Abstract
Different ways of calculating mortality during epidemics have yielded very different results, particularly during the current COVID-19 pandemic. For example, the “CFR” has been interchangeably called the case fatality ratio, case fatality rate, and case fatality risk, often without standard mathematical definitions. The most commonly used CFR is the case fatality ratio, typically constructed using the estimated number of deaths to date divided by the estimated total number of confirmed infected cases to date. How does this CFR relate to an infected individual’s probability of death? To explore such issues, we formulate both a survival probability model and an associated infection duration-dependent SIR model to define individual- and population-based estimates of dynamic mortality measures to show that neither of these are directly represented by the case fatality ratio. The key parameters that affect the dynamics of different mortality estimates are the incubation period and the time individuals were infected before confirmation of infection. Using data on the recent SARS-CoV-2 outbreaks, we estimate and compare the different dynamic mortality estimates and highlight their differences. Informed by our modeling, we propose more systematic methods to determine mortality during epidemic outbreaks and discuss sensitivity to confounding effects and uncertainties in the data arising from, e.g., undertesting and heterogeneous populations.
INTRODUCTION
Mortality metrics are key quantities describing the severity of a viral disease [1]. During an outbreak, these metrics typically evolve in time before converging to a constant value and can be defined in a number of ways. Commonly used metrics are the case fatality ratio, case fatality rate, and case fatality risk, which are all confusingly denoted “CFR” [2, 3]. Fatality rate implies a change in deaths per unit time, risk implies an individual probability, while ratio implies a fraction of two numbers, typically populations. CFR is most often defined as the ratio of the total estimated number of deaths to date, D(t), to the estimated number of all confirmed cases to date Nc(t) [1, 4–6]. These numbers are key to estimating disease severity. Usually, antibody [7] and reverse transcription-polymerase chain reaction (RT-PCR) testing [8] is used to confirm SARS-CoV-2-positive patients. To find D(t), the number of patients who actually die of COVID-19 must also be quantified. In Italy, deaths of patients with positive RT-PCR testing for SARS-CoV-2 are reported as COVID-19 deaths, but the criteria for COVID-19-related deaths are currently not clearly defined and may vary from region to region [9].
Some studies define CFR as the “case fatality risk” and associate it with the probability of death of an individual confirmed case within “a period of time” [10]. Yet others define case fatality ratio as simply “case fatality” and reserve the term case fatality ratio to mean the ratio of the case fatalities of two different diseases [3]. Infection fatality ratios (IFR), the number of deaths to date divided by the number of all infected individuals, have also been used [11–13] although the IFR = D(t)/N(t) requires an estimate of N(t), the number of total (including unconfirmed) infected individuals. Similarly, IFR has also been called the “infection fatality risk,” the probability of an individual dying conditioned on being infected. This individual-based definition of IFR is thus equivalent to the individual-based case fatality risk. However, in nearly all practical cases, both the CFR and IFR are estimated from aggregated population data from past outbreaks [4] as well as from those of the recent SARS-CoV-2 outbreaks [1, 11, 13–17].
Since case fatality ratio is the most commonly used, we henceforth define CFR = D(t)/Nc(t). We show examples of CFR curves (orange), which typically vary significantly both by region and in time, in Fig. 1 and in the Results and Discussion section. During the severe 2003 acute respiratory syndrome (SARS) outbreak in Hong Kong, the World Health Organization (WHO) also used the aforementioned estimate to obtain an initial CFR ~ 3% while the final values, after resolution of infections, approached 17.0% [18, 19] (see Fig. 1(a)). Another population-based mortality ratio is Mp(t) = D(t)/(D(t) + R(t)), the number of deaths divided by the sum of death and recovered cases (the number of resolved cases), up to time t is shown in blue in Figs. 1(a–b). In principle, Mp(t) should be a better measure of the likelihood of death, but it is underestimated by the CFR = D(t)/Nc(t). For example, Mp(t) is currently (as of April 25, 2020) 203, 164/(203, 164 + 836, 612) ≈ 20%, significantly higher than the April 25, 2020 CFR(t) = D(t)/Nc(t) = 203, 164/2, 919, 404 ≈ 7% estimate [22]. Despite this underestimation, the CFR is still commonly used by the WHO and other health officials, such as in the ongoing SARS-CoV-2 outbreaks [1, 12, 13, 15, 17] (see Table I). As shown in Fig. 1(c), the CFR would correspond to the mortality ratio only if all tested infected individuals recover. Such underestimations by CFRs may lead to insufficient countermeasures and a more severe epidemic [23, 24].
FIG. 1. Mortality estimates.
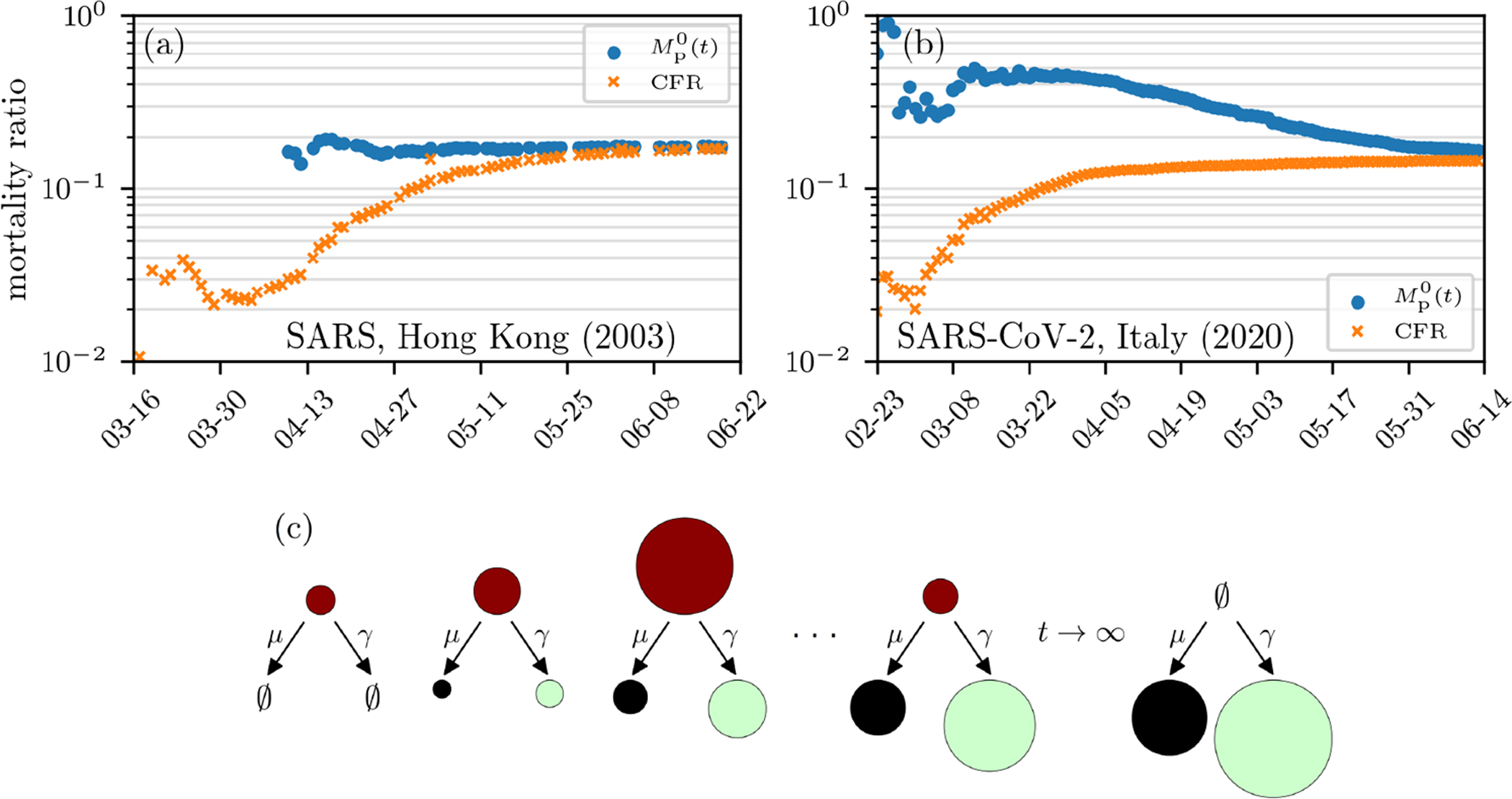
(a–b) Estimates of mortality ratios (see Eqs. (9) and (14)) of SARS-CoV infections in Hong Kong (2003) [20] and SARS-CoV-2 infections in Italy. (c) Evolution of the cumulative number of infected (red), death (black), and recovered (green) cases. The size of the circles indicates the number of cases in the respective compartments on a certain day. Note that CFR and have exhibited qualitatively similar behavior across different epidemics. The data are based on Ref. [21].
TABLE I.
Different CFR estimates of COVID-19.
Since meaningful and accurate mortality metrics are critical for assessing the risks associated with epidemic outbreaks, we first unambiguously define the probability M1(t) that a single, newly infected individual will die of the disease by a given time. This probability has also been called the case fatality risk, but without specifying its dependence on time after infection [10]. This intrinsic mortality or probability of death, can be identified as one minus the survival probability of a single infected individual. It should be an intrinsic property of the virus and the infected individual, depending on age, health, access to health care, etc., and not directly on the population-level dynamics of infected and recovered individuals. Whether this individual infects others does not directly affect his probability of eventually dying [31].
