Abstract
Current clinical assays for determining antibiotic susceptibility in Mycobacterium tuberculosis require many weeks to complete due to the slow growth of the bacilli. Here we demonstrate an extremely sensitive single-tube PCR assay that takes less than 3 h and reliably identifies rifampin-resistant M. tuberculosis in DNA extracted directly from sputum. Ninety-five percent of mutations associated with rifampin resistance occur in an 81-bp core region of the bacterial RNA polymerase gene, rpoB. All mutations that occur within this region result in rifampin resistance. The assay uses novel nucleic acid hybridization probes called molecular beacons. Five different probes are used in the same reaction, each perfectly complementary to a different target sequence within the rpoB gene of rifampin-susceptible bacilli and each labeled with a differently colored fluorophore. Together, their target sequences encompass the entire core region. The generation of all five fluorescent colors during PCR amplification indicates that rifampin-susceptible M. tuberculosis is present. The presence of any mutation in the core region prevents the binding of one of the molecular beacons, resulting in the absence of one of the five fluorescent colors. When 148 M. tuberculosis clinical isolates of known susceptibility to rifampin were tested, mutations associated with rifampin resistance were detected in 63 of the 65 rifampin-resistant isolates, and no mutations were found in any of the 83 rifampin-susceptible isolates. When DNA extracted directly from the sputum of 11 patients infected with rifampin-resistant tuberculosis was tested, mutations were detected in all of the samples. The use of this rapid assay should enable early detection and treatment of drug-resistant tuberculosis in clinical settings.
The worldwide increase in drug-resistant tuberculosis poses a major public health threat (16). First-line antituberculosis treatment often fails in patients with rifampin-monoresistant or multidrug-resistant tuberculosis (13). Inappropriate treatment can result in the development of resistance to additional antibiotics (3, 4) and in increased mortality (15, 26). Because Mycobacterium tuberculosis grows extremely slowly (5, 17), conventional susceptibility testing can require many weeks to complete (9). There is thus an urgent need to develop a rapid, simple, and accurate assay to assess drug resistance in M. tuberculosis (7).
Rifampin resistance is an excellent marker for multidrug-resistant tuberculosis because all of these strains are resistant to rifampin (23). Therefore, a screening assay does not need to test susceptibility to all antituberculosis drugs. Rifampin resistance is particularly amenable to detection by rapid genotypic assays because 95% of all rifampin-resistant strains contain mutations localized in an 81-bp region of the bacterial RNA polymerase gene, rpoB, which encodes the active site of the enzyme (14, 21). Moreover, all mutations that occur in this region result in rifampin resistance. By contrast, all rifampin-susceptible M. tuberculosis isolates have the same wild-type nucleotide sequence in this region (14, 22). Thus, it is only necessary to detect a mutation in the rpoB core region to know that the bacilli are rifampin resistant. A number of novel assays based on the PCR have been developed to detect these mutations (2, 24, 25). However, they are technically difficult and have been of limited clinical utility.
We have now developed a single-tube PCR assay that detects all mutations that occur in the M. tuberculosis rpoB core region in real time. The assay is simple, rapid, specific, and extremely sensitive. In less than 3 h, it reliably detects resistance mutations in DNA extracted directly from sputum. The assay is based on previous work of members of our group that demonstrated that fluorogenic nucleic acid hybridization probes, called molecular beacons (27), can be used to detect mutations in the M. tuberculosis rpoB gene (18, 19). The assay utilizes five differently colored molecular beacons, each of which binds to a different target segment within the rpoB core region. Together, the five probes interrogate the entire 81-bp core. Each molecular beacon was designed to be so specific that it does not bind to its target if the target sequence differs from the rifampin-susceptible sequence by as little as a single nucleotide substitution (12, 28). Since molecular beacons fluoresce only when they are bound to their targets, the absence of any one of the five colors in the assay indicates that the bacilli in the sample are rifampin resistant.
MATERIALS AND METHODS
Molecular beacons and primers.
Both conventional and wavelength-shifting molecular beacons were prepared by solid-phase DNA synthesis on an Applied Biosystems (Foster City, Calif.) 394 DNA/RNA synthesizer. The nucleotide sequences of the six molecular beacons that were synthesized were as follows: Probe A, 5′-Texas red-TTTTTT-fluorescein-CGAGCTCAGCTGGCTGGTGCGCTCG-dabcyl-3′; Probe B, 5′-tetrachlorofluorescein-GCTACGGAGCCAATTCATGGACCAGACGTAGC-dabcyl-3′; Probe C, 5′-tetramethylrhodamineTTTTTT-fluorescein-CCGACGCCGACAGCGGGTTGTTCGTCGG-dabcyl-3′; Probe D, 5′-rhodamine-TTTTTT-fluorescein-CCACGCTTGTGGGTCAACCCCCGTGG-dabcyl-3′; Probe E, 5′-fluorescein-CCTGCCGCCGACTGTCGGCGCTGGCAGG-dabcyl-3′; and a 16S probe, 5′-fluorescein-GCGCCCGCGGCCTATCAGCTTGTTGGTGGCGC-dabcyl-3′; underlines identify the arm sequences.
