Abstract
The recent discovery of Candida dubliniensis as a separate species that traditionally has been identified as Candida albicans has led to the development of a variety of biochemical and molecular methods for the differentiation of these two pathogenic yeasts. rRNA sequences are well-established phylogenetic markers, and probes targeting species-specific rRNA sequences have been used in diagnostic assays for the detection and identification of microorganisms. Peptide nucleic acid (PNA) is a DNA mimic with improved hybridization characteristics, and the neutral backbone of PNA probes offers significant advantages in whole-cell in situ hybridization assays. In this study, we developed PNA probes targeting the rRNAs of C. albicans and C. dubliniensis and applied them to a fluorescence in situ hybridization method (PNA FISH) for differentiation between C. albicans and C. dubliniensis. Liquid cultures were smeared onto microscope slides, heat fixed, and then hybridized for 30 min. Unhybridized PNA probe was removed by washing, and smears were examined by fluorescence microscopy. Evaluation of the PNA FISH method using smears of 79 C. dubliniensis and 70 C. albicans strains showed 100% sensitivity and 100% specificity for both PNA probes. We concluded that PNA FISH is a powerful tool for the differentiation of C. albicans and C. dubliniensis.
The discovery of Candida dubliniensis as a separate species traditionally identified as Candida albicans has necessitated the development of new methods to clearly and easily distinguish the two species. C. dubliniensis shares many phenotypic characteristics with C. albicans and is therefore incorrectly identified as C. albicans by current methods, such as germ tube analysis and commercially available carbon assimilation tests (23). C. albicans is responsible for the majority of all fungal infections worldwide; however, retrospective studies have recently shown that some previously diagnosed C. albicans infections were actually C. dubliniensis infections (4). The clinical importance of C. dubliniensis and the role of its drug resistance remain to be determined, although studies have shown that C. dubliniensis primarily has been associated with infections in the oral cavity of human immunodeficiency virus patients and that it has the ability to develop resistance to fluconazole in vitro (4, 23). Clinical studies to determine the role of C. dubliniensis in candidiasis rely on the availability of reliable laboratory methods to distinguish C. dubliniensis from C. albicans, and a number of different phenotypic methods have been described. These methods include fatty acid methyl ester analysis (14), PCR methods (2, 8, 12), DNA fingerprinting (5), β-d-glucosidase activity (19), growth at 42 and 45°C (16, 23, 25), and chlamydospore formation (10, 24, 25). Many of these methods, in particular, the phenotypic tests, produce results that are not 100% accurate.
Comparative analysis of ribosomal DNA (rDNA) sequences is a well-established method for phylogenetic analysis (3, 26) and has successfully been used to order and, in some cases, to reorder the current taxonomy of microorganisms. For yeasts, the D1-D2 region of 26S rDNA shows a high degree of species variation and has therefore been used not only for systematic studies (7) but also for the development of species-specific probes for identification (9, 20).
Peptide nucleic acid (PNA) is a DNA mimic with a polyamide backbone to which individual nucleobases are attached (11). This structure enables PNA probes to hybridize to complementary nucleic acid targets obeying Watson-Crick base-pairing rules with high specificity and rapid binding kinetics (1). These properties have been applied to a broad range of rapid microbiologic methods (22a). In particular, the relative hydrophobic character of PNA, which allows PNA probes to diffuse through the cell wall under conditions which do not lead to the disruption of cell morphology, has been exploited to develop simple and highly specific culture identification methods based on fluorescent in situ hybridization assays using PNA probes targeting species-specific rRNA sequences (PNA FISH) (15, 20, 21).
In this study, we designed specific PNA probes targeting the rRNAs of C. albicans and C. dubliniensis and used these two PNA probes in a PNA FISH format for differentiation between these two closely related species.
Clinical isolates and reference strains.