In the next section, we derive a survival probability model for M1(t) similar to that in Ghani et al. [32]. Importantly, our individual survival model incorporates the duration of infection (including an incubation period) before a patient tests positive at time t = 0. However, the CFR and other mortality measures are typically reported based on population data. Do these population-based measures, including CFR, provide reasonable measures of the probability of death of an individual? To address these and related issues, we develop an analogous population-based mortality metric based on a disease duration-structured SIR model. While population-based estimates of CFR are typically not a meaningful measure of individual mortality, under simplifying assumptions, the population-based mortality ratio Mp(t) is more closely related to the true probability of death M1(t) [32].
We will use the same rate parameters in our individual and population models to compute and compare the different mortality measures. By critically analyzing and comparing these estimates, the CFR, and a “delayed” case fatality ratio CFRd, we illustrate and interpret the differences among these measures and discuss how changes or uncertainty in the data affect them. In the Results and Discussion section, we identify a correction factor to transform population-level mortality estimates into individual mortality probabilities, and discuss the effects of other possible confounding factors such as heterogeneous populations and undertesting (unconfirmed cases).
MORTALITY MEASURES
In this section, we present different mortality measures for confirmed cases and outline their underlying mathematical models.
Intrinsic individual mortality rate
Consider an individual that, at the time of positive testing (t = 0), had been infected for a duration τ1. A “survival” probability density can be defined such that P(τ, t|τ1)dτ is the probability that the patient is still alive and infected (not recovered) at time t > 0 and has been infected for a duration between τ and τ + dτ. Since τ1 is unknown, it must be estimated or averaged over some distribution. The individual survival probability evolves according to [33].
| (1) |
where the death and recovery rates, μ(τ, t|τ1) and γ(τ, t|τ1), depend explicitly on the duration of infection at time t and can be further implicitly stratified according to patient age, gender, health condition, etc. [1, 34]. They may also depend explicitly on time t to reflect changes in clinical policy or available health care. For example, enhanced medical care may decrease the death rate μ, giving the individual’s intrinsic physiological processes a chance to cure the patient.
If we assume an initial condition of one individual having been infected for time τ1 at the time of confirmation, Eq. 1 can be solved using the method of characteristics shown in the Appendix. From the solution P(τ = t + τ1, t|τ1), one can derive the probabilities of death and recovery by time t as
| (2) |
The probability that an individual died before time t, conditioned on resolution (either death or recovery), is then defined as
| (3) |
Equations (2) and (3) also depend on all other relevant patient attributes such as age, accessibility to health care, etc. In the long-time limit, when resolution has occurred (Pd(∞|τ1) + Pr(∞|τ1) = 1), the individual mortality ratio is simply M1(∞|τ1) = Pd(∞|τ1). In order to capture the dependence of death and recovery rates on the time an individual has been infected, we propose a constant recovery rate γ and a piecewise constant death rate μ(τ|τ1) that is not explicitly a function of time t:
| (4) |
Here, τinc is the incubation time during which the patient is asymptomatic, has negligible chance of dying, but can recover by clearing the virus. In other words, some patients fully recover without ever developing serious symptoms.
For coronavirus infections, the incubation period appears to be highly variable with a mean of τinc ≈ 6.4 days [36]. We can estimate μ1 and γ using recent individual patient data from Singapore where 178 patients (mean age: 46 years) had been tracked from the date on which their first symptoms occurred until they recovered [35], on average, after 13.7 days. We show the recovery-time distribution in Fig. 2(a). Compared to other existing datasets, the Singapore COVID-19 dataset provides complete line lists for a large number of patients and is being updated regularly.
FIG. 2. Individual mortality.
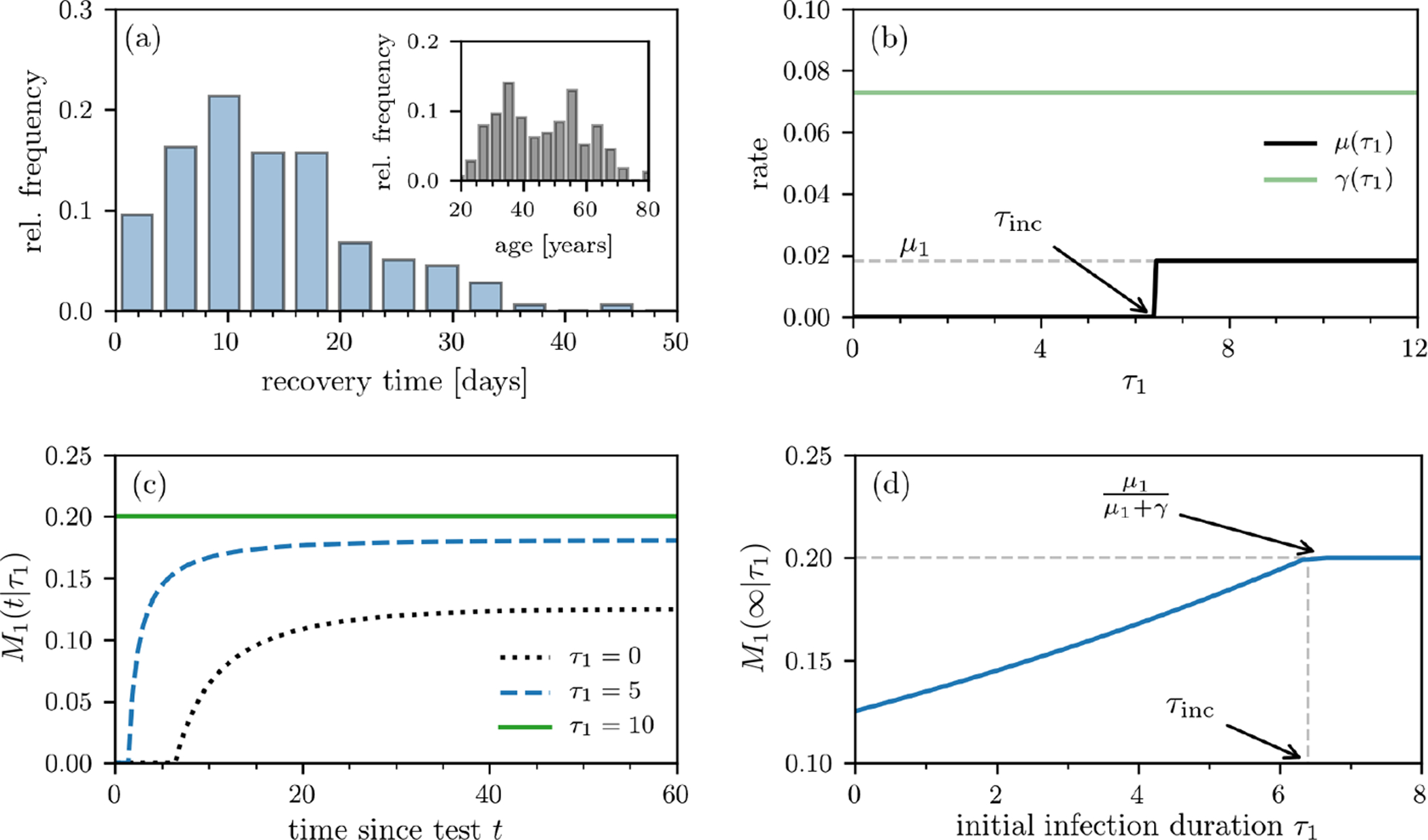
(a) Recovery time after first symptoms occurred based on individual data of 178 patients [35]. The inset shows the age distribution of these patients. (b) Death- and recovery rates as defined in Eq. (4). The death rate μ(τ1) approaches μ1 for τ1 > τinc, where τinc is the incubation period and τ1 is the time the patient has been infected before first being tested positive. (c) The individual mortality ratio M1(t|τ1) for τinc = 6.4 days at different values of τ1. Note that the individual death probability Pd(t|τ1) and M1(t|τ1) are nonzero only after t > τinc − τ1. (d) The asymptotic individual mortality ratio M1(∞) (see Eq. (3)) as a function of τ1.
We then use the global mortality of all resolved cases (≈ 20% [22]) to determine the dependence between μ1 and γ via μ1/(μ1 + γ) ≈ 1/5 (or γ/μ1 ≈ 4). The constant recovery and post incubation death rates [37] are thus
| (5) |
Using these numbers, the recovery and death rate functions γ(τ, t|τ1) and μ(τ|τ1) are plotted as functions of τ in Fig. 2(b). We show the evolution of M1(t|τ1) at different values of τ1 in Fig. 2(c). The corresponding long-time limit M1(∞|τ1) is readily apparent in Fig. 2(d): for τ1 ≥ τinc, M1(∞|τ1) = μ1/(μ1 + γ) ≈ 0.2, while M1(∞|τ1) < μ1/(μ1 +γ) when τ1 < τinc. The smaller expected mortality associated with early identification of infection arises from the remaining incubation time during which the patient has a chance to recover without possibility of death. When conditioned on testing positive at or after the incubation period, the patient immediately experiences a positive death rate, increasing his M1(∞|τ1).
In order to infer M1 (and also indirectly μ and γ) during an outbreak, a number of statistical issues must be considered. First, if the outbreak is ongoing, there may not be sufficient long-time cohort data. Second, τ1 is unknown. Since testing typically occurs at the onset of symptoms, most positive patients will have been infected a few days earlier. The uncertainty in τ1 can be represented by a probability density ρ(τ1) for the individual. The expected mortality can then be constructed as an average over ρ(τ1):
| (6) |
where and are the τ1-averaged probabilities death and cure probabilities.
Some properties of the distribution ρ(τ1) can be inferred from the behavior of patients. Before symptoms arise, only very few patients will know they have been infected, seek medical care, and get their case confirmed (i.e., ρ(τ1) ≈ 0 for τ1 ≈ 0). The majority of patients will seek care when they have been infected for approximately τinc. We choose the gamma distribution
| (7) |
with shape parameter n = 8 and rate parameter λ = 1.25/day so that the mean n/λ is equal to τinc = 6.4. Note that, independent of the distribution ρ, the average is bounded from above by M1(∞) = μ1/(μ1 + γ) for all times t.
