Abstract
Assembling peptides allow the creation of structurally complex materials, where amino acid selection influences resulting properties. We present a synergistic approach of experiments and simulations for examining the influence of natural and non-natural amino acid substitutions via incorporation of charged residues and a reactive handle on the thermal stability and assembly of multifunctional collagen mimetic peptides (CMPs). Experimentally, we observed inclusion of charged residues significantly decreased the melting temperature of CMP triple helices with further destabilization upon inclusion of the reactive handle. Atomistic simulations of a single CMP triple helix in explicit water showed increased residue-level and helical structural fluctuations caused by the inclusion of the reactive handle; however, these atomistic simulations cannot be used to predict changes in CMP melting transition. Coarse-grained (CG) simulations of CMPs at experimentally relevant solution conditions, showed, qualitatively, the same trends as experiments in CMP melting transition temperature with CMP design. These simulations show that when charged residues are included electrostatic repulsions significantly destabilize the CMP triple helix and that an additional inclusion of a reactive handle does not significantly change the melting transition. Based on findings from both experiments and simulations, the sequence design was refined for increased CMP triple helix thermal stability, and the reactive handle was utilized for the incorporation of the assembled CMPs within covalently crosslinked hydrogels. Overall, a unique approach was established for predicting stability of CMP triple helices for various sequences prior to synthesis, providing molecular insights for sequence design towards the creation of bulk nanostructured soft biomaterials.
Keywords: assembling peptides, collagen mimetic peptides, atomistic simulations, coarse-grained simulations, soft biomaterials
Graphical Abstract
Introduction
Assembling peptides continue to be useful building blocks for the design of materials with controlled and multiscale properties based on bottom up design using molecular engineering principles.1, 2 Depending on the application, peptide sequence design and assembly conditions can be modulated to control nano-, micro-, and macro-scale properties, from peptide conformations and higher ordered assemblies to bulk material properties.3–6 In particular, peptides mimetic of and inspired by the assembly of native collagen, known as collagen mimetic peptides (CMPs) or collagen like peptides (CLPs), have been designed to control the kinetics of assembly and the thermal stability and organization of the resulting assembled structures. These sequences are often comprised of an (X-Y-G)n repeat structure, where X and Y can be any amino acid except glycine. Most commonly in native human collagen X and Y are proline (P) and hydroxyproline (O), yielding the triplet (POG)n that forms triple helices in solution exhibiting melting temperatures (Tm) around or above physiological (≥ 37 °C) when n ≥ 7.7, 8 Nanostructures composed of triple helices to short fibrils on the order of ~10–100 nm long have been created with this approach and have been used for promoting selective integrin binding to hydrogels and targeting therapeutics to collagen-rich tissues.9 Achieving controlled higher ordered assembled structures from the nano- to micro- and macro-scales for the ultimate creation of bulk materials has been more challenging and often necessitates use of amino acid substitutions to the ‘canonical’ (POG) repeat unit.
Several types of amino acid substitutions have been integrated into CMPs, both natural and non-natural amino acids and functional groups, to control properties for specific applications, from the stabilization and modulation of properties in solution to the modification of surfaces and the formation of soft materials (e.g., hydrogels).10–14 For example, within CMP sequences built upon the canonical (POG) repeat unit, cysteines and lysines amongst other natural amino acids have been incorporated at the end and in the middle of sequences, respectively, for covalent modification of assembled structures.15, 16 Such substitutions have allowed the formation of disulfide bonds to crosslink strands for stabilization of the resulting triple helices and the functionalization of structures to gold nanoparticles for creating nanowires. Utilizing non-natural amino acids, a range of modifications have been made to modulate triple helix thermal stability, including integration of azaproline or azaglycine within the peptide backbone and click reactive handles as pendant groups along the peptide backbone.17–20 For the formation of long fibrils, both non-natural and natural amino acids also have been integrated to impart end-to-end physical interactions between triple helices, such as phenylalanine-pentafluorophenylalanine and charged amino acid pairs on the peptide termini.15, 21, 22 Excitingly for the creation of soft materials, the integration of select charged pairs (e.g., lysine [K] - aspartic acid [D]) on opposite ends of CMP sequences also leads to the formation of supramolecular structures and hydrogels at higher peptide concentrations in buffer; in these sequences, salt-bridged hydrogen bonds between K and D were found to promote end-to-end interactions between triple helices and the formation of fibrils reminiscent of native collagen.21 To integrate these robust assembled structures within biomaterials with tunable mechanical properties, we recently demonstrated the utility of integrating an orthogonal reactive handle, an allyloxycarbonyl-protected lysine (Kalloc), within the middle in addition to K and D substituted ends to enable light-triggered thiol–ene reactions and covalent crosslinking with multifunctional polymers.23 Given the utility of such multiple substitution sequences, important questions remain in how the placement, type, and number of specific natural and non-natural amino acids control triple helix thermal stability and higher ordered assembly to facilitate the fabrication of soft biomaterials with multiscale, thermally-responsive structures.
Molecular dynamics (MD) simulations are valuable predictive tools that can link the placement, type, and number of specific natural and non-natural amino acids to the thermal stability and self-assembly of peptides and peptide conjugates in general.24 Specifically for CMPs, during the past decade, there have been several MD studies focused on the effects of sequence length,25 amino acid substitutions,26 and amino acid stereochemistry27 on the thermal stability of the CMP triple helix. Many of these studies used atomistic models that quantified key atomic structural rearrangement and interactions, such as root mean square deviations (RMSD) of the peptide backbone and water-mediated hydrogen bonding. For example, Ramasami and coworkers studied (GPO)-based CMPs with different lengths from one to ten (GPO) repeat units using all-atom MD simulations, where (GPO) is the same repeat as (POG) with the repeat unit simply started at glycine rather than proline.25 They observed that a minimum of five repeat units were required to form a stable triple helix as evident by their RMSD, peptide-water radial distribution functions, and intermolecular hydrogen bonding analyses. While informative data can be gleaned from these unbiased atomistic simulations, they are limited to small system sizes (e.g., 1–2 triple helices) and cannot simulate melting of the CMP triple helix, owing to the intractable simulation times required for sampling rare-events during the triple helix melting.
Coarse-grained (CG) simulations are better suited than unbiased atomistic simulations to observe CMP melting at experimentally relevant concentrations because of their reduced degrees of freedom. For example, the CMP model of Condon and Jayaraman captures the directionality and specificity of hydrogen bonding interactions within the CMP triple helix, making it applicable for studying CMP melting due to breaking of hydrogen bonds with increasing temperature as well as CMP assembly at larger length and time scales.28 Other CG models developed by Buehler and coworkers29–31 based on MARTINI forcefield for proteins can also reproduce mechanical and structural properties of CMPs, such as persistence lengths and elastic moduli, but do not maintain the inter-strand N-H to C=O hydrogen bonding pattern found in collagen triple helical structures.30, 32
Besides the MD-based predictive tools discussed above, other computational methods involving energy scoring functions have been used to predict the stability of the CMP triple helix, specifically A:B:C heterotrimers, without conducting molecular simulations.33, 34 For example, Fallas and Hartgerink designed an A:B:C collagen heterotrimer consisting of (PKG), (PDG), (DOG), (KOG), and (POG) repeat units, where O represents hydroxyproline and all other letters are the standard 1 letter amino acid abbreviations. In this work they used a sequence-based energy scoring function that takes into account the stability conferred to collagen by axial lysine-aspartate salt bridges.34 Also, melting temperature (Tm) calculators and online tools35 can predict the melting temperatures for varying CMP designs but cannot be extended towards predicting larger scale structure from assembly of CMPs. Integration of atomistic and CG simulations, as done in this paper, can link the design of new peptides to CMP triple helix stability and also enable studies of larger scale assembly with CG simulations to compare to and guide experiments.
In this paper, we utilized a combination of atomistic MD simulations, CG MD simulations, and experiments to examine the effects of both natural and non-natural amino acid substitutions on CMP triple helix assembly and stability, and ultimately facilitate the design of soft biomaterials. Specifically, we investigated the thermal properties of CMPs with two different sets of substitutions to determine how changes in amino acid sequence (e.g., type and placement of substitutions) influenced the stability of the resulting CMP triple helices in comparison to a neutral ‘wild type’ (WT) control sequence (the canonical (POG)n). The first substitutions involved the inclusion of charged amino acids on both ends of the peptide that allow for stable triple helix formation while also encouraging fibril formation on larger size scales. This focus allowed benchmarking of simulation results to the literature, as well as our own experimental results, for new sequence design and refining elements of our established model. Second, we evaluated the incorporation and placement of a non-natural amino acid, a lysine with alkene pendant group (Kalloc), that allows light-triggered covalent reaction after assembly for modification and integration of the resulting hierarchical structures within soft biomaterials.23 The placement of this reactive handle, either at the end or middle of the sequence, was studied to determine which had the least impact on triple helix stability. Having validated our simulations versus experimental data, we then utilized our CG model to explore the effects of increased sequence length via the number of hydrogen bonding residues, while keeping the number of charged residues the same, for insights into relevant sequence designs for achieving desired thermal properties prior to synthesis. Finally, assembled structures formed with the designed sequence for increased thermal stability were integrated within covalently crosslinked hydrogels utilizing the non-natural reactive handle to fabricate bulk biomaterials with robust mechanical properties. This comprehensive approach that combines experimental results and insights from simulations could prove useful for the prediction and optimization of the thermal properties of not only CMP sequences, but other peptide sequences (e.g., long or challenging to synthesize) for the rapid design of innovative assembling peptides and soft materials with a range of thermal and structure properties of relevance for a variety of applications.
Experimental Section
Materials.
All general organic reagents were purchased from commercial sources, Thermo Fisher Scientific (Waltham, MA) and Millipore Sigma (St. Louis, MO), and used as received unless otherwise stated. Amino acids and 2-(1H-Benzotriazol-1-yl)-1,1,3,3-tetramethyluronium hexafluorophosphate (HBTU) were purchased from ChemPep (Wellington, FL) unless stated otherwise.
Peptide synthesis.
