Abstract
The increased infectivity and transmissibility of SARS‐CoV‐2 new variants were contributed largely by increase binding of receptor binding domain (RBD) domain of the Spike (S) protein to its cellular receptor ACE2 (Angiotensin-Converting Enzyme 2). Several studies have indicated that heparin and its derivatives interact to SARS-CoV-2 S-RBD and inhibits the binding of ACE2 which blocks the viral invasion. However, it is largely unclear how these SARS-CoV-2 variants affects ACE2 binding in the presence of heparin. Herein, using the molecular docking and interaction energy analysis, we showed that N501Y, L452R-E484Q, and E484K mutations bind strongly with heparin in the range of − 7.4 to − 8.0 kcal/mol. The triple mutations, K417N‐E484K‐N501Y, and K417T‐E484K‐N501Y displayed weaker binding affinity to heparin (−6.6 kcal/mol). Further, we showed that most of the RBD mutations increased the binding affinity of ACE2 in the absence of heparin, with the maximum increase observed for N501Y (−13.7 kcal/mol). Also, in the presence of heparin, ACE2 binds strongly to the mutant RBD as compared to WT RBD. The strong RBD/ACE2 interaction was observed in case of triple variants (−11.3 kcal/mol) whereas, N501Y showed weakest binding of RBD/ACE2 in the presence of heparin (−9.2 kcal/mol). The strong binding of ACE2 to RBD-heparin complex in these variants will leads to strong inhibition of their entry into host cells.
Keywords: SARS-CoV-2 variants, RBD mutations, Heparin, ACE2, Molecular docking, Binding affinity
Graphical Abstract
1. Introduction
Since the outbreak of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in China in December 2019, it is clear that the coronavirus disease 2019 (COVID-19) pandemic is going to persist for the foreseeable future [1]. Currently, it has spread worldwide, infecting more than 274 million people and resulting in over 5.3 million deaths. Across the world, multiple variants of SARS-CoV-2 have been reported recently. The recently emerging variants of SARS-CoV-2 have been categorized into different categories, variants of interest (VOI) and variants of concern (VOC). The variants including B.1.526, B.1.526.1, B.1.527, and P.2 lineages have been classified under VOI and the variants including B.1.1.7, B.1.351, B.1.427, B.1.429, B.1.617, and P.1 lineages have been classified under VOC. The newly variants have rapidly become dominant within their countries [2]. The variants, B.1.1.7 (501Y.V1, also known as Alpha variant), B.1.351 (501Y.V2, also known as Beta variant), B.1.1.28 (Gamma variant), B.1.525 (eta variant), B.1.617.1 (kappa variant), and B.1.427/ B.1.429 (epsilon variant) are responsible for increased transmissibility, infectivity, and disease severity. The B.1.1.7 variant carries 17 mutations in its genome, including N501Y in the Spike Receptor-Binding Domain (RBD) [3], [4]. The variant, B.1.351 [5] and B.1.1.28 [6] carries 9 and 11 mutations in the spike (S) protein, respectively, including three mutations in the receptor binding domain (RBD), K417N/T, E484K, and N501Y. B.1.427 and B.1.429 harbor L452R mutation in RBD [7]. The emergence of these variants has been associated to increased transmissibility, disease severity, and infectivity [8], [9]. Many studies have shown that these mutations increase the binding affinity of the RBD to the angiotensin-converting enzyme 2 (ACE2) receptor [10], [11], thus leads to increase in transmissibility and possibly pathogenicity. Very recently, a new variant of SARS-CoV-2, B.1.617 (Delta variant) discovered in India, carries two RBD mutations namely E484Q and L452R which is a double whammy. The Delta variant have invaded over 175 nations and becomes the most prevalent strain in the world. On November 26, 2021, the World Health Organization (WHO) reported the variant B.1.1.529, commonly known as Omicron, as a VOC. The Omicron variant has thirty mutations in the S protein, of which 15 mutations occur in the RBD of S protein, as well as the presence of three deletions and one insertion.
Many cell-based studies have reported that heparin, and its derivatives prevent the infection of many viruses, including Zika virus [12], herpes [13], human immunodeficiency virus [14], and SARS-associated coronavirus isolate HSR1 [15]. Recently, Clausen et al. [16] showed that heparan sulfate (HS) binds with the RBD and shifts the spike structure to an open conformation to enable ACE2 binding. The authors further showed that heparin binding to SARS-CoV-2 RBD inhibit viral invasion to host cells and thus can prevent the infection, that has been confirmed by many other works [17], [18], [19], [20]. These results suggest that heparin binds to the RBD at RBD/ACE2 interface and inhibits the RBD-ACE2 interaction which further prevents virus entry to host cell. However, how heparin modulates the RBD/ACE2 interaction in reference to new SARS-CoV-2 variants has not been studied in detail. Here, we sought to know whether S proteins mediated entry of the SARS-CoV-2 variants can be inhibited by heparin. Therefore, we study the structural basis behind heparin induced inhibition of RBD/ACE2 interaction using molecular docking and interaction energy analysis. The different binding affinity of ACE2 to RBD variants in the presence of heparin will help to understand the inhibitory role of heparin towards the newly emerged SARS‐CoV‐2 variants.
2. Methods
2.1. Structural modeling
We used the crystal structure of the SARS-CoV-2 S protein RBD-ACE-2 receptor complex protein for the study (PDB ID: 6M0J) [21]. The RBD (6M0J_ Chain E) was used to create the mutations in the protein. Different three-dimensional structures of the RBD variants namely, K417N, K417T, E484K, E484Q, L452R, N501Y, L452R-E484Q, K417N-E484K-N501Y, and K417T-E484K-N501Y were modeled using the BuildModel module of FoldX [22].
2.1.1. Molecular docking analysis of SARS-CoV-2 RBD-heparin tetrasaccharides
The molecular docking of heparin tetrasaccharides to the SARS-CoV-2 S-RBD structure (6M0J, Chain: E, residues 319–541) was achieved through the ClusPro webserver [23] which has an advanced option for heparin tetrasaccharides docking (http://cluspro.bu.edu/). The tool is based on a rigid body docking algorithm PIPER [24], created on the Fast Fourier Transform (FFT) correlation approach which generates and evaluates almost billion poses of a heparin tetrasaccharides probe. The docked structures were then clustered using pairwise root-mean-square deviations (RMSD) as the distance measure of 9 Å C-alpha RMSD radius. The server results in 10 different docked poses which were ranked according to the cluster size and lowest energy. Finally, the clusters with the highest protein-heparin contacts reliably predicts the heparin binding site. Both the wild type (WT) RBD and the mutant RBD were docked with heparin tetrasaccharides to investigate the effect of RBD variants on the protein-heparin interactions. The residues involved in the protein-heparin interactions in the selected docked models were then studied by Discovery Studio Visualizer (BIOVIA) and Ligplot+ [25].
The binding affinity for the WT RBD-heparin and mutant RBD-heparin complexes were then determined through the PROtein binDIng enerGY prediction (PRODIGY)-LIG webserver [26]. Prodigy-LIG predicts the binding affinity in protein-ligand complex using atomic contacts-based prediction method.
