Abstract
The rate of human intestinal infections with more than a single Campylobacter strain was determined and the genetic variabilities of Campylobacter strains throughout an infection episode were investigated by means of pulsed-field gel electrophoresis (PFGE) and enterobacterial repetitive intergenic consensus sequence PCR (ERIC-PCR). For 48 and 49 of 50 patients, all isolates from one sample showed identical patterns by PFGE and ERIC-PCR, respectively. Throughout an infection episode in 47 of 52 patients, the PFGE fingerprints of the isolates remained stable, while in 1 patient two different species were observed and in 4 patients different patterns were observed. Therefore, ERIC-PCR proved less discriminative than PFGE. These findings suggest that human infection with more than one Campylobacter strain is rare and should not significantly impair epidemiologic analyses. However, changes in the genetic fingerprint throughout an infection should be considered in the assessment of epidemiologic studies of Campylobacter spp.
The thermophilic Campylobacter species, mainly Campylobacter jejuni and Campylobacter coli, are recognized as important causative agents of acute human diarrheal disease worldwide. Dogs, cats, and other pets may be a source of human infection (1). However, Campylobacter enteritis is mainly a food-borne infection. Healthy food animals, especially poultry, are intestinal carriers of the organisms, and molecular typing studies have shown that individual birds can harbor up to seven different strains (H. Aarts, E. Bouw, and B. van Lith, COST Action Workshop on Pathogenic Microorganisms in Poultry and Eggs, Uppsala, Sweden, p. 14, 1997). Due to the highly automated process in modern poultry slaughterhouses, additional cross-contamination may occur, leading to the isolation of several different Campylobacter clones from one poultry meat product (5). These findings suggest that most cases of human food-borne Campylobacter infection might be caused by several bacterial strains instead of by only one bacterial strain.
In addition to these investigations, many studies have shown a high degree of genetic heterogeneity among Campylobacter isolates by techniques like arbitrarily primed PCR (10), enterobacterial repetitive intergenic consensus sequence PCR (ERIC-PCR) (6), restriction fragment length polymorphism (RFLP) analysis, mainly of the flagellin gene (12), and pulsed-field gel electrophoresis (PFGE) (14). Possible reasons for this heterogeneity are genetic rearrangements due to genomic instability that either occurs spontaneously or is induced by mobile elements (18); programmed DNA inversion, as described for Campylobacter fetus (3); horizontal gene transfer (17); and natural transformation (16). While some investigators have found evidence for genetic mosaicism or natural transformation during experimental colonization of chickens (8), other groups failed to do so (18). Additional phases that may be candidates for such rearrangements are during environmental transition or human infection. The study described here was performed to investigate (i) whether genetically distinct Campylobacter strains can be simultaneously isolated from stool samples of patients with Campylobacter enteritis and (ii) whether during an infection episode genetic rearrangements can be observed.
(The results described here were presented in part at the 9th International Workshop on Campylobacter, Helicobacter and Related Organisms, Cape Town, South Africa, 15 to 19 September 1997.)
Stool samples and bacterial strains.
Human stool samples were plated on blood-free Campylobacter agar (Oxoid, Basingstoke, United Kingdom) supplemented with 32 mg of cefoperazone per liter and 10 mg of amphotericin B per liter. Incubation in all experiments was carried out for 48 h at 37°C. Jars were partially evacuated and were refilled with a gas mixture that contained 5% O2, 10% CO2, and 85% N2 (final concentrations). Isolates were identified by standard biochemical tests. To avoid misidentification of hippurate-negative C. jejuni strains, all hippurate-negative isolates were subjected to previously described PCRs specific for C. jejuni and C. coli (4). Subcultures were performed on nonselective agar plates (yeast-cysteine agar) supplemented with 10% sheep erythrocytes.
