Abstract
The scenery of molecular diagnostics for infectious diseases is rapidly evolving to respond to the COVID-19 epidemic. The sensitivity and specificity of diagnostics, along with speed and accuracy, are crucial requirements for effective analytical tools to address the disease spreading around the world. Emerging diagnostic devices combine the latest trends in isothermal amplification methods for nucleic acids with state-of-the-art biosensing systems, intending to bypass roadblocks encountered in the last 2 years of the pandemic. Isothermal nucleic acid amplification is a simple procedure that quickly and efficiently accumulates nucleic acid sequences at a constant temperature, without the need for sophisticated equipment. The integration of isothermal amplification into portable biosensing devices confers high sensitivity and improves screening at the point of need in low-resource settings. This review reports the latest trends reached in this field with the latest examples of isothermal amplification-powered biosensors for detecting SARS-CoV-2, with different configurations, as well as their intrinsic advantages and disadvantages.
Keywords: Isothermal amplification, SARS-CoV-2 diagnostics, Sensitive and specific screening, High throughput, Biosensors
1. Introduction
In human history, numerous infectious diseases caused by viruses have threatened countries worldwide as the recent global events of COVID-19, caused by severe acute respiratory syndrome-coronavirus (SARS-CoV-2). This serious and epidemic disease has had an enormous social and economic impact on all sectors, alerting the population to the need to develop the right tools to tackle this pandemic and future ones. This necessity has been further exacerbated in the past months as, even with the spread of vaccines, COVID-19 pandemic is still persisting and new more contagious variants are occurring. To this aim, considering the global debate about the emerging and re-emerging of infectious diseases viruses related, and thus the requirement to face pandemic spread among the population, the scientific community has increased interest and commitment to developing novel effective and rapid diagnostics. Indeed, as stated by Vandenberg and colleagues “ending the pandemic involves the accurate application of diagnostic testing in high volumes and the rapid use of the results to help implement the appropriate therapy and prevent further spread (Vandenberg et al., 2021)”.
Many analytical systems have been described in the literature for virus detection in general and SARS-CoV-2 in particular, with very diverse biosensing configurations (Morales-Narváez and Dincer, 2020; Taleghani and Taghipour, 2021). COVID-19 diagnosis entails the analysis of antibodies, antigens, and nucleic acids. In particular, antibody IgG/IgM testing is rapid and cheap; however, a high false-positive rate occurs in such analyses. Antigen tests can detect an active infection with a fast response, although they have a low sensitivity and show the risk of false-negative results (Fabiani et al., 2021). Moreover, the RNA based SARS-CoV-2 detection needs to be first transcribed into its complementary DNA by reverse transcriptase and then amplified by PCR. In addition, COVID-19 testing in low-resource settings as well as underfunded rural clinics, or politically unstable environments, needs to establish more performant diagnostic conditions.
In this scenario, it is clear that each test shows pros and cons related to specificity, sensitivity, accuracy, costs, experimental settings, easiness of use, and cost-effectiveness. To this aim, coupling the newest biosensor technology with isothermal amplification systems provides a great opportunity to design novel diagnostics with real benefits in terms of sensitivity, targeting the globally addressed challenge of increasing population screening and reducing the spread of infectious diseases.
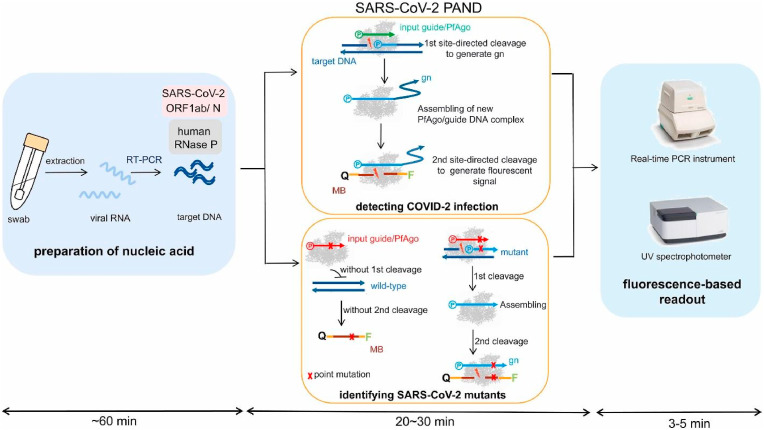
For example, Wang and colleagues (Wang et al., 2021) reported the design of a highly sensitive and accurate molecular diagnosis platform. This technique exploits Reverse Transcription Polymerase Chain Reaction (RT-PCR) to amplify conserved regions of the viral RNA extracted from nasopharyngeal or oropharyngeal swabs. In particular, the authors set up a novel protocol to shorten the amplification procedure to only 3–5 min per batch and thus consequently reduce the costs. However, the use of a laboratory set-up instrumentation hinders the use of biosensors for in-field population screening, since it is required an incubation step at 95 °C for 20–30 min and a total procedure time of about 1.5 h (Fig. 1 ).
Fig. 1.
Schematic of recommended SARS-CoV-2 PAND workflow. MB: molecular beacon; gn: newly generated guide; Q: quencher; F: fluorophore. Reprinted with permission fromWang et al. (2021)for Microalgal-bacterial consortia: PfAgo-based detection of SARS-CoV-2. Biosensors and Bioelectronics. Copyright (2021) Elsevier.
An additional relevant issue to consider is the development of virus variants, which can cause difficulties in molecular diagnostic protocols to identify the presence of SARS-CoV-2 (Janik et al., 2021). Indeed, throughout the pandemic, the viral genome, during the course of its replication, undergoes adaptive evolution, this results in genetic variation, as shown by the episodes that occurred during the SARS-CoV-2 pandemic. Actually, from the beginning of the pandemic, SARS-CoV-2 has undergone numerous mutations within its genome, giving rise to a series of functional variants originating from the RNA genome of the original Wuhan strain (Corey et al., 2021). Although most mutations in SARS-CoV-2 genome are silent, as they do not cause structural changes in the encoded proteins, mutations in surface proteins, usually, have a significant effect on the functionality of the virus. The SARS-CoV-2 genome encodes four structural proteins: Spike protein (S), Envelope protein (E), membrane protein (M), and nucleocapsid protein (N). The Centers for Disease Control and Prevention (CDC) and the World Health Organization (WHO) have classified some of these variants as variants of concern (VOC) based on conserved mutations primarily in the spike protein (e.g. D614G, L452R, P681R, T478K, E484K/Q, E156-F157del). Such mutations can affect viral functions, such as infectivity or the ability to evade vaccine-induced antibody responses. In particular, analyses regarding the protein S mutations are of particular importance as this protein is responsible for the recognition of the surface receptor of the host cell and the consequent cell fusion. Constant monitoring of the GISAID data revealed an increasing frequency of variants carrying mutations in the spike protein.
To date, in literature there is any pint-of-care (POC) test that can discriminate a COVID-19 variant. However, Zhao et al. (2021) demonstrated in 2021 that the electrochemical system integrating reconfigurable enzyme-DNA nanostructures (eSIREN) could be the only promising method for identifying different genetic loci of SARS-CoV-2 using various molecular nanostructures to form a cascading circuitry that bypasses all steps of conventional detection.
As above mentioned, the assay currently used for the diagnosis of SARS-CoV-2 is based on a molecular qRT-PCR test on respiratory samples (Böger et al., 2021). The primers used for the PCR analysis (about 20 bp) are designed on the basis of a reference genome (SARS-CoV-2, NC_045512.2 (Wu et al., 2020)). Any slight mutation within the annealing region of the primer can result in lower amplification efficiency and therefore may cause false negative for virus detection (Bru et al., 2008). Sequencing studies performed by GenBank and GISAID on samples from Germany and China showed that the mutations, in this case, were mainly related to ORF1ab (Farkas et al., 2020; Koyama et al., 2020). Surprisingly, a new study based on the analysis of 31421 samples showed that most of the mutations were observed within the primers used for the detection of the N32 gene by RT-PCR, therefore such mutations would jeopardize the reliability of the results (Wang et al., 2020).
Some studies conducted by Ziegler et al. (2020) demonstrate that single nucleotide polymorphism (SNP) in the SARS-CoV-2 N gene can interfere with detection in commonly used assays. However, the other probes used can still detect the presence of the virus in the samples analysed. These data underline that at least two key regions of the virus need to be considered for reliable detection. In addition, studies conducted on the alpha variant (B.1.1.7) have shown that it possesses a deletion (H69–V70) in the spike protein (S) that causes the failure of the amplification of the target gene S (SGTF), while the amplification of the gene N and ORF1ab were not affected. Based on these observations, a valid diagnostic test of RT-qPCR (TaqPath COVID-19 Combo Kit, Thermo Fisher) was created for the monitoring of the Alpha variant (Kidd et al., 2021). It is important to underline that the Omicron variant (B.1.1.529) also contains the deletion H69–V70, therefore, this kit is able to detect the latter as well. However, because of what has been observed, it is clear that to be reliable, the commercially available kits should be periodically remodelled in accordance with the new emerging variants (Kaden, 2020). In any case, currently, in combination with NGS (next generation sequencing) verification, RT-qPCR represents a valid method for the rapid identification of variants using characteristic mutations. Considering the dynamics of the mutation and the speed with which new variants emerge, such monitoring is essential for the early diagnosis of the new variants.
In addition to molecular analyses, antigen tests are also widely used; this type of tests detect the presence of viral antigens as parts of the S protein. As well as the molecular tests, also antigenic ones can be affected by viral mutations (Böger et al., 2021). During the execution of the validation study of the Abbott Panbio6 ™ COVID-19 Ag assay, it was observed that some samples analysed gave a negative result despite the high viral load obtained with the RT-PCR tests. Analysis of the sequence revealed the presence of multiple amino acid substitutions in a region containing an immunodominant epitope of the antigen N. A portion of the variants, not detected by the antigen assays, contained the A376T mutations coupled to M241I (Vecchio et al., 2021).
However, given the growing number of variants, it is easy to imagine the urgent need to develop new technologies capable of simultaneously detecting all variants of the SARS-CoV-2 virus quickly and efficiently. Indeed, variant testing is not the main focus of fighting the pandemic, rather it is necessary to make diagnostic tests for COVID-19 as rapid, sensitive, practical, and reliable as possible by targeting both sequences stored within the genome and antigenic regions in viral proteins.
Actually, antigenic tests are extremely rapid, practical, and friendly-use, but they show a rather variable sensitivity due to the absence of an amplification step, which causes false negatives in case of low viral load. Moreover, a low specificity is determined by the continuous rising of new variants. On the other hand, molecular diagnosis by PCR is much more sensitive, being based on nucleic acids amplification. Also, the emerging of new variants is largely obviated by PCR thanks to the use of various primers for the amplification steps. These features make it a very reliable technique in several respects but it is nevertheless slow and less practical, as it requires the use of specialized machines and qualified personnel.
In this context, isothermal in vitro amplification of target nucleic acids enables the production of genetic copies from a single fragment at a constant temperature without the need for a thermocycling machine. Thus, compared with PCR, such methods retain their advantages while reducing the complexity of analysis, thereby simplifying and speeding up the diagnosis process. Furthermore, isothermal amplification methods can also take advantage from the use of a variety of primers, as PCR, limiting the false negative detection of unidentified variants.
In addition, such amplification methods result in a high potential for simple integration into point-POC testing especially suitable in resource-limited conditions. In addition, they are easy to use, especially when lateral flow devices are used for detection. Finally, the outputs of these techniques can be coupled with both optical and electrochemical detection as well as lateral flow strip platforms to facilitate readout processes.
