Abstract
A large proportion of colorectal carcinomas (CRC) evolve from colorectal adenomas. However, not all individuals with colonic adenomas have a risk of CRC substantially higher than those of the general population. The aim of the study was to determine the differences or similarities of mutation profile among low- and high-grade adenomas and in situ carcinoma with detailed follow up. We have investigated the mutation spectrum of well-known genes involved in CRC (such as APC, BRAF, EGFR, NRAS, KRAS, PIK3CA, POLE, POLD1, SMAD4, PTEN, and TP53) in a large, well-defined series of 96 adenomas and in situ carcinomas using a high-throughput genotyping technique. Besides, the microsatellite instability and APC and MLH1 promoter methylation were studied as well. We observed a high frequency of pathogenic variants in the studied genes. The APC, KRAS and TP53 mutation frequencies were slightly lower in adenoma samples than in in situ carcinoma samples. Further, when we stratified mutation frequency based on the grade, the frequency distribution was as follows: low-grade adenoma—high-grade adenomas—in situ carcinoma: APC gene 42.9–56.0–54.5%; KRAS gene 32.7–32.0–45.5%; TP53 gene 8.2–20.0–18.2%. The occurrence of KRAS mutation was associated with the presence of villous histology and methylation of the APC promoter was significantly associated with the presence of POLE genetic variations. However, no association was noticed with the presence of any singular mutation and occurrence of subsequent adenoma or CRC. Our data supports the multistep model of gradual accumulation of mutations, especially in the driver genes, such as APC, TP53 and KRAS.
Subject terms: Cancer genetics, Cancer epigenetics
Introduction
Colorectal cancer (CRC), the third most common cancer and the fourth most frequent cause of cancer death worldwide1, represents an ideal model to investigate and dissect the genetic alterations involved in tumor initiation and progression. It has been known for some time that the majority of CRCs arises and progresses through a series of well-defined molecular and histopathological changes, the so-called adenoma-carcinoma sequence2,3, first described by Fearon and Vogelstein4.
The adenoma-carcinoma sequence was described as a gradual transformation of colorectal epithelium to adenomatous lesions and ultimately to an adenocarcinoma and a metastatic tumor. Even though most neoplastic adenomas will not give rise to cancer, it is well accepted that most colorectal carcinomas evolve from adenomatous polyps5. However, it cannot currently be predicted which of the early lesions will develop into cancer6. Molecular alterations that play a role in the initiation and progression of CRC suggest a heterogeneous adenoma-carcinoma sequence that comprises several distinct molecular pathways. These include chromosomal instability, microsatellite instability (MSI), and CpG island methylator phenotype (CIMP) pathways that all of which are responsible for genetic and epigenetic instability in CRC2. These genetic and epigenetic alterations affect different pathways that regulate multiple biological processes critical to cancer development7.
Genetic mutations enriched in both adenomas and carcinomas are likely to represent early driver events. Mutations present predominantly in the carcinomas may constitute later driver mutations involved in tumor progression. Genes mutated in adenomas and not mutated in associated carcinoma tissue comprise either random mutation events not important for cancer initiation, or rare events that were not identified in the carcinomas8.
The aim of the present study was to explore the genetic heterogeneity of adenomas and early carcinomas by analyzing the mutation spectrum of well-known genes involved in colorectal carcinogenesis (APC, BRAF, EGFR, NRAS, KRAS, PIK3CA, POLE, POLD1, SMAD4, PTEN, and TP53) in a large, well-defined series of adenomas and in situ carcinomas with follow up using a next generation sequencing approach.
In this context, we have analyzed each gene for the number and type of mutations present in the adenomas and in situ carcinomas and their specific relationship to MSI status. In addition, we have analyzed the levels of methylation of CRC-related genes by methylation-sensitive high-resolution melting (MS-HRM). We investigated the tumor suppressor adenomatous polyposis coli gene (APC), which encodes a key protein in the WNT signaling pathway and is indicated as an early event in carcinogenesis9, and MLH1 gene whose aberrant promoter methylation is responsible for the loss of mismatch repair activity.