2.2. Molecular docking of SARS-CoV-2 RBD-heparin complex to ACE2 and interaction analysis
To investigate the effect of heparin on RBD-ACE2 interaction, we performed the molecular docking of ACE-2 (PDB Id: 6M0J, Chain A) with WT or mutated RBD structures in the absence and presence of heparin using the HDOCK server [27], which is based on a hybrid algorithm of template-based modeling and ab initio free docking. The docked structures with the lowest RMSD and large docking score were considered for further study. For ACE-2/RBD-heparin complexes, a template-based docking without residue constraint was performed. The residues involved in protein-protein interactions in the selected docked structures were then investigated through PDBsum server (http://www.ebi.ac.uk/pdbsum). The bonded interactions between the protein-protein interactions were then compared among different RBD/ACE2 and RBD-heparin/ACE2 complexes.
2.2.1. Binding affinity and interaction energies of RBD-heparin/ACE2 complexes
The binding affinity of RBD/ACE2 and RBD-heparin/ACE2 complex structures was predicted using PRODIGY webserver that predicts the binding affinity and dissociation constant for different biological complexes [28]. Finally, the interaction energies of the docked complexes were computed through PPCheck web server [29]. The strength of a protein-protein interface was estimated by calculating the total stabilizing energy (ET) which includes the contribution from hydrogen bonding, electrostatic and van der Waals interactions, thus providing complete information on the qualitative and quantitative features of RBD/ACE2 interface in the absence and presence of heparin.
3. Results and discussion
Binding between SARS-CoV-2 S-RBD and ACE2 can be inhibited using small molecules [30], and heparin and its derivatives [16], [18], [31]. It is likely that heparin binding sites in SARS-CoV-2 RBD overlap with the ACE2 binding region, indicating that heparin and its derivatives might restricts the SARS-CoV-2 entry.
3.1. Molecular docking studies of SARS-CoV-2 RBD and heparin oligosaccharides
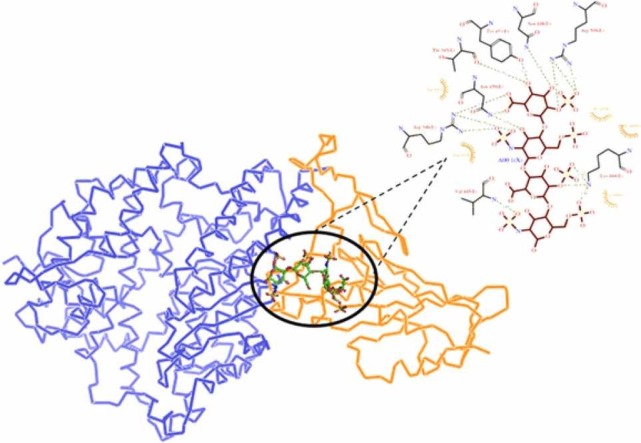
In order to determine how strongly SARS-CoV-2 RBD binds to heparin in its WT form and how the RBD mutations affect heparin binding, we here used the ClusPro docking webserver [23]. Cluspro scan the entire protein surface of SARS-CoV-2 RBD to search for heparin binding sites using the models of heparin tetrasaccharides as probes. The docking results in ten models based on the cluster size and low energy. We have identified two clusters with the highest protein-heparin contacts and through structural inspection of these top two models, we find two different modes of heparin binding to the RBD ( Fig. 1) which are in agreement with the previous studies [18], [31].
Fig. 1.
Molecular docking interactions between SARS-CoV-2 spike RBD (PDB ID:6M0J_Chain E) to heparin tetrasaccharides. The docked structures of Heparin-WT RBD complexes were obtained from Cluspro webserver. (A) Three-dimensional (3D) diagram of heparin-RBD complex of the model of cluster 0. (B) Two-dimensional (2D) representation of the interactions between RBD and heparin from cluster 0 model. (C) 3D model of heparin-RBD complex obtained from cluster 1 of Cluspro results. (D) 2D representation of intermolecular interactions of RBD-heparin complex.
To explore the key heparin binding residues in the SARS-CoV-2 RBD region, the intermolecular interactions between the heparin tetrasaccharides and the RBD residues were identified from Ligplot+ and shown in Fig. 1, Fig. 2. The Cluspro docking model from cluster 0 (Fig. 1A) indicates that the heparin binding site consists of residues T345, R346, F347, L441, D442, K444, V445, G446, G447, N448, Y449, N450, Y451, and R509 (site I) (Fig. 1B). The critical residues of RBD involved in hydrogen (H) bond formation in WT RBD-heparin complex was T345, K444, V445, N448, N450, Y451, and R509. The three salt bridges were shown to be formed by residues R346, K444, and R509 along with Vander Waal interactions with residues F347, L441, D442, V445, G446, and G447. Most of these heparin binding residues overlaps to the ACE2 binding regions as the residues 446–456 and 486–505 of RBD was predicted to participate in the RBD-ACE2 interaction [21], [31].
Fig. 2.
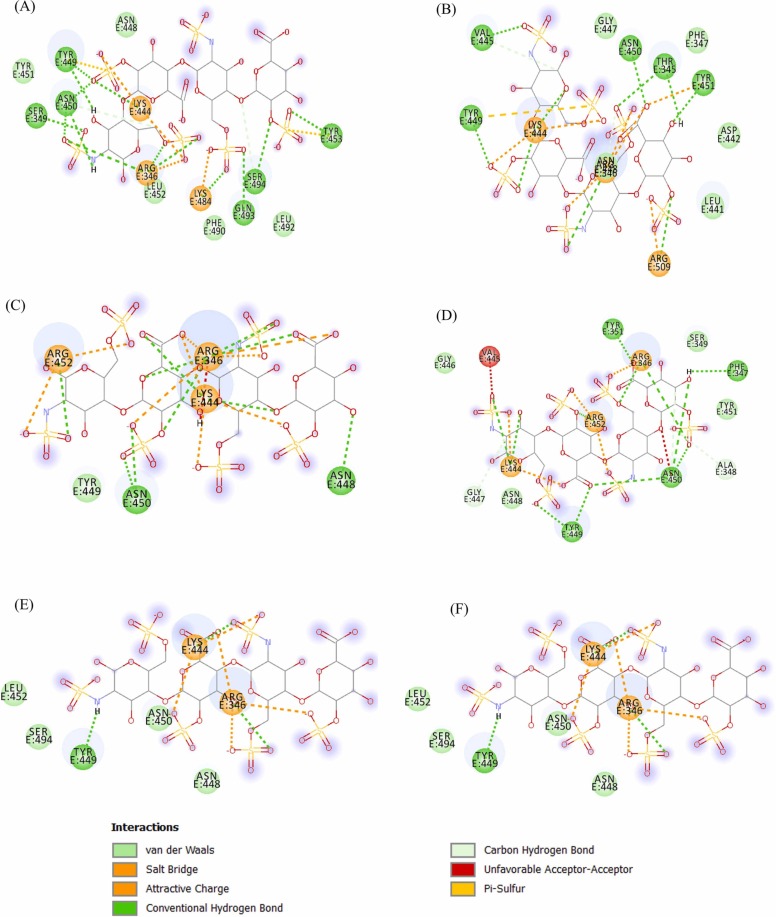
Molecular docking interactions of heparin to SARS-CoV-2 variants. The docked structures of Heparin-mutant RBD complexes were obtained from Cluspro webserver. Two-dimensional (2D) diagrams of heparin-RBD interactions for (A) E484K, (B) N501Y, (C) L452R, (D) E484Q, (E) L452R-E484Q, (F) K417N‐E484K‐N501Y, and (G) K417T‐E484K‐N501Y. The protein residues and interactions are colored accordingly and provided in the figure.