To detect the simultaneous presence of distinct Campylobacter clones in one stool sample, four individual colonies were picked from different areas of the original selective agar and were separately subcultured. Four colonies were chosen for standardization since more than four colonies can rarely be discriminated on primary agar plates due to contamination with other bacteria or fungi, swarming, or low colony counts. To study the stability of the genetic fingerprint throughout human infection, one colony per plate was subcultured and analyzed by molecular typing techniques.
Molecular typing.
All isolates were subjected to PFGE and ERIC-PCR. For PFGE the bacteria were suspended in 0.9% NaCl solution and were diluted to an optical density of 0.85 at 600 nm. A total of 1.5 ml of this suspension was centrifuged at 15,000 × g for 4 min. The pellet was washed three times and was resuspended in 0.5 ml of 0.9% NaCl solution. A mixture with 0.7 ml of 1.5% PFGE agarose (Sigma, St. Louis, Mo.) was cast in blocks, incubated in lysis buffer (0.5 mM EDTA [pH 9.5], 1% N-lauroyl-sarcosine, 1.8 mg of proteinase K per ml) overnight, and washed three times in TE buffer (10 mM Tris, 10 mM EDTA [pH 7.5]). If not otherwise stated, restriction was performed with 20 U of SmaI (New England Biolabs, Beverly, Mass.). Fragments were separated in a 1% PFGE agarose gel (Sigma) with 0.5× TBE (45 mM Tris, 45 mM boric acid, 1 mM EDTA) at 200 V and 12°C for 19 h with ramped pulse times from 10 to 30 s in a CHEF DR II system (Bio-Rad, Richmond, Calif.). In some experiments KpnI (20 U; New England Biolabs) was used as an additional enzyme, and in this case separation was performed for 12.7 h with ramped pulse times from 1 to 8 s. Bands were stained with ethidium bromide, and a 48.5-kb bacteriophage lambda ladder (Bio-Rad) served as a DNA size standard.
DNA for ERIC-PCR was prepared with an extraction kit (Qiagen, Hilden, Germany), according to the manufacturer's instructions. The amount of purified DNA was determined photometrically on a GeneQuant system (Pharmacia Biotech, Uppsala, Sweden) and was adjusted to a concentration of 20 μg/ml. For the reaction mixture (final volume, 50 μl), Taq DNA polymerase buffer (Pharmacia Biotech) with 1 mM MgCl2 and 0.4 μg of bovine serum albumin was used. Each primer, primer ERIC 1R and primer ERIC 2 (15), was used at a concentration of 25 pmol. A total of 100 ng of template DNA and 2 U of Taq DNA polymerase (Pharmacia) were added to each reaction mixture. PCR involved 30 cycles of consecutive denaturation (95°C, 1 min), primer annealing (40°C, 1 min), and chain extension (65°C, 8 min). Prior to cycling the samples were heated at 95°C for 7 min. Finally, an additional extension step (16 min, 65°C) was performed. A GeneAmp System 9600 thermocycler (Perkin-Elmer Cetus, Norwalk, Conn.) was used. PCR products were separated in 2% agarose gels and stained with ethidium bromide. C. jejuni NCTC 11351 was included as a reference strain in each experiment, PFGE, as well as ERIC-PCR. All gel images were recorded with an EASY Image Plus computer based video documentation system (Herolab, Wiesloch, Germany). All band patterns (PFGE and ERIC-PCR) were inspected and analyzed manually as well as with a computer. Automated analysis was performed by the unweighted pair group method with arithmetic averages and with GelCompar software (version 4.0; Applied Maths, Kortrijk, Belgium). For the analysis of similarity between the band patterns, the Dice coefficient with a position tolerance of 1.1% was used for the PFGE patterns and the Pearson coefficient was used for the more complex PCR patterns. The existence of double bands was evaluated by computer-based densitometry of the individual bands within a pattern. Throughout this study, all Campylobacter isolates were typeable by PFGE as well as by ERIC-PCR.