Results
Patient’s characteristics
The studied set included 96 patients, out of which 74 were patients with adenomas and 22 with in situ carcinoma. The clinic-pathological characteristics are presented in Table 1.
Table 1.
Patient´s clinical characteristics.
| All n = 96 (%) |
Adenomas n = 74 (%) |
In situ carcinomas n = 22 (%) |
p value (test) for difference between adenomas and in situ carcinomas |
||
|---|---|---|---|---|---|
| Age (mean ± SD) years | 65.6 ± 10.4 | 65.1 ± 10.6 | 67.0 ± 9.9 | 0.48 (t test) | |
| Sex | Men | 58 (60.4) | 46 (62.2) | 12 (54.5) | 0.62 (Fisher’s) |
| Women | 38 (39.6) | 28 (37.8) | 10 (45.5) | ||
| Lesion site | Colon | 54 (56.2) | 46 (62.2) | 8 (36.4) | 0.05 (Fisher’s) |
| Rectum | 42 (43.8) | 28 (37.8 | 14 (63.6) | ||
| Polyp type | Tubular | 46 (47.9) | 37 (50) | 9 (40.9) | 0.62 (Fisher’s) |
| Tubulo-villous | 38 (39.6) | 29 (39.2) | 9 (40.9) | ||
| Villous | 12 (12.5) | 8 (10.8) | 4 (18.2) | ||
| Grade* | Low | – | 49 (66.2) | – | – |
| High | – | 25 (33.8) | – | ||
*For adenoma patients only.
APC and MLH1 promoter methylation
The promoter methylation status of APC and MLH1 genes was studied by MS-HRM.
There was no remarkable difference in the distribution of promoter methylation in the APC gene (mean 9%): for in situ carcinoma, the mean promoter methylation in affected tissue was 8% and in adenomas it was 8.4% (9.1% for low grade and 6.8% for high grade dysplasia).
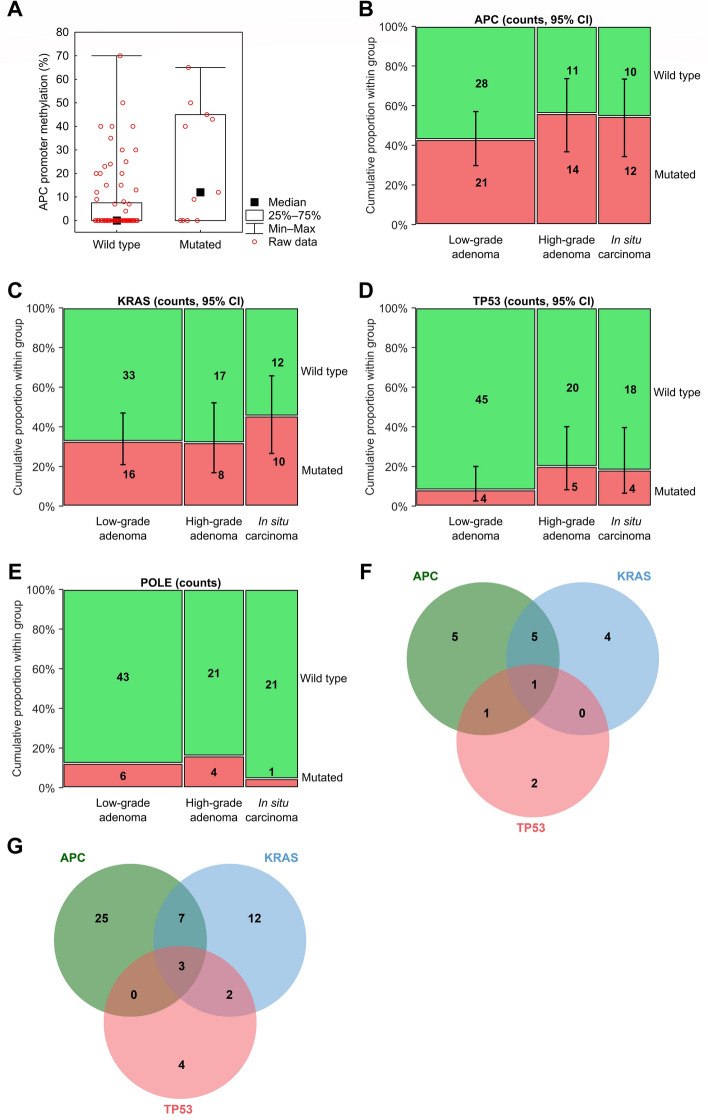
Interestingly, methylation of the APC promoter was significantly associated with the presence of POLE genetic variations (p = 0.02, Fig. 1A; n = 11).