Upon using the rates in Eqs. (4) and averaging over ρ(τ1), we derived expressions for , , and which are explicitly given in the Appendix. Using the values in Eq. (5) we find an expected individual mortality ratio (which are subsequently plotted in Fig. 3) and its asymptotic value (slightly less than M1(∞|τ1) due to averaging over ρ(τ1)). Of course, it is also possible to account for more complex time-dependent forms of γ and μ1 [38], but we will primarily use Eqs. (4) in our subsequent analyses. We stress that M1(t) tracks mortality of a cohort of individuals infected at about the same time, and does not include mortality of newly infected individuals. Thus, it can be trivially stratified according to different age groups and defined as the mortality M1(t|μ) of each age subpopulation with death rate μ.
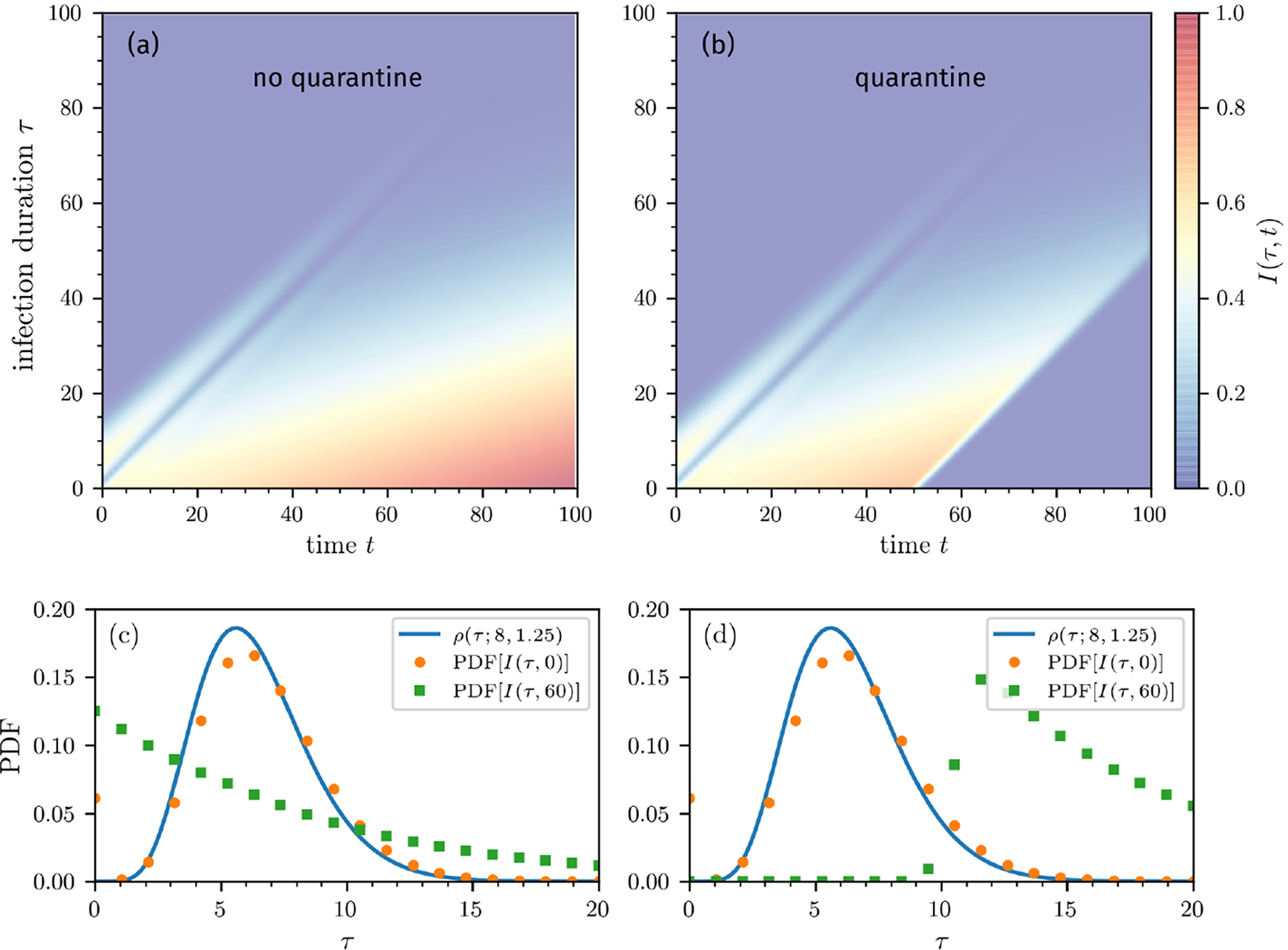
FIG. 3. Population-level mortality estimates.
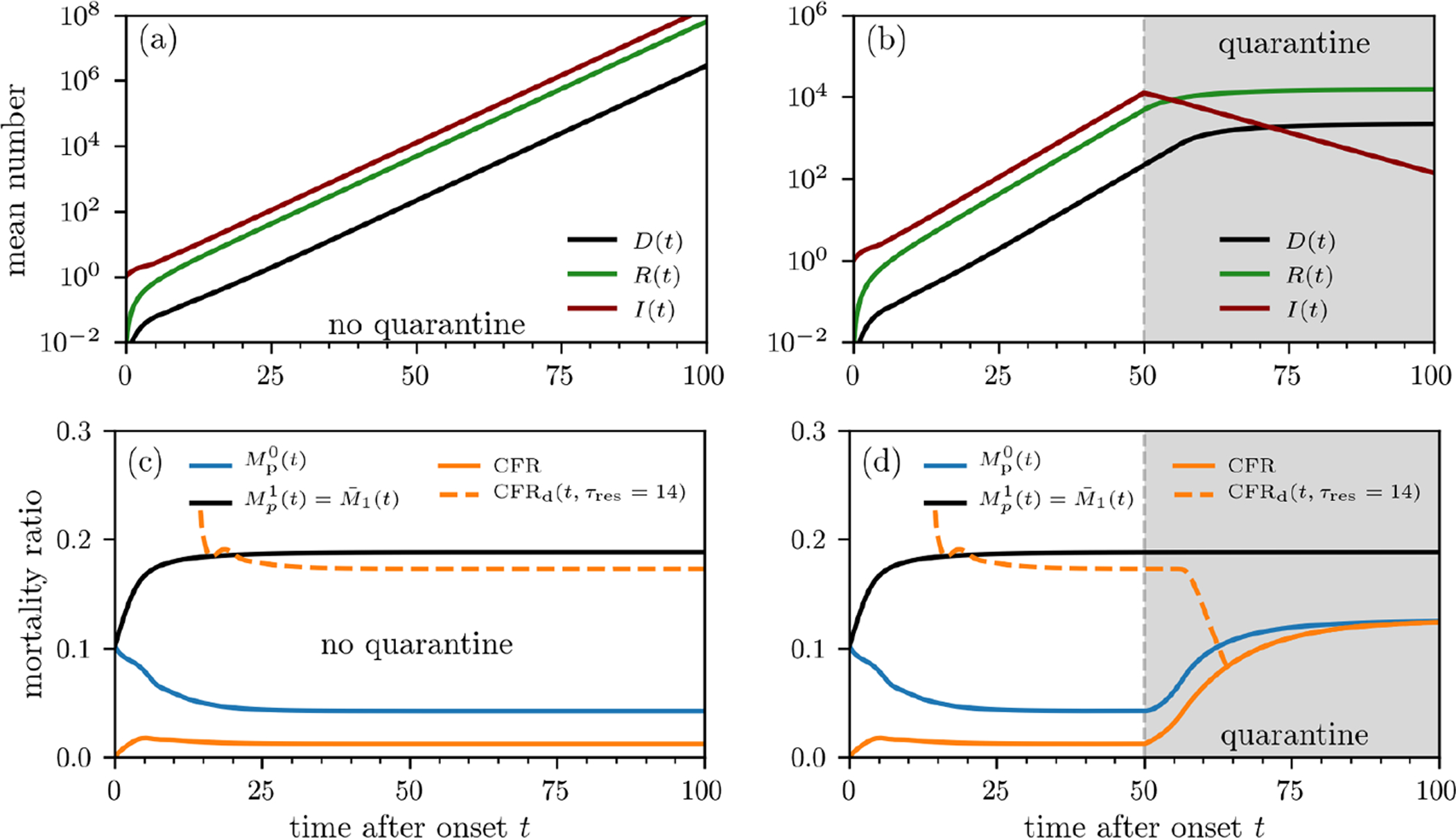
Outbreak evolution and mortality ratios without containment measures (a,c) and with quarantine (b,d). The curves are based on numerical solutions of Eqs. (10) using the initial condition I(τ, 0) = ρ(τ; 8, 1.25) (see Eq. (7)). The death and recovery rates are defined in Eqs. (4) and (5). We use an infection rate (Eq. (16)) defined by β0S0 = 4.64/day, which we estimated from the basic reproduction number of SARS-CoV-2 [36]. To model quarantine effects, we set β0S0 = 0 for t > 50 days. We show the mortality-ratio estimates and (see Eq. (14)) and CFRd(t, τres) (see Eqs. (8), (12), and (14)). CFRd(t, τres = 14 days) behaves very differently from CFR, initially decreasing for τres > 0 and significantly overestimating but providing a reasonable estimate of without quarantine. Note that under quarantine, CFR(∞), CFRd(∞), and approach the same value since they reflect the mortality ratio of the total cohort at the time of quarantine. On the other hand, reflects the ratio of the initial cohort at the start of the outbreak and remains unchanged from the no-quarantine case.