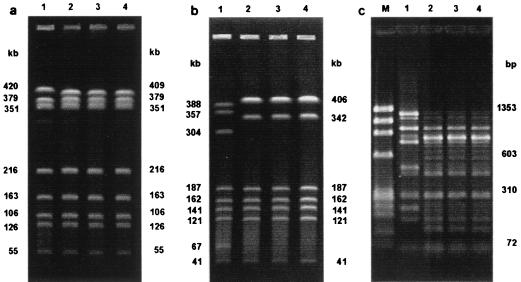
Four distinct colonies were picked from the first stool sample submitted from 50 consecutive patients with Campylobacter enteritis, leading to a total of 200 Campylobacter isolates. The isolates of 47 patients were identified as C. jejuni, and the isolates of 3 patients were identified as C. coli. A simultaneous infection with C. jejuni and C. coli was not detected in any patient. A total of 44 different band patterns were demonstrated by PFGE. In 48 of 50 patients the four isolates exhibited identical band patterns. Three isolates from one patient showed identical patterns, while for the fourth isolate a 409-kb band was replaced by a 420-kb band (Fig. 1a). However, when KpnI was used as the restriction enzyme, all isolates exhibited identical patterns (not shown). In another patient three isolates showed identical patterns, while one isolate differed from the others by seven bands when the band at 406 kb was considered a double band, as demonstrated by densitometry (data not shown) (Fig. 1b).
FIG. 1.
(a) PFGE patterns for four isolates (lanes 1 to 4, respectively) found in one stool sample of a patient with acute Campylobacter infection. In lane 1, a 409-kb band was replaced by a 420-kb band. (b) Four isolates (lanes 1 to 4, respectively) from one stool sample showing two different PFGE patterns. The pattern for the first isolate differed from those for the other isolates by seven bands (when the band at 406 kb is considered a double band, as determined by densitometry). (c) ERIC-PCR patterns for four isolates (lanes 1 to 4, respectively) found in one stool sample. The pattern for one isolate (lane 1) differed from the patterns for the other three isolates, which showed no significant differences. Lanes M, molecular size markers.
The analysis of the ERIC-PCR patterns showed a total of 44 different patterns. For 49 of 50 patients the four isolates showed identical patterns. For one patient the pattern for one isolate differed from the patterns for the other three isolates (Fig. 1c), thus reproducing the results found by PFGE. The similarity between those two different patterns was 51%, and the patterns for the isolates were the same those as shown in Fig. 1b.
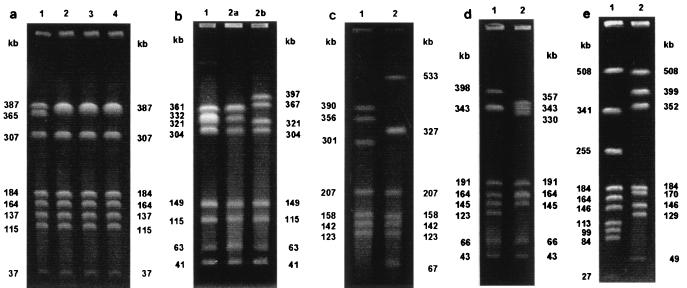
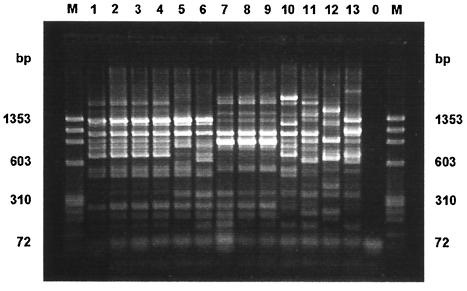
More than one stool sample from 52 patients was positive for Campylobacter (2 to 9 samples, with a mean of 2.4 samples) during the course of their Campylobacter infections. The total number of isolates was 124, and the interval between the time of recovery of the first and the last isolate from each patient was between 2 and 358 days (mean, 23 days; median, 15 days). All isolates from 49 patients were identified as C. jejuni, and all isolates from 2 patients were identified as C. coli. For one patient C. jejuni and C. coli (as proved by PCR) were detected at an interval of 25 days. A total of 55 different band patterns were demonstrated by PFGE. For 47 of 52 patients, the patterns for the isolates obtained by PFGE remained unchanged during the infection, while for 5 patients the band patterns for the isolates changed. In the case of a 27-year-old immunosuppressed patient, the initial isolate exhibited a band at 365 kb, while after 152, 314, and 358 days, a double band at approximately 387 kb was detected for the subsequently recovered isolates (Fig. 2a). To confirm this result an additional restriction with KpnI was performed, and it revealed identical patterns for isolates 1 to 3 from this patient and a pattern for isolate 4 that differed by one band from those for isolates 1 to 3 (data not shown). A second stool sample was obtained from a 