Abstract
Human intestinal spirochetosis, characterized by end-on attachment of densely packed spirochetes to the epithelial surface of the large intestines as a fringe has been associated with the weakly beta-hemolytic spirochetes Brachyspira aalborgi and Brachyspira (Serpulina) pilosicoli. In this study, fluorescent in situ hybridization with oligonucleotide probes targeting 16S or 23S rRNA of B. aalborgi, B. pilosicoli, and the genus Brachyspira was applied to 40 sections of formalin-fixed, paraffin-embedded intestinal biopsy specimens from 23 Danish and 15 Norwegian patients with histologic evidence of intestinal spirochetosis. Five biopsy specimens from patients without intestinal spirochetosis and three samples from pigs with experimental B. pilosicoli colitis were examined as well. In addition, the 16S ribosomal DNAs of two clinical isolates of B. aalborgi were sequenced, and a PCR procedure was developed for the identification of B. aalborgi in cultures. The genotypic characteristics of the two clinical isolates showed very high (99.5%) similarity with two existing isolates, the type strain of B. aalborgi and a Swedish isolate. Hybridization with the Brachyspira genus-specific probe revealed a brightly fluorescing fringe of spirochetes on the epithelia of 39 biopsy specimens, whereas 1 biopsy specimen was hybridization negative. The spirochetes in biopsy specimens from 13 Danish and 8 Norwegian patients (55.3%) were identified as B. aalborgi. The spirochetes in the biopsy specimens from the other 17 patients hybridized only with the Brachyspira probe, possibly demonstrating the involvement of as-yet-uncharacterized Brachyspira spirochetes in human intestinal spirochetosis.
Human intestinal spirochetosis (HIS) is a condition of the large intestinal mucosa microscopically characterized by colonization and extensive end-on attachment of densely packed spirochetes to the epithelial surface (16). The spirochetes are easily demonstrated in hematoxylin-eosin-stained biopsy sections as a diffuse blue fringe 3 to 6 μm thick. The colonization is usually associated with no or only minor morphologic and inflammatory reactions in the mucosa (16, 17, 23, 36). Ultrastructurally, spirochetes are attached perpendicular to the epithelial membrane between the microvilli, which appear shorter or depleted (11, 17, 23, 36). However, more severe lesions with spirochetal invasion of the epithelium and the adjacent lamina propria together with purulent discharge may occur (15, 27, 30, 42). HIS is a morphological description based on microscopic examination of biopsy samples. Hence, most reports have outlined histologic and electron microscopic findings, whereas there have been only a few reports of concurrent culturing of spirochetes from the same individuals (11, 15, 17, 36), and two species, Brachyspira aalborgi and Brachyspira (Serpulina) pilosicoli, have been identified (17, 36).
Recently, the phylogeny of intestinal spirochetes has been studied by multilocus enzyme electrophoresis (20, 21), polyacrylamide gel electrophoresis (7), pulsed-field gel electrophoresis (6, 29), and 16S rRNA gene sequencing (13); these studies have revealed considerable heterogeneity among human isolates. However, it seems that human intestinal spirochetes can be separated into at least two groups: a heterogeneous group closely related to or indistinguishable from B. pilosicoli and a second group consisting of the type strain of B. aalborgi (13, 20).
In 1982, Hovind-Hougen et al. (17) described the isolation of B. aalborgi from biopsy specimens from six patients with HIS in Aalborg, Denmark. Since then, most studies on HIS have assumed that the spirochetes involved are B. aalborgi. The second report on the isolation of B. aalborgi has only just been published (19). Distinction between intestinal spirochetes based on morphological criteria is possible using transmission electron microscopy, but the ultrastructural differences are subtle and may not be reliable diagnostic aids with biopsy samples. The involvement of B. aalborgi in HIS has recently been verified by molecular methods with biopsy specimens, with histological confirmation of spirochetal attachment (24).
B. pilosicoli is a known intestinal pathogen causing mild colitis and diarrhea in pigs (porcine spirochetal colitis) and other animals, including dogs and poultry (14, 33, 35, 40). Experimental infections in pigs are characterized by epithelial erosions and spirochetal invasion of the epithelium, while end-on attachment of spirochetes (intestinal spirochetosis [IS]) has only been found occasionally (18, 35, 40). Human isolates of B. pilosicoli are also capable of inducing colitis in pigs and chickens, like porcine isolates (37, 39). Although B. pilosicoli has previously been isolated from human stools, the involvement in HIS has just recently been documented by culturing of spirochetes from biopsy specimens from patients with HIS (36). Furthermore, B. pilosicoli has also been cultured from the blood of critically ill humans, some of whom had a history of gastrointestinal disorders (38). Thus, differentiation and specific in situ identification of the spirochetes observed in biopsy samples may be clinically important.
In addition to B. aalborgi and B. pilosicoli, other Brachyspira (Serpulina) species are Brachyspira alvinipulli, an intestinal pathogen of chickens (32); and the porcine species Brachyspira hyodysenteriae, the cause of swine dysentery, a severe mucohemorrhagic colitis (34), Brachyspira innocens, which is believed to be nonpathogenic, and Serpulina murdochii and Serpulina intermedia (31). The pathogenic potential of the latter remains unknown, whereas S. murdochii is considered to be nonpathogenic.
A number of diagnostic methods based on genotypes have been developed for the identification of Brachyspira spp. in cultures, including specific detection of B. pilosicoli by PCR targeting 16S ribosomal DNA (rDNA) (28) and specific detection of B. pilosicoli, B. hyodysenteriae, and S. intermedia by PCR targeting 23S rDNA (22). PCR methods using formalin-fixed paraffin-embedded human biopsy specimens and targeting the 16S rRNA and NADH oxidase (nox) genes of B. aalborgi and B. pilosicoli have also been described (24). While PCR assays are useful for the detection of bacteria in formalin-fixed paraffin-embedded tissue, they do not provide information on the in situ location, extension, and distribution of the organisms in the samples. On the other hand, in situ hybridization has been shown to be a reliable diagnostic tool for the specific detection of various pathogenic microorganisms in tissue samples. Detection of the genus Brachyspira, B. hyodysenteriae, and B. pilosicoli by fluorescent rRNA in situ hybridization with specific oligonucleotide probes has been developed and applied to formalin-fixed paraffin-embedded intestinal samples in studies of experimental as well as natural Brachyspira infections in pigs (9, 18; T. K. Jensen, K. Møller, G. E. Duhamel, K. K. Hansen, J. Szancer, and M. Boye, Abstr. Proc. 15th IPVS Congr., p. 58, 1998).
