Abstract
Artificial intelligence based interpretation of cardiac magnetic resonance (CMR) studies relies on the ability to accurately identify the imaging plane, which can be achieved by both deep learning (DL) and machine learning (ML) techniques. We aimed to compare the accuracy of ML and DL for CMR view classification and to identify potential pitfalls during training and testing of the algorithms. Both DL and ML algorithms accurately classified CMR images, but DL outperformed ML (95% and 90%) when classifying images with complex heart anatomy. Reaching this level of accuracy required training on a carefully curated cohort of studies.
Background:
Artificial intelligence is increasingly utilized to aid in the interpretation of cardiac magnetic resonance (CMR) studies. One of the first steps is the identification of the imaging plane depicted, which can be achieved by both deep learning (DL) and classical machine learning (ML) techniques without user input. We aimed to compare the accuracy of ML and DL for CMR view classification and to identify potential pitfalls during training and testing of the algorithms.
Methods:
To train our DL and ML algorithms, we first established datasets by retrospectively selecting 200 CMR cases. The models were trained using two different cohorts (passively and actively curated) and applied data augmentation to enhance training. Once trained, the models were validated on an external dataset, consisting of 20 cases acquired at another center. We then compared accuracy metrics and applied class activation mapping (CAM) to visualize DL model performance.
Results:
The DL and ML models trained with the passively-curated CMR cohort were 99.1% and 99.3% accurate on the validation set, respectively. However, when tested on the CMR cases with complex anatomy, both models performed poorly. After training and testing our models again on all 200 cases (active cohort), validation on the external dataset resulted in 95% and 90% accuracy, respectively. The CAM analysis depicted heat maps that demonstrated the importance of carefully curating the datasets to be used for training.
Conclusions:
Both DL and ML models can accurately classify CMR images, but DL outperformed ML when classifying images with complex heart anatomy.
Keywords: artificial intelligence, magnetic resonance imaging, automated diagnosis
Tweet:
Deep learning outperformed machine learning (95% vs 90%) when classifying images with complex heart anatomy. Reaching this level of accuracy required training on a carefully curated cohort of studies.
Introduction
Artificial intelligence (AI) is increasingly utilized to improve the way cardiac magnetic resonance (CMR) studies are interpreted. AI holds the promise of improving the efficiency, standardization, and types of quantification that can be obtained from CMR datasets[1, 2]. Regardless of the AI task being utilized, one of the first necessary steps in the analysis of CMR images is the identification of the imaging plane depicted. Deep learning (DL) and classical machine learning (ML) (two subtypes of AI) can potentially do it without user input. Given the important role that AI is expected to play in future clinical image analysis, it is important for the cardiac imager to have a foundational understanding of how an algorithm that accomplishes this initial task is developed and of the potential sources of error that may impact the performance of the AI algorithm.
Although the terms ML and DL are often used interchangeably, the techniques they refer to are not the same. ML techniques are a collection of mathematical and statistical concepts such as random forest, support vector machine, K-nearest neighbors (KNN), etc. DL algorithms, on the other hand, are specialized techniques that are a subset of ML. The most important difference between the two approaches is that ML algorithms require direct input from the developer to address any errors. DL algorithms have built-in mechanisms for assessing and addressing the root of any inaccuracies and do not require guidance[3]. In order to “interpret” an image, both DL and ML must deconstruct the image into specific features, such as sharpness, curvedness, brightness, etc., that can be numerically inputted into a computer algorithm. This feature extraction process differs between ML and DL algorithms. DL applies a range of convolution filters to an image to extract the features from the image[3, 4]. By changing the weight assigned to any given feature, the DL algorithm can be trained to identify a specific type of image. On the other hand, ML algorithms use simpler feature extraction methods, such as applying a transformation to the image matrix[3].
However, DL techniques can still be outperformed by ML techniques, such as linear regression, on certain tasks[5]. In addition, ML approaches may sometimes be more practical as they can be generally trained on less computationally demanding equipment and more quickly than DL techniques. The goals of this study were: (1) to determine whether DL is superior to ML for classifying different CMR views; (2) to identify strategies to develop a successful AI algorithm for identification of cardiac views; and (3) to gain insight into the aspects of a CMR image that are utilized by DL algorithms.