7-year-old child 28 days after isolation of C. jejuni from the first stool sample. Colonies with two different morphologies were noticed on the primary isolation medium, and the isolates in both colonies were identified as C. jejuni. The band pattern for the smaller colonies exhibited a double band at 361 kb and differed from the pattern for the previous isolate by two bands, while the band pattern for the larger colonies differed from that for the previous isolate by four bands. The patterns for the small and the large colonies differed by six bands (Fig. 2b). Two patterns for the C. jejuni isolates obtained from a 27-year-old man within an interval of 7 days exhibited a difference of seven bands when the band in lane 2 (Fig. 2c) at 327 kb is considered a double band, as demonstrated by densitometry (data not shown). Two isolates were obtained within an interval of 33 days from a child 21 months of age. The patterns for these isolates differed by six bands if the bands at 343 kb (lane 1) and 164 kb (lane 2) are considered double bands (Fig. 2d). For a 23-year-old patient, a second isolate that was identified as C. coli was isolated 25 days after the isolation of C. jejuni. The patterns for the isolates of the two species differed by 12 bands (Fig. 2e). All isolates from the five patients that exhibited different PFGE patterns during the course of infection were also typed by ERIC-PCR. By ERIC-PCR the isolates from one patient that differed by two bands by PFGE (Fig. 2b, lanes 1 and 2a, respectively) exhibited identical patterns. Also, the three isolates from one patient with different colony morphologies and with PFGE patterns that differed by two to six bands had identical PCR patterns. The three pairs of isolates whose patterns differed by 6 to 12 PFGE bands showed distinct PCR patterns, with similarities of 89, 56, and 38%, respectively (Fig. 3).
FIG. 2.
(a) PFGE patterns for four isolates from four different stool samples of a 27-year-old immunosuppressed male. Lanes 1 to 4 show the patterns for the isolates obtained at the first examination and 152, 314, and 358 days after the first examination, respectively. The pattern for the initial isolate exhibited a band at 365 kb, whereas the other isolates showed double bands at 387 kb. (b) Isolates collected from two samples of a 7-year-old child. Two isolates from the second sample with small and large colony morphologies (lanes 2a and 2b, respectively) were detected. The PFGE patterns for all three isolates differed from each other. (c) PFGE patterns for two isolates obtained from a 27-year-old male showing a difference of seven bands with a double band at 327 kb. (d) Isolates collected from a 21-month-old child. The PFGE patterns differed by six bands, with the bands at 343 kb (lane 1) and 164 kb (lane 2) considered double bands. (e) Different PFGE patterns for two isolates obtained from a 23-year-old male and identified as C. coli (lane 1) and C. jejuni (lane 2).
FIG. 3.
ERIC-PCR patterns for those isolates that exhibited different PFGE patterns during the course of infection. Weight markers (lane M) and the pattern for a negative control (lane 0) are also shown. All isolates with PFGE patterns that differed at 6 to 12 bands also had different PCR patterns. Lanes 5 and 6, lanes 10 and 11, and lanes 12 and 13 correspond to the lanes in Fig. 2d, c, and e, respectively. Isolates with PFGE patterns that differed by two to six bands had identical PCR patterns. The PCR patterns in lanes 1 to 4 correspond to the PFGE patterns in Fig. 2a, lanes 1 to 4, respectively. The PCR patterns in lanes 7 to 9 correspond to the PFGE patterns in the three lanes in Fig. 2b, respectively.
