproteinases (MMPs) and cathepsin K were also induced by BMP-2 cmRNA and rhBMP-2 (table S6). Of particular note were the high fold up-regulation values obtained for cathepsin K and Mmp-2, Mmp-13, and Mmp-16 at 4 weeks and Mmp-12 at 8 weeks after both treatments.
By far, the largest increases occurred in genes encoding various collagens, with type I being the most up-regulated, followed by types V and III (table S5). These data are consistent with type I collagen being the most abundant protein in bone, with type V collagen also being present. The type III collagen signal probably reflects its abundance in the endothelial linings of blood vessels in bone. These data, along with high expression of transcripts, such as Spp1, Sparc, Postn, related to bone mineralization, and the collagen chaperone Serpinh1, collectively reflect a highly active mineralized tissue environment in both cmRNA- and rhBMP-2–treated defects.
Differences between transcripts induced by BMP-2 cmRNA and rhBMP-2 were minor, apart from collagens associated with chondrogenesis. Both PCR arrays detected transcripts encoding type II and type X collagens in defects treated with BMP-2 cmRNA but not in defects treated with rhBMP-2 (Fig. 3 and table S5). Type II collagen signifies the presence of chondrocytes, and type X collagen is a marker of chondrocyte hypertrophy and endochondral ossification. Thus, these findings strongly suggest that there is a major difference in the biology of bone healing using these two osteogenic agents, with cmRNA provoking endochondral ossification and rhBMP-2 provoking intramembranous ossification.
Confirmation by histology
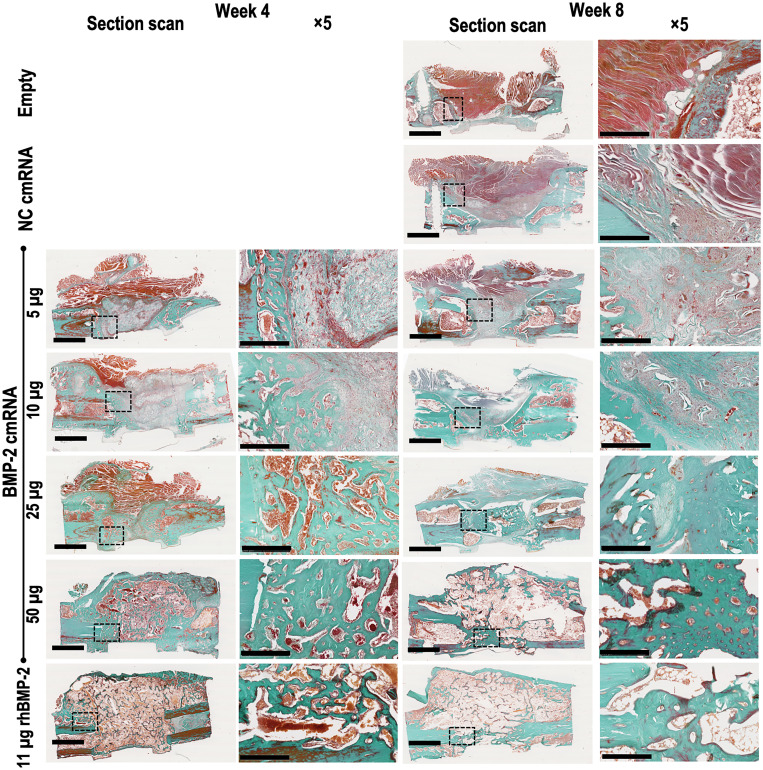
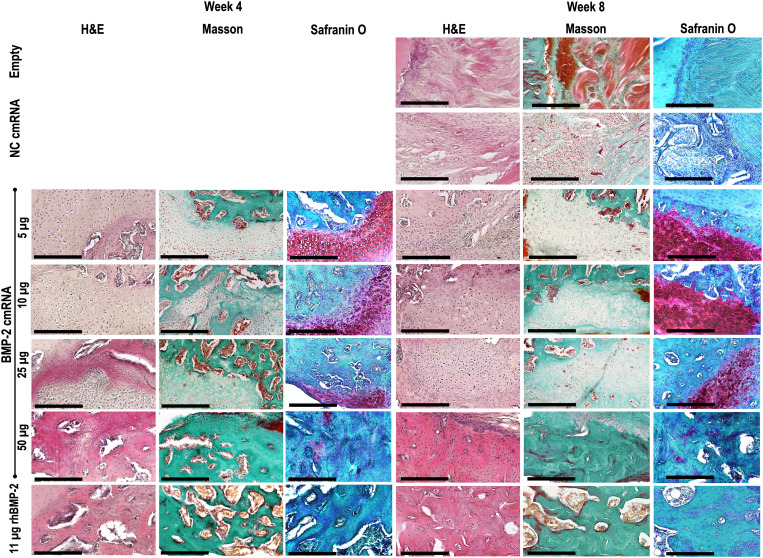
Histological examination confirmed that bone defects were bridged by osseous material at 4 and 8 weeks in the BMP-2 cmRNA (25 and 50 μg) and rhBMP-2 groups. Both Masson trichrome and hematoxylin and eosin (H&E) staining reveal newly formed bone (green in Masson trichrome) with cortication and the reestablishment of marrow (Fig. 4 and fig. S2). The bone fibrous matrix (osteoid) detected by Toluidine blue at 4 weeks seemed to be less prominent at 8 weeks, indicating mineralization of the osteoid over time (fig. S3). Restoration of the cortices seemed to be less pronounced with rhBMP-2, where a greater volume of callus tissue could be appreciated when compared to the 50-μg BMP-2 cmRNA. At lower concentrations of cmRNA producing partial bridging, areas of new bone were interspersed with fibrous and, in some specimens, abundant cartilaginous tissue (Figs. 4 and 5).
Fig. 4. BMP-2 cmRNA administration induces de novo bone deposition in the defect.
Masson-Goldner trichrome staining shows bone defects filled with newly formed bone (compact green) as a result of BMP-2 cmRNA treatment. Dose dependency is apparent; images corresponding to the higher BMP-2 cmRNA doses show deposits of a denser, green stained new matrix when compared to the lower doses. This was observed in fully healed animals. Staining was performed on harvested explants from all groups 4 and 8 weeks after surgery. From left to right, a section scan illustrates the entire defect area, while a ×5 magnification image shows the detailed appearance within the defect. Insets in section scans indicate the area where the ×5 magnification picture was taken. Representative images from n = 6 rats per group are shown. Scale bar on section scans, 2.5 mm. Scale bar on ×5 magnification images, 500 μm.
Fig. 5. Chondrocytic cells are only visible within defects administered BMP-2 cmRNA.
H&E, Masson-Goldner trichrome, and safranin O images show the morphology of the cells populating the newly formed tissue and its extracellular matrix. For the BMP-2 cmRNA groups, a clear chondrocytic morphology can be observed; using Masson-Goldner trichrome, cells appear embedded in a green-staining, collagen-rich matrix. Using safranin O, rounded, chondrocytic cells that are embedded in a proteoglycan-rich matrix appear (dark red). From left to right, the first three images correspond to week 4 after surgery, while the last three images were taken of specimens 8 weeks after surgery. Images were taken at ×20 magnification. Scale bars, 250 μm.