Methods
Dataset Building
Our dataset was composed of 200 unique CMR exams containing nearly 100,000 cine and late gadolinium enhancement images. The study was approved by the Institutional Review Board. To curate the dataset, we first retrospectively identified 100 consecutive patients, which represented a typical range of CMR indications (~15% stress testing, ~30% heart failure evaluation, ~30% arrhythmia evaluation, ~25% other). We refer to this first set the passively curated cohort since it was simply comprised of consecutive cases performed at our medical center. We then supplemented the dataset with 100 specifically selected and non-consecutive patients who had more complex cardiac anatomy or more challenging image quality. These additional 100 patients were selected to evenly represent 5 groups of challenging disease states: dilated cardiomyopathy, cardiac amyloidosis, atrial septal defect, coronary artery disease and patients with an implantable cardioverter defibrillator. The combination of the passively curated dataset plus the 100 specifically selected non-consecutive CMR cases are referred to as the actively curated cohort. Of the 200 total exams, 160 of these exams were used to train the algorithms, 20 were used for internal iterative testing, and the remaining 20 were set aside for validation. An additional 20 CMR exams acquired at a different institution were used for final validation of the algorithms on a dataset acquired elsewhere and referred to as the external validation cohort. A summation of the dataset is presented in Table 1. All images were acquired on a 1.5T scanner (Achieva, Philips Healthcare, Andover, Massachusetts) using a 5-Channel surface coil. A short-axis (SAX) stack covering the left ventricle and 2-, 3-, and 4-Chamber (2-Ch, 3-Ch, 4-Ch) views of the left ventricle were acquired using commercial steady state free precession and inversion recovery with a phase sensitive reconstruction pulse sequence. Each frame of each cine and the magnitude-, phase-, and phase-shifted images from each of late gadolinium enhancement images were labeled as either SAX, 2-Ch, 3-Ch or 4-Ch view. The 20 Cine-CMR cases from another institution (1.5T Ingenia, Philips; V5.1.7) were acquired using a 32-Channel array in the 2-, 3- and 4-Chamber views, and in 3 short-axis slices at the apical, mid, and basal ventricular levels using a steady state free precession pulse sequence. Since the images in the dataset only had these labels and no other details or annotations were utilized in training, the training of the algorithms was minimally supervised.
Table 1:
Dataset Description and Composition
| Cohort | SAX | 2-Ch | 3-Ch | 4-Ch |
|---|---|---|---|---|
| Passively Curated | 40157 (78.54%) | 3558 (6.96%) | 3562 (6.97%) | 3849 (7.53%) |
| Complex Anatomy | 35012 (78.76%) | 3210 (7.22%) | 3078 (6.92%) | 3154 (7.10%) |
| Actively Curated | 75169 (78.65%) | 6768 (7.08%) | 6640 (6.95%) | 7003 (7.32%) |
| Remote Validation | 80 (25%) | 80 (25%) | 80 (25%) | 80 (25%) |
The table describes the composition of the cohorts and the dataset used for training, testing and validation of the DL and ML algorithms.
Data Preprocessing
Prior to training our models, we needed to preprocess the CMR files in our datasets. One of the first steps of the process was converting the CMR DICOM files into JPEG files, which were then sorted into patient folders. Once the files were curated, we applied the preprocess_input function, the default preprocessing function from the Keras library. The function creates batches of images that can then be inputted for training the ML and DL models.
Machine Learning Algorithm: K-Nearest Neighbors
We trained and tested multiple ML-based classification algorithms, including random-forest classifiers, decision trees, and support vector machines. KNN notably outperformed the others in the preliminary testing, and accordingly, we decided to utilize this method as a representative model for ML-based classification. KNN is a clustering-based ML approach [6, 7]. The K value or “number of neighbors” was set to 3 and distance-weighted voting was used. ML algorithms such as KNN differ from DL because the weights in the DL evolve with each training iteration or epoch. Altering an inaccuracy for the KNN-approach requires programmer-directed changes to the algorithm or the training dataset.
Deep Learning Algorithm: VGG19 and Hyperparameters
For the DL algorithms, we trained and tested several neural networks, including VGG19, ResNet50, DenseNet, and Inception Net v3, which are some of the most popular neural networks used for image classification. Because the VGG19 neural network outperformed the other 3 neural networks, we used it as a representative for DL. VGG19 is a pre-trained neural network that was trained on the ImageNet dataset, a large dataset consisting of everyday objects. To repurpose the VGG19 network for our task, we removed the final classification layers of the VGG19 neural network and replaced them with a global average pooling layer and four fully convolutional layers. We then reprogrammed the last classification layers and trained them to only classify cardiac MRI views. None of the feature processing layers that preceded the classification layers were altered. By only training the last layer, we were able to reduce the training time and retain the feature processing abilities of an already effective neural network, which is a form of transfer learning.
When training the DL model, we conducted thorough experimentation to determine the most optimal values for the model hyperparameters, which can be altered to design models apt for our tasks. We set the number of epochs (training rounds) to 5 and the batch size to 32. The activation function set for all the feature extraction layers was the rectified linear unit, a ramp function, while the final layer of the DL model, the classification layer, utilized the softmax activation function, which is a generalization of the logistic function. During the training itself, we also applied the Adam optimizer and tracked loss using the categorical cross-entropy function.