The following sections provide a comprehensive overview of the diagnostics realized in the last two years based on the available methods for the isothermal amplification of SARS-CoV-2 nucleic acid.
2. Isothermal amplification-assisted diagnostics
Reverse Transcription Polymerase Quantitative Chain Reaction (RT-qPCR) represents the gold standard for RNA virus detection. However, this technique requires up to 4 h for the results as well as an expensive thermal cycler coupled with fluorimetry. To fulfil the demand for diagnoses during disease outbreaks, isothermal amplification-powered biosensors provide an excellent alternative thanks to their potential for rapid and sensitive analysis. In the following sections, the available types of isothermal nucleic acid amplification methods are described, including Loop-Mediated Isothermal Amplification (LAMP), Rolling Circle Amplification (RCA), Nucleic Acid Sequence-Based Amplification (NASBA), Multiple Cross Displacement Amplification (MCDA), Recombinase Polymerase Amplification (RPA), in conjunction with the ultimate diagnostics for SARS-CoV-2 detection.
2.1. Loop-mediated isothermal amplification (LAMP)-based diagnostics
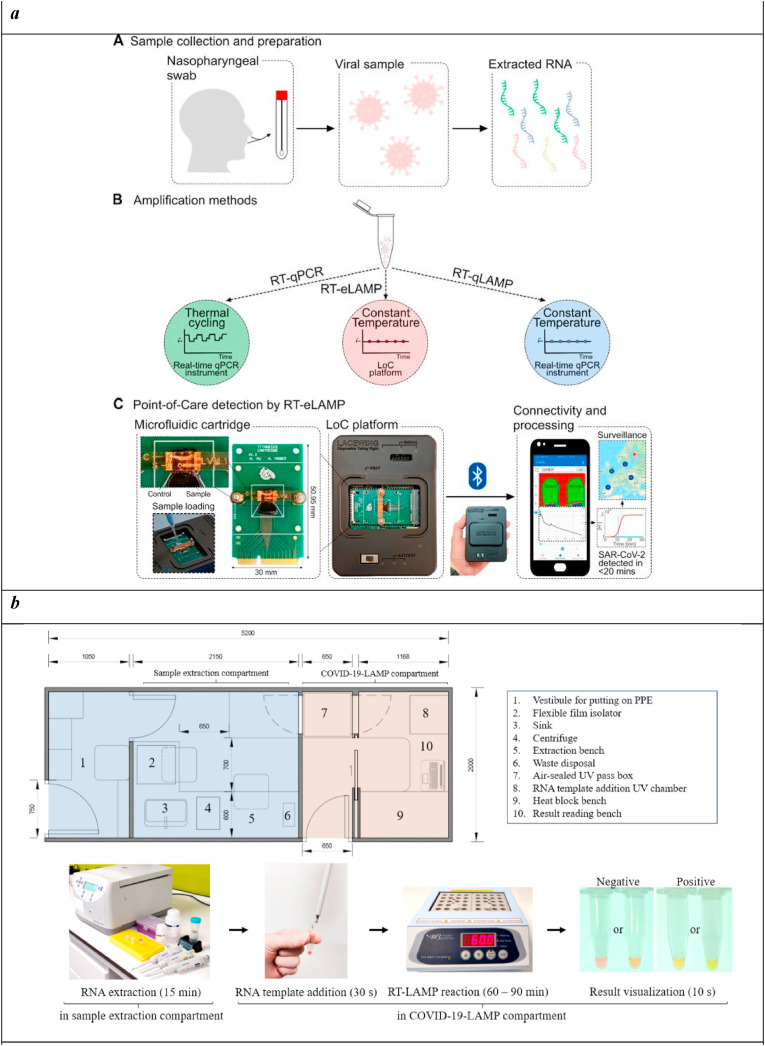
Loop-Mediated Isothermal Amplification (LAMP) technique amplifies target DNAs with high efficiency, specificity, and rapidity exploiting a DNA polymerase with high displacement strand activity. The reaction occurs under isothermal conditions (60–65 °C), whose choice is critical to allow the specific binding of primers specifically designed for the target region (Tomita et al., 2008). LAMP produces a large number of DNA copies within 1 h, with a yield 100 times higher than conventional PCR, thus improving biosensor sensitivity (Zhang et al., 2014). Diverse LAMP-assisted diagnostics have been realized for the detection of SARS-CoV-2 in the form of lab-on-chip and point-of-care devices suitable for effective, rapid, affordable monitoring also in low-resource laboratory settings. Indeed, this technique can be considered a trustworthy alternative for the precise analysis of nucleic acids low copy number, since it doesn't require a thermal cycler and it has a high amplification efficacy (Augustine et al., 2020). Moreover, LAMP has undergone numerous developments through the combination with other approaches, like reverse transcription and multiplex amplification, that made it an excellent candidate for the detection of infectious diseases caused by microorganisms in humans, livestock, and plants, including SARS-CoV-2. Reverse transcriptase loop-mediated isothermal amplification (RT-LAMP) was described for the development of a rapid test (<20 min) to detect SARS-CoV-2 from extracted RNA samples with a limit of detection of 10 RNA copies per reaction (Rodriguez-Manzano et al., 2021). A LAMP assay was designed and optimized on phylogenetic analysis of SARS-CoV-2 sequences and other related viruses, selecting N gene as the optimal target being conserved across available sequences and more resilient to emergent mutations. Both the amplification and the detection systems were integrated into a Complementary Metal Oxide Semiconductor (CMOS) Ion-Sensitive Field-Effect Transistor (ISFET) LOC platform equipped with a microfluidic cartridge for sample inlets (Fig. 2 a). This device was able to identify the voltage change as a result of pH variations related to the presence of the target. An algorithm produced a standard amplification curve, monitoring the proton concentration in real-time during the reaction and hence the increase in DNA copies. Moreover, the LOC platform was connected to a smartphone application to acquire and process the data, with a GPS geolocation to map the experiment. This device was validated on nasopharyngeal swab samples, showing 91% sensitivity and 100% specificity when compared to RT-qPCR, and providing a time-to-positive value ranging from 9 to 24 min and a limit of detection of 10 copies per reaction.
Fig. 2.
a) SARS-CoV-2 diagnostic workflow. A) Sample collection and preparation illustrating nasopharyngeal swab and RNA extraction. B) Nucleic acid amplification methods for SARS-CoV-2 RNA detection used in this study (RT-qPCR, RT-qLAMP, and RT-eLAMP). Thermal profiles are illustrated for comparison of the assays. C) Point-of-care diagnostics by RT-eLAMP showing the proposed handheld LOC platform including the microfluidic cartridge with control and sample inlets, and the smartphone-enabled application for geolocation and real-time visualization of results.
Reprinted with permission ofRodriguez-Manzano et al. (2021) for Handheld Point-of-Care System for Rapid Detection of SARS-CoV-2 Extracted RNA in under 20 min. ACS central science. Copyright (2021) American chemical society.
b) Illustration of a small van-sized mobile COVID-19-LAMP diagnostic unit. A drawn-to-scale layout of a van-sized mobile COVID-19-LAMP diagnostic unit, with sample processing and LAMP reactions compartments have been illustrated. A cargo van/lorry can be modified quickly to become a mobile diagnostic unit for rapid deployment in any region.
Reprinted with permission fromChow et al. (2020)for A Rapid, Simple, Inexpensive, and Mobile Colorimetric Assay COVID-19-LAMP for Mass On-Site Screening of COVID-19. Int. J. Mol. Sci. Copyright (2020) MDPI.
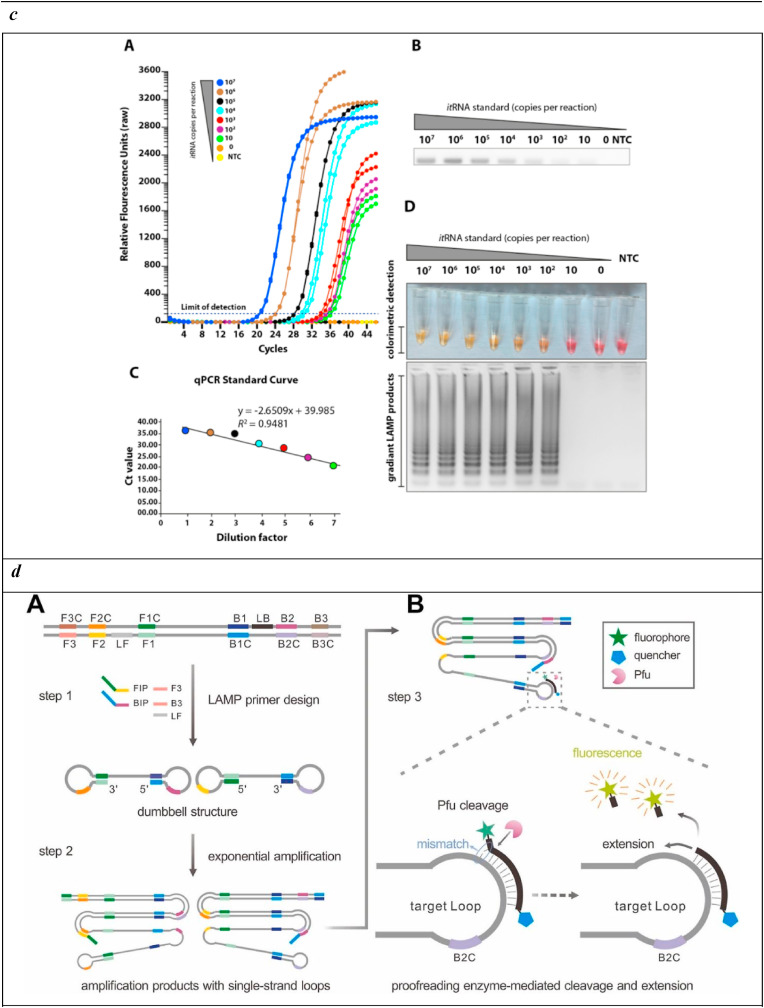
c) Sensitivities of the loop-mediated isothermal amplification (LAMP) assay. A) Seven different dilutions of in vitro transcribed RNA were run for quantitative measurement using qRT-PCR. Relative fluorescence units show a gradient decrease in signals. B) The corresponding PCR products on the electrophoresis gel. C) qRT-PCR standard curve based on the Ct value and dilution factor. D) The serially diluted synthetic RNAs were run in the LAMP assay and colour change represents positive (yellow) or negative (pink). The lower panel shows the LAMP gradient products.
Reprinted with permission from Rohaim et al. (2020) for Artificial Intelligence-Assisted Loop Mediated Isothermal Amplification (AI-LAMP) for Rapid Detection of SARS-CoV-2. Viruses. Copyright (2020) MDPI.