In the next subsection, we define population-based estimates for mortality ratios, Mp(t), and explore how they can be computed using SIR-type models. By comparing to Mp(t), we gain insight into whether population-based metrics are good proxies for individual mortality ratios.
Relation to infection duration-dependent SIR model
While individual mortalities can be estimated by tracking many individuals from infection to recovery or death, often, the available data are not resolved at the individual level and only total populations are given. Typically, one only has the total number of confirmed cases accumulated up to time t, Nc(t), the number of deaths to date D(t), and the number of cured/recovered patients to date R(t) (see Fig. 1). Note that Nc(t) includes unresolved cases and that Nc(t) ≥ R(t) + D(t). Resolution (death or recovery) of all patients, Nc(∞) = R(∞) + D(∞), occurs only well after the epidemic completely passes.
A variant of the CFR commonly used in the literature is the delayed CFR [5, 6]
| (8) |
which uses an earlier and smaller case number to compensate for underestimation by the standard CFR
| (9) |
The delay τres used is typically the time between the day symptoms first occurred and the day of death or recovery. To determine a realistic value of the delay time τres (which can be qualitatively interpreted as a resolution time), we use data on death/recovery periods of 36 tracked COVID-19 patients [39] and find that patients recover/die, on average, τres ≈ 2 weeks after first symptoms occurred. The delayed CFRd(t, τres > 0) also underestimates the individual mortality in previous epidemic outbreaks of SARS [18, 32] and Ebola [40], but is highly sensitive to τres. If the delay between the time of infection and time of resolution were vanishingly small, we can set τres = 0 and find that the CFRd and CFR are equivalent (see Eq. (9)).
Alternatively, a simple and interpretable population-level mortality is Mp(t) = D(t)/(R(t) + D(t)), the ratio of infected deaths to all resolved cases of confirmed infections. To provide a concrete model for D(t) and R(t), and hence Mp(t), we will use a variant of the standard infection duration-dependent susceptible-infected-recovered (SIR)-type model described by [41, 42]
| (10) |
and , where S(t) is the number of susceptibles, I(τ, t) is density of individuals at time t who have been infected for time τ, and R(t) is the number of recovered individuals. The rate at which an individual infected for time τ at time t infects susceptibles is denoted by β(τ, t)S(t). For simplicity, we assume only community spread and neglect immigration of infected individuals, which could be straightforwardly included [42].
Note that the equation for I(τ, t) is identical to the equation for the survival probability described by Eq. (1). It is also equivalent to McKendrick age-structured models [43, 44]. In both the individual model (Eq. (1)) and population model (Eq. (10)), the death and recovery rates are insensitive to changes in age a over the ≲ 1 year epidemic timescale. In this limit, we consider only infection-duration dependence in the population dynamics. However, in contrast to the individual survival probability, new infections of susceptibles are described by the boundary condition (or renewal equation)
| (11) |
which is similar to that used in age-structured models to represent birth [43]. The initial time t = 0 is arbitrary as long as the initial condition I(τ, 0) is defined. We use an initial condition corresponding to a single infected with the infection duration density given by Eq. (7): I(τ, 0) = ρ(τ; n = 8, λ = 1.25). Note that Eq. (11) assumes that all newly infected individuals are immediately identified; i.e., these newly infected individuals start with τ1 = 0. After solving for the infected population density, we find the total number of deaths, recoveries, and total cases to date,
| (12) |
and use D0(t) and N0(t) for D(t) and Nc(t) in definitions of CFR(t) and CFRd(t, τres) (Eq. (8)). In the definitions of D0(t), R0(t), and N0(t), we account for all possible death and recovery cases to date (see Appendix) and that newly infected individuals are immediately identified. We use these case numbers as approximations of the reported case numbers to study the evolution of mortality ratio estimates. Mortalities based on these numbers underestimate the actual individual mortality M1 (see the previous “Intrinsic individual mortality rate” subsection) since they involve individuals that have been infected for different durations τ, particularly recently infected individuals who have not yet died.
An alternative way to compute populations is to exclude new infections and consider only an initial cohort. The corresponding populations in this case are defined as
| (13) |
Since D1(t) and R1(t) do not include infected individuals with τ < t, they exclude the effect of newly infected individuals and may yield more meaningful mortalities as they would be based on an initial cohort of individuals in the distant past. It is superfluous to define CFR using D1(t)/Nc because the corresponding Nc of a cohort is a constant. The infections that occur after t = 0 contribute only to I(τ < t, t); thus, D1(t) and R1(t) do not depend on the transmission rate β, possible immigration of infected individuals, or the number of susceptibles S(t). Note that all the populations derived above implicitly average over ρ(τ1; n, γ) for the first cohort of identified infected individuals (but not subsequent infections). Moreover, the population density I(τ ≥ t, t) follows the same equation as provided the same ρ(τ1; n, λ) is used in their respective calculations.
The two different ways of partitioning populations (Eqs. (12) and (13)) lead to two different population-level mortality ratios
| (14) |
Since the populations D0(t) and R0(t), and hence , depend on disease transmission through β(τ, t) and S(t), we expect to carry a different interpretation from M1(t) and .
In the special case in which μ and γ are constants, the time-integrated populations and factor out of and , rendering them time-independent and
| (15) |
Thus, only in the special time-homogeneous case do both population-based mortality ratios become independent of the population (and transmission β) and coincide with the individual death probability.
To illustrate, in more general cases, the differences between M1(t), and CFRd(t, τres), we use the simple death and recovery rate functions given by Eqs. (4) in solving Eqs. (1) and (10). For β(τ, t) in Eq. (11), we use a recently inferred infectiousness profile [45] which is described by a gamma distribution
| (16) |
with a peak that occurs shortly before the onset of symptoms at time τinc and coincidentally has n = 8 and λ = 1.25/day as in the testing time distribution ρ(τ1) from a single infected (Eq. (7)). The constant dimensionless prefactor β0 sets the amplitude of the transmission rate. For the chosen parameters n and λ, the gamma distribution ρ(τ; n, λ) reaches a maximum at τ ≈ 5.6 days, about one day before τinc = 6.4 days [45]. Assuming that the susceptible pool is not appreciably depleted, S(t) ≈ S0 and Eq. (11) becomes . The amplitude β0S0 can be found by assuming a single infected for I(τ, t) in the renewal equation and using the estimated basic reproduction number. The basic reproduction number is the average number of secondary infections that result from any single infected individual before he dies or recovers [37]. There are two terms to consider when determining is the probability that an infection occurs in [τ, τ + dτ] and (ii) is the probability that a single infected individual has not died or recovered prior to time τ. If we integrate over the product of these quantities and multiply by the total susceptible population S0 (which is equivalent to the boundary condition (11) applied to a single infected individual), we obtain the average number of susceptibles infected by one infected individual, i.e., . Thus, upon using Eq. (16), β0S0 can be found by solving
| (17) |
Using the death and recovery rate functions given by Eqs. 4 and 5, we find β0S0 ≈ 4.64/day. Using this value, we numerically solve Eqs. (10) and (11) (see Appendix for further details) and use these solutions to compute D0,1(t), R0,1(t), and N0,1(t), which are then used in Eqs. (14) and CFRd(t, τres).
RESULTS AND DISCUSSION
Comparison of mortalities
Here, we evaluate and compare the different mortality metrics and show how some of them qualitatively resemble the measured mortality estimates shown in Fig. 1. In Fig. 3(a), we show the unbounded subpopulations I0(t), D0(t), and R0(t) computed using Eqs. (10), (11), and (12) when the susceptible population is assumed constant. Fig. 3(b) shows the populations when a strict quarantine (S(t > tq) = 0) is applied after tq = 50 days. The mortalities plotted in Figs. 3(c) show that approaches the individual mortality ratio given in the “Intrinsic individual mortality rate” subsection above. This occurs because the model for P(τ, t) and I(τ, t) are equivalent and we assumed the same initial distribution ρ(τ; 8, 1.25) for both quantities. However, the population-level mortality ratios CFRd(t, τres) and also take into account recently infected individuals who may recover before symptoms. This difference yields different mortality ratios because newly infected individuals are implicitly assumed to be detected immediately and all have τ1 = 0. Thus, the underlying infection-time distribution is not the same as that used to compute (see Appendix for further details). The mortality ratio should not be used to quantify the individual mortality probability of individuals who tested positive, while the accuracy of CFRd(t, τres) is sensitive to τres and quarantining. Moreover, due to evolution of the disease, D(t), R(t), and N (t) do not change with the same rates during an outbreak, the population-level mortality measures CFRd(t, τres) and reach their final steady state values only after sufficiently long times. Figs. 3(d) shows the corresponding mortalities with quarantining after tq = 50 days.
The population-level ratios and CFR(t) implicitly depend on new infections and the transmission rate β. Despite this confounding factor, and CFRd(t, τres) approach as t → ∞, where is the probability that no recovery occurred during the incubation time τinc. Based on these results, we can establish the following connection between the different mortality ratios for initial infection times with distribution ρ(τ1; n, λ) and mean :
| (18) |
According to Eq. (18), population-level mortality estimates (e.g., CFR and ), can be transformed, at least approximately, into individual mortality probabilities using the correction factor with . Although population-level quarantining does not directly affect the individual mortality M1(t|τ1) or , it can be easily incorporated into the SIR-type population dynamics equations through changes in β(τ, t)S(t). For example, we have set S(t > tq) = 0 to represent implementation of a perfect quarantine after tq = 50 days of the outbreak. After tq = 50 days, no new infections occur and the estimates CFR(t) and start to converge towards their common larger value (see Fig. 3(d)). In other words, without quarantining, the infected and recovered populations are continuously increasing, keeping CFR and low. Since the number of deaths decreases after the implementation of quarantine measures, the delayed CFRd(t, τres = 14 days) is first decreasing until t = tq + τres = 64 days. For t > 64 days, the CFRd(t, τres = 14 days) measures no new cases and is thus equal to the CFR.