The possible presence of chondrocytes in the unbridged or partially bridged BMP-2 cmRNA specimens is interesting given the expression of type II and X collagen noted earlier. Safranin O confirmed the presence of this cell type within an abundant anionic extracellular matrix (Fig. 5). Rounded, chondrocytic cells can be seen quite clearly embedded in a proteoglycan-rich matrix (stained dark red by safranin O) in the defects treated with 5, 10, and 25 μg of BMP-2 cmRNA (Fig. 5). Figure S4 depicts, in detail, the morphological features of the identified chondrocytic cells. At 4 weeks, the cells were rounded and organized in perpendicular columns. Later, at 8 weeks after treatment, these cells resembled hypertrophic chondrocytes deeply buried in the surrounding matrix. This morphologic change indicates a transition from a cartilaginous matrix to a more calcified one. This suggests that the lower dose of endogenously produced BMP-2 to which they were exposed stimulated chondrogenesis of mesenchymal precursors but failed to drive them all the way to endochondral ossification, despite enhanced expression of type X collagen (Fig. 3). Chondrocytic cells are also evident, but less extensively, in defects treated with 50 μg of BMP-2 cmRNA, where endochondral ossification proceeds to bone formation. No chondrocytic cells were visible in defects treated with rhBMP-2 (Fig. 5 and fig. S4). At neither the 4-week nor 8-week time point was there histological evidence of a local inflammatory response.
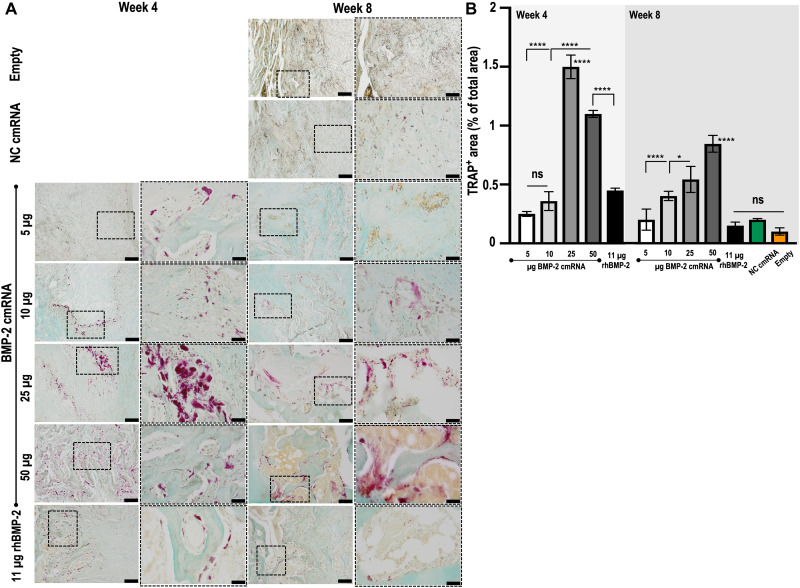
Tissue remodeling, indicated by cathepsin K expression (Fig. 3), was confirmed by tartrate-resistant acid phosphatase (TRAP) staining. In keeping with the PCR array data, TRAP activity was detected as early as 4 weeks after treatment and was clearly stronger in the 25- and 50-μg BMP-2 cmRNA–treated groups (Fig. 6A). In higher-magnification images (fig. S5), the presence of multinucleated osteoclast-like cells can be appreciated populating the newly formed tissue as a result of the BMP-2 cmRNA treatment. Semiquantitative analysis of the TRAP staining confirmed significantly higher percentage of TRAP-positive area in the 25- and 50-μg BMP-2 cmRNA–treated groups when compared to lower-dose cmRNA groups and to rhBMP-2 (P < 0.001; Fig. 6B). It is interesting that cmRNA treatment provoked more TRAP activity, indicative of more tissue remodeling, than its rhBMP-2 counterpart. This is further supported by the local increase in interleukin-17A (IL-17A) production, concomitant with elevated RANKL expression that was observed at 3 days after BMP-2 cmRNA treatment (P < 0.0001; fig. S6).
Fig. 6. TRAP activity is prominent in defects treated with higher doses of BMP-2 cmRNA but trivial in those treated with rhBMP-2.
TRAP staining was performed on harvested explants from all groups 4 and 8 weeks after surgery. (A) From left to right, a ×10 magnification image shows a general view within the region of interest (ROI = interface between the old bone and the newly formed tissue), while a ×40 magnification image shows detailed positive stained areas (magenta) within the ROI. Insets in ×10 magnification images are indicative of the area where the ×40 magnification picture was taken. Scale bar on ×10 magnification images, 200 μm. Scale bar on ×40 magnification images, 50 μm. Representative images from n = 6 rats per group are shown. (B) Quantification of TRAP-positive area by means of ImageJ. Results are reported as % of TRAP-positive area normalized to total area of the tissue section. ns, not significant.
Confirmation by immunohistochemistry of type II and X collagens in defects treated with cmRNA but not rhBMP-2
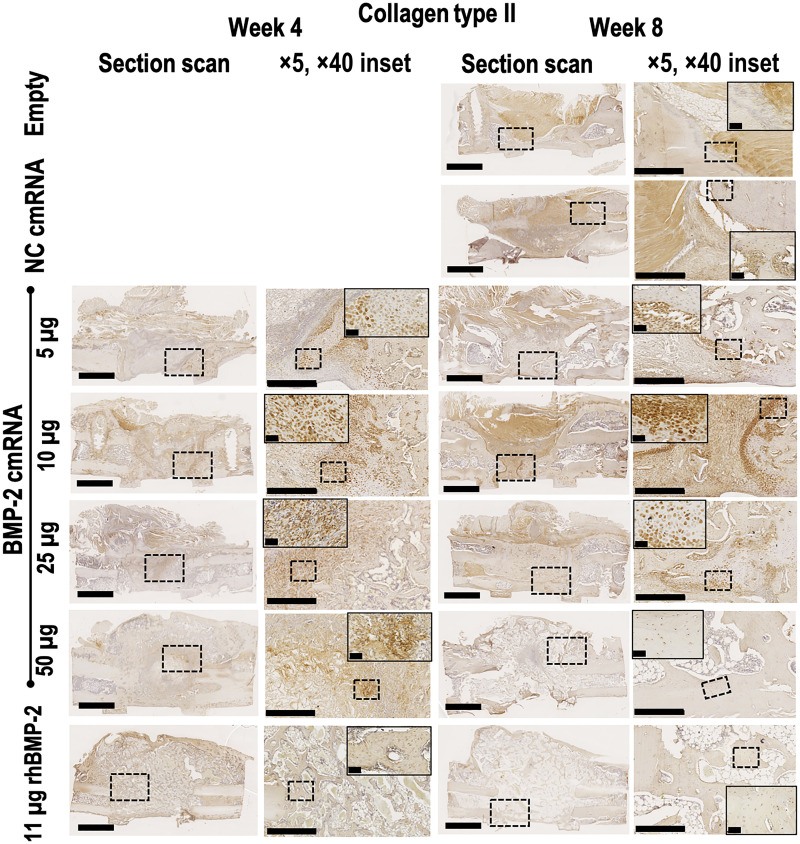
In keeping with the PCR array data (Fig. 3) and the histology images (Figs. 4 and 5), immunohistochemistry (IHC) detected type II (Fig. 7) and X (Fig. 8) collagens in defects treated with cmRNA but not in those treated with rhBMP-2. Consistent with the histology, expression of these collagens was strongest in those defects treated with sub-healing doses of BMP-2 cmRNA. In defects treated with 50 μg of cmRNA, type II collagen staining was visible at 4 weeks, but not at 8 weeks, presumably because endochondral ossification was advanced in the later animals.
