Abstract
Phenotypic susceptibility testing for clarithromycin by E-test and disk diffusion of 109 cultured Helicobacter pylori isolates was compared with the genotypic susceptibility determination by fluorescent in situ hybridization (FISH). No discrepancies were found between these three methods. However, FISH has the advantage of providing results after 3 h.
Clarithromycin is a key component of most treatment recommendations to eradicate Helicobacter pylori (1). Resistance of H. pylori to this antibiotic drug is the most important cause of treatment failure (2, 6, 7). In clinical H. pylori isolates, resistance to clarithromycin is caused predominantly by three distinct point mutations within the peptidyl transferase region of the 23S rRNA (A2143G, A2144G, and A2143C) (8, 10, 14). After culturing the pathogen from gastric biopsies, probably most laboratories use disk diffusion or E-test for the determination of macrolide resistance. Both methods require further subculturing for several days and cannot identify the type of point mutation present in the strain.
Our laboratory has demonstrated the application of fluorescent in situ hybridization (FISH) for simultaneous detection of H. pylori and identification of the 23S rRNA point mutations responsible for macrolide resistance directly in gastric biopsy specimens (13). In a more recent study, we compared FISH and conventional culturing for identification of H. pylori in gastric biopsies (9). The additional application of FISH to patients' biopsies requires a special preparation (e.g., fixation, embedding, and sectioning). For laboratories performing conventional culturing but not FISH directly from biopsy material, it might be an attractive alternative to use this technique for determination of clarithromycin resistance in cultured H. pylori strains, because FISH has the advantage of providing results within hours.
The aim of the present study was to directly compare FISH with E-test and disk diffusion regarding the practicality and reliability of clarithromycin susceptibility testing of cultured H. pylori. The pathogen was isolated from antral biopsies obtained from 109 dyspeptic patients with known H. pylori infection. All oligonucleotide probes used in this study have been previously described and evaluated (13). Briefly, probe Hpy-1 (5′-CACACCTGACTGACTATCCCG-3′) targeted to a 16S rRNA position was used to specifically identify H. pylori, whereas probes ClaR1 (A2143G) (5′-CGGGGTCTTCCCGTCTT-3′), ClaR2 (A2144G) (5′-CGGGGTCTCTCCGTCTT-3′), and ClaR3 (A2143C) (5′-CGGGGTCTTGCCGTCTT-3′) were designed to detect 23S rRNA point mutations responsible for clarithromycin resistance. In contrast, probe ClaWT (5′-CGGGGTCTTTCCGTCTT-3′) was used to identify clarithromycin-sensitive H. pylori strains which had not been detected by either ClaR1, ClaR2, or ClaR3. Oligonucleotide probes were 5′ labeled with the fluorochromes Cy3 (ClaR1, ClaR2, ClaR3, and ClaWT; red signal) or fluorescein isothiocyanate (Hpy-1; green signal). This set of hybridization probes is also commercially available in the creaFAST H. pylori Combi Kit (Oxoid Ltd., Basingstoke, United Kingdom). For hybridization of cultured H. pylori, bacteria were suspended in phosphate-buffered saline, fixed in 4% paraformaldehyde for 30 min at 4°C, subsequently spotted on five glass slides, and then dried for 15 min at 46°C. Dehydration steps were carried out in 50, 80, and 96% ethanol for 3 min each. Glass slides were hybridized with the respective oligonucleotides as previously described (9). Besides specific labeling with fluorescent probes, all samples were subsequently stained with DAPI (4′,6′-diamidino-2-phenylindole), which binds to bacterial DNA and thus helped to localize the microorganisms on glass slides during microscopy (12, 13).
For clarithromycin susceptibility testing of H. pylori by disk diffusion and E-test, colonies from agar plates were suspended into a 0.9% NaCl solution and swabbed onto Mueller-Hinton agar plates supplemented with 5% sheep erythrocytes. After applying 15-μg clarithromycin disks (Oxoid, Wesel, Germany) or clarithromycin E-test strips (AB Biodisk, Solna, Sweden) on the agar surface, plates were incubated in a microaerophilic environment at 37°C for 2 days. Thereafter, zone diameters and the MIC for each strain were determined. Each clarithromycin susceptibility testing was repeated twice.