Training and Testing
We utilized a total of 200 CMR cases, i.e. the entire cohort, to train and test our ML and DL algorithms. The cohort was organized into two different sets briefly described above.
Passively Curated Dataset:
This dataset was curated from 100 randomly selected CMR cases. We split the images in the passively curated cohort randomly into three categories: training (~60% of images), testing (~20% of images) and validation (~20% of images). Images included in the training category were used to determine the weights of the KNN and the Dense layers classifier of VGG19. The feature extraction layers of the two models were unchanged and used default ImageNet weights. The testing images were used to determine whether overfitting was occurring during training, and the validation images were used to determine the accuracy of the two models. Validation for the passively curated cohort was carried out in four different ways: 1. on the full validation dataset, which included all the images within the dataset generated from the passively curated cohort; 2. on a ‘balanced’ validation dataset, which was created by removing 90% of the SAX images to ensure that all views of the heart were equally represented, generated from the passively curated cohort; 3. on a full validation dataset which included all the images within the dataset generated from the images with complex anatomy; and 4. on a ‘balanced’ validation dataset, which was created by removing 90% of the SAX images to ensure that all views of the heart were equally represented, from the images with complex anatomy. The balanced datasets were created to avoid skewing by over-representation of the SAX images.
Actively Curated Dataset:
We trained another version of the DL and ML algorithms using the actively curated cohort which was a summation of the passively curated cohort and the above-described images with complex anatomy. In this case, the training set, containing 2000 images (500 images for each view), was created from 160 randomly selected MRI cases (80 from the passively curated cohort and 80 cases with abnormal cardiac anatomy) from the larger dataset of 200 MRI cases. The size of the training dataset was amplified through augmentation as described below, allowing the full cohort training dataset to have 10,000 images. The remaining 40 cases from the actively curated cohort were utilized to create the testing set consisting of 320 images, 80 from each view. The final validation of the algorithms trained and tested with the actively curated cohort was done on the 20 cases included in the external validation cohort.
We first used the passively curated dataset to measure the performance and “warm-up” the ML and DL models, which were then trained and tested on the actively curated dataset.
Data Augmentation
We utilized Keras, a module for designing and training neural networks, for data augmentation to amplify the full cohort training set size. Augmentations included rotating over 360°, flipping the image along the x and y axes, altering brightness and applying zero-form component analysis whitening. The goals of applying these augmentations were to (1) increase the size of the training set and (2) diversify the features available for training[8, 9].
Visualization with Class Activation Maps
In order to understand the region of a CMR image used by the VGG19 model to classify images, we implemented class activation maps (CAM), a set of algorithms that create heatmaps highlighting which parts of an image are primarily influencing classification decisions[10]. This type of visualization is specific to the DL models and cannot be used for the KNN-based classification.
Data analysis and Evaluation metrics
The McNemar test was used to compare the accuracy differences between the KNN-classifier and the VGG19-classifier[11]. A p-value <0.05 was considered statistically significant. In addition, for each test conducted with the models, we evaluated performance using accuracy, sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and the F1 score, a statistic that assesses the accuracy of classification using the harmonic mean of the PPV and sensitivity.
Results
Results from the passively curated cohort
The KNN and DL algorithms trained on the passively curated cohort achieved very high accuracy of image view classification, when tested on the full set of validation images selected from the passively curated cohort and on the ‘balanced’ dataset (Table 2). On the balanced validation set, the KNN algorithm had an accuracy of 99.3% while the VGG19 algorithm had an accuracy of 99.1%, a statistically significant but marginal difference.