Using a similar approach, we have designed an oligonucleotide probe for the diagnostic identification of B. aalborgi by fluorescent in situ hybridization and applied it, together with probes for the genus Brachyspira and B. pilosicoli, to formalin-fixed paraffin-embedded biopsy specimens from patients with histologically confirmed HIS in Denmark and Norway. In addition, we report the cultural and molecular characterization of two isolates of B. aalborgi from humans and the development of a specific PCR assay for the detection of B. aalborgi in cultures.
MATERIALS AND METHODS
Microbiologic procedures and PCR. (i) Isolates.
Isolates were obtained from two patients with HIS, as confirmed by histopathologic analysis (see specimens 1 and 2 in Tables 2 and 4). Primary isolation was performed by streaking fresh biopsy specimens on prereduced tryptose soy agar with 10% bovine blood, 400 μg of spectinomycin per ml, and 5 μg of polymyxin per ml as described by Hovind-Hougen et al. (17). Subculturing was performed on fastidious anaerobe agar (FAA) with 5% bovine blood as described by Moller et al. (25). The cultures were incubated anaerobically (80% N2, 10% CO2, 10% H2) at 37°C for 3 to 4 weeks.
TABLE 2.
Polymorphic positions of B. aalborgi 16S rDNA partial gene sequences (positions 57 to 1389) (E. coli numbering)
| Strain | Residue at the following position:
|
|||||
|---|---|---|---|---|---|---|
| 236 | 239 | 630 | 637 | 1246 | 1305 | |
| 513AT | G | C | A | C | T | G |
| W1 | G | C | G | C | C | G |
| Specimen 1 strain | G | T | G | T | C | G |
| Specimen 2 strain | A | C | G | C | C | A |
TABLE 4.
Application of fluorescent in situ hybridization to the detection and identification of spirochetes in various samples
| Specimena | Patient or pig data
|
Biopsy specimen location | Hybridization result forc:
|
Pathology, diagnosis, or symptomsd | |||
|---|---|---|---|---|---|---|---|
| Sexb | Age (yr) | Brachyspira (SER 1410) | B. aalborgi (Aalborgi183) | B. pilosicoli (Pilosi209) | |||
| 1 | M | 29 | Rectum | + | + | − | IS, diarrhea |
| 2 | F | 18 | Rectum | + | + | − | IS, diarrhea |
| 3 | M | 26 | Colon | + | + | − | IS, melanosis coli, diarrhea |
| 4 | M | 29 | Rectum | + | + | − | IS, NM, upper dyspepsia |
| 5 | M | 40 | Rectum | + | + | − | IS, NM, rectal bleeding (same patient for as specimen 9) |
| 6 | M | 52 | Rectum | + | + | − | IS, rectal adenocarcinoma, obstipation, diarrhea |
| 7 | M | 47 | Rectum | + | + | − | IS, NM, chronic diarrhea |
| 8 | M | 60 | Rectum | + | + | − | IS, NM, chronic diarrhea |
| 9 | M | 40 | Rectum | + | + | − | IS, NM, rectal bleeding (same patient as for specimen 5) |
| 10 | M | 65 | Rectum | + | + | − | IS, NM, chronic diarrhea |
| 11 | F | 86 | Colon | + | + | − | IS, adenoma, chronic diarrhea |
| 12 | M | 37 | Colon | + | + | − | IS, NM, obstipation, diarrhea |
| 13 | M | 71 | Rectum | + | + | − | IS, adenoma, rectal bleeding |
| 14 | M | 53 | Rectum | + | + | − | IS, NM, chronic diarrhea |
| 15 | F | 22 | Colon | + | − | − | IS; NM; bloody, mucinous stools (same patient as for specimen 18) |
| 16 | M | 60 | Rectum | + | − | − | IS, NM, anal fistula, purulent discharge |
| 17 | F | 33 | Colon | + | − | − | IS, NM, hemorrhagic proctitis, bloody stools |
| 18 | F | 22 | Rectum | + | − | − | IS, NM, bloody mucinous stools (same patient as for specimen 15) |
| 19 | M | 33 | Rectum | + | − | − | IS, hyperplastic polyp, irritable bowel |
| 20 | F | 21 | Rectum | + | − | − | IS, solitary rectal ulcer, bloody stools |
| 21 | F | 61 | Rectum | + | − | − | IS, NM, abdominal pain |
| 22 | M | 62 | Rectum | + | − | − | IS, hyperplastic polyp, diarrhea |
| 23 | F | 17 | Rectum | + | − | − | IS, NM, irritable bowel |
| 24 | M | 26 | Rectum | + | − | − | IS, NM, irritable bowel |
| 25 | F | 37 | Colon | + | − | − | IS, NM, diarrhea |
| 26 | M | 31 | Rectum | − | − | − | NIS, NM, negative control |
| 27 | M | 29 | Rectum | − | − | − | NIS, NM, negative control |
| 28 | F | 29 | Rectum | − | − | − | NIS, NM, negative control |
| 29 | M | 53 | Rectum | − | − | − | NIS, NM, negative control |
| 30 | F | 27 | Rectum | − | − | − | NIS, NM, negative control |
| 31 | M | 70 | Rectum | + | + | − | IS, NM, rectal adenocarcinoma |
| 32 | F | 55 | Colon | + | + | − | IS, NM, rectal adenocarcinoma |
| 33 | M | 44 | Rectum | + | + | − | IS, proctitis |
| 34 | M | 69 | Sigmoid | + | + | − | IS, tubular adenoma |
| 35 | M | 80 | Colon | + | + | − | IS, NM, diverticulitis with perforation |
| 36 | M | 65 | Colon | + | + | − | IS, NM, colonic adenocarcinoma |
| 37 | M | 29 | Colon | + | + | − | IS, NM, diarrhea |
| 38 | M | 70 | Rectum | + | + | − | IS, NM, rectal adenocarcinoma |
| 39 | F | 76 | Colon | + | − | − | IS, NM, colonic adenocarcinoma |
| 40 | F | 37 | Colon | − | − | − | IS, NM, colitis with perforation |
| 41 | M | 69 | Rectum | + | − | − | IS, NM, colitis |
| 42 | F | 33 | Appendix | + | − | − | IS, NM, mesenteric lymphadenopathy |
| 43 | F | 37 | Colon | + | − | − | IS, NM, ulcerous colitis |
| 44 | M | 36 | Colon | + | − | − | IS, NM, colitis, diarrhea |
| 45 | F | 71 | Colon | + | − | − | IS, NM, ulcerous colitis |
| 46 | Pig 1 | Colon | + | − | + | IS, spirochetal colitis, diarrhea | |
| 47 | Pig 2 | Colon | + | − | + | IS, spirochetal colitis, diarrhea | |
| 48 | Pig 3 | Colon | + | − | + | IS, spirochetal colitis, diarrhea | |
Biopsy specimens 1 to 30 were obtained from Danish patients, and specimens 31 to 45 were obtained from Norwegian patients. Specimens were from 38 HIS-positive patients, 5 HIS-negative patients, and 3 pigs with experimental B. pilosicoli IS.
