Summary
Background
Sensitivity has been a key issue for Enhancer of zeste homolog 2 (EZH2) inhibitors in cancer therapy. The EZH2 inhibitor EPZ-6438 was first approved by the US Food and Drug Administration (FDA) in 2020. However, its inadequate anti-cancer activity in solid tumors limits its clinical application. In this study, we utilized the multiple cancer cell lines, which are less sensitive to the EZH2 inhibitor GSK126, combining animal model and clinical data to investigate the underlying mechanism.
Methods
IncuCyte S3 was used to explore the difference in the responsiveness of hematological tumor cells and solid tumor cells to GSK126. Transcriptome and metabolome of B16F10 cells after GSK126 treatment were analyzed and the distinct changes in the metabolic profile were revealed. Real-time quantitative PCR and western blot experiments were used to further verify the multi-omics data. ChIP-qPCR was performed to detected H3K27me3 enrichment of target genes. Finally, the anti-tumor effects of combining GSK126 and lipid metabolism drugs were observed with IncuCyte S3 platform, CCK-8 and animal model respectively.
Findings
We found that although the proliferative phenotype did not show strong difference upon treatment with GSK126, the transcriptome and metabolome changed profoundly. GSK126 treatment led to broad shifts in glucose, amino acid, and lipid metabolism. Lipid synthesis was strengthened manifested by the increasing abundance of unsaturated fatty acids. SCD1 and ELOVL2 were regulated by H3K27me3 at gene regulatory region, and upregulated by EZH2 knockdown and inhibitors. SCD1 knockdown increased cellular sensitivity to GSK126. Based on the findings above, the application of the combination with SCD1 inhibitor significantly attenuated the proliferation of cancer and increased the sensitivity to GSK126 by suppressing desaturation of fatty acids.
Interpretation
Dysregulated lipid metabolism can blunt the sensitivity of cancer cells to GSK126. These characteristics shed light on the novel combination therapy strategies to combat tumor resistance.
Funding
National Natural Science Foundation of China (No. 81672091, No.91749107 and No. 81972966).
Keywords: EZH2, SCD1, Lipid metabolism, Combination treatment, GSK126
Abbreviations: EZH2, Enhancer of zeste homolog 2; EZH2i, EZH2 inhibitors; FA, fatty acid; MUFA, monounsaturated fatty acid; PRC2, polycomb repressive complex 2; PUFA, polyunsaturated fatty acid; SCD1, stearoyl-CoA desaturase 1
Research in context.
Evidence before this study
EZH2 has been found closely correlated with the majority of tumor genesis and progression. However, the effectiveness in previous clinical trials targeting EZH2 was not satisfactory, especially in solid tumors. Increasing evidence has suggested that EZH2 is involved in altering the metabolic profiles of tumor cells by multiple pathways, which cover glucose, lipid and amino acids metabolism. EZH2 methyltransferase activity can facilitate adipogenesis. Severe defects of adipogenesis in Ezh2−/− mouse primary preadipocytes have been identified. However, inhibition of EZH2 induces lipid accumulation in nonalcoholic fatty liver and certain cancer cells such as breast cancer. Cancer cells often activate de novo fatty acid (FA) synthesis to provide a constant supply of FAs for membrane biogenesis, energy production and protein modification. Currently, it is not clear how tumor lipid metabolism affects the efficacy of EZH2 inhibitors (EZH2i).
Added value of this study
This is the first time of describing the effect of EZH2i on the metabolic profile comprehensively in cancer cells. GSK126 enhances lipid synthesis and increases the content of unsaturated fatty acids which blunts its capacity in anti-tumor effect. In addition, we identified SCD1 and ELOVL2 as new target genes of EZH2 via H3K27me3 modification. Finally, we proposed a new strategy for enhancing the sensitivity of GSK126 by applying the SCD1 inhibitor MF-438, and it showed more promising therapeutic effect in vivo and in vitro experiments.
Implications of all the available evidence
This study uncovered the Achilles heel of EZH2i and found a complementary combination approach to enhance the power of EZH2i in cancer therapy by applying lipolysis agents. This combination therapy provides a novel strategy to combat the EZH2i drug resistance and shows the potential for future clinical applications.
Alt-text: Unlabelled box
Introduction
Enhancer of zeste homolog 2 (EZH2) is the catalytic subunit of the polycomb repressive complex 2 (PRC2) and functions as a histone methyltransferase. PRC2 catalyzes mono-, di-, and trimethylation of histone H3K27, leading to transcriptional silencing of target genes. EZH2 overexpression is frequently observed in multiple cancer subtypes and is correlated with tumorigenesis, metastasis, and prognosis.1 Therefore, numerous EZH2 inhibitors (EZH2i) have been developed as potential anticancer treatments. Among them, GSK126 and CPI1205 have been tested in clinical trials, and EPZ-6438 (tazemetostat) was approved by the FDA for the treatment of epithelioid sarcoma in 2020.2,3 However, the application of EZH2i is limited and the antitumor effect of EZH2i alone in solid tumors remains unsatisfactory.
Emerging evidence has revealed that EZH2 is involved in the regulation of various metabolism pathways.4 For example, EZH2 can facilitate tumor progression by promoting aerobic glycolysis.5 EZH2 knockdown diminished the expression level of fatty acid synthase (FASN) and reduced the abundance of intracellular fatty acids (FAs) in gliomas.6 However, inhibition of EZH2 induces lipid accumulation in adipocytes7 and breast cancer cells.8 EZH2 inactivation enhanced branched-chain amino acid (BCAA) metabolism by upregulating BCAA transferase 1 (BCAT1) expression in leukemia.9 Whether the alteration in the metabolic profile imposed by EZH2 or its inhibitors affects tumor genesis and progression is far from clear. More comprehensive studies are needed to investigate the effects of EZH2 on tumor metabolism in various tissues, which is beneficial for the accurate medication in the clinical practice of EZH2i.
Reprogramming of lipid metabolism in tumors are now frequently discussed and well recognized. The typical lipid metabolism characteristic of cancer cells is that excessive lipids and cholesterol are stored in lipid droplets (LDs), and high LDs and cholesterol ester content in tumors are now considered an indicator of cancer aggressiveness.10 Cancer cells often activate de novo FA synthesis to provide a constant supply of FAs for membrane biogenesis, energy production and protein modification.11 Increased lipogenesis is coordinated with upregulation of the key enzymes involved in the conversion of glucose into FAs, such as ATP citrate lyase (ACLY), acetyl-CoA carboxylase (ACAC), FASN, and stearoyl-CoA desaturase 1 (SCD1),etc.12 Fatty acid elongase such as ELOVL5 has been found overexpressed and regulated the oxidative stress in cancer.13 Given this fact, targeting metabolic regulation might pave the way for increasing the effectiveness of traditional chemotherapy.
Metabolism is a network that interacts with multiple metabolites and pathways. The overall pattern should be considered when focusing on a single metabolic pathway. Therefore, in this study, we first took a glance at the metabolism profile affected by EZH2i in a cancer model by multi-omics analysis. We found that FA biosynthesis was upregulated, accompanied with downregulation of the tricarboxylic acid (TCA) cycle and amino acid levels in EZH2i-treated cancer cells. SCD1 and ELOVL2 were identified as target genes of EZH2 via H3K27me3 modification. In terms of the prominent changes in lipid metabolism, we further combined the SCD1 inhibitor MF-438 with GSK126 and found it improved the inhibition effect of EZH2i on cancer cells significantly. Therefore, these findings suggest that dysregulated lipid metabolism in tumor cells could be the Achilles heel of EZH2i, while combining certain lipid-regulatory drugs may provide a potential strategy to optimize its effect for further clinical application.
Materials and methods
Cell culture
Human liver cancer cell lines SMMC7721 and Huh7, human melanoma cell line A375 and A875, human Burkitt lymphoma cell line Daudi, human monocytic leukemia cell line THP-1, human embryonic kidney (HEK) 293T cells, mouse melanoma cell line B16F10, and mouse colorectal cancer cell line MC38 were purchased from the Cell Resources Center of Peking Union Medical College (Beijing, China). Cell lines were grown in Dulbecco's modified Eagle's medium (DMEM, HyClone, Cat # SH30023.01) or RPMI-1640 (HyClone, Cat # SH30809.01) supplemented with 10% fetal bovine serum (FBS, GIBCO, Cat # 10099141), 50 IU/mL penicillin, and 50 mg/mL streptomycin (GIBCO, Cat # 15140122). All cells were cultured at 37 °C in a humidified incubator in the presence of 5% CO2.
All cell lines were validated by STR analysis at the Cell Resources Center of Peking Union Medical College and regularly tested for mycoplasma contamination (culture testing).
Cell proliferation analysis
IncuCyte S3 platform and CCK-8 assay
Cells were seeded in 96-well plates (about 5000 cells per well) and treated with GSK12614 or EPZ643815 after 24 h. The experiment was performed in quintuplicate. Then imaged cells using phase contrast channel in the IncuCyte S3 platform (Sartorius, Göttingen, Germany). Four sets of phase contrast images from distinct regions within each well were taken at intervals of 3 h using a 10X objective. IncuCyte S3 image analysis software was set to detect the edges of the cells and to determine their confluence in percentage.
Cell viability was also measured by CCK-8 assay. After culture in 96-well plates for 24, 48 or 72 h, 10 μL of CCK-8 (Dojindo Laboratories, Rockville, MA, USA) was added to each well, and the absorbance was measured at 450 nm one hour later using the microplate reader. The combination index (CI) was calculated by CompuSyn software. A CI < 0.9 indicates synergism and CI > 1.1 an antagonistic effect.
Preparation of palmitic acid and stearic acid
Palmitic acid (Sigma, Cat # P5585) and stearic acid (TRC, Cat # S686495) was conjugated to fatty acid–free (FFA) bovine serum albumin (BSA) (Sigma, Cat # A0281). Palmitic acid (51.2 mg) or stearic acid (56.9 mg) was dissolved in 100% ethanol (1 ml) to make 200 mM of stock solution. About 10% FFA low-endotoxin BSA was prepared in DMEM. 0.04 ml of 200 mM palmitate or stearic acid and 1.96 ml of 10% FFA BSA were mixed for at least 2 h to generate 4 mM palmitate stock solution.
Wound healing assay
Cells were seeded in 96-well plates. When cells growth reached 90–100% confluence, the IncuCyte® WoundMaker tool was used to create precise scratches within a cell monolayer. Wells were washed with PBS once, and the medium was changed to serum-free GSK126 treatment medium. Then imaged cells using phase contrast channel in the IncuCyte S3 platform (Sartorius, Göttingen, Germany). Two sets of phase contrast images from distinct regions within each well were taken at intervals of 3 h using a 10X objective. The scratched area was measured by Image J software.
Apoptosis assay and Annexin V staining assay
Fluorescein Isothiocyanate (FITC)-conjugated Annexin V (Biolegend, Cat #640906) and 7-amino-actinomycin D (7-AAD) kit (Biolegend, Cat #420403) was used to detect the apoptotic cells according to the manufacturer's instructions. After the treatment of 10 μM GSK126 for 48 h, the B16F10 cells were harvested with trypsin and washed twice with PBS. Cells were then resuspended in 1x binding buffer at a concentration of 1 × 106 cells/mL. Next, 100 μL of the solution (1 × 105 cells) was transfered to a 5 ml culture tube and 5 μL of FITC Annexin V and 5 μL 7-AAD were added into the solution. The mixture was incubated for 15 min at RT (25°C) in the dark. 200 μL of 1 x Binding Buffer was added to each tube and the apoptosis rate of the cells was analyzed by flow cytometry within 1 h.
