Summary
Hepatocellular carcinoma (HCC) is the most common liver cancer with a high rate of metastasis. However, the molecular mechanisms that drive metastasis remain unclear. We combined single-cell transcriptomic, proteomic, and chromatin accessibility data to investigate how heterogeneous phenotypes contribute to metastatic potential in five HCC cell lines. We confirmed that the prevalence of a mesenchymal state and levels of cell proliferation are linked to the metastatic potential. We also identified a rare hypoxic subtype that has a higher capacity for glycolysis and exhibits dormant, invasive, and malignant characteristics. This subtype has increased metastatic potential. We further identified a robust 14-gene panel representing this hypoxia signature and this hypoxia signature could serve as a prognostic index. Our data provide a valuable data resource, facilitate a deeper understanding of metastatic mechanisms, and may help diagnosis of metastatic potential in individual patients, thus supporting personalized medicine.
Subject areas: Cell biology, Omics, Transcriptomics
Graphical abstract

Highlights
-
•
Provide a high-resolution single-cell triple-omics data of five liver cancer cell lines
-
•
Identify a robust 14-gene set representing hypoxia signature
-
•
The hypoxia signature is associated with prognosis
-
•
Establish an assessment model to characterized metastasis ability
Cell biology; Omics; Transcriptomics
Introduction
Hepatocellular carcinoma (HCC), a neoplasm derived from hepatocytes, is the dominant form of primary liver cancer, and has a poor prognosis because of a high recurrence rate (Singal et al., 2020; Zheng et al., 2017b). The high metastasis rate is a major obstacle to treating HCC, but the mechanisms underlying this characteristic remain unclear (Fang et al., 2015; Li et al., 2020). HCC is also characterized by high heterogeneity (Marquardt and Thorgeirsson, 2014), which contributes to both tumor relapse and drug resistance (Liu et al., 2018). However, the relationship between the heterogeneous phenotypes and metastatic potential has not been systematically assessed. It is increasingly believed that epithelial-mesenchymal transition (EMT), a critical step in metastasis (Aiello et al., 2018), is not a binary process. Instead, a hybrid EMT state which is associated with an increased ability to migrate and invade tissues has been proposed to exist (Schliekelman et al., 2015; Hendrix et al., 1997). Aberrant cell proliferation is also one of the hallmarks of cancer, and operates both in early tumorigenesis and during tumor metastasis (Feitelson et al., 2015; Jarrett et al., 2018). Indeed, cell cycle progression is increased in patients with metastatic pancreatic ductal adenocarcinoma (PDAC) (Connor et al., 2019). Hypoxia, which is common in most tumors, is also linked to metastasis, tumor immune responses, and resistance to therapy (Nobre et al., 2018; Multhoff and Vaupel, 2020; Wigerup et al., 2016; Muz et al., 2015). It is still difficult to define hypoxia status in tumors because of the incredible diversity in hypoxia levels across tissues (Muz et al., 2015).
High throughput single-cell sequencing technologies make it possible to study tumor heterogeneity, and identify rare cell subtypes (Kieffer et al., 2020; Qian et al., 2020; Zhang et al., 2021). The human liver cell atlas revealed heterogeneous cell types (Aizarani et al., 2019) and diversity of cancer stem cell (CSC) subpopulations in HCC. This highlighted the role of CSC in tumor heterogeneity, prognosis, and hepatic CSC therapy (Zheng et al., 2018). Higher genomic complexity has also been related to a worse prognosis in HCC (Kwon et al., 2019). Single-cell genomics have revealed that genomic complexity results from mutation and copy number variations (CNVs), as well as diverse modes of clonal evolution in HBV-related HCC (Duan et al., 2018). Several studies have profiled the single-cell landscape of the tumor microenvironment, and elucidated the nature of the immune response in primary HCC as well as relapsed cases (Yang et al., 2020; Sun et al., 2021; Zheng et al., 2017a). A limitation of single-cell methods is that most studies focus on a single omics dimension. Multiomics approaches enable the integration of data from diverse omics platforms, providing multifaceted insights into the heterogeneity within a population of cells (Hou et al., 2016; Lareau et al., 2021; Mimitou et al., 2021). Combined quantification of the transcriptome and proteome, as well as chromatin accessibility, would give a more comprehensive view of cell states linked to metastatic potential in HCC.
Here, we gathered single-cell transcriptomic, proteomic, and epigenomic data from five HCC cell lines with different metastatic potentials. We thereby assessed the link between phenotypic heterogeneity and HCC metastatic potential, examining EMT capacity, cell proliferation, and hypoxia status. Through this analysis we identified a rare hypoxia subtype that expresses a robust hypoxia signature based on a 14-gene panel. We show that this hypoxia signature is applicable to clinical data. Our data provide deep insights into metastatic mechanisms in HCC, which may accelerate the development of tumor treatments.
Results
Single-cell transcriptomic, proteomic, and epigenetic landscapes in HCC cell lines
To systematically characterize the global transcriptomic, proteomic, and epigenetic landscapes of individual cells in HCC cell lines, we generated single-cell transcriptomic, proteomic, and chromatin accessibility data by applying cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) (Stoeckius et al., 2017) and single-cell assay for transposase-accessible chromatin sequencing (scATAC-seq) (Buenrostro et al., 2015) on DNBelab C Series Single-cell System (Liu et al., 2019). Five human-derived HCC cell lines with different metastatic potentials were used in the study (Figure 1A) (Aden et al., 1979; Hidekazu Nakabayashi et al., 1982; Li et al., 2001; Eun et al., 2014): HepG2 and Huh7 cells lack metastatic potential, MHCC97L cells have low metastatic potential, and MHCC97H and SK-HEP-1 cells have high metastatic potential. Given that CSC has been linked to tumorigenicity, invasiveness, and metastasis, the six markers of liver cancer stem cells (LCSC) were measured originating from antibody-derived tag (ADT) sequencing (Stoeckius et al., 2017), including CD44 (Yang et al., 2008; Zhu et al., 2010), EPCAM (Yamashita et al., 2007), CD133 (Yin et al., 1997), CD24 (Lee et al., 2011), CD90 (Yang et al., 2008), and CD54 (Liu et al., 2013). We obtained qualified and integrated single-cell data from 7311 (mean: 1462, range: 907–1833) transcriptomes and 17,736 (mean: 3547, range: 2735–4572) epigenomes from all five cell lines as well as 5683 (mean: 1421, range: 907–1833) proteomes from the Huh7, MHCC97L, MHCC97H, and SK-HEP-1 cell lines (Figures 1B, 1C, and S1A, and Table S1; see STAR Methods). Different cell lines showed distinct patterns in terms of gene expression levels or chromatin accessibility states.
Figure 1.
Single-cell multiomics landscape of five HCC cell lines
(A) Overview of study design and experimental pipeline for single-cell sequencing.
(B) Uniform manifold approximation and projection (UMAP) plot visualization of color-coded clustering of 7311 cells from five cell lines based on cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) data.
(C) UMAP plot visualization of color-coded clustering of 17,736 cells from five cell lines based on single-cell assay for transposase-accessible chromatin sequencing (scATAC-seq) data.
(D) UMAP plot of protein expression (top; centered log ratio (CLR) normalized), RNA (middle; log gene expression (Exp.)), and gene activity score (GA) (bottom; logGA) reflects signal from antibody panel (CD24, EPCAM, CD44, CD54, CD133, and CD90). See also Figure S1A and Table S1.
We compared the expression and gene activity score (GA) of six LCSC markers at the triple-omics level (Figure 1D). CD24 (CD24), CD44 (CD44), and CD90 (THY1) were consistent in five cell lines but showed variations in expression level and GA. For instance, CD24 (CD24) had a higher level of expression and GA in Huh7 and a lower level in MHCC97L, MHCC97H, and SK-HEP-1 cells. In contrast, CD44 (CD44) had a relatively high-level of expression and GA in MHCC97L, MHCC97H, and SK-HEP-1 cells, and a lower level in Huh7. Other markers including EPCAM (EPCAM), CD133 (PROM1), and CD54 (ICAM1) were not consistent at the triple-omics level. For example, EPCAM (EPCAM) which was highly expressed in Huh7 and CD133 (PROM1) was expressed at low levels across cell lines. However, GA for these examples was inconsistent with protein/gene expression levels. In addition to the inconsistency between GA and protein/gene expression levels, protein and gene expression levels were inconsistent in some cases. For example, CD54 (ICAM1) had relatively high protein expression and low gene expression in MHCC97H cells, indicating that gene expression levels are not sufficient to predict protein expression levels, consistent with previous reports (Rodriguez et al., 2019; Liu et al., 2016). Our data profiled heterogeneous landscapes of six LCSC markers at the molecular level in different HCC cell lines.