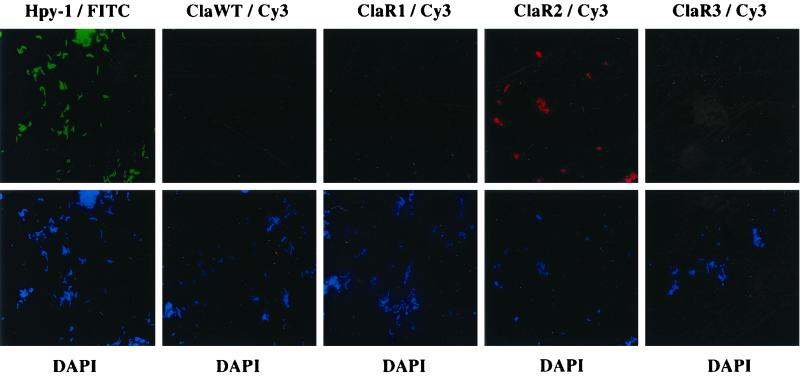
As shown in Table 1, all isolated strains bound to the H. pylori-specific probe Hpy-1. In 75 of these 109 isolates, binding of probe ClaWT but not of ClaR1, ClaR2, or ClaR3 was observed. In contrast, the remaining 34 isolates hybridized either with probe ClaR1, ClaR2, or ClaR3, thus identifying the corresponding point mutation responsible for clarithromycin resistance. Twelve strains harbored the point mutation A2143G, 20 strains carried transition A2144G, and only 2 strains with the transversion A2143C were found. Figure 1 shows typical images obtained from hybridization of probe ClaR2 with a clarithromycin-resistant H. pylori isolate harboring the A2144G point mutation. No cross-reactivity with probes ClaR1, ClaR3, or ClaWT was observed. By applying FISH to all 109 H. pylori strains examined in this study, a specific discrimination between single point mutations causing macrolide resistance and the corresponding wild-type 23S rRNA associated with clarithromycin sensitivity could be performed.
TABLE 1.
Determination of clarithromycin susceptibility of 109 clinical H. pylori isolates by FISH, disk diffusion, and E-test
| No. of H. pylori isolates | Result by:
|
||||||
|---|---|---|---|---|---|---|---|
| FISHa
|
Disk diffusionb | E-testc | |||||
| Hpy-1 | ClaWT | ClaR1 | ClaR2 | ClaR3 | |||
| 4 | Pos. | Pos. | Neg. | Neg. | Neg. | 64 | 0.016 |
| 1 | Pos. | Pos. | Neg. | Neg. | Neg. | 62 | 0.016 |
| 16 | Pos. | Pos. | Neg. | Neg. | Neg. | 60 | 0.016 |
| 6 | Pos. | Pos. | Neg. | Neg. | Neg. | 58 | 0.016 |
| 7 | Pos. | Pos. | Neg. | Neg. | Neg. | 56 | 0.016 |
| 5 | Pos. | Pos. | Neg. | Neg. | Neg. | 54 | 0.016 |
| 11 | Pos. | Pos. | Neg. | Neg. | Neg. | 52 | 0.016 |
| 6 | Pos. | Pos. | Neg. | Neg. | Neg. | 50 | 0.016 |
| 5 | Pos. | Pos. | Neg. | Neg. | Neg. | 48 | 0.016 |
| 1 | Pos. | Pos. | Neg. | Neg. | Neg. | 46 | 0.016 |
| 1 | Pos. | Pos. | Neg. | Neg. | Neg. | 50 | 0.023 |
| 1 | Pos. | Pos. | Neg. | Neg. | Neg. | 46 | 0.023 |
| 1 | Pos. | Pos. | Neg. | Neg. | Neg. | 56 | 0.032 |
| 1 | Pos. | Pos. | Neg. | Neg. | Neg. | 54 | 0.032 |
| 1 | Pos. | Pos. | Neg. | Neg. | Neg. | 52 | 0.032 |
| 1 | Pos. | Pos. | Neg. | Neg. | Neg. | 50 | 0.032 |
| 1 | Pos. | Pos. | Neg. | Neg. | Neg. | 48 | 0.032 |
| 1 | Pos. | Pos. | Neg. | Neg. | Neg. | 46 | 0.032 |
| 2 | Pos. | Pos. | Neg. | Neg. | Neg. | 50 | 0.047 |
| 2 | Pos. | Pos. | Neg. | Neg. | Neg. | 48 | 0.047 |
| 1 | Pos. | Pos. | Neg. | Neg. | Neg. | 44 | 0.094 |
| 12 | Pos. | Neg. | Pos. | Neg. | Neg. | 0 | >256 |
| 4 | Pos. | Neg. | Neg. | Pos. | Neg. | 0 | 16 |
| 1 | Pos. | Neg. | Neg. | Pos. | Neg. | 0 | 24 |
| 2 | Pos. | Neg. | Neg. | Pos. | Neg. | 0 | 32 |
| 2 | Pos. | Neg. | Neg. | Pos. | Neg. | 0 | 64 |
| 11 | Pos. | Neg. | Neg. | Pos. | Neg. | 0 | >256 |
| 2 | Pos. | Neg. | Neg. | Neg. | Pos. | 0 | >256 |
Probe Hpy-1 specifically identifies H. pylori. Probe ClaWT identifies clarithromycin-sensitive strains, whereas probes ClaR1 (A2143G), ClaR2 (A2144G), and ClaR3 (A2143C) identify point mutations responsible for clarithromycin resistance. Pos., positive; Neg., negative.