Fifteen C. albicans and 6 C. dubliniensis reference strains (Agricultural Research Service Culture Collection [NRRL], Peoria, Ill.) as well as 73 C. dubliniensis and 55 C. albicans clinical isolates (Institute of Medical Microbiology, University Hospital, Aachen, Germany) were used for this study. The C. dubliniensis clinical isolates were mainly from human immunodeficiency virus-positive patients (25) and from respiratory specimens from patients with cystic fibrosis (13). The clinical isolates of C. albicans were chosen to represent different strains, i.e., serotypes A and B, the biovar stellatoidea, and phenotypically aberrant strains such as a red-pigmented strain (6) and strains that failed to assimilate glucosamine and N-acetylglucosamine (17). All strains and isolates were identified by D1-D2 26S rDNA sequence analysis as previously described (7).
For PNA FISH analysis, reference strains and clinical isolates were inoculated into yeast-mold broth (Difco Laboratories, Detroit, Mich.) and incubated overnight at 35°C.
Preparation of smears.
One drop of phosphate-buffered saline was placed in the well of a Teflon-coated microscope slide (Clear Coat; Erie Scientific, Portsmouth, N.H.), and 10 μl of an overnight culture was added, mixed, and spread throughout the well. The smear was fixed either by placing the slide on an 80°C slide warmer for 2 h or by flame fixation by passing the slide through the blue cone of a Bunsen burner. The slide was subsequently immersed 95% ethanol for 1 to 2 min and allowed to air dry.
Selection of probe sequences.
Sequence processing was performed using computer software from DNASTAR (Madison, Wis.) and from the National Center for Biotechnology Information (www.ncbi.nlm.nih.gov/). Alignments of sequence data obtained either from the GenBank database (primarily 18S rRNA sequences) or from the most variable regions of the 26S rRNA were performed using the Megalign (version 4.03) program. From these alignments, PNA probes targeting the 18S rRNA of C. dubliniensis (TAGCCAGAAGAAAGG) and the 26S rRNA of C. albicans (ACAGCAGAAGCCGTG) were identified. The probes were selected to minimize any secondary structure in the probes using the PrimerSelect (version 4.03) program and to achieve Tm values within 68 to 76°C. Finally, each target sequence was checked for specificity against the GenBank database using both the GeneMan (version 3.30) program and an Advanced BLAST search of the GenBank database (www.ncbi.nlm.nih.gov/blast).
Synthesis of fluorescein-labeled PNA probes.
The PNA probes were synthesized at Boston Probes as previously described using an Expedite 8909 nucleic acid synthesis system with PNA option and reagents from Applied Biosystems, Foster City, Calif. (20).
PNA FISH.
PNA FISH was performed as described by Stender et al. (20) with minor modifications. Briefly, smears were covered with approximately 10 μl of hybridization solution, containing 10% (wt/vol) dextran sulfate (Sigma Chemical Co., St. Louis, Mo.), 10 mM NaCl, 30% (vol/vol) formamide (Sigma), 0.1% (wt/vol) sodium pyrophosphate (Sigma), 0.2% (wt/vol) polyvinylpyrrolidone (Sigma), 0.2% (wt/vol) Ficoll (Sigma), 5 mM disodium EDTA (Sigma), 0.1% (vol/vol) Triton X-100 (Aldrich), 50 mM Tris-HCl (pH 7.5), and 100 nM fluorescein-labeled PNA probe targeting C. albicans or 500 nM fluorescein-labeled PNA probe targeting C. dubliniensis. Coverslips were placed on the smears to ensure even coverage with hybridization solution, and the slides were placed on a slide warmer with a humidity chamber (Slidemoat, Boeckel, Germany) and incubated for 30 min at 50°C. Following hybridization, the coverslips were removed by submerging the slides in approximately 20 ml of prewarmed 5 mM Tris (pH 10)–15 mM NaCl–0.1% Triton X-100 per slide in a water bath at 50°C and the slides were washed for 30 min. The slides were then air dried. Each smear was finally mounted using 1 drop of IMAGEN mounting fluid (DAKO, Ely, United Kingdom) and covered with a coverslip. Microscopic examination was conducted using a fluorescence microscope (Optiphot; Nikon Corporation, Tokyo, Japan) equipped with a ×60 1.4 oil objective (Nikon), an HBO 100-W mercury lamp, and a fluorescein isothiocyanate-Texas Red dual-band filter set (Chroma Technology Corp., Brattleboro, Vt.). Images were obtained using a color charge-coupled device camera (Diagnostic Instruments, Inc., Sterling Heights, Mich.) connected to a computer system.