During synthesis, controlled-pore glass columns (Biosearch Technologies, Novato, Calif.) were used to incorporate dabcyl at the 3′ end of the oligodeoxyribonucleotides, fluorescein phosphoramidites (Glen Research, Sterling, Va.) were used to incorporate internal fluorescein moieties, and tetrachlorofluorescein phosphoramidites were used to incorporate tetrachlorofluorescein at the 5′ end. A thiolmodifier phosphoramidite (Glen Research) was incorporated at the 5′-terminal position when fluorescein or tetramethylrhodamine was used as a label, while an aminomodifier phosphoramidite (Glen Research) was incorporated at the 5′-terminal position when rhodamine or Texas red was used as a label. Iodoacetylated fluorescein or tetramethylrhodamine (Molecular Probes, Eugene, Ore.) was then coupled to the 5′-thiol groups, or succinimidyl esters of rhodamine or Texas red (Molecular Probes) were coupled to the 5′-amino groups. All probes were purified by gel exclusion chromatography through a NAP-5 Sephadex column (Amersham Pharmacia Biotech, Piscataway, N.J.), followed by high-pressure liquid chromatography on a Beckman Coulter (Fullerton, Calif.) System Gold chromatograph through a C-18 reverse-phase column (Waters Corporation, Milford, Mass.). A detailed protocol for molecular beacon synthesis is available (www.molecular-beacons.org).
The nucleotide sequences of the primers that were used to amplify a 189-bp segment of the rpoB gene that contains the entire core region were 5′-GGAGGCGATCACACCGCAGACGTT-3′ and 5′-ACCTCCAGCCCGGCACGCTCACGT-3′; and the nucleotide sequences of the primers that were used to amplify a 209-bp segment of the mycobacterial 16S rRNA gene that contains a conserved sequence were 5′-GAGATACTCGAGTGGCGAAC-3′ and 5′-GGCCGGCTACCCGTCGTC-3′.
Sample preparation.
One or two colonies of previously well-characterized M. tuberculosis clinical isolates that were grown on Löwenstein-Jensen slants were lysed by boiling in 1 ml of H2O for 20 min. Five microliters of each lysate was used as a template for each PCR assay.
Purified DNA from other Mycobacterium species was also used in the assay. The species were the following: M. africanum, M. asiaticum, M. avium, M. celatum, M. chelonae, M. flavescens, M. fortuitum, M. gastri, M. genavense, M. intracellulare, M. kansasii, M. leprae, M. lufu, M. malmoense, M. marinum, M. phlei, M. scrofulaceum, M. senegalense, M. simiae, M. smegmatis, M. szulgai, M. thermoresistibile, M. triviale, M. vaccae, and M. xenopi. Between 0.1 and 1 ng of DNA was used as a template for each PCR assay, to mimic the concentration expected to occur in sputum samples.
Expectorated sputum samples were collected from patients in Rio de Janeiro who had a previous diagnosis of pulmonary tuberculosis and who were smear positive for multidrug-resistant M. tuberculosis after 3 months of treatment. Drug resistance was confirmed by conventional mycobacterial culture and susceptibility testing (6). The sputum samples were lysed and detoxified with N-acetylcysteine and NaOH (9). One-milliliter samples from 11 different patients were brought to New York and spun in a Shelton Scientific VSB-14 microcentrifuge (Shelton, Conn.) at 13,000 rpm for 5 min, and each pellet was resuspended in 100 μl of 1 M NaOH in 2% Triton X-100. The samples were then boiled for 8 min and neutralized by the addition of 10 μl of 30% glacial acetic acid and 190 μl of Tris-HCl (pH 8.0). DNA was extracted from each sample using a Geneclean II kit (BIO 101, Carlsbad, Calif.), following the manufacturer's instructions, and was then eluted into 20 μl of H2O. Five microliters of eluant was used as a template for each PCR assay.
Assay conditions.
PCRs were performed in 96-well microtiter plates (Applied Biosystems). Each well contained a total of 50 μl of 1× PCR buffer (Applied Biosystems), 4 mM MgCl2, 250 μM dATP, 250 μM dCTP, 250 μM dGTP, 250 μM dTTP, 2.5 U of AmpliTaq Gold DNA polymerase (Applied Biosystems), a 500 nM concentration of each primer, a 200 nM concentration of each molecular beacon, and 5 μl of template DNA. The wells were hermetically sealed prior to amplification to prevent cross-contamination of untested samples. Amplification was performed in an Applied Biosystems 7700 Prism spectrofluorometric thermal cycler. The reaction mixtures were incubated for 10 min at 95°C to activate the DNA polymerase, followed by 40 to 50 cycles of 95°C denaturation for 30 s, 58°C annealing for 60 s, and 72°C extension for 30 s.