Epithelial-mesenchymal transition states are associated with the metastatic potential of HCC cell lines
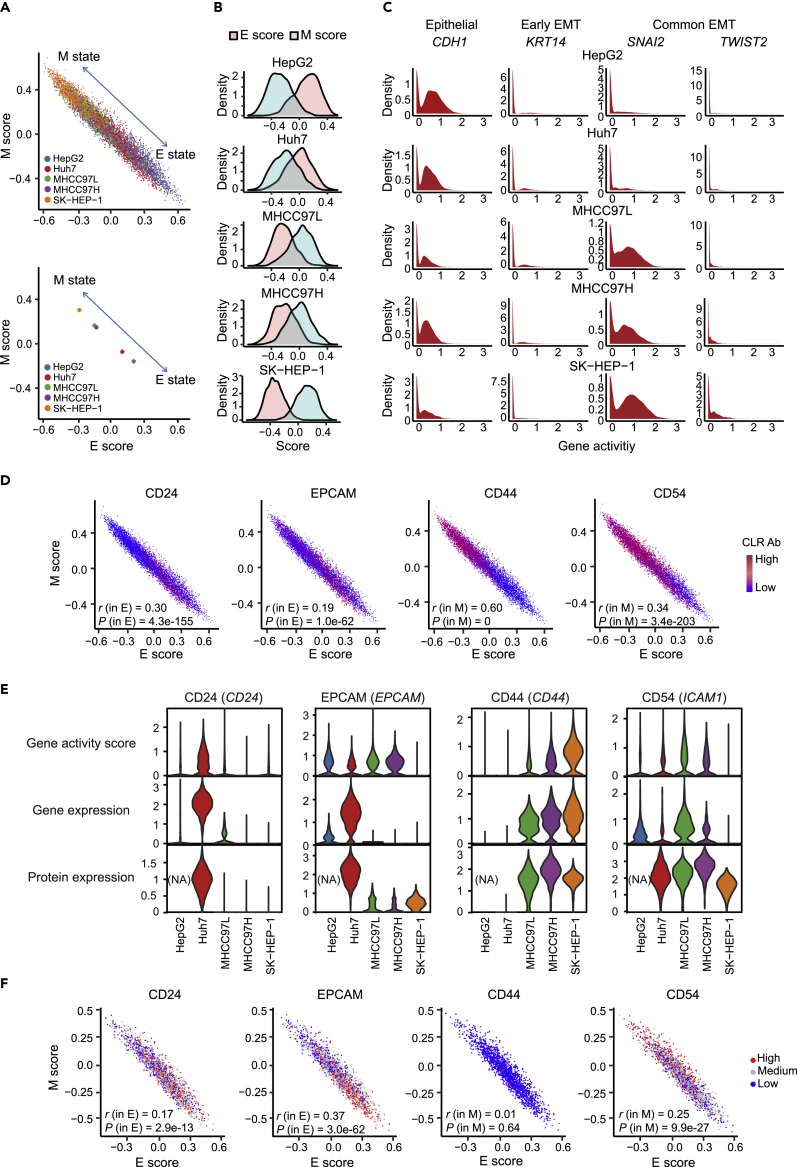
Using previously defined epithelial (E) and mesenchymal (M) related gene expression programs (Huang et al., 2013; Aiello et al., 2018) (Table S2), we calculated the E and M score of each cell in five cell lines using single-cell RNA sequencing (scRNA-seq) data (see STAR Methods). We observed increased M scores along with increased metastatic potential. MHCC97H had a similar M score to MHCC97L despite the former having a higher metastatic potential (Figure 2A). We further verified that HepG2 and Huh7 showed the dominant E state, whereas MHCC97L, MHCC97H, and SK-HEP-1 cells tended to show the M state (Figure 2B).
Figure 2.
Identification and characterization of different EMT states in HCC cell lines
(A) Scatter diagram of epithelial (E) and mesenchymal (M) states for five HCC cell lines at the single-cell level (top panel) and pseudo-bulk level (bottom panel) based on single-cell RNA sequencing (scRNA-seq) data. The top left of the arrow indicates M state, and the bottom right indicates E state. In the pseudo-bulk level analysis, the average scores of E and M in each cell line were plotted on the scatter diagram.
(B) Density distribution of E (pink) and M (light blue) scores in five HCC cell lines.
(C) Density map of normalized GA based on scATAC-seq data. CDH1, a classic epithelial gene. KRT14, an early EMT gene. SNAI2 and TWIST1, classical EMT transcription factors.
(D) EMT scatter diagram showing the relevance of adhesion and migration related surface markers (CD24, CD44, EPCAM, and CD54) with E or M state using normalized protein expression. p and r values of Pearson correlation coefficients analysis were compiled in Table S3.
(E) Violin plot depicting EMT state in each cell line using the normalized expression level of CD24, EPCAM, CD44, and CD54 in triple-omics. Wilcoxon rank-sum test was used to test the statistically significant differences. p values were compiled in Table S4.
(F) EMT scatter diagram showing the normalized protein expression level of CD24, EPCAM, CD44, and CD54 in Huh7. Cells with expression greater than the upper quartile were defined as “High”, less than the lower quartile were defined as “Low”, and others were defined as “Medium”. p and r values of Pearson correlation coefficients analysis were compiled in Table S3. See also Figures S1 and S2 and Tables S2–S4.
Data from scRNA-seq and scATAC-seq in terms of EMT showed the relative consistency (Figure S1B; see STAR Methods). The EMT state in each cell line was further investigated using chromatin accessibility of the characteristic epithelial marker CDH1 (Lamouille et al., 2014), early hybrid EMT state of the gene KRT14 (Pastushenko et al., 2018), and the classical EMT transcription factors SNAI2 and TWIST1 (Nieto et al., 2016) (Figures 2C, S1C, and S1D). CDH1 was more accessible and KRT14 was less accessible in HepG2 and Huh7 cells, consistent with a global E state in the two cell lines. KRT14 was more accessible in MHCC97L and MHCC97H cells than SK-HEP-1, whereas SNAI2 and TWIST1 were more accessible in SK-HEP-1 than MHCC97L and MHCC97H. The accessibility patterns confirmed that MHCC97L and MHCC97H exhibited an earlier hybrid EMT state (Pastushenko and Blanpain, 2019) compared with SK-HEP-1. The various EMT states of cell lines were highly correlated with their metastatic potential where M state showed a positive correlation with cancer metastasis.
Both the EMT program and LCSC markers, including CD24, EPCAM, CD44, and CD54, are relevant to cell adhesion and migration capacity (Nieto et al., 2016; Aiello et al., 2018; Ye and Weinberg, 2015). We next explored the potential relevance between the expression of LCSC markers and E or M state (Table S3; see STAR Methods). Notably, expression of the CD24 and EPCAM proteins were correlated to the E state, whereas CD44 and CD54 were correlated to the M state (Figure 2D, Table S3). Expression of CD24, EPCAM, CD44, and CD54 at the triple-omics levels were further visualized in each cell line, and the overall expression tendency supported the above conclusion (Figure 2E, Table S4). In addition, the expression of CD44 at gene and chromatin accessibility levels was increased in MHCC97L, MHCC97H, and SK-HEP-1 according to metastatic potential (Figure 2E, Table S4). It indicated the strongly underlying relevance between CD44 and M state (r = 0.60) (Figure 2D). All the results elucidated that CD24 and EPCAM characterized a more epithelial state, whereas CD44 and CD54 characterized a more mesenchymal state in HCC cell lines.
In addition to the overall differences of EMT states in cell lines, intercellular heterogeneity was explored. We verified the correlations of LCSC markers assessed above with E or M state within each cell line, and only Huh7 exhibited the significant correlations (Figure 2F and S2, Table S3). Cells were further classified into CD24, EPCAM, CD44, and CD54-high, CD54-medium, or CD54-low based on the expression of markers (Figures 2F and S2; see STAR Methods). EPCAMhigh and CD24high cells had higher E scores in Huh7 (p = 3.4e-24, p = 1.14e-05), whereas CD54high cells had higher M scores (p = 1.13e-27). CD44 showed no specific signal in the E (p = 0.26) or M state (p = 0.75), owing to the low proportion of HCC cells expressing this marker. In sum, our results indicated that expression levels of surface markers were capable of characterizing the nuances of EMT state in the HCC cell lines we tested.
Taken together, heterogeneous EMT states corresponded to metastatic potential in five HCC cell lines indicating that it works as a valuable signal for metastasis in HCC. Varying degrees in the M state hinted at its importance in EMT and cancer metastasis. In addition, LCSC markers were capable of characterizing E vs. M state and metastatic potential in HCC.
Proliferation capability correlates in part with the metastatic potential of cell lines
Cell proliferation drives tumor development and has been linked to metastasis (Hanahan and Weinberg, 2000). To define the cell cycle phase of each cell more precisely, we performed K-means analysis (Macosko et al., 2015; Wagstaff et al., 2001) using cell cycle gene sets (Macosko et al., 2015; Whitfield et al., 2002) (Table S5; see STAR Methods). We identified four prominent clusters (cluster C1-C4) (Figure 3A). Cluster C3 had the highest gene expression levels of G2/M and M phase signatures among clusters, and was assigned G2-M (Figures S3A, S3B, S3F, and S3G). Cluster C2 had the highest gene expression levels of G1/S and S phase signatures among clusters, and was assigned G1-S (Figures S3C, S3D, S3H, and S3I). Cluster C1 had the lowest cell cycle gene expression scores among clusters (Figures 3B, S3E), and showed low levels of the proliferation marker MKI67 (Sun and Kaufman, 2018; Sobecki et al., 2017) (Figure 3C), revealing C1 cells were in G0 phase. The expression pattern of cluster C4 was ambiguous; therefore, we assigned it to the intermediate period.
Figure 3.
Identification of cell cycle phases in HCC cell lines
(A) Classification of cell cycle phases inferred from scRNA-seq data by K-means clustering. Four color-coded clusters (C1-C4) of five HCC cell lines appeared in t-SNE plot and were further divided into four groups: C1, G0 phase; C2, G1-S phase; C3, G2-M phase; C4, intermediate.
(B) Violin plot visualizing the cycle scores in each K-means cluster. Student’s t test was used to test the statistically significant differences of cycle score between clusters of G0 phase (marked with red dotted line) and other clusters.
(C) t-SNE plot visualization of gene expression of MKI67 in all cells. The red dotted line showed gene expression of cells in the G0 phase.
(D) Proportional bar graphs showing distribution of the four cell cycle groups (G2-M, G1-S, intermediate, and G0) in each cell line. See also Figure S3 and Table S5.
The heterogeneity of cell proliferation in the five cell lines was explored by elucidating the composition of cells in different cell cycle phases. The proportion of cell cycle phases for each cell line was calculated, which indicated varying proliferation profiles for the different HCC lines (Figure 3D). The HCC cell lines could be ranked by the fraction of cells in G0 phase in decreasing order: HepG2 (0.53), MHCC97L (0.41), MHCC97H (0.16) to SK-HEP-1 (0.02). This suggested a gradual increase in proliferation capability in cell lines in the same order, which also aligned with increasing metastatic potential. This result was consistent with a previous study (O'Connor et al., 2021) where the population of G0 cells was significantly associated with less aggressive tumors. This held true in our study, except for the Huh7 cell line, which had a median proliferation rate (G0: 0.19), but had the lowest metastatic potential. In summary, though higher cell proliferation was associated with metastasis capacity, it was not exactly consistent with proliferation capability across all cell lines, indicating other factors acting on metastasis.