Annexin V staining assay was detected by IncuCyte S3 platform (Sartorius, Göttingen, Germany). Cells were seeded in 96-well plates and treated with GSK126 and appropriate FITC Annexin (with binding buffer) on the next day. Four sets of phase contrast images from distinct regions within each well were taken at intervals of 6 h using a 10X objective with phase and green image channels.
Real-time quantitative PCR (real-time qPCR)
Real-time qPCR was performed using the Applied Biosystems QuantStudio 5 system (Applied Biosystems, CA, USA). Actin beta (ACTB) was included as a house keeping gene control to normalize expression levels. The primers utilized for qPCR are presented in Table 1.
Table 1.
Sequence of primers used in RT-qPCR.
| Species | Gene | Primer sequence (5′-3′) |
|---|---|---|
| Human |
ACTB |
F:TGTTACAGGAAGTCCCTTGCC |
| R:ATGCTATCACCTCCCCTGTGTG | ||
|
EZH2 |
F:GTACACGGGGATAGAGAATGTGG |
|
| R:GGTGGGCGGCTTTCTTTATCA | ||
|
FASN |
F:ACAGCGGGGAATGGGTACT |
|
| R:GACTGGTACAACGAGCGGAT | ||
|
ACLY |
F:GCTCAGCCAGAACTTGGTAGTC |
|
| R:CTTGCCAACTGTGGCTTCCTG | ||
|
ACACA |
F:CCTGAAGACCTTAAAGCCAATGC |
|
| R:AGCCCACACTGCTTGTACTG | ||
|
ACACB |
F:GGGTCATCGAGAAGGTGCTTAT |
|
| R:GATGTACTCTGCGTTGGCCTTA | ||
|
SCD |
F:TCCAGAGGAGGTACTACAAACCT |
|
| R:CCGGGGGCTAATGTTCTTGT | ||
|
ABCA1 |
F:CTCCACAAGGTATTTTTGCAAGGC |
|
| R:AGTTTAGTGTTCTCAGGATTGGCT | ||
|
ACSL6 |
F:AAGATTCAGGCCAGTCTTGGTG |
|
| R:TAACCTTCATAAACCTGGCACCC | ||
|
ELOVL2 |
F:ATGTTTGGACCGCGAGATTCT |
|
| R:CCCAGCCATATTGAGAGCAGATA | ||
| Mouse |
Actb |
F:AGGGTGTGATGGTGGGAATG |
| R:CCAGTTGGTAACAATGCCATGT | ||
|
Ezh2 |
F:GCATGGTGACAGAGAATGTGG |
|
| R:ATTTCCGAGGTGGGCAAGTT | ||
|
Acsl6 |
F:AAGAAGTGGAGGACGGTGGT |
|
| R:GGGTCCATTCCCTGAGATGCTA | ||
|
Abca1 |
F:ACATGAGTGCCACTTTCCGAAT |
|
| R:GCGAGACACGATGGACTTGTT | ||
|
Slc27a1 |
F:GGCATGGATGATCGGCTGTTT |
|
| R:TGATGTTCCCTGCAGAGTGGTA | ||
|
Fasn |
F:GACCTCAGGCTGCAGTGAAT |
|
| R:TGGTTCCCGGATCACCTTCT | ||
|
Scd1 |
F:TCCCTCCGGAAATGAACGAG |
|
| R:GTAGAAAATCCCGAAGAGGCAGG | ||
| Elovl2 | F:CCTGCTCTCGATATGGCTGG |
|
| R:AAGAAGTGTGATTGCGAGGTTAT | ||
Data were analyzed by ΔΔCt method and gene expression was normalized to ACTB or Actb.
Western blot analysis
Western blot was performed using the cell lysate, according to standard protocols. Briefly, proteins were separated by SDS-PAGE and transferred to nitrocellulose filter membranes; subjected to immunoblotting with antibodies. The following antibodies were used: α-tubulin (Cell Signaling Technology, Cat # 3873S, RRID:AB_1904178, 1:5000), ACLY (Abcam, Cat # ab40793, RRID:AB_722533, 1:1000), SCD1 (Abcam, Cat # ab236868, 1:1000), FASN (Cell Signaling Technology, Cat # 3180S, RRID:AB_2100796, 1:1000), ELOVL2 (Abcam, Cat #ab176327, 1:1000), H3 (Proteintech, Cat # 17168-1, RRID:AB_2716755, 1:1000), H3K27me3 (Abcam, Cat # ab6002, RRID:AB_305237, 1:1000), β-ACTIN (Proteintech, Cat # 66009-1, RRID:AB_2687938, 1:2000), EZH2(CST, Cat # 5246S, RRID:AB_10694683, 1:2000). Secondary antibodies conjugated to LI-COR IRDye were obtained from LICOR Biosciences (Lincoln, Cat # 925-68071, RRID:AB_2721181, Cat # 925-68072, RRID:AB_2814912, 1:10000). The membranes were visualized using Odyssey Imager (LICOR Biosciences). All antibodies were validated by the commercial vendor.
Sample preparation, RNA sequencing and data processing
After treated with DMSO or GSK126 for 48 h, B16F10 cells were harvested, and total RNA was isolated using RNeasy Mini Kit (Qiagen). Paired-end libraries were synthesized by using the TruSeq ® RNA Sample Preparation Kit (Illumina, USA) following TruSeq ® RNA Sample Preparation Guide. Briefly, the poly-A containing mRNA molecules were purified using poly-T oligo-attached magnetic beads. The purified mRNA was used to construct cDNA library and quantified by Qubit® 2.0 Fluorometer (Life Technologies, USA) and validated by Agilent 2100 bioanalyzer (Agilent Technologies, USA) to confirm the insert size and calculate the mole concentration. cDNA libraries were sequenced on the Illumina Novaseq 6000 (Illumina, USA) at Shanghai Biotechnology Corporation. Basecalls was performed using CASAVA (version 1.8). Reads were aligned to the GRCm38.p4 (mm10) genome assembly using Hisat2 (version:2.0.4). Uniquely mapped reads with mapping quality >=20 were retrieved for downstream analysis. Reads Per Kilobase of exon per Megabase of library size (RPKM) were calculated. In short, exons from all isoforms of a gene were merged to create one meta-transcript. The number of reads falling in the exons of this meta-transcript were counted and normalized by the size of the meta-transcript and by the size of the library. Principal component analysis was performed by R software (version: 4.0.2). The differential gene expression analysis was based on the negative-binomial statistical model of read counts as implemented in the edgeR package. Differential expressions with FDR-adjusted p-values less than 0.05 and log2 fold change more than 1.0 were considered to be statistically significant. Gene enrichment analysis of Gene Ontology and KEGG pathways was conducted on gene set enrichment analysis software (http://www.broadinstitute.org/gsea/index.jsp) to identify a priori-defined gene sets that show statistically significant differences between control and GSK126-treated cells.
The accession number for the RNA-seq data reported in this paper is NCBI GEO: GSE163078.
Sample preparation and targeted metabolic profiling
After treated with DMSO or GSK126 for 48 h, cells were harvested and mixed with 10 pre-chilled zirconium oxide beads and 20 μL of deionized water in an Eppendorf Safelock microcentrifuge tube. The sample was homogenated for 3 min and 150 μL of Methanol containing internal standard was added to extract the metabolites. The sample was homogenated for another 3 min and then centrifuged at 18,000 g for 20 min. Then the supernatant was transferred to a 96-well plate. The following procedures were performed on a Biomek 4000 workstation (Biomek 4000, Beckman Coulter, Inc., Brea, California, USA). 20 μL of freshly prepared derivative reagents was added to each well. The plate was sealed and the derivatization was carried out at 30°C for 60 min. After derivatization, the sample was evaporated for 2 h. 330 μL of ice-cold 50% methanol solution was added to reconstitute the sample. Then the plate was stored at -20°C for 20 min and followed by 4000 g centrifugation at 4 °C for 30 min. 135 μL of supernatant was transferred to a new 96-well plate with 10 μL internal standards in each well. Serial dilutions of derivatized stock standards were added to the left wells. Finally, the plate was sealed for LC-MS analysis. All of the standards were obtained from Sigma-Aldrich, Steraloids Inc. (Newport, RI, USA) And TRC Chemicals (Toronto, ON, Canada). An ultra-performance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS) system (ACQUITY UPLC-Xevo TQ-S, Waters Corp., Milford, MA, USA) was used to quantitate all targeted metabolite s in this project.
Metabolome analysis
The raw data files generated by UPLC-MS/MS were processed using the MassLynx software (v4.1, Waters, Milford, MA, USA) to perform peak integration, calibration, and quantitation for each metabolite. was performed by R software (version: 4.0.2). SIMCA-P 14.0 (Umetrics, Umea, Sweden) software was used to conduct principal component analysis (PCA) and orthogonal partial least square discriminant analysis (OPLS-DA). To validate the model against over-fitting, a 7-round cross-validation and response permutation testing was carried out. Cumulative values of R2X, R2Y and Q2Y close to 1.0 indicate an excellent model with a reliable predictive ability. Those variables with variable influence on projection (VIP) greater than 1.0 are considered significantly different between classes. Metabolic pathway enrichment analysis was conducted on the MetaboAnalyst website (http://www.metaboanalyst.ca).
Plasmids and cell transfection
siRNA of negative control (si-NC) and EZH2 (si-EZH2#2-3, mouse) were purchased from Ribobio Biotech (Guangzhou, China). The plasmids sh-NC, sh-EZH2#1-3 (human), sh-Scd1#1-3 (mouse) were purchased from Shanghai Genechem (Shanghai, China). All plasmids were transfected into B16F10 and A375 cells with Lipofectamine 3000 (Thermofisher, Cat # L3000001). Tumor cells were seeded into the 6-well plates (about 3 × 105 cells per well) and left overnight. 4 μg plasmids and 10 μL lipofectamine reagent per well were used to perform cell transfection according to the manufacturer's instruction manual. To obtain stable cell lines, the lentiviral expression vector (sh-Scd1) and packaging vectors (pMD2.G and psPAX) were transfected into HEK293T cells to produce virus. After collecting the viral supernatant fractions at 48 h and 72 h, B16F10 cells were infected and screened with puromycin.
Chromatin immunoprecipitation (ChIP) assay
A375 cells plated in 10 cm dishes were treated with DMSO or 10 μM GSK126 for 48 h respectively. Then cells were fixed with 1% formaldehyde for 10 min at room temperature. To stop the reaction, glycine was added to a final concentration of 0.125 M at room temperature for 5 min. Cells were scraped into cold PBS with proteinase inhibitor and transferred with contents of each group to a 15 mL tube. ChIP assay was performed using SimpleChIP Plus Enzymatic Chromatin IP Kit (Magnetic Beads) (Cell Signaling Technology, #9005) and anti-Histone H3K27me3 (Cell Signaling Technology, #9733) according to the procedures provided by the manufacturer. The final ChIP DNA samples were then used as templates in qPCR reactions. Primers were designed according to our previous ChIP-seq results.16 The sequences of the primers were as follows:
| Primers | Sequences (5’–3’) | Position from TSS |
| ELOVL2 peak20688-F | GAGGCCACCGTTCTGTTCA | -701 ∼ -576 |
| ELOVL2 peak20688-R | GCGGATCAGTTCGGATAACG | |
| ELOVL2 peak8265-F1 | AGCGGCTGGGTTTCTATCAG | 679 ∼ 763 |
| ELOVL2 peak8265-R1 | GGGAAATCGGGCAGAGAGAG | |
| ELOVL2 peak8265-F2 | GTCAAGCTCTTGCCCCTCTC | 580 ∼ 712 |
| ELOVL2 peak8265-R2 | TCCGAGCCCAGACACTGATA | |
| SCD1 peak3138-F1 | AGTGGCCAGTGACAAACACA | -19422 ∼ -19311 |
| SCD1 peak3138-R1 | TGCAAGCCCTCTAGGAAAGC | |
| SCD1 peak3138-F2 | AGCTTTCCTAGAGGGCTTGC | -19331 ∼ -19182 |
| SCD1 peak3138-R2 | TAGGTGTTCTAGGCTCCGCA | |
| SCD1 peak1160-F | CCTTCCTTGCCCATCACCTT | -17807 ∼ -17691 |
| SCD1 peak1160-R | CTCTACAAGCCAGGGCCTTC | |
In vivo experiments in mice
Ethics statement
All experiments were performed in accordance with the “Guide for the Care and Use of Laboratory Animals” issued by the National Institutes of Health (NIH publication no. 85–23, revised 1996). The experimental protocols approved by the Peking University Third Hospital Medical Science Research Ethics Committee (permission No. S2021094). All of mice were fed and housed in the Peking University Health Science Center Department of Laboratory Animal Science.