Table 2:
Algorithm performance for different training datasets
| F1 Score | PPV | Sensitivity | Specificity | NPV | Accuracy | |
|---|---|---|---|---|---|---|
| Models Trained on Passively Curated Cohort | ||||||
| Validation on Full Dataset | ||||||
| Deep Learning | 0.988 | 0.988 | 0.988 | 0.996 | 0.996 | 0.988* |
| Machine Learning | 0.995 | 0.995 | 0.995 | 0.998 | 0.998 | 0.995 |
| Validation on Balanced Dataset | ||||||
| Deep Learning | 0.991 | 0.991 | 0.991 | 0.997 | 0.997 | 0.991* |
| Machine Learning | 0.993 | 0.993 | 0.993 | 0.998 | 0.998 | 0.993 |
| Validation on Full Dataset of Images with Complex Anatomy | ||||||
| Deep Learning | 0.70 | 0.90 | 0.64 | 0.88 | 0.88 | 0.64* |
| Machine Learning | 0.82 | 0.90 | 0.79 | 0.79 | 0.93 | 0.80 |
| Validation on Balanced Dataset of Images with Complex Anatomy | ||||||
| Deep Learning | 0.81 | 0.85 | 0.81 | 0.94 | 0.94 | 0.81* |
| Machine Learning | 0.83 | 0.84 | 0.83 | 0.94 | 0.94 | 0.83 |
| Model Trained on Actively Curated Cohort | ||||||
| Deep Learning | 0.93 | 0.93 | 0.93 | 0.98 | 0.98 | 0.93* |
| Machine Learning | 0.87 | 0.87 | 0.87 | 0.96 | 0.96 | 0.87 |
| Remote Cohort Validation of Actively Curated Cohort | ||||||
| Deep Learning | 0.95 | 0.96 | 0.95 | 1.0 | 0.98 | 0.95 |
| Machine Learning | 0.90 | 0.91 | 0.90 | 0.97 | 0.97 | 0.90 |
Statistically significant difference between DL & ML (p-value < 0.05)
When tested on the images with complex and abnormal anatomy, both the DL and the KNN models performed significantly worse (Table 2). The DL model had the lowest accuracy for classifying SAX images, at 57.2%, while the KNN model had the lowest accuracy for 4-Ch images, at 76.8% (Figure 1).
Figure 1:
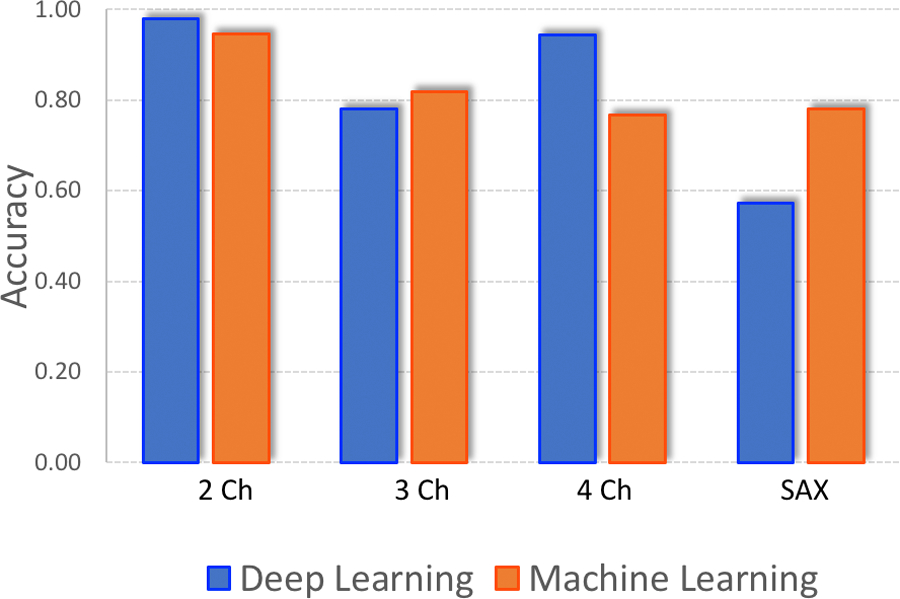
DL v ML Accuracy (Complex Anatomy Cases).
The figure depicts the accuracy of the passively trained ML and DL algorithms validated on CMR images with complex, abnormal anatomy. ML and DL performance varied more and the difference in classification accuracy was the greatest for short-axis images.
Results from the Actively Curated Cohort
The ML and DL algorithms trained using the actively curated cohort produced more uniform results across the four image views (Figure 2). The DL and ML algorithms showed more notable differences in diagnostic performance. Compared to the accuracy values achieved through training/testing on the passively curated dataset, the DL accuracy was nearly 93% while the ML accuracy was 87% following training on the actively curated cohort (Table 2).