M, male; F, female.
Probes specific for the organism are shown in parentheses. +, hybridization; −, no hybridization.
NM, no mucosal lesions in the examined section; NIS, no signs of IS.
(ii) Sequencing.
Partial 16S rDNA sequences (from positions 57 to 1389; E. coli numbering) of B. aalborgi 513AT and of each of the two colony types of the two clinical isolates were obtained as previously described (5). These sequences were aligned with the sequence of B. aalborgi 513AT (GenBank accession number Z22781) and the sequence described by Kraaz et al. (19) as W1 (GenBank accession number AF200693).
(iii) PCR.
Prior to PCR, B. aalborgi cells were lysed by boiling. Approximately 50 colonies were transferred to 100 μl of phosphate-buffered saline, boiled for 10 min, and centrifuged at 10,000 × g for 20 s; the supernatant was diluted 1:100 in Milli-Q-water (Millipore, Molsheim, France). Primer sequences used to specifically detect the presence of the 16S rDNA gene of B. aalborgi were identified on the basis of the GenBank sequence database and the sequences obtained in this study. Primers 5′GCATATACTCTTGACGCTA′3 and 3′TGTTCTTCTCGATATCTATA′5, obtained from DNA Technology (Aarhus, Denmark), were used in a PCR assay generating a 520-bp fragment. The amplification mixtures (50 μl) consisted of 0.2 μM each primer, 100 μM each nucleotide, 2.5 mM MgCl2, 0.5 U of Taq polymerase (Bethesda Research Laboratories), and 2 μl of boiled and diluted specimen. The thermocycling program was as follows: denaturation at 94°C for 3 min; 35 cycles at 94°C for 1 min, 52°C for 1 min, and 72°C for 1 min; and extension at 72°C for 10 min. The PCR products were electrophoresed on agarose gels and visualized by ethidium bromide staining.
In situ hybridization. (i) Oligonucleotide probes.
A specific oligonucleotide probe for B. aalborgi was selected by using the function Probe Design in ARB software (http://www.mikro.biologie.tu-muenchen.de).
The oligonucleotide probes used in this study are listed in Table 1. The probes, synthesized by standard phosphoramidite chemistry, were purchased from Hobolth DNA Syntese, Hillerod, Denmark. The oligonucleotides were 5′ labeled with an aminohexyl linker, conjugated to fluorescein isothiocyanate (Peninsula Laboratories, Inc., Belmont, Calif.), and purified by reverse-phase chromatography.
TABLE 1.
Sequences, target sites (Escherichia coli numbering), and specificities of rRNA-targeted oligonucleotide probes used for in situ hybridization in this study
| Probe name | Systematic namea | Probe sequence (5′→3′) | Target (positions) | Specificity |
|---|---|---|---|---|
| Aalborgi183 | S-S-B.aalb-0183-a-A-18 | CTACGCTTTAGCGTCAAG | 16S (183–200) | B. aalborgi |
| EUB338b | S-D-bact-0338-a-A-18 | GCTGCCTCCCGTAGGAGT | 16S (338–355) | Domain Bacteria |
| SER1410c | L-G-Serp-1410-a-A-19 | GTCATTCCATCGAAACATA | 23S (1410–1428) | Genus Brachyspira |
| Hyo1210c | L-S-S.hyo-1210-a-A-19 | CTCACGATGAACCTTCGAC | 23S (1210–1228) | B. hyodysenteriae |
| Pilosi209c | S-S-S.pilo-0209-a-A-18 | GCTCATCGTGAAGCGAAA | 16S (209–226) | B. pilosicoli |
| NON-EUB338b | S-∗-non-0338-a-S-18 | CGACGGAGGGCATCCTCA | Nonspecific control probe |
(ii) Biopsy specimens.
Formalin-fixed paraffin-embedded biopsy specimens (n = 25) of the large intestine (colon and rectum) from 23 patients with a histopathologic diagnosis of HIS (presence of the characteristic, 3- to 6-μm-thick bluish fringe in hematoxylin-eosin-stained sections) were obtained from the files of the Institute of Pathology, Aalborg Hospital, Aalborg, Denmark, the hospital from which B. aalborgi was originally reported. Similarly, biopsy specimens from five patients without the characteristic bluish fringe on the large intestinal epithelium were included as HIS-negative controls. Biopsy specimens 1 and 2 were taken from the same patients as the two isolates of B. aalborgi.
Additionally, 15 biopsy specimens from patients with a histopathologic diagnosis of HIS in Vest-Agder Sentralsykehus, Kristiansand, Norway, were included (see Table 4 for sex, age, symptoms, and histopathologic diagnosis). Five of these biopsy specimens (31, 32, 35, 39, and 40) had previously been examined by PCR for the presence of B. aalborgi and B. pilosicoli by Mikosza et al. (24).
For comparison, colon samples from three pigs with experimental B. pilosicoli IS were included (T. K. Jensen, unpublished data). The pigs had been inoculated with B. pilosicoli as described previously (Jensen et al., 15th IPVS Congr.). Sections (3 μm thick) of the samples were mounted on Super Frost∗/plus slides (Menzel-Gläser, Braunschweig, Germany) and kept at 4°C until use. After epifluorescence microscopy, the sections were stained with hematoxylin-eosin and reexamined by light microscopy.
(iii) Hybridization of formalin-fixed tissue sections.
Prior to hybridization, paraffin was removed from the sections by use of xylene, and the sections were transferred to 96% ethanol and kept there for 10 min. Before the hybridization solution was applied, the sections were circumscribed with a hydrophobic PAP-pen (Daido Sangyo Co. Ltd., Tokyo, Japan). Hybridization was carried out at 37°C with 20 μl of hybridization buffer (100 mM Tris [pH 7.2], 0.9 M NaCl, 0.1% sodium dodecyl sulfate) and 100 ng of probe for 16 h in a moisture chamber. The samples were washed in 100 ml of prewarmed (37°C) hybridization buffer for 15 min and subsequently in 100 ml of prewarmed (37°C) washing solution (100 mM Tris [pH 7.2], 0.9 M NaCl) for 15 min. The samples were rinsed in water, air dried, and mounted in Vectashield (Vector Laboratories Inc., Burlingame, Calif.) for fluorescence microscopy.