Figure 1.
The mutated gene signature of colorectal adenomas and in situ carcinomas. (A) The APC promoter methylation distribution with POLE genetic variations, (B) The mutation distribution of APC gene between low-, high-grade adenomas and in situ carcinomas, (C) The mutation distribution of KRAS gene between low-, high-grade adenomas and in situ carcinomas, (D) The mutation distribution of TP53 gene between low-, high-grade adenomas and in situ carcinomas, (E) The mutation distribution of POLE gene between low-, high-grade adenomas and in situ carcinomas, (F) The Venn diagram of mutations of APC, TP53, and KRAS genes in in situ carcinomas, (G) The Venn diagram of mutations of APC, TP53, KRAS, and POLE genes in adenomas.
The only one hypermethylated MLH1 promoter corresponded to the only sample with MSI-H status.
MSI status
MSI status of all adenomas and in situ carcinomas was tested. In our set, only 3 samples had MSI instability: 2 of them with MSI-L and one with MSI-H status. The MSI-H status was observed in an in situ carcinoma located in the right colon while MSI-L was noticed in two low grade dysplasia samples located in the left colon.
Mutation spectrum
In total, 96 adenomas and in situ carcinomas were analyzed for mutations in 11 genes (APC, BRAF, EGFR, NRAS, KRAS, PIK3CA, POLE, POLD1, SMAD4, PTEN, and TP53). The mutation hotspots in KRAS, NRAS, BRAF, PIK3CA, EGFR, SMAD4 genes as well as exonuclease domain for POLE and POLD1 genes were studied. Concerning the APC, PTEN and TP53 genes, we have focused on the entire open reading frame (ORF).
Of all 96 samples, 21 (18 adenomas and 3 in situ carcinomas) of them did not bear any mutation in the studied genes. Out of the remaining 75 samples with mutation(s), 56 were adenomas and 19 carcinomas (Fig. 2). We did not observe any significant differences in mutation distribution between adenomas and in situ carcinomas.