Interpretation of test results.
Two PNA probes in parallel hybridization reactions served as complementary controls such that identification as C. albicans or C. dubliniensis was based on a positive reaction with one PNA probe complemented by a negative reaction with the other PNA probe. Positive reactions were determined as bright green fluorescent cells, whereas negative reactions were determined as nonfluorescent cells with a reddish appearance. The results for double-negative or double-positive samples were inconclusive and were reported as “not identified.”
Fifteen C. albicans and 6 C. dubliniensis reference strains were tested with PNA FISH (Table 1). Of the 15 C. albicans strains, 15 (100%) produced a positive result with the C. albicans probe and a negative result with the C. dubliniensis probe. Of the six C. dubliniensis strains, 6 (100%) produced a negative result with the C. albicans probe and a positive result with the C. dubliniensis probe. In a blind study, 128 clinical isolates representing 73 C. dubliniensis and 55 C. albicans isolates were tested with PNA FISH. The results proved the 100% accuracy, as the 73 C. dubliniensis and 55 C. albicans isolates were correctly identified. As predicted, the C. albicans PNA probe did not cross-hybridize with any of the C. dubliniensis isolates and the C. dubliniensis PNA probe did not cross-hybridize with any of the C. albicans isolates.
TABLE 1.
Results for reference strains analyzed by PNA FISH with C. albicans and C. dubliniensis PNA probes
| Organism | Strain | Resulta obtained with the following PNA probe:
|
|
|---|---|---|---|
| C. albicans | C. dubliniensis | ||
| C. albicans | NRRL Y-79 | + | − |
| C. albicans | NRRL Y-81 | + | − |
| C. albicans | NRRL Y-82 | + | − |
| C. albicans | NRRL Y-107 | + | − |
| C. albicans | NRRL Y-302 | + | − |
| C. albicans | NRRL Y-477 | + | − |
| C. albicans | NRRL Y-6359 | + | − |
| C. albicans | NRRL Y-6943 | + | − |
| C. albicans | NRRL Y-12983 | + | − |
| C. albicans | NRRL Y-17967 | + | − |
| C. albicans | NRRL Y-17968 | + | − |
| C. albicans | NRRL Y-17974 | + | − |
| C. albicans | NRRL Y-17975 | + | − |
| C. albicans | NRRL Y-17976 | + | − |
| C. albicans | NRRL YB-3898 | + | − |
| C. dubliniensis | NRRL Y-17841 | − | + |
| C. dubliniensis | NRRL Y-17512 | − | + |
| C. dubliniensis | NRRL Y-17969 | − | + |
| C. dubliniensis | NRRL Y-17971 | − | + |
| C. dubliniensis | NRRL Y-17972 | − | + |
| C. dubliniensis | NRRL Y-17973 | − | + |
+, positive; −, negative.