Values are clarithromycin zone diameters (in millimeters).
Values are clarithromycin MICs (in micrograms per milliliter).
FIG. 1.
Genotypic detection of H. pylori clarithromycin resistance by FISH. (Upper panels) Fluorescein isothiocyanate-labeled oligonucleotide Hpy-1 specifically identifies H. pylori. Cy3-labeled probe ClaR2 detects the A2144G transition responsible for macrolide resistance, whereas oligonucleotides ClaWT/Cy3, ClaR1/Cy3, and ClaR3/Cy3 do not hybridize to the 23S rRNA of the respective strains. (Lower panels) Identification of microorganisms on glass slides by staining of bacterial DNA with DAPI. The microscopic fields correspond to the respective fields of the upper panel.
As determined by FISH, all 75 clarithromycin-sensitive H. pylori isolates underwent phenotypic testing of susceptibility to the macrolide drug using disk diffusion and E-test. Table 1 demonstrates a complete correlation between all three test methods provided that isolates were considered to be resistant when the MICs of clarithromycin were >2 μg/ml. In the disk diffusion test, all 34 clarithromycin-resistant isolates grew right up to the margin of the disk (Table 1). Apparently, in all 14 strains reactive with oligonucleotide probes ClaR1 or ClaR3, the corresponding point mutations at position 2143 correlated with a high MIC of clarithromycin (>256 μg/ml). In contrast, for 20 H. pylori isolates carrying the point mutation at position 2144, the MICs ranged from 16 μg/ml (4 strains), 24 μg/ml (1 strain), 32 μg/ml (2 strains), 64 μg/ml (2 strains), to >256 μg/ml (11 strains).
Methods for the detection of point mutations in H. pylori at the molecular level are mainly based on PCR and subsequent restriction length polymorphism or colorimetric hybridization of amplified DNA fragments (3, 4, 7, 8, 10, 11, 14). Analysis of amplicons with restriction enzymes BsaI or BbsI specifically detects the A→G transition at positions 2143 and 2144 but not the A2143C point mutation of the 23 rRNA (7, 14). In contrast, all three relevant mutations within the peptidyl transferase region are detectable by the application of a PCR-oligonucleotide ligation assay (10) or a PCR hybridization approach (8). In a recent study, a new and rapid method of detecting A2143G and A2144G point mutations in the 23S rRNA gene of H. pylori by using a LightCycler has been reported (5).
The detection of pathogen-specific DNA and genotypic determination of H. pylori macrolide resistance directly in gastric biopsies are useful tools in the microbiology laboratory. Nevertheless, conventional culturing of H. pylori still appears to be mandatory because isolated strains should be used for susceptibility testing with all relevant drugs and for epidemiological studies. Clearly, the disk diffusion test is the most cost-effective and simplest method for the screening of clarithromycin resistance in H. pylori. By applying the more expensive E-test to cultured bacteria, an accurate MIC of the macrolide can be determined. Results from disk diffusion and E-test are available after 2 days of incubation. In contrast, the application of FISH to cultured H. pylori and the microscopic examination of hybridized slides are performed in approximately 3 h and are very easy to carry out. In summary, FISH is a rapid and accurate but also cost-effective method for detection of macrolide resistance in cultured H. pylori, thereby contributing to facilitating the choice of an appropriate treatment regimen for the individual patient.
Acknowledgments
H.R. was supported by the AIDS-Stipendienprogramm from the Bundesministerium für Bildung, Wissenschaft, Forschung und Technologie of Germany.