Histopathological examination, lipid accumulation evaluation in mice
Six to eight-week-old female C57BL/6 mice were purchased from and raised in the Peking University Health Science Center Department of Laboratory Animal Science. GSK126 (Selleck, Cat # S7061) or vehicle was administered intraperitoneally at a dose of 100 mg/kg. B16F10 cells were implanted subcutaneously in female C57BL/mice (n = 4–6 mice/group). Then GSK126 treatment (three times/four days) was initiated once tumor volume was approximately 50 mm3. Mice were sacrificed after 12 days of drug treatment. Liver samples were excised and embedded in Tissue-Tek OCT compound for histopathological analysis. The OCT-embedded samples were serially sectioned at 10 μm. Oil Red O staining was performed for the evaluation of lipid accumulation, and results were photographed under microscope. The tissue sections were also subjected to hematoxylin and eosin (H&E) staining, and the results were scanned by Nano Zoomer SQ (Hamamatsu, Japan). The positive or negative area of staining were measured by Image J software. Collected retro-orbital blood samples were and send them to Peking University Health Science Center Department of Laboratory Animal Science for biochemical analysis (TG etc.).
Combination treatment in vivo
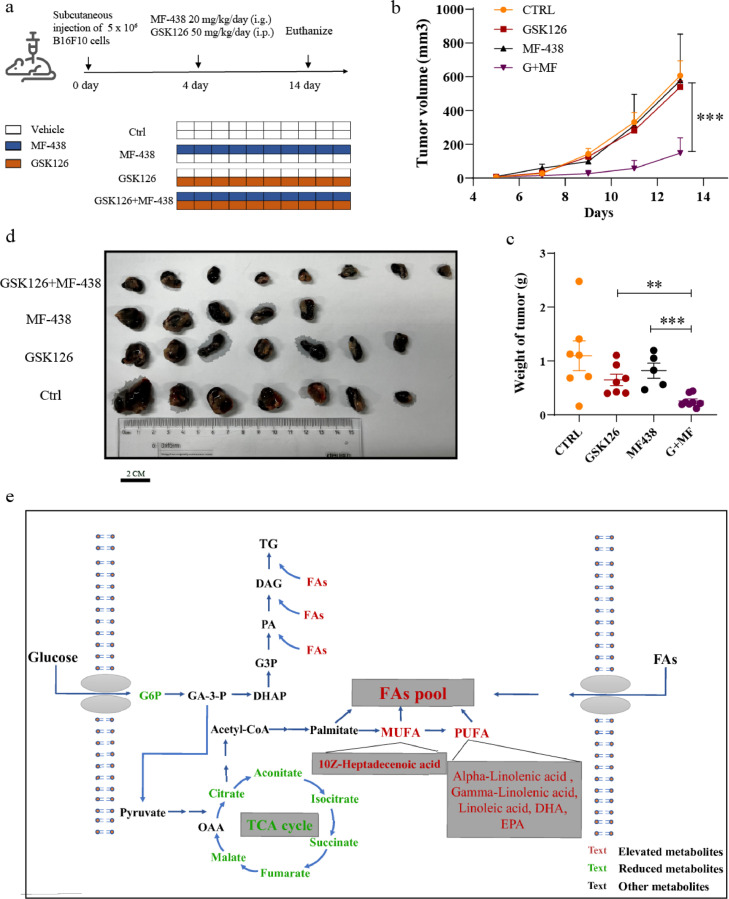
To explore the therapeutic effect of combination treatment in vivo, six to eight-week-old female C57BL/6 mice (6-8 weeks of age, female) were applied. At the weight of 20–22 g, mice were subcutaneously injected in the right flank with 5 × 105 B16F10 cells in 100 μL PBS. When tumors reached an average volume of 50 mm3, mice were allocated to experimental groups (n = 5-8 mice/group) and treated with vehicle or drugs (Figure 6a). Intragastric administration of MF-43817 (20 mg/kg, MCE, Cat # HY-15822) per day. GSK126 (50 mg/kg, Selleck, Cat # S7061) was injected intraperitoneally per day. Fenofibrate18 or GSK126 was dissolved in vehicle (10% DMSO + 40% PEG300 + 5% Tween-80 + 45% saline). The combination treatment group used the equal dose of MF-438 (20 mg/kg) and GSK126 (50 mg/kg) per day. After 2 weeks, mice were sacrificed and the subcutaneous tumor tissues were removed. Tumor volume was monitored each two days using a digital caliper. Tumor volume = Length*Width* Width/2.
Figure 6.
Combination treatment of GSK126 and MF-438 reduced the growth of B16F10 cells in vivo. (a) Schematic diagram of tumor bearing experiment in mice. (b, c) Tumor growth curve (b) and tumor weight (c) for mice carrying B16F10.The tumor volume was measured with a vernier caliper every 2 days. Tumor volume = Length*Width*Width)/2. (d) Visual pictures showing representative tumor volumes of each treatment group. Scale bars are marked in individual images. (n=5–8) (e) GSK126 was speculated to promote the flow of glucose metabolism to lipid metabolism. Eventually, the level of glycolysis and TCA intermediate metabolites declined, and the level of desaturated fatty acids raised, which provided raw materials for TG synthesis. (b, c) analyzed by one-way ANOVA with Tukey multiple comparison posttest. *P < 0.05, **P < 0.01, ***P < 0.001.
The method for combination treatment of GSK126 and MF-438 in pre-B16F10 tumor implantation in vivo. Before subcutaneous implantation, B16F10 cells were treated for 48h according to the following groups: 1) DMSO, 2) GSK126 (10 μM, Selleck, Cat # S7061), 3) MF-438 (6 μM, MCE, Cat # HY-15822), and 4) GSK126 (10 μM) and MF-438 (6 μM). Mice were allocated to four experimental groups (n = 6 mice/group) as above shown (Fig. S6a). Tumor volume was monitored each two days using a digital caliper. Tumor volume = Length*Width* Width/2.
Glucose consumption assay
Collected the culture medium of B16F10 cells before and after GSK126 treatment, and utilized the glucose oxidase assay kit (Applygen, Cat# E1010) to detect the glucose concentration in the culture medium. The glucose consumption levels were calculated as the difference between the initial and the remaining glucose levels in the cell culture media of B16F10 cells subjected to GSK126 treatment. In the meanwhile, cell confluence was monitored in IncuCyte S3 platform (Sartorius, Göttingen, Germany). All glucose consumption levels were normalized to cell confluence.
Statistical analysis
All data were expressed as the mean ± standard error of the mean (S.E.M.). Differences between groups were evaluated using an unpaired Student's t-test. When more than two groups of samples were compared concurrently, one-way ANOVA with Tukey multiple comparison posttests was used. A Kaplan-Meier curve was used to illustrate cumulative survival Significance levels were indicated as suggested by Prism Software: P < 0.05 was considered statistically significant.
Online databases
ONCOMINE database: https://www.oncomine.org/19
GEO database: https://www.ncbi.nlm.nih.gov/gds/20
The cancer genome atlas: http://www.tcga.org/21
The Human Protein Atlas: https://www.proteinatlas.org/22
Role of funding source
This work was funded by The National Natural Science Foundation of China (No. 81672091, No.91749107 and No.81972966). The funders have no roles in study design, data collection, data analysis, interpretation or the writing of this research.
Results
Solid tumor cell lines are less responsive to the EZH2 inhibitor GSK126
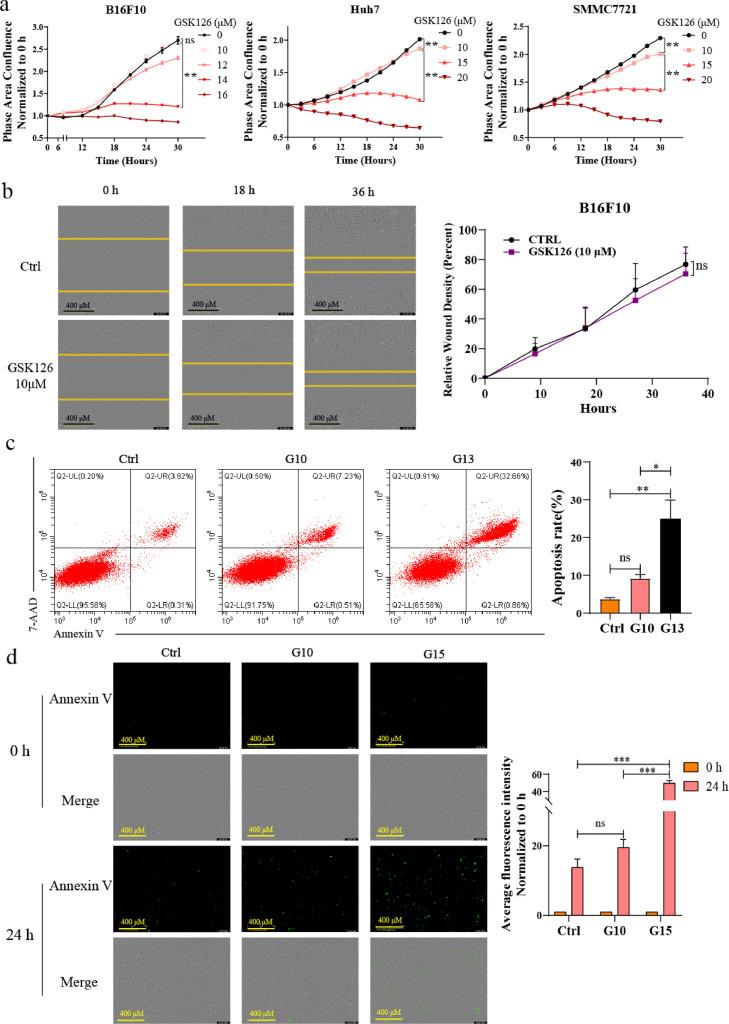
To evaluate the response of solid tumor cells to EZH2 inhibition accurately, we first detected the effect of GSK126 of various concentrations on proliferation of B16F10 melanoma cells, hepatocellular carcinoma (HCC) cells Huh7 and SMMC7721 and colorectal cancer (CRC) cells MC38 in vitro. The results showed that GSK126 inhibited cell proliferation in a dose-dependent manner, and 10 μM of GSK126 only had a weak effect (Figure. 1a and S1a). Under the same conditions, the proliferation of hematological cancer cell Daudi and THP-1 cells was significantly inhibited after 10 μM GSK126 treatment for 24 h and 48 h. Especially Daudi Burkitt's lymphoma cell was already extremely sensitive to GSK126 at 1 μM (Fig. S1b). Consistent with other studies, we found that solid tumor cell lines are generally not sensitive to EZH2i (Fig. S1c), while many studies have shown IC50s (half-maximal inhibitory concentration) of GSK126 below 1 μM in most hematological cancer cell lines.23 We also observed that GSK126 has significantly down-regulated the H3K27me3 level of B16F10, A375 and Daudi cells at 6 μM or 10 μM, which showed the sufficient H3K27me3 inhibitory activity of EZH2i (Fig. S1d, e), but it is not enabled to inhibit the proliferation of cancer cells at 10 μM effectively (Figure. 1a and S1c). Next, we conducted a wound-healing assay in B16F10 melanoma cells. Similar to the cell proliferation experiment, after culturing for 36 h, GSK126 did not effectively inhibit cell migration at 10 μM compared with the control group (Figure 1b). Furthermore, to evaluate the pro-apoptosis effect of GSK126, we treated B16F10 cells with GSK126 at different concentrations and analyzed the apoptosis rate of the cells using flow cytometry (Figure 1c) and fluorescence live-cell imaging (Figure 1d). The results showed that GSK126 had a limited ability to induce apoptosis in B16F10 cells at 10 μM. These results indicated that the proliferation of solid tumor cells B16F10, Huh7, SMMC7721 and MC38 and the migration and survival of B16F10 cells were not significantly inhibited after treatment with high concentrations of GSK126 (10 μM).