Fluorescence was measured in every well during each annealing step throughout the course of each reaction. The spectral data were automatically analyzed by the computer program controlling the spectrofluorometric thermal cycler to determine the fluorescence intensity contributed by each of the differently colored molecular beacons. The background fluorescence of each probe (calculated from the readings that were taken between the 8th and the 15th thermal cycles) was then subtracted. These fluorescence intensities were then normalized to correct for intrinsic well-to-well variation by multiplying all of the fluorescence readings in each well by a factor that adjusted the maximum fluorescence intensity in each well to the same arbitrary value. The threshold cycle was automatically determined by the computer program controlling the spectrofluorometric thermal cycler to be the number of thermal cycles required before the intensity of the fluorescence signal exceeded six times the standard deviation of the background.
RESULTS
Design of the assay.
Molecular beacons (27) are oligonucleotides that contain a probe sequence embedded within “arm” sequences that are unrelated to the probe. The arms are chosen to be complementary to each other, so that under assay conditions they hybridize to form a stem-and-loop secondary structure in which the probe sequence is located in the loop. A fluorophore is covalently linked to the end of one arm, and a nonfluorescent quencher is covalently linked to the end of the other arm. The fluorophore and the quencher are so close together in the stem helix that fluorescence is suppressed (28). However, when the probe binds to its complementary target, it forms a probe-target helix that is longer and stronger than the stem helix. Since double-stranded DNA is relatively rigid, it is topologically impossible for the probe-target helix and the stem helix to coexist. Consequently, the molecular beacon undergoes a conformational reorganization that causes the arms to unwind. This separates the fluorophore from the quencher, and the probe fluoresces brightly.
In order to detect any deletion, insertion, or nucleotide substitution that may occur in the rpoB core region, it was necessary to design the molecular beacons so that they each could hybridize only to the wild-type sequence. This was accomplished by choosing short probe sequences that form probe-target helices that are just strong enough to overcome the strength of the stem helix. The presence of a mutation in the target creates a mismatched base pair that weakens the probe-target helix, favoring the retention of the stem helix. Because molecular beacons can exist in two different stable states (probe-target hybrid or stem-and-loop structure), they are “finicky” and form a probe-target hybrid only if the target is perfectly complementary to the probe (12, 28). Furthermore, it does not matter where the mismatch occurs within the probe-target hybrid. The melting temperature of a mismatched hybrid is significantly lower than the melting temperature of a perfectly complementary hybrid, irrespective of the identity or location of the mismatch (1). Thus, the presence of a mutation anywhere within the rpoB core region will prevent the binding of one of the molecular beacons, and the characteristic fluorescence of that molecular beacon will not occur.
Five different molecular beacons were synthesized, each perfectly complementary to a different segment of the wild-type rpoB core. Together, the five molecular beacons covered the entire core sequence. In order to carry out the assay in a single tube, each of the molecular beacons was labeled with a differently colored fluorophore. Because five different colors needed to be detected in the same assay, it was highly desirable that each type of fluorophore emit a signal of comparable strength. However, we used a spectrofluorometric thermal cycler to carry out the assays that employs a blue argon-ion laser to stimulate fluorescence. Although energy from blue light is well absorbed by fluorophores that emit green or yellow fluorescence, this energy is not efficiently absorbed by fluorophores that emit orange or red fluorescence, resulting in relatively weak signals. We therefore used two conventional molecular beacons to generate a green signal and a yellow signal, and we used three “wavelength-shifting” molecular beacons to generate differently colored signals in the orange-red portion of the spectrum (29). Figure 1 compares wavelength-shifting and conventional molecular beacons. In conventional molecular beacons, a single fluorophore carries out the dual functions of absorbing energy from the stimulating light and then emitting that energy as fluorescent light of a longer wavelength. In wavelength-shifting molecular beacons, two fluorophores are present on one arm of the probe. A “harvester” fluorophore efficiently absorbs energy from the blue laser light, and that energy is then rapidly transferred to an “emitter” fluorophore (by fluorescence resonance energy transfer), which then fluoresces brightly at its own characteristic (orange or red) wavelength. Figure 2 shows the two complementary strands of the rpoB core region and identifies the target sequence for each of the five molecular beacons.
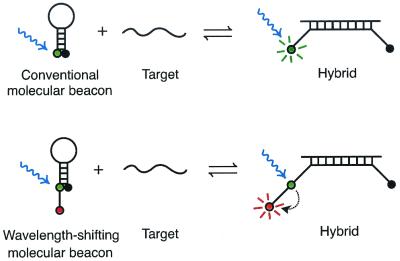
FIG. 1.