Figure 2:
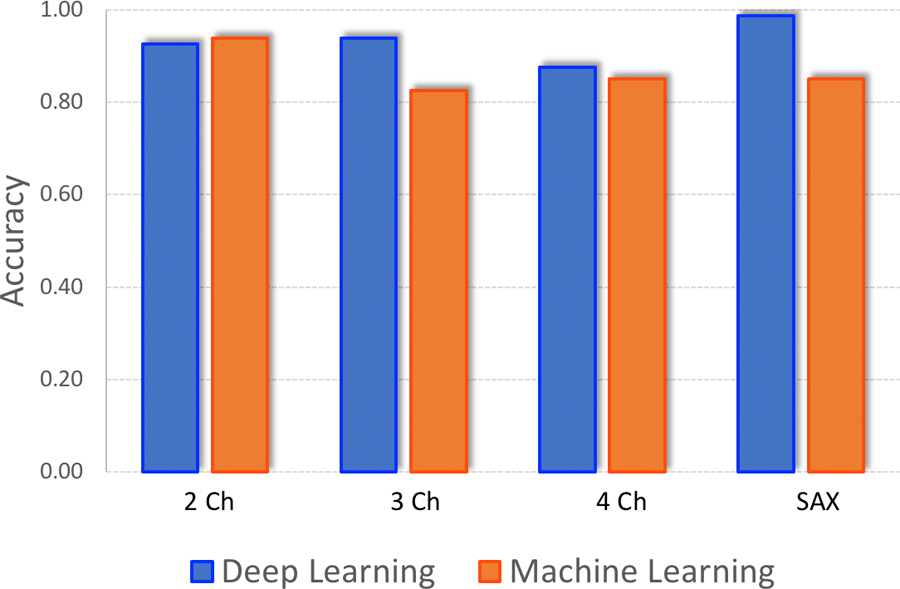
DL v ML Accuracy (Actively Curated Cohort)
After training the ML and DL algorithms with the active cohort, ML and DL performance improved. The DL model also demonstrated an edge over the ML model, indicating that DL might be the preferable algorithm for datasets with complex cases.
In addition, we noted that images misclassified by the ML and DL algorithms had recurring patterns. For images mislabeled by both the ML and DL algorithms, the models misclassified the images with the same incorrect label in 71% of the cases. More than half the misclassified cases were of 4-Ch views being labelled as 3-Chamber.
We also assessed errors specific to the ML and DL models. The KNN algorithm performed most poorly in patients with implanted cardiac devices. On the other hand, the VGG19 frequently confused 2-Ch views for SAX views and 4-Ch views for 3-Ch views (Figure 3).
Figure 3:
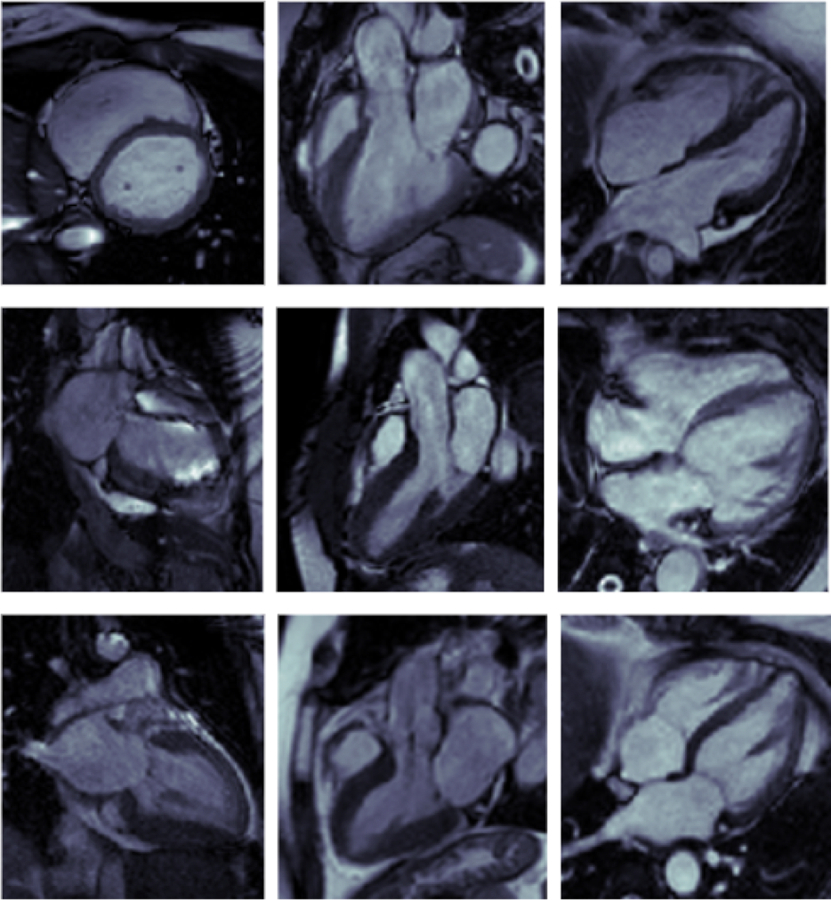
Sample of misclassified cardiac MRI images.