Whereas, the structural model from cluster 1 (Fig. 1C) indicates the interacting residues were W353, N354, R355, K356, R357, N394, Y396, R466, and L518 (site II) (Fig. 1D). The H bond forming residues in this heparin binding site was N354, R355, K356, R357, N394, Y396, and R466. The four salt bridges reported were from residues R355, K356, R357, and R466 along with Vander Waal interactions mediated by W353, and L518.
Next, to evaluate the effect of RBD mutations on the heparin binding, we have selected structural model from cluster 0 for further study because of its large size and lowest energy. The E484K substitution results in stronger binding of heparin to RBD as indicated by the formation of H-bond from a different set of residues including S349, K444, Y453, K484 Q493, and S494, along with different salt bridge forming residues K484 and Y453 (Fig. 2A). Thus, the substituted residue Lys484 form an extra salt bridge and an additional H-bond which would result in increased binding affinity of heparin to RBD. Moreover, the substitution leads to complete loss of WT binding pocket, hence is largely affecting the interaction of RBD-heparin complex. The E484K mutation binds to heparin at a site which is not overlapping to the ACE2 binding region. This could lead to the inability of heparin to inhibit the RBD-ACE2 interaction in case of E484K variant. The E484K variant leads to high transmissibility and infectivity of SARS-CoV-2.
The mutation N501Y (Alpha variant) also increases the infectivity and has increased the COVID-19 cases. The intermolecular interactions of N501Y RBD-heparin complex revealed increase in number of H-bonds and salt bridges as compared to WT RBD (Fig. 2B). The additional two H-bonds were formed by K444 and Y449 along with an increase in salt bridge formation by Y451 and N448, indicating the strong affinity to heparin. The L452R reported in epsilon variant (B.1.427and B.1.429) exhibited a similar extent of heparin binding as in N501Y. The L452R substitution is evident with the formation of an extra H-bond and two salt bridges from the substituted residue R452. The docking study thus revealed that L452R variant has stronger heparin binding affinity compared to WT RBD. However, a significant loss of Van der Waal interactions was observed (Fig. 2C). The E484Q substitution behaves overall similar to WT RBD except that Y449 form two H-bonds and a loss of H-bond from V445 (Fig. 2D). In addition, K444 forms an additional H-bond in E484Q variant along with an unfavorable acceptor-acceptor interaction by T345.
To investigate the heparin binding to L452R-E484Q RBD, we generated the structural model of L452R-E484Q mutant and studied its docking to heparin (Fig. 2E). Compared to WT RBD, the L452R-E484Q mutant complex form unique H-bonds with residues F347, Y351, Y449, and R452 along with the loss of H-bond from residues T345, V445, N448, Y451, and R509. In addition, two unfavorable acceptor-acceptor interactions were formed by V445 and N450 along with loss of salt bridges. The salt bridges were formed only by R346, K444, and R452. The overall interactions suggest that the L452R-E484Q binds to heparin with a similar extent to WT RBD.
The triple variants, K417N‐E484K‐N501Y (Fig. 2F) and K417T‐E484K‐N501Y (Fig. 2G) showed significant loss of intermolecular interactions between RBD-heparin complex. In both the variants, the H-bonds were formed only by R346, K444, and Y449 along with the salt bridge through R346, and K444. Residues T345, V445, N448, N450, Y451, and R509 failed to form any interactions in these variants. These results thus indicate that heparin will bind very weakly to the RBD of both the Beta and Gamma SARS-CoV-2 variants. The lesser binding affinity of heparin suggest that heparin mediated inhibition of SARS-CoV-2 invasion might not be observed in the case of triple variants or it required higher concentration of heparin which could not be feasible for human use.
In case of K417N substitution, Y451 failed to form H-bond but form an extra salt bridge and an extra H-bond formed by Y449 and R346 (Supporting information, Fig. S1A). Similarly, in case of K417T, R346 failed to form H-bond but form an extra salt bridge (Supporting information, Fig. S1B). Rest of the interactions in case of K417 substitution to N/T were very similar to that of WT RBD-heparin complex.
Interestingly, for the structural model of cluster 1 obtained from Cluspro, the intermolecular interactions in RBD-heparin complexes are similar to WT RBD for the variants N501Y, L452R, E484Q, K417N, and K417T (Supporting information, Fig. S2 A-E). However, the triple variants, K417N‐E484K‐N501Y (Supporting information, Fig. S3A) and K417T‐E484K‐N501Y (Supporting information, Fig. S3B) showed different binding pocket consisting of residues R346, K444, Y449, N448, N450, K484, Q493, and S494 involved in H-bonding. For L452R-E484Q mutant, the binding pocket has residues involved in H-bonding are R346, S349, Y351, K444, G447, Y449, N450, and R452 (Supporting information, Fig. S3C), and is very much similar to the binding pocket observed in case of structure model of cluster 0. Similarly, the binding pocket of E484K is also very much similar to the pocket obtained from other structure model (Supporting information, Fig. S3D). These results thus clearly indicate that the different SARS-CoV-2 variants displayed different binding pockets and thus leads to the complete loss of WT RBD-heparin binding pocket.
Next, we used Prodigy-LIG server [26] to predict the binding affinities of the WT RBD-heparin and mutant RBD-heparin complexes ( Table 1). Here, the binding affinity obtained depends on the number of contacts formed by the protein-ligand complex. The WT RBD-heparin complex showed a binding affinity (ΔG) of − 7.7 kcal/mol, whereas the mutant RBD showed comparable binding affinity towards heparin, with the strongest binding affinity exhibited by E484K, N501Y and L452R-E484Q (Table 1). The decrease in heparin binding affinity was observed for triple variants K417N/T-E484K-N501Y. We observed that the higher binding affinity was due to increased numbers of atomic contacts.
Table 1.