Figure 2.
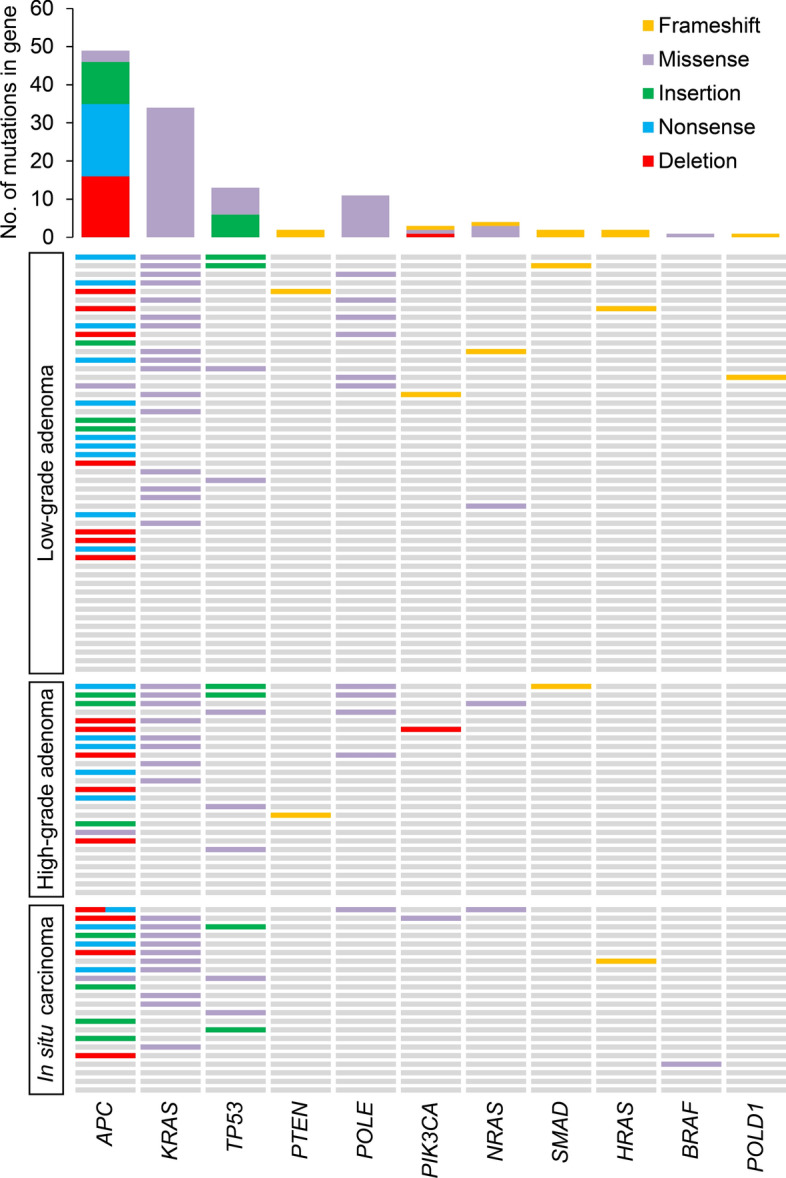
The distribution of genetic alterations detected in low-grade, high-grade adenomas, and in situ carcinomas. Each row represents a patient, and each column represents a gene. Different mutation types are indicated by different colors. The bar chart on the top shows the total number of the given gene’s mutations observed in the sample.
APC
In the present study, we have focused on the entire ORF of the APC gene as it harbors most of the mutations described in the APC gene. In both the low- and high-grade adenoma samples (Fig. 1B), 36 deleterious (according to HGMD guidelines and pathogenic according to ACMG guidelines; Supplementary Table 1) mutations in 35 individuals were observed: 12 deletion (p.E1268fs, p.1275_1275del, p.D1279fs, p.T1283fs, p.1289_1291del, p.1302_1304del, p.1344_1350del, p.P1406fs, p.1431_1432del, p.T1469fs, p.P1479fs, and p.1561_1562del), 7 insertion (p.Q1226fs, p.S1316fs, p.V1359fs, p.P1391fs, p.T1469fs, p.G1481fs, and p.E1536fs), 15 nonsense (three times p.Q1273X, twice p.Q1349X, twice p.E1379X, twice p.R1432X, p.Q1276X, p.E1288X, p.S1297X, p.E1304X, p.E1335X, and p.Q1349X), and 2 missense (p.T1274M and p.E1299Q) mutations. While in carcinoma samples, 13 mutations in 12 patients were detected: 4 deletion (p.1284_1286del, p.1454_1455del, p.T1469fs, and p.E1542fs), 4 insertion (p.Q1226fs, p.M1365fs, p.S1377fs, and p.T1478fs), 4 nonsense (p.E1198X, p.K1292X, p.E1361X, and p.E1379X), and 1 missense (p.L1493I) mutations. Interestingly, one patient with low grade adenoma and one patient with carcinoma had two concurrent APC deleterious mutations (2 insertions (p.G1481fs and p.E1536fs) in the adenoma sample and 1 deletion (p.E1542fs) and 1 nonsense (p.E1379X) type of APC mutation in the carcinoma sample).