Figure 1.
Solid tumor cell lines are less responsive to the EZH2 inhibitor GSK126. (a) Cell proliferation of B16F10, Huh7 and SMMC7721 cells in GSK126 (10-20 μM) treatment monitored by IncuCyte S3. Mean ± SEM is shown (n = 6). (b) Wound-healing assay showing the effect of GSK126 (10 μM) treatment on the migration ability of B16F10 cell lines. Mean ± SEM is shown (n = 3). Scale bars are marked in individual images. (c, d) The apoptosis rate of B16F10 cells in the presence of GSK126 (10, 13, 15 μM) was validated by flow cytometry (c) and Annexin V staining assay (d). Scale bars are marked in individual images. Mean ± SEM is shown (n = 3). One-way ANOVA with Tukey multiple comparison posttest (a, c, d) and two-tailed unpaired Student t test (b) were used to evaluate statistical significance (*P < 0.05, **P < 0.001, ***P < 0.0001). ns, nonsignificant.
Characterization of the metabolic-related genes expression profile of B16F10 cells under GSK126 treatment
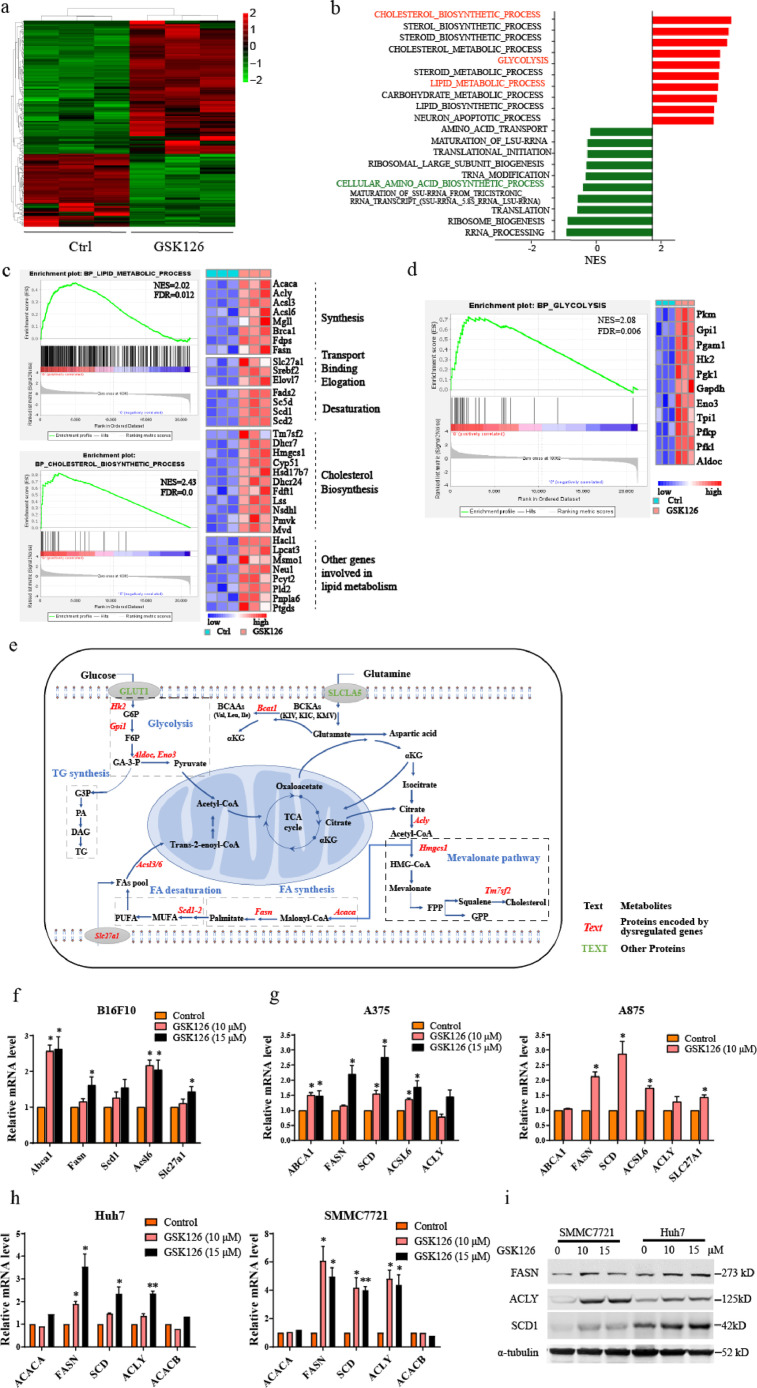
To further investigate the underlying mechanism of EZH2 inhibition, we performed RNA-sequencing (RNA-seq) on B16F10 cells treated with 10 μM GSK126 for 48 h. The RNA-seq data detected a total of 21,279 RNA transcripts, and principal component analysis showed a clear separation between these two groups (Fig. S2a). A total of 209 transcripts including 135 upregulated and 74 downregulated transcripts were found to be significantly dysregulated in GSK126-treated cells compared with the control (Figure. 2a and S2b). Gene set enrichment analysis (GSEA) revealed that GSK126 dysregulated gene expression were involved in cholesterol biosynthetic process, lipid metabolic process, glycolysis, and cellular amino acid biosynthetic process (Figure. 2b–d, and S2c, d). These results indicate that GSK126 profoundly affects the whole metabolic pattern and plays an important role in lipid, glucose, and amino acid metabolism.
Figure 2.
Transcriptional profiling in mouse B16F10 melanoma cells under DMSO (Ctrl) or GSK126 treatment (GSK126-treated). (a) Heat map of the differentially expressed genes between Ctrl and GSK126-treated cells (n = 3). (b) Gene set enrichment analysis (GSEA) of the top 20 gene sets enrichment in biological process between Ctrl and GSK126-treated cells. (c) GSEA of the lipid metabolic process, cholesterol biosynthetic process (left) and hit genes expression in heat map (right). C: Ctrl, G: GSK126-treated. (d) GSEA of the glycolysis(left) and hit genes expression in heat map (right). C: Ctrl, G: GSK126-treated. (e) Schematic representation of metabolic enzymes and metabolic processes that fluctuate after GSK126 treatment in B16F10 cells. (f, g) The mRNA level of fatty acids synthesis genes in murine melanoma cells B16F10 (f) and human melanoma cells A375, A875 (g) which treated with GSK126 detected by real-time qPCR. (h, i) The mRNA (h) and protein (i) level of fatty acids synthesis genes in HCC cells treated with GSK126 detected by real-time qPCR and western blot. Error bars represent mean ± SEM, n = 3 replicates for (f–i). (f–i) analyzed by unpaired two-tailed t test, statistical comparisons are to control. *P < 0.05, **P < 0.01.
GSK126 upregulated genes for lipid and cholesterol biosynthesis, such as Slc27a1, Acly, Acaca, Fasn, Scd1/2, Acsl6, and Tm7sf2 (Figure 2c). As shown in the schematic diagram (Figure 2e), they are involved in FA transport, synthesis, desaturation, and cholesterol synthesis respectively. For example, Acly, Acaca and Fasn catalyze the generation of palmitate from citrate, and oxygen consuming enzymes Scd1/2 participate in the conversion of saturated FAs into monounsaturated FAs. Acyl-CoA synthetase long-chain family member 6 (Acsl6) catalyzes the conversion of long-chain FAs to their active form acyl-CoA for the synthesis of cellular lipids.24,25 Transmembrane 7 superfamily member 2 (Tm7sf2) catalyzes the reduction of the C14-unsaturated bond of lanosterol, as part of the metabolic pathway leading to cholesterol biosynthesis.
The up-regulation of a panel of genes related lipid synthesis was confirmed by real-time qPCR in B16F10 cells treated with GSK126 for 48 h (Figure 2f). Abca1 expression was also upregulated, which functions in cholesterol efflux and regulating cholesterol homeostasis. We managed the same verification in human melanoma cells A375 and A875 to investigate the universality of the role in up-regulating lipid synthesis. It was found that the lipid synthesis genes including ACLY, FASN, SCD1, ACSL6 were up-regulated to varying degrees under the treatment of GSK126 at 10 μM and 15 μM (Figure 2g).
The liver plays an important role in the metabolism of lipids, including fatty acid oxidation and synthesis. We further detected the expression of FA synthesis-related genes in HCC cell lines Huh7 and SMMC7721. As shown in Figure 2h, i, the mRNA and protein levels of ACLY, FASN, and SCD1 were significantly elevated under GSK126 treatment, which is consistent with the RNA-seq data. These lipogenic enzymes, which contribute to increased lipogenesis, display strong activity and high expression in tumor cells.12 According to the Oncomine database,19 the expression of FASN and ACLY was upregulated in liver cancer and melanoma (Fig. S2e). The high expression of ACLY and FASN was correlated with poor prognosis in patients with liver cancer and melanoma, respectively, which were validated using data from the TCGA: LIHC19 and GEO: GSE840120 datasets (Fig. S2f). This suggests that the enzymes involved in lipid biosynthesis play important roles in the development and progression of cancer.
Cancer cells frequently undergo aerobic glycolysis and produce extensive intermediate metabolites and lactic acids, by a phenomenon known as the “Warburg effect”. In addition to lipid metabolism, GSK126 also upregulated genes involved in glycolysis, e.g., Aldoc, Eno3, Gpi1, and Pgk1 (Figure 2d and 2e). They are involved in regulating the key steps of glycolysis. For example, Gpi1 catalyzes the conversion of glucose-6-phosphate to fructose-6-phosphate, and Aldoc is involved in the synthesis of pyruvate from D-glyceraldehyde 3-phosphate. These data indicate that, rather than inhibiting the Warburg effect, GSK126 at 10 μM boosted glycolysis, which favors cancer cells.