Hypoxia clusters have a characteristic profile
Hypoxia is a common feature of most tumors and hypoxic cells are more aggressive and invasive with a greater ability to metastasize (Muz et al., 2015). To explore the underlying link between hypoxia and metastatic potential, hypoxia-related genes in Hallmark gene sets (Liberzon et al., 2015) were analyzed in five cell lines by dividing cells into two groups representing nonmetastatic ability (HepG2, Huh7) and metastatic ability (MHCC97L, MHCC97H, and SK-HEP-1) (Figure 4A). Hypoxia-related genes were expressed at higher levels in cell lines with higher metastasis potential. Each cell line was separated into four to seven subgroups by an unsupervised clustering method using transcriptomic data, and a total of 29 clusters were identified (Figures S4A–S4E). We next calculated pairwise correlation coefficients of shared differentially expressed genes (DEGs) from 29 clusters and thereby identified three common subtypes among the cell lines (Group 1–3) (Figure 4B; DEGs were compiled in Table S6; see STAR Methods). Applying functional enrichment analysis, Group 1 was mainly enriched in mitosis (M phase) related pathways, and Group 3 in interphase associated cell cycle related pathways (Figure S4F). Remarkably, we found a hypoxia group (Group 2) was enriched in “Response to oxygen levels”, “Response to hypoxia”, and “Response to decreased oxygen levels”, indicating a hypoxia phenotype which included three hypoxia (HY) clusters from three cell lines (Huh7, MHCC97L, and MHCC97H) (Figure 4C).
Figure 4.
Profiling of hypoxia clusters with metabolic, dormant, invasive, and malignant characteristics
(A) Average expression of hypoxia related genes originating from Hallmark gene sets in cells with metastatic ability (MHCC97L, MHCC97H, and SK-HEP-1) and cells with no metastatic ability (HepG2 and Huh7). Wilcoxon rank-sum test was used to test the statistically significant differences.
(B) Heatmap depicting pairwise correlations of 29 clusters identified by unsupervised clustering. Three groups showed coherent gene expression patterns across cell lines. Groups and related clusters were marked in red boxes.
(C) Functional enrichment analysis of highly expressed DEGs (log fold change (FC) > 0.25) in Group 2 highlighting hypoxia-related processes. Hypergeometric test was used to test the statistically significant differences.
(D) Differences in glycolytic metabolism between hypoxia clusters and other cells. Violin plot depicting tricarboxylic acid (TCA) scores (light blue dots) and glycolysis scores (pink dots) of hypoxia (HY) clusters compared to other cells in Huh7, MHCC97L, and MHCC97H clusters. Each box in the graphs showed the median and IQR (IQR) of the group data, with the lower quartile (bottom) and upper quartile (top). The whiskers extended from the hinge to the smallest or largest value within 1.5 times the IQR from the box boundaries; outliers outside the whiskers range were also presented. Wilcoxon rank-sum test was used to test the statistically significant differences.
(E) t-SNE plot showing the distribution of cells from hypoxia clusters. These cells were shown in red and cells in G0_HY were marked by a blue dotted box. Chi-Squared test was used to test the statistically significant differences.
(F) Proportional bar graphs of G0 phase cells and cycling cells in hypoxia clusters (HY) and all cells in Huh7, MHCC97L, and MHCC97H.
(G) Violin plots indicating CASP8, CDKN1B, EGFR, and VEGFA gene expression in the three groups (G0_HY, G0_NonHY, and Others). Student's t-test was used to test the statistical differences of gene expression between G0_HY and G0_NonHY or Others. See also Figure S4 and Tables S2, S6.
The Warburg effect fuels tumor metastasis and describes increased glycolysis in cancer cells despite the availability of oxygen (Person, 1957; Warburg, 1925; Lu, 2019). Gene expression characteristic of hypoxia in these clusters may originate from metabolic reprogramming leading to the Warburg effect. We analyzed expression of glycolysis and tricarboxylic acid (TCA) cycle related genes to compare the glycolytic capacity of HY clusters with other cells in Huh7, MHCC97L, and MHCC97H (PathCards: https://pathcards.genecards.org/) (Belinky et al., 2015) (Figure 4D, Table S2). Our analysis showed that HY clusters indeed exhibited a higher glycolytic index indicating a higher glycolytic capacity. Accordingly, HY clusters were considered to be a specific glycometabolic subpopulation with features of Warburg effect. The results supported metabolic heterogeneity within cancer cell lines which might enhance cancer cell survival when encountering hypoxic environments.
To profile the cell cycle phase of cells in HY clusters, we projected them onto the cell cycle scatter diagram showing enrichment of cells in G0 (p < 2.2e-16) (Figure 4E). We compared the proportion of G0 cells in HY clusters and corresponding cell lines, which further confirmed enrichment in this cell cycle phase (Figure 4F). We then merged all data from Huh7, MHCC97L, and MHCC97H cell lines and divided them into three groups: G0 phase of HY (G0_HY), G0 phase of Non-HY (G0_NonHY), and Others. Expression of the classic apoptosis gene CASP8 was low, whereas the dormancy gene CDKN1B (Sosa et al., 2015; Fluegen et al., 2017) was upregulated in G0_HY (Figure 4G). This suggested a no-apoptotic and dormant state of cells in G0_HY. In addition, we highlighted EGFR (Giannelli et al., 2008) (tumor-promoting) and VEGFA (Senger et al., 1983; Cao et al., 2004) (pro-angiogenesis), both of which exhibited specific elevated expression in G0_HY (Figure 4G). The results revealed enhanced invasiveness and malignancy of cells in the G0_HY cluster.
In conclusion, we found rare HY clusters which had higher glycolytic capacity and exhibited significant enrichment in the G0 phase. In addition, cells in G0_HY exhibited dormant, invasive, and malignant characteristics. The specific metabolic pattern and characteristics are all associated with metastatic potential.
Hypoxia signature may provide a prognosis index in clinical studies
To better determine the hypoxia status of tumors, we derived a hypoxia signature from HY clusters based on common gene expression features. DEGs (fold-change (FC) > 2) of the HY clusters intersected and 14 genes (CA9, ENO2, SLC6A8, BNIP3, FAM162A, BNIP3L, INSIG2, NDRG1, LDHA, PLOD2, ALDOC, ANGPTL4, ZNF395, and HILPDA) were shared by HY cells across HCC cells lines (Figures 5A, S5A–S5C). We calculated hypoxia scores based on the 14 genes using data from a previous study (Ye et al., 2019) and found that the hypoxia score was increased with the hypoxic treatment time, confirming the robustness of the 14-gene signature (Figure 5B).
Figure 5.
Identification and verification of 14-gene hypoxia signature
(A) Venn diagram showing intersection of DEGs (FC > 2) in hypoxia clusters.
(B) Hypoxia scores of three cancer cell lines (HepG2, MDA-MB231, and U87) under normoxic and hypoxic conditions. The treatment time of hypoxia was 4, 8, and 12 h, respectively. Sample size for each condition was 3 and Student's t-test was used to assess the statistical differences. Each box showed the median, the lower quartile (bottom) and upper quartile (top) in different treatment time groups. The whiskers extended from the hinge to the smallest or largest value within 1.5 times the IQR from the box boundaries.
(C) Density distribution diagram showing the average expression of 14 genes in HCC tissue samples using scRNA-seq data (Sun et al., 2021). Small peaks indicated by red arrowheads represent a small set of cells with a higher expression of 14 genes.
(D) Kaplan-Meier analysis showing the survival probability of HCC patients from The Cancer Genome Atlas (TCGA) dataset. The numbers of patients and the classification are indicated in the figure. Log rank test was used to test the statistically significant differences.
(E) Relapse-free survival curve of HCC patients using published data (Sun et al., 2021). The numbers of patients and the classification are indicated in the figure. Log rank test was used to test the statistically significant differences. See also Figure S5.
To verify whether the signature we found in the cell lines could be applied to tissue samples, scRNA-seq data from HCC clinical tissues (Sun et al., 2021) were utilized to analyze the presence of the hypoxia signature (Figure 5C). A small peak indicated a small set of cells which had a higher expression of 14-gene in Patient 10 (P10) and Patient 18 (P18), but not in Patient 09 (P09). This hinted that the 14-gene hypoxia signature was able to profile characteristics of hypoxia in tissue samples. It also reflected differences in tissue samples which may indicate that the signature is useful for analysis of patient-specific prognosis, and may allow personalized treatment protocols to be developed. We further assessed the clinical relevance of the 14-gene signature. Survival analysis indicated that HCC patients in The Cancer Genome Atlas (TCGA) dataset with high hypoxia scores had a worse prognosis (Figure 5D; see STAR Methods). Relapse-free survival analysis indicated that HCC patients (Sun et al., 2021) with high hypoxia scores had a higher recurrence rate (Figure 5E; see STAR Methods). These results confirmed that the 14-gene signature might serve as a valuable prognosis index.
In short, a robust 14-gene panel serves as hypoxia signature, which may be useful for evaluating the hypoxia status in clinical samples from patients with HCC. The signature might work as a prognosis index and predict the likelihood of relapse in a clinical context. It might also aid targeted therapy aimed at the hypoxic state.
Establishing a metastasis assessment model and extracting a metastasis feature gene set
To better decipher the contributions of the feature factors above to examine HCC metastatic potential, we developed an assessment model using multiple linear regression as follows (Table 1; see STAR Methods).
Table 1.
Coefficients and p values of the metastasis assessment model
| Model | EMT capacity | Cell proliferation | Hypoxia | Intercept | |
|---|---|---|---|---|---|
| p value | 1.8e-05 | 0.31244 | 0.00917 | 0.00205 | 0.00018 |
| Coefficients | N/A | 0.3957 | 8.3690 | 12.5696 | −6.6205 |
Student’s t-test was used to test the statistically significant differences.
Related to Figure 6.
The model considered the contribution of EMT capacity (EM), cell proliferation (P), and hypoxia status (H) to metastatic potential. The predicted metastatic potential scores were consistent with the actual metastatic potential of the five cell lines (Figure 6A). We performed survival analysis using HCC data from the TCGA, which showed this model outperformed other models that only applied a single factor (Figures 6B and 6C). These analyses verified the reliability of our model. Combined with the metastasis-related factors we characterized, the model could be used for evaluating the likelihood of metastasis and may provide referable insights into metastatic mechanisms. Further analysis, beyond the five cell lines used here for model construction, could ultimately develop a more precise model.
Figure 6.
Verification of the metastatic assessment model and extraction of metastasis feature gene set
(A) Histogram showing metastatic potential scores of one-fifth of cells for each cell line to verify reliability of the model.