Top: Images Misclassed by both DL and ML; Left: SAX image misclassed as 3-Ch by DL and 4-Ch by ML; Center: 3-Ch image misclassed as SAX by DL and 2-Ch by ML; Right: 4-Ch image misclassed as 3-Ch by both DL and ML
Middle: Images Misclassed by DL only; Left: 2-Ch image misclassed as SAX; Center: 3-Ch image misclassed as SAX; Right: 4-Ch image misclassed as 3-Ch
Bottom: Images Misclassed by ML only; Left: 2-Ch image misclassed as SAX; Center: 3-Ch image misclassed as SAX; Right: 4-Ch image misclassed as 3-Ch
Results from the Remote Cohort Validation
Validation on the remote cohort yielded accuracy values over 90% for both the ML and the VGG19-classifier models. The DL method was more accurate than the ML method, but the difference, according to the McNemar test, was not statistically significant (p = 0.25). It is also worth noting that the DL method and the ML method varied in their misclassifications. The DL method primarily missed 2-Ch images, while the ML method missed 3-Ch and 4-Ch images.
Visualization by Class Activation Maps
The CAM analysis depicted regions of focus for the DL algorithm and showed that each of the 4 labels had a unique pattern (Figure 4). In 2-Ch images, the features extracted from the heart, as well as the superior anatomical structures, such as the collar bones, had more influence on the algorithm. For 3-Ch images, features extracted from the aortic root had the most weight, while for 4-Ch images features extracted from the left atrium, left ventricle, and the liver had greater importance in classification. However, for SAX images, the CAM indicated that the DL algorithm decision seems to be based more on the features extracted from the anatomic structures adjacent to the heart rather than the heart itself. Misclassifications by the DL algorithm seem to occur because the algorithm extracted features in a pattern that resembled those of another label.
Figure 4:
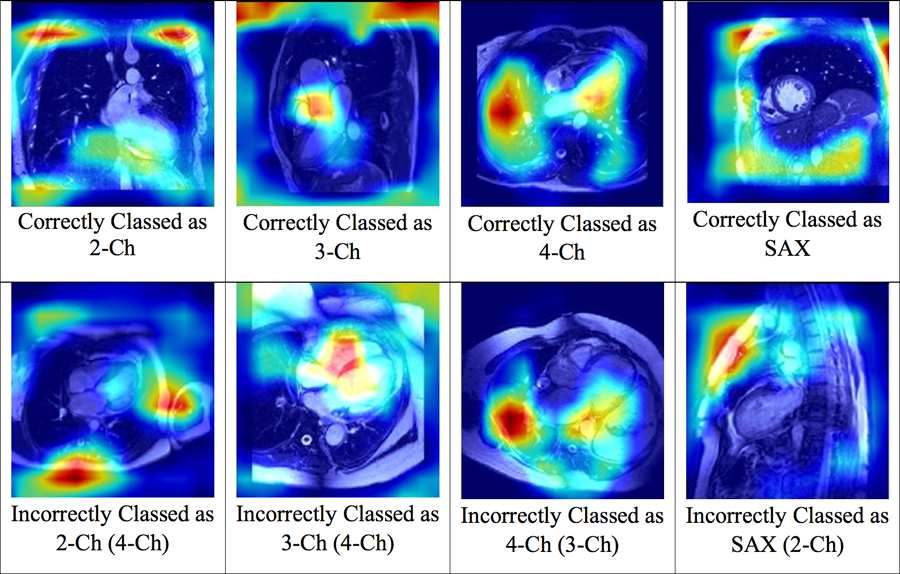
Class Activation Map Patterns Generated for the DL Algorithm.
The figure above depicts class activation maps for the images processed by the DL algorithms. Each image view has a distinct CAM pattern and images misclassified by the DL model demonstrate the pattern of the misclassified image view.
Discussion
In this study, we used a minimally supervised training approach to show that: (1) DL was superior to ML for classifying cardiac imaging planes from CMR images as the complexity of the cohort increased; (2) the importance of an actively curated training dataset, which includes a wide range of disease states and image qualities to develop robust DL and ML algorithms; (3) the need to compose well-balanced validation datasets to determine the accuracy of an algorithm; and (4) the use of CAM patterns to understand the image features that are utilized by the algorithm to classify an image. We specifically chose the task of CMR view classification despite it being a relatively straightforward challenge, as view classification must precede most other complex operations, such as segmentation of the heart chambers. In addition, since one of the main goals of this study was to compare two different algorithms, we sought a problem that would not be too computationally challenging and something that is the first step of CMR image analysis.
There are many different AI approaches that can be utilized to achieve a specific task. Selecting the correct approach requires a deep understanding of the task that the algorithm needs to solve. Accordingly, prior to initiating our study, we performed preliminary testing on several candidate ML and DL algorithms that had the potential to correctly classify CMR images. Interestingly, in our assessment of DL algorithms for the specific task of CMR view classification, we discovered that ResNet50, despite being the premier neural network utilized for classification tasks, was inferior to VGG19. We ultimately selected KNN and VGG19 to formally test one of our main hypotheses, namely whether ML can outperform DL in CMR image classification, or vice versa. We found that both ML and DL algorithms could accurately categorize cardiac CMR views. However, the DL algorithm performed better as the complexity of cases increased and when tested on images acquired at another center. This suggests that the DL algorithm may be better suited for classifying a larger variety of images. This trend has been observed in general image classification as well[12]. In the 2012 ImageNet Large-Scale Visual Recognition Challenge (ILSVRC), a competition for classifying everyday images from nearly 1000 categories, the convolutional neural network AlexNet outperformed all ML algorithms. Its top-5 error rate was 15.3 percent, nearly 10.8% less than the runner up[4]. Since 2012, DL models have been consistently the champions of the ILSVRC competitions.