All peptides, except for the (POG)12 sequence, were synthesized by solid phase peptide synthesis (SPPS) using standard Fmoc chemistry using a Tribute peptide synthesizer (Gyros Protein Technologies, Tuscon, AZ). Briefly, Fmoc protected amino acids and HBTU were loaded into cartridges at 4x molar excess for coupling on Tentagel R RAM Resin Rink-type (Peptides International; 0.19 meq/g). The Fmoc group was deprotected using 20% piperidine in N,N-dimethylformamide (DMF) for 30 minutes. Amino acids were activated using 0.4 M 4-methylmorpholine (4-MMP) and coupled on resin for 1 hour. All amino acids were double coupled and a final deprotection of the Fmoc group on the final amino acid was performed on the instrument.
In the case of the (POG)12 sequence, a modified approach was used to ensure successful synthesis and purification of the desired peptide sequence, after observing amino acid deletions in sequences with more than 7 (POG) repeat units when made strictly on the automated peptide synthesizer where these deletions were difficult to mitigate or subsequently separate. Moroder and coworkers and Chenoweth and coworkers, amongst others, previously have shown how use of a fragment condensation approach improves yields for CMPs by reducing the number of amino acid deletions.36, 37 In addition to improving crude peptide yield, this approach also makes the desired peptide easier to separate from undesired products owing to the increased disparity in their chemical structures and molecular weights (e.g., deletions of (POG) rather than a single P, O, or G). Here, we adapted this approach to synthesize CMPs with a combination of automated SPPS and manual fragment condensation. Specifically, Fmoc-GPO(tBu)-OH fragments were synthesized following a previously published protocol.23, 36, 38 (POG)7 was synthesized on resin via automated SPPS as described above using a Tribute peptide synthesizer (Gyros Protein Technologies, Tucson, AZ). After removal of the peptide on resin from the automated peptide synthesizer, a standard Fmoc-G-OH amino acid was coupled manually using 2 molar equivalents of the amino acid, 1.9 molar equivalents of HBTU, and 2 molar equivalents of diisopropylethylamine (DIPEA) in DMF and stirred for 30 minutes followed by Fmoc removal. Subsequently, three Fmoc-GPO(tBu)-OH fragments were coupled manually using similar molar equivalents as described above, followed by manual coupling of a standard Fmoc-O-OH and Fmoc-P-OH, resulting in (POG)12. A Kaiser test for primary amines was used to monitor coupling progress, and final deprotections were performed using 20% piperidine in DMF for 30 minutes each, repeated 3x to ensure complete Fmoc removal.
After synthesis the peptides were cleaved from the resin using a cleavage cocktail (95% trifluoroacetic acid (TFA), 2.5% deionized water, and 2.5% triisopropylsilane (TIPS) v/v) for 2 hours. Peptides within the cleavage solution were precipitated in cold ethyl ether at 10x excess in a 50 mL conical tube and refrigerated overnight. The next day the peptides were centrifuged at 4400 RPM at 4 °C for 10 minutes and washed 3x with fresh cold ethyl ether, resuspending and centrifuging after each wash. Peptides were then dried in the hood at room temperature overnight and then placed in a desiccator for at least 1 hour to remove any excess solvent. Peptides were purified by reverse phase high pressure liquid chromatography (HPLC) where a linear gradient was used (initially 95:5 water:acetonitrile with increasing acetonitrile of 0.5% per minute for (POG)12, for improved separation of this sequence with >7 (POG) repeats, and 1.1% per minute for charged sequences with <7 (POG) repeats; XBridge C18 OBD 5 μm column) (Waters Corporation, Milford, MA). The collected fraction was frozen at −80 °C and lyophilized (FreeZone 4.5 Plus, Labconco, Kansas City, MO). The mass of each peptide was confirmed through electrospray ionization (ESI+) mass spectrometry (ESI-MS, LCQ Advantage Thermo Scientific, Waltham, MA; Acquity UPLC H-class/SQD2, Waters Corporation, Milford, MA) (Figures S1–S6).
General peptide sample procedure.
Sodium phosphate buffer (10 mM) was prepared with monobasic and dibasic sodium phosphate (Sigma Aldrich, St. Louis, MO) utilizing an online buffer calculator (http://serge.engi.tripod.com/MolBio/Buffer_cal.html): specifically, 81.94 mg of dibasic sodium phosphate (Na2HPO4) and 50.75 mg of monobasic sodium phosphate (NaH2PO4) were dissolved in 100 mL of deionized water, and after mixing, the pH verified to be 7. Peptide samples were prepared in deionized water or 10 mM sodium phosphate buffer at pH 7, which were sterile filtered prior to use in dissolving the peptide. Specifically, the neutral (POG)12 sequence was prepared in deionized water, and sequences with charged residues were prepared in 10 mM sodium phosphate buffer for salt-bridge formation to facilitate comparison with published data in the literature. Samples were prepared in dilute solution (0.1–0.3 mM) to focus on the formation of triple helices, rather than higher ordered structures that may be formed at higher CMP concentrations, and analysis of triple helix thermal stability; note, triple helix formation/melting is reversible within the temperature ranges probed here (4 to 85 °C).21, 22, 39, 40 Accordingly, samples were heated to 85 °C on a temperature block for 15 minutes, as the standard for melting any triple helices, and allowed to slowly cool to room temperature and assemble for 48 hours prior to characterization of the resulting assembled structures.
Circular dichroism (CD).
Triple helix formation and melting curves were measured using a CD spectropolarimeter (J-810 JASCO Corporation, Easton, MD). For wavelength scans, CMP samples were prepared at a concentration of 0.1 mM peptide, and for temperature scans, the samples were prepared at 0.3 mM. Measurements were performed in a quartz cuvette with a 1 mm path length. Wavelength scans were measured from 195 nm to 250 nm at desired temperatures using a scan rate of 20 nm/minute and averaged over 3 scans. The millidegree values were collected every nanometer and the data were converted to mean residue ellipticity [θ] (deg cm2 dmol−1) using the following formula:
where θ is the measured ellipticity in millidegrees; L is the pathlength in cm; c is the concentration of the peptide solution in M; and N is the number of amino acids in the peptide. Similarly, to determine the melting temperature of the CMPs, the value of the ellipticity at the 225 nm peak was measured between the temperatures of 4 °C and 80 °C, where data points were taken every degree with a heating rate of 10 °C per hour. To determine the specific melting temperature (Tm) of assembled triple helices for each peptide design, defined as the temperature at which 50% of the peptide strands are assembled and disassembled, the data were fit to a Boltzmann distribution, and the first derivative of the fit was taken to determine the inflection point thereby giving a value for Tm. Each data set was fit between an approximate 40 °C window around the melting temperature to determine the Tm. The melting temperature of each peptide was measured 3 times, and the Tm values from the Boltzmann fit were averaged.
Atomistic CMP simulations.
We conducted all atomistic CMP simulations using the GROMACS 4.6.5 package to investigate the effects of natural and non-natural amino acid substitutions to the canonical (POG)n on the structural transitions of the CMP triple helix.41 We used the gencollagen package42 to generate initial configurations of CMPs with sequences, (XYG)12, where X and Y are proline (P) and 4’-hydroxyproline (O), respectively, with the backbone dihedrals of glycine, proline, and hydroxyproline set to φ = −74°, −75°, −75°; ψ = 170°, 168°, 153°; ω = 180°, 180°, 180°, respectively, in order to reproduce the native structure of the CMP triple helix. The N-terminus of each CMP chain presented an NH2 group while the C-terminus presented a CO-NH2 group, consistent with the peptides that were prepared experimentally on a Rink Amide resin. CMP sequences with natural and non-natural amino acid substitutions were then built using the mutagenesis plugin of PyMol.43 For example, hydroxyproline residues (O) in (POG) repeat units of (POG)12 were replaced with positively charged residues (K) and proline residues (P) in (POG) repeat units of (POG)12 were replaced with negatively charged residues (D) to obtain (PKG)4(POG)4(DOG)4. For CMPs containing Kalloc residue (Ka), either the lysine residue in the fourth (PKG) triplet of (PKG)4(POG)4(DOG)4 was replaced with a Kalloc residue to obtain (PKG)3(PKaG)(POG)4(DOG)4 or a Kalloc residue was also placed at the N-terminus of (PKG)4(POG)4(DOG)4 to obtain Ka(PKG)4(POG)4(DOG)4.
All initial configurations were constructed with a one residue stagger between CMP strands, in agreement with previously reported (POG)n triple helical structures,44 to achieve a ‘blunt-ended’ configuration rather than the alternative, proposed ‘sticky-ended’ configuration.21, 45 We note that the experimental work by Hartgerink and co-workers has shown that CMP sequences in which charged blocks are located at both termini can assemble to form fibrillar structures with either a blunt-ended design or sticky-ended design.45 However, we assume a blunt- ended design for all CMP sequences in this study to enable a fair comparison between the charged CMP sequences and (POG)n that are expected to be ‘blunt-ended’.44 Our assumption of blunt-ended design for all CMP sequences is justified at the dilute CMP concentrations as the presence of sticky ends would only impact the higher order (e.g., fibrillar) assembly at high CMP concentrations.44 As a result, we do not expect our choice of a blunt-ended design to impact the results of this study.
For each CMP sequence, a single CMP triple helix was solvated in water and an appropriate number of Na+ and Cl- counterions were added for charged sequences to achieve charge neutral systems. The box size was chosen such that the nearest distance between any peptide atom and the edge of the box was at least 1 nm to avoid artifacts due to periodic boundary conditions. Details of the number of atoms, system size, and number of counterions for a single CMP triple helix formed with each CMP sequence are shown in Table 1, where Ka represents a Kalloc within the sequence.