Interestingly, several identical APC mutations have been observed both in high-grade adenomas and in situ carcinomas, namely: deletion (p.T1469fs), insertion (p.Q1226fs) and nonsense (p.E1379X) mutations.
KRAS
For the KRAS gene, several missense mutations were successfully identified in 34 individuals [24 individuals with either low- or high-grade adenoma and 10 with in situ carcinoma, (Fig. 1C)] at codon 12: c.34G > C/A/T, c.35G > A/T/C, codon 13: c.38G > A/C/T, codon 61: c. 182A > G, c.183A > C: and 146: c.437C > T. Both adenomas and carcinomas were most frequently mutated at codon 12, position 35 (22 cases out of 34: in particular, in 16 adenomas and 6 carcinomas, 73% and 27%, respectively) and a G > A transition was the most common nucleotide for both phenotypes (14 cases out of 34: in particular, 10 in adenomas and 4 in carcinomas, 71% and 29%, respectively). The presence of KRAS mutation was significantly associated with the presence of villous histology (p = 0.02).
TP53
Concerning the TP53 gene, we have focused on the entire ORF. Nine patients with adenomas and 4 carcinoma patients bore mutations in the TP53 gene (Fig. 1D). In the adenoma samples (either grade), 5 of them had missense (p.R43H, p.P45S, p.H47R, p.R49C, and p.R116Q) mutations and 4 had insertion (twice p.H46fs and twice p.C143fs). On the other hand, 4 mutations were identified in carcinoma samples: 2 missense (p.R43H and p.I122V) and 2 insertion (p.P59fs and p.V71fs). None of the analyzed patients had more than one TP53 mutation. The p.R43H missense mutation was observed both in high-grade adenoma and in in situ carcinoma tissues.
APC, KRAS and TP53 co-mutations
Only 4 out of 96 samples had all 3 APC, KRAS and TP53 genes mutated. Interestingly, 1 sample was a carcinoma, 2 samples were high grade dysplasia adenomas, and 1 sample a low grade dysplasia adenoma (Fig. 1F,G). All above samples had an MSS status and were located in the right colon. The individual with low grade dysplasia later developed another adenoma in the bowel.
Simultaneously, when we compared the frequency of mutations in these three genes between both low- and high-grade adenomas and in situ carcinoma, the APC mutation frequency was lower in adenoma samples than in carcinoma samples (47.3% vs. 54.5%, p = 0.63). The KRAS mutation frequency was again lower in adenoma than in carcinoma samples (32.4% vs. 45.5%, 0.31), while the TP53 mutations frequency was the lower in adenomas and the highest in carcinoma samples (12.2% vs. 18.2%, p = 0.27). These distribution differences among adenomas and in situ carcinomas were not significant. Further, when we stratified mutation frequency for low- and high-grade adenoma, the distribution was as follows: low-grade adenoma—high-grade adenomas- in situ carcinoma: APC gene 42.9–56.0–54.5%; KRAS gene 32.7–32.0–45.5%; TP53 gene 8.2–20.0–18.2%.
Other mutations
Four NRAS mutations were found, two missense (twice c.181A (p.Q61K) in the right colon and a c.38A (p.G13R)) mutations and insertion (c.430dupA:p.T144fs) in the left colon. The first two mentioned missense mutations have recently been described in adenomas7. The second mentioned NRAS mutation was observed in in situ carcinoma in our set of patients.
In the present study, BRAF mutation c.1799 T > A (p.V600E) was only observed in a single carcinoma sample that also displayed MSI-H status. Further, three PIK3CA mutations were detected, namely a hotspot missense mutation (c.1624G > A, p.E542K) in an in situ carcinoma and two deletions ((twice c.3114delT:p.Y1038fs), one in a low-grade and one in a high-grade dysplasia sample). SMAD4 (c.941dupT:p.I314fs in high-grade dysplasia and c.353_354insA:p.A118fs in low-grade dysplasia) and PTEN (twice c.908dupT:p.I303fs in low- and high-grade dysplasia) mutations were also recorded.