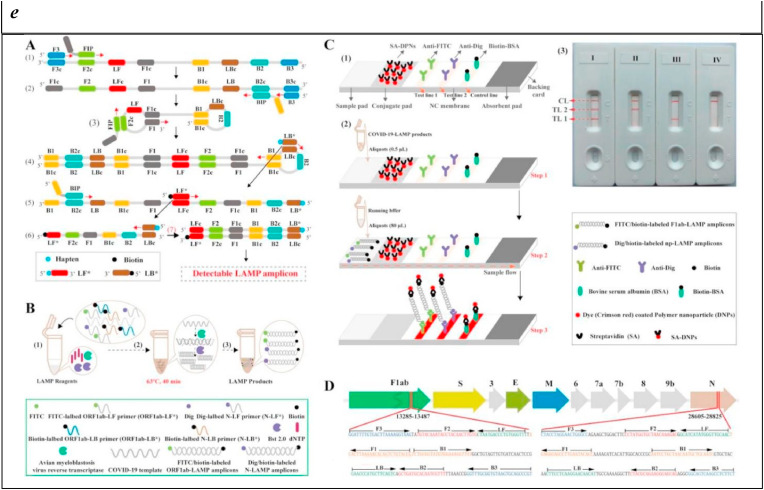
d) The schematic of specific detection of LAMP products using the Proofman probe. A) LAMP reaction process including the initial amplification phase and exponential amplification phase. Primer design was based on the target sequence: FIP and BIP were the inner primers; F3 and B3 were the outer primers; LF was the loop primer. B) The principle of sequence-specific detection using the Proofman probe. The Proofman probe was designed based on the target sequence and a deliberate mismatch at the 3′ end of the probe was needed to trigger the cleavage activity of the proofreading enzyme (Pfu). Once the Proofman probe binds to the target loop of LAMP products, the Pfu can cleave the probe at the mismatching nucleotide, releasing the fluorophore. Then, the cleaved probe serves as a loop primer to enhance the amplification efficiency.
Reprinted with permission from Ding et al. (2021) for Sequence-specific and multiplex detection of COVID-19 virus (SARS-CoV-2) using proofreading enzyme-mediated probe cleavage coupled with isothermal amplification. Biosensors and Bioelectronics. Copyright (2021) Elsevier.
e) Outline of COVID-19 RT-LAMP-LFB design. A) Outline of LAMP assay with LF* and LB*. B) Mechanistic description of the COVID-19 RT-LAMP-LFB assay. C) The principle of LFB for visualization of COVID-19 RT-LAMP products. D) Primer design of COVID-19 mRT-MCDA-LFB assay. Up row, SARS-CoV-2 genome organization (GenBank: MN908947, Wuhan-Hu-1).
Reprinted with permission from Zhu et al. (2020) for Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosensors and Bioelectronics. Copyright (2020) Elsevier.
RT-LAMP was associated also with optical biosensors (Park et al., 2020). A simple and affordable equipment was developed for the detection of SARS-CoV-2 using respiratory samples from patients with COVID-19 (n = 223) and other respiratory virus infections (n = 143); it is based on a one-step colorimetric RT-LAMP assay easily interpreted by naked eyes (Chow et al., 2020). In detail, the assay combines reverse transcriptase, DNA polymerase, pH indicator, and six primers to amplify RNA templates, causing a drop in pH and, thus, a colour change from pink to yellow. SARS-CoV-2 RNA was detected by this assay with a detection limit of 42 copies/reaction, within a time response from 60 to 90 min, a 95.07% sensitivity on spiked samples, 96.88% on nasopharyngeal swabs, 94.03% on sputum/deep throat saliva samples, and 93.33% on throat swab samples. In addition, any of the 143 samples with other respiratory viruses resulted positive, displaying a 100% specificity. On balance, this system showed high sensitivity and specificity, rapidity, and capability to perform unlimited high-throughput mass screening within mobile diagnostic units operating in resource-limiting areas (Fig. 2b).
Colorimetric, fluorometric, and lateral dipstick readout have been also coupled to RT-LAMP in an integrated Palm Germ-Radar (PaGeR) device (Ge et al., 2022) for the rapid and simple detection of SARS-CoV-2 down to 1 copy/μL from extracted patient swab specimens sample RNA. The results showed that 15 out of 17 COVID-19 patients were diagnosed as positive while all 55 normal samples were diagnosed as negative.
Orf1ab and spike gene have been also selected as targets for the monitoring of SARS-CoV-2 using RT-LAMP for their amplification (Yan et al., 2020). A primer set was designed for a rapid reaction run for 18–20 min at 63 °C. The detection was assessed by naked eye, depending on turbidity or colour change from orange to green. This assay provided sensitivities of 2 × 101 copies and 2 × 102 copies for Orf1ab and spike gene, respectively. Any cross-reactivity was obtained with 60 other respiratory pathogens. To test the method, 130 specimens from patients with clinically suspected SARS-CoV-2 infection were analysed, and 58 were confirmed to be positive while 72 were negative by RT-LAMP, showing a 100% sensitivity and 100% specificity.
To further improve the biosensor performance and avoid operator bias in result interpretation, RT-LAMP was connected to automated image acquisition and processing algorithms capable of data analysis using artificial intelligence (AI) (Rohaim et al., 2020). This configuration, based on colorimetric detection targeting the RNA-dependent RNA polymerase gene, provided SARS-CoV-2 analysis with high sensitivity and specificity on ∼200 CoVID-19 suspected patient samples. Extracted RNA was subjected to LAMP reactions using the commercial WarmStartTM Colorimetric LAMP 2X Master Mix. Once amplified, the target was detected by colour change using pH-sensitive dyes able to sense pH variations resulting from proton accumulation due to dNTP incorporation. Colour change was thus observed directly by the naked eye (Fig. 2c) or through AI image processing, determining a detection limit of 10 copies.
Commercial kits have been also exploited for the design of SARS-CoV-2 diagnostics, as the Loopamp® 2019-SARSCoV-2 Detection Reagent Kit, which enables cDNA synthesis from RNA using reverse transcriptase, followed by DNA amplification under isothermal conditions in one step (Kitagawa et al., 2020). A comparison with the results obtained by RT-qPCR on 76 nasopharyngeal swab samples underlined a 100% sensitivity and a 97.6% specificity. In addition, a detection sensitivity of 1.0 × 101 copies/μL was achieved with a fast response of 35 min. Conversely, this system can deliver a limited number of sample analyses as well as it lacks validation of cross-reactivity with other respiratory pathogens.
However, RT-LAMP shows high potential, not only for highly sensitive analysis but also because of the easy performance and the option to bypass RNA extraction. Indeed, some problems can occur if RNA isolation procedures are inadequate. Exploiting RNA variplex™ RT-LAMP, SARS-CoV-2 was detected on unprocessed respiratory secretions samples, bypassing RNA purification because of the robustness of the polymerase (Rödel et al., 2020). The variplex™ SARS-CoV-2 is a qualitative assay based on a 6-oligonucleotide primer mix targeting the membrane protein (M) gene; this kit was used for the target amplification that was successively tested in parallel with the Allplex™ and VIASURE BD MAX RT-PCRs, commercial kits for SARS-CoV-2 detection. A panel of pharyngeal swabs was analysed within a response time from 15 to 24 min and compared to RT-PCR, showing a lower sensitivity of 75% but no false-positive results were observed. In conclusion, this assay can be considered an easy-to-perform and rapid molecular test, but its main limitation regards the small samples to be processed, which hinders its use for high sample throughputs.
To further enhance the specificity and, thus, avoid non-specific detection of LAMP products, a sequence-specific LAMP assay was described using proofreading enzyme-mediated probe cleavage (Proofman), which could realize real-time and visual detection without uncapping (Ding et al., 2021). This assay combines a proofreading enzyme and the fluorogenic probe to RT-LAMP, to specifically detect SARS-CoV-2 RNA with a detection limit of 100 copies (Fig. 2d). In addition to the real-time analysis, the assay is capable of endpoint visualization under a transilluminator within 50 min, providing a convenient reporting manner under the setting of a POC format. Exploiting different fluorophores, a one-pot multiplex platform was realized to detect multiple SARS-CoV-2 targets simultaneously, becoming highly specific and versatile for rapid in-field screening.
A multiplex RT-LAMP was also described for diagnosing COVID-19, coupled with a nanoparticle-based lateral flow biosensor (Zhu et al., 2020). A primer set, based on the ORF1ab (opening reading frame 1a/b) and N (nucleoprotein) genes of SARS-CoV-2, was used for the single-tube amplification reaction. In presence of FITC (fluorescein)-/digoxin- and biotin-labeled primers, mRT-LAMP produced numerous FITC-/digoxin- and biotin-attached duplex amplicons, successively detected by immunoreactions (FITC/digoxin on the duplex and anti-FITC/digoxin on the test line of LFB) and biotin/streptavidin interactions (biotin on the duplex and streptavidin on the polymerase nanoparticles). The accumulation of nanoparticles led to a characteristic crimson band, enabling multiplex analysis of ORF1ab and N gene without instrumentation (Fig. 2e). This system provided in less than 1 h a limit of detection of 12 copies (for each detection target) per reaction, with any cross-reactivity from non-SARS-CoV-2 templates, a 100% sensitivity (33/33 oropharynx swab samples collected from COVID-19 patients), and a 100% specificity (96/96 oropharynx swab samples collected from non-COVID-19 patients). As stated by the authors, these results highlighted the high potential of such tools “for diagnosing SARS-CoV-2 infections in frontline public health field and clinical laboratories, especially from resource-poor regions”.
2.2. Rolling circle amplification (RCA)-based diagnostics
Rolling Circle Amplification (RCA) is a DNA amplification technique based on in vivo rolling circle replication (Fire and Xu, 1995; Liu et al., 1996). This method can be used for a wide number of applications: genomics (Schweitzer and Kingsmore, 2001), proteomics (Kingsmore and Patel, 2003), nanotechnologies (Lin et al., 2006), and functional nucleic acids (Navani and Li, 2006). Circular DNA is the predominant template for this amplification technique and numerous rounds of amplification are required to obtain the extension of short primers by the action of a DNA/RNA polymerase. Usually, the reaction is carried out at 30–37 °C and the amplification takes from several hours to several days. This very simple mechanism has a very high yield and specificity, being capable to distinguish single-base mismatches and single-nucleotide polymorphisms (Qi et al., 2001). Moreover, RCA can be carried out in most liquids (like blood and urine), solid (like cells and tissues), and clinical samples, representing a great approach for target in situ detection (Mignardi et al., 2015; Qi et al., 2019). For these reasons, RCA represents a powerful amplification technique to be exploited for sensitive diagnostics, in association with both optical and electrochemical biosensors.