PRODIGY-LIG derived binding affinity ΔGnoelec (kcal/mol) and PPCheck-derived total stabilizing energy (kJ/mol) for the WT RBD-heparin complex and mutant RBD-heparin complexes.
| RBD-Heparin complex | ΔGnoelec (kcal/mol) | Total Stabilizing Energy (kJ/mol) |
|---|---|---|
| WT RBD-Heparin | -7.7 | -129.55 |
| K417N- Heparin | -7.8 | -130.19 |
| K417T- Heparin | -7.7 | -133.75 |
| L452R- Heparin | -7.4 | -95.64 |
| E484K- Heparin | -7.9 | -141.53 |
| E484Q- Heparin | -7.6 | -122.30 |
| N501Y- Heparin | -8.1 | -152.04 |
| K417N-E484K-N501Y- Heparin | -6.6 | -73.20 |
| K417T-E484K-N501Y- Heparin | -6.6 | -72.94 |
| L452R-E484Q- Heparin | -8.0 | -148.46 |
PPCheck was further used to calculate the total stabilization energy (Es) for the RBD-heparin complexes. The Es for the RBD-heparin interaction was largely contributed by Vander Waals interactions. As can be seen, the Es for WT RBD-heparin complex was − 129.55 kJ/mol which was much higher than L452R and K417N/T-E484K-N501Y variants (Table 1). The other RBD variants exhibited higher Es than WT RBD, with the maximum value was seen in L452R-E484Q, E484K, and N501Y. Overall, the results indicated that the increasing affinity of heparin to different RBD mutations in SARS-CoV-2 variants is arranged in the following manner:
N501Y> >L452R-E484Q>E484K>K417N~WT~417 T > E484Q>L452R> >K417N-E484K-N501Y~ K417N-E484K-N501Y.
3.2. Binding affinity and interaction energy of the RBD-Heparin/ACE2 complexes
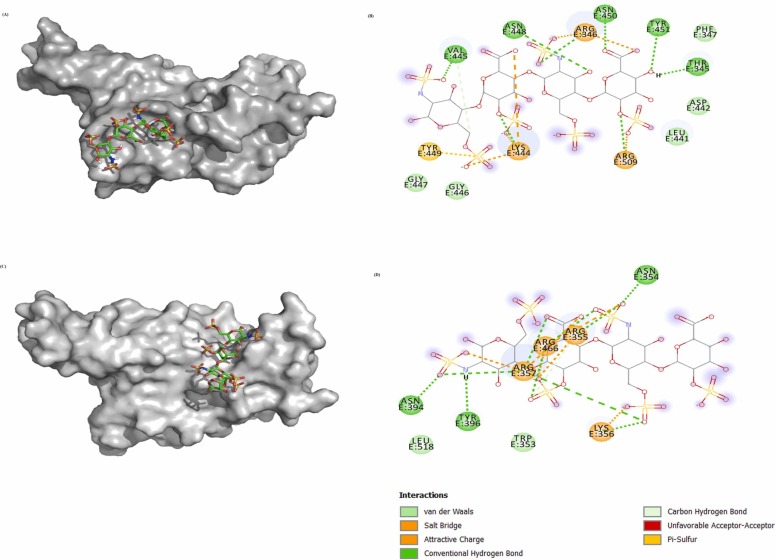
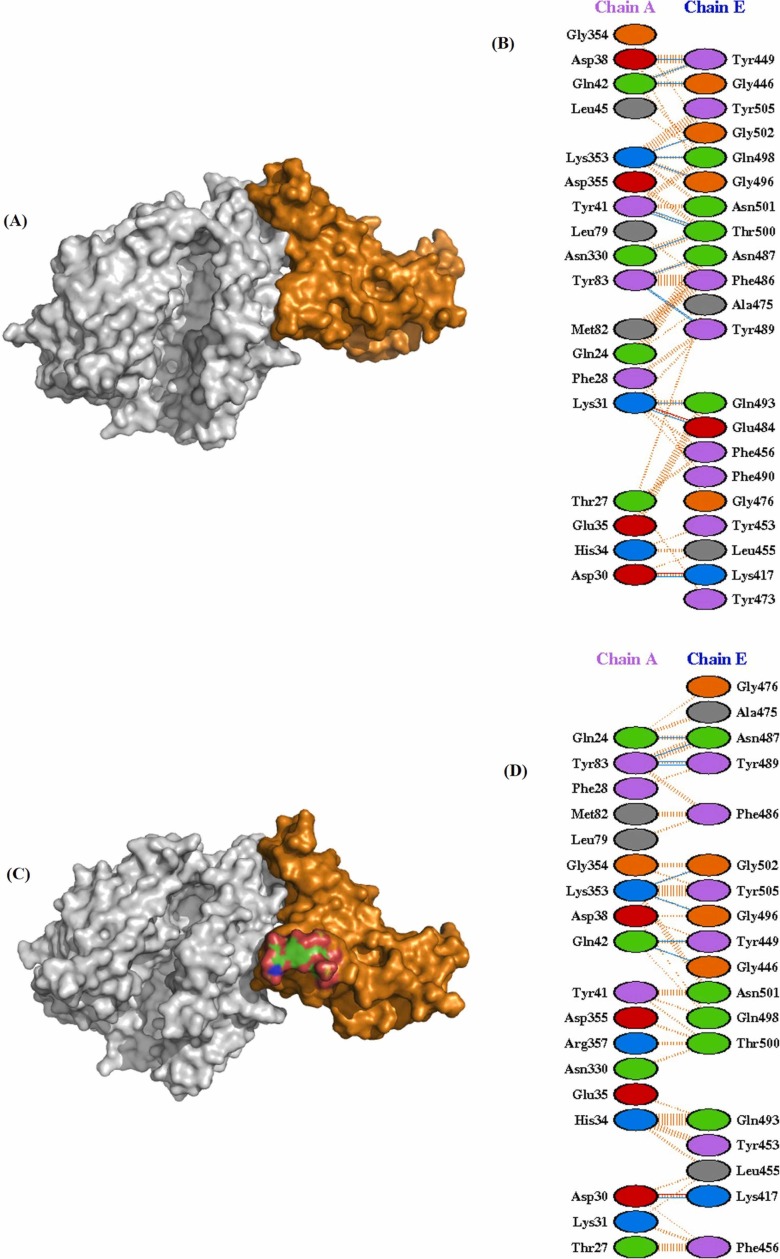
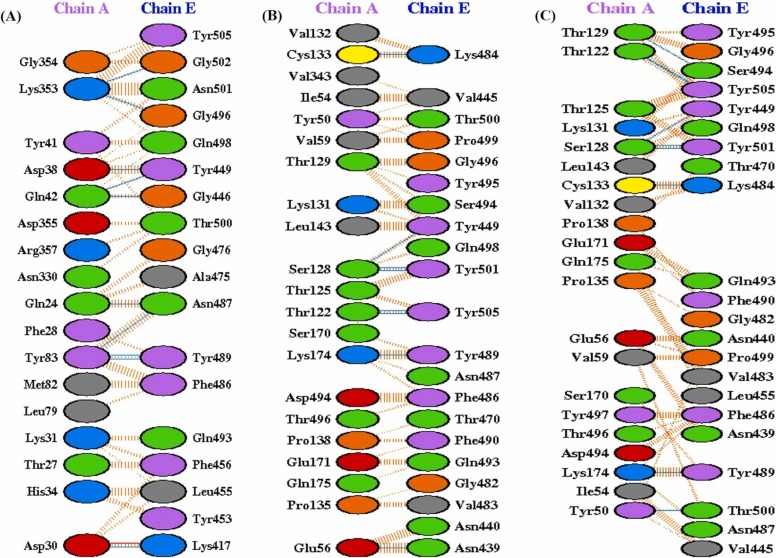
Molecular docking of WT RBD/ACE2 (6M0J: Chain E/Chain A) showed that RBD binds strongly to ACE2 with a docking score of − 310.19 kcal/mol. Interestingly, heparin bound RBD interact with a lesser affinity to ACE2 with the docking score of − 269.41 kcal/mol, which confirmed that bound heparin inhibited the interfacial interactions between the RBD-ACE2 complex. Further, to investigate the interactions in the WT RBD/ACE2 complex in both the absence and presence of heparin, representative structures were subjected to PDBsum analysis to identify the interfacial contacts. Interaction analysis revealed that WT RBD and ACE2 strongly interact in the absence of heparin by forming 2 salt bridges, 15 H-bonds, and 176 nonbonded interactions. The H-bonds formed by the RBD-ACE2 includes K417-E30, G446-Q42, Y449-D38, Y449-Q42, E484-K31, N487-Y83, Y489-Y83, Q493-K31, G496-K353, Q498-K353, T500-Y41, T500-N330, and G502-K353 ( Fig. 3A and B). The two salt bridges were formed between K417-D30, and E484-K31.