It is also known that EZH2 can influence the transport of amino acids and the cycle of methionine,4 so we inspected changes in amino acid-related gene expression. Several genes important for the cellular amino acid biosynthetic process, such as Asns, Bcat1, and Psph (Figs. S2c and 2e), were downregulated by GSK126 treatment. Among them, Bcat1 catalyzes the first reaction in the catabolism of the essential branched-chain amino acids leucine, isoleucine, and valine.
Metabolic signatures of B16F10 cells under GSK126 treatment
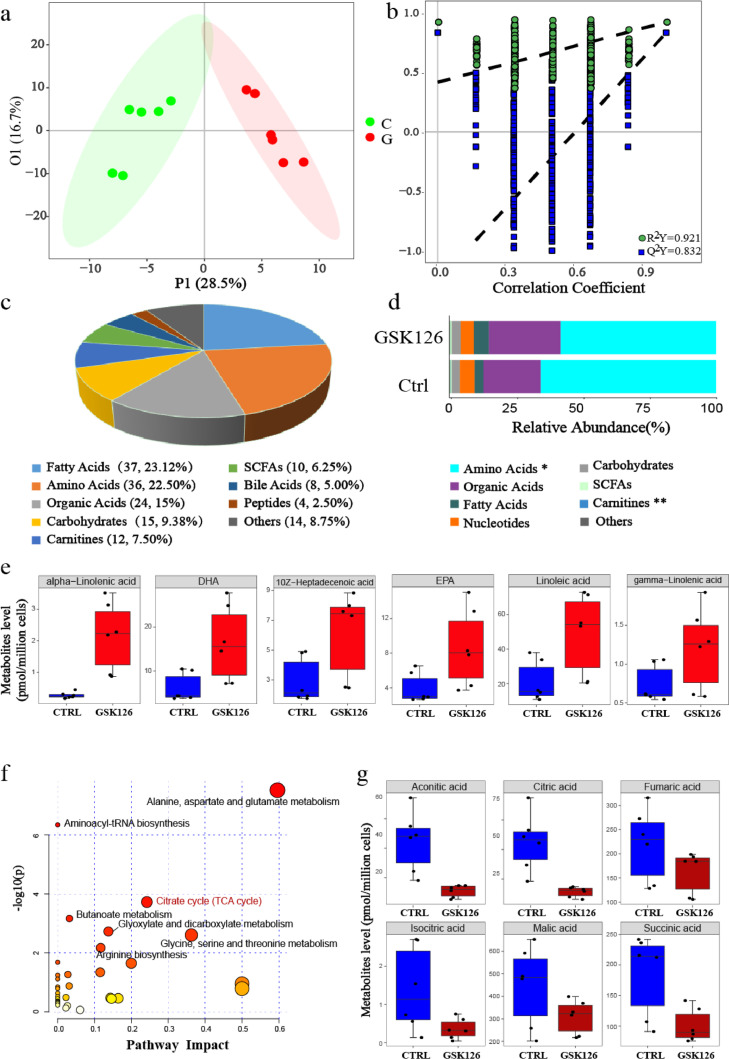
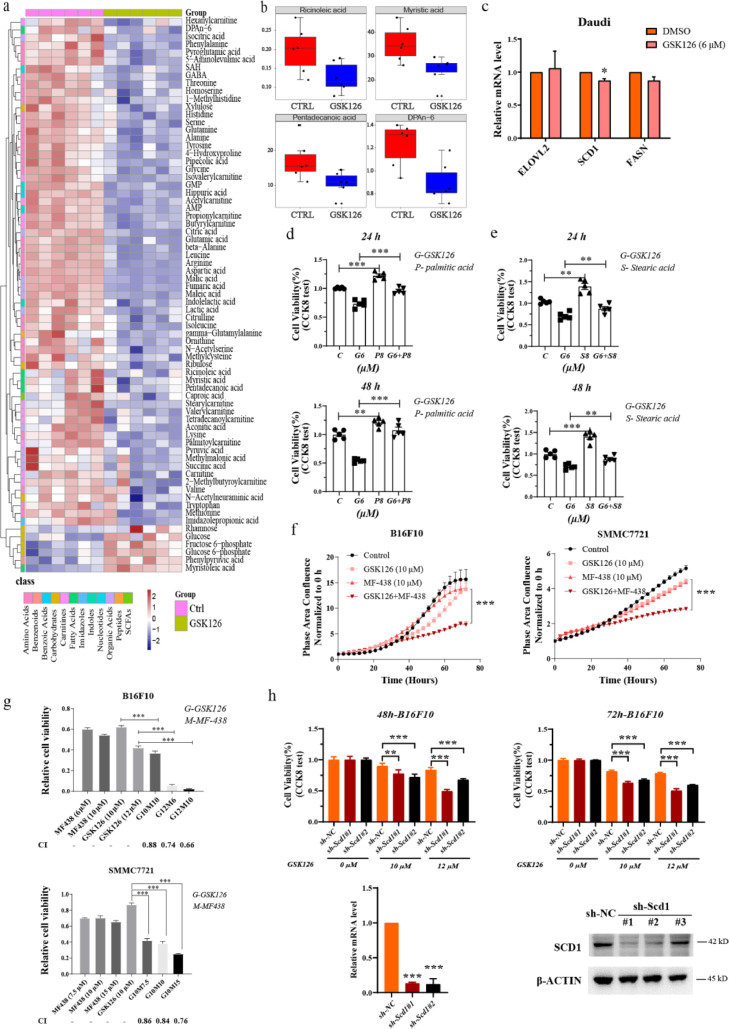
Since RNA-seq results indicated the involvement of metabolic pathways, we further investigated the metabolic changes induced by GSK126. We analyzed the metabolic profiles of B16F10 cells treated with GSK126 or DMSO using ultra-performance liquid chromatography coupled to tandem mass spectrometry (UPLC-MS/MS) system, which refers to determining the substance concentration of a sample corresponding to known concentrations in standard samples. This method can be used for the absolute quantification of more than 300 metabolites. In total, 160 target metabolites were detected after quality control. To investigate the metabolic distinction between these two groups, we performed orthogonal partial least square discriminant analysis (OPLS-DA), and the discrimination between control and GSK126-treated cells was obvious (Figure 3a), and the permutation test showed that the model was reliable and had no overfitting (Figure 3b). The cumulative values of R2Y, and Q2Y indicated an excellent model with a reliable predictive ability. Among the 160 identified metabolites, 70.0% were amino acids, FAs, organic acids, and carbohydrates (Figure 3c and 3d), indicating that GSK126 has a global effect on metabolic patterns. Among these, 65 metabolites with variable influence on projection (VIP) greater than 1.0, were significantly different (Fig. S3a, b, Table S1), with 7 upregulated and 58 downregulated metabolites in GSK126-treated cells compared with the control.
Figure 3.
Metabolic profiling in mouse B16F10 melanoma cells under DMSO (Ctrl) or GSK126 treatment (GSK126-treated). (a) Orthogonal partial least squares discriminant analysis (OPLS-DA) score plot based on the metabolic profiling (n=6). C: Ctrl, G: GSK126-treated. (b) Validate plot of the OPLS-DA model (R2Y = 0.921; Q2Y = 0.832). (c) The classes and composition of metabolites. (d) The relative abundance of each metabolite class in different groups. Amino acids and carnitines were analyzed by Wilcoxon test, and others were analysed by Student's t test, *P < 0.05, **P < 0.01. (e) Six significantly elevated fatty acids in GSK126-treated cells. (f) Overview of the pathway enrichment analysis based on decreased metabolites. (g) Significantly decreased levels of metabolites involved in TCA cycle with GSK126 treatment.
Metabolic alterations in FA biosynthesis and TCA cycle induced by GSK126
Among the seven significantly elevated metabolites in GSK126-treated cells, there are five PUFAs (including alpha-linolenic acid, DHA, EPA, linoleic acid, and gamma-linolenic acid) and one monounsaturated fatty acid (MUFA, including 10Z-Heptadecenoic acid) (Figure. 3e and S3a). This indicated that GSK126 could elevate FA abundance, which was partially supported by the metabolic pathway analysis based on RNA sequencing. These PUFAs can be used as substrates in lipid synthesis to form membranes, but inhibit cell growth and induce apoptosis when excessively accumulated.26
To systematically investigate the metabolic alterations associated with GSK126 treatment, we performed pathway enrichment analysis utilizing metabolites that were present at reduced levels in GSK126-treated cells (Figure 3f, Table S2). One of the decreased pathways was the TCA cycle, which plays a central role in ATP production through the oxidation of acetyl-CoA. Six TCA cycle metabolites were decreased, including aconitic, citric, isocitric, succinic, malic, and fumaric acids. In particular, aconitic acid, citric acid, and isocitric acid levels were markedly reduced (>1.8-fold) in GSK126-treated cells (Figure 3g). This may be related to increased conversion of citric acid to FAs.
Amino acid metabolism is also downregulated with GSK126 treatment. Notably, some amino acids, such as glycine and tryptophan, and the intermediate product in amino acid metabolic processes, such as pipecolic acid, homocitrulline, 2-hydroxyglutaric acid, and dimethylglycine, decreased significantly in the GSK126-treated cells (VIP>1.0) (Fig. S3a and S3b). Serine releases a one-carbon unit when converted to glycine. Glycine and tryptophan could also contribute to the one-carbon pool and are involved in methylation.27 The decrease in glycine and tryptophan indicates that one-carbon metabolism was interrupted by GSK126, which is an S-adenosyl-L-methionine (SAM) competitive inhibitor of EZH2; thus, the methylation and enzymatic activity of EZH2 was impaired. This indicates that GSK126 effectively influences amino acid metabolism and may further block protein synthesis.
Since various FAs were upregulated in B16F10 cells treated with GSK126, we explored the alteration of lipid metabolism in tumor-bearing mice. Hematoxylin and eosin and Oil Red O staining of liver sections showed that fat vacuoles and lipid deposition increased significantly in the GSK126 treatment group (Fig. S3c). The level of triglycerides (TG) in the blood was also elevated (Fig. S3d). These results suggest that GSK126 could regulate lipid metabolism in the liver and the whole body.