Fig. 7. Collagen type II deposition and chondrocytic cells are only visible within defects administered BMP-2 cmRNA.
Collagen type II immunohistochemical staining was performed on harvested explants from all groups 4 and 8 weeks after surgery. From left to right, a section scan illustrates the entire defect area, while a ×5 magnification image shows detail. Images taken at ×40 magnification show the appearance of cells populating the newly formed tissue. Insets in section scans indicate the area where the ×5 magnification picture was taken. Insets in ×5 magnification pictures indicate the area where the ×40 magnification image was taken. Representative images from n = 6 rats per group are shown. Scale bar on section scans, 2.5 mm. Scale bar on ×5 magnification images, 500 μm. Scale bar on ×40 magnification images, 50 μm.
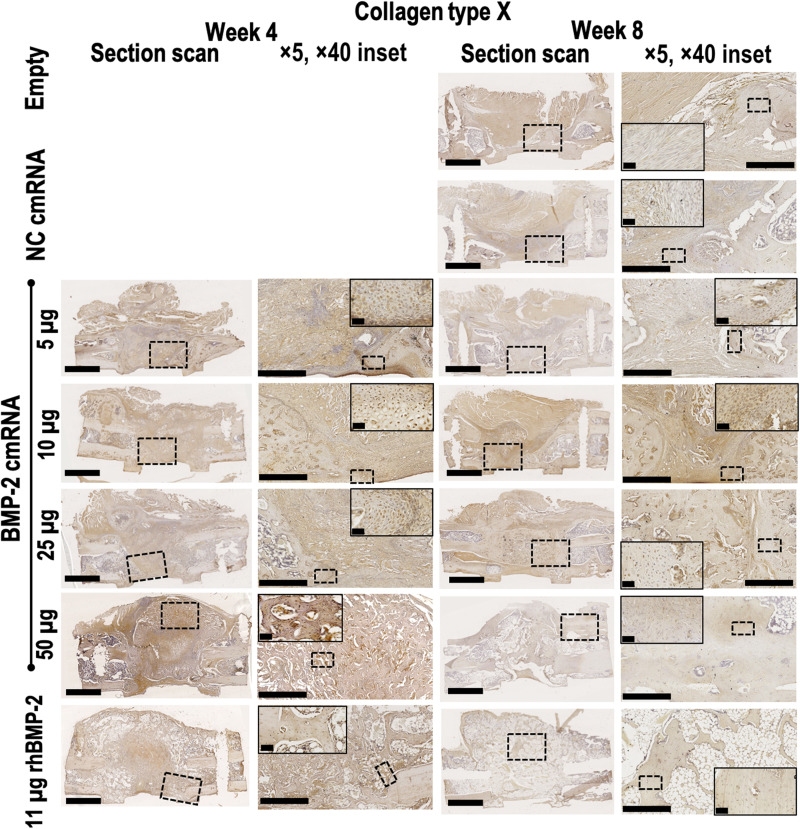
Fig. 8. Collagen type X is clearly visible 4 weeks after BMP-2 cmRNA treatment but decreases at 8 weeks.
Hypertrophic chondrocytes are embedded within the newly formed tissue. Collagen type X immunohistochemical staining was performed on explants 4 and 8 weeks after surgery. From left to right, a section scan illustrates the entire defect area, while a ×5 magnification image shows detail. Images taken at ×40 magnification show the appearance of cells populating the newly formed tissue. Insets in section scans indicate the area where the ×5 magnification picture was taken. Insets in ×5 magnification pictures indicate the area where the ×40 magnification image was taken. Representative images from n = 6 rats per group are shown. Scale bar on section scans, 2.5 mm. Scale bar on ×5 magnification images, 500 μm. Scale bar on ×40 magnification images, 50 μm.
Expression of type X collagen was weaker than that of type II collagen in defects treated with suboptimal amounts of cmRNA and had almost disappeared by 8 weeks. This presumably reflects failed endochondral ossification. Defects treated with 50 μg of cmRNA had less distinctive staining for type X collagen at 4 weeks, reflecting the advanced stage of repair at this time. No type X collagen was detected by IHC at 8 weeks, in agreement with the PCR array data.
Circulating anti- and pro-inflammatory cytokines were induced by cmRNA
Concentrations of a variety of inflammatory [interferon-γ (IFN-γ), IL-1α, IL-1β, IL-12p70, IL-17A, IL-2, IL-5, IL-6, and tumor necrosis factor–α (TNF-α)] and anti-inflammatory (IL-4, IL-10, and IL-13) cytokines were measured in the peripheral blood at various times after surgery and implantation of BMP-2 cmRNA or rhBMP-2 (fig. S7 and table S8). No increase in any of these cytokines was noted when comparing levels in rats with empty defects and those receiving rhBMP-2. In contrast, cytokine levels were increased in animals receiving cmRNA. In most cases, noncoding cmRNA alone elevated cytokine production and this was further augmented when BMP-2 cmRNA was applied. Induction of the pro-inflammatory cytokine IL-1α was particularly strong, with circulating levels of several hundred picograms per milliliter in rats receiving BMP-2 cmRNA. A similar but slightly milder scenario was observed with the anti-inflammatory cytokine IL-10. In other instances, as in the case of IL-1β, IL-2, IL-6, IL-13, IL-17A, and TNF-α, NC cmRNA induced levels that were either very low or similar to those of the empty and rhBMP-2 groups for days 1 and 10, but not for day 2. In all cases, the increase in cytokines was maximal by 1 day after surgery and remained elevated for at least 10 days.
DISCUSSION
With two modified mRNA-based COVID-19 vaccines currently approved for clinical use, the potential of this technology has been validated (14, 15). In addition, modified mRNA has been used in the treatment of various diseases caused, for example, by single-gene defects (16, 17). A less explored therapeutic application of this technology is to stimulate tissue regeneration. Tissue restoration is a particularly appropriate target for mRNA, because it may not need repetitive or systemic administration. Transient, local protein expression may be sufficient to initiate the sustained regeneration of diverse tissues.
Recent investigations have shown mRNA therapy to be applicable in liver (18), cardiac tissue [reviewed in (19)], and skin regeneration (20). The last of these progressed into clinical trials for patients with type 2 diabetes, where it improved blood flow in patients’ skin (20). These studies demonstrate that, unlike its recombinant protein counterpart, mRNA can be administered at a low dose while achieving a successful therapeutic outcome without side effects. mRNA therapeutics are generally considered to be safe and affordable (21, 22). However, turning mRNA into drugs may be hampered by two main problems: poor pharmacology (rapid degradation) and high immunogenicity. Decades of research have developed several strategies to overcome these obstacles, one being to modify the 5′ cap, the 5′ and 3′UTRs, and the poly(A) tail among other mRNA structural elements (22); more than 100 nucleotide modifications have been investigated (10, 23).