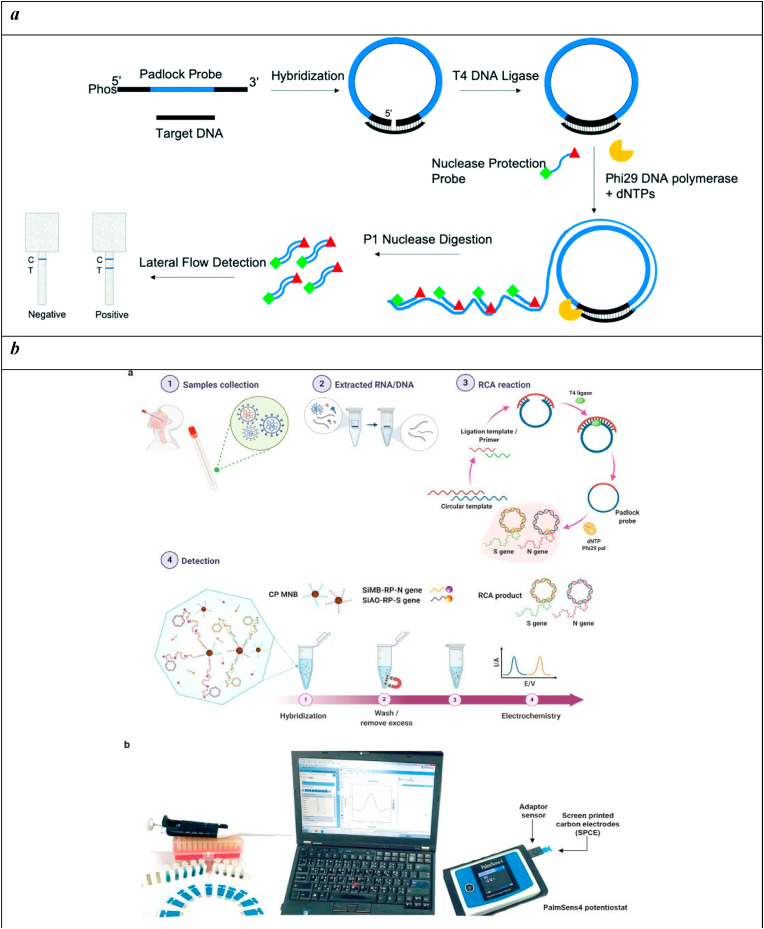
A central example of an RCA-assisted colorimetric test for SARS-CoV-2 was described based on padlock probe rolling circle amplification, nuclease protection, and a paper-based lateral flow detection (Jain et al., 2021). The reaction requires a “padlock” probe that first needs to be closed to form a circular template that can go through isothermal circle amplification. Thus, the reaction starts with a sequence-specific ligation of the probe which, once circularized and in the presence of primers, can be amplified under the catalysis of a DNA polymerase; this process gives rise to a long, linear concatenated ssDNA that includes the binding sites for the labeled probe along its sequence. The single-strand DNA is selectively cleaved by the action of a single-strand DNA nuclease while the double-stranded DNA is detected in a lateral flow immunoassay format to provide a visual, colorimetric readout of results. This principle was exploited for the development of an RCA-powered lateral flow immunoassay for SARS-CoV-2 (Fig. 3 a). This configuration allowed for reaching a limit of detection of 1.1 pM target DNA (or 1.3 × 106 copies per reaction) also in complex matrices as saliva, plant tissue extract, and bacterial culture without any sample pre-treatment.
Fig. 3.
b) Overview of the detection platform. a Detection workflow of SARS-CoV-2 from clinical samples using the electrochemical biosensor with RCA of the N and S genes. b The detection setup for electrochemical analysis by using a portable potentiostat device connected to a laptop. Reprinted with permission fromChaibun et al. (2021)for Rapid electrochemical detection of coronavirus SARS-CoV-2. Nature Communications. Copyright (2021) Nature.c) Schematic illustration of homogeneous circle-to-circle amplification. In the first round of RCA, polymerases act tandemly to generate intermediate amplicons. Intermediate amplicons anneal to CT2 for the second round of RCA, generating amplicon coils that lead to the assembly of MNPs. After a ligation step, all processes of amplification, hybridization, and detection take place simultaneously on-chip at 37 °C.
An example of an electrochemical biosensor based on RCA was reported involving the hybridization of the amplicons with probes functionalized with redox-active labels electrochemically detectable (Chaibun et al., 2021). This technique can be performed in two different ways: stepwise hybridization and one-step hybridization, although no significant differences were observed between the two resulting signals. Therefore, the one-step hybridization is preferentially used for optimized assays since it results easier and faster. The stepwise hybridization consists of the sequential hybridization of the probe-conjugated magnetic bead particle (CP-MNB) to the amplified target. The use of magnetic beads simplifies the isolation and purification of the nucleic acid target. The resulting CP-MNB-target was bound to the silica-reporter probe (Si-RP) with three washing steps after each hybridization. On the contrary, the one-step strategy requires a single hybridization step mixing CP-MNB, Si-RP, and the target simultaneously, followed by a single washing step. This technique has become a useful approach because of its low cost, compatibility with liquid samples, and high surface area for hybridization (Fig. 3b). This one-step sandwich hybridization test was able to detect as low as 1 copy/μL of N and S genes, in less than 2 h, and was challenged on 106 clinical samples showing 100% accordance with RT-qPCR.
Circle-to-circle amplification (C2CA) is a particular amplification method based on padlock probe ligation and rolling circle amplification. It shows a high amplification efficiency with a low risk of false-positives, but it contains several step-by-step operation processes. This method was combined with an optomagnetic analysis based on a magnetic nanoparticle assembly (Tian et al., 2020). In detail, a homogeneous circle-to-circle amplification was proposed to avoid the need for additional monomerization and ligation steps after the first round of RCA, which combines two amplification rounds in a one-pot reaction. A second RCA round produces amplicon coils able to anneal to detection probes grafted onto magnetic nanoparticles that are detected in real-time using an optomagnetic sensor (Fig. 3c). This methodology enabled the detection of a synthetic complementary DNA of SARS-CoV-2 RNA-dependent RNA polymerase coding sequence, with a detection limit of 0.4 fM in a total assay time of 100 min.
Reprinted with permission from Tian et al. (2020) for Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosensors and Bioelectronics. Copyright (2020) Elsevier.
2.3. Nucleic Acid Sequence-Based Amplification (NASBA)-based diagnostics
Nucleic Acid Sequence-Based Amplification (NASBA) has been specifically designed for the amplification of single-stranded RNA or DNA sequences (Compton, 1991) and takes inspiration from the retroviral RNA replication by cDNA intermediates. The reaction is carried out at 41 °C and utilizes two RNA target-specific primers and three enzymes: avian myeloblastosis virus reverse transcriptase (AMV), T7 DNA-dependent RNA polymerase (DdRp), and RNase H. NASBA shows many advantages such as its utility in RNA virus diagnosis and the low temperature of the reaction. However, it also possesses some drawbacks. Indeed, this technique cannot be considered truly isothermal because denaturation steps are required and the enzymes used in the reaction are thermolabile, so they need to be added separately. Another important limitation is that only short oligonucleotides (120–250 bp) can be efficiently amplified (Piepenburg et al., 2006).
A conformational switch biosensor was realized based on a selective toehold RNA biosensor scaffold and NASBA isothermal amplification for the sensitive and specific colour read-out of SARS-CoV-2 RNA fragments (Chakravarthy et al., 2021). Toehold RNA scaffolds are synthetic switches that control RNA translation when placed in tandem. The classical arrangement of a toehold switch is formed by a variable region, a ribosome binding site (RBS), a translational start sequence (AUG), and a 3′ overhang containing a reporter gene. The 5’ overhang variable sequence is designed to specifically bind with a trigger RNA in trans and to partially anneal to the central stem. The binding of the trigger RNA breaks the central stem structure inducing the exposure of RBS and the start codon and this conformational change causes a switch in the sensor and, consequently, the translation of a reporter protein (for example lacZ) that can be easily revealed with a chromogenic substrate (Fig. 4 ), with sensitivity down to attomolar (100 copies of) SARS-CoV-2 RNA.
Fig. 4.
A) Schematic of Toehold switches. Toehold RNA switches consist of a central stem loop structure that harbours a ribosome binding site (RBS, blue) and a translation start site (AUG, pink) with a downstream reporter gene (such as lacZ, grey). A variable region with the toehold (green) are designed to specifically base-pair with a trigger RNA (dark green). In the absence of trigger RNA (left), the RBS and AUG are sequestered within the sensor structure and inaccessible to the ribosome. Presence of the trigger RNA (right) induces intermolecular interactions between the toehold and the trigger RNA, resulting in an alternate conformation wherein the RBS and AUG are accessible to the ribosome, enabling translation of the downstream LacZ enzyme. Production of LacZ is easily monitored with colour, using a chromogenic substrate. The concept is modular and allows the use of alternate reporter genes and modes of detection. B) Schematic showing our assay development pipeline. RNA extracted from viral particles is amplified isothermally using NASBA (Nucleic Acid Sequence-Based Amplification) and detected with specifically designed toehold-based biosensors in an in vitro transcription-translation (IVTT) assay. The NASBA coupled IVTT assay leads to production of colour that can be easily visualized by eye or with cell phone cameras or luminescence that can be quantified by luminometry. Our assay development pipeline focused on identifying targetable regions of the SARS-CoV-2 genome, design of specific biosensors, optimized primers for efficient NASBA and overall sensitivity and response of the assay. Reprinted with permission fromChakravarthy et al. (2021)for Engineered RNA biosensors enable ultrasensitive SARS-CoV-2 detection in a simple colour and luminescence assay. Copyright (2021) medRxiv.
2.4. Multiple Cross Displacement Amplification (MCDA)-based diagnostics
Multiple Cross Displacement Amplification (MCDA) is a new isothermal amplification strategy developed in 2015, based on the filament displacement polymerization reaction (Wang et al., 2015). This low-cost method is highly specific and sensitive thanks to the high primer concentration. The reaction is carried out between 60 and 67 °C for 40 min, making it also a high-speed technique. In brief, MCDA assay encompasses the use of ten primers, each complementary to a target DNA region, and a DNA polymerase with strand-displacement activity to amplify the target.
An MCDA assay was designed for targeting SARS-CoV-2 N gene and ORF1ab and can be compared to LAMP and RT-PCR for its sensitivity and rapidity (Luu et al., 2021). The amplification reactions were accomplished using the WarmStart LAMP (DNA and RNA) kit (NEB) based on a warmstart RTx reverse transcriptase and Bst2.0 polymerase. N gene amplification by MCDA provided faster response but lower sensitivity, with a detection limit of 100 copies/μL with respect to 500 copies/μL of LAMP and 10 copies/μL of RT-PCR. ORF1ab amplification by MCDA showed similar speed to LAMP but this latter was more sensitive with 50 copies/μL compared to MCDA (500 copies/μL). In light of these findings, it is clear that each technique has pros and cons to be thoroughly considered when choosing the most suitable diagnostics based on needs, such as sensitivity, speed, and portability.
Alternative isothermal amplification assays are also available to address the speed and portability of conventional laboratory set-up diagnostics. A reverse transcription multiple cross-displacement amplification (RT-MCDA) was described in combination with a nanoparticle-based biosensor assay for SARS-CoV-2 detection. The monitoring of multiple clinical specimens has demonstrated that this method is an attractive, rapid, reliable, low-cost, and easy-to-use tool for primary care in resource-poor settings (Li et al., 2020). This technique requires two primer sets designed to bind the open reading frame ORF1ab and SARS-CoV-2 N gene. The instrument used for this assay was able to maintain a constant reaction temperature of 64 °C for only 35 min. This test detected down to five copies of target sequences within 1 h, and was challenged on 65 clinical samples from faeces, nasal, pharyngeal, and anal swabs of COVID-19 infected patients, obtaining 22 (33.8%) positive results, while RT-qPCR assay only detected 20 (30.7%) positive results in these samples. Any positive results were achieved from clinical samples without infections.
2.5. Recombinase polymerase amplification (RPA)-based diagnostics
Recombinase Polymerase Amplification (RPA) technology is a low temperature (37 °C) isothermal amplification method that amplifies a target DNA sequence with a recombinase, a single-stranded DNA-binding protein (SSB), and a strand-displacing DNA polymerase (Yonesaki and Minagawa, 1985). The recombinase, together with the single-strand binding protein, unwinds the complementary strands. For this reason, RPA does not require the use of heat to separate double-stranded DNA. Thanks to this enormous advantage, this amplification technique is very often associated with lateral flow biosensors for in-field, rapid, and simple analysis. The lateral flow assisted by RPA amplification uses biotinylated reverse primers and an internal fluorophore-labeled probe. When the amplified products rise to the top of the strip, its fluorophore-labeled probe can interact with anti-fluorophore antibody-labeled gold particles present in the strip. Since the amplified DNA contains also a biotinylated reverse primer it can be immobilized on the lateral flow strip by the anti-biotin antibodies, making the test line visible. The only products that result visible with this technique are those that contain both, biotinylated primers and fluorophores. The reaction is confirmed when a control band appears in the upper part of the strip.