REFERENCES
- 1.European Helicobacter Pylori Study Group. Current European concepts in the management of Helicobacter pylori infection. The Maastricht Consensus Report. Gut. 1997;41:8–13. doi: 10.1136/gut.41.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Graham D Y. Antibiotic resistance in Helicobacter pylori: implications for therapy. Gastroenterology. 1998;115:1272–1277. doi: 10.1016/s0016-5085(98)70100-3. [DOI] [PubMed] [Google Scholar]
- 3.Maeda S, Yoshida H, Matsunaga H, Ogura K, Kawamata O, Shiratori Y, Omata M. Detection of clarithromycin-resistant Helicobacter pylori strains by a preferential homoduplex formation assay. J Clin Microbiol. 2000;38:210–214. doi: 10.1128/jcm.38.1.210-214.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Maeda S, Yoshida H, Ogura K, Kanai F, Shiratori Y, Omata M. Helicobacter pylori specific nested PCR assay for the detection of 23S rRNA mutation associated with clarithromycin resistance. Gut. 1998;43:317–321. doi: 10.1136/gut.43.3.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Matsumura M, Hikiba Y, Ogura K, Togo G, Tsukuda I, Ushikawa K, Shiratori Y, Omata M. Rapid detection of mutations in the 23S rRNA gene of Helicobacter pylori that confers resistance to clarithromycin treatment to the bacterium. J Clin Microbiol. 2001;39:691–695. doi: 10.1128/JCM.39.2.691-695.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Megraud F. Antibiotic resistance in Helicobacter pylori infection. Br Med Bull. 1998;54:207–216. doi: 10.1093/oxfordjournals.bmb.a011671. [DOI] [PubMed] [Google Scholar]
- 7.Occhialini A, Urdaci M, Doucet-Populaire F, Bebear C M, Lamouliatte H, Megraud F. Macrolide resistance in Helicobacter pylori: rapid detection of point mutations and assays of macrolide binding to ribosomes. Antimicrob Agents Chemother. 1997;41:2724–2728. doi: 10.1128/aac.41.12.2724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pina M, Occhialini A, Monteiro L, Doermann H P, Megraud F. Detection of point mutations associated with resistance of Helicobacter pylori to clarithromycin by hybridization in liquid phase. J Clin Microbiol. 1998;36:3285–3290. doi: 10.1128/jcm.36.11.3285-3290.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rüssmann H, Kempf V A J, Koletzko S, Heesemann J, Autenrieth I B. Comparison of fluorescent in situ hybridization and conventional culturing for detection of Helicobacter pylori in gastric biopsy specimens. J Clin Microbiol. 2001;39:304–308. doi: 10.1128/JCM.39.1.304-308.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stone G G, Shortridge D, Versalovic J, Beyer J, Flamm R K, Graham D Y, Ghoneim A T, Tanaka S K. A PCR-oligonucleotide ligation assay to determine the prevalence of 23S rRNA gene mutations in clarithromycin-resistant Helicobacter pylori. Antimicrob Agents Chemother. 1997;41:712–714. doi: 10.1128/aac.41.3.712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Szczebara F, Dhaenens L, Vincent P, Husson M O. Evaluation of rapid molecular methods for detection of clarithromycin resistance in Helicobacter pylori. Eur J Clin Microbiol Infect Dis. 1997;16:162–164. doi: 10.1007/BF01709478. [DOI] [PubMed] [Google Scholar]
- 12.Trebesius K, Harmsen D, Rakin A, Schmelz J, Heesemann J. Development of rRNA-targeted PCR and in situ hybridization with fluorescently labeled oligonucleotides for detection of Yersinia species. J Clin Microbiol. 1998;36:2557–2564. doi: 10.1128/jcm.36.9.2557-2564.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Trebesius K, Panthel K, Strobel S, Vogt K, Faller G, Kirchner T, Kist M, Heesemann J, Haas R. Rapid and specific detection of Helicobacter pylori macrolide resistance in gastric tissue by fluorescent in situ hybridisation. Gut. 2000;46:608–614. doi: 10.1136/gut.46.5.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Versalovic J, Shortridge D, Kibler K, Griffy M V, Beyer J, Flamm R K, Tanaka S K, Graham D Y, Go M F. Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob Agents Chemother. 1996;40:477–480. doi: 10.1128/aac.40.2.477. [DOI] [PMC free article] [PubMed] [Google Scholar]