Additionally, mutations in POLD1 and POLE genes were studied. For the POLD1 gene, a c.1263 dupG:p.L421fs mutation was observed in low grade dysplasia. Interestingly, in 12 samples (11 adenomas and 1 in situ carcinoma, (Fig. 1E), POLE (c.T1166C:p.F389S) mutation was identified. However, no record about this particular mutation was published in COSMIC10, LOVD11 or HGMD12. For this reason, we rather assessed this genetic variant as variant of uncertain significance (VUS) rather than pathogenic mutation.
There was no significant overlap between these mutations identified in our set of samples.
Follow up
For 70 out of 96 patients included into the study, follow up data were available. Out of these, 21 developed a subsequent adenoma. Seventeen of them had previously adenoma (7 high-grade and 10 low-grade adenoma) and four in situ carcinoma (Table 2). The only common feature of these patients is that their first samples included in our study were all of an MSS status (apart one low grade adenomas that was MSI-L). Unfortunately, we were not able to obtain the following adenoma tissue for analysis. Apart from one low-grade and one high-grade adenoma, all of them had previously at least one mutation in the high-risk genes, such as APC, KRAS, or TP53, however no association was observed in relation of the presence of any singular mutation and occurrence of subsequent adenoma. Concerning the histology and subsequent adenoma, patients with mixed histology (tubule-villous) were less prone to develop further adenomas than those with villous or tubular histology alone (p = 0.02). Besides, patients with higher age also develop subsequent adenomas rather than younger patients (p = 0.04).
Table 2.
Follow up of patients included into the study.
| Normal findings | Recurrence of adenoma | Occurrence of CRC | |
|---|---|---|---|
| Low grade adenomas | 39 | 10 | 2 |
| High grade adenomas | 18 | 7 | 1 |
| In situ carcinomas | 12 | 4 | 6 |
In all patients with in situ carcinoma, the tumor was surgically removed, and patients were regularly monitored by colonoscopy procedure.
Furthermore, out of 22 patients with in situ carcinoma, 4 developed adenoma (as stated above) and 6 even invasive carcinoma within few years after the first in situ carcinoma. The proportion of patients developing subsequent invasive carcinoma depended significantly on the type of their primary lesion (p = 0.009), being by far the highest for in situ carcinomas (27.3%) in comparison to low-grade and high-grade adenomas (4.1% and 4.0%, respectively).
Most of the patients with in situ carcinoma who underwent a clinical follow up had further developed CRC in about the same location as the previous findings quite soon or within two years.
For the patient with adenoma and subsequently developed CRC, the situation is a bit different. In a few years, the emerging CRC was located in a completely different colon segment than the previous adenoma. One patient with low grade adenoma developed liver metastases very quickly.
Discussion
The progression from adenoma into cancer can take as long as 20 years and is not usually affected by a single pathway5,13,14. This transition represents a multistep process that is characterized by chromosomal instability (CIN), MSI, and CIMP. Effects of all these pathways may combine and are responsible for genetic instability in an adenoma that underlies malignant transformation15.
Despite this, little is known about the mutation profiles of advanced adenomas and in situ carcinomas. It is not certain whether they share the same genetic background, or the driver mutations are more abundant in in situ carcinomas compared to adenomas. As these two forms of neoplasia are described as subsequent stages of carcinogenesis in the Vogelstein model, the cascade of low grade—high grade- adenoma and in situ carcinoma represents a suitable model for the analysis of CRC development. Up to now, the research of the transformation of colon adenoma into cancer has mostly focused on association studies assessing the cancer risk or advanced colorectal carcinomas.
The aim of the present study was to use a well-defined series of adenomas and in situ carcinomas to perform a parallel investigation of the mutation status of 11 genes known to be involved in CRC, as hypothesized by Vogelstein. The studied genes are involved in several different signal transduction pathways.