In a recent study, a two-step reaction was assembled, requiring a separate cDNA synthesis step before the RPA reaction and lateral flow readout (Shelite et al., 2021). This assay provided a limit of detection of 35.4 viral cDNA nucleocapsid (N) gene copies/μL. 37 nasopharyngeal samples were also evaluated in comparison with the reference test, obtaining 100% concordance between RPA and RT-qPCR.
A microfluidic-integrated lateral flow was also described as powered with RPA for the rapid and sensitive detection of SARS-CoV-2, based on a universal dipstick detection system integrated into a single microfluidic chip (Liu et al., 2021). RPA reaction occurs in a single chamber to deliver to the detection strips biotin- and FAM-labeled amplified sequences. Samples containing COVID-19 target genes are loaded in a small reservoir (I and A) so first isothermally reverse transcribed, then amplified, and lastly labeled with biotin and FAM to bind to anti-FAM antibody. The samples are mixed (III) with a running buffer that comes from a large chamber (II) and allows for the capillary diffusion of the samples to pass through the detection bands contained in the lateral flow chamber (IV). Therefore, only the amplified gold-labeled products bind onto the biotin-ligand molecules at the test band, whereas the free gold-labeled anti-FAM antibodies bind the control band as a positive control. Testing required 30 min in total procedure consisting of nucleic acid extraction, loading, and incubation, and furnished a limit of detection of 1 copy per μL, or 30 copies per sample. This portable and affordable system was tested on clinically diagnosed COVID-19 cases and revealed a sensitivity of 97% and specificity of 100%.
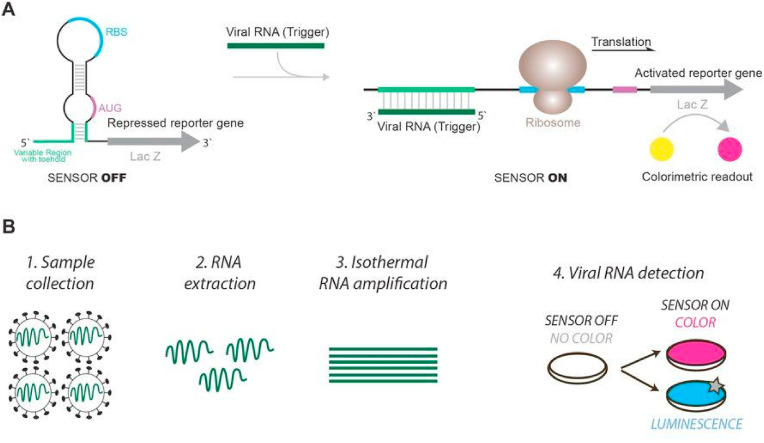
A similar dipstick was developed as a multiplexed, 1–2 step, fast (20–30 min) molecular test that can simultaneously detect both SARS-CoV-2 E and RdRP genes (Cherkaoui et al., 2021). A One-pot RPA amplification was carried out at a constant temperature (37–39 °C) using sequences of the primers/probe sets specific for SARS-CoV-2 E and RdRP genes.
Real-time fluorescence was detected after nuclease cleavage at the THF site of the probes, which causes the release of fluorophore from its quencher (Fig. 5 a). The sensitivity for the dipstick method was 130 (95%) RNA copies per reaction and the specificity was assessed towards SARS-CoV and MERS-CoV, underlining high potential as “point-of-care testing in decentralised settings, with minimal or equipment-free incubation methods and a user-friendly prototype smartphone application”, as stated by the authors.
Fig. 5.
a) Schematic representation of the rapid and multiplex RT-RPA assay with real-time fluorescence and dipstick detection. A) One-pot RT-RPA assay including reverse transcription of the viral RNA and amplification by RPA at constant temperature (37–39 °C). B) Sequences of the primers/probe sets used for SARS-CoV-2 E gene and RdRP gene in the multiplex RT-RPA assay with real-time detection (blue) and sequences of the modified primers used for the multiplex dipstick detection (orange). C) Real-time fluorescence detection by exonuclease cleavage of the probes for E gene and RdRP gene at their THF residue. D) Design of the dipstick for multiplexed detection of the E gene and the RdRP gene. Reprinted with permission fromCherkaoui et al. (2021)for Harnessing recombinase polymerase amplification for rapid multi-gene detection of SARS-CoV-2 in resource-limited settings. Biosensors and Bioelectronics. Copyright (2021) Elsevier.
b) Combining RPA with a rkDNA-GO system for the detection of COVID-19. Reprinted with permission fromChoi et al. (2021)for Combined recombinase polymerase amplification/rkDNA–graphene oxide probing system for detection of SARS-CoV-2. Analytica Chimica Acta. Copyright (2021) Elsevier.
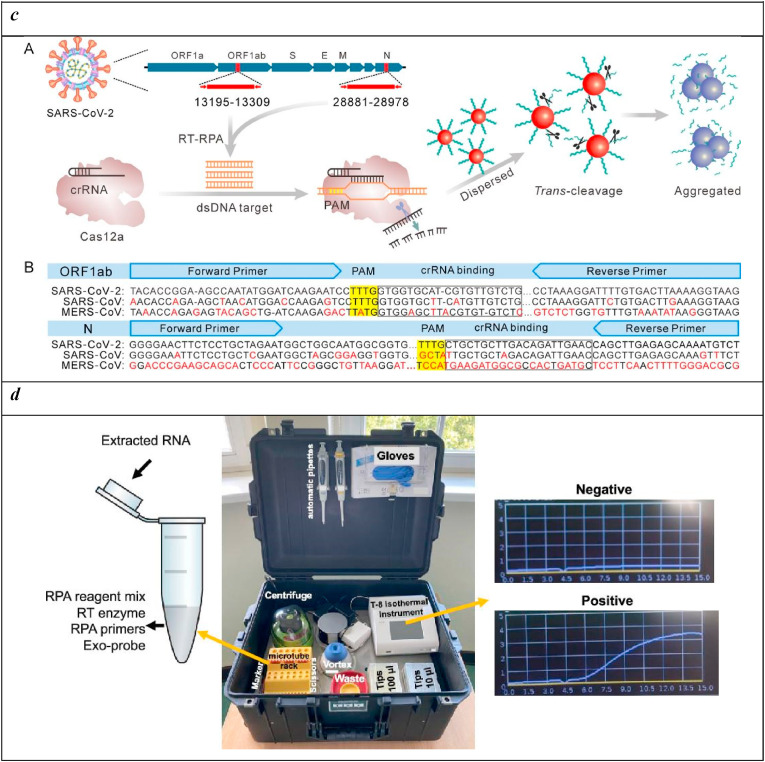
c) RT-RPA-Coupled Cas12a for Colorimetric Detection of SARS-CoV-2; (A) Schematic Illustration of the Strategy Design. The Whole Process Consists of Three Steps: RT-RPA of the Selected SARS-CoV-2 Genome Region, Cas12a Activation and Colorimetric Detection; (B) SARS-CoV-2 Genome Alignment of the Selected Target Region in the ORF1ab Gene and the N Protein gene; The Accession Numbers of SARS-CoV-2, SARS-CoV, and MERS-CoV Genomes Were NC_045512.2, AY278741.1, and NC_019843.3, Respectively. Reprinted with permission fromZhang et al. (2021). Reverse Transcription Recombinase Polymerase Amplification Coupled with CRISPR-Cas12a for Facile and Highly Sensitive Colorimetric SARS-CoV-2 Detection. Copyright (2021) American Chemical Society.
d) All-inclusive portable suitcase lab for deployment at the point of need. Reprinted with permission fromEl Wahed et al. (2021). Reverse Transcription Recombinase Polymerase Amplification Coupled with CRISPR-Cas12a for Suitcase Lab for Rapid Detection of SARS-CoV-2 Based on Recombinase Polymerase Amplification Assay. Copyright (2021) American Chemical Society.
Another lateral flow strip was realized using RPA amplification and SYBR Green I for the colorimetric detection of the amplification reaction and direct visualization (Lau et al., 2021). A 98% sensitivity and 100% specificity were obtained on 10-fold serial diluted synthetic RNA and genomic RNA of similar viruses, as well as 78 positive and 35 negative nasopharyngeal samples, reaching a detection limit of 7.659 copies/μL RNA, with no cross-reactivity. This data demonstrated how such combined systems can be considered as a valid alternative to RT-qPCR for the detection of SARS-CoV-2, especially in areas with limited infrastructure.
More recently, a customized RT-RPA-assisted colorimetric lateral flow strip was designed to allow for the rapid and simultaneous visual screening of SARS-CoV-2 and Flu viruses (Flu A and Flu B) without cross reactivity, false positives, and false negatives (Sun et al., 2022). This biosensor was capable to reduce the detection time (1 h) and improve sensitivity, detecting as low as 10 copies of the viral RNA, with 100% accuracy for evaluating 15 SARS-CoV-2-positive clinical samples, 10 Flu viruses-positive clinical samples, and 5 negative clinical samples, pre-confirmed by qRT-PCR.
Nanotechnology further helped in the last months to enhance the performances of similar diagnostics, highlighting their high potential to augment the analytical sensitivity and specificity. This was the case of a rkDNA-graphene oxide (GO) probe system to allow for the rapid detection of SARS-CoV-2 with high sensitivity and selectivity (Choi et al., 2021). An rkDNA probe was enzymatically synthesized complementary to an RPA-amplified sequence of the target N-gene of SARS-CoV-2. In the graphene oxide (GO) probe system the fluorescence of rkDNA is quenched, then, when the target RPA-amplified sequence is present in the mixture, the fluorescence is immediately recovered and detected (Fig. 5b). This combined system displayed high selectivity (discrimination factor: 17.2) and sensitivity (LOD = 6.0 aM), within a total processing time of 1.6 h, all crucial features for an efficient and simple point-of-care diagnostic suitable for rapid and in-field screening.
At the same strength of nanotechnology, biotechnology likewise furnished valid support in the development of efficient diagnostics for SARS-CoV-2. The design of a colorimetric assay for the SARS-CoV-2 detection was obtained by the combination of RPA amplification with nanomaterials, such as the ultimate CRISPR-Cas technology (Zhang et al., 2021). In detail, DNA-modified gold nanoparticles (AuNPs) were exploited as a universal colorimetric readout to specifically target ORF1ab and N regions of the SARS-CoV-2 genome. These regions are amplified by RPA and recognized by Cas12a. This interaction activates the nucleolitic enzyme that cleaves both the target DNA (cis) and the capped DNA substrate from AuNPs (trans). The aggregation of AuNPs particles causes a change in surface plasmon resonance (SPR) that is monitored by UV–vis absorbance spectroscopy and naked eye observation (Fig. 5c). This system allowed for the detection of 1 copy of viral genome sequence per test, with any false positive events thanks to the dual action deriving from the isothermal amplification and Cas12a activation process.
Thanks to the convergence of cross-cutting technologies, including microfluidics, nanotechnology, and biotechnology, many portable and cost-effective diagnostics have been realized for point-of-care in-field screening. A central and very recent example comes from the international cooperation of diverse research groups that produced a portable suitcase lab that combines nucleic acid extraction, RPA amplification, and detection at the point of need (El Wahed et al., 2021). As reported in Fig. 5d, this portable, simple, and all-inclusive equipment allowed for rapid (15 min), sensitive, and specific detection of pathogenic viruses with no cross-reactivity also in a low-resource setting as in developing countries.