The overall time-evolution of some of the mortalities in Fig. 3 qualitatively resembles the behavior of the mortality estimates in Fig. 1. As shown in Fig. 1, the CFR is increasing over time whereas provides a more stable mortality estimate for the SARS-CoV outbreak in Hong Kong (2003) and seems to follow a similar behavior in the current SARS-CoV-2 outbreak in Italy. In Fig. 4, we show additional examples of mortality-ratio estimates for China, South Korea, Spain, Germany, Switzerland, and the United Kingdom. After an initial transient, the CFR, in most cases, increases to a new asymptote after the epidemic passes. As in Fig. 1, we observe, consistent with their definitions, that the population-based mortality ratio is larger than the corresponding CFR in all cases. also appears to be a temporally more stable metric. Differences in the evolution of mortality ratios in different regions could result from changing practices in data collection or from explicitly time-inhomogeneous parameters μ(τ, t), γ(τ, t), and/or β(τ, t).
FIG. 4. Mortality estimates in different countries.
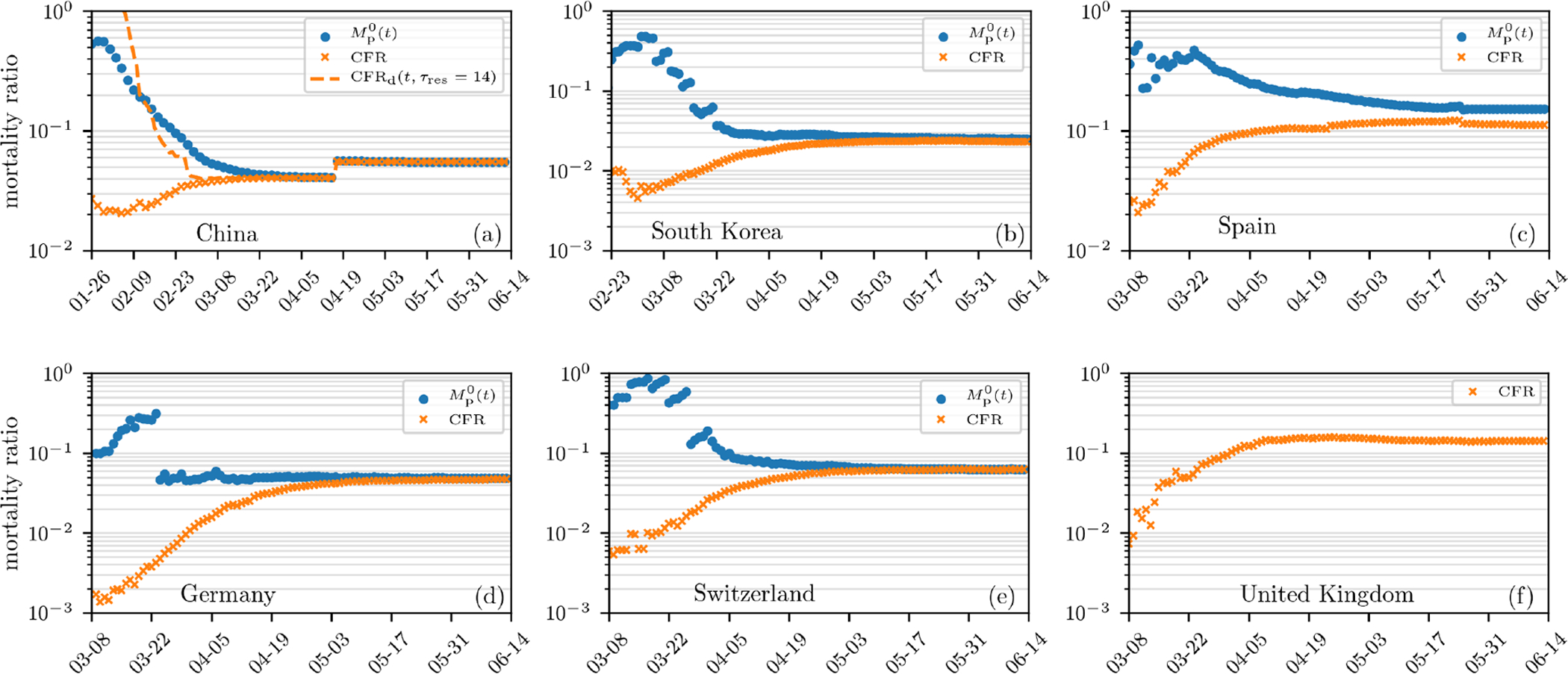
Estimates of mortality ratios (see Eqs. (8) and (14)) of SARS-CoV-2 infections in different countries. The data are derived from Ref. [21]. The case fatality rate, CFR, corresponds to the number of deaths to date divided by the total number of cases to date. The “delayed” mortality-ratio estimate CFRd corresponds to the number of deaths to date divided by total number of cases at time t − τres is also shown for China. The population-based mortality ratios Mp(t) are also shown, except for the UK which has reported an inexplicable .
In addition to the mathematical differences between M1(t), and , and CFR, estimating and CFR(t) from aggregate populations implicitly incorporate a number of confounding factors that contribute to their variability. On April 25, 2020, the value of in Sweden, Belgium, France, the US, and Italy are 2, 194/(2, 194 + 1, 005) ≈ 69%, 7, 094/(7, 094 + 10, 785) ≈ 40%, 22, 856/(22, 856 + 44, 903) ≈ 34%, 54, 941/(54, 941 + 118, 633) ≈ 32%, and 26, 384/(26, 384 + 63, 120) ≈ 29%, respectively. These have slowly decreased (see Fig. 4) as patients resolve even if current conditions (e.g., treatment methods, age group proportion of infected individuals, etc.) have not changed. By comparison, on April 25, 2020, and 6% in Germany and China, respectively. These differences result from delays and inaccuracies in reporting, varied guidelines for assigning cause of death, differing medical treatment strategies, and demographic heterogeneity among different countries.
Heterogeneous populations
Differences in demographics can easily be a source of variability in mortality rates measured across different regions. Older patients and those with underlying medical conditions typically have a higher death rate μ(τ, t) and/or lower recovery rate γ. Since we focus on mortality, the different subpopulations within the confirmed population matter only through their differences in μ and/or γ. For the and metrics, no new infections are used in their determination. Thus, these metrics are associated with the mean death and recovery rates of the original group of infected individuals, i.e., the ratios and refer to the mortality ratios of each subpopulation or individual described by μ and γ. The effective over the entire confirmed population can be trivially constructed by population-averaging D1(t|μ, γ) and R1(t|μ, γ) over μ and γ before constructing .
For the other confirmed mortalities and CFR(t), new infections are taken into account and subpopulations with different death and recovery rates can infect each other. Suppose there are two subpopulations “a” and “b” (e.g., young and old) with associated death and recovery rates μa,b and γa,b, respectively. The equations for each subpopulation are
| (19) |
indicating that each subpopulation follows their own dynamics for τ > 0. However, the subpopulations interact with each other through the coupled boundary conditions
| (20) |
that describe cross-infections between the “a” and “b” subpopulations. Thus, the infection levels in each subpopulation also depend on the transmission rates βaa, βab, and βbb. To compute the overall confirmed mortality or CFR(t) of the entire population, we must solve Eqs. (19) and (20) for Ia and Ib, and hence Da(t), Db(t), and D(t) = Da(t) + Db(t).
In Fig. 5, we show the evolution of for two age groups representing young and old individuals with different mortality and infection rates. The behavior of for the entire population is qualitatively similar to, but falls in between those of each age group (see Fig. 3). Whether the overall mortality is closer to that of the young or old population depends on the relative populations of young and old infecteds, their death and recovery rates, and their cross transmission rates βab. For age-stratified case data, the subpopulation model outlined above, or other approaches such as scaling approximations [46] may be useful for capturing age-dependent variations in .
FIG. 5. Population-level mortality estimate for two age groups.

The mortality ratio without containment measures (a) and under quarantining (b). The curves are based on numerical solutions of Eqs. (19) and (20) assuming constant S(t) ≈ S0 and using the initial condition Ia(τ, 0) = Ib(τ, 0) = ρ(τ; 8, 1.25)/2 (see Eq. (7)), where the subscripts “a” and “b” denote the young and old age group, respectively. The death and recovery rates for the younger age group are defined in Eqs. (4) and (5). For the older age group, we set μb = 4μa and γb = γa. We use an infection rate (Eq. (16)) defined by βaaS0 = 4.64/day, which we estimated from the basic reproduction number of SARS-CoV-2 [36]. The remaining infection rates are defined via . To model quarantine effects, we set β0S0 = 0 for t > 50 days in (b).
Undertesting and unconfirmed cases
Another important confounding factor is the large number of untested and often asymptomatic infected individuals. The mortality rate often quoted in the literature ranges from < 1 – 3%, which is much smaller than the resolved mortality ratios we have used for illustration. Our estimates of and CFRd(t, τres) using I(τ, t) actually describe the mortality of the population conditioned on being tested positive. Since we used Eqs. (10) to compute infected populations, we implicitly assumed that all infected individuals have been tested/confirmed. However, the total infected population is comprised of tested and untested individuals, which may or may not carry different death and recovery rates. Typically, only a small fraction f of the total number of infected individuals might be tested and confirmed positive.
Our confirmed mortalities (derived from only the positively tested population) can be extended to the entire population, tested or untested. The “true” and the fatality ratio conditioned on having been infected (the IFR) would typically be much smaller than the and CFR calculated using only confirmed cases. How the testing fraction f < 1 might qualitatively affect the “true” underlying mortality measures (the mortality conditioned on simply being infected) is illustrated in Fig. 6.