Comparison of conventional molecular beacons (top diagrams) to wavelength-shifting molecular beacons (lower diagrams). The fluorescence of both conventional and wavelength-shifting molecular beacons is well quenched when the probes are free in solution (left diagrams), yet both types of probes undergo a conformational reorganization and fluoresce brightly when they bind to their target (right diagrams). When a conventional molecular beacon is bound to a target, its fluorophore absorbs energy from the stimulating light, stores the energy for a few nanoseconds, and then emits that energy as bright fluorescent light of a longer wavelength. However, if the particular fluorophore that is chosen for a conventional molecular beacon does not efficiently absorb energy from the stimulating light, then its fluorescence signal will be weak (for example, when blue laser light is used to stimulate the fluorescence of a red fluorophore). In wavelength-shifting molecular beacons, however, the harvester fluorophore is chosen because it efficiently absorbs energy from the stimulating light (for example, fluorescein efficiently absorbs energy from blue light), and the emitter fluorophore (usually orange or red) is chosen because it is able to efficiently accept energy from the harvester fluorophore, store the energy for a few nanoseconds, and then emit that energy as bright fluorescent light at its own characteristic wavelength.
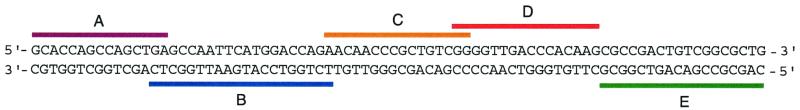
FIG. 2.
Location of the target sequence for each of the five molecular beacons on the complementary strands of the 81-bp M. tuberculosis rpoB core region. Probe B (labeled with tetrachlorofluorescein) and Probe E (labeled with fluorescein) were conventional molecular beacons, while Probe A (labeled with Texas red), Probe C (labeled with tetramethylrhodamine), and Probe D (labeled with rhodamine) were wavelength-shifting molecular beacons.
The assay was designed to work as follows: DNA from a clinical sample is amplified in the presence of the five differently colored molecular beacons. If all five fluorescent colors occur, then rifampin-susceptible M. tuberculosis is present in the sample; if one or more probes fail to fluoresce, then rifampin-resistant M. tuberculosis is present; and if no fluorescence occurs, then M. tuberculosis is not present. In addition, the assay was designed to be sufficiently sensitive to detect even a single bacillus in a sputum sample; fluorescence measurements are taken throughout the course of the amplification, in order to precisely determine the number of bacilli in the sample.
Detection of rifampin resistance.
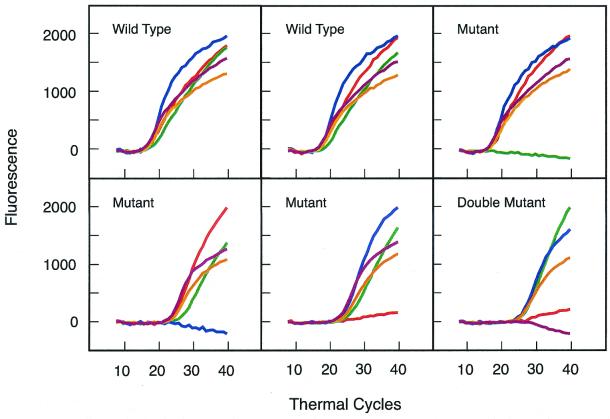
Genomic DNA from 148 previously well-characterized M. tuberculosis clinical isolates was tested. Conventional culture-based analysis had identified 83 of these isolates as being rifampin susceptible and 65 as being rifampin resistant. Fluorescence signals developed in all 148 assays. All five colors appeared in each of the 83 assays carried out with DNA from the rifampin-susceptible isolates. Of the 65 rifampin-resistant isolates that were tested, one color failed to develop in 59 isolates, two colors failed to develop in four isolates (indicating the presence of two different mutations), and two isolates developed all five colors, despite being rifampin resistant. Subsequent nucleotide sequence analysis of these two isolates showed that there were no mutations in their rpoB core region. Thus, they were among the 5% of rifampin-resistant clones whose resistance is due to a mutation outside of the core region. The development of a multicolored signal confirms that rpoB amplicons were synthesized. No fluorescence developed in control reactions that did not contain template DNA. Figure 3 shows representative results obtained from two rifampin-susceptible isolates and four rifampin-resistant isolates (one of which possessed two mutations).
FIG. 3.
Detection of mutations that cause rifampin resistance in M. tuberculosis by the amplification of the rpoB core region in the presence of five differently colored molecular beacons. All of the probes hybridized to the amplicons and generated strong fluorescence signals when rifampin-susceptible strains were tested. However, one or two molecular beacons failed to provide a fluorescence signal when rifampin-resistant strains were tested. In all, 148 different M. tuberculosis clinical isolates were tested. The results obtained from six of these isolates are shown.
Species specificity.