3. Integration into point-of-care devices and market entry
The diffusion of the COVID-19 pandemic and its penetration in all strata of society, accounting for 356.955.803 cases with 5.610.291 deaths globally (World Health Organization, 2022), has prompted for the creation of commercial solutions, based on research-derived COVID-19 sensing platforms, capable to diagnose the infection even in asymptomatic subjects and exploitable outside clinical labs.
In this optic, isothermal amplification techniques targeted to SARS-CoV-2 genes have demonstrated high potential in providing sensitive as well as compact and rapid systems for COVID-19 diagnosis. Taking advantage from the literature, with crucial examples reported above, several companies have tried to integrate the isothermal amplification techniques into instruments able to detect SARS-CoV-2 in different human fluids, aiming to:
-
i)
lower as much as possible the detection limit,
-
ii)
increase the specificity of the method,
-
iii)
minimize the discomfort on the patient by targeting easy-to-extract bodily fluid, such as sputum or direct nasal secretion.
However, most of the current available products present on the market require a nasal swab to be performed to provide reliable results. Some examples of developed commercial products, requiring trained personnel to be used but still useful for on-the-field testing, are reported in Table 1 .
Table 1.
Some examples of commercially available instrumentations for outside-lab detection of SARS-CoV-2 (white background: products based on isothermal replication; grey background: product utilizing other detection strategy. Taken from (Johns Hopkins Center for Health Security, 2022).
| Test | Manufacturer | Type of test | Sensitivity | Specificity | Sample needed for the test | Target gene | Limit of detection (LOD) | Point of application |
|---|---|---|---|---|---|---|---|---|
| Sherlock CRISPR SARS-CoV-2 Kit | Sherlock Biosciences, Inc | CRISPR,RT-LAMP | 100% | 100% | nasal swabs, nasopharyngeal swabs, oropharyngeal swabs, nasopharyngeal wash/aspirate or nasal aspirate, and bronchoalveolar lavage | ORF1ab and N genes | 6750 copies/mL | Clinical laboratories |
| ID NOW COVID-19 | Abbott Diagnostics Scarborough | Isothermal amplification | 100% | 100% | direct nasal secretions, nasopharyngeal or throat swabs | RdRP | 125 copies/mL (3000 copies/mL as measured by the FDA reference panel) | Clinical laboratories, Point-of-care |
| Cue COVID-19 Test | Cue Health Inc. | Isothermal amplification | 100% | 92,0% | nasal swab | N gene | 1300 copies/mL | Clinical laboratories, Point-of-care |
| iAMP COVID-19 Detection Kit | Atila Biosystems, Inc | Isothermal amplification (OMEGA), patented | 100% | 100% | nasal, nasopharyngeal and/or oropharyngeal swabs | N and ORF1ab genes | 60 copies/reaction of 25 uL | Clinical laboratories |
| Cue COVID-19 Test for Home and Over The Counter (OTC) Use | Cue Health Inc. | Isothermal amplification, | 96% (symptomatic), 100% (asymptomatic) | 98% (symptomatic), 100% (asymptomatic) | anterior nares swabs | N gene | 2700 copies/mL | At-home test |
| Solana SARS-CoV-2 Assay | Quidel Corporation | Molecular isothermal Reverse Transcriptase – Helicase-Dependent Amplification (RT-HDA) | 90,30% | 97,8% | nasopharyngeal (NP) and nasal (NS) swab specimens | pp1ab gene | 1.16 x 10^4 copies/mL | Clinical laboratories |
| Talis One COVID-19 Test System | Talis Biomedical Corporation | NAAT (reverse transcriptase isothermal amplification) | 100,0% | 100,00% | nasal mid-turbinate swabs | ORF1ab gene, N gene | 500 copies/mL | Clinical laboratories, Point-of-care |
| DETECTR BOOST SARS-CoV-2 Reagent Kit | Mammoth Biosciences, Inc. | RT and isothermal amplification test | 95,7% | 100,00% | nasopharyngeal, anterior nasal, mid-turbinate nasal, or oropharyngeal swabs | N gene | 100 copies/mL | Clinical laboratories |
| Detect Covid-19 Test | Detect, Inc. | RT-LAMP | 90,9% | 97,50% | anterior nasal swabs | ORF1ab | 800 copies/mL | At-home test |
| MobileDetect Bio BCC19 Test Kit | Detectachem Inc | RT-LAMP | 98% | 100% | nasopharyngeal, anterior nasal swabs, mid-turbinate nasal swabs, and oropharyngeal swab | N and E genes | 75000 copies/mL | Clinical laboratories |
| Lucira COVID-19 All-In-One Test Kit | Lucira Health, Inc | RT-LAMP | 94% | 98% | Nasal swab (self collection), nasal swab (healthcare collection) | N gene (two regions) | 2700 copies/swab (or 900 copies/mL if the sample is 100% lysed and eluted) | At-home test |
| SARS-CoV-2 DETECTR Reagent Kit | Mammoth Biosciences, Inc | RT-LAMP | 95% | 100% | nasopharyngeal swabs, oropharyngeal (throat) swabs, mid-turbinate nasal swabs, anterior nasal swabs, nasopharyngeal wash/aspirate or nasal aspirate | N gene | 20,000 copies/mL | Clinical laboratories |
| AQ-TOP COVID-19 Rapid Detection Kit PLUS | Seasun Biomaterials, Inc | RT-LAMP | 100% | 100% | oropharyngeal and nasopharyngeal swabs, anterior nasal and mid-turbinate nasal swabs, nasopharyngeal washes/aspirates or nasal aspirates as well as bronchoalveolar lavage | ORF1ab and N genes | 1000 copies/mL | Clinical laboratories |
| AQ-TOP™ COVID-19 Rapid Detection Kit | Seasun Biomaterials, Inc. | RT-LAMP | 100% | 100% | oropharyngeal and nasopharyngeal swab specimens, anterior nasal and mid-turbinate nasal swabs, nasopharyngeal wash/aspirate or nasal aspirate specimens, BAL and sputum | ORF1ab | 7000 copies/mL | Clinical laboratories |
| SynergyDx SARS-CoV-2 RNA Test | Synergy Diagnostic Laboratory, Inc., DBA SynergyDx | rRT-PCR | 100% | 100% | anterior nasal swab | ORF1ab and N genes | 20.8 copies/mL | Clinical laboratories |
| LumiraDx SARS-CoV-2 RNA STAR Complete | LumiraDx UK Ltd. | RT-qSTAR amplification | 97,8% | 98,5% | nasal, mid-turbinate, nasopharyngeal, and oropharyngeal swabs | ORF1a | 7500 copies/mL | Clinical laboratories |
| Procleix SARS-CoV-2 Assay | Grifols Diagnostic Solutions Inc | transcription-mediated nucleic acid amplification (TMA) | 100% | 100% | anterior nasal and mid-turbinate nasal swabs, nasopharyngeal and oropharyngeal swabs, nasopharyngeal washes/aspirates or nasal aspirates, and bronchoalveolar lavage specimens | Not stated | 60 copies/mL | Clinical laboratories |
As demonstrated by the large number of biosensing configurations, numerous methods based on isothermal amplifications have found a commercial outlet. Indeed, considering Table 1 (reporting commercial products having received the Food and Drug Administration's Emergency Use Authorization), it is clear that isothermal amplification technique has been used to produce instrument suitable for fast laboratory analysis, testing by trained personal for on-field applications, and even test for autonomous use by common users for at-home testing. It is interesting to underline that, while the at-home testing products report slightly lower sensitivity and specificity values and higher LODs, they are still quite competitive with the more complex commercial products designed for professional use. Finally, by comparing the commercial product based on isothermal amplification (white cells in Table 1) with some examples of commercial product employing different SARS-CoV-2 recognition techniques (grey cells in Table 1), it is clear that isothermal amplification is truly competitive as detection technique to realize commercial solutions for COVID-19 diagnosis, from clinical laboratories to autonomous testing.
4. Conclusions and future perspectives
The events related to the COVID-19 pandemic of the past two years have highlighted the need for surveillance and diagnosis of diseases as crucial services for public health and the economy. Accurate and rapid diagnostic strategies have proved decisive for minimizing or eradicating the spread of infectious diseases. The sensitivity and specificity of diagnostics are also vital characteristics, as are the portability, affordability, and simplicity to provide suitable handheld devices at the point of need. Such diagnostics could have a huge impact on the success of high-throughput screening and disease control. Nucleic acid isothermal amplification has been confirmed to be an effective technology for powering biosensors, being able to provide numerous benefits, including sensitivity and specificity. Furthermore, such combined systems obviate the need for a thermal cycler thus simplifying the diagnosis procedures, especially in low-resource settings. Diverse isothermal amplification methods are available, with intrinsic pros and cons (Table 2 ). LAMP relies on strand displacement DNA polymerase and primers to amplify specific DNA sequences of pathogens. It possesses a very high specificity; indeed, it can amplify a specific gene by discriminating a single nucleotide difference (Tavares et al., 2011). Moreover, this method has also the great advantage of checking the amplification products by SYBR Green I dye mediated naked eye visualization and by real-time monitoring (Nakamura et al., 2007). Although LAMP has the advantages described above, it also has some disadvantages. One of the major drawbacks is related to the primers that, if not correctly designed, may deliver primer-primer interactions (Torres et al., 2011). RCA utilizes highly processive strand displacement DNA polymerase and circularizable oligonucleotide probes for detecting single strand DNA or RNA. This method has a simple reaction scheme only requiring a single primer and a productive enzyme and generates long chains of tandem repeats. RCA has a wide range of applications, from genomics to proteomics, nanotechnology, and functional nucleic acids (Li and Macdonald, 2015), and the products can be visualized on a gel, or be monitored in real-time. Although RCA is considered a powerful technique, it has the clear inconvenience of occurring only linearly over time. NASBA can amplify more than 109 copies of nucleic acid sequences in just 90 min. It produces ssRNA amplicons without an additional reverse transcription (RT) step; the amplicons can be used directly in the next round of amplification without denaturation steps. That is why it is especially suitable for the detection of Retroviruses. As for the other techniques, NASBA also presents some disadvantages. As for RT-qPCR and other RNA amplification procedures, one of the major drawbacks is RNA integrity. Moreover, although it is considered an isothermal technique (41 °C), it requires a melting step before the amplification reaction to allow the annealing of the primers to the target region. Lastly, the amplified RNA target sequence should be 120–250 nucleotides long, shorter or longer regions would be amplified less efficiently. MCDA is the fastest method for SARS-CoV-2 detection: 5 min could be sufficient even if the amplification is performed for 20 min to ensure reliable results for negative samples. MCDA can amplify SARS-CoV-2 RNA using boiling as an extraction procedure, reducing time and costs (Luu et al., 2021), which are further reduced by the use of a single enzyme. On the other hand, the use of several primers ensures a high recognition specificity of the target sequences (Wang et al., 2015) but, at the same time, it has a disadvantage, due to the possible primer-primer annealing, as already described for LAMP. The detection limit is about 100 copies/μL, better than LAMP but lower than PCR. RPA has a simple reaction scheme since it occurs at a low and constant temperature, and does not require an initial denaturation step or the use of multiple primers. Moreover, RPA has high sensitivity and extreme rapidity; indeed, it is capable of amplifying 1–10 DNA target copies in less than 20 min. However, RPA is inhibited by high genomic DNA concentrations in whole blood samples (20–100 ng/μL). Another disadvantage occurs when the products are detected by agarose gel electrophoresis or lateral flow; in these cases, a purification/protein digestion, after the amplification process, is required to avoid smearing or impaired flow (Lobato and O'Sullivan, 2018).