Fig. 3.
Molecular docking of SARS-CoV-2 spike RBD (PDB ID:6M0J_Chain E) to ACE2 (PDB ID:6M0J_Chain A) in the absence (A-B) and presence of heparin (C-D). (A) Three-dimensional (3D) diagram of ACE2-RBD complex (B) Two-dimensional (2D) representation of the interactions between ACE2 and RBD. (C) 3D model of RBD-heparin /ACE2 complex. The docked structure of heparin-RBD complex was obtained from Cluspro webserver. (D) 2D representation of intermolecular interactions of RBD-heparin/ACE2 complex.
In case of WT RBD-ACE2 complex in the presence of heparin, most of the intermolecular interactions between RBD and ACE2 lost significantly. In the presence of heparin, WT RBD binds to ACE2 through 9 H-bonds, 1 salt bridge, and 153 non-bonded contacts. Total six H-bonding (Y449-D38, E484-K31, E493-K31, Q498-K353, T500-Y41, and T500-N330) and one salt bridge (E484-K31) interactions were lost due to heparin binding to the RBD (Fig. 3C and D).
Protein-protein docking of mutant RBD/ACE2 in the absence and presence of heparin were then analyzed and the docking score of RBD/ACE2 complexes are shown in Table 2. The docking results indicates that most of the SARS-CoV-2 variants binds strongly to ACE2 in the absence of heparin as revealed by increase in the docking score (Table 2). Among the RBD variants, the increasing affinity of ACE2 to RBD in the absence of heparin is arranged as:
Table 2.
Binding results of ACE2 to SARS-CoV-2 RBD variants in the absence and presence of heparin obtained from in-silico investigations using HDOCK, PRODIGY, and PPCheck webservers.
| – Heparin |
+ Heparin |
|||||||
|---|---|---|---|---|---|---|---|---|
| RBD /ACE2 Complex | Docking Score | ΔG | Kd (M) at 25.0 ℃ | Total stabilization energy (kJ/mol) | Docking Score | ΔG | Kd (M) at 25.0 ℃ | Total stabilization energy (kJ/mol) |
| (kcal/ mol) |
(kcal/ mol) |
|||||||
| RBD-ACE2 | -310.19 | -12.6 | 6.0E-10 | -174.93 | -290.56 | -8.9 | 3.1E-07 | -120.78 |
| K417T /ACE2 | -339.90 | -13.7 | 8.7E-11 | -219.07 | -277.02 | -13.0 | 2.8E-10 | -63.67 |
| K417N /ACE2 | -325.91 | -12.7 | 5.1E-10 | -227.30 | -275.07 | -14.1 | 4.6E-11 | -160.27 |
| L452R/ACE2 | -314.81 | -13.2 | 2.0E-10 | -218.67 | -284.50 | -10.7 | 1.4E-08 | -135.72 |
| E484K/ACE2 | -325.91 | -13.1 | 2.3E-10 | -210.76 | -281.99 | -10.8 | 1.3E-08 | -118.41 |
| E484Q /ACE2 | -331.06 | -12.7 | 5.1E-10 | -203.51 | -286.98 | -10.6 | 1.5E-08 | -132.84 |
| N501Y/ACE2 | -363.41 | -13.7 | 8.6E-11 | -258.88 | -275.11 | -9.2 | 1.8E-07 | -74.87 |
| K417T-E484K-N501Y/ACE2 | -364.08 | -13.5 | 1.2E-10 | -211.68 | -295.93 | -11.3 | 4.9E-09 | -168.91 |
| K417N-E484K-N501Y/ACE2 | -311.75 | -12.7 | 4.6E-10 | -182.15 | -293.04 | -11.4 | 4.4E-09 | -182.88 |
| L452R-E484Q /ACE2 | -316.27 | -12.9 | 3.5E-10 | -208.67 | -297.18 | -11.1 | 6.7E-09 | -158.78 |
N501Y> K417T-E484K-N501Y>L452R-E484Q> K417N-E484K-N501Y> E484K > L452R> K417N > E484Q> K417T >WT RBD (Table 2).
However, heparin binding reduced the ACE2 binding affinity to RBD as indicated by decrease in the docking energy scores (Table 2). In the presence of heparin, overall binding affinity towards ACE2 reduced and the binding affinity among the variants are arranged in decreasing order as:
WT< N501Y< E484Q < L452R < E484K< L452R-E484Q< K417T-E484K-N501Y< K417N-E484K-N501Y< K417T< K417N (Table 2).
Interestingly, in the presence of heparin the ACE2 binds weakly to N501YRBD-heparin complex with a docking score of − 275.11 kcal/mol. The other three single RBD mutants, E484Q, L452R, and E484K also displayed reduced binding affinity to ACE2 in the presence of heparin. The L452R-E484Q mutant SARS-CoV-2 variants binds strongly with ACE2 (−297.18 kcal/mol) as compared to the docking scores of triple mutants, K417T-E484K-N501Y (−295.93 kcal/mol), and K417N-E484K-N501Y (−293.04 kcal/mol). Whereas, K417N and K417T showed maximum binding affinity to ACE2 in the presence of heparin.
The lesser binding of ACE2 to N501Y in the presence of heparin was explored through the PDBsum analysis of intermolecular interactions of the mutant RBD and ACE2 structures. The UK variant N501Y reported to form 5 H-bonds, 1 salt bridge, and 192 non-bonded contacts with the ACE2 in the presence of heparin ( Fig. 4A), suggesting a weaker binding affinity. The H bonds forming residues were G416-D615, Y421-D615, Y501-S170, G504-D494, and Y505-P492. The interaction clearly suggests that N501Y binds to ACE2 in a completely different binding pocket where all the H-bond interactions observed in case of WT/ACE2 interactions were lost. This could further explain by the fact that heparin binds more strongly to N501Y mutants as compared to other variants. This increased strength of heparin binding induces a large conformational change in the RBD which might allow the less binding to its cognate ACE2 receptor.