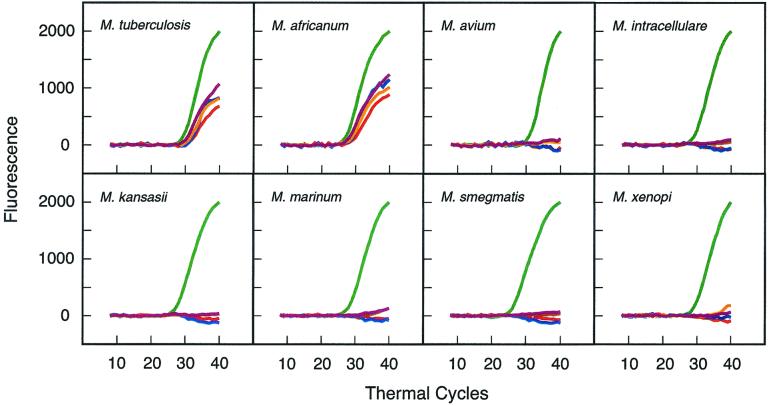
To determine whether the assay is specific for M. tuberculosis, we tested DNA that was isolated from 26 different mycobacterial species. The results (Fig. 4) show that only M. tuberculosis and the closely related M. africanum (a member of the M. tuberculosis group, all of which cause tuberculosis) elicited a positive response from each of the differently colored rpoB molecular beacons that were present (the fluorescein-labeled rpoB molecular beacon was purposely omitted). Significantly, none of the rpoB molecular beacons gave a positive signal with the other 24 species. To confirm that the absence of these fluorescence signals was due to species-specific differences in the rpoB sequence, rather than being due to PCR failure, each reaction was designed to generate an additional signal that served as an amplification control. The reactions contained a second set of primers specific for a conserved region of the 16S rRNA gene of mycobacteria and a fluorescein-labeled molecular beacon to detect the resulting amplicon. This molecular beacon gave a signal with all 26 species tested. Taken together, the results imply that positive signals will occur only when M. tuberculosis or other members of the M. tuberculosis group are present.
FIG. 4.
Determination of the specificity of the assay. Twenty-six mycobacterial species were tested (representative results from eight of the species are shown). Only M. tuberculosis (strain H37Rv) and the closely related M. africanum (which is a member of the M. tuberculosis group) elicited fluorescence from the four differently colored rpoB probes that were present (a fifth rpoB probe, labeled with fluorescein, was omitted). The reactions also contained primers for the amplification of a region of the 16S rRNA gene that is conserved in all mycobacteria and a fluorescein-labeled molecular beacon that was complementary to that region. The presence of fluorescence from the fluorescein-labeled probe (plotted in green) served as a control signal that confirmed that the absence of the other four colors was due to significant differences between the rpoB sequence of M. tuberculosis and the rpoB sequence in each of the other species and not due to PCR failure.
Sensitivity of the assay.
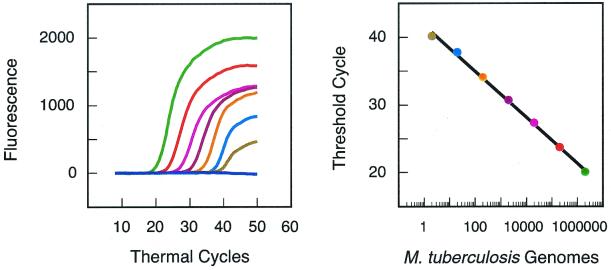
To determine the ability of the assay to provide quantitative results, we prepared samples containing 10-fold serial dilutions of M. tuberculosis genomic DNA. The most concentrated sample contained an amount of DNA equivalent to 2,000,000 bacilli, and the least concentrated sample contained an amount of DNA equivalent to two bacilli. The lower the initial DNA concentration, the greater the number of thermal cycles required to generate a detectable fluorescence signal (the threshold cycle). The results demonstrate (Fig. 5) that the logarithm of the number of target DNA molecules initially present is inversely proportional to the threshold cycle (8). The assay is thus quantitative over an extremely wide range of target concentrations, and it is sufficiently sensitive to detect the DNA from two bacilli.
FIG. 5.
Determination of the sensitivity of the assay. Eight PCR assays were initiated with different quantities of DNA obtained from the M. tuberculosis laboratory strain, H37Rv. The amount of DNA added as a template to each of the eight reactions was calculated to be equivalent to the amount of genomic DNA contained in 0, 2, 20, 200, 2,000, 20,000, 200,000, and 2,000,000 bacilli, respectively. Primers were present to amplify the rpoB core region, and probe E was used to detect the amplicons in real time. The results (shown in the left panel) demonstrate that the number of amplification cycles required to generate a detectable fluorescence signal decreases as the number of target molecules initially present in a reaction increases. A control reaction, which did not contain any template DNA, did not give a signal. The results (shown in the right panel) demonstrate that: (i) the threshold cycle is inversely proportional to the logarithm of the number of target molecules initially present, (ii) quantitative results can be obtained over a wide range of target concentrations, and (iii) the assay is sufficiently sensitive to detect as few as two bacilli.
When preparing samples by serial dilution, it is difficult to obtain a sample containing exactly two genomic DNA equivalents. A test reaction can, by chance, contain a greater or smaller number of DNA molecules. We therefore prepared 10 different samples designed to contain two genomic DNA equivalents and found that 8 of the 10 samples gave positive signals. In addition, we prepared 10 different samples designed to contain two-tenths of a genomic DNA equivalent and found that 1 of the 10 samples gave a positive signal. These results imply that the assay can detect a single target molecule if it is present in the sample.
Determination of rifampin susceptibility in sputum samples.