Table 1:
Number of atoms, system size, and number of counterions for atomistic CMPs
| System Description | # of Atoms (CMP and water) | System size (units in nm) | # of Counterions |
|---|---|---|---|
| (POG)12 | 138,954 | 14.1 × 10.0 × 10.0 | 0 |
| (PKG)4(POG)4(DOG)4 | 238,305 | 13.8 × 13.5 × 13.5 | 0 |
| Ka(PKG)4(POG)4(DOG)4 | 251,640 | 15.1 × 13.1 × 13.1 | 0 |
| (PKG)3 (PKaG)(POG)4(DOG)4 | 201,519 | 14.1 × 12.1 × 12.1 | 3 |
MD simulations were performed using the Gromos53a6 forcefield46 and the SPC water model.47 All non-bonded Lennard Jones interactions were truncated at 1 nm with long-tail dispersion corrections treated analytically and the group neighbor list was updated every 10 fs. The electrostatic interactions were evaluated using the particle-mesh Ewald (PME) method, using fourth-order interpolation and a Fourier grid spacing of 0.12 nm. A simulation time step of 2 fs was used with all bond lengths constrained via the LINCS algorithm.48 First, each system was energy minimized using the steepest descent algorithm, followed by equilibration for 200 ps at 300 K in the NVT ensemble and then for 2 ns in the NPT ensemble at 300 K and 1 bar. Next, each production run was performed for 100 ns at 300 K and 1 bar. The Berendsen thermostat and barostat were used with a damping coefficient of 1 ps-1.49 Configurations were stored every 5 ps and only the last 30 ns of each production simulation was used for the analysis.
Atomistic Simulations Analysis.
To quantify the structural integrity of the entire CMP triple helix, we calculated the root mean square deviation (RMSD) and the diameter of the triple helix. The RMSD was defined as the average change in the displacement of the backbone atoms (Cα-NH-CO) during the simulation, relative to the initial configuration as shown in equation 1.
| (1) |
Next, the local (i.e., residue-level) fluctuation of the collagen triple helix was captured by calculation of the helix diameter at each residue (from the three chains) along the triple helix. The diameter of the triple helix was calculated using the diameter of the circle encompassing the three Cα atoms of all three chains as shown in equation 2.
| (2) |
where a, b, and c are the distances between Cα atoms. The average diameter of the triple helix was then calculated using equation 3.
| (3) |
where Di represents the diameter of a circle encompassed by the ith set of the three Cα atoms in the three CMP chains.
Coarse-Grained (CG) CMP simulations.
We used the CG CMP model of Condon and Jayaraman28 where each CMP strand involving (POG) repeats was represented as a chain of CG beads – a proline backbone (PB) bead, a proline H-bond (PH) acceptor bead, a hydroxyproline backbone bead (OB), a glycine backbone (GB) bead, and a glycine H-bond (GH) donor bead. The PH and GH beads were incorporated to capture directional hydrogen bonding between the residues on the CMP strands that keep the triple helix intact at low temperature and destabilize at high temperature. All backbone (BB) beads had a diameter of 1.0 σ and a mass of 3.0 m while all H-bond beads had a diameter of 0.3 σ and a mass of 1.0 m; all distances were defined in terms of and all masses in terms of an arbitrary m as the simulations were not intended to capture the correct dynamics of these CMP systems. In other words, the bead masses do not need to reproduce the exact masses of the residues they represent. For CMP sequences involving (PKG) repeats, lysine residues (K) were modeled using a single BB (KB) bead, which has the same characteristics as the hydroxyproline (OB) bead along with an additional +1 charge. Similarly, for CMP sequences involving (DOG) repeats, aspartic acid residues (D) were modeled using a backbone (DB) bead which has a charge of −1 but also had a H-bond (DB) bead which had the same characteristics as the proline H-bond acceptor bead (PB). Monovalent counterions (IN) beads that had either a +1 charge or −1 charge were included to maintain charge neutrality. These IN beads had a diameter of 0.7 σ and a mass of 1.0 m. In this paper, extending the original model of Condon and Jayaraman, we modeled the Kalloc residue with two CG beads: the KB backbone bead and an alloc side chain bead (AB). This AB bead has a diameter of 0.86 σ and a mass of 3.0 m. The diameters of the KB and AB beads were chosen to reproduce the ratio of van der Waals volumes of the alloc group to its respective “parent” lysine residue determined using the modeling package, Marvinsketch.50
In each CMP chain/strand, adjacent BB beads were connected via a harmonic bond potential with a bond length of 0.5 σ and force constant of 1000 ε/σ2 and each H-bond bead was connected to its “parent” BB bead via a harmonic bond potential with a bond length of 0.37 σ and force constant of 1000 ε/σ2. In the Kalloc residue the AB bead is connected to its parent KB bead via a harmonic bond with a bond length of 0.93 σ and force constant of 1000 ε/σ2. All energy terms were defined in terms of . The SHAKE algorithm was used to constrain all BB-HB bond lengths.51 To account for the rigidity of the PPII conformation of CMP,7 we included a harmonic angle potential between three adjacent BB beads with the angle constant set at 20 ε/rad2 and equilibrium bond angle set at 180°. To ensure that H-bond formation occurs perpendicular to the backbone, a H-bond bead-parent BB bead-adjacent BB bead angle potential was introduced with a force constant of 300 ε/rad2 and equilibrium angle of 90°. To ensure the directionality of H-bond beads with respect to neighboring H-bond beads along the same strand, we included two H-bond bead—BB bead—BB bead—H-bond bead dihedral angle potentials,
| (4) |
in which the dihedral constant kd was set at 15 ε, and the reference dihedral, φ0, was set at 120° for (PH-PB-GB-GH) dihedrals and –120° for (GH-GB-PB-PH) dihedrals.
Non-bonded H-bond donor (D) - acceptor (A) interactions were represented using the Lennard-Jones (LJ) potential,52
| (5) |
where is the interaction strength between D and A beads and was set at 50.4 ε, and σHB is the diameter of a H-bond bead. All other pairwise interactions (besides the H-bond donor-acceptor interactions) for CMP beads were modeled using the purely repulsive Weeks-Chandler-Andersen (WCA) potential.53
| (6) |
In Equation (5) σij is the arithmetic mean of the diameters of beads i and j, and εij, the interaction strength, was set at 1.0 ε. For donor-donor and acceptor-acceptor interactions, σii was set at 0.7 σ instead of 0.3 σ to ensure that a donor HB bead only forms an H-bond with a single HB acceptor bead, and vice versa. The interactions involving alloc side chain (AB) beads were strictly repulsive and were modeled using the WCA potential with εij set to 1.0 ε and σij is the arithmetic mean of the diameters of an AB bead and bead j. Lastly, we modeled all electrostatic interactions using the Coulomb potential.
| (7) |
Where qi is the charge valency of bead i; qj is the charge valency of bead j; ε0 is the permittivity of free space; and εr was set at 80 to match the dielectric constant of water at room temperature.
We performed Langevin dynamics simulations in the NVT ensemble using the LAMMPS simulation package.54 For all systems, the initial configuration was created by randomly placing 10 intact CMP triple helices in a cubic simulation box of size 110 σ with periodic boundary conditions, achieving a concentration of approximately 0.3 mM. For systems containing charged amino acids, monovalent counterion (IN) beads were added such that a counterion of the opposite charge was added to the simulation box for every charged CMP backbone bead in order to achieve net charge neutrality. We also inserted an additional 10 mM of monovalent salt only for charged systems to mimic experimental conditions where salts are added. Moreover, in all Langevin simulations the friction coefficient was set to 10 τ (where 0.001 τ is approximately 6 fs) as in Ghobadi and Jayaraman.55 A two-level RESPA56 integrator was also used so that non-bonded and bonded interactions were integrated with a time step of 0.001 τ and 0.0005 τ, respectively. All systems were first equilibrated for 108 time steps followed by 107 time steps long production run in which data was collected every 100,000 time steps.
Coarse-Grained (CG) Simulation Analyses.
To quantify the hybridization state of the CMP triple helix, we defined the ensemble average fraction of inter-strand glycine (G) triplets that are intact, fintact. An inter-strand G triplet refers to three G BB beads that are all part of separate strands and are located at the same relative position along the triple helix. For example, the first G BB bead in each CMP strand would constitute the first G triplet. Next, the triplet is counted as intact if the maximum distance between any pair of G BB beads is less than 3 σ. Therefore, a plot of fintact vs. reduced temperature, T*, would describe the melting transition of a given sequence, with the melting temperature defined as . We also measured the diameter of the triple helix at every triplet of BB beads along the triple helix. The diameter was obtained by calculating the center of mass (COM) of the triplet of BB beads (each part of different strands), and the maximum pairwise distance between the COM and each BB bead was defined as the radius of the triple helix. For all results, error bars show the standard deviation obtained from three trials in which the average value was first calculated for each trial, then the standard deviation of the three average values were reported.
Formation and characterization of hydrogels containing designed CMPs.
Hydrogels integrating designed CMPs ((PKG)4(POG)6(DOG)4 or (PKG)3(PKaG)(POG)6(DOG)4) were formed by photoinitiated thiol–ene polymerization with multifunctional reactive monomers. Briefly, each CMP was assembled as described earlier now using a higher concentration in a physiologically-relevant buffer (20 mM CMP in phosphate buffered saline (PBS)) to facilitate hydrogel preparation. The assembled peptide was then diluted to 5 mM in a solution containing a thiolated 4-arm poly(ethylene glycol) (PEG-SH, Mn ~ 20 kDa) monomer, non-assembling peptide linker (KKaGDQGIAGFKaK), and photoinitiator (lithium phenyl-2,4,6-trimethylbenzoylphosphinate, LAP, 2.2 mM) in PBS, synthesized as previously described.23, 57 These hydrogel precursor solutions were prepared at 10 wt% PEG-SH (20 mM SH) along with an equivalent stoichiometric concentration of Kalloc groups coming from a combination of the non-assembling peptide linker and the CMP. Specifically, the total SH to Kalloc ratio remained 1:1, and, in the conditions that contained 5 mM CMP presenting Kalloc, the peptide linker concentration was reduced to achieve 15 mM Kalloc, resulting in a total of 20 mM Kalloc to match the 20 mM thiol. The precursor solutions were gently mixed with a pipette and then placed on the bottom quartz plate of UV-vis attachment on a rheometer (TA AR-G2, TA Instruments, New Castle, DE) connected by a liquid-filled light guide to a filtered mercury arc lamp (Exfo Omnicure Series 2000 light with 365 nm bandpass filter, Excelitas Technologies Corp, Waltham, MA). An 8 mm flat plate geometry was then lowered on to the sample to fill the gap. Samples were photopolymerized to form crosslinked hydrogels (10 mW/cm2 at 365 nm for 4 minutes) while the storage (G′) and loss (G′′) moduli were measured at 1% strain and a frequency of 6 rad s−1, which is within the linear viscoelastic regime for these materials. The modulus values reported represent the mean ± standard error for 4 independent samples after in situ photopolymerization. Statistical significance was determined using a One-Way Analysis Of Variance (ANOVA) with a Tukey’s honestly significant difference (HSD) post hoc test.