GSK126 upregulated lipid synthesis genes via EZH2 mediated H3K27me3 modification
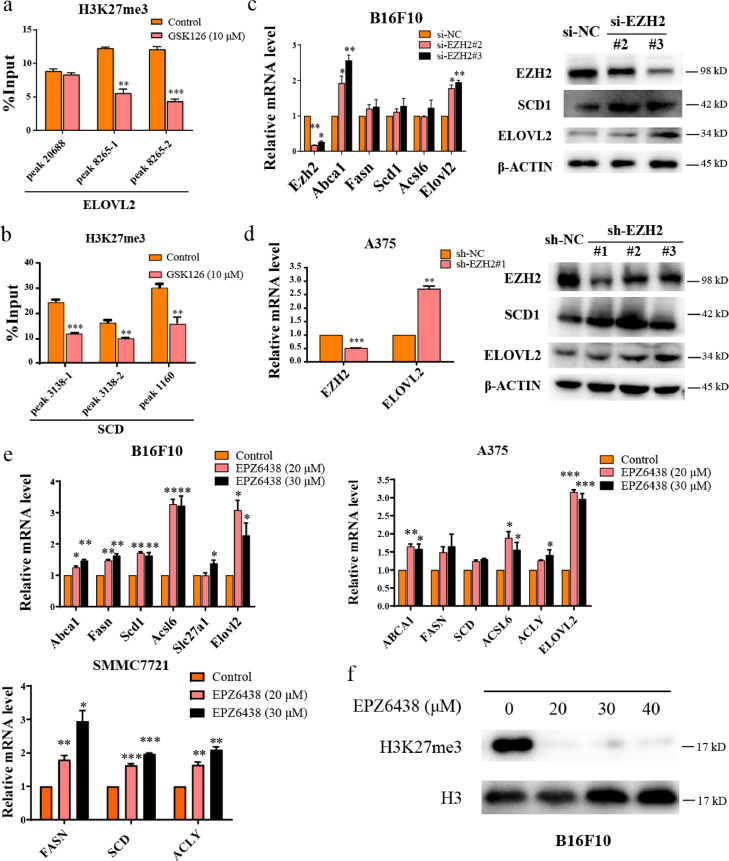
In our previous study, the ChIP-seq and RNA-seq data of EZH2-knockout ovarian cancer cells showed that there were genes related to lipid metabolism directly regulated by EZH2, such as SCD (Scd1) and ELOVL2. ELOVL2 functions in PUFA elongation. As shown in Fig. S4a, the level of H3K27me3 occupied on ELOVL2 promoter and SCD (Scd1) regulatory region was significantly decreased after knockout of EZH2 in SKOV3 cells. The RNA-seq data showed that the pathway of biosynthesis of unsaturated fatty acid was significantly altered,16 and ELOVL2 and SCD expression were elevated in EZH2-KO SKOV3 cells (Fig. S4b). We validated the ChIP-seq results in melanoma cell A375 by ChIP-qPCR, and found that GSK126 treatment reduced the H3K27me3 level on ELOVL2 promoter and SCD regulatory region (Figure 4a, b), and upregulated ELOVL2 and SCD (Scd1) expression (Figs. S4c, 2c, 2f, g). These results indicated that GSK126 elevated ELOVL2 and SCD1 expression by inhibiting the histone methyltransferase activity of EZH2.
Figure 4.
GSK126 inhibited H3K27me3 of lipid synthesis genes. (a-b) The H3K27me3 level of ELOVL2 (a) and SCD1 (b) in control group and GSK126 treated A375 cells detected by ChIP-qPCR. (c) mRNA and protein level of lipid metabolism genes in EZH2-knockdown B16F10 cells by siRNA detected by real-time qPCR and western blot. (d) mRNA and protein level of lipid metabolism genes in EZH2-knockdown A375 cells by shRNA detected by real-time qPCR and western blot. (e) mRNA level of lipid metabolism genes in EPZ6438 treated B16F10, A375 and SMMC7721 cells detected by real-time qPCR. (f) The total H3K27me3 level of B16F10 cells treated by EPZ6438 detected by western blot. (a-e) analyzed by unpaired two-tailed t test, statistical comparisons are to control. *P < 0.05, **P < 0.01, ***P < 0.001.
To further confirm these findings, we knocked down EZH2 expression by siRNA or shRNA. EZH2 knockdown caused upregulation of ELOVL2 and SCD expression in B16F10 and A375 cells (Figure 4c, d). We also treated cancer cells with another EZH2i EPZ6438, to confirm the universality of EZH2i in up-regulation of lipid metabolism genes. The expression of ELOVL2, SCD (Scd1) or other genes was up-regulated to varying degrees after EPZ6438 treatment in B16F10, A375 and SMMC7721 cells (Figure 4e), and total H3K27me3 in B16F10 cells was inhibited (Figure 4f), confirming the regulation role of EZH2i on fatty acid synthesis.
Regulating lipid metabolism enhances the inhibitory effect of GSK126 on cancer cells
To demonstrate whether lipid metabolism of cancer cells was the key factor that affected the sensitivity to EZH2, the metabolomics analysis in Daudi cells treated with GSK126 was conducted using UPLC-MS/MS system, with concentration of 6 μM, close to IC50. To investigate the metabolic distinction between these two groups, we performed OPLS-DA, and the discrimination between control and GSK126-treated cells was obvious (Fig. S5a, b). Seventy-one metabolites with VIP greater than 1.0 were significantly different, with 65 upregulated and 6 downregulated metabolites in GSK126-treated cells compared with the control (Figure. 5a, S5c). Among these significantly different metabolites, we found four decreased fatty acids in GSK126-treated cells, including one MUFA (ricinoleic acid) and one PUFA (DPA n-6) (Figure 5b). According to The Human Protein Atlas database, the positive rate of SCD1 protein expression in lymphoma was lower than that in other solid tumors (Fig. S5d), which fits with our results. Consistent with the metabolomics result, GSK126 did not upregulate the expression of ELOVL2, SCD1 and FASN (Figure 5c). We also detected whether supplement of fatty acid could decrease the sensitivity to GSK126 in Daudi cells. Cell proliferation was promoted moderately by adding palmitic acid and stearic acid (Fig. S5e), and the supplement of fatty acids rescued the cell viability inhibited by GSK126 to varying degrees (Figure 5d, e). These results suggested that the level of fatty acids in cancer cells could weaken tumor inhibition effect of EZH2i.
Figure 5.
Combination with SCD1 inhibitor enhanced the inhibitory effects of GSK126 on cancer cell. (a) Differential metabolites between Ctrl and GSK126-treated Daudi cells. Heat map shows the scaled abundance of 71 differential metabolites with VIP (variable importance in the projection) value greater than 1. The class of metabolites shows in different colors. (b) Four significantly decreased fatty acids in GSK126-treated Daudi cells. (c) mRNA level of lipid metabolism genes in GSK126 treated Daudi cells detected by real-time qPCR (n=3). (d) Cell viability of GSK126 and palmitic acid treated Daudi cells analyzed by CCK-8 kit (n=5). (e) Cell viability of GSK126 and stearic acid treated Daudi cells analyzed by CCK-8 kit (n=5). (f) Cell growth of GSK126 and MF-438 treated B16F10 and SMMC7721 cells monitored by IncuCyte S3. Mean ± SEM is shown (n=5). (g) Cell viability of GSK126 and MF-438 treated B16F10 and SMMC7721 cells detected by CCK-8 kit. CI was calculated by CompuSyn software. A CI < 0.9 indicates synergism and CI > 1.1 an antagonistic effect. (h) The effect of Scd1 knockdown to GSK126 treatment on B16F10 cells activity measured by CCK-8 assay. Mean ± SEM is shown (n=5). Scd1 mRNA and protein level after knockdown were detected by real-time qPCR and western blot. (d-f) analyzed by one-way ANOVA with Tukey multiple comparison posttest. (c, g, h) analyzed by unpaired two-tailed t test, statistical comparisons are to control. *P < 0.05, **P < 0.01, ***P < 0.001.
According to our results, ELOVL2 and SCD1 were directly regulated by EZH2 and H3K27me3 on the promoter, which indicated that ELOVL2 or SCD1 inhibition may enhance cell sensitivity to EZH2i. Since SCD1 desaturates saturated fatty acids, upstream of PUFA elongation, we use SCD1 inhibitor MF-438 for combination treatment. The combination of GSK126 and MF-438 significantly inhibited the proliferation of B16F10 and SMMC7721 cells compared to GSK126 or MF-438 single treatment for 72 h (Figure 5f). The CI analyzed by CCK-8 test and CompuSyn software showed the synergism of GSK126 and MF-438 at various concentration in both cells (Figure 5g). To verify whether SCD1 inhibition could make resistant cells more sensitive to EZH2 inhibitors, the CCK-8 assay was performed, which showed that knockdown of Scd1 enhanced the inhibitory effect of GSK126 on tumor cell activity (Figure 5h). In vivo tests showed that the combination treatment of GSK126 and MF-438 in either pre- or post- B16F10 tumor implantation in mice, the volume of tumors was significantly reduced (Figure. 6a–d and S6a–d). The results above suggested that SCD1 inhibition could enhance the anti-cancer effect of EZH2i.
The anti-cancer effect of SCD1 inhibitors are still studied in preclinical stage, so we further explored whether the clinical lipid-lowering drug fenofibrate (FF) could enhance the inhibitory effect of GSK126 on cancer cells. FF activates peroxisome proliferator-activated receptor α (PPARα), leading to the activation of target gene transcription of lipid metabolism regulation genes.18 We combined GSK126 with FF to treat melanoma cells B16F10, HCC cells SMMC7721, and found that the combination with FF significantly enhanced the inhibitory effect on cell proliferation (Fig. S6e). Simultaneously, we also confirmed that the drug combination can down-regulate the elevated level of lipid metabolism caused by GSK126 treatment. It was found that lipid metabolism genes including Abca1 and Acsl6 of B16F10 were significantly reduced in the combined treatment group compared to GSK126 treatment (Fig. S6f). In SMMC7712 cells, FA synthesis genes including ACLY, FASN, and SCD1 were significantly declined in the combination therapy group (Fig. S6f).
Discussion
In recent decades, a series of EZH2i have been discovered and tested in clinical trials which include tazemetostat, GSK126, CPI-1205, PF-06821497, and SHR2554. In early 2020, tazemetostat was approved by the US Food and Drug Administration (FDA) for use in treatment of adults and adolescents with locally advanced or metastatic epithelioid sarcoma. However, the therapeutic effect of EZH2i on solid tumors is still very limited. Previous studies on the antitumor effects of EZH2i have focused on the epigenetic regulation of target genes, but less on its effect on metabolism, one of the most important characteristics of tumors. Especially the reason of its inefficacy or limitation is far from being uncovered. In this study, we comprehensively described the effect of an EZH2i on metabolism and found that its effect on lipid metabolism may be an important factor that impairs its efficacy.
We found that EZH2 inhibition affected metabolic phenotypes in all respects. At the transcriptional level, GSEA of RNA-seq data revealed that gene expression in various metabolic pathways was altered. In GSK126-treated cells, genes involved in cholesterol biosynthesis, lipid metabolic processes, and lipid biosynthesis were significantly upregulated, while genes related to protein synthesis, such as amino acid transport, translational initiation, and ribosome biosynthesis, were downregulated (Figure 2b). In line with this pattern, the key enzyme-coding genes in these processes showed characteristic changes.
EZH2 inhibition increase the expression of FA synthesis genes. For example, ACLY is upregulated, which cleaves citrate into acetyl-CoA to serve as a building block for FAs (Figure 2c). It is upregulated in liver cancer patients with a poor prognosis (Fig. S2e, f). FASN activation is an early and near-universal hallmark of most human carcinomas and their precursor lesions.28 At both the transcriptional level and fatty acid proportion, elevation of FASN and FA levels were pronounced upon GSK126 treatment (Figure. 2c, 2f–i, and 3d). In addition, SCD1 inhibitors suppressed the formation of tumors that rely on de novo FA synthesis and are more potent in suppressing cancer cell proliferation compared to ACC or FASN inhibitors.29 From our data, Scd1, Scd2, and Scd5 were all upregulated upon GSK126 treatment (Figure 2c and 2f–i). Therefore, from the original substrates, long-chain FA synthesis to the desaturation process, the de novo lipid synthesis process becomes prominent consistently.
The acyl-CoA synthetase long-chain family (ACSL) participates in lipid droplet biogenesis as well as the formation of TG and cholesteryl esters (CE).24,25 This is particularly relevant in cancer because mounting evidence supports the fact that the number and size of lipid droplets are associated with cancer aggressiveness and resistance to therapy.30, 31, 32, 33 ACSL3 was upregulated in melanoma, and its levels correlate with a worse patient prognosis.34 We first found that ACSL3 and ACSL6 were upregulated upon GSK126 treatment (Figure 2c). These evidences strongly indicated that upregulated lipid metabolism can weaken the anticancer effect of GSK126 via metabolic pathways.