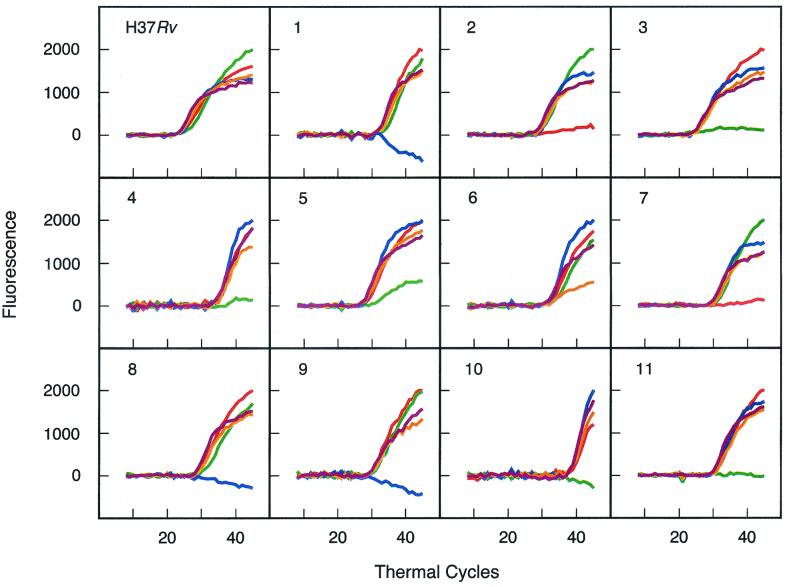
DNA was extracted from 11 smear-positive sputum samples obtained from patients infected with rifampin-resistant M. tuberculosis and was used as a template in the assay. A control sample containing DNA extracted from a rifampin-susceptible M. tuberculosis laboratory strain was evaluated in parallel. All five differently colored molecular beacons gave a fluorescence signal with the rifampin-susceptible control, while one of the five differently colored molecular beacons failed to give a signal with each of the 11 rifampin-resistant samples (Fig. 6). Subsequent nucleotide sequence analysis of the rpoB core region of the amplicons generated from each sputum sample showed that a mutation was present within the target sequence of whichever molecular beacon failed to produce a fluorescence signal.
FIG. 6.
Determination of the rifampin susceptibility of M. tuberculosis found in sputum samples. Eleven PCR assays were carried out with DNA extracted from sputum obtained from smear-positive patients infected with rifampin-resistant M. tuberculosis. A control reaction contained DNA from the rifampin-susceptible M. tuberculosis laboratory strain, H37Rv. Although all five differently colored molecular beacons gave a fluorescence signal with the rifampin-susceptible control, one of the five fluorescent colors failed to develop in each of the 11 rifampin-resistant samples.
DISCUSSION
The results demonstrate that rifampin-resistant M. tuberculosis can be detected in DNA isolated from sputum samples in a single-tube assay that takes less than 3 h to perform. The assay is extremely specific and extraordinarily sensitive. Moreover, the assay is simple to perform and readily automatable for high-throughput screening. The results that are obtained from the assay indicate whether a patient is infected with M. tuberculosis, what concentration of bacilli is present in the sample, and whether the bacilli are rifampin resistant. Because all multidrug-resistant M. tuberculosis strains are rifampin resistant (23), the results of the assay enable an immediate decision to be made as to whether to prescribe a more rigorous course of antibiotic treatment. Furthermore, the assay provides quantitative results, which could eliminate the need to perform sputum microscopy for routine tuberculosis screening.
We utilized wavelength-shifting molecular beacons to provide strong fluorescence signals in the orange and red portion of the visible spectrum. This was necessitated by our use of the Applied Biosystems 7700 spectrofluorometric thermal cycler, which employs a blue argon-ion laser to stimulate fluorescence. However, a number of other instruments are capable of carrying out multiplex real-time gene amplification assays utilizing multicolored light sources for stimulating fluorescence (for example, the Bio-Rad iCycler iQ, the Cepheid Smart Cycler, and the Stratagene Mx4000). All of the probes used in the assay can be conventional molecular beacons if one of these instruments is used. Although the spectrofluorometric instruments used to carry out these assays are relatively expensive, they are becoming common in clinical diagnostic laboratories as a result of an increasing awareness of the broad applicability of closed-tube gene amplification techniques (20, 25, 30).
It should also be possible to replace the multitemperature PCR that is used in the assay with an isothermal gene amplification protocol. For example, molecular beacons provide strong signals with amplicons produced during nucleic acid sequence-based amplification (10), rolling-circle amplification (11, 32), and strand-displacement amplification (31).
Commercial versions of the assay should include a sixth uniquely colored probe that provides a positive signal to confirm that gene amplification is taking place. Ultimately, these assays will enable more rapid diagnosis, earlier treatment, and prompt implementation of infection control procedures to reduce the morbidity, mortality, and spread of drug-resistant tuberculosis.