Fig. 4.
Molecular docking interactions of ACE2 to SARS-CoV-2 RBD variants in the presence of heparin. Two-dimensional (2D) diagrams of RBD/ACE2 interactions in the presence of heparin for (A) N501Y, (B) L452R (C) E484Q, and (D) E484K. The protein residues and interactions are colored accordingly and provided in figure.(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
Both the L452R and E484Q variant formed 9 H-bonds and 1 salt bridge with the similar interfacial contacts between WT RBD/ACE2 in the presence of heparin (Fig. 4B-C). The intermolecular non-bonded contacts in L452R and E484Q mutant complex were 169 and 159, respectively. The E484K RBD form 8 hydrogen bonds, 1 salt bridge, and 120 nonbonded contacts with ACE2 in the presence of heparin (Fig. 4D). The H-bonds includes the key RBD residues K417, G446, Y449, N487, Y489, G496, and G502. The only salt bridge involves K417-D30. The substituted residue 484 K leads to the loss of one H-bond normally formed between E484-K31 of ACE2.
The higher binding affinity of ACE2 to L452R-E484Q mutant can be explained by the presence of 10 H-bonds, 1 salt bridge, and 180 non-bonded contacts in the RBD/ACE2 interface. The H-bond between Y449 (RBD) and D38 (ACE2) is the one extra H-bond formed as compared to WT RBD/ACE2 complex ( Fig. 5A). All other H-bonds are K417-E30, G446-Q42, Y449-Q42, N487-Q24, N487-Y83, Y489-Y83, G496-K353, and G502-K353 along with a salt bridge K417-D30.
Fig. 5.
Molecular docking interactions of ACE2 to SARS-CoV-2 RBD variants in the presence of heparin. Two-dimensional (2D) diagrams of RBD/ACE2 interactions in the presence of heparin for (A) L452R-E484Q, (B) K417N‐E484K‐N501Y, and (C) K417T‐E484K‐N501Y. The protein residues and interactions are colored accordingly and provided in figure.(For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
A total of 9 H-bonds, and 145 non-bonded contacts were reported in triple variant K417N-E484K-N501Y (Fig. 5B). All of the H-bonds observed in the WT RBD/ACE2 interface were lost and the formation of H-bonds from the different residues, K484, S494, T500, Y501, and Y505 not observed in the WT were appeared. A total of 8 H-bonds with 136 non-bonded contacts were observed in K417T-E484K-N501Y variant (Fig. 5C). The H-bonds forming residues were also different in this variant, indicating the presence of new ACE2 binding pocket in these triple variants. Among the hydrogen bonds, Y449-S128, K484-C133, Y489-K174, Y501-S128, and Y505-T122 are common in both the variants. Thus, the substituted residues, 484 K and 501Y were involved in the formation of H-bonds indicating a strong and stable binding to ACE2.
The K417 single substitution variant, K417N and K417T formed 8 H-bonds and 153 non-bonded contacts with similar interfacial residues (Supporting information, Fig. S4A–B).
Next, we calculated the binding affinities (ΔG) and dissociation constants (Kd) of the docked structures using the PRODIGY server [18] (Table 2). The WT RBD-ACE2 complex displayed ΔG of − 12.6 kcal/mol and Kd of 6.0E-10 M in the absence of heparin. Interestingly, the ACE2 binding to RBD/heparin complex significantly decreased with ΔG of − 8.9 kcal/mol and Kd of 3.1E-07 M. The binding of ACE2 to RBD-heparin complex was largely due to increased charged-apolar, polar-apolar, and apolar-apolar intermolecular contacts. Moreover, the results also showed that the ACE2 binds to SARS-CoV-2 RBD variants with a strong affinity in the absence of heparin as compared to the presence of heparin. However, among the SARS-CoV-2 variants, in the presence of heparin, ACE2 binds to RBD in the following fashion:
K417N>K417T> K417N-E484K-N501Y~ K417T-E484K-N501Y> L452R-E484Q > E484K~ L452R ~E484Q > N501Y >WT RBD (Table 2).
Overall, the results reflect the stronger binding of the SARS-CoV-2 variants to ACE2 in the presence of heparin than WT. The triple variants displayed strongest binding to ACE2 in the presence of heparin. Also, the higher docking score and binding affinity displayed by L452R-E484Q indicates that it could possess the similar infectivity potential as the triple variants. Notably, the similar binding affinity of WT and N501Y variant to ACE2 in the presence of heparin suggests that the N501Y behave similarly to the WT and the already developed COVID-19 vaccines might work against the N501Y Alpha variant.
These findings are consistent with the results of interaction energies obtained from PPCheck server. The total stabilization energy (ET) was computed to estimate the strength of a protein-protein interface. The results indicated that the RBD/ACE2 interface has significant stabilization energy with the value of − 174.93 kJ/mol as compared to RBD/ACE2 interface in the presence of heparin (−120.78 kJ/mol). The results also revealed the presence of stabilized interfaces between the mutant RBD-heparin/ACE2 complex. The maximum ET for the K417N-E484K-N501Y, K417T-E484K-N501Y, and L452R-E484Q RBD/ACE2 complex in the presence of heparin was − 182.88, − 168.91, and − 158.78 kJ/mol, respectively (Table 2). A closer look for the interactions and energy components revealed that the binding of heparin to RBD/ACE2 complex in both the WT and mutant variants are largely governed by Van der Waals and electrostatic energies.
The COVID-19 pandemic has taken a major toll on human health and prosperity. The increased transmissibility for the recently emerged SARS-CoV-2 variants raises the possibility that these variants might display altered host cell interactions. The results of this study suggest that SARS-CoV-2 variants has higher affinity toward ACE2 and therefore likely increased transmissibility and pathogenicity. The different binding affinity of the RBD variants to ACE2 revealed that the Alpha and Delta variants are more deadly than the others. Heparin binds to the interface of the viral RBD and host cell-surface receptors, ACE2 thus blocking the virus entry to host cells. A few studies on heparin binding to the SARS-CoV-2 RBD have shown that the specific residues were involved in the binding [31], [32]; however, no study has been conducted on the binding of RBD to ACE2 in the presence of heparin. In keeping with these findings, we here performed protein–protein docking experiments in the absence and presence of heparin which gives clear insight into molecular interactions between RBD and ACE2 and how these interactions were blocked by heparin.