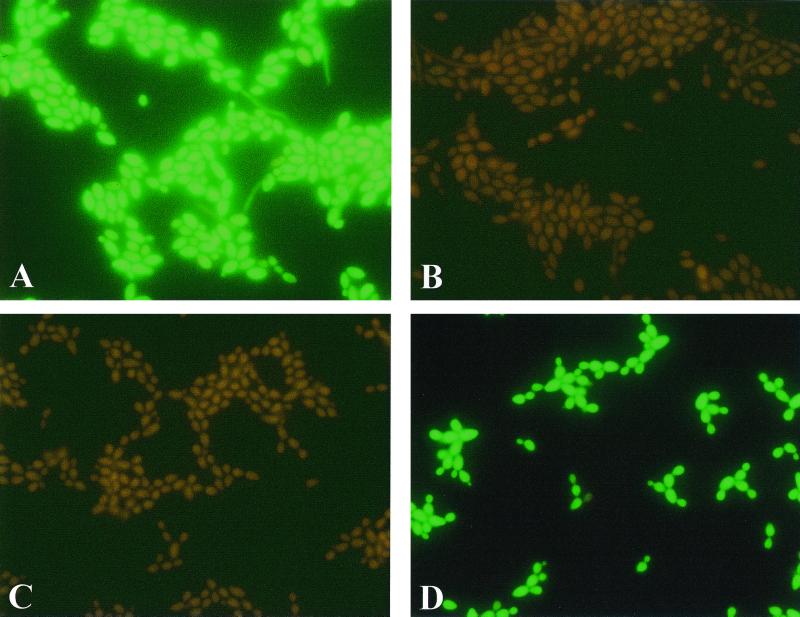
Representative images of assay results are shown in Fig. 1. Negative results were observed as reddish cells, whereas positive results were seen as bright green fluorescent yeasts. In some instances, variable fluorescence was observed between individual yeast cells. This result could have been due to various amounts of rRNA in cells or because of variable permeability of the cell walls.
FIG. 1.
Microscope images of C. albicans analyzed by PNA FISH using the C. albicans PNA probe (A) and the C. dubliniensis PNA probe (B) and C. dubliniensis analyzed by PNA FISH with the C. albicans PNA probe (C) and the C. dubliniensis PNA probe (D).
Conclusion.
We have shown that PNA FISH using PNA probes targeting the rRNAs of C. dubliniensis and C. albicans is a 100% accurate method for the differentiation of C. albicans and C. dubliniensis. The excellent sensitivity and specificity of the assay are typical of other culture identification methods based on PNA FISH (21, 22). The test is performed on smears of cultures, and interpretation of results is conducted by microscopy, such that the PNA FISH procedure simply adds the high specificity of PNA probes to standard microbiological staining procedures to provide definitive identification. These attributes make this method easily adaptable by typical clinical microbiology laboratories. Another benefit of the assay is the use of two PNA probes such that identification is based on the combination of a positive reaction with one PNA probe and a negative reaction with the other PNA probe. This format reduces the risk of false identification, as that would require incorrect reactions by both PNA probes.
The PNA FISH method is intended for differentiation between C. dubliniensis and C. albicans presumptively identified as C. albicans by standard yeast identification methods, such as germ tube analysis or carbon assimilation methods. It is likely that this method can be applied directly to patient specimens, as has been described for PNA FISH of Mycobacterium tuberculosis in sputum smears and biopsy specimens (22, 28). This application is currently being investigated and has the potential to determine the clinical significance of C. dubliniensis. Finally, besides using the described probes to identify mixed cultures upon primary isolation, this method could be complemented by PNA probes targeting other medically important Candida species and other yeasts to provide a general diagnostic tool for definitive identification of yeasts, essential for the optimal selection of antifungal therapy (18).
Acknowledgments
We thank the Chemistry Group at Boston Probes for the synthesis of PNA probes and S. van Oy for excellent assistance.