Our study also demonstrated the importance of actively curating a training dataset in order to generate a robust algorithm to accomplish the goal at hand. We compared the use of an actively curated dataset, containing a diverse set of hand-picked images, to the use of a passively curated dataset that included consecutively performed CMR cases. On initial testing of an algorithm developed using this passively curated dataset, it was over 99% accurate for identifying the correct CMR imaging plane; however, as the algorithm was tested on more complex images, its performance clearly deteriorated. On the other hand, an actively curated training dataset that was generated by experts that proactively identified diverse types of cases encountered in clinical practice performed considerably better. This finding highlights the continued role of subject matter experts when developing an AI algorithm[13, 14].
In clinical research, it is well established that the sensitivity and specificity of a test are highly dependent on the prevalence of a disease state within a population. The same holds true when validating an artificial intelligence algorithm. As can be seen in Table 2, when the algorithms were trained and tested on the “fully passive dataset” of images, the ML algorithm seemed to outperform the DL algorithm; however, both algorithms performed similarly when compared using a more “balanced passive dataset.” The reason for this is that a CMR exam typically has many more short-axis images of the left ventricle than long-axis images. Because the ML algorithm tended to classify images as short-axis regardless of the actual view and because there were more short-axis images in the dataset, it appeared to perform better than it really did. Imbalanced data has been an issue that DL researchers across disciplines have faced, and applying data-level methods to balance classes is important in order to get an accurate picture of the algorithm’s performance[14].
An important part of validating an image analysis algorithm is to test it on a dataset acquired at an independent center. In our study, the DL algorithm marginally outperforms the ML algorithm for classifying CMR images on the external validation dataset. The importance of re-validating an algorithm on an external dataset is to avoid any imaging center-specific settings that are not actually related to the underlying anatomy itself. For example, perhaps an imaging center might systemically use a larger field of view when acquiring short-axis views of the heart and a smaller field of view when acquiring long-axis views of the heart. An algorithm might simply learn to recognize the field of view and use that finding to classify an image even though the field of view does not actually have anything to do with the underlying anatomy. Arnaout et al., voiced a similar sentiment and recommended using images with variations in quality and from a range of ages, hemodynamics and sizes to train DL models for medical image classification[15].
One of the challenges in the adoption of DL algorithms in clinical practice is the perception that the clinician does not know what an algorithm is “looking at” to make its decision. The lack of understanding of what features an algorithm utilizes is one of the factors that make clinicians wary of extensively using AI algorithms [16]. In this study, we used CAMs to identify the structures that our DL algorithm was using to classify images[12]. These CAMs have the potential to help developers and clinicians understand when and how an algorithm is failing and may be used to refine the algorithm. Misclassifications occur when CAM patterns resemble that of another label, suggesting that more detailed annotations on training datasets may improve algorithm development. Additionally, a CAM can show the features being used by a DL algorithm to make the decision. This information may be provided to the clinician as a mechanism of oversight of the algorithm. Such oversight by physicians may allow DL algorithms to be more readily adopted into clinical practice.
Limitations
First, the algorithms we tested have been developed using relatively small sample sizes acquired at a single imaging center. It is possible, that the use of multi-center data to train the algorithm might result in somewhat different findings. Second, the task selected was fairly narrow in scope, i.e. classifying CMR images by imaging plane; however, identifying the cardiac imaging plane is a fundamental step needed before any further analysis of the images can occur. Future studies are needed to compare algorithms for performing other more complex tasks, such as identifying specific disease states or segmenting the heart.
In addition, the CMR views may not always be clear cut. Situations in which the imaging view is imperfectly acquired may represent a mixture of two different views. For instance, a 4-Ch image may look like a 3-Ch image, if the plane was slightly off-axis. Such a mixed view would likely be somewhat arbitrarily assigned to one view or another by an expert. An AI algorithm would similarly have to arbitrarily categorize it as one view or the other.
Conclusions
In this study, we showed that DL algorithms outperform ML algorithms for classifying CMR images especially as the complexity of a case increased. We highlighted the importance of actively curating a training dataset, which includes a wide range of disease states. We also underscored the need for creating balanced validation datasets to avoid biases when testing the performance of an algorithm. Finally, CAMs may be a useful tool to understand potential sources of error made by a DL algorithm when classifying an image.