FIG. 6. Fractional testing.
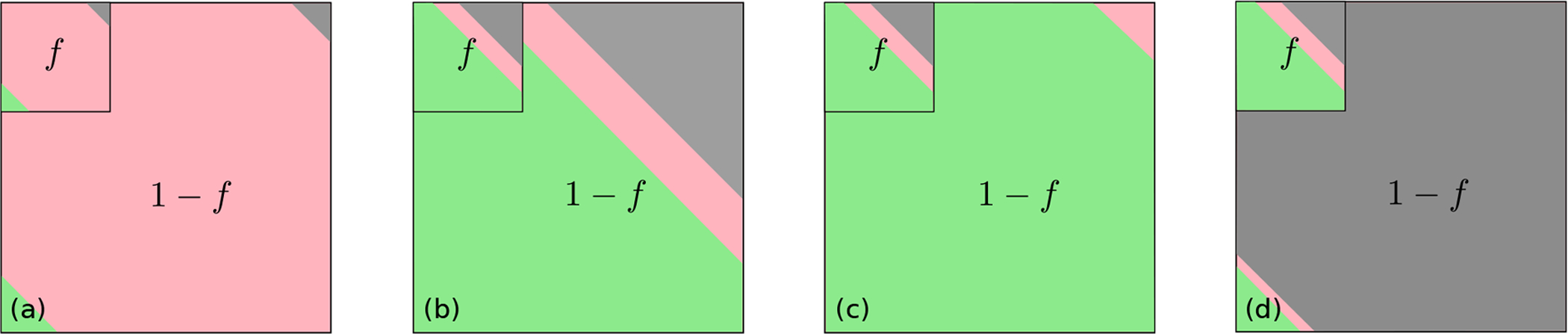
An example of fractional testing in which a fixed fraction f of the real total infected population is assumed to be tested. The remaining 1 − f proportion of infected individuals are untested. Equivalently, if the total tested fraction has unit population, then the fraction of the population that remains untested is 1/f − 1. (a) At short times after an outbreak, most of the infected patients, tested and untested, have not yet resolved (red). Only a small number have died (gray) or have recovered (green). (b) At later times, if the untested population dies at the same rate as the tested population, Mp(t) and CFR remain accurate estimates for the entire infected population. (c) If the untested population is, say, asymptomatic and rarely dies, the true mortality can be significantly overestimated by the tested mortality . (d) Finally, in a scenario in which untested infected individuals die at a higher rate than tested ones, and CFR based on the tested fraction underestimate the true mortality .
Estimates for SARS-CoV-2 show that f is small (e.g., f ≈ 14% in China before January 23, 2020) [47]. At early times (Fig. 6(a)) most patients, tested or untested, have not yet resolved. A reported/tested fraction f < 1 would not directly affect or alter the CFRs or mortality ratios if the unreported/untested population dies and recovers in the same proportion as those tested, as depicted in Fig. 6(b). That is, undertesting would still provide a good estimate of the true mortality if the entire population were homogeneous in death and recovery rates. However, if the untested (presumably mildly or asymptomatic infected) are less likely to die than the tested infected individuals, undertesting would give rise to and CFR(t) that overestimate the true mortality and the infection fatality ratio (IFR). If untested infected individuals do not die at all, as depicted in Fig. 6(c), the true long-time mortality . In the unlikely scenario in which untested individuals do not receive medical care and hence die at a faster rate (Fig. 6(d)), and CFR based on the tested fraction would underestimate the true long-time mortality and IFR, respectively.
To quantitatively estimate the underlying mortality of the population conditioned simply on being infected, we have to quantify the number of confirmed and untested infected individuals, Ic(τ, t) and Iu(τ, t), which can be further divided into subpopulations with intrinsically different transmission, death, and recovery rates. The act of confirmation itself may change behavior and/or treatment, further changing transmission, death, and recovery parameters.
By constructing the accumulated deaths and recoveries associated with Ic(τ, t) and Iu(τ, t), and , respectively, we can define true, whole population mortality ratios as listed in Table II. For example,
| (21) |
where μc,u and γc,u are the death and recovery rates associated with infected individuals who are confirmed and untested, respectively. Analogous expressions arise for and . If the confirmed and untested populations are further subdivided, the μc,uIc,u and γc,uIc,u integrands would be replaced by a Hamdamard (i.e., element-wise) product of two vectors representing subpopulations and their corresponding rates. The populations Ic,u themselves can be found from a specific disease transmission model that also includes a testing process that converts Iu to Ic.
TABLE II. Definitions of the main metrics.
The superscript “0” and “1” denote quantities that are based on the total population (including new infections) and a cohort (excluding new infections), respectively. Quantities with subscript “c” and “u” denote confirmed and untested pools (for example, is the total number of untested individuals at time t) that must be inferred using other measurements such as random testing. We have suppressed the time dependences for notational simplicity.
| Subpopulation╲Metric | Fatality Ratios | Resolved Mortality w/inf | Resolved Mortality w/o inf | Individual Risk |
|---|---|---|---|---|
| confirmed (tested) | ||||
| total (tested+untested) | not defined |
SUMMARY AND CONCLUSIONS
The CFR has been predominantly used but appears to evolve in qualitatively similar ways as epidemics evolve. Although CFRd(t, τres) is based on a delay reflecting the timescales for recovery, in general, there is no clear mechanistic interpretation for using the CFR or IFR as mortality ratios.
Here, we stress that more mechanistically meaningful and interpretable metrics can be readily defined and just as easily estimated from population data as CFRs are. Our proposed mortality ratios for viral epidemics are defined in terms of (i) individual survival probabilities and (ii) population ratios using numbers of deaths and recovered individuals. Both of these measures are based on the within-host evolution of the disease, and in the case of , the population-level transmission dynamics. On a single patient level, is the metric of interest. However, to estimate this, one needs accurate cohort data, for which few exist for coronavirus. Nonetheless, cumulative population-based mortalities can provide insight.
Among the metrics we describe, is structurally closest to the individual mortality in that both are independent of disease transmission since new infections are not counted. Both of these mortality ratios converge after an incubation time τinc to a value smaller than or equal to μ1/(μ1 + γ) and are best interpreted as approximately the mortality probability conditioned on being tested positive. The most accurate estimates of can be obtained if we keep track of the fate of cohorts who were confirmed within a small time window in the past. By following only these individuals, one can track how many of them die as a function of time. As more cases arise, one should stratify them according to their estimated times since infection to obtain better statistics for M1(∞). With the further spread of SARS-CoV-2 in different countries, data on more individual cases of death and recovery can also be more easily stratified according to other central factors in COVID-19 mortality: age, sex, health condition. Population heterogeneity and uncertainty in intrinsic disease parameters such as the incubation period and the time τ1 a patient had been infected before confirmation can affect the mortality measures.
Besides demographic heterogeneity and the highly variable estimates of COVID-19 deaths due to different clinical protocols for assigning cause of death, undertesting also confounds accurate estimation of the true underlying mortality. Infected individuals in the population at large who are untested comprise an unknown population Iu which contributes to deaths and recovery, and need to be factored into the “true” mortalities or the IFR.
These untested/unconfirmed populations can, in principle, be computed from a multicompartment mathematical model for disease transmission and testing. The relevant expressions for are listed in Table II. Even though Mp(t) typically overestimates the true mortality, tracking or of an initially confirmed cohort can still provide a reasonable estimate of the mortality ratio, especially if untested infected individuals die at the same rate as confirmed individuals.
ACKNOWLEDGEMENTS
LB acknowledges financial support from the SNF Early Postdoc.Mobility fellowship on “Multispecies interacting stochastic systems in biology”. The authors also acknowledge financial support from the Army Research Office (W911NF-18-1-0345), the NIH (R01HL146552), and the National Science Foundation (DMS-1814364). TC also acknowledges the hospitality of the Beijing Computational Science Research Center.
APPENDIX
Numerical scheme
To numerically solve Eqs. (10) and (11), we use a uniform discretization τk = kΔτ, k = 0, 1, …, K. A backward difference operator [I(τk, t) − I(τk−1, t)] /(Δτ) is used to approximate ∂τ I(τ, t) and a predictor-corrector Euler scheme is used to advance time [48]. Setting the cut-offs I(−Δτ, t) ≡ 0 and I(KΔτ, t) ≡ 0, the resulting discretized equations for the full SIR model are
| (A1) |
where is the initial predicted guess, and the last term proportional to δk,0 encodes the boundary condition Eq. (11). Note that we use to indicate the numerical evaluation of . Quadrature methods such as Simpson’s rule and the trapezoidal rule can be used to approximate the integral more efficiently.
The total number of dead, recovered, and infected individuals at time t are found by
with analogous expressions for D1(mΔt) and R1(mΔt). To obtain a stable integration scheme, the time steps Δt and Δτ have to satisfy Δt/(2Δτ) < 1. In all of our numerical computations, we thus set Δt = 0.002, Δτ = 0.02, and K = 104. In the next section, we show additional plots of the magnitude of I(τ, t) in the t − τ plane.
Solutions for τ1-averaged probabilities
Using the method of characteristics, we find the formal solution to Eq. (1):
| (A2) |
which can be used to construct the death and cure probabilities
| (A3) |
If we now invoke the functional forms of μ and γ given in Eq. (4), we find explicitly
| (A4) |
and
| (A5) |
FIG. A1. Phase plot for P (τ > t, t) and I(τ > t, t).