Results and Discussion
Collagen mimetic peptide designs
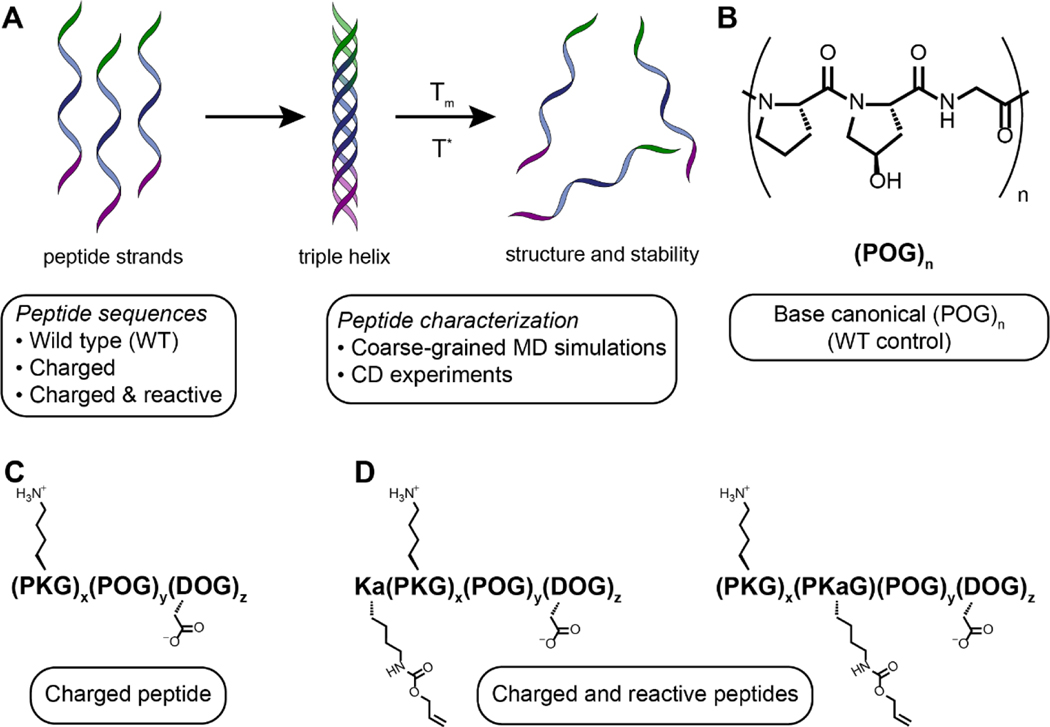
Different collagen mimetic peptide designs were studied using a combination of experiments and simulations to determine the effects of different types of amino acid substitutions on triple helix assembly and stability (Figure 1A). A base sequence of canonical (POG)n served as the ‘WT’ control sequence in all studies (Figure 1B) because this repeat is the most commonly found repeating subunit in native human collagen. The goal of the first set of substitutions to this WT sequence was to incorporate charged amino acids on the termini of the peptides, similar to previously reported work, for systematically understanding their impact on triple helix formation and thermal properties along with establishing and validating our approaches.21, 22 Specifically, a positively charged lysine residue was substituted for the hydroxyproline residue to create a (PKG) block on the N-terminus, and a negatively charged aspartic acid residue was substituted for the proline residue to create a (DOG) block on the C-terminus, where salt-bridged hydrogen bonds between these residues previously have been shown to support end-to-end interactions between triple helices and the formation of fibrils reminiscent of native collagen in seminal studies by Hartgerink and coworkers.21 Melting temperatures of triple helices formed in dilute solution were measured using circular dichroism (CD), where the significant impact of these substitutions on triple helix stability was clear. Atomistic and CG simulations subsequently were used to probe underlying molecular interactions. Experimental and simulations results then were compared to each other, as well as benchmarked against published literature.
Figure 1.
Sequences and important functional groups of designed peptides. (A) Collagen mimetic peptide assembly includes three individual peptide strands interacting via hydrogen bonding to form triple helices, as well as higher ordered structures, and when heated these triple helices disassemble or ‘melt’ allowing for the characterization of the melting temperature and stability of the triple helix. We utilized a combination of atomistic MD simulations, coarse-grained MD simulations, and experiments in this work to examine the effects of both natural and non-natural amino acid substitutions on CMP triple helix assembly and stability for facilitating the design of soft biomaterials. Three classes of peptide designs were examined to probe the effects of amino acid type, placement, and number: (B) a canonical (POG)n base sequence (‘WT’ control), (C) a charged sequence where charged natural amino acid substitutions were made on either end of the peptide to facilitate long range assembly, and (D) a charged and reactive sequence which included an alkene functional handle that enabled post-assembly modification of the assembled peptides.
We subsequently performed a second set of substitutions, expanding upon these previously established sequences, integrating a reactive handle orthogonal to assembly within the charged peptide sequences for covalent modifications if and as desired. Here, we used an allyloxycarbonyl protected lysine residue (Kalloc, Ka), which provides an alkene reactive handle that can be modified using radically-initiated thiol–ene click chemistry and that we have recently shown can be used to integrate these thermally-responsive, assembled structures within covalently crosslinked hydrogels.23 In the new study presented here, not only were we interested in investigating at a molecular level how incorporation of such non-natural amino acid reactive handles influenced peptide assembly, but also how the position for substitution of the reactive handle within the CMP sequence (e.g., substitution at N-terminus versus middle of the sequence) and the length of the hydrogen bonding block influenced assembly and thermal stability (Figure 1C, D).
Experimental examination of the influence of substitutions on triple helix formation and melting
All sequences were synthesized, either wholly or in part, by SPPS techniques, and CD was used to confirm formation of the triple helix and monitor its disassembly upon increasing temperature to determine the melting temperature (Tm) associated with each sequence. The Tm is defined as the point at which half of the triple helices are assembled and disassembled, a measure of the stability of the resulting assembled structures and a property that often is tuned for use of sequences in specific applications (e.g., deployment under physiological conditions). Synthesis of sequences with long repeats of (POG)n for production of bulk materials can face challenges owing, in part, to steric hinderance that can lead to amino acid deletions.36, 37, 58 Consequently, we utilized a combination of automated SPPS and fragment condensation techniques to synthesize the (POG)12 sequence, fabricating on resin (POG)7 using automated SPPS and extending to (POG)12 using manual coupling of the Fmoc-GPO(tBu)-OH fragment and related amino acids, as detailed in Experimental Section.
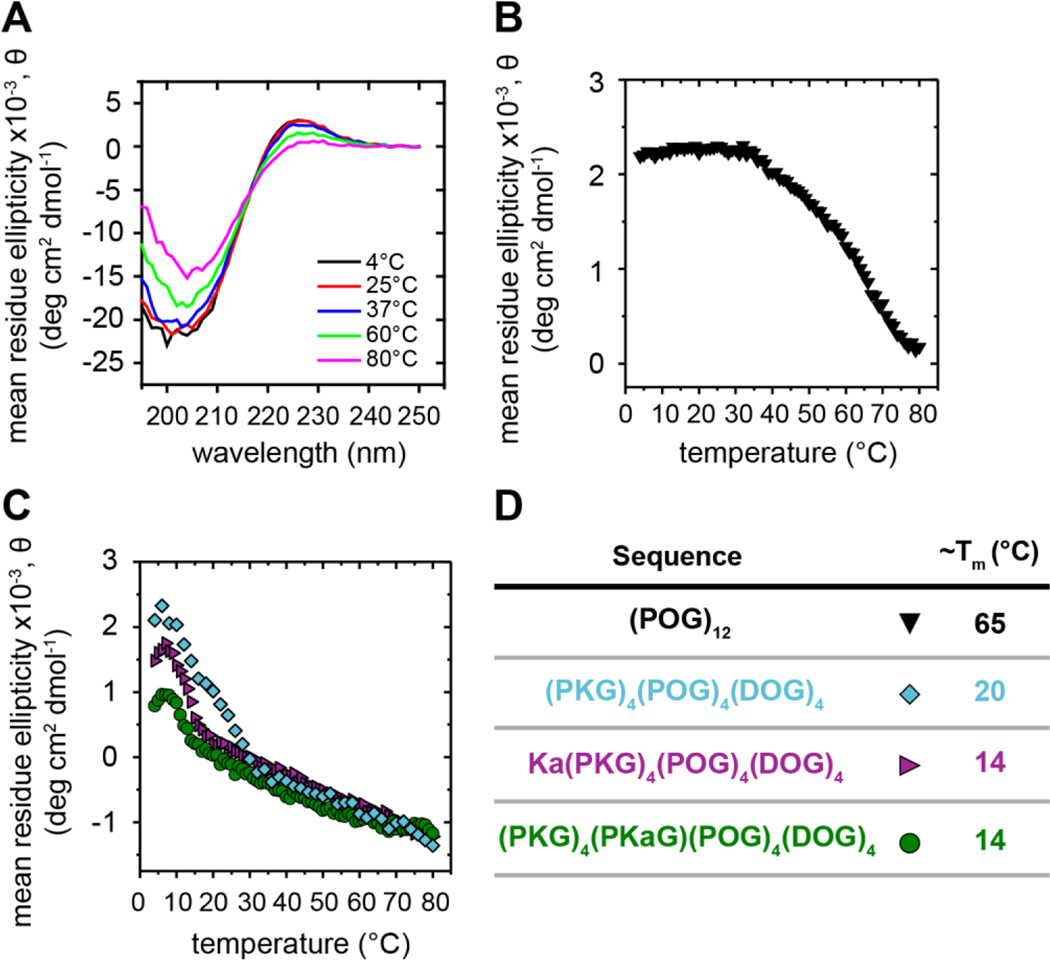
Stable triple helices formed in solution for the WT sequence (POG)12, as determined by the characteristic peak observed in the wavelength scans at approximately 225 nm (Figure 2A), which was then further tracked upon increasing temperature to produce temperature scans (Figure 2B). The Tm recorded for this sequence was approximately 65 °C, similar in magnitude to literature reports.59 Initial studies with peptides containing charged residue substitutions were carried out using sequences with four of each triplet type, (PKG)4(POG)4(DOG)4, which has been established previously to produce large fibrils in dilute solution in aqueous buffer.21, 22 While triple helices were observed in solution for this sequence (Figure S8), notably, a melting temperature of approximately room temperature was observed (Figure 2C), corresponding to a decrease of ~45 °C compared to (POG)12. Further, when the Kalloc residue was substituted into the charged peptide on the N-terminus, the Tm decreased to ~14 °C, corresponding to a decrease of ~51 °C relative to (POG)12. A similar Tm of ~14 °C was observed when the Kalloc residue was substituted within the middle of the sequence, replacing a standard lysine residue in the (PKG) triplet closest to the (POG) triplets. Interestingly, on a per residue basis, a single Kalloc substitution to the charged sequence (9 total substitutions) resulted in the same shift in Tm on a per residue basis as the 8 charged amino acids substitutions relative to (POG)12, corresponding to a ~6 °C reduction per residue substitution. Based on these observations, substitutions involving either charged residues or charged residues and the reactive handle had a significant impact on the stability of the triple helix, as measured by melting temperature. Simulations provided an opportunity to understand these observations at the molecular level.