(iv) Fixation of whole bacterial cells and whole-cell hybridization.
Cultured cells were fixed in 4% paraformaldehyde as described earlier (2). Fixed cells were stored at −20°C until use. Before hybridization, cells were bound to six-well poly-l-lysine (Sigma Chemical Co., St. Louis, Mo.) Teflon-coated slides (NovaKemi AB, Enskede, Sweden) and dried by sequential washes in 50, 80, and 100% ethanol (3 min each). Hybridization was carried out as described above for in situ hybridization of tissue sections using 10 μl of hybridization buffer and 50 ng of probe per well.
(v) Epifluorescence microscopy.
An Axioplan 2 epifluorescence microscope (Carl Zeiss, Oberkochen, Germany) equipped for epifluorescence with a 75-W xenon lamp and filter set XF23 (Omega Optical, Brattleboro, Vt.) was used to visualize fluorescein. For obtaining a more differentiated background, we used double-filter set XF53 (Omega Optical) for simultaneous detection of red and green fluorescence. Micrographs were taken with a Carl Zeiss MC200 camera using Kodak Elite Ektachrome 400 film.
Nucleotide sequence accession numbers.
The partial 16S rDNA sequences of the two clinical isolates (from specimens 1 and 2) have been deposited in GenBank under accession numbers AF395882 and AF395883.
RESULTS
Culturing of spirochetes.
After 2 to 3 weeks of incubation of tryptase soy agar plates, a thin haze of pinpoint colonies could be seen mixed with other bacterial colonies. The pinpoint colonies were transferred to FAA plates every 2 to 3 weeks until pure. As previously described by Hovind-Hougen et al. (17), two types of colonies were observed after three or four subcultures. Type A consisted of flat, rough-edged colonies with weak hemolytic activity, and type B consisted of smooth-edged, pinpoint colonies without hemolytic activity. The colony types remained stable on FAA plates. Types A and B were examined by dark-field microscopy, and both types showed highly motile, comma-shaped, helical cells.
PCR and sequence analysis of the 16S rDNA gene.
PCR products of the expected size of 520 bp were obtained from type A and B colonies of the two clinical isolates. In contrast, no amplification product was obtained from B78T (B. hyodysenteriae), P43T (B. pilosicoli), 256T (B. innocens), or 56-150T (S. murdochii).
The partial 16S rDNA sequences showed total identity between type A and B colonies from the same patient. Six polymorphic positions were identified among isolates from specimens 1 and 2, 513AT, and W1, corresponding to more than 99.5% nucleotide similarity (Table 2). By sequencing 513AT, we found guanosine residues at positions 1089, 1094, and 1388 (E. coli numbering), where information formerly has been lacking. We found a G at position 1099. According to GenBank, 513AT has a C at this position.
In situ hybridization.
A specific oligonucleotide probe was designed for B. aalborgi and given the systematic name S-S- B.aalb-0183-a-A-18, according to Oligonucleotide Probe Database nomenclature (1), but for simplicity the probe was named Aalborgi183. It has a perfect match to the four 16S rRNA sequences available for B. aalborgi. The probe sequence and its comparison with sequences of other members of the genus Brachyspira are given in Table 3. The probe Aalborgi183 has eight mismatches to other species of the genus and at least four mismatches to all other bacteria in the ARB database (currently 10,073 species).
TABLE 3.
Sequence comparisons (E. coli numbering 183–200)a
| Probe or strain | Sequence |
|---|---|
| Aalborgi183 | 3′-GAACTGCGATTTCGCATC-5′ |
| B. aalborgi ATCC 43994 | 5′-CTTGACGCTAAAGCGTAG-3′ |
| B. hyodysenteriae ATCC 27164 | ....CTA.AT...TAG.. |
| S. intermedia ATCC 51140 | ....CTA.AT...TAG.. |
| S. murdochii ATCC 51284 | ....CTA.AC...TAG.. |
| B. innocens ATCC 29796 | ....CTA.AC...TAG.. |
| B. pilosicoli ATCC 51139 | ....CTA.AT...TAG.. |
Pairing is represented by dots, and mismatches within the target site are indicated.
Probe Aalborgi183 was tested with the two clinical isolates of B. aalborgi as well as the reference strain B. aalborgi ATCC 43994; it was able to bind to the 16S rRNA of B. aalborgi, and the ribosomal content in the cells was sufficient to result in a reasonable signal. Similarly, probe SER1410 gave a positive signal for the clinical isolates and the reference strain. Additionally, it also gave a positive signal when hybridized with the B. alvinipulli type strain (M. Boye and T. K. Jensen, unpublished data), verifying that the probe hybridizes with all known species of the genus Brachyspira (9). The probes specific for B. pilosicoli and B. hyodysenteriae, Pilosi209 and Hyo1210, respectively, gave a negative signal when hybridized with the same isolates.
We applied the different probes to 40 biopsy specimens from patients with HIS, 5 specimens from patients without IS, and colon samples from three pigs with experimental B. pilosicoli infection. Fluorescent in situ hybridization using general bacterial probe EUB338 showed morphologically different microorganisms in all the biopsy specimens, with one exception; biopsy specimen 40 failed to hybridize with any of the applied probes, although a fringe of spirochetes was observed on the epithelial surface (the negative hybridization could have been caused by a processing error before paraffin embedding). The hybridization results for the genus Brachyspira, B. aalborgi, and B. pilosicoli are summarized in Table 4. Biopsy specimens 5 and 9 were taken from the same patient at an interval of 5 months. Similarly, biopsy specimens 15 and 18 were taken from the same patient at an interval of 8 weeks. Application of the Brachyspira-specific probe SER1410 gave a positive signal for all IS-positive biopsy specimens (except for biopsy specimen 40). The positive reaction revealed the spirochetes as a bright green fringe on the epithelial surface and occasionally a little further into the crypts (Fig. 1A). The fringe, 3 to 5 μm high, appeared uniform and without any distinction of a single spirochete, despite being observed at high magnifications. In most cases, the fringe covered the whole surface epithelium of the biopsy specimen, leaving empty goblet cells unaffected. However, single and sparsely packed end-on-attached spirochetes were seen between areas with IS in some biopsy specimens. The epithelial cells colonized by spirochetes and the underlying mucosa usually appeared normal, without any morphological signs of damage or cellular inflammation.