(B) Survival analysis using EMT capacity, hypoxia status, and cell proliferation as indicators, respectively. The numbers of patients and the classification are indicated in the figure. Log rank test was used to perform significance differences.
(C) Survival analysis using metastatic potential score as indicator. The numbers of patients and the classification are indicated in the figure. Log rank test was used to perform significance differences.
(D) Survival analysis using the metastasis feature gene set as indicator. The numbers of patients and the classification are indicated in the figure. Log rank test was used to test the statistically significant differences.
(E) Functional enrichment analysis of metastasis feature gene set referring to Hallmark gene set. Permutation test was used to test the statistically significant differences. See also Table S7.
We further extracted a gene set which was highly correlated with metastatic potential (see STAR Methods). 114 genes were found (Table S7) and survival analysis using HCC data from the TCGA dataset was carried out to verify the reliability (Figure 6D). Functional enrichment analysis showed the gene set was mainly enriched in “MYC targets v1”, “mTORC1 signaling”, “Reactive oxygen species pathway”, “Glycolysis”, and “Epithelial mesenchymal transition” gene sets (Figure 6F). These pathways were also mainly related to cell proliferation, hypoxia, and EMT, as we used to build the model. Though we had pointed out the important role of EMT in metastatic potential among five cell lines, we found that both the model and feature extraction indicated EMT capacity had a relatively low weight. We speculated that the potential connections between E or M states and metastatic potential might not be applicable to all cell lines. In summary, both the model and feature extraction analysis assessed the contribution of EMT capacity, cell proliferation, and hypoxia to the metastatic potential at a global level which might accelerate the research for tumor mechanisms.
Discussion
HCC cells exhibit extensive heterogeneity in terms of a variety of molecular and phenotypic features. In this study, we generated a multiomics single-cell resource of five liver cancer cell lines with different metastatic potential. Molecular heterogeneity was observed among cell lines and associated with phenotypic heterogeneity including EMT capacity, cell proliferation, and hypoxia status. We performed a comprehensive assessment of how heterogeneous phenotypes track with HCC metastatic potential using this multiomics single-cell resource. At the single-cell resolution, we found that LCSC markers characterize E or M state in HCC, and also identified specific HY clusters. In addition, we uncovered a robust gene panel that represented a hypoxia signature, validated this in published clinical data, and found that this hypoxia signature was associated with HCC prognosis. Our data provide a valuable resource, facilitate a deeper understanding of metastatic mechanisms, and provide clinical evidence that may be a basis for personalized treatment depending on the presence of the hypoxia signature.
The nonbinary process of EMT was also profiled at the molecular level. Prominent E or M state was responsible for EMT heterogeneity in each cell line and corresponded with metastatic potential. Furthermore, in cell populations with a hybrid EMT state, a prominent M state was an important factor for metastatic potential. As for proliferation capacity, it gradually increased in cell lines consistent with metastatic potential in the following order: HepG2, MHCC97L, MHCC97H, and SK-HEP-1. MHCC97L and MHCC97H are two cell lines with a very similar genetic background but different metastatic potential. In these cells, proliferation seems to play a major role in metastatic differences. However, Huh7 had a relationship between proliferation capacity and metastatic potential that was inconsistent with that seen in the other HCC cell lines. This result suggests that cell proliferation is not a factor that is essential to metastatic potential for Huh7. HY clusters had a higher glycolytic capacity. Cells in G0_HY exhibited dormancy, invasiveness, and malignance characteristics, which were linked to metastatic potential. Cells in the G0_HY cluster might be specific to hypoxia-related promotion of metastasis. We calculated hypoxia scores based on a 14-gene panel in HCC patients. Higher hypoxia scores, indicating a poorer prognosis and an increased chance of relapse, might highlight the contribution of hypoxia to metastatic potential. Our metastasis assessment model systematically depicted the contribution of three factors to metastatic potential and confirmed the model built by combining three factors outperforms those based on a single factor. Metastasis feature gene set further assessed the contribution of EMT capacity, cell proliferation, and hypoxia to the metastatic potential at a global level from the data mining perspective, which made up for the incomplete consideration of variable factors in model construction.
At present, liver transplantation and resection are effective therapeutic options at early stages of disease, but only 20–30% of all HCC patients are eligible for these treatment methods (She and Chok, 2015). Sorafenib (anti-AKT) and regorafenib (anti-MEK) are the only two FDA-approved drugs used for HCC treatment, but it turns out they have limited benefits in terms of survival (Caruso et al., 2019; Forner et al., 2018). One of the reasons for poor efficacy in HCC might be the heterogeneous expression profile of drug target genes (Figure S6). For example, in HepG2 cluster 4, the FGFR4/FGF19 signaling genes, FGFR3 and FGFR4 were downregulated or even lacking compared to other HepG2 clusters. In contrast, MET and genes encoding enzymes that removed histone modifications, HDAC1 and HDAC3, were specifically upregulated in cluster 4 of HepG2. Therefore, some drug targets are expressed heterogeneously, reducing the broad efficacy of small molecule inhibitors in treating HCC.
Our data indicate that MHCC97L and MHCC97H may be more sensitive to drugs such as PHA-665752 and JUJ-38877605 for their high expression level of MET (Figure S6). TOP2A, involved in DNA replication and AURKB, which regulates mitosis, were upregulated in cell cycle Groups 1 and 3, indicating that they may be sensitive to Doxorubicin acting on DNA replication and Alisertib acting on mitosis. More effective drugs that can target cells through the gene clusters we have uncovered may exist. Meanwhile, instead of targeting the cell cycle pathway, cells in the G0_HY cluster might be sensitive to drugs targeting EGFR (a tyrosine kinase). These results suggest that HCC cells may have different drug responses depending on their intratumoral vs. extratumoral location. We speculate that a multi-target drug combination will be more effective than monotherapy, and our study suggests the important role of personalized medicine in cancer treatment.
In summary, our results advance understanding of metastasis in HCC, and have practical implications for the clinic. The data and analysis will hopefully provide a basis for clinical treatment and improve the outcomes of cancer therapy.
Limitations of the study
In this work, we systematically assessed the pro-metastasis contributors in HCC cell lines involving EMT, cell proliferation, and hypoxia. Nevertheless, it is hard to deny that metastasis is a very complex process that encompasses multiple aspects, especially in vivo microenvironment. Because employed HCC cell lines that were cultured in vitro, we paid more attention to the characteristics of HCC cells in metastasis rather than the interaction between HCC cells and microenvironment which is crucial for metastasis. Collecting more clinical samples will support further investigations. Besides, though we proved the reliability of the metastatic assessment model, a larger sample size is required for large-scale use of the model. In this context, combining multiple aspects analysis on large-scale sample size will allow for more thorough integration and a more precise model.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| PE Mouse Anti-Human CD24 | BD | Cat# 555428; RRID: AB_395822 |
| CD133/1 Antibody, Anti-human | Miltenyi Biotec | Cat# 130-113-668; RRID: AB_2726210 |
| CD326 (EpCAM) Antibody, Anti-mouse | Miltenyi Biotec | Cat# 130-118-075; RRID: AB_2751452 |
| PE-Cy7 Mouse Anti-Human CD90 | BD | Cat# 561558; RRID: AB_10714644 |
| BB515 Mouse Anti-Human CD54 | BD | Cat# 564685; RRID: AB_2738892 |
| APC Mouse Anti-Human CD44 | BD | Cat# 559942; RRID: AB_398683 |
| Chemicals, peptides, and recombinant proteins | ||
| Dulbecco's modified Eagle's medium (DMEM) | Gibco | Cat# 11-995-040 |
| Fetal bovine serum (FBS) | Gibco | Cat# 26010066 |
| Penicillin-Streptomycin | Gibco | Cat# 15140122; CAS: 8025-06-7 |
| Trypsin (0.25%) | Gibco | Cat# 25200056; CAS: 9002-07- |
| FcR Blocking Reagent | Miltenyi Biotec | Cat# 130-059-901; RRID: AB_2892112 |
| AMPure XP | Beckman Coulter | Cat# A63882 |
| Transposase | BGI | Cat# BGE005 |
| Streptavidin kit | Bio-Rad | Cat# LNK163STR |
| EZ-link Sulpho-NHS S-S Biotin | Thermo Fisher Scientific | Cat# 21328 |
| Tris-HCl pH7.5 | Thermo Fisher Scientific | Cat# 15567027 |
| 5 M NaCl | Thermo Fisher Scientific | Cat# AM9760G; CAS: 7647-14-5 |
| 1 M MgCl2 | Thermo Fisher Scientific | Cat# AM9530G; CAS: 7786-30-3 |
| 0.1% Tween-20 | Sigma | Cat# P9416; CAS: 9005-64-5 |
| 0.1% NP-40 | Roche | Cat# 11754599001; CAS: 123359-41-1 |
| 0.01% Digitonin | Sigma | Cat# D141-100MG; CAS: 59033-71-5 |
| 1% bovine serum albumin (BSA) | BBI | Cat# A600332-0005; CAS: 9048-46-8 |
| Deposited data | ||
| Raw and analyzed data | This paper | CNSA: CNP0001350 |
| Expression profiling of hypoxic HepG2 hepatoma, U87 glioma, and MDA-MB231 breast cancer cells | Ye et al. (2019) | GEO: GSE18494 |
| Single-cell RNA-seq data from HCC clinical tissues | Sun et al. (2021) | CNSA: CNP0000650 |
| Expression profiling of HCC patients | TCGA | TCGA-LIHC |
| Experimental models: Cell lines | ||
| HepG2 | BGI technology services center | RRID: CVCL_0027 |
| MHCC97H | BeNa Culture Collection | RRID: CVCL_4972 |
| Huh7 | Zhongshan Hospital, Fudan University | RRID: CVCL_0336 |
| MHCC97L | Zhongshan Hospital, Fudan University | RRID: CVCL_4973 |
| SK-HEP-1 | ATCC | RRID: CVCL_0027 |
| NIH/3T3 | ATCC | RRID: CVCL_0594 |
| Oligonucleotides | ||
| Oligonucleotides for CITE-seq | This paper | Table S8 |
| Software and algorithms | ||
| Seurat (version 3.1) | Stuart et al. (2019) | RRID: SCR 016341; https://github.com/satijalab/seurat |
| ArchR (version 0.9.5) | Granja et al. (2021) | RRID: SCR_020982; https://github.com/GreenleafLab/ArchR |
| clusterProfiler (v3.16.0) | Yu et al. (2012) | RRID: SCR_016884; https://github.com/YuLab-SMU/clusterProfiler |
| PISA (version 0.3) | N/A | https://github.com/shiquan/PISA |
| STAR (version 2.5) | Dobin et al. (2012) | RRID: SCR_004463; https://github.com/alexdobin/STAR |
| GSEA | Mootha et al. (2003); Subramanian et al. (2005) | RRID: SCR_003199; http://www.gsea-msigdb.org/gsea/index.jsp |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Liang Wu (wuliang@genomics.cn).