Figure 2.
Experimental observations of triple helices and related melting temperature of CMPs with different natural and non-natural amino acid substitutions in solution. (A) Example wavelength scans for (POG)12 at various temperatures examining the presence of triple helices (peak at 225 nm). Similar profiles were observed for (PKG)4(POG)4(DOG)4, Ka(PKG)4(POG)4(DOG)4, (PKG)3(PKaG)(POG)4(DOG)4 (Figure S8). Temperature scans were taken for all sequences, monitoring the mean residue ellipticity at 225 nm while increasing the temperature: (B) (POG)12 in black and (C) (PKG)4(POG)4(DOG)4 in cyan, Ka(PKG)4(POG)4(DOG)4 in purple, and (PKG)3(PKaG)(POG)4(DOG)4 in green. The data were fit with a Boltzmann distribution; the first derivative of the fit was taken; and the inflection point, which occurred at the minimum of the first derivative, was used to determine the Tm. (D) Summary table of experimentally measured Tm values for all sequences.
Computational examination of the influence of substitutions on triple helix formation and melting
To complement the experimental measurements and to understand the impact of amino acid substitutions on triple helix stability and structure at the atomistic level, we first used atomistic MD simulations of a single triple helix in explicit water (Figure S7A). Even though the experimentally relevant question of how Tm changes with CMP design cannot be investigated in these unbiased atomistic MD simulations, we briefly summarize the observed atomistic effects of placement of the Kalloc residue on the local and global structure and fluctuations of a single triple helix. These simulations showed that the root mean square deviations (RMSD) from the initial configuration of the peptides for (PKG)3(PKaG)(POG)4(DOG)4 is larger than the RMSD of Ka(PKG)4(POG)4(DOG)4 at multiple time steps, indicating that the placement of the Kalloc residue imparts increased instability to the entire CMP triple helix (Figure S9A). We also calculated the diameter of the CMP triple helix at every triplet of amino acids, where a triplet is defined as three residues that are each part of separate CMP strands and are located at the same relative position along the triple helix, to show if the helix exhibited larger triple helix diameters due to the Kalloc residue placement (Figure S9B). When the Kalloc residue was placed at the end of the CMP sequence (Ka(PKG)4(POG)4(DOG)4), the value of the diameter at the 3rd and 4th triplet were significantly larger than the corresponding values in the CMP sequence with the Kalloc residue located at the center of the sequence ((PKG)3(PKaG)(POG)4(DOG)4). Similarly, these simulations also showed the average end-to-end distance (<Ree>) for (PKG)3(PKaG)(POG)4(DOG)4 is larger than that of Ka(PKG)4(POG)4(DOG)4 (Figure S9). We observed visually that the residues at the ends of the Ka(PKG)4(POG)4(DOG)4 exhibit splaying which in turn decreased the (<Ree>) of that sequence. Overall, the larger RMSD, larger diameters of certain triplets, and smaller (<Ree>) due to splaying at the ends of the CMP triple helix imply that the placement of the Kalloc at the end of the CMP sequence led to larger fluctuations and structural destabilization of the triple helix. Based on this observation, our subsequent investigations focused on CMP sequences with the Kalloc placed in the middle of the sequence.
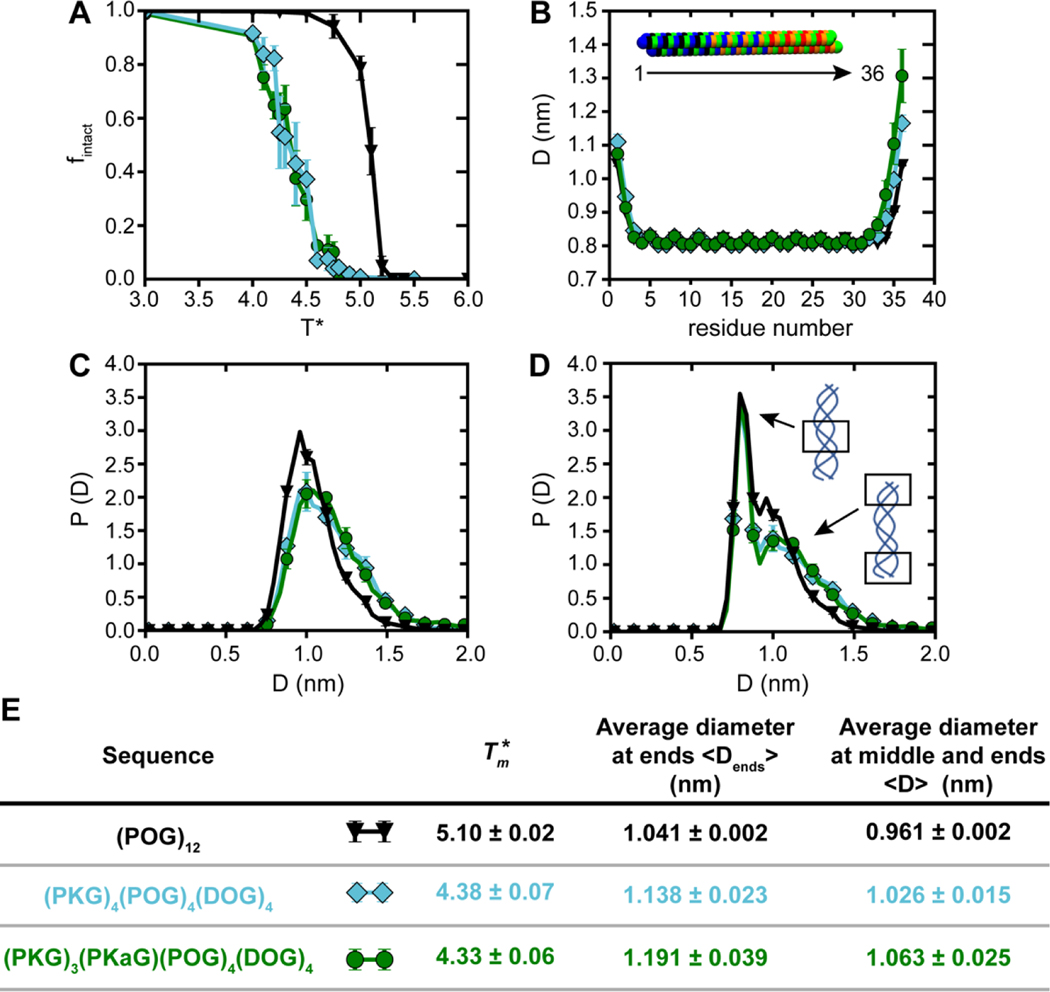
As stated before, the atomistic MD simulations capture local instabilities within a single triple helix but do not predict melting of helices. To predict the melting thermodynamics of CMP triple helices, with and without charged and Kalloc residues, at experimentally relevant concentrations we used CG MD simulations (Figure S7B–D). The results of our CG MD simulations showed that the computational melting temperature () of (POG)12 (black curve) was higher than the of (PKG)4(POG)4(DOG)4 (cyan) and (PKG)3(PKaG)(POG)4(DOG)4 (green), mirroring our experimental results (Figure 3A). The smaller difference in Tm between (PKG)4(POG)4(DOG)4 and (PKG)3(PKaG)(POG)4(DOG)4 seen in experiments was not observed in our CG MD simulations. When comparing (POG)12 (black curve) to (PKG)4(POG)4(DOG)4 (cyan) and (PKG)3(PKaG)(POG)4(DOG)4 (green), we observed a decrease in despite all three sequences having a similar number of H-bonds at the same temperature; thus the decrease in is attributed to the electrostatic repulsion experienced in (PKG)4(POG)4(DOG)4 (cyan) and (PKG)3(PKaG)(POG)4(DOG)4 (green) CMP triple helices (Figure S11).
Figure 3.
Computational melting curves and triple helix diameters obtained from CG MD simulations. (A) Ensemble average fraction of intact glycine triplets (fintact) as a function of computational melting temperature () for (POG)12 (black), (PKG)4(POG)4(DOG)4 (cyan), and (PKG)3(PKaG)(POG)4(DOG)4 (green). (B) Triple helix diameter measured at each residue along the triple helix, where residue 1 corresponds to the (PKG) end (N-terminus) of the triple helix while residue 36 corresponds to the (DOG) end (C-terminus). (C) Probability distributions of diameters measured only at the two ends (and not in the middle) of the triple helix. (D) Probability distributions of diameters measured at both the middle and ends of the triple helix. (E) A table summarizing values of and average diameters at the ends and middle for all sequences.