According to the recent study by Lee-Six et al.16, mutations in APC, KRAS and TP53 genes are common in CRC (accounting for 56% of base-substitution and indel driver mutations) while being rare among unaffected colonic crypts. The authors suggested that mutations in these genes confer higher likelihoods of conversion of normal epithelium to adenoma and carcinoma. However, in our study, only 4 out of 96 individuals had all APC, KRAS and TP53 genes mutated concurrently suggesting that further alterations in mutational frequency and spectrum may occur along with CRC progression.
In our study, we did not observe any significant differences in the distribution frequency of mutations in APC, KRAS and TP53 genes between adenoma and in situ carcinomas. Without the stratification for low- and high-grade adenoma, the mutation frequencies were rather similar. The APC mutation frequency was moderately lower in adenoma samples (47.3%) than in carcinoma samples (54.5%). The KRAS mutation frequency was again lower in adenoma than in carcinoma samples (32.4% and 45.5%), as were the TP53 mutation frequency (12.2% and 20%). Only 4 out of 96 individuals had all APC, KRAS and TP53 genes mutated concurrently. Further, when we stratified mutation frequencies for low- and high-grade adenoma, the distribution frequencies of mutations in the APC and TP53 genes had an increasing tendency towards in situ carcinoma, while the frequency of mutations in the KRAS gene in the low- and high-grade adenoma was lower than that in in situ carcinoma. This may point to the fact that in situ carcinoma still carries the mutation profile of the adenoma and only during further progression the mutation frequencies change, or, alternatively vice versa that high-grade adenomas are already approaching in situ carcinomas with their mutation profile. Therefore, a more thorough examination and assessment of the risk of CRC in people with adenomas is needed. Nonetheless, the observed higher mutation frequency in in situ carcinomas resembles the generally accepted Fearon and Vogelstein model of carcinogenesis4. However, additional studies are warranted to track the dynamics of mutational in relation to the disease heterogeneity.
The rather higher KRAS mutation frequency than expected might be explained by the fact that the current study investigated KRAS mutations in codons 12, 13, 61 and 146 covering most of all reported mutations for KRAS in CRC, while earlier studies investigated mostly codon 12 and/or 13 of the KRAS gene only17,18.
In our study we have observed the presence of KRAS mutation associated with the presence of villous histology. Similar outcomes were obtained by Zauber et al.19. The authors hypothesized that non-mucinous and MSS CRC with wild-type KRAS gene may have had a mutation in the KRAS gene during their earlier stages, however the mutation was lost during further growth.
Over the last years, many studies have shown that germline mutations in the proofreading domains of POLD1 and POLE predispose to CRC and other malignancies20,21. These mutations arise early in oncogenesis and serve as gatekeeper mutations—conferring a growth advantage to cellular subpopulations and driving tumor growth. Interestingly, in 12 samples (11 adenomas and 1 in situ carcinoma), POLE (c.T1166C:p.F389S) VUS was identified. However, no record about this identified VUS was published in public databases yet, this VUS was observed in oral squamous cell carcinoma22 and small cell lung cancer23 and thus it deserves further attention. Besides this, APC promoter methylation was significantly associated with the presence of POLE VUS. Although inactivating framshift not likely leads to a hypermutation profile, Poulos et al.24 observed the presence of coding mutation hotspots in POLE-mutant cancers at highly-methylated CpGs in the tumor-suppressor genes APC and TP53. This finding points to the links between methylation and mutations and DNA repair, and these mechanisms define a key part of the mutational background of cancer genomes.
While BRAF mutations are more common in MSI colorectal cancers, they are less prevalent in adenomas25. In our study, we have observed BRAF mutation only in one in situ carcinoma sample that simultaneously possessed the MSI-H phenotype. BRAF and KRAS mutations are mutually exclusive26, as was observed in our set of samples.