C. jejuni NCTC 11351 was included in each experiment as a reference strain. In all experiments the patterns for this strain clustered with similarities of at least 84 and 80% with the patterns in the PFGE and PCR databases, respectively. Therefore, in the computer-based analysis all PFGE and ERIC-PCR patterns that clustered with similarities higher than 84 and 80%, respectively, were considered identical. Those clusters of patterns considered identical by computer-based analysis were additionally checked by visual comparison. Since ERIC-PCR amplicons smaller than 300 bp and larger than 1,700 bp had poor reproducibilities while amplicons within this size range had high reproducibilities, only bands between 300 and 1,700 bp were analyzed.
Tracking the sources and routes of infection in outbreak situations and, even more interestingly, in sporadic cases of infection is based to a large extent on analysis of genetic markers and molecular fingerprinting methods. All these strategies depend on two major preconditions: (i) the exclusive presence of the investigated clone of a certain species in the putative source as well as in the infected individual and (ii) the stability of the distinctive molecular characteristics used as epidemiologic markers in a defined bacterial clone. For epidemiologic studies of human Campylobacter infections, neither of these two conditions has yet been unequivocally proven. In contrast, the isolation of multiple Campylobacter strains from single sources of infection (Aarts et al., COST Action Workshop on Pathogenic Microorganisms in Poultry and Eggs) and the presence of genotypic variation within single Campylobacter clones, as demonstrated by RFLP analysis of the flagellin gene (9) and as detected even more reliably by PFGE (17, 18), have raised substantial doubts concerning these preconditions (13). The appearance of 55 and 44 different band patterns by PFGE and ERIC-PCR, respectively, in the Campylobacter isolates from a total of 50 patients from a restricted geographic area proved the high discriminatory powers of both of these methods for the typing of Campylobacter spp. For isolates from 96% of the patients, a single band pattern was exclusively obtained by both typing techniques. For the isolate from one patient (Fig. 1a), a difference in the PFGE pattern of only two bands at least indicated a strong genetic relation between the isolates. Even more, the difference might be interpreted as the mobility shift of one band for identical isolates. This assumption was supported by restriction with KpnI and ERIC-PCR, which did not discriminate between the two isolates. The PFGE band patterns from various isolates from one stool sample from only one patient differed by seven bands, indicating genetic diversity. This finding suggests that human Campylobacter infection most frequently is due to a single bacterial clone. This finding is somewhat surprising in light of the previously described contamination of putative sources, i.e., poultry, with more than one clone (5). A possible reason for this discrepancy might be differences in the pathogenicities of food-contaminating strains, with only certain strains being able to establish infections in humans. Furthermore, different growth demands could lead to the overgrowth of a single strain. The preparation of food could also result in the selection of one strain. Improper cooking of chicken meat might lead to killing of superficially contaminating bacteria, while a strain in the inner parts of the food would be exposed to lower temperatures, thus being able to survive. Finally, those results could be explained by overestimation of poultry meat as a source of infection since simultaneous contamination with different strains has been demonstrated to a much lesser extent for other sources like raw milk and drinking water (7, 11). However, this subject has not been studied in detail.