Interestingly, even though the CG MD simulations predicted similar Tm for (PKG)4(POG)4(DOG)4 and (PKG)3(PKaG)(POG)4(DOG)4 (Figure 3A), the CG MD simulations captured a residue-level destabilizing effect of the Kalloc residue on the structure as shown by the increased diameter at the end of the triple helix for (PKG)3(PKaG)(POG)4(DOG)4 versus (PKG)4(POG)4(DOG)4 (Figure 3B). For all three CMP sequences there are oscillations in the diameter of the CMP triple helix for the triplets in the central portion of the triple helix. As each triplet corresponds to three residues from three different CMP strands, for triplets comprised of residues with hydrogen bonding sites, the hydrogen bonding interactions as modeled in our CG model facilitates closer contacts between neighboring strands than for triplets comprised of residues without hydrogen bonding.
Next, to understand how the inclusion of charged amino acids impacted the dimensions of the triple helix, we obtained probability distributions of triple helix diameters for (POG)12, (PKG)4(POG)4(DOG)4, and (PKG)3(PKaG)(POG)4(DOG)4, where the diameter was measured at the ends of the triple helix. We observed that the probability distribution of helix diameters at the ends of the triple helix (Figure 3C) was broader and shifted to larger diameters for charged CMP sequences compared to neutral CMP sequences, which meant that charged CMP triple helices were more structurally unstable relative to the tightly wound, neutral CMP triple helices (Figure 3C, black, cyan, and green curves). Furthermore, we hypothesized that the shift to larger diameters for charged CMP sequences is due to the electrostatic repulsion between charged residues leading to an increase in both the average helical diameter as well as the breadth of the probability distributions of helix diameters for charged CMP sequences. This is confirmed by the probability distributions of the diameter all along the triple helix (i.e., both in the middle and at the ends of the triple helix) (Figure 3D). For all CMP sequences, we observed a bimodal distribution with a single peak followed by a shoulder-like appearance at larger diameters. The first peak in each distribution corresponded to the diameter measured at the center of the helix comprised of POG repeats where the helix is intact and stable, and the shoulder at larger diameters corresponds to the splay near the ends of the helix. For the charged CMP sequences, this shoulder is shifted to larger distances as compared to the uncharged WT CMP sequence, confirming that the charged residues present at the ends of the CMP sequence create a structural destabilization.
Overall, these simulations allowed us to gain insight at the molecular level into the origins of differences between triple helix stability for different sequences designs. In both atomistic and CG MD simulations increased fluctuations were observed when charged groups were incorporated due to electrostatic repulsions. CG MD simulations confirm that these electrostatic repulsions are significant enough to reduce the melting transition of the CMP compared to the neutral sequences. The presence and absence of the reactive handle (Kalloc) in the CMP sequence exhibited significant differences in fluctuations within a single helix in the atomistic MD simulations but exhibited insignificant changes in melting transition temperature at experimentally relevant concentrations explored with the CG MD simulations. This understanding of key interactions allowed us to then propose and simulate new sequence designs prior to synthesis and confirm stability a priori.
CMP sequence designs guided by model predictions
After studying sequences with the same number of triplets, which resulted in melting temperatures lower than those needed for many biological applications, we set out to use our developed model to predict melting trends and triple helix stability for two additional sequences where the number of hydrogen bonding units was increased. Specifically, based on insights from our prior experimental and simulations results, we hypothesized that the Tm of the sequences would increase upon the addition of more (POG)n repeats to offset the effects of charge repulsion and steric hinderance introduced by the natural and non-natural amino acid substitutions. Here, (PKG)4(POG)6(DOG)4 (red) and (PKG)3(PKaG)(POG)6(DOG)4 (yellow) were simulated, with an increase to six POG repeats compared to the four POG repeats in the previous section.
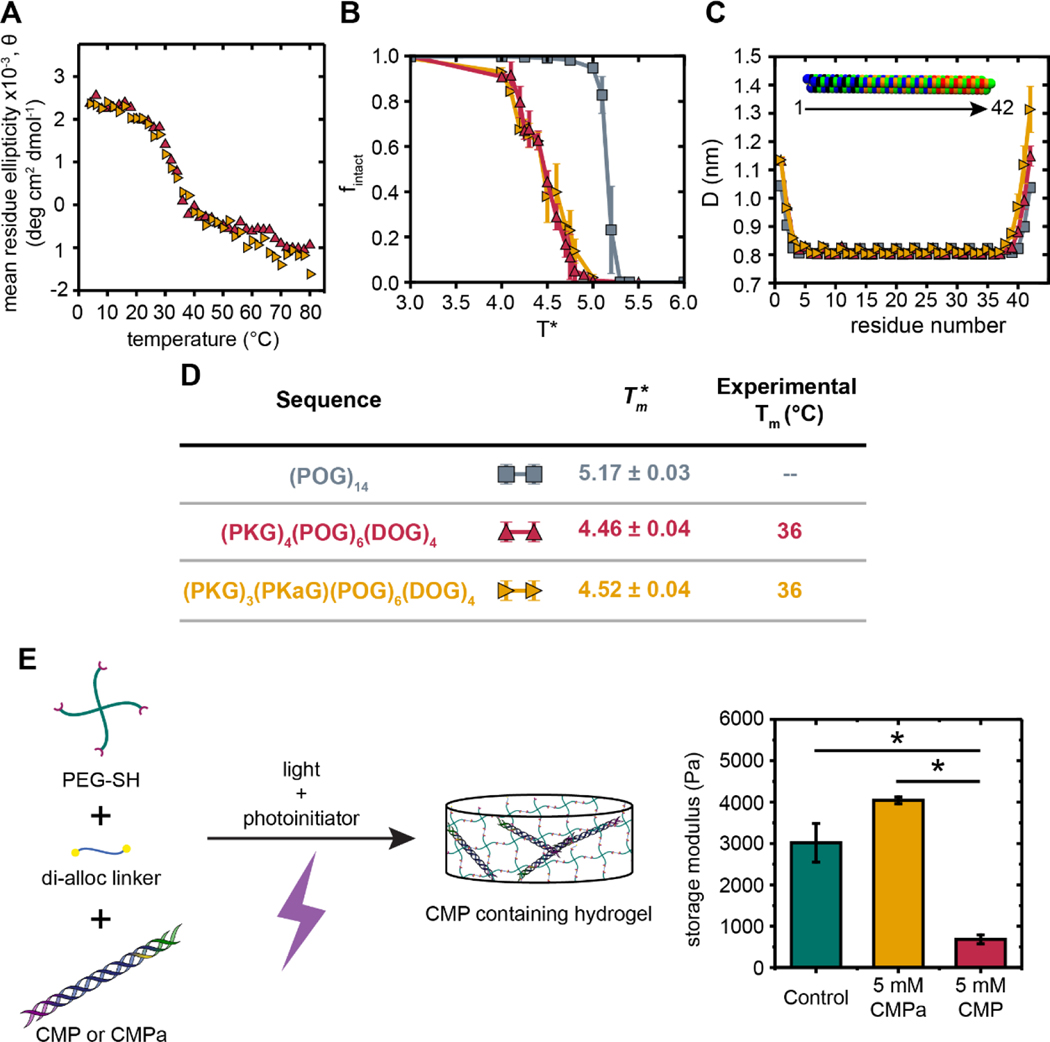
Our CG MD simulations supported this hypothesis by showing an increase in the number of hydrogen bonds with an increasing number of POG units for uncharged (POG12 vs. POG14), charged CMPs without reactive handles ((PKG)4(POG)4(DOG)4 vs. (PKG)4(POG)6(DOG)4), and charged CMPs with a reactive handle ((PKG)3(PKaG)(POG)4(DOG)4 vs. (PKG)3(PKaG)(POG)6(DOG)4) (Figure S12). This increased number of hydrogen bonds with an increasing number of POG units leads to higher melting temperatures for CMPs in Figure 4 as compared to Figure 3. Besides the increase in melting temperature going from (POG)12 to (POG)14, (PKG)4(POG)4(DOG)4 to (PKG)4(POG)6(DOG)4, and from (PKG)3(PKaG)(POG)4(DOG)4 to (PKG)3(PKaG)(POG)6(DOG)4, all trends seen before in Figure 3 are repeated in Figure 4. For example, the charged CMP sequences have lower melting temperature compared to the neutral CMP sequences. Further, the charged CMP sequences with and without the reactive handle exhibit similar values. The impact of charged residues on the dimensions of longer CMP triple helices (Figure 4C) also were similar to what was observed for the shorter sequences (Figure 3B–D). Specifically, we observed that the locations of the splay (i.e., the last three residues near the C-terminus and last three residues near the N-terminus) were similar for both the long (Figure 4C) and short CMP sequences (Figure 3B) shown above, despite the former having larger Tm. Larger diameters at the C-terminus of the peptide emerged when the charged groups were added, which increased even more when the Kalloc residue was integrated in the middle of the CMP sequence from 1.15 nm to approximately 1.3 nm.
Figure 4.
Comparison of experimental and computational melting curves and triple helix diameters for sequences designed for integration within soft biomaterials. (A) Temperature scans for designed CMPs in solution were obtained by monitoring the CD value at a wavelength of 225 nm while increasing temperature from 4 °C to 80 °C. (B) Plot of fintact as a function for (PKG)3(PKaG)(POG)6(DOG)4 (CMPa, yellow), (PKG)4(POG)6(DOG)4 (CMP, red), and (POG)14 (grey). (C) Triple helix diameter measured at each residue along the triple helix where residue 1 corresponds to the (PKG) end of the triple helix (N-terminus) while residue 42 corresponds to the (DOG) end (C-terminus). (D) A summary table of and Tm values for lengthened sequences. (E) Designed peptides (CMP or CMPa) were assembled (20 mM in PBS) and diluted (5 mM) into reactive monomer solution for formation of a covalently crosslinked hydrogel via light-triggered thiol–ene reaction: 10 wt% 4-arm PEG-SH, non-assembling di-alloc peptide linker, and either i) CMP (red), ii) CMPa (yellow), or iii) no CMP (control hydrogel, blue-green) (1:1 SH:alloc). The mechanical properties of the resulting hydrogels were measured by in situ rheometry. Note, schematic not drawn to scale. The data shown here illustrate the mean with error bars showing the standard error (n = 4, *p < 0.05). Statistical significance was determined by a One-way ANOVA with a Tukey’s HSD post hoc test.