Materials availability
This study did not generate new unique reagents
Experimental model and subject details
Liver cancer cell lines and NIH/3T3 culture
This study was approved by the Institutional Review Board on Ethics Committee of BGI (permit no. BGI- IRB20200811003). Cell lines including HepG2 (BGI technology services center, China), Huh7, MHCC97L, MHCC97H (BeNa Culture Collection, China), SK-HEP-1 (ATCC, USA), and NIH/3T3 (ATCC, USA) were employed in the experiment. Huh7 and MHCC97L were kindly provided by Zhongshan Hospital, Fudan University. Cells were cultured in high glucose Dulbecco's modified Eagle's medium (DMEM, Gibco, USA) containing 10% fetal bovine serum (FBS, Gibco, USA) and 1% Penicillin-Streptomycin (Gibco, USA). Trypsin (0.25%) (Gibco, USA) was used to dissociate cells which were then resuspended in DMEM cell culture medium. All cell lines are male. HepG2 was authenticated by short tandem repeats (STR). The remaining cell lines were not authenticated.
Method details
Single-cell preparation and cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq)
Oligonucleotides with a 5′amine modification were synthesized (see Table S8 for oligonucleotides information). Antibodies used included CD24, CD44, CD54, and CD90 from BD (USA) and CD133, EPCAM from Miltenyi Biotec (Germany). To exclude nonspecific antibody binding, we spiked 5% NIH/3T3 into HCC cells as a negative control before cell staining. Species-specific mouse cells are easily distinguished from HCC cells and can be used to calibrate background noise. Oligonucleotides were biotinylated with EZ-link Sulpho-NHS S-S Biotin (Thermo Fisher Scientific, USA) and antibodies were linked to oligonucleotides using streptavidin kit (Bio-Rad, USA). Pooled cells from NIH/3T3 and each cell line were incubated with FcR Blocking Reagent (Miltenyi Biotec, Germany) for 10 min at 4°C and then incubated with antibody-oligo complexes for 30 min at 4°C. We followed manufacturer’s instructions (Countstar, China) to count cells and their viability. All cells employed in the experiment had a viability >91%. We then resuspended cells in cell resuspension buffer at a concentration of 1,000 cells/μL and DNBelab C Series Single-cell System helped construct the single-cell library (Liu et al., 2019). Antibody derived tags (ADT) and cDNA were separated using AMPure XP (Beckman Coulter, USA) and libraries were constructed. The final sequencing library comprised 10% ADT and 90% cDNA library and was sequenced using a BGISEQ500 sequencer with the paired-end (PE) model. Reads 1 and reads 2 included 100-bp cDNA sequences and 41-bp barcode/unique molecular identifier (UMI) sequences respectively.
Single-nucleus preparation and single-cell assay for transposase-accessible chromatin sequencing (scATAC-seq)
Cells were collected in 1.5 mL centrifuge tube and added lysis buffer which consisted of 10 mM Tris-HCl pH7.5 (Thermo Fisher Scientific, USA), 10 mM NaCl (Thermo Fisher Scientific, USA), 3 mM MgCl2 (Thermo Fisher Scientific, USA), 0.1% Tween-20 (Sigma, USA), 0.1% NP-40 (Roche, Switzerland), 0.01% Digitonin (Sigma, USA), and 1% bovine serum albumin (BSA, BBI, UK). Then the lysates were centrifuged at 500 g for 5 min at 4°C and the supernatant was discarded. The obtained nuclei were then washed three times with ATAC wash buffer (10 mM Tris-HCl pH7.5 (Thermo Fisher Scientific, USA), 10 mM NaCl (Thermo Fisher Scientific, USA), 3 mM MgCl2 (Thermo Fisher Scientific, USA), 0.1% Tween-20 (Sigma, USA), 1% bovine serum albumin (BSA, BBI, UK)). Nuclei were then transposed using transposase (BGI, China) and were resuspended in nuclear resuspension buffer. Transposed single-nucleus suspensions were converted to barcoded scATAC-seq libraries, through procedures including droplet encapsulation, pre-amplification, emulsion breakage, capture beads collection, DNA amplification and purification. The libraries were sequenced on the ultra-high-throughput DIPSEQ T1 sequencer with PE 50-bp read length.
CITE-seq data processing and filtering
For human-mouse mixed data, raw scRNA-seq reads were aligned to the human reference genome (GRCh38) and mouse genome (mm10) using STAR (version 2.5) (Dobin et al., 2012). We removed cells with less than 500 UMI mapping to the human or mouse genome. For the reads with same cell barcode, if more than 90% of UMI counts were aligned to human genome, cell corresponding to the reads was determined from human. If it was less than 10% of UMI counts were aligned to human genome, cell corresponding to the reads was determined from mouse. Cells with reads in between 10% UMI counts and 90% UMI counts mapped to human genome were considered mixed species and were removed. Cell versus gene UMI count matrix was generated with PISA (version 0.3): https://github.com/shiquan/PISA.
CITE-seq ADT data processing and filtering
CITE-seq ADT data included the protein expression of six surface markers. Antibody barcodes and cell barcodes were directly extracted from the reads in the sequencing data files. ADT was assigned to individual cell according to cell barcodes and assigned to antibodies according to antibody barcodes. Cells with less than 10 ADT UMI counts were removed from the subsequent analysis. Cell versus ADT UMI count matrix was generated with PISA (version 0.3): https://github.com/shiquan/PISA. The six dimensions ADT count data were normalized and applied the centered log ratio (CLR) transformation (Stoeckius et al., 2017). Then we defined the species-independent cutoff using the median of normalized ADT counts from mouse cells. The value of each ADT in HCC cells was calculated by subtracting the median value in mouse cells.
Single-cell RNA sequencing (scRNA-seq) cell clustering and differential gene expression analysis
Clustering analysis of the cell lines dataset was performed using Seurat (version 3.1) (Stuart et al., 2019) in the R program. Parameters used in each function were manually curated to portray the optimal clustering of cells. In preprocessing, the data of cell lines was treated separately. Cells were filtered based on the criteria of expressing a minimum of 200 genes and less than 5% mitochondrial UMI. Data per gene were expressed using a minimum of 3 cells. Filtered data were ln (counts per million (CPM)/100 + 1) transformed.
In the process of clustering with each cell line, top 2000 highly variable genes were chosen according to their average expression and dispersion. Each gene was scaled using the default option. Dimension reduction analysis started with principal component analysis (PCA), and the number of principal components used for UMAP depended on the importance of embeddings. The Louvain method was then used to detect subgroups of cells. Top 16 principal components (PCs) were used to build the K-NN graph by setting the number of neighbors K as 20. Clusters were identified using a resolution of 0.5. Differentially expressed gene (DEGs) analysis in each cell line was performed using the FindAllMarkers function of the Seurat package (version 3.1).
In the process of clustering with five merged cell lines, filtered data in each cell line were used and then the procedures of merged data were identical to those for a single cell line, except that the top 15 PCs were used to build the K-NN graph and clusters were identified using a resolution of 0.7.
ScATAC-seq data processing and clustering
Raw sequencing reads from DIPSEQ-T1 were filtered and demultiplexed using PISA (version 0.3): https://github.com/shiquan/PISA. Cells with a low fragment (<1000) and transcription start site (TSS) proportion (<0.1) were removed. The filtered data were imported into R and dimensionality was reduced by latent semantic indexing. We analyzed scATAC-seq data using ArchR (version 0.9.5) (Granja et al., 2021). Peak calling was performed using MACS2 (version 2.1.2) (Feng et al., 2012) with options set to -f BAM -B -q 0.01 –nomodel. The cell versus peak reads count matrix was generated using a custom script. The gene activity score matrix was calculated by ArchR.
Integrating analysis of scATAC-seq and scRNA-seq datasets
FindTransferAnchors function was used to get the anchors between scATAC-seq and scRNA-seq datasets in Seurat, and then all data were co-embedded by the TransferData function of Seurat. Top 30 PCs were used in RunUMAP function for obtaining cluster coordinates.
Epithelial and mesenchymal analysis
We examined epithelial (E) and mesenchymal (M) related gene expression programs in five HCC cell lines by calculating the E score and M score based on scRNA-seq data. The E score and M score of each cell was calculated with gsva() in the R package GSVA (Gene Set Variation Analysis) using E and M gene sets collected from previous research (Table S2) (Huang et al., 2013; Aiello et al., 2018). Position of cells were decided according to the two scores on EMT scatter diagram and cell lines' information was projected on the plot. At the pseudo-bulk level, the average scores of E and M in each cell line were plotted on the scatter diagram. Density distribution was displayed for each cell line based on E score and M score.
In data consistency analysis, we aggregated single-cell level counts/gene activity score into “pseudo-bulk” data at the level of cell line in scRNA-seq or scATAC-seq, then normalized the “pseudo-bulk” data. Data consistency between the two omics in terms of EMT were assessed using Pearson correlation coefficient analysis referring to the E and M gene set (Table S2).
In the relevance analysis of LCSC markers with the EMT program, the protein expression of CD24, EPCAM, CD44, and CD54 was projected on the scatter diagram of E and M states where all cells were merged based on scRNA-seq data.
Pearson correlation coefficient analysis was used to assess the correlation of LCSC markers with E or M state. The protein expression of CD24, EPCAM, CD44, and CD54 was used to calculate the correlation with E or M score. Those who satisfied with the conditions of | r | > 0.1 and p < 0.05 on both the E and M scales were considered to be correlated.
In intercellular EMT state heterogeneity analysis, the protein expression of each cell line of CD24, EPCAM, CD44, and CD54 was projected on the scatter diagram of E and M states (cell lines were not merged). First, all cells were divided into 3 categories according to protein expression. Those greater than the upper quartile were defined as “High”, then those lower than the lower quartile were defined as “Low”, and others were defined as “Medium”.