The second part of the study focused on a further prerequisite of epidemiologic investigations, namely, the stability of the genetic fingerprint of Campylobacter. For 90% of the patients, a single bacterial genetic fingerprint was detected by PFGE throughout the course of a Campylobacter infection. For 10% of the patients, however, the band patterns for the isolates changed during the infection episode. In one patient (Fig. 2e) the difference was probably due to a coinfection or a reinfection since the two isolates detected within an interval of more than 3 weeks were identified as C. jejuni and C. coli, respectively. Underlining the lack of genetic relatedness of these two isolates, the PFGE patterns differed by as many as 12 bands. For isolates from four patients (Fig. 2a to d), two to seven different bands in the patterns for the Campylobacter isolates obtained during the course of infection could be observed by PFGE. For one patient (Fig. 2a) an additional difference was detected by the use of a second restriction enzyme. Interestingly, the isolates that differed by less than six bands by PFGE could not be discriminated by ERIC-PCR, while isolates that differed by more than six bands by PFGE also showed distinct ERIC-PCR patterns. These findings underline the high degree of discriminatory power of PFGE for the typing of Campylobacter spp. and support the recommendation that more than a single typing technique should be used for epidemiologic studies (2). Due to the design of this study, it cannot definitely be determined whether the genetic differences in Campylobacter isolates obtained during the course of an infection were due to genetic changes or to a coinfection or a reinfection. With regard to the first part of the investigation, in which the simultaneous detection of distinct genotypes was very rare, however, genetic instability is more probable. This instability was associated with the acquisition of DNA of approximately 22 kb (Fig. 2a), 40 kb, and 70 kb (Fig. 2b). For the isolates from two patients with differences of six (Fig. 2d) or seven (Fig. 2c) bands, the mechanisms of acquisition or loss of DNA and the simultaneous rearrangement or acquisition of point mutations are much more complex, and several causative genetic events would be possible. To exclude the possibility that these strains had high-grade genetic instability, they were serially passaged in vitro 45 times on nonselective medium. The PFGE patterns, however, remained unchanged (data not shown).
In conclusion, simultaneous infection with more than one Campylobacter strain seems to be a rare event and therefore does not remarkably impair genotypic analysis of epidemiologic patterns. However, during an infection episode the causative Campylobacter strain can undergo substantial genetic changes. This must be taken into account for future epidemiologic investigations. Otherwise, misinterpretation of data by underestimation of epidemiologic relations might be the consequence.
Acknowledgments
We thank Marianne Vetter-Knoll and colleagues of the stool laboratory at the Institut für Medizinische Mikrobiologie und Hygiene for skillful assistance.
REFERENCES
- 1.Bottone R J, Hanna B, Hong T, Inzana T J, Namdari H, Qureshi M N, Weyant R S. Cumitech 27, Laboratory diagnosis of zoonotic infections: bacterial infections obtained from companion and laboratory animals. 1996. Coordinating ed., T. J. Inzana. American Society for Microbiology, Washington, D.C. [Google Scholar]
- 2.de Boer P, Duim B, Rigter A, van der Plas J, Jacobs-Reitsma W F, Wagenaar J A. Computer-assisted analysis and epidemiological value of genotyping methods for Campylobacter jejuni and Campylobacter coli. J Clin Microbiol. 2000;38:1940–1946. doi: 10.1128/jcm.38.5.1940-1946.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dworkin J, Blaser M J. Molecular mechanisms of Campylobacter fetus surface layer protein expression. Mol Microbiol. 1997;26:433–440. doi: 10.1046/j.1365-2958.1997.6151958.x. [DOI] [PubMed] [Google Scholar]