We previously reported the combination of a translation initiator of short UTRs (TISU) and 5-iodo–modified pyrimidine nucleotides (12) as a strategy for improving translational efficiency while reducing inflammatory responses to mRNA (fig. S1A). When coding for BMP-2, cmRNA containing TISU in conjunction with 5-iodinated pyrimidines was shown to be osteogenic in a dose-dependent manner (12).
Here, we provide convincing evidence that the optimized cmRNA we developed is able to heal large segmental defects in the rat when delivered by a nonviral, lipid vector. The BMP-2 cmRNA:lipoplex particles were able to induce robust and timely expression of BMP-2. The time frame of detectable protein production was just 5 days yet sufficient to heal the defects. BMP-2 protein production remained intralesional, and almost no drug was detected in blood, lung, spleen, liver, or kidney at relevant times after administration. No relevant side effects were associated with BMP-2 cmRNA administration, consistent with the transient and anatomically restricted expression of BMP-2.
The transcriptomic analyses provided important insights into osteogenic mechanisms in operation after 4 and 8 weeks. Various components of the extracellular matrix, including osteopontin (spp1), osteonectin (sparc), and several types of collagen particularly types I and V and, with cmRNA, types II and X were highly up-regulated, as were various MMPs, particularly MMP2 and MMP13. Coupled to the high expression of cathepsin K and TRAP, these data suggest robust remodeling of both the mineral and organic components of the extracellular matrix of the newly formed bone. The up-regulation of BMP-1, BMP-2, and BMP-3, as well as the BMP receptor 1a, indicates the importance of endogenous BMPs in regulating these processes. There was also a robust induction of TGF-β3, TGF-β receptors 1 and 2, and fibroblast growth factor (FGF) receptor 1.
The cmRNA used here was engineered to reduce induction of inflammatory cytokines. We have previously shown that induction of IL-1β, IL-6, TNF-α, and IFN-γ production by human peripheral blood mononuclear cells is greatly reduced by these modifications (12). It is therefore unexpected that circulating cytokine levels, especially IL-1α, were elevated in rats receiving BMP-2 cmRNA. It is also interesting that NC cmRNA alone generally did not do this, suggesting the importance of BMP-2, although rhBMP-2 did not produce these changes. Intracellular expression of BMP-2 in conjunction with a cmRNA triggers effects that are not observed with rhBMP-2. A certain amount of early inflammation seems required for bone healing, and it is possible that the residual inflammatory properties of the cmRNA matched this requirement. In this context, it is interesting that BMP2 cmRNA induced more IL-1β than did rhBMP-2 at early time points (fig. S6). Moreover, RANKL, which is induced by IL-1, was higher at 3 days in the cmRNA group. Induction of IL-17A, on the other hand, was much lower and did not differ statistically between BMP-2 cmRNA and rhBMP-2. Additional research is required to investigate these matters.
Our data suggest that BMP-2 delivered by cmRNA heals bone by the endochondral route, whereas rhBMP-2 does so intramembraneously. Long bones form via endochondral ossification during development and heal fractures by the same route, unless absolute stability is achieved using compression hardware, which is not possible when dealing with a large segmental defect. This suggests that part of the superiority of BMP-2 cmRNA over rhBMP-2 reflects the former’s ability to trigger the native endochondral ossification pathway to bone formation in a segmental defect model. It is interesting that doses of cmRNA too low to heal these segmental defects nevertheless generated cartilage, as gauged histologically and by detection of type II collagen expression. Although type X collagen expression was detected by PCR array in the 50-μg cmRNA groups and by IHC in all cmRNA groups, these cells failed to proceed to endochondral ossification at lower doses. No type II or type X collagen was detected at any time in the defects receiving rhBMP-2.
Bone remodeling, a process essential for bone healing, was also visibly superior in the BMP-2 cmRNA group as indicated by the high TRAP activity observed in the animals treated with 25 and 50 μg of BMP-2 cmRNA. This may help account for the observation that with BMP-2 cmRNA healing lacks the massive, ectopic callus formation that has been described for rhBMP-2 (24) and confirmed here. The de novo tissue formed with BMP-2 cmRNA was localized and constrained to the defect area. Correspondingly, newly formed tissue as a consequence of BMP-2 cmRNA treatment showed earlier biomechanical improvement and remained superior 8 weeks after treatment.
Overall, the data summarized in this study are the first demonstration that cmRNA can heal large, critical-sized, segmental osseous defects of long bones in a superior fashion to its recombinant protein counterpart. This technology promises to couple affordability with efficacy, and clinical expediency with safety, while presenting fewer barriers than traditional gene therapy to clinical translation. Remaining challenges for this technology include the development of mRNA-specific carrier matrices. These three-dimensional carriers may provide further protection to the cmRNA from degradation while ensuring optimal cmRNA delivery (25). In addition, such a matrix might also prolong the cmRNA action time; this may be interesting for complex tissues and tissue interfaces such as cartilage and the osteochondral unit.
Although convincing, this study does have limitations. Young, healthy male rats were used for the experimental model; this may be relevant to certain types of trauma patients but not, for example, to patients whose segmental defects were created by tumor resection and whose surrounding soft tissue may have been subjected to irradiation. Before contemplating clinical translation of this technology, it will be necessary to confirm that similar results are obtained for female rats. A second requirement is to confirm that the technology is effective in a large animal model; sheep, pigs, and goats are frequently used for this purpose (26). Translation will be facilitated by the widespread, effective, and safe use of cmRNA in vaccines as well as the fact that BMP-2, the protein product of our cmRNA, is already approved by the U.S. Food and Drug Administration (FDA) for certain applications in bone healing.
MATERIALS AND METHODS
Study design
The goal of this study was to determine the efficacy, potency, and safety of an optimized cmRNA encoding BMP-2 in healing a critical-sized, diaphyseal, femoral defect in the rat. Its properties in these regards were compared to those of rhBMP-2, the current clinical gold standard. We used a validated rat model in which a 5-mm mid-femoral osteotomy was performed and stabilized with a custom-made plate. Various doses of cmRNA encoding BMP-2 were placed in the defect on a collagen sponge. Noncoding cmRNA was used as a negative control, and the clinically equivalent dose of rhBMP-2 was included for comparison. A collagen sponge was used as a carrier to secure a valid comparison of cmRNA and rhBMP-2 under identical settings, matching the current clinical practice for rhBMP-2 delivery. In addition, using the well-known collagen sponge removed potential complications from unknown effects related to an alternative carrier. Furthermore, using an already approved collagen carrier should facilitate translation into clinical use because the regulatory authorities are very familiar with, and have already approved, the collagen sponge. Biodistribution of cmRNA and its expression were assessed by reverse transcription PCR (RT-PCR) and in vivo imaging in conjunction with cmRNA encoding luciferase. Bone healing was monitored by weekly x-ray and analyzed at 4 and 8 weeks by μCT, mechanical testing, and histology. Mechanistic data were obtained from gene expression analysis using PCR arrays and by IHC.
cmRNA production and formulation
cmRNA encoding BMP-2 and FLuc, as well as an NC construct, was synthesized by T7 RNA polymerase in vitro transcription (IVT). BMP-2 coding sequence (NM_001200.2) was used in combination with a translation initiator of short 5′UTR element, TISU (fig. S1A). In the plasmid template for the NC cmRNA, the Kozak element (GCCACC) was scrambled (CGCACC) and all in-frame ATGs were converted to stop codons (TGA). For the FLuc cmRNA, the FLuc coding sequence (amplified from pGL4.10 plasmid; Promega, USA) was used in combination with a minimal 5′UTR sequence. Regarding ribonucleotide substitution, 35% of uridine residues were replaced with 5-iodo-uridine and 7.5% of cytidine residues were replaced with 5-iodo-cytidine. Details of the IVT reaction and later purification can be found elsewhere (12). The concentration and quality of all cmRNAs were measured on NanoDrop 2000C (Thermo Fisher Scientific, USA). The purity and size were confirmed by automated capillary electrophoresis (Fragment Analyzer; Advanced Analytical, USA). Nucleotide analysis was performed by high-performance liquid chromatography (Agilent 1260 Infinity, Agilent Technologies, USA).