To confirm the computationally predicted melting trends experimentally, the two designed CMP sequences (PKG)4(POG)6(DOG)4 and (PKG)3(PKaG)(POG)6(DOG)4 were synthesized and purified, using SPPS and HPLC, and assembled in 10 mM sodium phosphate buffer. Temperature scans of both new sequences, with and without the Kalloc, showed that both sequences had melting temperatures of approximately 36 °C, and that the corresponding melting curves heavily overlapped as predicted by the CG simulations. These Tm values were higher, and closer to physiologically relevant values, than those of the previous sequences that only contained four (POG) repeat units, which is of particular interest for biological applications.
An important facet of the designed sequences is the combination of non-natural and natural amino acids: the reactive handle presented by the non-natural Kalloc provides opportunities for modifying assembled structures with designed thermal properties for the construction of nanostructured biomaterials. To confirm that the Kalloc reactive handle allowed integration of the resulting assembled structures within robust biomaterials (Figure 4E), each designed sequence ((PKG)4(POG)6(DOG)4 (denoted as CMP) or (PKG)3(PKaG)(POG)6(DOG)4 (denoted as CMPa)) was assembled (20 mM) and then briefly diluted (5 mM) into a solution with reactive monomers (10 wt% 4-arm PEG-SH with non-assembling dialloc peptide linker; 1:1 SH:alloc) for light-triggered hydrogel formation. Covalently crosslinked hydrogels incorporating i) the assembled CMP, ii) the assembled and reactive CMPa, or iii) no CMP (control hydrogel) then rapidly were formed by photoinitiated thiol–ene click chemistry in situ on a rheometer for facile monitoring of the resulting hydrogel properties, as detailed in the Methods. When the assembled and reactive CMPa was incorporated, hydrogels formed with statistically the same moduli as control hydrogels without any CMP, indicating integration of the assembled nanostructures within the covalently crosslinked hydrogel network. However, when the non-reactive CMP was incorporated, hydrogels formed with a statistical decrease in the modulus compared to both the control and the CMPa hydrogel conditions. Overall, these observations show the utility of incorporating the reactive handle within computationally-designed CMPs for the formation of robust hydrogels that integrate assembled structures.
In summary, the above computational and experimental results for the various CMP sequences show that, even though the presence of the non-natural Kalloc residue induces increased structural destabilization, it does not alter the number of hydrogen bonds formed within the triple helix. As a result, upon inclusion of the Kalloc residue, the CMP triple helix melting transition is either not altered (for longer CMP sequences) or exhibits small changes (for shorter CMP sequences). Incorporation of natural charged residues in the short and long CMP sequences led to increased electrostatic repulsion causing structural and thermal destabilization, in both experiments and simulations. Impacts of these substitutions on the melting temperature of the CMP could be offset through inclusion of additional hydrogen bonding units, where additional (POG) repeats were used here. Broadly, these results highlight the usefulness of this approach and platform for capturing structure-property trends for different peptide designs relevant for the fabrication of soft biomaterials without having to iterate using only experimental approaches that can be resource and time intensive. Opportunities for future investigations include the design of new sequences that integrate different types, positions, and numbers of both natural and non-natural functionalities (e.g., multiple reactive handles within a single sequence, end and pendant functional groups, number of repeats), where the CG model versatility will allow probing the effects of these different functional groups on triple helix stability. Further, integration of these different CMP designs within hydrogels may prove useful in both biological applications and more fundamental materials studies, where the interplay between Tm and crosslink density can be investigated for mimicry of and comparison to naturally-derived systems.60
Conclusion
In this work, we have established a robust approach for the molecular design, prediction, and characterization of the properties of multifunctional CMPs. First, we synthesized peptides with both natural and non-natural amino acid residues of relevance for fabrication of hydrogels to determine the impact of substitutions and their placement on the stability of the CMP triple helix relative to a WT canonical (POG)n sequence. In parallel, atomistic and CG models were used to both understand and predict trends in structural and thermal stability of triple helices when substitutions were made. Having established the validity of model predictions versus experimental data, the CG models subsequently were used to predict behavior of newly designed sequences prior to synthesis for achieving desired thermal stability and integrating these assembled structures within hydrogels. Overall, these multiscale simulations gave insight and understanding into how specific molecular interactions within such engineered sequences influenced their structure and stability. This combination of experimental and molecular simulations provides a robust platform for informing peptide design and synthesis based on predicted thermal properties. Furthermore, the validated coarse-grained CMP model can be extended to study thermal stability and self-assembly of peptides containing other natural and non-natural amino acids as well as reactive handles; this CG model is being made available to the public on an open source simulation platform (www.mosdef.org). This ability to study and design multifunctional CMPs with natural and non-natural amino acid substitutions, or potentially other assembling sequences, could allow evaluation of the effects of a variety of substitutions and predicting the resulting properties prior to their synthesis. This synergy offers a large phase space for innovation in which this platform for rational design of assembling peptides tailored to a specific application could be used to study amino acid substitutions and resulting stability and structures.
Supplementary Material
Acknowledgements
The computational work in this paper and the ongoing work on coarse grained CMP model dissemination via mosdef.org were financially supported by National Science Foundation (NSF) Grants 1703402 and 1835613, respectively. The computational work in this paper was also supported by the information technologies resources at the University of Delaware, specifically in the form of the Farber high-performance computing resources. The experimental work in this paper was supported in part by grants on related work from the National Science Foundation DMR BMAT (1253906), the Pew Charitable Trusts (26178), and National Institutes of Health (NIH) Director’s New Innovator Award (DP2 HL152424–01). The authors would like to acknowledge the University of Delaware Mass Spectrometry Core facilities, supported by the Delaware COBRE programs with grants from the National Institute of General Medical Sciences (NIGMS) (P20GM104316 and P30GM110758) from the NIH, and the Millicent Sullivan group for use of equipment. The authors would like to thank Dr. Bryan Sutherland for helpful feedback and discussions on drafts of this manuscript.
Footnotes
Supporting Information. Includes CD measurements for charged sequences, atomistic simulation results, simulation snapshots, additional hydrogen bond analyses, and mass spectrometry of all peptides.
Competing interests: The authors declare no competing interests.
References
- 1.Ryadnov MG; Woolfson DN, Engineering the morphology of a self-assembling protein fibre. Nature Materials 2003, 2, 329. [DOI] [PubMed] [Google Scholar]
- 2.Song Z; Fu H; Wang R; Pacheco LA; Wang X; Lin Y; Cheng J, Secondary structures in synthetic polypeptides from N-carboxyanhydrides: design, modulation, association, and material applications. Chemical Society Reviews 2018, 47 (19), 7401–7425. [DOI] [PubMed] [Google Scholar]
- 3.Zhang HV; Polzer F; Haider MJ; Tian Y; Villegas JA; Kiick KL; Pochan DJ; Saven JG, Computationally designed peptides for self-assembly of nanostructured lattices. Science Advances 2016, 2 (9), e1600307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cui H; Webber MJ; Stupp SI, Self-assembly of peptide amphiphiles: from molecules to nanostructures to biomaterials. Biopolymers 2010, 94, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hartgerink JD; Beniash E; Stupp SI, Self-Assembly and Mineralization of Peptide-Amphiphile Nanofibers. Science 2001, 294 (5547), 1684. [DOI] [PubMed] [Google Scholar]
- 6.Tang JD; Mura C; Lampe KJ, Stimuli-Responsive, Pentapeptide, Nanofiber Hydrogel for Tissue Engineering. Journal of the American Chemical Society 2019, 141 (12), 4886–4899. [DOI] [PubMed] [Google Scholar]
- 7.Shoulders MD; Raines RT, Collagen structure and stability. Annual Review of Biochemistry 2009, 78, 929–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Holmgren S; Taylor K; Bretscher L; Raines RT, Code for collagen’s stability deciphered. Nature 1998, 392, 666–667. [DOI] [PubMed] [Google Scholar]
- 9.Hosoyama K; Lazurko C; Muñoz M; McTiernan CD; Alarcon EI, Peptide-Based Functional Biomaterials for Soft-Tissue Repair. Frontiers in Bioengineering and Biotechnology 2019, 7, 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yu SM; Li Y; Kim D, Collagen mimetic peptides: progress towards functional applications. Soft Matter 2011, 7, 7927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Przybyla DE; Chmielewski J, Metal-Triggered Radial Self-Assembly of Collagen Peptide Fibers. Journal of the American Chemical Society 2008, 130 (38), 12610–12611. [DOI] [PubMed] [Google Scholar]
- 12.Pires MM; Przybyla DE; Chmielewski J, A metal-collagen peptide framework for three-dimensional cell culture. Angewandte Chemie International Edition 2009, 48, 7813–7. [DOI] [PubMed] [Google Scholar]