ACKNOWLEDGMENTS
We thank Richard Chaisson of Johns Hopkins University in Baltimore, Md., and Fernanda Mello and Afranio Kritski of the University of Rio de Janeiro in Brazil for providing infected sputum samples, Amalio Telenti of the Centre Hospitalier Universitaire Vaudois in Switzerland and Michael Levi of Montefiore Medical Center for providing cultured M. tuberculosis isolates, and Pablo Bifani and Barry Kreiswirth of the Public Health Research Institute for providing purified DNA from other mycobacterial species. We also thank Cindy Fung for her expert technical assistance.
This work was supported by National Institutes of Health grants AI-07501, AI-43268, AI-46669, and HL-43521.
REFERENCES
- 1.Bonnet G, Tyagi S, Libchaber A, Kramer F R. Thermodynamic basis of the enhanced specificity of structured DNA probes. Proc Natl Acad Sci USA. 1999;96:6171–6176. doi: 10.1073/pnas.96.11.6171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.De Beenhouwer H, Lhiang Z, Jannes G, Mijs W, Machtelinckx L, Rossau R, Traore H, Portaels F. Rapid detection of rifampin resistance in sputum and biopsy specimens from tuberculosis patients by PCR and line probe assay. Tuber Lung Dis. 1995;76:425–430. doi: 10.1016/0962-8479(95)90009-8. [DOI] [PubMed] [Google Scholar]
- 3.Fischl M A, Uttamchandani R B, Daikos G L, Poblete R B, Moreno J N, Reyes R R, Boota A M, Thompson L M, Cleary T J, Lai S. An outbreak of tuberculosis caused by multiple-drug-resistant tubercle bacilli among patients with HIV infection. Ann Intern Med. 1992;117:177–183. doi: 10.7326/0003-4819-117-3-177. [DOI] [PubMed] [Google Scholar]
- 4.Goble M, Iseman M D, Madsen L A, Waite D, Ackerson L, Horsburgh C R., Jr Treatment of 171 patients with pulmonary tuberculosis resistant to isoniazid and rifampin. N Engl J Med. 1993;328:527–532. doi: 10.1056/NEJM199302253280802. [DOI] [PubMed] [Google Scholar]
- 5.Harris G, Rayner A, Blair J, Watt B. Comparison of three isolation systems for the culture of mycobacteria from respiratory and non-respiratory samples. J Clin Pathol. 2000;53:615–618. doi: 10.1136/jcp.53.8.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heifets L B. Rapid automated methods (BACTEC system) in clinical mycobacteriology. Semin Respir Infect. 1986;1:242–249. [PubMed] [Google Scholar]
- 7.Heifets L B, Cangelosi G A. Drug susceptibility testing of Mycobacterium tuberculosis: a neglected problem at the turn of the century. Int J Tuberc Lung Dis. 1999;3:564–581. [PubMed] [Google Scholar]
- 8.Higuchi R, Fockler C, Dillinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Bio/Technology. 1993;11:1026–1030. doi: 10.1038/nbt0993-1026. [DOI] [PubMed] [Google Scholar]
- 9.Kent P R, Kubica G P. Public health mycobacteriology: a guide for the level III laboratory. U.S. Washington, D.C.: Department of Health and Human Services; 1985. [Google Scholar]
- 10.Leone G, van Schijndel H, van Gemen B, Kramer F R, Schoen C D. Molecular beacon probes combined with amplification by NASBA enable homogeneous, real-time detection of RNA. Nucleic Acids Res. 1998;26:2150–2155. doi: 10.1093/nar/26.9.2150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lizardi P M, Huang X, Zhu Z, Bray-Ward P, Thomas D C, Ward D C. Mutation detection and single-molecule counting using isothermal rolling-circle amplification. Nat Genet. 1998;19:225–232. doi: 10.1038/898. [DOI] [PubMed] [Google Scholar]
- 12.Marras S A E, Kramer F R, Tyagi S. Multiplex detection of single-nucleotide variations using molecular beacons. Genet Anal. 1999;14:151–156. doi: 10.1016/s1050-3862(98)00018-7. [DOI] [PubMed] [Google Scholar]
- 13.Mitchison D A, Nunn A J. Influence of initial drug resistance on the response to short-course chemotherapy of pulmonary tuberculosis. Am Rev Respir Dis. 1986;133:423–430. doi: 10.1164/arrd.1986.133.3.423. [DOI] [PubMed] [Google Scholar]
- 14.Musser J M. Antimicrobial agent resistance in mycobacteria: molecular genetic insights. Clin Microbiol Rev. 1995;8:496–514. doi: 10.1128/cmr.8.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pablos-Mendez A, Sterling T R, Frieden T R. The relationship between delayed or incomplete treatment and all-cause mortality in patients with tuberculosis. JAMA. 1996;276:1223–1228. doi: 10.1001/jama.1996.03540150025026. [DOI] [PubMed] [Google Scholar]