For in vivo application, lipoplexes were formed using a nonviral lipidoid vector. This vector is based on a cationic lipidoid with dipalmitoyl phosphatidylcholine (DPPC), cholesterol helper lipids, and 1,2-dimyristoyl-rac-glycero-3-methylpolyoxyethylene PEGylated lipid (DMG-PEG) (27). This vector has been shown to be biocompatible in mice (27, 28) and rats (12). A solvent exchange methodology was followed to obtain the lipoplexes (29). cmRNAs were mixed with the lipid at an N/P ratio of 8 (molar ratio of polymer amine (N) to nucleic acid phosphate (P) groups), ensuring a final cmRNA concentration of 1 mg/ml. Lipoplexes thus formed had particle sizes of 60 to 80 nm and a positive zeta potential of approximately 20 mV. The encapsulation efficiency, as determined by RiboGreen, was found to be >90% and similar for all three cmRNAs.
Treatment of rats with critical-sized femoral defects
cmRNA lipoplexes were loaded onto an FDA-approved collagen sponge (sterile, sized 0.6 × 1.27 cm, Helistat, Integra LifeSciences, USA) 20 min before implantation into the rat bone defect. Doses of 5, 10, 25, and 50 μg of BMP-2 cmRNA per sponge were used. For NC cmRNA and FLuc cmRNA, 50 μg was loaded per sponge. As a clinically relevant control, 11 μg of rhBMP-2 (INFUSE, Medtronic, USA) was loaded to the collagen sponges following the same procedure as used for the cmRNAs. This dose of rhBMP-2 is equivalent to the one used clinically for bone formation in humans.
Animal care and experimental protocols were approved by Mayo Clinic’s Institutional Animal Care and Use Committee (no. A00004410-19). F344 rats (n = 190) at 18 weeks of age were used for surgery as described before (30). Briefly, a critical-sized, mid-diaphyseal, 5-mm defect was created in the right femur of each rat (fig. S1B). Rats were anesthetized with isoflurane and placed in the left lateral decubitus position with the right hind limb protruding through a sterile fenestrated drape. The skin incision was made on the posterolateral thigh, and the lateral intermuscular septum was dissected to expose the femoral diaphysis. Care was taken to avoid damaging the surrounding soft tissues. Using a polyacetal plate (Special Designs, USA), four holes were drilled along the mid-diaphysis using a 0.79-mm drill bit (RISystem AG, Switzerland). The plate was then secured carefully to the femur using four hand-driven 0.9-mm threaded K-wires (MicroAire Surgical Instruments, USA), which allowed the construct to act as a locked plate. Creation of a 5-mm ostectomy was performed using a 0.44-mm Gigli wire saw (RISystem AG). The 5-mm bone fragment was removed, and the site was irrigated with normal saline. The defect site either was left empty or received a collagen sponge containing one of the test substances (fig. S1C). A soft tissue pouch was created with the deep thigh muscles to contain the implants within the defect. The wound was closed in layers with 4-0 Vicryl, and the incision site was closed using 9-mm wound clips. Animals were followed for up to 8 weeks after surgery, at which time the treated femora and the contralateral, nontreated femora were harvested for analysis. The timeline as for this study is depicted in fig. S1D.
Radiographic monitoring
In vivo bone healing was monitored with radiographic images of the right femur using a digital MX-20 x-ray cabinet system (Faxitron Bioptics, USA) under general anesthesia with isoflurane. Rats were ventrally positioned inside the cabinet with the hind limbs abducted at a 90° angle from the body. Radiographs were obtained at 42-kV energy and 10-s exposure time on day 10 and weeks 4, 6, and 8 after surgery. Two observers analyzed the images for fractures, failure of fixation, bone formation, and bridging of the defect.
In vivo bioluminescence
Rats (n = 6 per group) underwent bioluminescence imaging using an IVIS Spectrum system (PerkinElmer, USA) at days 1 to 3 after surgery. d-Luciferin (GoldBio, USA) diluted in phosphate-buffered saline (PBS) was administered via intraperitoneal injection at a dose of 150 mg/kg. Rats were anesthetized with isoflurane, and fur was shaved from the operated limb, right lumbar region, and lower right abdomen to reduce the luminescent signal from the white fur. Rats were placed inside the IVIS chamber in the left lateral decubitus position so that the right hind limb was facing the charge-coupled device camera. Photon emissions from the operated limb were collected 30, 35, 40, and 45 min after injection using a 1-min exposure time. Bioluminescence signal was superimposed over a photograph of the rat, and the flux within a circular region of interest comprising the length of the femur was quantified using the Living Image software (PerkinElmer). At day 3, rats were euthanized after in vivo imaging and the femora were explanted, rinsed in PBS, and fixed in 10% neutral buffered formalin for immunohistological analysis.
Blood draws and organ harvest
Blood was drawn from the animals at days 1, 2, and 10 after surgery using microtainer K2EDTA tubes (BD, USA). Six animals per group were used. A 250-μl sample of whole blood was transferred to ribonuclease/deoxyribonuclease-free sterile tubes containing 1 ml of RNAlater (Thermo Fisher Scientific, USA). After an overnight incubation at 4°C, samples were transferred to −20°C until further PCR analysis. Microtainer tubes containing 250 μl of blood were incubated on ice for 15 min and then centrifuged at 1500g for 10 min. Plasma was carefully collected and stored at −80°C for further multiplex assay. At weeks 4 and 8 after surgery, rats were euthanized, and lung, spleen, liver, and kidney were surgically removed. The same portion of harvested organs was used for all the animals (entire spleen, right kidney, right caudate lobes of the liver, and right lower lobe of the lung). The organs were collected using a 5× volume of RNAlater (Thermo Fisher Scientific) and incubated overnight at 4°C. Tissue homogenization was performed using TRIzol (Thermo Fisher Scientific) in Qiagen TissueLyser II (two cycles of 2 min at 30 Hz; Germany). Samples were stored at −80°C until further PCR analysis.
ELISA for BMP-2
Groups of animals (n = 6 per group) were euthanized at days 1, 2, and 5 after surgery to evaluate in vivo BMP-2 production in the femoral defects, following a previously reported procedure (31). Collagen implants were collected and processed in stainless steel grinding jars using TissueLyser II (Qiagen, Germany). Care was taken to preserve all cellular material and/or tissue deposited within the harvested implant. The resulting homogenate was extracted with T-PER tissue protein extraction solution (Thermo Fisher Scientific) containing protease inhibitors to release protein. The supernatant was collected for BMP-2 determination using the commercial DuoSet ELISA Development System (R&D Systems, USA).