- 4.Eyers M, Chapelle S, Van Camp G, Goossens H, De Wachter R. Discrimination among thermophilic Campylobacter species by polymerase chain reaction amplification of 23S rRNA gene fragments. J Clin Microbiol. 1993;31:3340–3343. doi: 10.1128/jcm.31.12.3340-3343.1993. . (Erratum, 32:623, 1994.) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geilhausen B, Schuett-Gerowitt H, Aleksic S, Koenen R, Mauff G, Pulverer G. Campylobacter and Salmonella contaminating fresh chicken meat. Zentbl Bakteriol Parasitenkd Infektkrankh Hyg Abt 1 Orig. 1996;284:241–245. doi: 10.1016/s0934-8840(96)80099-5. [DOI] [PubMed] [Google Scholar]
- 6.Giesendorf B A J, Goossens H, Niesters H G M, van Belkum A, Koeken A, Endtz H P, Stegemann H, Quint W G V. Polymerase chain reaction-mediated DNA fingerprinting for epidemiological studies on Campylobacter spp. J Med Microbiol. 1994;40:141–147. doi: 10.1099/00222615-40-2-141. [DOI] [PubMed] [Google Scholar]
- 7.Hanninen M L, Niskanen M, Korhonen L. Water as a reservoir for Campylobacter jejuni infection in cows studied by serotyping and pulsed-field gel electrophoresis (PFGE) Zentbl Veterinärmed. 1998;45:37–42. doi: 10.1111/j.1439-0450.1998.tb00764.x. [DOI] [PubMed] [Google Scholar]
- 8.Hanninen M L, Hakkinen M, Rautelin H. Stability of related human and chicken Campylobacter jejuni genotypes after passage through chick intestine studied by pulsed-field gel electrophoresis. Appl Environ Microbiol. 1999;65:2272–2275. doi: 10.1128/aem.65.5.2272-2275.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harrington C S, Thomson-Carter F M, Carter P E. Evidence for recombination in the flagellin locus of Campylobacter jejuni: implications for the flagellin gene typing scheme. J Clin Microbiol. 1997;38:2386–2392. doi: 10.1128/jcm.35.9.2386-2392.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez J, Fayos A, Ferrus M A, Owen R J. Random amplified polymorphic DNA fingerprinting of Campylobacter jejuni and Campylobacter coli isolated from human faeces, seawater and poultry products. Res Microbiol. 1995;146:685–696. doi: 10.1016/0923-2508(96)81065-5. [DOI] [PubMed] [Google Scholar]
- 11.Hudson J A, Nicol C, Wright J, Hasell S K. Seasonal variation of Campylobacter types from human cases, veterinary cases, raw chicken, milk and water. J Appl Microbiol. 1999;87:115–124. doi: 10.1046/j.1365-2672.1999.00806.x. [DOI] [PubMed] [Google Scholar]
- 12.Nachamkin I, Ung H, Patton C M. Analysis of HL and O serotypes of Campylobacter strains by the flagellin gene typing system. J Clin Microbiol. 1996;34:277–281. doi: 10.1128/jcm.34.2.277-281.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.On S L. In vitro genotypic variation of Campylobacter coli documented by pulsed-field gel electrophoretic DNA profiling: implications for epidemiological studies. FEMS Microbiol Lett. 1998;165:341–346. doi: 10.1111/j.1574-6968.1998.tb13167.x. [DOI] [PubMed] [Google Scholar]
- 14.Owen R J, Sutherland K, Fitzgerald C, Gibson J, Borman P, Stanley J. Molecular subtyping scheme for serotypes HS1 and HS4 of Campylobacter jejuni. J Clin Microbiol. 1995;33:872–877. doi: 10.1128/jcm.33.4.872-877.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Versalovic J, Koeuth T, Lupski J R. Distribution of repetitive DNA sequences in eubacteria and application to fingerprinting of bacterial genomes. Nucleic Acids Res. 1991;19:6823–6831. doi: 10.1093/nar/19.24.6823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang Y, Taylor D. Natural transformation in Campylobacter species. J Bacteriol. 1990;172:949–955. doi: 10.1128/jb.172.2.949-955.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wassenaar T M, Fry B N, van der Zeijst B A M. Variation of flagellin gene locus of Campylobacter jejuni by recombination and horizontal gene transfer. Microbiology. 1995;141:95–101. doi: 10.1099/00221287-141-1-95. [DOI] [PubMed] [Google Scholar]
- 18.Wassenaar T M, Geilhausen B, Newell D G. Evidence of genomic instability in Campylobacter jejuni isolated from poultry. Appl Environ Microbiol. 1998;64:1816–1821. doi: 10.1128/aem.64.5.1816-1821.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]