Table 2.
Advantages and drawbacks of isothermal amplification techniques.
| Advantages | Drawbacks | |
|---|---|---|
| Rt-qPCR | Specific Sensitive Simultaneous detection and quantification |
High cost Specific machine required Absolute quantification relies on standard curve Temperature of reaction |
| LAMP | Highly specific Can be detected by a cheap turbidimeter No initial heating step Speed (within 1 h) |
Complex primers design High temperature (60–65 °C) Possible primer-primer annealing Detection limit |
| RCA | Simple mechanism Easy primer design Temperature of reaction (30–37 °C) Easily detectable |
Numerous rounds of amplifications RNA amplification is complex Works only with circular templates Occurs only linearly over time |
| NASBA | Specifically designed to detect RNA and in turn RNA viruses Temperature of reaction (41 °C) Fast |
Denaturation step Less efficient in amplifying RNA targets out of the range 120–250 bp RNA integrity Pre-heating step |
| MCDA | Fastest method No initial heating step Speed (40 min) Sensitivity, specificity Low cost |
Complex primers design High temperature (60–67 °C) |
| RPA | High sensitivity Temperature of reaction (37 °C) Simple primers design Extremely quick (20 min) No initial heating step |
Non-specific background Possible primer-primer annealing Inhibition by high genomic DNA concentrations Detection limit |
It is clear that each method shows its advantages and disadvantages; however, nearly all methods reported at least 90% specificity and 90% sensitivity, as well as most of them do not need any sample heating. Additional characteristics to be considered for effective diagnostics are repeatability, lack of homogeneity among the different tests developed, unpredictable viral load in the specimen, and sample preparation efficiency. Therefore, more comprehensive research is needed to develop a universal, high-quality multiplex biosensor device for point-of-care implementation that is essential to fight pandemics.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgment
This work was supported by the Project POR FESR PdR Lazio E-CROME.
References
- Augustine R., Hasan A., Das S., Ahmed R., Mori Y., Notomi T., Kevadiya B.D., Thakor A.S. Loop-mediated isothermal amplification (LAMP): a rapid, sensitive, specific, and cost-effective point-of-care test for coronaviruses in the context of COVID-19 pandemic. Biology. 2020;9:182. doi: 10.3390/biology9080182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Böger B., Fachi M.M., Vilhena R.O., Cobre A.F., Tonin F.S., Pontarolo R. Systematic review with meta-analysis of the accuracy of diagnostic tests for COVID-19. Am. J. Infect. Control. 2021;49:21–29. doi: 10.1016/j.ajic.2020.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bru D., Martin-Laurent F., Philippot L. Quantification of the detrimental effect of a single primer-template mismatch by real-time PCR using the 16S rRNA gene as an example. Appl. Environ. Microbiol. 2008 doi: 10.1128/AEM.02403-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaibun T., Puenpa J., Ngamdee T., Boonapatcharoen N., Athamanolap P., O'Mullane A.P., Vongpunsawad S., Poovorawan Y., Lee S.Y., Lertanantawong B. Rapid electrochemical detection of coronavirus SARS-CoV-2. Nat. Commun. 2021;12:802. doi: 10.1038/s41467-021-21121-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy A., Nandakumar A., George G., Ranganathan S., Umashankar S., Shettigar N., Palakodeti D., Gulyani A., Ramesh A. Engineered RNA biosensors enable ultrasensitive SARS-CoV-2 detection in a simple color and luminescence assay. Life Sci. Alliance. 2021;4 doi: 10.26508/lsa.202101213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherkaoui D., Huang D., Miller B.S., Turbé V., McKendry R.A. Harnessing recombinase polymerase amplification for rapid multi-gene detection of SARS-CoV-2 in resource-limited settings. Biosens. Bioelectron. 2021;189:113328. doi: 10.1016/j.bios.2021.113328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi M.H., Lee J., Seo Y.J. Combined recombinase polymerase amplification/rkDNA–graphene oxide probing system for detection of SARS-CoV-2. Anal. Chim. Acta. 2021;1158:338390. doi: 10.1016/j.aca.2021.338390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chow F.W.-N., Chan T.T.-Y., Tam A.R., Zhao S., Yao W., Fung J., Cheng F.K.-K., Lo G.C.-S., Chu S., Aw-Yong K.L., Tang J.Y.-M., Tsang C.-C., Luk H.K.-H., Wong A.C.-P., Li K.S.-M., Zhu L., He Z., Tam E.W.T., Chung T.W.-H., Wong S.C.Y., Que T.-L., Fung K.S.-C., Lung D.C., Wu A.K.-L., Hung I.F.-N., Woo P.C.-Y., Lau S.K.-P. A rapid, simple, inexpensive, and mobile colorimetric assay COVID-19-LAMP for mass on-site screening of COVID-19. Int. J. Mol. Sci. 2020;21:5380. doi: 10.3390/ijms21155380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Compton J. Nucleic acid sequence-based amplification. Nature. 1991;350:91–92. doi: 10.1038/350091a0. [DOI] [PubMed] [Google Scholar]
- Corey L., Beyrer C., Cohen M.S., Michael N.L., Bedford T., Rolland M. SARS-CoV-2 variants in patients with immunosuppression. N. Engl. J. Med. 2021;385:562–566. doi: 10.1056/NEJMsb2104756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding S., Chen G., Wei Y., Dong J., Du F., Cui X., Huang X., Tang Z. Sequence-specific and multiplex detection of COVID-19 virus (SARS-CoV-2) using proofreading enzyme-mediated probe cleavage coupled with isothermal amplification. Biosens. Bioelectron. 2021;178:113041. doi: 10.1016/j.bios.2021.113041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Wahed A.A., Patel P., Maier M., Pietsch C., Rüster D., Böhlken-Fascher S., Kissenkötter J., Behrmann O., Frimpong M., Diagne M.M., Faye M., Dia N., Shalaby M.A., Amer H., Elgamal M., Zaki A., Ismail G., Kaiser M., Corman V.M., Niedrig M., Landt O., Faye O., Sall A.A., Hufert F.T., Truyen U., Liebert U.G., Weidmann M. Suitcase lab for rapid detection of SARS-CoV-2 based on recombinase polymerase amplification assay. Anal. Chem. 2021;93:2627–2634. doi: 10.1021/acs.analchem.0c04779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabiani L., Caratelli V., Fiore L., Scognamiglio V., Antonacci A., Fillo S., De Santis R., Monte A., Bortone M., Moscone D., Lista F., Arduini F. State of the art on the SARS-CoV-2 toolkit for antigen detection: one year later. Biosensors. 2021;11:310. doi: 10.3390/bios11090310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas C., Fuentes-Villalobos F., Garrido J.L., Haigh J., Barría M.I. Insights on early mutational events in SARS-CoV-2 virus reveal founder effects across geographical regions. PeerJ. 2020;8 doi: 10.7717/peerj.9255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fire A., Xu S.Q. Rolling replication of short DNA circles. Proc. Natl. Acad. Sci. Unit. States Am. 1995;92:4641–4645. doi: 10.1073/pnas.92.10.4641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge A., Liu F., Teng X., Cui C., Wu F., Liu W., Liu Y., Chen X., Xu J., Ma B. A Palm Germ-Radar (PaGeR) for rapid and simple COVID-19 detection by reverse transcription loop-mediated isothermal amplification (RT-LAMP) Biosens. Bioelectron. 2022;200:113925. doi: 10.1016/j.bios.2021.113925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain S., Dandy S., Geiss D.J., Henry C B.S. Padlock probe-based rolling circle amplification lateral flow assay for point-of-need nucleic acid detection. Analyst. 2021;146:4340–4347. doi: 10.1039/D1AN00399B. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janik E., Niemcewicz M., Podogrocki M., Majsterek I., Bijak M. The emerging concern and interest SARS-CoV-2 variants. Pathogens. 2021;10:633. doi: 10.3390/pathogens10060633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johns Hopkins Center for Health Security . 2022. Antigen and Molecular Tests for COVID-19.https://www.centerforhealthsecurity.org/covid-19TestingToolkit/molecular-based-tests/current-molecular-and-antigen-tests.html [WWW Document]. COVID-19 Testing Toolkit. URL. [Google Scholar]
- Kaden R. Early phylogenetic diversification of SARS-CoV-2: determination of variants and the effect on epidemiology, immunology, and diagnostics. J. Clin. Med. 2020;9:1615. doi: 10.3390/jcm9061615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kidd M., Richter A., Best A., Cumley N., Mirza J., Percival B., Mayhew M., Megram O., Ashford F., White T., Moles-Garcia E., Crawford L., Bosworth A., Atabani S.F., Plant T., McNally A. S-variant SARS-CoV-2 lineage B1.1.7 is associated with significantly higher viral load in samples tested by TaqPath polymerase chain reaction. J. Infect. Dis. 2021;223:1666–1670. doi: 10.1093/infdis/jiab082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kingsmore S.F., Patel D.D. Multiplexed protein profiling on antibody-based microarrays by rolling circle amplification. Curr. Opin. Biotechnol. 2003;14:74–81. doi: 10.1016/S0958-1669(02)00019-8. [DOI] [PubMed] [Google Scholar]
- Kitagawa Y., Orihara Y., Kawamura R., Imai K., Sakai J., Tarumoto N., Matsuoka M., Takeuchi S., Maesaki S., Maeda T. Evaluation of rapid diagnosis of novel coronavirus disease (COVID-19) using loop-mediated isothermal amplification. J. Clin. Virol. 2020;129:104446. doi: 10.1016/j.jcv.2020.104446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koyama T., Platt D., Parida L. Variant analysis of SARS-CoV-2 genomes. Bull. World Health Organ. 2020;98:495–504. doi: 10.2471/BLT.20.253591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau Y.L., Ismail I. binti, Mustapa N.I. binti, Lai M.Y., Soh T.S.T., Hassan A.H., Peariasamy K.M., Lee Y.L., Kahar M.K.B.A., Chong J., Goh P.P. Development of a reverse transcription recombinase polymerase amplification assay for rapid and direct visual detection of Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) PLoS One. 2021;16 doi: 10.1371/journal.pone.0245164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Macdonald J. Advances in isothermal amplification: novel strategies inspired by biological processes. Biosens. Bioelectron. 2015;64:196–211. doi: 10.1016/j.bios.2014.08.069. [DOI] [PubMed] [Google Scholar]
- Li S., Jiang W., Huang J., Liu Y., Ren L., Zhuang L., Zheng Q., Wang M., Yang R., Zeng Y., Wang Y. Highly sensitive and specific diagnosis of COVID-19 by reverse transcription multiple cross-displacement amplification-labelled nanoparticles biosensor. Eur. Respir. J. 2020;56 doi: 10.1183/13993003.02060-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C., Xie M., Chen J.J.L., Liu Y., Yan H. Rolling-circle amplification of a DNA nanojunction. Angew. Chem. Int. Ed. 2006;45:7537–7539. doi: 10.1002/anie.200602113. [DOI] [PubMed] [Google Scholar]