Biomechanical testing
After euthanasia at weeks 4 and 8 after surgery, the femora (n = 8 per group) were explanted, cleaned of soft tissue, wrapped in gauze soaked in normal saline, and frozen at −20°C until used. Frozen femora were allowed to thaw at 4°C for 24 hours before testing. Biomechanical properties of the healed segmental defects were determined by torsional testing to failure. The member of our research team (C.V.N.) who conducted the testing was blinded to the treatments. Eight specimens (n = 8) were included per group. Before testing, the callus size was measured using a high-precision digital digimatic absolute caliper (Mitutoyo, USA) and the ends of the femora were potted in polymethylmethacrylate within a square metal fixture. A custom-made material testing system for small animals (Mayo Clinic, USA) was used to complete the torsional testing (movie S1 for native bone sample and movie S2 for explanted test bone sample). This machine interfaces with a LabVIEW (National Instruments, USA) software designed to output displacement and load (model RTS-200, Transducer Techniques LLC, USA). Bones were tested to failure under deformation control at a constant deformation rate of 5 rad/min. The torque and displacement were used to calculate failure strength and torsional stiffness.
μCT analysis
Bone explants were scanned by using a SkyScan 1176 apparatus (Bruker, Belgium) at 65 kV, 385 mA, and a voxel size of 9 mm. Image reconstruction and analysis were performed by using NRecon and CTAn Bruker software, respectively. Trabecular parameters such as separation (Tb.Sp; mm), number (Tb.N; 1/mm), and thickness (Tb.Th; mm) were determined. In addition, the ratio BV/TV (%) was calculated for each sample. Six samples per group were scanned for analysis.
cmRNA localization by qPCR
Total RNA was isolated from blood and organs using the RiboPure RNA Purification Kit (Thermo Fisher Scientific). Total RNA was reverse-transcribed into cDNA by using the First-Strand cDNA Synthesis Kit (Thermo Fisher Scientific). In both cases, the instructions of the manufacturer were carefully followed. Real-time quantitative PCR (qPCR) was performed with SsoFast Eva Green Supermix (Bio-Rad, USA) on a Bio-Rad CFX96 thermal cycler (Bio-Rad). The sequences of primers used to amplify BMP-2 cmRNA and rat tubulin beta 2A class IIa (Tubb2a) can be found in the Dataverse repository (https://dataverse.nl/privateurl.xhtml?token=73db97df-5749-44aa-83d6-e8b0a123aa9e). Spike-in controls were used for all samples. The percentage of recovery of the BMP-2 cmRNA used as spike-in control was >96.1 ± 3.4% for all tissue samples analyzed. Absolute quantification was performed using a standard curve, and cmRNA values were calculated by interpolation. Data are reported as picograms of BMP-2 cmRNA.
Extracellular matrix and osteogenesis PCR array
Total RNA was isolated from the defect sites of explanted femora 4 and 8 weeks after surgery. Frozen samples were homogenized with steel beads in TRIzol (Thermo Fisher Scientific) using TissueLyser II (Qiagen; 3 min at 30 Hz). Samples were then immediately processed following a standard phenol/chloroform method with subsequent purification using the Monarch RNA Cleanup Kit (New England BioLabs, USA). cDNA was synthesized using the RT2 First Strand Kit (Qiagen) following the instructions provided. For the PCR array, RT2 SYBR Green Master Mix was used as recommended by Qiagen. RT2 Profiler PCR Array Rat Extracellular Matrix and Adhesion Molecules (GeneGlobe Id: PARN-013ZD-2) and Rat osteogenesis (GeneGlobe Id: PARN-026Z) were used following the instructions provided for the Bio-Rad CFX96 thermal cycler (Bio-Rad). Gene list for both arrays can be found in table S9. Overall, 148 genes that are relevant during skeletal development and for bone mineral metabolism were analyzed. Expression of relevant growth factor, transcription factor, extracellular matrix, and cell adhesion molecule genes was also measured. Five housekeeping genes were included in each array. Data analysis was performed by uploading the obtained Ct values table (Excel file) on to the data analysis web portal at www.qiagen.com/geneglobe. Samples were assigned to controls (i.e., native bone) and test groups (i.e., 50 μg of BMP-2 cmRNA/4 weeks, 50 μg of BMP-2 cmRNA/8 weeks, 11 μg of rhBMP-2/4 weeks, and 11 μg of rhBMP-2/8 weeks). The Ct cutoff was set to 35 for analysis. Data are reported as fold change/regulation. The ΔΔCt method was used. For this, ΔCt was calculated between gene of interest and an average of all reference genes; ΔΔCt calculations were performed using the following equation: ΔCt (Test Group) − ΔCt (Control Group). Fold change was then calculated using the 2-ΔΔCt formula.
Histology
Femoral explants were decalcified in 10% buffered EDTA (Sigma-Aldrich, USA), dehydrated in ethanol, and embedded in paraffin. Longitudinal cross sections with a thickness of 7 μm were stained with H&E and Masson-Goldner trichrome kits (Carl Roth GmbH, Germany) using the protocol provided by the manufacturer. In addition, staining with safranin O and toluidine blue (both from Sigma-Aldrich) was performed following standard protocols described elsewhere (32). For TRAP staining, an l-tartaric acid solution (12 g/liter) was prepared in sodium acetate medium (pH adjusted to 5). Next, 120 mg of fast red violet LB salt and 1 ml of naphthol AS-MX phosphate solution [ethylene glycol monoethyl ether (20 mg/ml)] were mixed with 200 ml of the l-tartaric acid solution to obtain the TRAP staining solution mix. Deparaffinized slides were rehydrated with distilled water and placed inside a prewarmed TRAP staining solution mix. Samples were incubated at 37°C for 30 min, rinsed with distilled water, and then counterstained with 0.02% fast green. All reagents used for TRAP staining were purchased from Sigma-Aldrich. Six samples per group were used for each staining. Images of the complete section were taken using a Slide scanner (NanoZoomer 2.0, Hamamatsu Photonics, Japan). NDP.view 2 software was used to process the images. Higher-magnification images were taken using an Olympus BX43 microscope equipped with an Olympus camera SC50 (Olympus, Japan). The member of our research team (W.Z.) who conducted histological processing was blinded to the treatments. A semiquantitative analysis of the TRAP staining was performed by ImageJ v1.53k (National Institutes of Health, MD, USA). TRAP images were converted to 8-bit grayscale images. Next, the TRAP-positive area was selected by applying a black and white threshold (minimum, 95; maximum, 170). The total sample area and TRAP-positive area were calculated in pixels by using the ImageJ “measure” command. The TRAP-positive area was normalized to the total area analyzed. Two members of our research team (C.J.P.S. and E.R.B.) performed the ImageJ analysis independently, and results were averaged.