ELOVL2 is an enzyme that elongates long-chain omega-3 and omega-6 polyunsaturated fatty acids (LC-PUFAs) and participates in the synthesis of DPA and DHA.35 According to our data, ELOVL2 was directly regulated by H3K27me3 of the promoter. EZH2i caused inhibition of H3K27me3 and increase of ELOVL2 expression, which could contribute to the upregulation of PUFA synthesis. Notably, some PUFAs, such as EPA and DHA, the essential FAs linoleic acid and α-linolenic acid, increased significantly (Figure 3e). Regarding the role of PUFAs in carcinogenesis and progression, there are some contradictory notions. On one hand, erythrocyte levels of EPA and DHA were reported to be inversely associated with breast cancer risk in a case-control study.36 On the other hand, high levels of ω-3 PUFAs in the blood/tissue may exert carcinogenic effects in humans.37 Highly unsaturated PUFAs oxidize to aldehydic products that form DNA adducts and have adverse effects on genome stability.38 In our study, there were no additional essential FAs in the medium; the increased quantity of PUFAs should come from the serum. This data may reflect the increased proportion in the cellular membrane of EPA and DHA. Further identification of the specific PUFA (ω-3, ω-6), MUFA, and various phosphatidyl choline and phosphatidyl ethanolamine components, which are imposed by EZH2i, should be carried out for comprehensive lipid-target metabolism assays.
Although the overall tendency of GSK126 in lipid metabolism is to promote the synthesis of fatty acids, which most likely favor tumor progression, the alteration in amino acids seems to play the opposite role. EZH2, a methyltransferase, is involved in the methylation cycle in which methionine is converted into SAM. The elevation of some methylation related metabolites including SAM were found in the urine and colon of colon cancer mouse models and patients with colon cancer.39 GSK126 is a highly selective, SAM-competitive inhibitor of EZH2 methyltransferase activity. Among the amino acids, we found that glycine and tryptophan decreased upon GSK126 treatment (Fig. S3b), which accounts for the presence of methyl- for tetrahydrofolic acid (FH4). In addition to the methyl-transfer function, the amino acids mainly act as essential materials for cell proliferation. The effect of decreasing amino acids indicates that GSK126 can inhibit tumor cell expansion by downregulating the production of amino acids, which leads to further impaired protein synthesis.
Therefore, although lipid metabolism was significantly upregulated, it is necessary to consider the effect on metabolism as a whole when evaluating the effect of EZH2i. GSK126 interrupted the balance between glucose conversion and lipid metabolism in B16F10 cells. It was speculated that GSK126 may promote the flow of glucose metabolism to lipid metabolism without changing the glucose consumption of B16F10 cells (Fig. S7). After GSK126 treatment, glycolysis, FA synthesis, and desaturation are over-activated (Figure 2b–d), which may cause the intermediate metabolites of glycolysis to flow into lipid metabolism through the following two pathways: 1) Glyceraldehyde triphosphate is converted to glycerol triphosphate into the diacylglycerol pathway, and 2) pyruvate is converted into citric acid through the TCA cycle, which is cleaved into acetyl-CoA to provide the raw material for FA synthesis. Eventually, the levels of TCA intermediate metabolites declined, and the level of desaturated FAs increased, which provided raw materials for TG synthesis (Figure. 3 and 6e).
In this study, the differences of metabolic responses and resistance to EZH2i between hematologic malignancies and solid tumor cell lines were discovered. In contrast to the B16F10 cells, GSK126 treatment caused the reduction of fatty acids in the more sensitive Daudi cells (Figure 5b), and the expression of key lipid metabolism genes was not elevated (Figure 5c), which indicated that lipid metabolism may contribute to the resistance to EZH2i. It has been reported that H3K27ac gain and oncogenic reprogramming limit EZH2i response in solid tumors, compared with hematologic malignancies.23 The relationship between lipid synthesis upregulation and H3K27ac gain remains to be explored. A study in multiple myeloma reported that downregulation of methionine cycling-associated genes was the marker of EZH2i responsive cells, and fatty acid synthesis pathway was not focused.40 Actually the microenvironment of hematologic malignancies and solid tumor exists obvious difference, which may contribute to the distinct metabolic pattern. For example, the positive rate of SCD1 protein expression in lymphoma was lower than that in other solid tumors according to The Human Protein Atlas database (Fig. S5d), indicating that solid tumor cells are more dependent on fatty acid desaturation. Therefore, the upregulation of SCD1 by EZH2 inhibitors may weaken its anticancer effect in solid tumors.
SCD1 is a promising anti-cancer target in the field of inhibiting lipid synthesis. SCD1 catalyzes the conversion from saturated fatty acids (SFAs) into △9-MUFAs, playing an important role in the de novo synthesis of FAs. Higher levels of MUFAs were found in cancer cell and tissue and were related to tumorigenic pathways regulation. The increase of SCD1 expression is positively correlated with cancer aggressiveness and poor patient prognosis in multiple solid tumors, such as liver, thyroid, prostate, pancreatic, kidney, skin, and breast cancer. Various SCD1 inhibitors have been tested in preclinical studies and exhibited anti-cancer effect.41 In this study, we discovered that SCD1 can be regulated by H3K27me3 and EZH2 in the gene remote regulatory region (Figure 4b), and EZH2i caused the elevation of SCD1 expression (Figure. 2g, h and 4e). SCD1 inhibitor MF-438 inhibited cell growth of melanoma and HCC to a certain extent, and the combination of MF-438 and GSK126 showed synergism and stronger inhibitory effects in vitro and in vivo (Figure. 5f, g, 6a–d, S6a–d). These indicated that correcting the upregulation of MUFA synthesis caused by EZH2 inhibitors may be a new combination therapy strategy.
Because SCD1 inhibitors are not yet clinically available, we also selected a clinical agent that regulate lipid metabolism for combination. As a PPARα agonist, the lipid-lowering drug fenofibrate (FF) exhibits anticancer activities against various tumors. The related effects are involved in apoptosis, cell cycle arrest, invasion, and migration. FF induced transcriptional activation of the FA β-oxidation machinery and switched energy metabolism from glucose to FA utilization, resulting in decreased ATP levels, activated AMP-activated protein kinase (AMPK), and increased reactive oxygen species.18 Our results showed that the combination with FF significantly enhanced the inhibitory effects on cancer cell proliferation compared to treatment with GSK126 alone (Fig. S6e). This suggests that the combination of EZH2i and lipid-lowering drugs could exhibit stronger anticancer effects and provide a possible strategy for cancer treatment.
Meanwhile, one important factor restricting the clinical application of GSK126 is its toxic side effects. The results of phase I study of GSK126 showed that forty-one patients (21 solid tumors, 20 lymphoma) who received treatment experienced ≥1 adverse event. Dose-limiting elevated liver transaminases occurred in 2 of 7 patients receiving 3,000 mg of GSK126.42 This clinical study can contact to our results, we found that GSK126 caused liver lipid accumulation (Fig. S3c and S3d). When we dissected the mice treated with GSK126, we found that the liver and peritoneum were adhered severely. Adverse events such as mesenteric adhesions and flatulence also occurred. Other studies have also reported that in the high-fat diet (HFD)-induced mouse obesity model, it was found that down-regulation of EZH2 in hepatocytes led to cholesterol accumulation. HFD up-regulates TRAF6 and ubiquitinates EZH2 to promote the miR-429-dependent inhibition of PPARα, leading to cholesterol accumulation in liver and the occurrence of fatty liver.43 Besides the side effect of EZH2i, this evidence provides a theoretical basis for the combination of PPARα agonists, such as fenofibrate with EZH2i once again. PPARα agonists may ameliorate the drawback of EZH2i on liver function.
Mechanistically, EZH2i decreased H3K27me3 levels globally and reactivated silenced PRC2 target genes without affecting EZH2 protein levels. Regarding the alteration in lipid metabolism, it was reported that EZH2 was involved in adipocyte differentiation by inducing H3K27me3 of Wnt gene promoters.44 In addition, EZH2 inhibition induces the accumulation of cytoplasmic lipid droplets in human breast cancer8 and upregulates genes related to cholesterol homeostasis by activating sterol regulatory element-binding protein 2 (SREBP2) in HCC cells.45 In this study, ELOVL2 and SCD1 were found as a target genes regulated by H3K27me3 and EZH2. It is the first time to find that GSK126 can increase TG and PUFAs accumulation as well as promote FA synthesis. This will provide essential evidence for selecting targeted patients or a given type of cancer administered EZH2i. For instance, cancer patients who also suffer from lipid dysfunction, such as high TG and/or CE, will not be suitable for EZH2i treatment. Meanwhile, tumors that originate in the liver or organs with active lipid synthesis should also be taken into account.
In conclusion, this study revealed that EZH2 inhibition altered the metabolic profile of cancer cells globally, with upregulated lipid synthesis and downregulated TCA cycle and amino acid metabolism. This metabolic alteration may reduce the efficacy of EZH2i. SCD1 and ELOVL2 expression were regulated by EZH2 mediated H3K27me3 modification. To overcome this issue, combining with SCD1 inhibitor or lipid-lowering drugs, such as FF, can be a very promising approach to sharpen the weapon of EZH2i as well as reshaping the tumor microenvironment. Our research further illustrated the interaction between epigenetics and metabolism, and provided a drug combination strategy for future cancer treatment and the evidence for choosing the target patients precisely in EZH2i clinical practice in the future.
Data sharing statement
RNA-seq data are available indefinitely at NCBI GEO: GSE163078 (http://www.ncbi.nlm.nih.gov/geo).
Contributors
Tengrui Zhang and Zhengyang Guo contributed equally to this work.
Lixiang Xue and Changtao Jiang participated in the idea of the article. Tengrui Zhang, Zhengyang Guo, Xiao Huo, Yueqing Gong, Chen Li, Jiaqi Huang, Hao Feng, Xiaojuan Ma and Yan Wang conducted the experiments. Qianqian Yin conducted the bioinformation analysis. Tengrui Zhang, Zhengyang Guo, and Qianqian Yin performed the statistical analysis and wrote the manuscript. Lixiang Xue and Changtao Jiang revised the manuscript and supervised the project. All authors read and approved the final manuscript. Lixiang Xue and Qianqian Yin have verified the underlying data.
Declaration of interests
The authors declare that they have no competing interests.
Funding
This work was funded by The National Natural Science Foundation of China (No. 81672091, No.91749107 and No. 81972966).
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.ebiom.2022.103872.
Contributor Information
Tengrui Zhang, Email: zhangtengrui@hsc.pku.edu.cn.
Zhengyang Guo, Email: guozhengyang@bjmu.edu.cn.
Xiao Huo, Email: biohuoxiao@163.com.
Yueqing Gong, Email: bnu_gyq1988@126.com.
Chen Li, Email: 1911210521@pku.edu.cn.
Jiaqi Huang, Email: huangjq1993@163.com.
Yan Wang, Email: yanwang2019@bjmu.edu.cn.
Hao Feng, Email: hao.feng@mail.mcgill.ca.
Xiaojuan Ma, Email: maxiaojuans@163.com.
Changtao Jiang, Email: jiangchangtao@bjmu.edu.cn.
Qianqian Yin, Email: yinqianqian@bjmu.edu.cn.
Lixiang Xue, Email: lixiangxue@hsc.pku.edu.cn.