In Huh7, Wilcoxon rank-sum test was used to assess statistically significant differences in E or M score between "High" and the remaining cells.
Cell cycle phase confirmation in five cell lines
Gene sets reflecting five phases of the HeLa cell cycle (G1/S, S, G2/M, M, and M/G1) were taken from previous studies (Whitfield et al., 2002; Macosko et al., 2015). We removed the influence of genes that were previously detected in HeLa cells but did not appear in HCC cell lines data. Genes that were highly correlated (Spearman’s rank correlation coefficient, r > 0.3) with the average signature of the respective cell cycle phase (before excluding genes) were used to define cell cycle signatures (Table S5). The cell cycle score of each cell was defined as mean gene expression of the five cell cycle phase genes. Five cell cycle signature scores were generated for each cell, using averaged normalized expression levels (ln (CPM/100 + 1)) of genes in each set. Based on five cell cycle signatures, we first constructed a low dimensional embedding by t-SNE method, then inferred the cell cycle pattern of each cell based the t-SNE embedding by K-means clustering (with K = 4) (Wagstaff et al., 2001). Student’s t-test was used to assess statistically significant differences in this section.
Cluster correlation and functional enrichment analysis
In cluster correlation analysis, we identified the DEGs in each cluster relative to the other 28 clusters. DEGs that were shared by two clusters were used to calculate pairwise correlations which were defined as the value of the intersection of DEGs divided by the union of DEGs. Functional enrichment analysis using highly expressed DEGs (log FC > 0.25 and shared in at least two clusters) was performed in clusterProfiler (v3.16.0) (Yu et al., 2012) package referring to gene ontology (GO) term enrichment analysis of biological process (BP).
In metastasis feature gene set analysis, we performed the enrichment analysis of 114 genes on the Gene Set Enrichment Analysis (GSEA): http://www.gsea-msigdb.org/gsea/index.jsp (Mootha et al., 2003; Subramanian et al., 2005) using the Hallmark gene set.
Establishment and verification of metastasis assessment model
E and M gene sets in Table S2 were used and the average gene expression of E and M genes was calculated. Then the ratios of M/E were chosen to represent EMT capacity, and the average expression of G2/M gene set in Table S5 was used to represent cell proliferation capacity of cell lines. The average gene expression of Hallmark hypoxia genes was used to represent the hypoxia status of cell lines. According to the actual metastatic potential of cell lines, 1, 2, and 3 were set as the initial value of three levels of metastatic potential. ScRNA-seq data in each cell line were randomly divided into five equal parts. Four parts of the data were used to establish the metastasis assessment model by multiple linear regression with lm() function in R, and the last part was used for subsequent model verification by evaluating the Pearson correlation coefficients between metastasis potential score and initial value of metastasis potential score of five cell lines. Student’s t-test was used to test the statistically significant differences. We verified the result through five-fold crossover, and then selected the model with the highest correlation as the final version.
The Cancer Genome Atlas (TCGA) data of HCC patients were used to further assess the reliability of the model in a Kaplan-Meier survival analysis. We performed survival analysis for EMT capacity, cell proliferation capacity, hypoxia status, and metastasis score respectively. p values were used to confirm the best indicator.
Metastasis feature gene set analysis
The initial metastatic potential value of each cell was set as 1, 2, and 3 according to the actual metastatic potential of cell lines, then we calculated the Pearson correlation coefficients between the value and gene expression in each cell. Those genes with r > 0.5 and p < 0.05 were further verified using univariate Cox hazard analysis, and the genes (log rank p < 0.05) were retained as the candidate genes.
Survival analysis
The mean gene expression of 14 genes was used to define the hypoxia score. In survival analysis using TCGA data, the median value was chosen as a cutoff to classify HCC patients into two groups according to hypoxia score. The Kaplan-Meier survival analysis was performed to assess the difference in overall survival (OS) between the high hypoxia score group and low hypoxia score group. In a relapse-free survival analysis, samples were divided into high hypoxia score and low hypoxia score groups based on the hypoxia score as determined using the R function surv_cutpoint. Kaplan-Meier survival analysis was carried out to assess the difference in relapse-free survival ratios between the high-score and low-score group.
To verify the reliability of the model, the median values of the metastasis score, EMT capacity, cell proliferation capacity, and hypoxia status were used as the cutoff to classify patients into two groups respectively. Kaplan-Meier survival analysis was carried out to assess the difference in OS between the high-score and low-score group.
To verify the reliability of the metastasis feature gene set, the average expression of 114 genes was calculated and the median value was used as the cutoff to classify patients into two groups. Kaplan-Meier survival analysis was carried out to assess the difference in OS between the high-risk and low-risk group.
Quantification and statistical analysis
All analyses were performed on the R 3.6.0 framework. In the analysis of violin plot depicting expression level of CD24, EPCAM, CD44, and CD54 in triple-omics, E or M score assessment between "High" and the remaining cells in Huh7, hypoxia related genes (Hallmark gene sets) expression levels in cells with metastatic ability or none metastatic ability, and tricarboxylic acid (TCA) scores and glycolysis scores between hypoxia (HY) clusters and other cells, Wilcoxon rank-sum test was used to test the statistically significant differences. In the analysis of cell cycle phases scores among clusters, genes (CASP8, CDKN1B, VEGFA, and EGFR) expression levels among groups, metastasis assessment model establishing, and hypoxia score among different treatment time groups, Student's t-test was used to assess the statistical differences. In the enrichment analysis of HY clusters' cells in G0 phase, Chi-Squared test was used to test the statistically significant differences. In the survival analysis and metastasis feature gene set analysis, log rank test was used to assess the statistical differences. In the functional enrichment analysis of Group 1-3, hypergeometric test was used to test the statistically significant differences. In the functional enrichment analysis of metastasis feature gene set, permutation test was used to test the statistically significant differences.
In the analysis of correlation between LCSC markers and E or M state, metastasis assessment model choosing, and metastasis feature gene set extracting, Pearson correlation coefficient was used to evaluate the correlation. In the filtering analysis of cell cycle related genes, Spearman’s rank correlation coefficient was used to evaluate the correlation.
Acknowledgments
We thank the China National GeneBank for supporting this project. This project was supported by grants from Shenzhen Key Laboratory of Single-Cell Omics (NO. ZDSYS20190902093613831), Guangdong-Hong Kong Joint Laboratory on Immunological and Genetic Kidney Diseases (NO. 2019B121205005), and Guangdong Basic and Applied Basic Research Foundation (NO. 2021A1515110832). We thank L. Xu for suggestions on CITE-seq experimental design, Y. Huang and C. Liu for experimental support.
Author contributions
Conceptualization and methodology, L.W., S.W., and J.X.; Data generating, S.W., J.X., and Z.W.; Formal analysis, X.Z., T.P., and Z.Z.; Resources, Y.Y., X.W., and L.L.; Visualization, X.Z., T.P., Y.Z., and X.Z.; Writing Original Draft, S.W. and J.X.; Revising the manuscript, S.W., J.X., Q.Y., X.Z., L.W., R.L., Y.L., and J.Y.; Supervision, L.W., S.L., and H.Y.
Declaration of interests
The authors declare no competing interests.
Published: March 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103857.
Supplemental information
Data and code availability
-
•
Single-cell sequencing data have been deposited at the CNGB Sequence Archive (CNSA) (Guo et al., 2020) of the China National GeneBank DataBase (CNGBdb) (Chen et al., 2020) with accession number: CNP0001350.