Immunohistochemistry
Luciferase and collagen types II and X were stained by IHC. Samples used for luciferase staining were collected at day 3, while samples used for collagen analysis were harvested at weeks 4 and 8 after surgery. Dewaxed and rehydrated sections were incubated first in 3% hydrogen peroxide (Sigma-Aldrich) for 15 min and then with proteinase K (Dako, Denmark) for 7 min at room temperature. Slides used for collagen staining were blocked with 2% bovine serum albumin (Sigma-Aldrich) for 60 min, and the sections were incubated with the primary antibody solutions in blocking buffer overnight (4°C). Collagen primary antibodies were purchased from Abcam (Cambridge, United Kingdom). Anti–collagen type II (ab34712) was used at a dilution of 1:200, and anti–collagen type X (ab49945) was used at a dilution of 1:1000. Rabbit isotype control immunoglobulin G (IgG) (ab27478) was used at a dilution of 1:200. Subsequently, slides were incubated with EnVision Dual Link System-HRP Rabbit/Mouse (Dako) for 1 hour at room temperature. Color development was performed with liquid diaminobenzidine chromogen (Dako), and sections were counterstained with Mayer’s hematoxylin (Carl Roth GmbH). Sections used for luciferase detection were blocked with 2.5% donkey serum for 1 hour at room temperature. Next, an overnight incubation at 4°C was performed with primary anti-luciferase antibody (G7451, Promega, USA) diluted 1:200 in PBS supplemented with 0.3% Triton X-100. Tissue sections were washed in PBS and incubated with secondary donkey anti-goat IgG HRP (horseradish peroxidase) (V805A, Promega) for 1 hour at room temperature. Sections were counterstained with Mayer’s hematoxylin (Carl Roth GmbH). Goat anti-rabbit IgG from Abcam (ab6702, 1:200 dilution) was used as isotype control. Images of the complete sections were taken using a Slide scanner (NanoZoomer 2.0) and processed with the NDP.view 2 software. The member of our research team (W.Z.) who conducted IHC processing was blinded to the treatments.
Multiplex ELISA
Plasma samples (n = 6 per group) were used to detect levels of pro- and anti-inflammatory cytokines including granulocyte colony-stimulating factor (G-CSF)/CSF-3, granulocyte-macrophage CSF (GM-CSF), IFN-γ, IL-1α, IL-1β, IL-10, IL-12p70, IL-13, IL-17A, IL-2, IL-4, IL-5, IL-6, and TNF-α. ProcartaPlex Rat Th Complete Panel 14-plex (Invitrogen by Thermo Fisher Scientific, USA) was used carefully following the manufacturer’s instructions. Samples were allowed to thaw at room temperature and centrifuged at 1000g for 10 min immediately before assay. Data were acquired in a Luminex system (Luminex Co., USA) that was calibrated (MAGPIX Calibration kit, Luminex Co.) before each assay.
Statistical analysis
Analysis of the data was performed using GraphPad Prism software (version 9.1.0, GraphPad, USA). Data are presented as means ± SD. Statistical analysis was done using one- or two-way analysis of variance (ANOVA). When two groups were analyzed over time, a multiple t test was used. Correction for multiple comparisons was performed, following GraphPad Prism recommendation, using either Tukey’s, Dunnett’s, or Holm-Sidak corrections and 95% confidence intervals. The sample size (n = number of animals) was a priori determined to be the minimal necessary for statistical significance. Therefore, n varied between experiments. Six samples were used for IVIS, μCT, histology, and IHC, as well as BMP-2 enzyme-linked immunosorbent assays (ELISAs); eight samples were needed for PCR arrays, quantitative RT-PCR, and biomechanics. Blood was drawn from six animals for cytokine determination by multiplex ELISA. All operated animals were radiographically examined (x-ray of n = 190) each at relevant time of observation. The exact methodology followed for statistical analysis has been described in Materials and Methods and in the legend of each figure, along with the n used in each case. P > 0.05 was considered not significant, and P values were reported using the GraphPad Prism style (*P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, and ****P ≤ 0.0001).
Acknowledgments
We acknowledge the support provided by C. Dohmen during the nonviral lipidoid vector production and by V. Joris and N. Roumans during the protein quantification and ELISA assays.
Funding: This project was supported by NIH (R01 AR074395 from NIAMS to C.H.E. and E.R.B.) and the John and Posy Krehbiel Professorship in Orthopedics (C.H.E.). Partial support was received by NIAMS (T32AR56950 to C.V.N.).
Author contributions: Conceptualization: C.H.E. and E.R.B. Methodology: R.E.D.L.V., M.v.G., and E.R.B. Investigation: R.E.D.L.V., M.v.G., W.Z., M.J.C., C.V.N., J.A.P., C.J.P.S., J.G., and E.R.B. Formal analysis: R.E.D.L.V., C.V.N., and E.R.B. Visualization: E.R.B. Resources: M.v.G., C.P., C.H.E., and E.R.B. Funding acquisition: C.H.E. and E.R.B. Project administration: C.H.E. and E.R.B. Supervision: M.v.G., C.P., C.H.E., and E.R.B. Writing—original draft: R.E.D.L.V., C.H.E., and E.R.B. Writing—review and editing: R.E.D.L.V., M.v.G., C.H.E., and E.R.B.
Competing interests: C.P. and E.R.B. are inventors on a patent related to this work filed by ETHRIS GMBH (no. WO/2016/075154, filed on 10 November 2015, published on 19 May 2016). C.P., W.Z., and J.G. are employees of ETHRIS GMBH. C.P. is also a shareholder of ETHRIS GMBH. The authors declare no other competing interests.
Data and materials availability: A complete dataset supporting the findings of this study is present in the paper and the provided Supplementary Materials. In addition, cmRNA and primer sequences have been deposited in the Dataverse repository (https://dataverse.nl/privateurl.xhtml?token=73db97df-5749-44aa-83d6-e8b0a123aa9e).