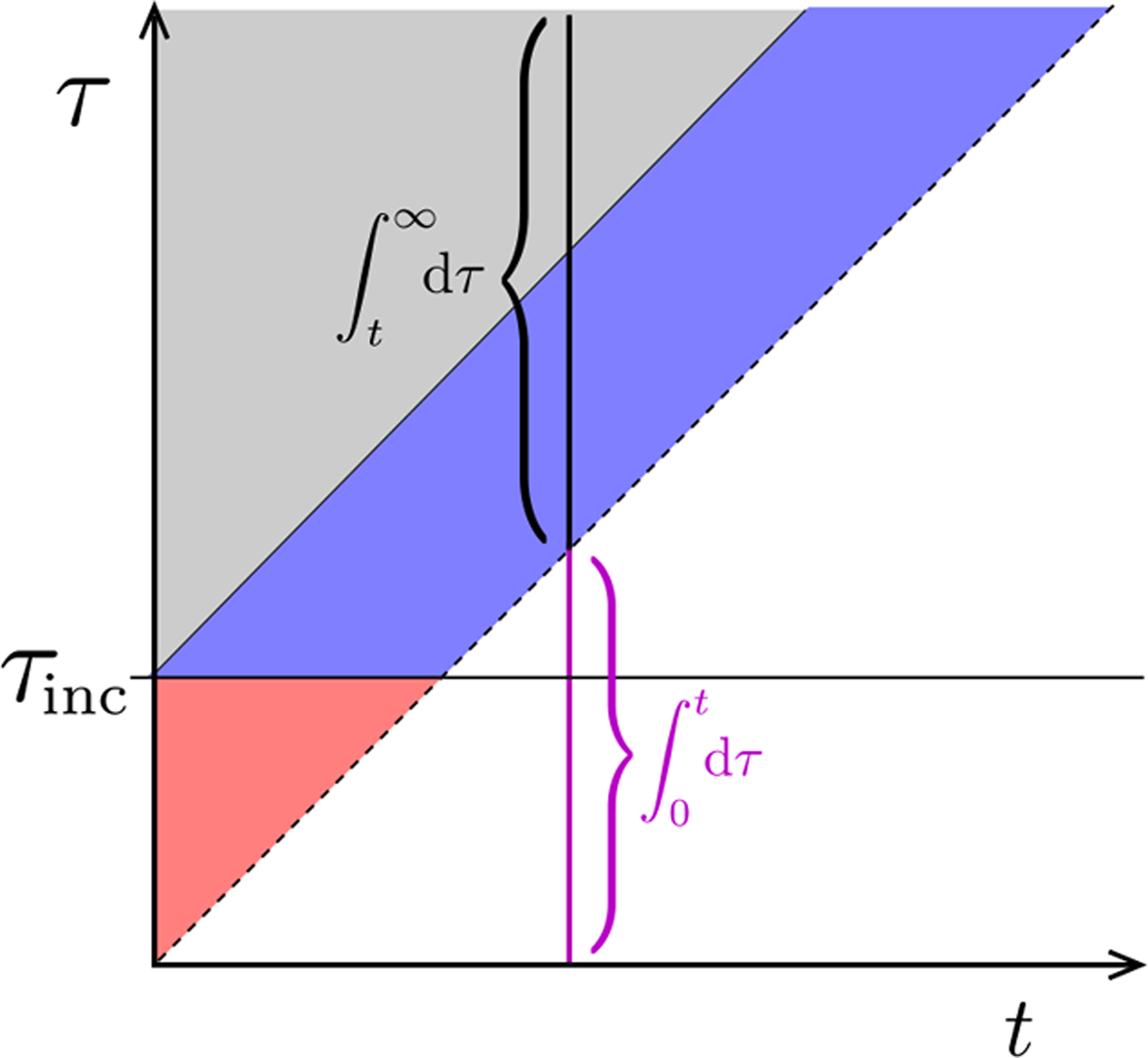
The regions delineate the different forms for the solution (Eq. (A6)). Here, we have included an incubation time τinc before which no death occurs. The solution for or I(τ, t) in the τ < t region must be self-consistently solved using the boundary condition Eq. (11). At any fixed time, the integral of I(τ, t) over t < τ ≤ ∞ captures only the initial population, excludes newly infected individuals, and is used to compute D1(t), R1(t), and . To compute D0(t), R0(t), and , we integrate across all infected individuals (including the integral over t > τ ≥ 0 shown in magenta).
Finally, we can also find the τ1-averaged probabilities for τ ≥ t by weighting over ρ(τ1; n, λ). For example,
| (A6) |
These solutions hold for the different regions shown in the phase plot of Fig. A1 and are equivalent to those for I(τ > t, t). Corresponding expressions for and can be found and used to construct . Fig. A2(a) shows the magnitude of I(τ, t) in the t − τ plane when we use Eq. (16), set S(t) = S0 constant (so that the first equation in Eq. (A1) does not apply) and assign β0S0 = 4.64/day. In this case, the epidemic continues to grow in time, but the mortality rates nonetheless converge as t → ∞. In Fig. A2(b), we set β0S0 = 0 for t > tq to model strict quarantining after tq = 50 days. We observe no new infection after the onset of strict quarantine measures. In both cases (quarantine and no quarantine), we use ρ(τ; n = 8, λ = 1.25) (see Eq. (7) in the main text) to describe the initial distribution of infection times τ. As time progresses, more of the distribution of τ moves towards smaller values until quarantine measures take effect (see Fig. A2(c) and (d)).
FIG. A2. Density plots of I(τ, t) in the t − τ plane.
Numerical solution of the equation for I(τ, t) in Eqs. (10) under the assumption of a fixed susceptible size and β0S0 = 4.64/day. (a) The density without quarantine monotonically grows with time t in the region τ < t as an unlimited number of susceptibles continually produces infections. (b) With quarantining after tq = 50 days, we set β0S0 = 0 for t > tq, which shuts off new infections. Both plots were generated using the same initial density ρ(τ1) defined in Eq. (7). In both cases, the density I(τ > t) is identical to P(τ > t) if the same ρ(τ1) is used and is independent of disease transmission, susceptible dynamics, etc. (c-d) Probability-density functions (PDFs) of the number of infected individuals I(τ, t) for t = 0, 60 days (b) without and (c) with quarantine. The blue solid line corresponds to the initial distribution ρ(τ; n = 8, λ = 1.25) (see Eq. (7)).
References
- [1].Verity Robert, Okell Lucy C., Dorigatti Ilaria, Winskill Peter, Whittaker Charles, Imai Natsuko, Cuomo-Dannenburg Gina, Thompson Hayley, Walker Patrick G. T., Fu Han, Dighe Amy, Griffin Jamie T, Baguelin Marc, Bhatia Sangeeta, Boonyasiri Adhiratha, Cori Anne, Cucunubá Zulma, FitzJohn Rich, Gaythorpe Katy, Green Will, Hamlet Arran, Hinsley Wes, Laydon Daniel, Nedjati-Gilani Gemma, Riley Steven, van Elsland Sabine, Volz Erik, Wang Haowei, Wang Yuanrong, Xi Xiaoyue, Donnelly Christl A., Ghani Azra C., and Ferguson Neil M., “Estimates of the severity of coronavirus disease 2019: a model-based analysis,” The Lancet Infectious Diseases (2020), 10.1016/S1473-3099(20)30243-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Kelly Heath and Cowling Benjamin J., “Case Fatality: Rate, Ratio, or Risk?” Epidemiology 24, 622–623 (2013). [DOI] [PubMed] [Google Scholar]
- [3].European Centre for Disease Prevention and Control, “Field Epidemiology Manual Wiki,” https://wiki.ecdc.europa.eu/fem/Pages/Attack%20rates%20and%20case%20fatality.aspx (2020), accessed: 2020-04-21.
- [4].Garske Tini, Legrand Judith, Donnelly Christl A, Ward Helen, Cauchemez Simon, Fraser Christophe, Ferguson Neil M, and Ghani Azra C, “Assessing the severity of the novel influenza A/H1N1 pandemic,” BMJ 339 (2009), 10.1136/bmj.b2840, https://www.bmj.com/content. [DOI] [PubMed] [Google Scholar]
- [5].Xu Zhe, Shi Lei, Wang Yijin, Zhang Jiyuan, Huang Lei, Zhang Chao, Liu Shuhong, Zhao Peng, Liu Hongxia, Zhu Li, et al. , “Pathological findings of COVID-19 associated with acute respiratory distress syndrome,” Lancet Resp. Med (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Wu Zunyou and McGoogan Jennifer M, “Characteristics of and Important Lessons From the Coronavirus Disease 2019 (COVID-19) Outbreak in China: Summary of a Report of 72314 Cases From the Chinese Center for Disease Control and Prevention,” JAMA (2020). [DOI] [PubMed] [Google Scholar]
- [7].“CDC Viral Test for COVID-19,” https://www.cdc.gov/coronavirus/2019-ncov/php/testing.html (2020), accessed: 2020-05-13.
- [8].“Research Use Only 2019-Novel Coronavirus (2019-nCoV) Real-time RT-PCR Primer and Probe Information,” https://www.cdc.gov/coronavirus/2019-ncov/lab/rt-pcr-panel-primer-probes.html (2020), accessed: 2020-05-13.
- [9].Onder Graziano, Rezza Giovanni, and Brusaferro Silvio, “Case-fatality rate and characteristics of patients dying in relation to covid-19 in italy,” Jama (2020). [DOI] [PubMed] [Google Scholar]
- [10].Lipsitch Marc, Donnelly Christl A., Fraser Christophe, Blake Isobel M., Cori Anne, Dorigatti Ilaria, Ferguson Neil M., Garske Tini, Mills Harriet L., Riley Steven, Van Kerkhove Maria D., and Hernán Miguel A., “Potential Biases in Estimating Absolute and Relative Case-Fatality Risks during Outbreaks,” PLOS Neglected Tropical Diseases 9, 1–16 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Jung Sung-mok, Akhmetzhanov Andrei R., Hayashi Katsuma, Linton Natalie M., Yang Yichi, Yuan Baoyin, Kobayashi Tetsuro, Kinoshita Ryo, and Nishiura Hiroshi, “Real-Time Estimation of the Risk of Death from Novel Coronavirus (COVID-19) Infection: Inference Using Exported Cases,” Journal of Clinical Medicine 9 (2020), 10.3390/jcm9020523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Famulare Mike, “2019-nCoV: preliminary estimates of the confirmed-case-fatality-ratio and infection-fatality-ratio, and initial pandemic risk assessment,” https://institutefordiseasemodeling.github.io/nCoV-public/analyses/first_adjusted_mortality_estimates_and_risk_assessment/2019-nCoV-preliminary_age_and_time_adjusted_mortality_rates_and_pandemic_risk_assessment.html (2020), accessed: 2020-03-31.