REFERENCES
- 1.Egholm M, Buchard O, Christensen L, Behrens C, Freier S M, Driver D A, Berg R H, Kim S K, Norden B, Nielsen P E. PNA hybridizes to complementary oligonucleotides obeying the Watson-Crick hydrogen bonding rules. Nature. 1993;365:556–568. doi: 10.1038/365566a0. [DOI] [PubMed] [Google Scholar]
- 2.Elie C M, Lott T J, Reiss E, Morrison C J. Rapid identification of Candida species with species-specific DNA probes. J Clin Microbiol. 1998;36:3260–3265. doi: 10.1128/jcm.36.11.3260-3265.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fox G E, Stackebrandt E, Hespell R B, Gibson J, Maniloff J, Dyer T A, Wolfe R S, Black W E, Tanner R S, Magrum L J, Zablen L B, Blakemore R, Gupta R, Bonen L, Lewis B J, Stahl S A, Luehrsen K R, Chen K N, Woese C R. The phylogeny of prokaryotes. Science. 1980;209:457–463. doi: 10.1126/science.6771870. [DOI] [PubMed] [Google Scholar]
- 4.Jabra-Rizk M A, Falkler W A, Jr, Merz W G, Baqul A A M A, Kelley J I, Meiller T F. Retrospective identification and characterization of Candida dubliniensis isolates among Candida albicans clinical laboratory isolates from human immunodeficiency virus (HIV)-infected and non-HIV-infected individuals. J Clin Microbiol. 2000;38:2423–2426. doi: 10.1128/jcm.38.6.2423-2426.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joly S, Pujol C, Rysk M, Vargas K, Soll D R. Development and characterization of complex DNA fingerprinting probes for the infectious yeast Candida dubliniensis. J Clin Microbiol. 1999;37:1035–1044. doi: 10.1128/jcm.37.4.1035-1044.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kerkmann M L, Schuppler M, Paul K D, Schoenian G, Smith M T. Red-pigmented Candida albicans in patients with cystic fibrosis. J Clin Microbiol. 1999;37:278. doi: 10.1128/jcm.37.1.278-278.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kurtzman C P, Robnett C J. Identification and phylogeny of ascomycetous yeasts from analysis of nuclear large subunit (26S) ribosomal DNA partial sequences. Antonie Leeuwenhoek. 1998;73:331–371. doi: 10.1023/a:1001761008817. [DOI] [PubMed] [Google Scholar]
- 8.Kurzai O, Heinz W J, Sullivan D J, Coleman D C, Frosch M, Mühlschlegel F A. Rapid PCR test for discriminating between Candida albicans and Candida dubliniensis isolates using primers derived from the pH-regulated PHR1 and PHR2 genes of C. albicans. J Clin Microbiol. 1999;37:1587–1590. doi: 10.1128/jcm.37.5.1587-1590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mannarelli B M, Kurtzman C P. Rapid identification of Candida albicans and other human pathogenic yeasts using short oligonucleotides in a PCR. J Clin Microbiol. 1998;36:1634–1641. doi: 10.1128/jcm.36.6.1634-1641.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mosaid A A, Sullivan D, Salkin I F, Shanley D, Coleman D. Differentiation of Candida dubliniensis from Candida albicans on staib agar and caffeic acid-ferric citrate agar. J Clin Microbiol. 2001;39:323–327. doi: 10.1128/JCM.39.1.323-327.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nielsen P E, Egholm M, Buchard O. Peptide nucleic acids (PNA). A DNA mimick with a peptide backbone. Bioconjugate Chem. 1994;5:3–7. doi: 10.1021/bc00025a001. [DOI] [PubMed] [Google Scholar]
- 12.Park S, Wong M, Marras S A E, Cross E W, Kiehn T E, Chaturvedi V, Tyagi S, Perlin D S. Rapid identification of Candida dubliniensis using a species-specific molecular beacon. J Clin Microbiol. 2000;38:2829–2836. doi: 10.1128/jcm.38.8.2829-2836.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Peltroche-Llacsahuanga, H., H. Döhmen, and G. Haase. Recovery of Candida dubliniensis from sputum of cystic fibrosis patients. Mycoses, in press. [PubMed]
- 14.Peltroche-Llacsahuanga H, Schmidt S, Seilbold M, Lütticken R, Haase G. Differentiation between Candida dubliniensis and Candida albicans by fatty acid methyl ester analysis using gas-liquid chromatography. J Clin Microbiol. 2000;38:3696–3704. doi: 10.1128/jcm.38.10.3696-3704.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Perry-O'Keefe, H., S. Rigby, K. Oliveira, D. S. Sørensen, H. Stender, J. Coull, and J. J. Hyldig-Nielsen. Identification of indicator micro-organisms using a standardized PNA FISH method. J. Microbiol. Methods, in press. [DOI] [PubMed]