Appendix. Supplementary materials
References
- 1.Kim K.H., Roberts C.W. Targeting EZH2 in cancer. Nat Med. 2016;22(2):128–134. doi: 10.1038/nm.4036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gulati N., Beguelin W., Giulino-Roth L. Enhancer of zeste homolog 2 (EZH2) inhibitors. Leuk Lymphoma. 2018;59(7):1574–1585. doi: 10.1080/10428194.2018.1430795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hoy S.M. Tazemetostat: first approval. Drugs. 2020;80(5):513–521. doi: 10.1007/s40265-020-01288-x. [DOI] [PubMed] [Google Scholar]
- 4.Zhang T., Gong Y., Meng H., Li C., Xue L. Symphony of epigenetic and metabolic regulation-interaction between the histone methyltransferase EZH2 and metabolism of tumor. Clin Epigenet. 2020;12(1):72. doi: 10.1186/s13148-020-00862-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zheng M., Cao M.X., Luo X.J., et al. EZH2 promotes invasion and tumour glycolysis by regulating STAT3 and FoxO1 signalling in human OSCC cells. J Cell Mol Med. 2019;23(10):6942–6954. doi: 10.1111/jcmm.14579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ahmad F., Patrick S., Sheikh T., et al. Telomerase reverse transcriptase (TERT) - enhancer of zeste homolog 2 (EZH2) network regulates lipid metabolism and DNA damage responses in glioblastoma. J Neurochem. 2017;143(6):671–683. doi: 10.1111/jnc.14152. [DOI] [PubMed] [Google Scholar]
- 7.Yiew N.K.H., Greenway C., Zarzour A., et al. Enhancer of zeste homolog 2 (EZH2) regulates adipocyte lipid metabolism independent of adipogenic differentiation: role of apolipoprotein E. J Biol Chem. 2019;294(21):8577–8591. doi: 10.1074/jbc.RA118.006871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hayden A., Johnson P.W., Packham G., Crabb S.J. S-adenosylhomocysteine hydrolase inhibition by 3-deazaneplanocin A analogues induces anti-cancer effects in breast cancer cell lines and synergy with both histone deacetylase and HER2 inhibition. Breast Cancer Res Treat. 2011;127(1):109–119. doi: 10.1007/s10549-010-0982-0. [DOI] [PubMed] [Google Scholar]
- 9.Gu Z., Liu Y., Cai F., et al. Loss of EZH2 reprograms BCAA metabolism to drive leukemic transformation. Cancer Discov. 2019;9(9):1228–1247. doi: 10.1158/2159-8290.CD-19-0152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Beloribi-Djefaflia S., Vasseur S., Guillaumond F. Lipid metabolic reprogramming in cancer cells. Oncogenesis. 2016;5(1):e189. doi: 10.1038/oncsis.2015.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Röhrig F., Schulze A. The multifaceted roles of fatty acid synthesis in cancer. Nat Rev Cancer. 2016;16(11):732–749. doi: 10.1038/nrc.2016.89. [DOI] [PubMed] [Google Scholar]
- 12.Che L., Paliogiannis P., Cigliano A., Pilo M.G., Chen X., Calvisi D.F. Pathogenetic, prognostic, and therapeutic role of fatty acid synthase in human hepatocellular carcinoma. Front Oncol. 2019;9:1412. doi: 10.3389/fonc.2019.01412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Centenera M.M., Scott J.S., Machiels J., et al. ELOVL5 is a critical and targetable fatty acid elongase in prostate cancer. Cancer Res. 2021;81(7):1704–1718. doi: 10.1158/0008-5472.CAN-20-2511. [DOI] [PubMed] [Google Scholar]
- 14.McCabe M.T., Ott H.M., Ganji G., et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–112. doi: 10.1038/nature11606. [DOI] [PubMed] [Google Scholar]
- 15.Knutson S.K., Warholic N.M., Wigle T.J., et al. Durable tumor regression in genetically altered malignant rhabdoid tumors by inhibition of methyltransferase EZH2. Proc Nat Acad Sci USA. 2013;110(19):7922–7927. doi: 10.1073/pnas.1303800110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huo X., Sun H., Qian Q., et al. CYP27B1 downregulation: a new molecular mechanism regulating EZH2 in ovarian cancer tumorigenicity. Front Cell Dev Biol. 2020;8 doi: 10.3389/fcell.2020.561804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Léger S., Black W.C., Deschenes D., et al. Synthesis and biological activity of a potent and orally bioavailable SCD inhibitor (MF-438) Bioorg Med Chem Lett. 2010;20(2):499–502. doi: 10.1016/j.bmcl.2009.11.111. [DOI] [PubMed] [Google Scholar]
- 18.Lian X., Wang G., Zhou H., Zheng Z., Fu Y., Cai L. Anticancer properties of fenofibrate: a repurposing use. J Cancer. 2018;9(9):1527–1537. doi: 10.7150/jca.24488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rhodes D.R., Yu J., Shanker K., et al. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6(1):1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xu L., Shen S.S., Hoshida Y., et al. Gene expression changes in an animal melanoma model correlate with aggressiveness of human melanoma metastases. Mol Cancer Res. 2008;6(5):760–769. doi: 10.1158/1541-7786.MCR-07-0344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tomczak K., Czerwińska P., Wiznerowicz M. The cancer genome atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol. 2015;19(1a):A68–A77. doi: 10.5114/wo.2014.47136. (Poznan, Poland) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Uhlén M., Fagerberg L., Hallström B.M., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220) doi: 10.1126/science.1260419. [DOI] [PubMed] [Google Scholar]
- 23.Huang X., Yan J., Zhang M., et al. Targeting epigenetic crosstalk as a therapeutic strategy for EZH2-aberrant solid tumors. Cell. 2018;175(1) doi: 10.1016/j.cell.2018.08.058. 186-199.e119. [DOI] [PubMed] [Google Scholar]
- 24.Grevengoed T.J., Klett E.L., Coleman R.A. Acyl-CoA metabolism and partitioning. Annu Rev Nutr. 2014;34:1–30. doi: 10.1146/annurev-nutr-071813-105541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kassan A., Herms A., Fernandez-Vidal A., et al. Acyl-CoA synthetase 3 promotes lipid droplet biogenesis in ER microdomains. J Cell Biol. 2013;203(6):985–1001. doi: 10.1083/jcb.201305142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Serini S., Ottes Vasconcelos R., Fasano E., Calviello G. How plausible is the use of dietary n-3 PUFA in the adjuvant therapy of cancer? Nutr Res Rev. 2016;29(1):102–125. doi: 10.1017/S0954422416000044. [DOI] [PubMed] [Google Scholar]
- 27.Newman A.C., Maddocks O.D.K. One-carbon metabolism in cancer. Br J Cancer. 2017;116(12):1499–1504. doi: 10.1038/bjc.2017.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Esslimani-Sahla M., Thezenas S., Simony-Lafontaine J., et al. Increased expression of fatty acid synthase and progesterone receptor in early steps of human mammary carcinogenesis. Int J Cancer. 2007;120(2):224–229. doi: 10.1002/ijc.22202. [DOI] [PubMed] [Google Scholar]
- 29.Peck B., Schulze A. Lipid desaturation-the next step in targeting lipogenesis in cancer? FEBS J. 2016;283(15):2767–2778. doi: 10.1111/febs.13681. [DOI] [PubMed] [Google Scholar]
- 30.Yue S., Li J., Lee S.Y., et al. Cholesteryl ester accumulation induced by PTEN loss and PI3K/AKT activation underlies human prostate cancer aggressiveness. Cell Metab. 2014;19(3):393–406. doi: 10.1016/j.cmet.2014.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Petan T., Jarc E., Jusovic M. Lipid droplets in cancer: guardians of fat in a stressful world. Molecules. 2018;23(8):1941–1982. doi: 10.3390/molecules23081941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ackerman D., Tumanov S., Qiu B., et al. Triglycerides promote lipid homeostasis during hypoxic stress by balancing fatty acid saturation. Cell Rep. 2018;24(10):2596–2605. doi: 10.1016/j.celrep.2018.08.015. e2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bensaad K., Favaro E., Lewis C.A., et al. Fatty acid uptake and lipid storage induced by HIF-1alpha contribute to cell growth and survival after hypoxia-reoxygenation. Cell Rep. 2014;9(1):349–365. doi: 10.1016/j.celrep.2014.08.056. [DOI] [PubMed] [Google Scholar]
- 34.Chen W.C., Wang C.Y., Hung Y.H., Weng T.Y., Yen M.C., Lai M.D. Systematic analysis of gene expression alterations and clinical outcomes for long-chain acyl-coenzyme a synthetase family in cancer. PLoS One. 2016;11(5) doi: 10.1371/journal.pone.0155660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baker E.J., Miles E.A., Burdge G.C., Yaqoob P., Calder P.C. Metabolism and functional effects of plant-derived omega-3 fatty acids in humans. Prog Lipid Res. 2016;64:30–56. doi: 10.1016/j.plipres.2016.07.002. [DOI] [PubMed] [Google Scholar]
- 36.Kuriki K., Hirose K., Wakai K., et al. Breast cancer risk and erythrocyte compositions of n-3 highly unsaturated fatty acids in Japanese. Int J Cancer. 2007;121(2):377–385. doi: 10.1002/ijc.22682. [DOI] [PubMed] [Google Scholar]
- 37.Serini S., Calviello G. Long-chain omega-3 fatty acids and cancer: any cause for concern? Curr Opin Clin Nutr Metab Care. 2018;21(2):83–89. doi: 10.1097/MCO.0000000000000439. [DOI] [PubMed] [Google Scholar]
- 38.Gruz P., Shimizu M., Sugiyama K.I., Honma M. Mutagenicity of omega-3 fatty acid peroxidation products in the Ames test. Mutat Res. 2017;819:14–19. doi: 10.1016/j.mrgentox.2017.05.004. [DOI] [PubMed] [Google Scholar]
- 39.Manna S.K., Tanaka N., Krausz K.W., et al. Biomarkers of coordinate metabolic reprogramming in colorectal tumors in mice and humans. Gastroenterology. 2014;146(5):1313–1324. doi: 10.1053/j.gastro.2014.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nylund P., Atienza Párraga A., Haglöf J., et al. A distinct metabolic response characterizes sensitivity to EZH2 inhibition in multiple myeloma. Cell Death Dis. 2021;12(2):167. doi: 10.1038/s41419-021-03447-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tracz-Gaszewska Z., Dobrzyn P. Stearoyl-CoA desaturase 1 as a therapeutic target for the treatment of cancer. Cancers (Basel) 2019;11(7):948–973. doi: 10.3390/cancers11070948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yap T.A., Winter J.N., Giulino-Roth L., et al. Phase I study of the novel enhancer of zeste homolog 2 (EZH2) inhibitor GSK2816126 in patients with advanced hematologic and solid tumors. Clin Cancer Res. 2019;25(24):7331–7339. doi: 10.1158/1078-0432.CCR-18-4121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Z., Wen H., Peng B., Weng J., Zeng F. High-fat diet-induced TRAF6 upregulation promotes liver cholesterol accumulation and fatty liver development through EZH2-mediated miR-429/PPARα axis. Mol Ther Nucleic Acids. 2021:711–727. doi: 10.1016/j.omtn.2021.01.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L., Jin Q., Lee J.E., Su I.H., Ge K. Histone H3K27 methyltransferase Ezh2 represses Wnt genes to facilitate adipogenesis. Proc Natl Acad Sci USA. 2010;107(16):7317–7322. doi: 10.1073/pnas.1000031107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang P.M., Hong Y.H., Hsu K.C., Liu T.P. p38alpha/S1P/SREBP2 activation by the SAM-competitive EZH2 inhibitor GSK343 limits its anticancer activity but creates a druggable vulnerability in hepatocellular carcinoma. Am J Cancer Res. 2019;9(10):2120–2139. [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.