FIG. 1.
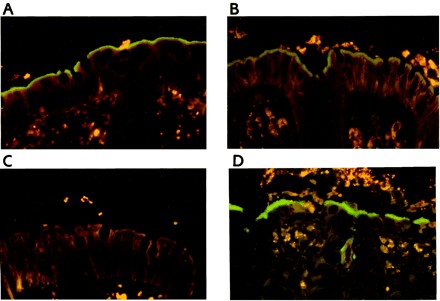
Fluorescent in situ hybridization for identification of intestinal spirochetes in formalin-fixed biopsy specimens. (A) Colon biopsy specimen from a case of HIS. Spirochetes (green) are visualized as a fringe on the luminal epithelium. Hybridization was done with the genus Brachyspira-specific probe SER1410 (fluorescein labeled) and biopsy specimen 15 (negative for B. aalborgi). The mucosal tissue is stained dark orange, whereas erythrocytes and leukocytes are bright yellow due to autofluorescence. Magnification, ×380 (original magnification, ×400, red-green filter set). (B) Specific identification of B. aalborgi in a colon biopsy specimen with HIS. The B. aalborgi organisms are seen as a green fringe on the luminal epithelium and further into the crypt opening. The dark profiles in the epithelium represent goblet cells. Fluorescein-labeled probe Aalborgi183 was used with biopsy specimen 37. Magnification, ×380 (original magnification, ×400, red-green filter set). (C) Negative staining of biopsy specimen 15 (same as panel A) from a case of HIS after hybridization with the B. aalborgi-specific probe Aalborgi183. The spirochetes are seen as a weakly autofluorescing fringe on the luminal epithelium. Magnification, ×380 (original magnification, ×400, red-green filter set). (D) Specific identification of B. pilosicoli in a colon specimen from a pig with experimental IS. The B. pilosicoli organisms are visualized as a green fringe on the luminal epithelium and in the crypts. Fluorescein-labeled probe Pilosi209 was used with biopsy specimen 47. Magnification, ×380 (original magnification, ×400, red-green filter set).
A similar positive reaction with visualization of a bright green fringe was observed with the probe specific for B. aalborgi (Fig. 1B) in 22 of the IS-positive biopsy specimens, whereas the remaining 17 biopsy specimens were all negative. Morphological differences in spirochetal colonization between B. aalborgi-positive and -negative samples were not observed. Application of the probe specific for B. pilosicoli, Pilosi209, to the biopsy specimens failed to give any positive results. Likewise, hybridization results with the B. hyodysenteriae-specific probe were negative. The presence of spirochetes in the B. aalborgi hybridization-negative biopsy sections was confirmed by a shadow of a fringe on the surface of the epithelium for all IS cases (Fig. 1C). In the IS-negative control biopsy specimens, spirochetes were not revealed by any of the probes. Control probing with the nonspecific probe EUB338 did not result in any specific hybridization signal, indicating that there was no nonspecific staining of microorganisms or tissue.
Application of the probes SER1410 and Pilosi209 to the colon samples from the pigs with experimental B. pilosicoli infection gave similar positive reactions, whereas both probes Aalborgi183 and Hyo1210 gave negative results. B. pilosicoli cells were found colonizing and invading the surface epithelium and the crypts. IS, represented by a fringe 4 to 6 μm thick, was evident in multiple areas on the luminal surface and occasionally on the crypt epithelium (Fig. 1D). HE-stained sections of the large intestines of three pigs were characterized by colitis, with multifocal erosions and sloughing of epithelial cells, slight edema, and an infiltrate of mononuclear cells in the lamina propria. Although the epithelial surface of the human mucosa was covered by IS, the underlying mucosa usually appeared normal in HE-sections, without any morphological signs of damage or cellular infiltration that could be associated with the spirochetes. A few cases showed a mononuclear infiltrate in the lamina propria.
DISCUSSION
This study is the second one since B. aalborgi was originally reported in 1982 describing reisolation of the organism. The new isolates were cultured from two patients at the same Danish regional hospital as in the original report by Hovind-Hougen et al. (17). Characterization of the two isolates confirmed the phenotypic and genotypic properties of the species B. aalborgi (the type strain and the newly described Swedish isolate, W1) (17, 19, 26). The identity of the 16S rDNA sequences between colony types A and B indicates the existence of a substantial heterogeneity among B. aalborgi isolates that might not be recognized by sequencing the ribosomal genes. Several other Brachyspira species show only minimal differences between 16S and 23S rDNA genes (22). Thus, to detect the extent of heterogeneity among types A and B, other genotyping methods such as DNA-DNA hybridization should be applied.
By aligning sequences of partial 16S rDNA of the two clinical isolates, the type strain (513AT), and the sequence of W1 from GenBank, a total of six polymorphic positions were identified (Table 2). Only A-G or C-T substitutions were recognized. Primer positions for amplification of parts of the 16S rDNA in two former publications (19, 24) as well as primer and probe positions in this study have all been chosen in areas outside these polymorphic sites.
As phenotypic characterization of intestinal spirochetes is a tedious procedure we designed specific PCR primers based on sequencing of the 16S rDNA of B. aalborgi. The PCR was tested on extracted DNA from cultures of selected Brachyspira species and was found to be specific for B. aalborgi.
In addition, this study is the first to demonstrate the applicability of fluorescent in situ hybridization for diagnostic identification of spirochaetes involved in HIS. The probe, Aalborgi183 targeting 16S rRNA of B. aalborgi hybridized with the type strain, the two new isolates verified genotypically, and spirochaetes in 22 of 40 formalin-fixed biopsies with HIS, including the biopsies from the 2 patients from which the new isolates were cultured. The remaining 17 biopsies showed no reaction with either Aalborgi183 or Pilosi209 probe. A similar prevalence (62%) of B. aalborgi in formalin-fixed biopsies with HIS has been found by PCR amplification of 16S rRNA and NADH oxidase genes (24). Mikosza et al. (24) failed to return a positive B. aalborgi PCR in 6 out of 16 biopsies. This could indicate involvement of as-yet-uncharacterized spirochaetes in HIS. However, a negative PCR result could also be explained by insufficient amount or probably too damaged specific DNA or due to a simple lack of the organism in the sample. Five of the Norwegian biopsies from the study of Mikosza et al. (24) were also examined in this study. Two of them (31 and 35) were positive for B. aalborgi, the other two were negative (39 and 40) while one (32) was positive for B. aalborgi by fluorescent in situ hybridization but negative by PCR. The negative B. aalborgi PCR in the latter biopsy specimen could probably be due to insufficient amount of DNA since the proportion of spirochete-infected mucosa compared to the total section size was very small.