-
•
All original code has been deposited at Github and is publicly available as of the date of publication (GitHub: https://github.com/xuanxuanzou/Hepatoma-cell-line).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Aden D.P., Fogel A., Plotkin S., Damjanov I., Knowles B.B. Controlled synthesis of HBsAg in a differentiated human liver carcinoma-derived cell line. Nature. 1979;282:615–616. doi: 10.1038/282615a0. https://pubmed.ncbi.nlm.nih.gov/?term=Fogel+A&cauthor_id=233137 [DOI] [PubMed] [Google Scholar]
- Aiello N.M., Maddipati R., Norgard R.J., Balli D., Li J., Yuan S., Yamazoe T., Black T., Sahmoud A., Furth E.E., et al. EMT subtype influences epithelial plasticity and mode of cell migration. Dev. Cell. 2018;45:681–695 e4. doi: 10.1016/j.devcel.2018.05.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aizarani N., Saviano A., Sagar, Mailly L., Durand S., Herman J.S., Pessaux P., Baumert T.F., Grun D. A human liver cell atlas reveals heterogeneity and epithelial progenitors. Nature. 2019;572:199–204. doi: 10.1038/s41586-019-1373-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belinky F., Nativ N., Stelzer G., Zimmerman S., Iny Stein T., Safran M., Lancet D. PathCards: multi-source consolidation of human biological pathways. Database. 2015;2015:bav006. doi: 10.1093/database/bav006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buenrostro J.D., Wu B., Litzenburger U.M., Ruff D., Gonzales M.L., Snyder M.P., Chang H.Y., Greenleaf W.J. Single-cell chromatin accessibility reveals principles of regulatory variation. Nature. 2015;523:486–490. doi: 10.1038/nature14590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao R., Eriksson A., Kubo H., Alitalo K., Cao Y., Thyberg J. Comparative evaluation of FGF-2-, VEGF-A-, and VEGF-C-induced angiogenesis, lymphangiogenesis, vascular fenestrations, and permeability. Circ. Res. 2004;94:664–670. doi: 10.1161/01.RES.0000118600.91698.BB. [DOI] [PubMed] [Google Scholar]
- Caruso S., Calatayud A.L., Pilet J., La Bella T., Rekik S., Imbeaud S., Letouzé E., Meunier L., Bayard Q., Rohr-Udilova N., et al. Analysis of liver cancer cell lines identifies agents with likely efficacy against hepatocellular carcinoma and markers of response. Gastroenterology. 2019;157:760–776. doi: 10.1053/j.gastro.2019.05.001. [DOI] [PubMed] [Google Scholar]
- Chen F.Z., You L.J., Yang F., Wang L.N., Guo X.Q., Gao F., Hua C., Tan C., Fang L., Shan R.Q., et al. CNGBdb: China National GeneBank database. Yi Chuan. 2020;42:799–809. doi: 10.16288/j.yczz.20-080. [DOI] [PubMed] [Google Scholar]
- Connor A., Denroche R., Jang G., Lemire M., Zhang A., Chan-Seng-Yue M., Wilson G., Grant R., Merico D., Lungu I., et al. Integration of genomic and transcriptional features in pancreatic cancer reveals increased cell cycle progression in metastases. Cancer Cell. 2019;35:267–282.e7. doi: 10.1016/j.ccell.2018.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2012;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan M., Hao J., Cui S., Worthley D.L., Zhang S., Wang Z., Shi J., Liu L., Wang X., Ke A., et al. Diverse modes of clonal evolution in HBV-related hepatocellular carcinoma revealed by single-cell genome sequencing. Cell Res. 2018;28:359–373. doi: 10.1038/cr.2018.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eun J.R., Jung Y.J., Zhang Y., Zhang Y., Tschudy-Seney B., Ramsamooj R., Wan Y.J., Theise N.D., Zern M.A., Duan Y. Hepatoma SK Hep-1 cells exhibit characteristics of oncogenic mesenchymal stem cells with highly metastatic capacity. PLoS One. 2014;9:e110744. doi: 10.1371/journal.pone.0110744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang J.H., Zhou H.C., Zhang C., Shang L.R., Zhang L., Xu J., Zheng L., Yuan Y., Guo R.P., Jia W.H., et al. A novel vascular pattern promotes metastasis of hepatocellular carcinoma in an epithelial-mesenchymal transition-independent manner. Hepatology. 2015;62:452–465. doi: 10.1002/hep.27760. [DOI] [PubMed] [Google Scholar]
- Feitelson M.A., Arzumanyan A., Kulathinal R.J., Blain S.W., Holcombe R.F., Mahajna J., Marino M., Martinez-Chantar M.L., Nawroth R., Sanchez-Garcia I., et al. Sustained proliferation in cancer: mechanisms and novel therapeutic targets. Semin. Cancer Biol. 2015;35:S25–S54. doi: 10.1016/j.semcancer.2015.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J., Liu T., Qin B., Zhang Y., Liu X.S. Identifying ChIP-seq enrichment using MACS. Nat. Protoc. 2012;7:1728–1740. doi: 10.1038/nprot.2012.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fluegen G., Avivar-Valderas A., Wang Y., Padgen M.R., Williams J.K., Nobre A.R., Calvo V., Cheung J.F., Bravo-Cordero J.J., Entenberg D., et al. Phenotypic heterogeneity of disseminated tumour cells is preset by primary tumour hypoxic microenvironments. Nat. Cell Biol. 2017;19:120–132. doi: 10.1038/ncb3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forner A., Reig M., Bruix J. Hepatocellular carcinoma. Lancet. 2018;391:1301–1314. doi: 10.1016/S0140-6736(18)30010-2. [DOI] [PubMed] [Google Scholar]
- Giannelli G., Sgarra C., Porcelli L., Azzariti A., Antonaci S., Paradiso A. EGFR and VEGFR as potential target for biological therapies in HCC cells. Cancer Lett. 2008;262:257–264. doi: 10.1016/j.canlet.2007.12.001. [DOI] [PubMed] [Google Scholar]
- Granja J.M., Corces M.R., Pierce S.E., Bagdatli S.T., Choudhry H., Chang H.Y., Greenleaf W.J. ArchR is a scalable software package for integrative single-cell chromatin accessibility analysis. Nat. Genet. 2021;53:403–411. doi: 10.1038/s41588-021-00790-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo X., Chen F., Gao F., Li L., Liu K., You L., Hua C., Yang F., Liu W., Peng C., et al. CNSA: a data repository for archiving omics data. Database. 2020;2020:baaa055. doi: 10.1093/database/baaa055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D., Weinberg R.A. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- Hendrix M.J., Seftor E.A., Seftor R.E., Trevor K.T. Experimental co-expression of vimentin and keratin intermediate filaments in human breast cancer cells results in phenotypic interconversion and increased invasive behavior. Am. J. Pathol. 1997;150:483–495. [PMC free article] [PubMed] [Google Scholar]
- Hidekazu Nakabayashi K.T., Miyano K., Yamane T., Sato J. Growth of human hepatoma cell lines with differentiated functions in chemically defined medium. Cancer Res. 1982;42:3858–3863. [PubMed] [Google Scholar]
- Hou Y., Guo H., Cao C., Li X., Hu B., Zhu P., Wu X., Wen L., Tang F., Huang Y., et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016;26:304–319. doi: 10.1038/cr.2016.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang R.Y., Wong M.K., Tan T.Z., Kuay K.T., Ng A.H., Chung V.Y., Chu Y.S., Matsumura N., Lai H.C., Lee Y.F., et al. An EMT spectrum defines an anoikis-resistant and spheroidogenic intermediate mesenchymal state that is sensitive to e-cadherin restoration by a src-kinase inhibitor, saracatinib (AZD0530) Cell Death Dis. 2013;4:e915. doi: 10.1038/cddis.2013.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jarrett A.M., Lima E., Hormuth D.A., 2nd, Mckenna M.T., Feng X., Ekrut D.A., Resende A.C.M., Brock A., Yankeelov T.E. Mathematical models of tumor cell proliferation: a review of the literature. Expert Rev. Anticancer Ther. 2018;18:1271–1286. doi: 10.1080/14737140.2018.1527689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kieffer Y., Hocine H.R., Gentric G., Pelon F., Bernard C., Bourachot B., Lameiras S., Albergante L., Bonneau C., Guyard A., et al. Single-cell analysis reveals fibroblast clusters linked to immunotherapy resistance in cancer. Cancer Discov. 2020;10:1330–1351. doi: 10.1158/2159-8290.CD-19-1384. [DOI] [PubMed] [Google Scholar]
- Kwon S.M., Budhu A., Woo H.G., Chaisaingmongkol J., Dang H., Forgues M., Harris C.C., Zhang G., Auslander N., Ruppin E., et al. Functional genomic complexity defines intratumor heterogeneity and tumor aggressiveness in liver cancer. Sci. Rep. 2019;9:16930. doi: 10.1038/s41598-019-52578-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamouille S., Xu J., Derynck R. Molecular mechanisms of epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2014;15:178–196. doi: 10.1038/nrm3758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lareau C.A., Ludwig L.S., Muus C., Gohil S.H., Zhao T., Chiang Z., Pelka K., Verboon J.M., Luo W., Christian E., et al. Massively parallel single-cell mitochondrial DNA genotyping and chromatin profiling. Nat. Biotechnol. 2021;39:451–461. doi: 10.1038/s41587-020-0645-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee T.K., Castilho A., Cheung V.C., Tang K.H., Ma S., Ng I.O. CD24(+) liver tumor-initiating cells drive self-renewal and tumor initiation through STAT3-mediated NANOG regulation. Cell Stem Cell. 2011;9:50–63. doi: 10.1016/j.stem.2011.06.005. [DOI] [PubMed] [Google Scholar]
- Li X., Hu J., Gu B., Paul M.E., Wang B., Yu Y., Feng Z., Ma Y., Wang X., Chen H. Animal model of intrahepatic metastasis of hepatocellular carcinoma: establishment and characteristic. Sci. Rep. 2020;10:15199. doi: 10.1038/s41598-020-72110-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y., Tang Z.-Y., Ye S.L., Liu Y.-K., Chen J., Xue Q., Chen J., Gao D.M., Bao W.-H. Establishment of cell clones with different metastatic potential from the metastatic hepatocellular carcinoma cell line MHCC97. World J. Gastroenterol. 2001;7:630–636. doi: 10.3748/wjg.v7.i5.630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liberzon A., Birger C., Thorvaldsdóttir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1:417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Wu T., Fan F., Liu Y., Wu L., Junkin M., Wang Z., Yu Y., Wang W., Wei W., et al. A portable and cost-effective microfluidic system for massively parallel single-cell transcriptome profiling. Preprint at bioRxiv. 2019 doi: 10.1101/818450. [DOI] [Google Scholar]
- Liu J., Dang H., Wang X.W. The significance of intertumor and intratumor heterogeneity in liver cancer. Exp. Mol. Med. 2018;50:e416. doi: 10.1038/emm.2017.165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S., Li N., Yu X., Xiao X., Cheng K., Hu J., Wang J., Zhang D., Cheng S., Liu S. Expression of intercellular adhesion molecule 1 by hepatocellular carcinoma stem cells and circulating tumor cells. Gastroenterology. 2013;144:1031–1041.e10. doi: 10.1053/j.gastro.2013.01.046. [DOI] [PubMed] [Google Scholar]
- Liu Y., Beyer A., Aebersold R. On the dependency of cellular protein levels on mRNA abundance. Cell. 2016;165:535–550. doi: 10.1016/j.cell.2016.03.014. [DOI] [PubMed] [Google Scholar]