- 13.Stahl PJ; Romano NH; Wirtz D; Yu SM, PEG-based hydrogels with collagen mimetic peptide-mediated and tunable physical cross-links. Biomacromolecules 2010, 11, 2336–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hulgan SAH; Jalan AA; Li IC; Walker DR; Miller MD; Kosgei AJ; Xu W; Phillips GN; Hartgerink JD, Covalent Capture of Collagen Triple Helices Using Lysine–Aspartate and Lysine–Glutamate Pairs. Biomacromolecules 2020. [DOI] [PubMed] [Google Scholar]
- 15.Gottlieb D; Morin SA; Jin S; Raines RT, Self-Assembled Collagen-like Peptide Fibers as Templates for Metallic Nanowires. Journal of Materials Chemistry 2008, 18, 3865–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krishna OD; Kiick KL, Supramolecular assembly of electrostatically stabilized, hydroxyproline-lacking collagen-mimetic peptides. Biomacromolecules 2009, 10, 2626–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kasznel AJ; Zhang Y; Hai Y; Chenoweth DM, Structural Basis for Aza-Glycine Stabilization of Collagen. Journal of the American Chemical Society 2017, 139 (28), 9427–9430. [DOI] [PubMed] [Google Scholar]
- 18.Melton SD; Chenoweth DM, Variation in the Yaa position of collagen peptides containing azaGlycine. Chemical Communications 2018, 54 (84), 11937–11940. [DOI] [PubMed] [Google Scholar]
- 19.Aronoff MR; Egli J; Menichelli M; Wennemers H, γ-Azaproline Confers pH Responsiveness and Functionalizability on Collagen Triple Helices. Angewandte Chemie International Edition 2019, 58 (10), 3143–3146. [DOI] [PubMed] [Google Scholar]
- 20.Hentzen NB; Smeenk LEJ; Witek J; Riniker S; Wennemers H, Cross-Linked Collagen Triple Helices by Oxime Ligation. Journal of the American Chemical Society 2017, 139 (36), 12815–12820. [DOI] [PubMed] [Google Scholar]
- 21.O’Leary LER; Fallas JA; Bakota EL; Kang MK; Hartgerink JD, Multi-hierarchical self-assembly of a collagen mimetic peptide from triple helix to nanofibre and hydrogel. Nature Chemistry 2011, 3, 821–8. [DOI] [PubMed] [Google Scholar]
- 22.Rele S; Song Y; Apkarian RP; Qu Z; Conticello VP; Chaikof EL, D-Periodic Collagen-Mimetic Microfibers. Journal of the American Chemical Society 2007, 129 (47), 14780–14787. [DOI] [PubMed] [Google Scholar]
- 23.Hilderbrand AM; Ford EM; Guo C; Sloppy JD; Kloxin AM, Hierarchically structured hydrogels utilizing multifunctional assembling peptides for 3D cell culture. Biomaterials Science 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Taylor PA; Jayaraman A, Molecular Modeling and Simulations of Peptide–Polymer Conjugates. Annual Review of Chemical and Biomolecular Engineering 2020, 11 (1), 257–276. [DOI] [PubMed] [Google Scholar]
- 25.Raman SS; Parthasarathi R; Subramanian Y; Ramasami T, Role of length-dependent stability of collagen-like peptides. Journal of Physical Chemistry B 2008, 112 (5), 1533–1539. [DOI] [PubMed] [Google Scholar]
- 26.Sundar Raman S; Parthasarathi R; Subramanian V; Ramasami T, Role of aspartic acid in collagen structure and stability: A molecular dynamics investigation. Journal of Physical Chemistry B 2006, 110 (41), 20678–20685. [DOI] [PubMed] [Google Scholar]
- 27.Punitha V; Raman SS; Parthasarathi R; Subramanian V; Rao JR; Nair BU; Ramasamr T, Molecular dynamics investigations on the effect of D amino acid substitution in a triple-helix structure and the stability of collagen. Journal of Physical Chemistry B 2009, 113 (26), 8983–8992. [DOI] [PubMed] [Google Scholar]
- 28.Condon JE; Jayaraman A, Development of a Coarse-Grained Model of Collagen-Like Peptide (CLP) for Studies of CLP Triple Helix Melting. The Journal of Physical Chemistry B 2018, 122 (6), 1929–1939. [DOI] [PubMed] [Google Scholar]
- 29.Buehler MJ, Atomistic and continuum modeling of mechanical properties of collagen: Elasticity, fracture, and self-assembly. Journal of Materials Research 2006, 21 (2), 1947–1961. [Google Scholar]
- 30.Buehler MJ, Nature designs tough collagen: Explaining the nanostructure of collagen fibrils. Proceedings of the National Academy of Sciences 2006, 103 (33), 12285–12290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gautieri A; Russo A; Vesentini S; Redaelli A; Buehler MJ, Coarse-grained model of collagen molecules using an extended MARTINI force field. Journal of Chemical Theory and Computation 2010, 6 (4), 1210–1218. [Google Scholar]
- 32.Monticelli L; Kandasamy SK; Periole X; Larson RG; Tieleman DP; Marrink SJ, The MARTINI coarse grained force field: extension to proteins. J. Chem. Theory Comput. 2008, 4 (5), 819–834. [DOI] [PubMed] [Google Scholar]
- 33.Xu F; Zahid S; Silva T; Nanda V, Computational design of a collagen A:B:C-type heterotrimer. J Am Chem Soc 2011, 133 (39), 15260–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fallas JA; Hartgerink JD, Computational design of self-assembling register-specific collagen heterotrimers. Nat Commun 2012, 3, 1087. [DOI] [PubMed] [Google Scholar]
- 35.Persikov AV; Ramshaw JA; Brodsky B, Prediction of collagen stability from amino acid sequence. Journal of Biological Chemistry 2005, 280 (19), 19343–19349. [DOI] [PubMed] [Google Scholar]
- 36.Ottl J; Jürgen Musiol H; Moroder L, Heterotrimeric collagen peptides containing functional epitopes. Synthesis of single-stranded collagen type I peptides related to the collagenase cleavage site. Journal of Peptide Science 1999, 5 (2), 103–110. [DOI] [PubMed] [Google Scholar]
- 37.Melton SD; Smith MS; Chenoweth DM, Incorporation of Aza-Glycine into Collagen Peptides. The Journal of Organic Chemistry 2020, 85 (3), 1706–1711. [DOI] [PubMed] [Google Scholar]
- 38.Dai N; Wang XJ; Etzkorn FA, The Effect of a Trans-Locked Gly-Pro Alkene Isostere on Collagen Triple Helix Stability. Journal of the American Chemical Society 2008, 130 (16), 5396–5397. [DOI] [PubMed] [Google Scholar]
- 39.Kar K; Amin P; Bryan MA; Persikov AV; Mohs A; Wang Y-H; Brodsky B, Self-association of collagen triple helic peptides into higher order structures. The Journal of Biological Chemistry 2006, 281, 33283–90. [DOI] [PubMed] [Google Scholar]
- 40.Li Y; Foss C. a.; Summerfield DD; Doyle JJ; Torok CM; Dietz HC; Pomper MG; Yu SM, Targeting collagen strands by photo-triggered triple-helix hybridization. Proceedings of the National Academy of Sciences of the United States of America 2012, 109, 14767–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abraham MJ; Murtola T; Schulz R; Páll S; Smith JC; Hess B; Lindah E, Gromacs: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 2015, 1–2, 19–25. [Google Scholar]
- 42.Huang C; Couch G; Pettersen E; Ferrin T; Howard A; Klein T. In The object technology framework: an object-oriented interface to molecular data and its application to collagen, Pacific Symposium on Biocomputing. Pacific Symposium on Biocomputing, 1998; pp 349–361. [PubMed] [Google Scholar]
- 43.DeLano WL, The PyMOL Molecular Graphics System. 2002. [Google Scholar]
- 44.Shoulders MD; Raines RT, Collagen structure and stability. Annual review of biochemistry 2009, 78, 929–958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarkar B; O’Leary LE; Hartgerink JD, Self-assembly of fiber-forming collagen mimetic peptides controlled by triple-helical nucleation. Journal of the American Chemical Society 2014, 136 (41), 14417–14424. [DOI] [PubMed] [Google Scholar]
- 46.Oostenbrink C; Soares TA; van der Vegt NFA; van Gunsteren WF, Validation of the 53A6 GROMOS force field. European Biophysics Journal 2005, 34 (4), 273–284. [DOI] [PubMed] [Google Scholar]
- 47.Mark P; Nilsson L, Structure and Dynamics of the TIP3P, SPC, and SPC/E Water Models at 298 K. The Journal of Physical Chemistry A 2001, 105 (43), 9954–9960. [Google Scholar]
- 48.Hess B; Bekker H; Berendsen HJC; Fraaije JGEM, LINCS: A linear constraint solver for molecular simulations. Journal of Computational Chemistry 1997, 18 (12), 1463–1472. [Google Scholar]
- 49.Berendsen HJC; Postma JPM; van Gunsteren WF; DiNola A; Haak JR, Molecular dynamics with coupling to an external bath. The Journal of Chemical Physics 1984, 81 (8), 3684–3690. [Google Scholar]
- 50.Csizmadia P, MarvinSketch and MarvinView: molecule applets for the World Wide Web. 1999. [Google Scholar]
- 51.Andersen HC, Rattle: A “velocity” version of the shake algorithm for molecular dynamics calculations. Journal of Computational Physics 1983, 52 (1), 24–34. [Google Scholar]
- 52.Lennard-Jones JE, On the Determination o f Molecular Fields. — II. From the Equation o f State o f a Gas. Proc. R. Soc. Lond. A 1924, 106 (738), 463–477. [Google Scholar]
- 53.Weeks JD; Chandler D; Andersen HC, Role of repulsive forces in determining the equilibrium structure of simple liquids. The Journal of Chemical Physics 1971, 54 (12), 5237–5247. [Google Scholar]
- 54.Plimpton S, Fast Parallel Algorithms for Short-Range Molecular Dynamics. Journal of Computational Physics 1995, 117 (1), 1–19. [Google Scholar]
- 55.Ghobadi AF; Jayaraman A, Effect of backbone chemistry on hybridization thermodynamics of oligonucleic acids: a coarse-grained molecular dynamics simulation study. Soft matter 2016, 12 (8), 2276–2287. [DOI] [PubMed] [Google Scholar]
- 56.Tuckerman M; Berne BJ; Martyna GJ, Reversible multiple time scale molecular dynamics. The Journal of Chemical Physics 1992, 97 (3), 1990–2001. [Google Scholar]
- 57.Fairbanks BD; Schwartz MP; Bowman CN; Anseth KS, Photoinitiated polymerization of PEG-diacrylate with lithium phenyl-2,4,6-trimethylbenzoylphosphinate: polymerization rate and cytocompatibility. Biomaterials 2009, 30, 6702–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jenkins CL; Vasbinder MM; Miller SJ; Raines RT, Peptide Bond Isosteres: Ester or (E)-Alkene in the Backbone of the Collagen Triple Helix. Organic Letters 2005, 7 (13), 2619–2622. [DOI] [PubMed] [Google Scholar]
- 59.Persikov AV; Ramshaw JAM; Brodsky B, Prediction of Collagen Stability from Amino Acid Sequence. Journal of Biological Chemistry 2005, 280 (19), 19343–19349. [DOI] [PubMed] [Google Scholar]
- 60.Brad Bennett C; Kruczek J; Rabson DA; Garrett Matthews W; Pandit SA, The effect of cross-link distributions in axially-ordered, cross-linked networks. Journal of Physics: Condensed Matter 2013, 25 (28), 285101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.