Weak orange autofluorescence confirmed the presence of spirochetes in the B. aalborgi negative sections when hybridized with Aalborgi183. The autofluorescence was clearly distinct from the specific signal. The B. aalborgi negative spirochetes, however, hybridized with the probe SER1410 as intensively as the B. aalborgi positive biopsy specimens indicating that the failure of hybridization with Aalborgi183 was not due to insufficient amounts of ribosomes in the spirochaetes or inaccessibility of the probe but reflected nucleotide differenties in the target site.
The probe SER1410, designed to target 23S rRNA of the porcine Brachyspira spp. (9), also hybridized with B. aalborgi and the avian spirochete B. alvinipulli indicating that the probe hybridized with all known members of the genus Brachyspira. Thus, we believe that the uncharacterized spirochetes belong to the genus Brachyspira and would like to propose the name Brachyspira christiani (after the Danish-Norwegian King Christian 4, founder of the city Kristiansand). The spirochetes, however, also belong to the species B. aalborgi or B. pilosicoli but differ in the rRNA sequence in the target area of the two probes. While only four sequences and three isolates of B. aalborgi were available for testing the probe Aalborgi183, the probe Pilosi209 has been tested on many samples of B. pilosicoli (9).
Since the uncharacterized spirochetes seem to be difficult to culture direct PCR amplification of DNA from biopsy specimens for sequencing may be included in future studies for identification of the organisms.
We were unable to morphologically differentiate between B. aalborgi and the uncharacterized spirochetes by light microscopy. Even ultrastructurally the differentiation between intestinal spirochetes is difficult e.g. B. aalborgi, is 2 to 7 μm long and has 4 to 5 periplasmic flagella inserted at each end (17, 26) whereas B. pilosicoli is 5 to 10 μm and has 5 to 7 periplasmic flagella (26, 40). Several isolates of human intestinal spirochetes assigned to be B. pilosicoli have, however, been reported to possess only four to six periplasmic flagella (21, 36). This could further support the hypothesis that species of spirochaetes other than B. aalborgi and B. pilosicoli may be involved in HIS.
Fluorescent in situ hybridization of whole cells with rRNA-targeted oligonucleotide probes has become a highly valuable tool for specific detection and identification of microorganisms without cultivation (3, 12). Nonculturable bacteria have also been identified in their natural environments as endosymbionts or intestinal pathogens in pigs and in activated sludge (4, 8, 41). RNA is naturally amplified in growing cells e.g., an exponentially growing E. coli cell contains 104 to 105 copies of 5S, 16S, and 23S rRNAs per cell (10). This means that a considerable increase in sensitivity can be achieved by targeting rRNA instead of DNA.
Due to its isolation from human stools, colonization of the gastrointestinal tract by B. pilosicoli has been known for some time (20), although it has not been associated with HIS until recently. Trivett-Moore et al. (36) cultured B. pilosicoli from 11 out of 22 rectal biopsy specimens with microscopically confirmed HIS from homosexual men attending a sexual clinic in Sydney, Australia. The biopsies were characterized by the absence of cellular inflammatory reactions and spirochetal invasion of the epithelium. Only a mild loss of microvilli was observed by transmission electron microscopy, and invasive capacities of the spirochetes were not revealed. Morphologic differences between the spirochetes in the culture positive and the culture-negative biopsy specimens were not reported. Thus, the spirochetes in the culture-negative biopsy specimens could have been other than B. pilosicoli. As shown in the present study, B. pilosicoli is clearly visualized by fluorescent in situ hybridization in formalin-fixed tissue. The presence of intestinal spirochaetosis in pigs due to both human and porcine derived B. pilosicoli is associated with evident colitis with epithelial erosion (18, 35, 37, 40). Similarly, the IS in the experimentally B. pilosicoli infected pigs was accompanied by evident colitis. Comparison of the spirochetes in humans and pigs showed that the fringe of B. pilosicoli in pigs was taller than that caused in humans by B. aalborgi as well as the other spirochetes. Furthermore, B. pilosicoli appeared to be invasive in the pigs whereas we were not able to reveal spirochetal invasion in the human biopsies by fluorescent in situ hybridization. Epithelial and subepithelial invasion by single spirochaetes, however, was revealed in the biopsy specimens 1 and 2 by immunohistochemistry (P. S. Teglbjærg, unpublished data).
In the present study we detected only one spirochete in each biopsy, hence species differentiation in the sections was not essential. Simultaneous demonstration of coexisting spirochetal infections is, however, possible by concurrent hybridizing with two different fluorochrome (fluorescein and CY3)-labeled probes (Jensen et al., 15th IPVS Congr.).
The study by Mikosza et al. (24) included B. aalborgi-positive biopsy specimens from Norway, the United States, and Australia, suggesting a wide distribution of the organism. A similar distribution is suggested for the uncharacterized spirochetes reported from Denmark, Norway, and possibly Australia.
Although the occurrence of HIS has been associated with a variety of gastrointestinal disorders such as longstanding diarrhea, the physiologic and pathologic significance of the spirochetal attachment is uncertain. In the survey by Lindboe et al. (23), 8 out of 30 patients revealed HIS in biopsy specimens from multiple locations indicating that the spirochetes were widely distributed throughout the large intestines. Thus, in such cases a fringe of densely packed spirochetes covering large areas of the large intestines may simply act as a physical barrier preventing the absorption of liquid and thereby inducing diarrhea. Depletion or loss of microvilli has commonly been observed in HIS by transmission electron microscopy that could further decrease the large intestinal absorption capacity.
Reexamination of two patients with a few weeks interval and identification of the same spirochete as in the first biopsy, suggests that persistent infection may occur with both B. aalborgi and the uncharacterized spirochete.
In conclusion, this study shows the applicability of fluorescent in situ hybridization with oligonucleotide probes for diagnostic detection and identification of pathogens in their natural environment. Hybridization with probes targeting 16S or 23S rRNA of B. aalborgi, B. pilosicoli, and genus Brachyspira was applied on 40 formalin-fixed, paraffin-embedded intestinal biopsy specimens from Danish and Norwegian patients with histologic evidence of HIS. B. aalborgi was identified in 22 (55%) of the biopsy specimens. The spirochetes in the biopsy specimens from 17 other patients were negative for B. pilosicoli but hybridized with the genus Brachyspira-specific probe, suggesting involvement of as-yet-uncharacterized Brachyspira spirochetes in HIS.