- Lu J. The Warburg metabolism fuels tumor metastasis. Cancer Metastasis Rev. 2019;38:157–164. doi: 10.1007/s10555-019-09794-5. [DOI] [PubMed] [Google Scholar]
- Macosko E.Z., Basu A., Satija R., Nemesh J., Shekhar K., Goldman M., Tirosh I., Bialas A.R., Kamitaki N., Martersteck E.M., et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. 2015;161:1202–1214. doi: 10.1016/j.cell.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marquardt J.U., Thorgeirsson S.S. SnapShot: hepatocellular carcinoma. Cancer Cell. 2014;25:550–e1. doi: 10.1016/j.ccr.2014.04.002. [DOI] [PubMed] [Google Scholar]
- Mimitou E.P., Lareau C.A., Chen K.Y., Zorzetto-Fernandes A.L., Hao Y., Takeshima Y., Luo W., Huang T.S., Yeung B.Z., Papalexi E., et al. Scalable, multimodal profiling of chromatin accessibility, gene expression and protein levels in single cells. Nat. Biotechnol. 2021;39:1246–1258. doi: 10.1038/s41587-021-00927-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mootha V.K., Lindgren C.M., Eriksson K.-F., Subramanian A., Sihag S., Lehar J., Puigserver P., Carlsson E., Ridderstråle M., Laurila E., et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Multhoff G., Vaupel P. Hypoxia compromises anti-cancer immune responses. Adv. Exp. Med. Biol. 2020;1232:131–143. doi: 10.1007/978-3-030-34461-0_18. [DOI] [PubMed] [Google Scholar]
- Muz B., De La Puente P., Azab F., Azab A.K. The role of hypoxia in cancer progression, angiogenesis, metastasis, and resistance to therapy. Hypoxia (Auckl) 2015;3:83–92. doi: 10.2147/HP.S93413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieto M.A., Huang R.Y., Jackson R.A., Thiery J.P. EMT: 2016. Cell. 2016;166:21–45. doi: 10.1016/j.cell.2016.06.028. [DOI] [PubMed] [Google Scholar]
- Nobre A.R., Entenberg D., Wang Y., Condeelis J., Aguirre-Ghiso J.A. The different routes to metastasis via hypoxia-regulated programs. Trends Cell Biol. 2018;28:941–956. doi: 10.1016/j.tcb.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor S.A., Feldman H.M., Arora S., Hoellerbauer P., Toledo C.M., Corrin P., Carter L., Kufeld M., Bolouri H., Basom R., et al. Neural G0: a quiescent-like state found in neuroepithelial-derived cells and glioma. Mol. Syst. Biol. 2021;17:e9522. doi: 10.15252/msb.20209522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastushenko I., Blanpain C. EMT transition states during tumor progression and metastasis. Trends Cell Biol. 2019;29:212–226. doi: 10.1016/j.tcb.2018.12.001. [DOI] [PubMed] [Google Scholar]
- Pastushenko I., Brisebarre A., Sifrim A., Fioramonti M., Revenco T., Boumahdi S., Van Keymeulen A., Brown D., Moers V., Lemaire S., et al. Identification of the tumour transition states occurring during EMT. Nature. 2018;556:463–468. doi: 10.1038/s41586-018-0040-3. [DOI] [PubMed] [Google Scholar]
- Person P. Otto warburg: "on the origin of cancer cells. Oral Surg. Oral Med. Oral Pathol. 1957;10:412–421. [Google Scholar]
- Qian J., Olbrecht S., Boeckx B., Vos H., Laoui D., Etlioglu E., Wauters E., Pomella V., Verbandt S., Busschaert P., et al. A pan-cancer blueprint of the heterogeneous tumor microenvironment revealed by single-cell profiling. Cell Res. 2020;30:745–762. doi: 10.1038/s41422-020-0355-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez J., Ren G., Day C.R., Zhao K., Chow C.C., Larson D.R. Intrinsic dynamics of a human gene reveal the basis of expression heterogeneity. Cell. 2019;176:213–226.e18. doi: 10.1016/j.cell.2018.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schliekelman M.J., Taguchi A., Zhu J., Dai X., Rodriguez J., Celiktas M., Zhang Q., Chin A., Wong C.H., Wang H., et al. Molecular portraits of epithelial, mesenchymal, and hybrid States in lung adenocarcinoma and their relevance to survival. Cancer Res. 2015;75:1789–1800. doi: 10.1158/0008-5472.CAN-14-2535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senger D.R., Galli S.J., Dvorak A.M., Perruzzi C.A., Harvey V.S., Dvorak H.F. Tumor cells secrete a vascular permeability factor that promotes accumulation of ascites fluid. Science. 1983;219:983–985. doi: 10.1126/science.6823562. [DOI] [PubMed] [Google Scholar]
- She W.H., Chok K. Strategies to increase the resectability of hepatocellular carcinoma. World J. Hepatol. 2015;7:2147–2154. doi: 10.4254/wjh.v7.i18.2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singal A.G., Lampertico P., Nahon P. Epidemiology and surveillance for hepatocellular carcinoma: new trends. J. Hepatol. 2020;72:250–261. doi: 10.1016/j.jhep.2019.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sobecki M., Mrouj K., Colinge J., Gerbe F., Jay P., Krasinska L., Dulic V., Fisher D. Cell-cycle regulation accounts for variability in Ki-67 expression levels. Cancer Res. 2017;77:2722–2734. doi: 10.1158/0008-5472.CAN-16-0707. [DOI] [PubMed] [Google Scholar]
- Sosa M.S., Parikh F., Maia A.G., Estrada Y., Bosch A., Bragado P., Ekpin E., George A., Zheng Y., Lam H.M., et al. NR2F1 controls tumour cell dormancy via SOX9- and RARβ-driven quiescence programmes. Nat. Commun. 2015;6:6170. doi: 10.1038/ncomms7170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoeckius M., Hafemeister C., Stephenson W., Houck-Loomis B., Chattopadhyay P.K., Swerdlow H., Satija R., Smibert P. Simultaneous epitope and transcriptome measurement in single cells. Nat. Methods. 2017;14:865–868. doi: 10.1038/nmeth.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart T., Butler A., Hoffman P., Hafemeister C., Papalexi E., Mauck W.M., 3rd, Hao Y., Stoeckius M., Smibert P., Satija R. Comprehensive integration of single-cell data. Cell. 2019;177:1888–1902.e21. doi: 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., Paulovich A., Pomeroy S.L., Golub T.R., Lander E.S., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U S A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun X., Kaufman P.D. Ki-67: more than a proliferation marker. Chromosoma. 2018;127:175–186. doi: 10.1007/s00412-018-0659-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y., Wu L., Zhong Y., Zhou K., Hou Y., Wang Z., Zhang Z., Xie J., Wang C., Chen D., et al. Single-cell landscape of the ecosystem in early-relapse hepatocellular carcinoma. Cell. 2021;184:404–421 e16. doi: 10.1016/j.cell.2020.11.041. [DOI] [PubMed] [Google Scholar]
- Wagstaff K., Cardie C., Rogers S., Schrödl S. Eighteenth International Conference on Machine Learning. Morgan Kaufmann Publishers Inc; 2001. Constrained K-means clustering with background knowledge; pp. 577–584. [Google Scholar]
- Warburg O. The metabolism of carcinoma cells. J. Cancer Res. 1925;9:148–163. [Google Scholar]
- Whitfield M.L., Sherlock G., Saldanha A.J., Murray J.I., Ball C.A., Alexander K.E., Matese J.C., Perou C.M., Hurt M.M., Brown P.O., et al. Identification of genes periodically expressed in the human cell cycle and their expression in tumors. Mol. Biol. Cell. 2002;13:1977–2000. doi: 10.1091/mbc.02-02-0030.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigerup C., Påhlman S., Bexell D. Therapeutic targeting of hypoxia and hypoxia-inducible factors in cancer. Pharmacol. Ther. 2016;164:152–169. doi: 10.1016/j.pharmthera.2016.04.009. [DOI] [PubMed] [Google Scholar]
- Yamashita T., Budhu A., Forgues M., Wang X.W. Activation of hepatic stem cell marker EpCAM by Wnt-beta-catenin signaling in hepatocellular carcinoma. Cancer Res. 2007;67:10831–10839. doi: 10.1158/0008-5472.CAN-07-0908. [DOI] [PubMed] [Google Scholar]
- Yang Y., Liu F., Liu W., Ma M., Gao J., Lu Y., Huang L.H., Li X., Shi Y., Wang X., et al. Analysis of single-cell RNAseq identifies transitional states of T cells associated with hepatocellular carcinoma. Clin. Transl Med. 2020;10:e133. doi: 10.1002/ctm2.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z.F., Ho D.W., Ng M.N., Lau C.K., Yu W.C., Ngai P., Chu P.W., Lam C.T., Poon R.T., Fan S.T. Significance of CD90+ cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–166. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- Ye X., Weinberg R.A. Epithelial-mesenchymal plasticity: a central regulator of cancer progression. Trends Cell Biol. 2015;25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye Y., Hu Q., Chen H., Liang K., Yuan Y., Xiang Y., Ruan H., Zhang Z., Song A., Zhang H., et al. Characterization of hypoxia-associated molecular features to aid hypoxia-targeted therapy. Nat. Metab. 2019;1:431–444. doi: 10.1038/s42255-019-0045-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin A.H., Miraglia S., Zanjani E.D., Almeida-Porada G., Ogawa M., Leary A.G., Olweus J., Kearney J., Buck D.W. AC133, a novel marker for human hematopoietic stem and progenitor cells. Blood. 1997;90:5002–5012. [PubMed] [Google Scholar]
- Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16:284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M., Hu S., Min M., Ni Y., Lu Z., Sun X., Wu J., Liu B., Ying X., Liu Y. Dissecting transcriptional heterogeneity in primary gastric adenocarcinoma by single cell RNA sequencing. Gut. 2021;70:464–475. doi: 10.1136/gutjnl-2019-320368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng C., Zheng L., Yoo J.K., Guo H., Zhang Y., Guo X., Kang B., Hu R., Huang J.Y., Zhang Q., et al. Landscape of infiltrating T cells in liver cancer revealed by single-cell sequencing. Cell. 2017;169:1342–1356.e16. doi: 10.1016/j.cell.2017.05.035. [DOI] [PubMed] [Google Scholar]
- Zheng H., Pomyen Y., Hernandez M.O., Li C., Livak F., Tang W., Dang H., Greten T.F., Davis J.L., Zhao Y., et al. Single-cell analysis reveals cancer stem cell heterogeneity in hepatocellular carcinoma. Hepatology. 2018;68:127–140. doi: 10.1002/hep.29778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng J., Kuk D., Gönen M., Balachandran V.P., Kingham T.P., Allen P.J., D'Angelica M.I., Jarnagin W.R., Dematteo R.P. Actual 10-year survivors after resection of hepatocellular carcinoma. Ann. Surg. Oncol. 2017;24:1358–1366. doi: 10.1245/s10434-016-5713-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Z., Hao X., Yan M., Yao M., Ge C., Gu J., Li J. Cancer stem/progenitor cells are highly enriched in CD133+CD44+ population in hepatocellular carcinoma. Int. J. Cancer. 2010;126:2067–2078. doi: 10.1002/ijc.24868. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
Single-cell sequencing data have been deposited at the CNGB Sequence Archive (CNSA) (Guo et al., 2020) of the China National GeneBank DataBase (CNGBdb) (Chen et al., 2020) with accession number: CNP0001350.
-
•
All original code has been deposited at Github and is publicly available as of the date of publication (GitHub: https://github.com/xuanxuanzou/Hepatoma-cell-line).
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.