- Liu D., Daubendiek S.L., Zillman M.A., Ryan K., Kool E.T. Rolling circle DNA synthesis: small circular oligonucleotides as efficient templates for DNA polymerases. J. Am. Chem. Soc. 1996;118:1587–1594. doi: 10.1021/ja952786k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Shen H., Zhang Y., Shen D., Zhu M., Song Y., Zhu Z., Yang C. A microfluidic-integrated lateral flow recombinase polymerase amplification (MI-IF-RPA) assay for rapid COVID-19 detection. Lab Chip. 2021;21:2019–2026. doi: 10.1039/D0LC01222J. [DOI] [PubMed] [Google Scholar]
- Lobato I.M., O'Sullivan C.K. Recombinase polymerase amplification: basics, applications and recent advances. Trac. Trends Anal. Chem. 2018;98:19–35. doi: 10.1016/j.trac.2017.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luu L.D.W., Payne M., Zhang X., Luo L., Lan R. Development and comparison of novel multiple cross displacement amplification (MCDA) assays with other nucleic acid amplification methods for SARS-CoV-2 detection. Sci. Rep. 2021;11:1873. doi: 10.1038/s41598-021-81518-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignardi M., Mezger A., Qian X., La Fleur L., Botling J., Larsson C., Nilsson M. Oligonucleotide gap-fill ligation for mutation detection and sequencing in situ. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gkv772. e151–e151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morales-Narváez E., Dincer C. The impact of biosensing in a pandemic outbreak: COVID-19. Biosens. Bioelectron. 2020;163:112274. doi: 10.1016/j.bios.2020.112274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura N., Ito K., Takahashi M., Hashimoto K., Kawamoto M., Yamanaka M., Taniguchi A., Kamatani N., Gemma N. Detection of six single-nucleotide polymorphisms associated with rheumatoid arthritis by a loop-mediated isothermal amplification method and an electrochemical DNA chip. Anal. Chem. 2007;79:9484–9493. doi: 10.1021/ac0715468. [DOI] [PubMed] [Google Scholar]
- Navani N.K., Li Y. Nucleic acid aptamers and enzymes as sensors. Curr. Opin. Chem. Biol. Combin. Chem. Mol. Divers. 2006;10:272–281. doi: 10.1016/j.cbpa.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Park G.-S., Ku K., Baek S.-H., Kim S.-J., Kim S.I., Kim B.-T., Maeng J.-S. Development of reverse transcription loop-mediated isothermal amplification assays targeting severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) J. Mol. Diagn. 2020;22:729–735. doi: 10.1016/j.jmoldx.2020.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. DNA detection using recombination proteins. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi L., Hu Q., Kang Q., Bi Y., Jiang Y., Yu L. Detection of biomarkers in blood using liquid crystals assisted with aptamer-target recognition triggered in situ rolling circle amplification on magnetic beads. Anal. Chem. 2019;91:11653–11660. doi: 10.1021/acs.analchem.9b02186. [DOI] [PubMed] [Google Scholar]
- Qi X., Bakht S., Devos K.M., Gale M.D., Osbourn A. L-RCA (ligation-rolling circle amplification): a general method for genotyping of single nucleotide polymorphisms (SNPs) Nucleic Acids Res. 2001;29 doi: 10.1093/nar/29.22.e116. e116–e116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rödel J., Egerer R., Suleyman A., Sommer-Schmid B., Baier M., Henke A., Edel B., Löffler B. Use of the variplexTM SARS-CoV-2 RT-LAMP as a rapid molecular assay to complement RT-PCR for COVID-19 diagnosis. J. Clin. Virol. 2020;132:104616. doi: 10.1016/j.jcv.2020.104616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Manzano J., Malpartida-Cardenas K., Moser N., Pennisi I., Cavuto M., Miglietta L., Moniri A., Penn R., Satta G., Randell P., Davies F., Bolt F., Barclay W., Holmes A., Georgiou P. Handheld point-of-care system for rapid detection of SARS-CoV-2 extracted RNA in under 20 min. ACS Cent. Sci. 2021;7:307–317. doi: 10.1021/acscentsci.0c01288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohaim M.A., Clayton E., Sahin I., Vilela J., Khalifa M.E., Al-Natour M.Q., Bayoumi M., Poirier A.C., Branavan M., Tharmakulasingam M., Chaudhry N.S., Sodi R., Brown A., Burkhart P., Hacking W., Botham J., Boyce J., Wilkinson H., Williams C., Whittingham-Dowd J., Shaw E., Hodges M., Butler L., Bates M.D., La Ragione R., Balachandran W., Fernando A., Munir M. Artificial intelligence-assisted loop mediated isothermal amplification (AI-LAMP) for rapid detection of SARS-CoV-2. Viruses. 2020;12:972. doi: 10.3390/v12090972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schweitzer B., Kingsmore S. Combining nucleic acid amplification and detection. Curr. Opin. Biotechnol. 2001;12:21–27. doi: 10.1016/S0958-1669(00)00172-5. [DOI] [PubMed] [Google Scholar]
- Shelite T.R., Uscanga-Palomeque A.C., Castellanos-Gonzalez A., Melby P.C., Travi B.L. Isothermal recombinase polymerase amplification-lateral flow detection of SARS-CoV-2, the etiological agent of COVID-19. J. Virol Methods. 2021;296:114227. doi: 10.1016/j.jviromet.2021.114227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Qin P., He J., Li Weiwei, Shi Y., Xu J., Wu Q., Chen Q., Li Weidong, Wang X., Liu G., Chen W. Rapid and simultaneous visual screening of SARS-CoV-2 and influenza viruses with customized isothermal amplification integrated lateral flow strip. Biosens. Bioelectron. 2022;197:113771. doi: 10.1016/j.bios.2021.113771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taleghani N., Taghipour F. Diagnosis of COVID-19 for controlling the pandemic: a review of the state-of-the-art. Biosens. Bioelectron. 2021;174:112830. doi: 10.1016/j.bios.2020.112830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tavares R.G., Staggemeier R., Borges A.L.P., Rodrigues M.T., Castelan L.A., Vasconcelos J., Anschau M.E., Spalding S.M. Molecular techniques for the study and diagnosis of parasite infection. J. Venom. Anim. Toxins Incl. Trop. Dis. 2011;17:239–248. doi: 10.1590/S1678-91992011000300003. [DOI] [Google Scholar]
- Tian B., Gao F., Fock J., Dufva M., Hansen M.F. Homogeneous circle-to-circle amplification for real-time optomagnetic detection of SARS-CoV-2 RdRp coding sequence. Biosens. Bioelectron. 2020;165:112356. doi: 10.1016/j.bios.2020.112356. [DOI] [PubMed] [Google Scholar]
- Tomita N., Mori Y., Kanda H., Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nat. Protoc. 2008;3:877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- Torres C., Vitalis E.A., Baker B.R., Gardner S.N., Torres M.W., Dzenitis J.M. LAVA: an open-source approach to designing LAMP (Loop-Mediated isothermal amplification) DNA signatures. BMC Bioinf. 2011;12:240. doi: 10.1186/1471-2105-12-240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenberg O., Martiny D., Rochas O., van Belkum A., Kozlakidis Z. Considerations for diagnostic COVID-19 tests. Nat. Rev. Microbiol. 2021;19:171–183. doi: 10.1038/s41579-020-00461-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vecchio C.D., Brancaccio G., Brazzale A.R., Lavezzo E., Onelia F., Franchin E., Manuto L., Bianca F., Cianci V., Cattelan A., Toppo S., Crisanti A. 2021. Emergence of N Antigen SARS-CoV-2 Genetic Variants Escaping Detection of Antigenic Tests. [DOI] [Google Scholar]
- Wang F., Yang J., He R., Yu X., Chen S., Liu Y., Wang L., Li A., Liu L., Zhai C., Ma L. PfAgo-based detection of SARS-CoV-2. Biosens. Bioelectron. 2021;177:112932. doi: 10.1016/j.bios.2020.112932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Hozumi Y., Yin C., Wei G.-W. Mutations on COVID-19 diagnostic targets. Genomics. 2020;112:5204–5213. doi: 10.1016/j.ygeno.2020.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Yi, Wang Yan, Ma A.-J., Li D.-X., Luo L.-J., Liu D.-X., Jin D., Liu K., Ye C.-Y. Rapid and sensitive isothermal detection of nucleic-acid sequence by multiple cross displacement amplification. Sci. Rep. 2015;5:11902. doi: 10.1038/srep11902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Health Organization WHO coronavirus (COVID-19) dashboard [WWW document] 2022. https://covid19.who.int WHO Coronavirus (COVID-19) Dashboard. URL.
- Wu F., Zhao S., Yu B., Chen Y.-M., Wang W., Song Z.-G., Hu Y., Tao Z.-W., Tian J.-H., Pei Y.-Y., Yuan M.-L., Zhang Y.-L., Dai F.-H., Liu Y., Wang Q.-M., Zheng J.-J., Xu L., Holmes E.C., Zhang Y.-Z. A new coronavirus associated with human respiratory disease in China. Nature. 2020;579:265–269. doi: 10.1038/s41586-020-2008-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan C., Cui J., Huang L., Du B., Chen L., Xue G., Li S., Zhang W., Zhao L., Sun Y., Yao H., Li N., Zhao H., Feng Y., Liu S., Zhang Q., Liu D., Yuan J. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020;26:773–779. doi: 10.1016/j.cmi.2020.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yonesaki T., Minagawa T. T4 phage gene uvsX product catalyzes homologous DNA pairing. EMBO J. 1985;4:3321–3327. doi: 10.1002/j.1460-2075.1985.tb04083.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang S.-Q., Tan B., Li P., Wang F.-X., Guo L., Yang Y., Sun N., Zhu H.-W., Wen Y.-J., Cheng S.-P. Comparison of conventional RT-PCR, reverse-transcription loop-mediated isothermal amplification, and SYBR green I-based real-time RT-PCR in the rapid detection of bovine viral diarrhea virus nucleotide in contaminated commercial bovine sera batches. J. Virol. Methods. 2014;207:204–209. doi: 10.1016/j.jviromet.2014.05.020. [DOI] [PubMed] [Google Scholar]
- Zhang W.S., Pan J., Li F., Zhu M., Xu M., Zhu H., Yu Y., Su G. Reverse transcription recombinase polymerase amplification coupled with CRISPR-cas12a for facile and highly sensitive colorimetric SARS-CoV-2 detection. Anal. Chem. 2021;93:4126–4133. doi: 10.1021/acs.analchem.1c00013. [DOI] [PubMed] [Google Scholar]
- Zhao H., Zhang Y., Chen Y., Ho N.R.Y., Sundah N.R., Natalia A., Liu Y., Miow Q.H., Wang Y., Tambyah P.A., Ong C.W.M., Shao H. Accessible detection of SARS-CoV-2 through molecular nanostructures and automated microfluidics. Biosens. Bioelectron. 2021;194:113629. doi: 10.1016/j.bios.2021.113629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X., Wang X., Han L., Chen T., Wang L., Li H., Li S., He L., Fu X., Chen S., Xing M., Chen H., Wang Y. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens. Bioelectron. 2020;166:112437. doi: 10.1016/j.bios.2020.112437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziegler K., Steininger P., Ziegler R., Steinmann J., Korn K., Ensser A. SARS-CoV-2 samples may escape detection because of a single point mutation in the N gene. Euro Surveill. 2020;25:2001650. doi: 10.2807/1560-7917.ES.2020.25.39.2001650. [DOI] [PMC free article] [PubMed] [Google Scholar]