REFERENCES
- 1.Alm E W, Oerther D B, Larsen N, Stahl D A, Raskin L. The oligonucleotide probe database. Appl Environ Microbiol. 1996;62:3557–3559. doi: 10.1128/aem.62.10.3557-3559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amann R I, Binder B J, Olson R J, Chrisholn S W, Devereux R, Stahl D A. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl Environ Microbiol. 1990;56:1919–1925. doi: 10.1128/aem.56.6.1919-1925.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amann R I, Ludwig W, Schleifer K H. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev. 1995;59:143–169. doi: 10.1128/mr.59.1.143-169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Amann R I, Springer N, Ludwig W, Görtz H D, Schleifer K H. Identification in situ and phylogeny of uncultured bacterial endosymbionts. Nature. 1991;351:161–164. doi: 10.1038/351161a0. [DOI] [PubMed] [Google Scholar]
- 5.Angen o, Ahrens P, Tegtmeier C. Development of a species-specific PCR test for identification of [Haemophilus] somnus in pure and mixed cultures. Vet Microbiol. 1998;63:39–48. doi: 10.1016/s0378-1135(98)00222-3. [DOI] [PubMed] [Google Scholar]
- 6.Atyeo R F, Oxberry S L, Hampson D J. Pulsed-field gel electrophoresis for sub-specific differentiation of Serpulina pilosicoli (formerly “Anguillina coli”) FEMS Microbiol Lett. 1996;141:77–81. doi: 10.1111/j.1574-6968.1996.tb08366.x. [DOI] [PubMed] [Google Scholar]
- 7.Barrett S P, Holton J J, Hookey J V, Costas M, Ganner M, Mundy R, Wright D J. Heterogenecity of human intestinal spirochaetes demonstrated by one-dimensional polyacrylamide gel electrophoresis of proteins visualised by (35)S-methionine labelling and Coomassie blue staining. J Med Microbiol. 1996;45:6–9. doi: 10.1099/00222615-45-1-6. [DOI] [PubMed] [Google Scholar]
- 8.Boye M, Jensen T K, Moller K, Leser T D, Jorsal S E. Specific detection of Lawsonia intracellularis in porcine enteropathy inferred from fluorescent rRNA in situ hybridization. Vet Pathol. 1998;35:153–156. doi: 10.1177/030098589803500212. [DOI] [PubMed] [Google Scholar]
- 9.Boye M, Jensen T K, Moller K, Leser T D, Jorsal S E. Specific detection of the genus Serpulina, Serpulina hyodysenteriae and Serpulina pilosicoli in porcine intestines by fluorescent rRNA in situ hybridization. Mol Cell Probes. 1998;12:323–330. doi: 10.1006/mcpr.1998.0193. [DOI] [PubMed] [Google Scholar]
- 10.Bremer H, Dennis P P. Modulation of chemical composition and other parameters of the cell by growth rate. In: Neidhardt F C, Ingraham J L, Low K B, Magasanik B, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella typhimurium: cellular and molecular biology. Washington, D.C.: American Society for Microbiology; 1987. pp. 1527–1542. [Google Scholar]
- 11.Cooper C, Cotton D W K, Hudson M J, Kirkham N, Wilmott F E. Rectal spirochaetosis in homosexual men: characterization of the organism and pathophysiology. Genitourin Med. 1986;62:47–52. doi: 10.1136/sti.62.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.DeLong E F, Wickham G S, Pace N R. Phylogenetic strains: ribosomal RNA-based probes for the identification of single cells. Science. 1989;243:1360–1363. doi: 10.1126/science.2466341. [DOI] [PubMed] [Google Scholar]
- 13.De Smet K A L, Worth D E, Barrett S P. Variation among human isolates of Brachyspira (Serpulina) pilosicoli based on biochemical characterization and 16S rRNA gene sequencing. Int J Syst Bacteriol. 1998;48:1257–1263. doi: 10.1099/00207713-48-4-1257. [DOI] [PubMed] [Google Scholar]
- 14.Duhamel G E, Trott D J, Muniappa N, Mathiesen M R, Tarasuik K, Lee J I, Hampson D J. Canine intestinal spirochetes consist of Serpulina pilosicoli and a newly identified group provisionally designated “Serpulina canis” sp. nov. J Clin Microbiol. 1998;36:2264–2270. doi: 10.1128/jcm.36.8.2264-2270.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gebbers J O, Marder H P. Unusual in vitro formation of cyst-like structures associated with human intestinal spirochaetosis. Eur J Clin Microbiol Infect Dis. 1989;8:302–306. doi: 10.1007/BF01963456. [DOI] [PubMed] [Google Scholar]
- 16.Harland W A, Lee F D. Intestinal spirochaetosis. Br Med J. 1967;3:708–709. doi: 10.1136/bmj.3.5567.718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hovind-Hougen K, Birch-Andersen A, Henrik-Nielsen R, Orholm M, Pedersen J O, Teglbjærg P S, Thaysen E H. Intestinal spirochetosis: morphological characterization and cultivation of the spirochete Brachyspira aalborgi gen. nov., sp. nov. J Clin Microbiol. 1982;16:1127–1136. doi: 10.1128/jcm.16.6.1127-1136.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jensen T K, Moller K, Boye M, Leser T D, Jorsal S E. Scanning electron microscopy and fluorescent in situ hybridization of experimental Brachyspira pilosicoli infection in growing pigs. Vet Pathol. 2000;37:22–32. doi: 10.1354/vp.37-1-22. [DOI] [PubMed] [Google Scholar]
- 19.Kraaz W, Petterson B, Thunberg U, Engstrand L, Fellström C. Brachyspira alborgi infection diagnosed by culture and 16S ribosomal DNA sequencing using human colonic biopsy specimens. J Clin Microbiol. 2000;38:3555–3560. doi: 10.1128/jcm.38.10.3555-3560.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee J I, Hampson D J. Genetic characterisation of intestinal spirochaetes and their association with disease. J Med Microbiol. 1994;40:365–371. doi: 10.1099/00222615-40-5-365. [DOI] [PubMed] [Google Scholar]
- 21.Lee J I, McLaren A J, Lymbery A J, Hampson D J. Human intestinal spirochetes are distinct from Serpulina hyodysenteriae. J Clin Microbiol. 1993;31:16–21. doi: 10.1128/jcm.31.1.16-21.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leser T D, Moller K, Jensen T K, Jorsal S E. Specific detection of Serpulina hyodysenteriae and pathogenic weakly β-hemolytic porcine intestinal spirochetes by polymerase chain reaction targeting 23S rDNA. Mol Cell Probes. 1997;11:363–372. doi: 10.1006/mcpr.1997.0129. [DOI] [PubMed] [Google Scholar]