- 16.Pablos-Mendez A, Raviglione M C, Laszlo A, Binkin N, Rieder H L, Bustreo F, Cohn D L, Lambregts-van Weezenbeek C S, Kim S J, Chaulet P, Nunn P. Global surveillance for antituberculosis-drug resistance, 1994–1997. N Engl J Med. 1998;338:1641–1649. doi: 10.1056/NEJM199806043382301. [DOI] [PubMed] [Google Scholar]
- 17.Palacios J J, Ferro J, Ruiz Palma N, García J M, Villar H, Rodríguez J, Macías M D, Prendes P. Fully automated liquid culture system compared with Löwenstein-Jensen solid medium for rapid recovery of mycobacteria from clinical samples. Eur J Clin Microbiol Infect Dis. 1999;18:265–273. doi: 10.1007/s100960050275. [DOI] [PubMed] [Google Scholar]
- 18.Piatek A S, Tyagi S, Pol A C, Telenti A, Miller L P, Kramer F R, Alland D. Molecular beacon sequence analysis for detecting drug resistance in Mycobacterium tuberculosis. Nat Biotechnol. 1998;16:359–363. doi: 10.1038/nbt0498-359. [DOI] [PubMed] [Google Scholar]
- 19.Piatek A S, Telenti A, Murray M R, El-Hajj H, Jacobs W R, Jr, Kramer F R, Alland D. Genotypic analysis of Mycobacterium tuberculosis in two distinct populations using molecular beacons: implications for rapid susceptibility testing. Antimicrob Agents Chemother. 2000;44:103–110. doi: 10.1128/aac.44.1.103-110.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pierce K E, Rice J E, Sanchez J A, Brenner C, Wangh L J. Real-time PCR using molecular beacons for accurate detection of the Y chromosome in single human blastomeres. Mol Hum Reprod. 2000;6:1155–1164. doi: 10.1093/molehr/6.12.1155. [DOI] [PubMed] [Google Scholar]
- 21.Riska P F, Jacobs W R, Jr, Alland D. Molecular determinates of drug resistance in tuberculosis. Int J Tuberc Lung Dis. 2000;4:S4–S10. [PubMed] [Google Scholar]
- 22.Sreevatsan S, Pan X, Stockbauer K E, Connell N D, Kreiswirth B N, Whittam T S, Musser J M. Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA. 1997;94:9869–9874. doi: 10.1073/pnas.94.18.9869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Telenti A, Imboden P, Marchesi F, Lowrie D, Cole S, Colston M J, Matter L, Schopfer K, Bodmer T. Detection of rifampin-resistance mutations in Mycobacterium tuberculosis. Lancet. 1993;341:647–650. doi: 10.1016/0140-6736(93)90417-f. [DOI] [PubMed] [Google Scholar]
- 24.Telenti A, Honore N, Bernasconi C, March J, Ortega A, Heym B, Takiff H E, Cole S T. Genotypic assessment of isoniazid and rifampin resistance in Mycobacterium tuberculosis: a blind study at a reference laboratory level. J Clin Microbiol. 1997;35:719–723. doi: 10.1128/jcm.35.3.719-723.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Torres M J, Criado A, Palomares J C, Aznar J. Use of real-time PCR and fluorimetry for rapid detection of rifampin and isoniazid resistance-associated mutations in Mycobacterium tuberculosis. J Clin Microbiol. 2000;38:3194–3199. doi: 10.1128/jcm.38.9.3194-3199.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Turett G S, Telzac E E, Torian L V, Blum S, Alland D, Weisfuse I, Fazal B A. Improved outcomes for patients with multidrug-resistant tuberculosis. Clin Infect Dis. 1995;21:1238–1244. doi: 10.1093/clinids/21.5.1238. [DOI] [PubMed] [Google Scholar]
- 27.Tyagi S, Kramer F R. Molecular beacons: probes that fluoresce upon hybridization. Nat Biotechnol. 1996;14:303–308. doi: 10.1038/nbt0396-303. [DOI] [PubMed] [Google Scholar]
- 28.Tyagi S, Bratu D P, Kramer F R. Multicolor molecular beacons for allele discrimination. Nat Biotechnol. 1998;16:49–53. doi: 10.1038/nbt0198-49. [DOI] [PubMed] [Google Scholar]
- 29.Tyagi S, Marras S A E, Kramer F R. Wavelength-shifting molecular beacons. Nat Biotechnol. 2000;18:1191–1196. doi: 10.1038/81192. [DOI] [PubMed] [Google Scholar]
- 30.Vet J A M, Majithia A R, Marras S A E, Tyagi S, Dube S, Poiesz B, Kramer F R. Multiplex detection of four pathogenic retroviruses using molecular beacons. Proc Natl Acad Sci USA. 1999;96:6394–6399. doi: 10.1073/pnas.96.11.6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walker G T, Fraiser M S, Schram J L, Little M C, Nadeau J G, Malinowski D P. Strand displacement amplification—an isothermal, in vitro DNA amplification technique. Nucleic Acids Res. 1992;20:1691–1696. doi: 10.1093/nar/20.7.1691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang D Y, Brandwein M, Hsuih T C, Li H. Amplification of target-specific, ligation-dependent circular probe. Gene. 1998;211:277–285. doi: 10.1016/s0378-1119(98)00113-9. [DOI] [PubMed] [Google Scholar]