- 16.Pinjon E, Sullivan D, Salkin I, Shanley D, Coleman D. Simple, inexpensive, reliable method for differentiation of Candida dubliniensis from Candida albicans. J Clin Microbiol. 1998;36:2093–2095. doi: 10.1128/jcm.36.7.2093-2095.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinto de Andrade M, Schönian G, Forche A, Rosado L, Costa I, Muller M, Presber W, Mitchell T G, Tietz H J. Assessment of genetic relatedness of vaginal isolates of Candida albicans from different geographical origins. Int J Med Microbiol. 2000;290:97–104. doi: 10.1016/s1438-4221(00)80112-5. [DOI] [PubMed] [Google Scholar]
- 18.Rowen J L, Tate J M, Nordoff N, Passarell L, McGinnis M R. Candida isolates from neonates: frequency of misidentification and reduced fluconazole susceptibility. J Clin Microbiol. 1999;37:3725–3737. doi: 10.1128/jcm.37.11.3735-3737.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schoofs A, Odds F C, Colebunders R, Ieven M, Goossens H. Use of specialised isolation media for recognition and identification of Candida dubliniensis isolates from HIV-infected patients. Eur J Clin Microbiol Infect Dis. 1997;16:296–300. doi: 10.1007/BF01695634. [DOI] [PubMed] [Google Scholar]
- 20.Stender H, Kurtzman C, Hyldig-Nielsen J J, Sørensen D, Broomer A, Oliveira K, Perry-O'Keefe H, Sage A, Young B, Coull J. Identification of Brettanomyces (Dekkera bruxellensis) from wine by fluorescence in site hybridization using peptide nucleic acid probes. Appl Environ Microbiol. 2001;67:938–941. doi: 10.1128/AEM.67.2.938-941.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stender H, Lund K, Petersen K H, Rasmussen O F, Hongmanee P, Miörner H, Godtfredsen S E. Fluorescence in situ hybridization assay using peptide nucleic acid probes for differentiation between tuberculous and nontuberculous Mycobacterium species in smears of Mycobacterium cultures. J Clin Microbiol. 1999;37:2760–2765. doi: 10.1128/jcm.37.9.2760-2765.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Stender H, Mollerup T A, Lund K, Petersen K H, Hongmanee P, Godtfredsen S E. Direct detection and identification of Mycobacterium tuberculosis in smear-positive sputum samples by fluorescence in situ hybridization (FISH) using peptide nucleic acid (PNA) probes. Int J Tuberc Lung Dis. 1999;3:830–837. [PubMed] [Google Scholar]
- 22a.Stender, H., M. Fiandaca, J. J. Hyldig-Nielsen, and J. Coull. PNA for rapid microbiology. J. Microbiol. Methods, in press. [DOI] [PubMed]
- 23.Sullivan D, Coleman D. Candida dubliniensis: characteristics and identification. J Clin Microbiol. 1998;36:329–334. doi: 10.1128/jcm.36.2.329-334.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sullivan D, Westerneng T J, Haynes K A, Bennett D E, Coleman D C. Candida dubliniensis sp. nov.: phenotypic and molecular characterization of a novel species associated with oral candidosis in HIV-infected individuals. Microbiology. 1995;141:1507–1521. doi: 10.1099/13500872-141-7-1507. [DOI] [PubMed] [Google Scholar]
- 25.Tintelnot K, Haase G, Seibold M, Bergmann F, Staemmler M, Franz T, Naumann D. Evaluation of phenotypic markers for selection and identification of Candida dubliniensis. J Clin Microbiol. 1999;38:1599–1608. doi: 10.1128/jcm.38.4.1599-1608.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Woese C R. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zerbi, P., A. Schønau, S. Bonetto, A. Gari, G. Costanzi, P. Luca, and L. Vago. Amplified in situ hybridization with peptide nucleic acid probes for differentiation of mycobacterium tuberculosis complex and nontuberculosis Mycobacterium species on formalin-fixed, paraffin-embedded archival biopsy and autopsy samples. Am. J. Clin. Pathol., in press. [DOI] [PubMed]