Acknowledgement.
The authors would like to thank Dr. Mubeena Abdulkarim who helped label the cardiac planes on some of the images.
Dr. Patel has received research grant and/ or support from Philips, General Electric, Arterys, CircleCVI, and Neosoft. This project was supported by the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health (NIH) through Grant Number 5UL1TR002389-02 that funds the Institute for Translational Medicine (ITM), and K25 HL141634.
Abbreviations:
- AI
Artificial Intelligence
- ASD
Atrial Septal Defect
- CAM
Class Activation Map
- CMR
Cardiac Magnetic Resonance
- DL
Deep Learning
- KNN
K-Nearest Neighbors
- ML
Machine Learning
- NPV
Negative Predictive Value
- PPV
Positive Predictive Value
- 2-Ch
2 Chamber
- 3-Ch
3 Chamber
- 4-Ch
4 Chamber
- SAX
Short Axis
References
- 1.Petersen S, Abdulkareem M, Leiner T (2019) Artificial Intelligence Will Transform Cardiac Imaging-Opportunities and Challenges. Frontiers in Cardiovascular Medicine. 10.3389/fcvm.2019.00133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Thrall J, Li X, Li Q (2018) Artificial Intelligence and Machine Learning in Radiology: Opportunities, Challenges, Pitfalls, and Criteria for Success. Journal of the American College of Radiology. 10.1016/j.jacr.2017.12.026 [DOI] [PubMed] [Google Scholar]
- 3.Shinde P, Shah S (2019) A Review of Machine Learning and Deep Learning Applications [Google Scholar]
- 4.Krizhevsky A, Sutskever I, Hinton G (2017) ImageNet classification with deep convolutional neural networks. Communications of the ACM 60:. 10.1145/3065386 [DOI] [Google Scholar]
- 5.Jiao S, Gao Y, Feng J, et al. (2020) Does deep learning always outperform simple linear regression in optical imaging? Optics Express 28:3717–3731. 10.1364/oe.382319 [DOI] [PubMed] [Google Scholar]
- 6.Li C, Zhang S, Zhang H, et al. (2012) Using the K-nearest neighbor algorithm for the classification of lymph node metastasis in gastric cancer. Computational and Mathematical Methods in Medicine. 10.1155/2012/876545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kramer O (2013) K-Nearest Neighbors. In Dimensionality Reduction with Unsupervised Nearest Neighbors., 1st ed. Springer-Verlag Berlin Heidelberg [Google Scholar]
- 8.Lundervold A, Lundervold A (2019) An overview of deep learning in medical imaging focusing on MRI. Zeitschrift Für Medizinische Physik 29:102–127. 10.1016/j.zemedi.2018.11.002 10. [DOI] [PubMed] [Google Scholar]
- 9.Wong S, Gatt A, Stamatescu V, McDonnell M (2016) Understanding Data Augmentation for Classification: When to Warp? IEEE, Gold Coast, QLD, pp 1–6 [Google Scholar]
- 10.Zhou B, Khosla A, Lapedriza A, et al. (2016) Learning Deep Features for Discriminative Localization. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2921–29. 10.1109/CVPR.2016.319 [DOI] [Google Scholar]
- 11.McNemar Q (1947) Note on the sampling error of the difference between correlated proportions or percentages. Psychometrika. 10.1007/BF02295996 [DOI] [PubMed] [Google Scholar]
- 12.O’Mahony N, Campbell S, Carvalho A, et al. (2019) Deep Learning vs. Traditional Computer Vision. Advances in Computer Vision 128–44. 10.1007/978-3-030-17795-9_10 [DOI] [Google Scholar]
- 13.Foody G, Mcculloch M, Yates W (1995) The Effect of Training Set Size and Composition on Artificial Neural Network Classification. International Journal of Remote Sensing 16:1707–1723. 10.1080/01431169508954507 [DOI] [Google Scholar]
- 14.Johnson J, Khoshgoftaar T (2019) Survey on Deep Learning with Class Imbalance. Journal of Big Data 6:. 10.1186/s40537-019-0192-5 [DOI] [Google Scholar]
- 15.Madani A, Arnaout R, Mofrad M, Arnaout R (2018) Fast and accurate view classification of echocardiograms using deep learning. npj Digital Medicine 1:. 10.1038/s41746-017-0013-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Reyes M, Meier R, Pereira S, et al. (2020) On the Interpretability of Artificial Intelligence in Radiology: Challenges and Opportunities. Radiology: Artificial Intelligence 2:. 10.1148/ryai.2020190043 [DOI] [PMC free article] [PubMed] [Google Scholar]