Our docking analyses revealed that heparin binding leads to a significant decrease in the binding affinity between ACE2 and RBD in both the WT and RBD variants, indicating that heparin destabilizes the RBD/ACE2 interface by decreasing the favorable interactions between RBD and ACE2. The significant decrease in binding affinity of ACE2 to RBD decrease in case of N501Y, indicating the more protective behavior of heparin. Whereas, the ACE2 binds strongly to different SARS-CoV-2 variants both in the absence and presence of heparin, suggesting the higher infectivity of these variants. The study of H-bonds revealed that most of the mutant-RBD/ACE2 complexes form lesser H-bond interactions than the WT-RBD/ACE2 complex both in the absence and presence of heparin. The detailed decomposition of energy components showed that the heparin binding to RBD or RBD/ACE2 is governed primarily through the van der Waals and electrostatic interactions. In the absence of any heparin-bound structure of the RBD/ACE2 complex, the results of this study suggests that heparin binding to the RBD leads to the structural destabilization, thus preventing the binding to host cell receptors and inhibits the viral invasion. Heparin is considered as an interesting compound to be considered as a repurposable drug that can be evaluated as a treatment against COVID-19. In the present in-silico based molecular docking study, we showed that heparin has a sub-nanomolar binding affinity to RBD and thus has a potential to block RBD of SARS-CoV-2, therefore indicating its potential as a transmission-blocking compound.
4. Conclusion
In conclusion, the present study explored the binding of the SARS-CoV-2 S-RBD variants with the host ACE2 in the presence and absence of heparin and revealed the differences in the binding of the WT and recently emerged SARS-CoV-2 variants. From the heparin-protein docking results, it was observed that heparin have stronger tendency towards most of the variants. The maximum heparin binding affinity is observed in Alpha variant, N501Y. In contrast, the epsilon and gamma variants showed weaker binding affinity to heparin suggesting its lethality than the other variants. The lesser binding affinity of heparin in case of triple variants also suggest that higher concentration of heparin is needed to prevent the invasion of these variants. Moreover, the RBD/ACE2 binding in the presence of heparin indicates that heparin exerts its viral entry inhibitory activity significantly to Alpha variant while the epsilon, gamma, and delta variants appeared to least inhibited and thus more lethal than the other variants. The current findings thus emphasize the possible beneficial use of heparin and stress the need to evaluate the antiviral activity and dosage of heparin in the clinic against COVID-19.
CRediT authorship contribution statement
V.K. designed the study, N.A. and V.K. performed the study and prepared figures of the results. R.K., A.F.A, and V.K. analysed the data and wrote the manuscript.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
The authors are thankful to the Researchers supporting project number (RSP-2021/335), King Saud University, Riyadh, Saudi Arabia. The author sincerely thanks the Amity University, Noida for providing facilities.
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.procbio.2022.02.012.
Appendix A. Supplementary material
Supplementary material
.
References
- 1.Zhou P., Yang X.L., Wang X.G., Hu B., Zhang L., Zhang W., Si H.R., Zhu Y., Li B., Huang C.L., Chen H.D., Chen J., Luo Y., Guo H., Jiang R.D., Liu M.Q., Chen Y., Shen X.R., Wang X., Zheng X.S., Zhao K., Chen Q.J., Deng F., Liu L.L., Yan B., Zhan F.X., Wang Y.Y., Xiao G.F., Shi Z.L. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature. 2020;579(7798):270–273. doi: 10.1038/s41586-020-2012-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pango lineages. Global report investigating novel coronavirus haplotypes., https://cov-lineages.org/global_report, 2021.
- 3.Kidd M., Richter A., Best A., Cumley N., Mirza J., Percival B., Mayhew M., Megram O., Ashford F., White T., Moles-Garcia E., Crawford L., Bosworth A., Atabani S.F., Plant T., McNally A. S-variant SARS-CoV-2 lineage B1.1.7 is associated with significantly higher viral loads in samples tested by ThermoFisher TaqPath RT-qPCR. J. Infect. Dis. 2021 doi: 10.1093/infdis/jiab082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leung K., Shum M.H., Leung G.M., Lam T.T., Wu J.T. Early transmissibility assessment of the N501Y mutant strains of SARS-CoV-2 in the United Kingdom, October to November 2020. Eur. Surveill. 2021;26(1) doi: 10.2807/1560-7917.ES.2020.26.1.2002106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mwenda M., Saasa N., Sinyange N., Busby G., Chipimo P.J., Hendry J., Kapona O., Yingst S., Hines J.Z., Minchella P., Simulundu E., Changula K., Nalubamba K.S., Sawa H., Kajihara M., Yamagishi J., Kapin’a M., Kapata N., Fwoloshi S., Zulu P., Mulenga L.B., Agolory S., Mukonka V., Bridges D.J. Detection of B.1.351 SARS-CoV-2 variant strain - Zambia, December 2020. MMWR Morb. Mortal. Wkly. Rep. 2021;70(8):280–282. doi: 10.15585/mmwr.mm7008e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Francisco R.D.S., Jr., Benites L.F., Lamarca A.P., de Almeida L.G.P., Hansen A.W., Gularte J.S., Demoliner M., Gerber A.L., de C.G.A.P., Antunes A.K.E., Heldt F.H., Mallmann L., Hermann B., Ziulkoski A.L., Goes V., Schallenberger K., Fillipi M., Pereira F., Weber M.N., de Almeida P.R., Fleck J.D., Vasconcelos A.T.R., Spilki F.R. Pervasive transmission of E484K and emergence of VUI-NP13L with evidence of SARS-CoV-2 co-infection events by two different lineages in Rio Grande do Sul, Brazil. Virus Res. 2021;296 doi: 10.1016/j.virusres.2021.198345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang W., Davis B.D., Chen S.S., Sincuir Martinez J.M., Plummer J.T., Vail E. Emergence of a novel SARS-CoV-2 variant in Southern California. JAMA. 2021;325(13):1324–1326. doi: 10.1001/jama.2021.1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abdool Karim S.S., de Oliveira T. New SARS-CoV-2 variants - clinical, public health, and vaccine implications. N. Engl. J. Med. 2021 doi: 10.1056/NEJMc2100362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Burki T. Understanding variants of SARS-CoV-2. Lancet. 