Supplementary Materials
This PDF file includes:
Supplementary Materials and Methods
Figs. S1 to S7
Other Supplementary Material for this manuscript includes the following:
Tables S1 to S9
Movies S1 and S2
REFERENCES AND NOTES
- 1.James A. W., LaChaud G., Shen J., Asatrian G., Nguyen V., Zhang X., Ting K., Soo C., A review of the clinical side effects of bone morphogenetic protein-2. Tissue Eng. Part B Rev. 22, 284–297 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Witte T. M., Fratila-Apachitei L. E., Zadpoor A. A., Peppas N. A., Bone tissue engineering via growth factor delivery: From scaffolds to complex matrices. Regen. Biomater. 5, 197–211 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.De la Vega R. E., Atasoy-Zeybek A., Panos J. A., Griensven M. V., Evans C. H., Balmayor E. R., Gene therapy for bone healing: Lessons learned and new approaches. Transl. Res. 236, 1–16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Evans C., De la Vega R. E., Evans C. H., van Griensven M., Balmayor E. R., Healing with RNA. Injury 50, 625–626 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sahin U., Kariko K., Tureci O., mRNA-based therapeutics—Developing a new class of drugs. Nat. Rev. Drug Discov. 13, 759–780 (2014). [DOI] [PubMed] [Google Scholar]
- 6.Carlsson L., Clarke J. C., Yen C., Gregoire F., Albery T., Billger M., Egnell A. C., Gan L. M., Jennbacken K., Johansson E., Linhardt G., Martinsson S., Sadiq M. W., Witman N., Wang Q. D., Chen C. H., Wang Y. P., Lin S., Ticho B., Hsieh P. C. H., Chien K. R., Fritsche-Danielson R., Biocompatible, purified VEGF-A mRNA improves cardiac function after intracardiac injection 1 week post-myocardial infarction in swine. Mol. Ther. Methods Clin. Dev. 9, 330–346 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ferizi M., Aneja M. K., Balmayor E. R., Badieyan Z. S., Mykhaylyk O., Rudolph C., Plank C., Human cellular CYBA UTR sequences increase mRNA translation without affecting the half-life of recombinant RNA transcripts. Sci. Rep. 6, 39149 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oh S., Kessler J. A., Design, assembly, production, and transfection of synthetic modified mRNA. Methods 133, 29–43 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trepotec Z., Aneja M. K., Geiger J., Hasenpusch G., Plank C., Rudolph C., Maximizing the translational yield of mRNA therapeutics by minimizing 5′-UTRs. Tissue Eng. Part A 25, 69–79 (2019). [DOI] [PubMed] [Google Scholar]
- 10.Kariko K., Buckstein M., Ni H., Weissman D., Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005). [DOI] [PubMed] [Google Scholar]
- 11.Kormann M. S., Hasenpusch G., Aneja M. K., Nica G., Flemmer A. W., Herber-Jonat S., Huppmann M., Mays L. E., Illenyi M., Schams A., Griese M., Bittmann I., Handgretinger R., Hartl D., Rosenecker J., Rudolph C., Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat. Biotechnol. 29, 154–157 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Zhang W., De La Vega R. E., Coenen M. J., Muller S. A., Peniche Silva C. J., Aneja M. K., Plank C., van Griensven M., Evans C. H., Balmayor E. R., An improved, chemically modified RNA encoding BMP-2 enhances osteogenesis in vitro and in vivo. Tissue Eng. Part A 25, 131–144 (2019). [DOI] [PubMed] [Google Scholar]
- 13.Balmayor E. R., Evans C. H., RNA therapeutics for tissue engineering. Tissue Eng. Part A 25, 9–11 (2019). [DOI] [PubMed] [Google Scholar]
- 14.Dolgin E., How COVID unlocked the power of RNA vaccines. Nature 589, 189–191 (2021). [DOI] [PubMed] [Google Scholar]
- 15.Wang F., Zuroske T., Watts J. K., RNA therapeutics on the rise. Nat. Rev. Drug Discov. 19, 441–442 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Jiang L., Park J. S., Yin L., Laureano R., Jacquinet E., Yang J., Liang S., Frassetto A., Zhuo J., Yan X., Zhu X., Fortucci S., Hoar K., Mihai C., Tunkey C., Presnyak V., Benenato K. E., Lukacs C. M., Martini P. G. V., Guey L. T., Dual mRNA therapy restores metabolic function in long-term studies in mice with propionic acidemia. Nat. Commun. 11, 5339 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.T. Michel, H.-P. Wendel, S. Krajewski, Next-generation therapeutics: mRNA as a novel therapeutic option for single-gene disorders, in Modern Tools for Genetic Engineering, M. Kormann, Ed. (IntechOpen, 2016). [Google Scholar]
- 18.Rizvi F., Everton E., Smith A. R., Liu H., Osota E., Beattie M., Tam Y., Pardi N., Weissman D., Gouon-Evans V., Murine liver repair via transient activation of regenerative pathways in hepatocytes using lipid nanoparticle-complexed nucleoside-modified mRNA. Nat. Commun. 12, 613 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaur K., Zangi L., Modified mRNA as a therapeutic tool for the heart. Cardiovasc. Drugs Ther. 34, 871–880 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gan L. M., Lagerstrom-Fermer M., Carlsson L. G., Arfvidsson C., Egnell A. C., Rudvik A., Kjaer M., Collen A., Thompson J. D., Joyal J., Chialda L., Koernicke T., Fuhr R., Chien K. R., Fritsche-Danielson R., Intradermal delivery of modified mRNA encoding VEGF-A in patients with type 2 diabetes. Nat. Commun. 10, 871 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lieberman J., Tapping the RNA world for therapeutics. Nat. Struct. Mol. Biol. 25, 357–364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Weissman D., mRNA transcript therapy. Expert Rev. Vaccines 14, 265–281 (2015). [DOI] [PubMed] [Google Scholar]
- 23.Gilbert W. V., Bell T. A., Schaening C., Messenger RNA modifications: Form, distribution, and function. Science 352, 1408–1412 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zara J. N., Siu R. K., Zhang X., Shen J., Ngo R., Lee M., Li W., Chiang M., Chung J., Kwak J., Wu B. M., Ting K., Soo C., High doses of bone morphogenetic protein 2 induce structurally abnormal bone and inflammation in vivo. Tissue Eng. Part A 17, 1389–1399 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Balmayor E. R., Synthetic mRNA—Emerging new class of drug for tissue regeneration. Curr. Opin. Biotechnol. 74, 8–14 (2021). [DOI] [PubMed] [Google Scholar]
- 26.Evans C. H., Stoddart M. J., Why does bone have TERM limits? Injury 47, 1159–1161 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Jarzebinska A., Pasewald T., Lambrecht J., Mykhaylyk O., Kummerling L., Beck P., Hasenpusch G., Rudolph C., Plank C., Dohmen C., A single methylene group in oligoalkylamine-based cationic polymers and lipids promotes enhanced mRNA delivery. Angew. Chem. Int. Ed. Engl. 55, 9591–9595 (2016). [DOI] [PubMed] [Google Scholar]
- 28.Schrom E., Huber M., Aneja M., Dohmen C., Emrich D., Geiger J., Hasenpusch G., Herrmann-Janson A., Kretzschmann V., Mykhailyk O., Pasewald T., Oak P., Hilgendorff A., Wohlleber D., Hoymann H. G., Schaudien D., Plank C., Rudolph C., Kubisch-Dohmen R., Translation of angiotensin-converting enzyme 2 upon liver- and lung-targeted delivery of optimized chemically modified mRNA. Mol. Ther. Nucleic Acids 7, 350–365 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jesorka A., Orwar O., Liposomes: Technologies and analytical applications. Annu. Rev. Anal. Chem. 1, 801–832 (2008). [DOI] [PubMed] [Google Scholar]
- 30.De La Vega R. E., De Padilla C. L., Trujillo M., Quirk N., Porter R. M., Evans C. H., Ferreira E., Contribution of implanted, genetically modified muscle progenitor cells expressing BMP-2 to new bone formation in a rat osseous defect. Mol. Ther. 26, 208–218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Liu F., Ferreira E., Porter R. M., Glatt V., Schinhan M., Shen Z., Randolph M. A., Kirker-Head C. A., Wehling C., Vrahas M. S., Evans C. H., Wells J. W., Rapid and reliable healing of critical size bone defects with genetically modified sheep muscle. Eur. Cell. Mater. 30, 118–131 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmitz N., Laverty S., Kraus V. B., Aigner T., Basic methods in histopathology of joint tissues. Osteoarthr. Cartil. 18 (suppl. 3), S113–S116 (2010). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials and Methods
Figs. S1 to S7
Tables S1 to S9
Movies S1 and S2