- [13].Oke Jason and Heneghan Carl, “Global Covid-19 Case Fatality Rates: Oxford COVID-19 Evidence Service,” (2020), accessed: 2020-03-27.
- [14].Angelopoulos Anastasios Nikolas, Pathak Reese, Varma Rohit, and Jordan Michael I., “Identifying and Correcting Bias from Time- and Severity- Dependent Reporting Rates in the Estimation of the COVID-19 Case Fatality Rate,” arXiv:2003.08592v2 (2020).
- [15].Mizumoto K and Chowell G, “Estimating risk for death from 2019 novel coronavirus disease, China, January-February 2020,” Emerging Infectious Diseases (2009), 10.3201/eid2606.200233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Ruan Shigui, “Likelihood of survival of coronavirus disease 2019,” The Lancet Infectious Diseases (2020), 10.1016/S1473-3099(20)30257-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Spychalski Piotr, Bayska-Spychalska Agata, and Kobiela Jarek, “Estimating case fatality rates of covid-19,” The Lancet Infectious Diseases (2020), 10.1016/S1473-3099(20)30246-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Paul SF Yip, Lam KF, Lau Eric HY, Chau Pui-Hing, Tsang Kenneth W, and Chao Anne, “A comparison study of realtime fatality rates: severe acute respiratory syndrome in Hong Kong, Singapore, Taiwan, Toronto and Beijing, China,” J. Roy. Stat. Soc. A 168, 233–243 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Paul SF Yip, Eric HY Lau, Lam KF, and Huggins Richard M, “A chain multinomial model for estimating the real-time fatality rate of a disease, with an application to severe acute respiratory syndrome,” Am. J. Epidemiol 161, 700–706 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].World Health Organization, “Cumulative Number of Reported Probable Cases of Severe Acute Respiratory Syndrome (SARS),” https://www.who.int/csr/sars/country/en/ (2020), accessed: 2020-03-30.
- [21].Dong Ensheng, Du Hongru, and Gardner Lauren, “An interactive web-based dashboard to track COVID-19 in real time,” The Lancet Infectious Diseases (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].“COVID-19 statistics,” https://www.worldometers.info/coronavirus/ (2020), accessed: 2020-02-26.
- [23].Böttcher Lucas, Olivia Woolley-Meza, Araújo Nuno AM, Herrmann Hans J, and Helbing Dirk, “Disease-induced resource constraints can trigger explosive epidemics,” Sci. Rep 5, 1–11 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Böttcher Lucas, Olivia Woolley-Meza, Goles Eric, Helbing Dirk, and Herrmann Hans J, “Connectivity disruption sparks explosive epidemic spreading,” Phys. Rev. E 93, 042315 (2016). [DOI] [PubMed] [Google Scholar]
- [25].Xu Zhou, Li Shu, Tian Shen, Li Hao, and Kong Ling-quan, “Full spectrum of covid-19 severity still being depicted,” The Lancet (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Mahase Elisabeth, “Coronavirus: COVID-19 has killed more people than SARS and MERS combined, despite lower case fatality rate,” (2020). [DOI] [PubMed]
- [27].World Health Organization, “WHO Director-General’s opening remarks at the media briefing on COVID-19 – 24 February 2020,” https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---24-february-2020 (2020), accessed: 2020-02-28.
- [28].World Health Organization, “WHO Director-General’s opening remarks at the media briefing on COVID-19 – 3 March 2020,” https://www.who.int/dg/speeches/detail/who-director-general-s-opening-remarks-at-the-media-briefing-on-covid-19---3-march-2020 (2020), accessed: 2020-03-05.
- [29].Porcheddu R, Serra C, Kelvin D, Kelvin N, and Rubino S, “Similarity in Case Fatality Rates (CFR) of COVID-19/SARS-COV-2 in Italy and China,” The Journal of Infection in Developing Countries 14, 125–128 (2020). [DOI] [PubMed] [Google Scholar]
- [30].Peeri Noah C, Shrestha Nistha, Rahman Md Siddikur, Zaki Rafdzah, Tan Zhengqi, Bibi Saana, Baghbanzadeh Mahdi, Aghamohammadi Nasrin, Zhang Wenyi, and Haque Ubydul, “The SARS, MERS and novel coronavirus (COVID-19) epidemics, the newest and biggest global health threats: what lessons have we learned?” International Journal of Epidemiology (2020), 10.1093/ije/dyaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31]. Of course, at the population level, if there are many deaths, medical facilities may be stressed, which can indirectly lead to an increase in death rates.
- [32].Ghani AC, Donnelly CA, Cox DR, Griffin JT, Fraser C, Lam TH, Ho LM, Chan WS, Anderson RM, Hedley AJ, et al. , “Methods for estimating the case fatality ratio for a novel, emerging infectious disease,” Am. J. Epidemiology 162, 479–486 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33]. Since the disease timescale is much shorter than the timescale over which aging appreciably affects death or recovery, the age-dependent transport terms ∂P/∂a can be neglected.
- [34].Riou Julien, Hauser Anthony, Counotte Michel J, and Althaus Christian L, “Adjusted age-specific case fatality ratio during the COVID-19 epidemic in Hubei, China, January and February 2020,” medRxiv (2020), 10.1101/2020.03.04.20031104. [DOI] [Google Scholar]
- [35].covid19 SG, “Dashboard of the COVID-19 Virus Outbreak in Singapore,” https://co.vid19.sg/cases (2020), accessed: 2020-04-04.
- [36].Lai Chih-Cheng, Shih Tzu-Ping, Ko Wen-Chien, Tang Hung-Jen, and Hsueh Po-Ren, “Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) and corona virus disease-2019 (COVID-19): the epidemic and the challenges,” Int. J. Antimicrob. Agents, 105924 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Keeling Matt J and Rohani Pejman, Modeling infectious diseases in humans and animals (Princeton University Press, 2011). [Google Scholar]
- [38].Böttcher Lucas and Antulov-Fantulin Nino, “Unifying susceptible-infected-recovered processes on networks,” arXiv preprint arXiv:2002.11765 (2020).
- [39].nCoV 2019 Data Working Group, “Epidemiological Data from the nCoV-2019 Outbreak: Early Descriptions from Publicly Available Data,” http://virological.org/t/epidemiological-data-from-the-ncov-2019-outbreak-early-descriptions-from-publicly-available-data/337 (2020), accessed: 2020-02-26.
- [40].Atkins Katherine E, Wenzel Natasha S, Ndeffo-Mbah Martial, Altice Frederick L, Townsend Jeffrey P, and Galvani Alison P, “Under-reporting and case fatality estimates for emerging epidemics,” BMJ 350, h1115 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Webb GF, “Population models structured by age, size, and spatial position,” in Structured Population Models in Biology and Epidemiology, edited by Magal Pierre and Ruan Shigui (Springer Berlin Heidelberg, Berlin, Heidelberg, 2008) pp. 1–49. [Google Scholar]
- [42].Wu Joseph T., Leung Kathy, Bushman Mary, Kishore Nishant, Niehus Rene, de Salazar Pablo M., Cowling Benjamin J., Lipsitch Marc, and Leung Gabriel M., “Estimating clinical severity of COVID-19 from the transmission dynamics in Wuhan, China,” Nature Medicine (2020), 10.1038/s41591-020-0822-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].McKendrick AG, “Applications of mathematics to medical problems,” Proc. Edinburgh Math. Soc 44, 98–130 (1926). [Google Scholar]
- [44].Chou T and Greenman CD, “A Hierarchical Kinetic Theory of Birth, Death and Fission in Age-Structured Interacting Populations,” Journal of Statistical Physics 164, 49–76 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].He Xi, Lau Eric H. Y., Wu Peng, Deng Xilong, Wang Jian, Hao Xinxin, Yiu Chung Lau Jessica Y. Wong, Guan Yujuan, Tan Xinghua, Mo Xiaoneng, Chen Yanqing, Liao Baolin, Chen Weilie, Hu Fengyu, Zhang Qing, Zhong Mingqiu, Wu Yanrong, Zhao Lingzhai, Zhang Fuchun, Cowling Benjamin J., Li Fang, and Leung Gabriel M., “Temporal dynamics in viral shedding and transmissibility of COVID-19,” Nature Medicine (2020), 10.1038/s41591-020-0869-5. [DOI] [PubMed] [Google Scholar]
- [46].Seoane Beatriz, “A scaling approach to estimate the covid-19 infection fatality ratio from incomplete data,” (2020), arXiv:2006.02757 [q-bio.PE]. [DOI] [PMC free article] [PubMed]
- [47].Li Ruiyun, Pei Sen, Chen Bin, Song Yimeng, Zhang Tao, Yang Wan, and Shaman Jeffrey, “Substantial undocumented infection facilitates the rapid dissemination of novel coronavirus (SARS-CoV2),” Science (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Press WH, Teukolsky SA, Vetterling WT, and Flannery BP, Numerical Recipes: The Art of Scientific Computing (Cambridge University Press, 2007). [Google Scholar]