2021;397(10273):462. doi: 10.1016/S0140-6736(21)00298-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williams A.H., Zhan C.G. Fast prediction of binding affinities of the SARS-CoV-2 spike protein mutant N501Y (UK variant) with ACE2 and miniprotein drug candidates. J. Phys. Chem. B. 2021 doi: 10.1021/acs.jpcb.1c00869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Luan B., Wang H., Huynh T. Enhanced binding of the N501Y-mutated SARS-CoV-2 spike protein to the human ACE2 receptor: insights from molecular dynamics simulations. FEBS Lett. 2021 doi: 10.1002/1873-3468.14076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ghezzi S., Cooper L., Rubio A., Pagani I., Capobianchi M.R., Ippolito G., Pelletier J., Meneghetti M.C.Z., Lima M.A., Skidmore M.A., Broccoli V., Yates E.A., Vicenzi E. Heparin prevents Zika virus induced-cytopathic effects in human neural progenitor cells. Antivir. Res. 2017;140:13–17. doi: 10.1016/j.antiviral.2016.12.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.WuDunn D., Spear P.G. Initial interaction of herpes simplex virus with cells is binding to heparan sulfate. J. Virol. 1989;63(1):52–58. doi: 10.1128/jvi.63.1.52-58.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rusnati M., Coltrini D., Oreste P., Zoppetti G., Albini A., Noonan D., d’Adda di Fagagna F., Giacca M., Presta M. Interaction of HIV-1 Tat protein with heparin. Role of the backbone structure, sulfation, and size. J. Biol. Chem. 1997;272(17):11313–11320. doi: 10.1074/jbc.272.17.11313. [DOI] [PubMed] [Google Scholar]
- 15.Vicenzi E., Canducci F., Pinna D., Mancini N., Carletti S., Lazzarin A., Bordignon C., Poli G., Clementi M. Coronaviridae and SARS-associated coronavirus strain HSR1. Emerg. Infect. Dis. 2004;10(3):413–418. doi: 10.3201/eid1003.030683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Clausen T.M., Sandoval D.R., Spliid C.B., Pihl J., Perrett H.R., Painter C.D., Narayanan A., Majowicz S.A., Kwong E.M., McVicar R.N., Thacker B.E., Glass C.A., Yang Z., Torres J.L., Golden G.J., Bartels P.L., Porell R.N., Garretson A.F., Laubach L., Feldman J., Yin X., Pu Y., Hauser B.M., Caradonna T.M., Kellman B.P., Martino C., Gordts P., Chanda S.K., Schmidt A.G., Godula K., Leibel S.L., Jose J., Corbett K.D., Ward A.B., Carlin A.F., Esko J.D. SARS-CoV-2 infection depends on cellular heparan sulfate and ACE2. Cell. 2020;183(4):1043–1057. doi: 10.1016/j.cell.2020.09.033. e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tandon R., Sharp J.S., Zhang F., Pomin V.H., Ashpole N.M., Mitra D., McCandless M.G., Jin W., Liu H., Sharma P., Linhardt R.J. Effective Inhibition of SARS-CoV-2 entry by heparin and enoxaparin derivatives. J. Virol. 2021;95(3) doi: 10.1128/JVI.01987-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kwon P.S., Oh H., Kwon S.J., Jin W., Zhang F., Fraser K., Hong J.J., Linhardt R.J., Dordick J.S. Sulfated polysaccharides effectively inhibit SARS-CoV-2 in vitro. Cell Disco. 2020;6:50. doi: 10.1038/s41421-020-00192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi X., Liu Y., Wang J., Fallowfield J.A., Li X., Shi J., Pan H., Zou S., Zhang H., Chen Z., Li F., Luo Y., Mei M., Liu H., Wang Z., Li J., Yang H., Xiang H., Liu T., Zheng M.H., Liu C., Huang Y., Xu D., Kang N., He Q., Gu Y., Zhang G., Shao C., Liu D., Zhang L., Kawada N., Jiang Z., Wang F., Xiong B., Takehara T., Rockey D.C. Clinical course and risk factors for mortality of COVID-19 patients with pre-existing cirrhosis: a multicentre cohort study. Gut. 2020 doi: 10.1136/gutjnl-2020-321666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim S.Y., Jin W., Sood A., Montgomery D.W., Grant O.C., Fuster M.M., Fu L., Dordick J.S., Woods R.J., Zhang F., Linhardt R.J. Characterization of heparin and severe acute respiratory syndrome-related coronavirus 2 (SARS-CoV-2) spike glycoprotein binding interactions. Antivir. Res. 2020;181 doi: 10.1016/j.antiviral.2020.104873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lan J., Ge J., Yu J., Shan S., Zhou H., Fan S., Zhang Q., Shi X., Wang Q., Zhang L., Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020;581(7807):215–220. doi: 10.1038/s41586-020-2180-5. [DOI] [PubMed] [Google Scholar]
- 22.Schymkowitz J., Borg J., Stricher F., Nys R., Rousseau F., Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mottarella S.E., Beglov D., Beglova N., Nugent M.A., Kozakov D., Vajda S. Docking server for the identification of heparin binding sites on proteins. J. Chem. Inf. Model. 2014;54(7):2068–2078. doi: 10.1021/ci500115j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kozakov D., Brenke R., Comeau S.R., Vajda S. PIPER: an FFT-based protein docking program with pairwise potentials. Proteins. 2006;65(2):392–406. doi: 10.1002/prot.21117. [DOI] [PubMed] [Google Scholar]
- 25.Laskowski R.A., Swindells M.B. LigPlot+: multiple ligand-protein interaction diagrams for drug discovery. J. Chem. Inf. Model. 2011;51(10):2778–2786. doi: 10.1021/ci200227u. [DOI] [PubMed] [Google Scholar]
- 26.Vangone A., Schaarschmidt J., Koukos P., Geng C., Citro N., Trellet M.E., Xue L.C., Bonvin A. Large-scale prediction of binding affinity in protein-small ligand complexes: the PRODIGY-LIG web server. Bioinformatics. 2019;35(9):1585–1587. doi: 10.1093/bioinformatics/bty816. [DOI] [PubMed] [Google Scholar]
- 27.Yan Y., Zhang D., Zhou P., Li B., Huang S.Y. HDOCK: a web server for protein-protein and protein-DNA/RNA docking based on a hybrid strategy. Nucleic Acids Res. 2017;45(W1):W365–W373. doi: 10.1093/nar/gkx407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xue L.C., Rodrigues J.P., Kastritis P.L., Bonvin A.M., Vangone A. PRODIGY: a web server for predicting the binding affinity of protein-protein complexes. Bioinformatics. 2016;32(23):3676–3678. doi: 10.1093/bioinformatics/btw514. [DOI] [PubMed] [Google Scholar]
- 29.Sukhwal A., Sowdhamini R. Oligomerisation status and evolutionary conservation of interfaces of protein structural domain superfamilies. Mol. Biosyst. 2013;9(7):1652–1661. doi: 10.1039/c3mb25484d. [DOI] [PubMed] [Google Scholar]
- 30.Zhou Y., Hou Y., Shen J., Huang Y., Martin W., Cheng F. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Disco. 2020;6(1):14. doi: 10.1038/s41421-020-0153-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mycroft-West C.J., Su D., Pagani I., Rudd T.R., Elli S., Gandhi N.S., Guimond S.E., Miller G.J., Meneghetti M.C.Z., Nader H.B., Li Y., Nunes Q.M., Procter P., Mancini N., Clementi M., Bisio A., Forsyth N.R., Ferro V., Turnbull J.E., Guerrini M., Fernig D.G., Vicenzi E., Yates E.A., Lima M.A., Skidmore M.A. Heparin inhibits cellular invasion by SARS-CoV-2: structural dependence of the interaction of the spike S1 receptor-binding domain with heparin. Thromb. Haemost. 2020;120(12):1700–1715. doi: 10.1055/s-0040-1721319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gupta Y., Maciorowski D., Zak S.E., Kulkarni C.V., Herbert A.S., Durvasula R., Fareed J., Dye J.M., Kempaiah P. Heparin: a simplistic repurposing to prevent SARS-CoV-2 transmission in light of its in-vitro nanomolar efficacy. Int. J. Biol. Macromol. 2021;183:203–212. doi: 10.1016/j.ijbiomac.2021.04.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material