- 23.Lindboe C F, Tostrup N E, Nersund R, Rekkavik G. Human intestinal spirochaetosis in mid-Norway. APMIS. 1993;101:858–864. [PubMed] [Google Scholar]
- 24.Mikosza A S J, La T, Brooke C J, Lindboe C F, Ward P B, Heine R G, Guccion J G, de Boer W B, Hampson D J. PCR amplification from fixed tissue indicates frequent involvement of Brachyspira aalborgi in human intestinal spirochetosis. J Clin Microbiol. 1999;37:2093–2098. doi: 10.1128/jcm.37.6.2093-2098.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Moller K, Jensen T K, Jorsal S E, Leser T D, Carstensen B. Detection of Lawsonia intracellularis, Serpulina hyodysenteriae, weakly beta-haemolytic spirochaetes, Salmonella enterica, and Escherichia coli from swine herds with and without diarrhea in growing pigs. Vet Microbiol. 1998;62:59–72. doi: 10.1016/s0378-1135(98)00199-0. [DOI] [PubMed] [Google Scholar]
- 26.Ochiai S, Adachi Y, Mori K. Unification of the genera Serpulina and Brachyspira, and proposals of Brachyspira hyodysenteriae comb. nov., Brachyspira innocens comb. nov., and Brachyspira pilosicoli comb. nov. Microbiol Immunol. 1997;41:445–452. doi: 10.1111/j.1348-0421.1997.tb01877.x. [DOI] [PubMed] [Google Scholar]
- 27.Padmanabhan V, Dahlstrom J, Maxwell L, Kaye G, Clarke A, Barratt P J. Invasive intestinal spirochetosis: a report of three cases. Pathology. 1996;28:283–286. doi: 10.1080/00313029600169174. [DOI] [PubMed] [Google Scholar]
- 28.Park N Y, Chung C Y, McLaren A J, Atyeo R F, Hampson D J. Polymerase chain reaction for identification of human and porcine spirochaetes recovered from cases of spirochaetosis. FEMS Microbiol Lett. 1995;125:225–230. doi: 10.1111/j.1574-6968.1995.tb07362.x. [DOI] [PubMed] [Google Scholar]
- 29.Rayment S J, Barrett S P, Livesley M A. Sub-specific differentiation of intestinal spirochaetes isolates by macrorestriction fragment profiling. Microbiology. 1997;143:2923–2929. doi: 10.1099/00221287-143-9-2923. [DOI] [PubMed] [Google Scholar]
- 30.Rodgers F G, Rodgers C, Shelton A P, Hawkey C J. Proposed pathogenic mechanism for the diarrhea associated with human intestinal spirochetes. Am J Clin Pathol. 1986;86:679–682. doi: 10.1093/ajcp/86.5.679. [DOI] [PubMed] [Google Scholar]
- 31.Stanton T B. Physiology of ruminal and intestinal spirochaetes. In: Hampson D J, Stanton T B, editors. Intestinal spirochaetes in domestic animals and humans. Wallingford, United Kingdom: CAB International; 1997. pp. 7–46. [Google Scholar]
- 32.Stanton T B, Postic D, Jensen N S. Serpulina alvinipulli sp.nov., a new Serpulina species that is enteropathogenic for chickens. Int J Syst Bacteriol. 1998;48:669–676. doi: 10.1099/00207713-48-3-669. [DOI] [PubMed] [Google Scholar]
- 33.Swayne D E, McLaren A J. Avian intestinal spirochaetes and avian intestinal spirochaetosis. In: Hampson D J, Stanton T B, editors. Intestinal spirochaetes in domestic animals and humans. Wallingford, United Kingdom: CAB International; 1997. pp. 267–300. [Google Scholar]
- 34.Taylor D J, Alexander T J L. The production of dysentery in swine by feeding cultures containing a spirochaete. Br Vet J. 1971;127:58–61. doi: 10.1016/s0007-1935(17)37282-2. [DOI] [PubMed] [Google Scholar]
- 35.Taylor D J, Simmons J R, Laird H M. Production of diarrhea and dysentery in pigs by feeding pure cultures of a spirochaete differing from Treponema hyodysenteriae. Vet Rec. 1980;106:324–332. doi: 10.1136/vr.106.15.326. [DOI] [PubMed] [Google Scholar]
- 36.Trivett-Moore N L, Gilbert G L, Law C L H, Trott D J, Hampson D J. Isolation of Serpulina pilosicoli from rectal biopsy specimens showing evidence of intestinal spirochetosis. J Clin Microbiol. 1998;36:261–265. doi: 10.1128/jcm.36.1.261-265.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trott D J, Huxtable C R, Hampson D J. Experimental infection of newly weaned pigs with human and porcine strains of Serpulina pilosicoli. Infect Immun. 1996;64:4648–4654. doi: 10.1128/iai.64.11.4648-4654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Trott D J, Jensen N S, Saint Girons I, Oxberry S, Stanton T B, Lindquist D, Hampson D J. Identification and characterization of Serpulina pilosicoli isolates recovered from the blood of critically ill patients. J Clin Microbiol. 1997;35:482–485. doi: 10.1128/jcm.35.2.482-485.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Trott D J, McLaren A J, Hampson D J. Pathogenicity of human and porcine intestinal spirochetes in one-day-old specific-pathogen-free chicks: an animal model of intestinal spirochetosis. Infect Immun. 1995;63:3705–3710. doi: 10.1128/iai.63.9.3705-3710.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trott D J, Stanton T B, Jensen N S, Duhamel G E, Johnson J L, Hampson D J. Serpulina pilosicoli sp. nov., the agent of porcine intestinal spirochetosis. Int J Syst Bacteriol. 1996;46:206–215. doi: 10.1099/00207713-46-1-206. [DOI] [PubMed] [Google Scholar]
- 41.Wagner M, Erhart R, Manz W, Amann R I, Lemmer H, Wedi D, Schleifer K H. Development of an rRNA-targeted oligonucleotide probe specific for the genus Acinobacter and its application for in situ monitoring in activated sludge. Appl Environ Microbiol. 1994;60:792–800. doi: 10.1128/aem.60.3.792-800.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.White J, Roche D, Chan Y, Mitchell E A. Intestinal spirochetosis in children: report of two cases. Pediatr Pathol. 1994;14:191–199. doi: 10.3109/15513819409024252. [DOI] [PubMed] [Google Scholar]