In the current study, three mutations were found in the PI3KCA and two mutations in the negative regulator of the PI3K-AKT pathway, the PTEN gene. According to the literature, the PIK3CA mutation frequency in CRC ranges between 10–30%27. However, PIK3CA mutations are less common in colorectal adenomas, around 3%, indicating that mutations in PIK3CA would generally arise later during the adenoma-carcinoma transition. Although it may seem that our results are not in complete agreement with these observations, we have detected a hotspot mutation (c.1624G > A, p.E542K) in an in situ carcinoma and two deletion (c.3114delT:p.Y1038fs), one in a low-grade, and one in a high-grade dysplasia sample. However, we focused only on in situ carcinomas and not on invasive CRC and the mutation frequency rate in adenomas is in concordance with other studies, being around 3%.
A further aim of the study was to regularly follow up all patients included into study. Unfortunately, we were not able to follow many of the patients for various reasons: patients did not attend the regular check-ups, moved away, died for other than gastrointestinal reasons, etc. Follow-up data were recorded by clinicians to determine if patients had, a) clear colonoscopy findings, b) recurrence of adenoma, and c) occurrence of CRC. Clinical follow-up of 70 patients out of 96 was performed within this study. Several patients developed a subsequent adenoma or even CRC. The only common feature of these patients is that their first samples included in our study were all of an MSS status and all of them previously had at least one mutation in the high-risk genes, such as APC, KRAS, or TP53.
Concerning the histology and subsequent adenoma, patients with mixed histology (tubule-villous) were less prone to develop further adenomas than those with tubular or villous histology (p = 0.02). The fact that patients with higher age developed subsequent adenomas rather than younger patients might be a result of more regular colonoscopy controls advised among older people.
Besides, out of 22 patients with in situ carcinoma, 4 further developed adenoma and 6 even invasive carcinoma within few years after the first in situ carcinoma diagnosis. The occurrence of in situ carcinoma is in itself a clear evidence of a malignant reversal in the body and therefore these conclusions are not so unexpected.
As it was stated earlier, mutations identified in both adenomas and in situ carcinomas are likely to represent early driver events. However, mutations present predominantly in carcinomas may indicate later driver mutations involved in tumor progression. Genes mutated in adenomas and not mutated in cancer tissue can illustrate either random mutation events that are not as important for cancer onset or rare events that have not been identified in cancers. So how do we know that the adenoma is no longer embarking on transformation into cancer and, conversely, that the cancer does not carry even the historical "adenoma" mutations? Or can it be stated that once the adenoma tissue has accumulated all the necessary mutations, it will switch to carcinoma so quickly that we do not have much chance of catching it in this transitional phase?
Of course, it is still more likely to find those driver mutations in carcinomas and rare events in adenomas, but depending on how fast such a transformation takes place, the classification of genes as potential early and late driving events can help dissect pathways involved in both tumor initiation and progression.
The weaknesses of the present study include the still limited number of patients and incomplete clinical follow-up, partly responsible for the impossibility of comparing the mutational profile of initial lesion and following lesion. Also, the used method detected the presence of the mutation in the one region of the lesion, thus the results do not reflect possible heterogeneity of the non-invasive colorectal lesions. Furthermore, only adenoma tissue was analyzed in this study. Unfortunately, due to Ethical reason, the adjacent unaffected tissue was not collected and analyzed. For this reason, we cannot say with certainty whether we have identified exclusively somatic variants.
The early cancer biomarkers are strongly needed and are taking advantage of rapid progress in molecular biology. These biomarkers should be able to distinguish healthy people from patients with adenomas and subjects with early-stage CRC (stage Tis, I or II) with relative ease and low cost, and to be minimally invasive with aim to increase screening acceptability. With this respect, circulating nucleic acid-based biomarkers (or so-called “liquid biopsy”) are currently extensively studied in cancer research. Circulating cell-free DNA (cfDNA) is probably the most promising tool among all components of liquid biopsy.
Pathogenic mutations in the KRAS, BRAF, APC, and TP53 genes have been predominantly analyzed in the cfDNA isolated from CRC patients and less in patients with adenoma. The concordance of the mutations found in these genes in tumor tissue and plasmatic cfDNA was 100% (reviewed in28). Recently, the study by Cervena et al.29 proved the clinical relevance of APC and TP53 genes especially in the light of longitudinal monitoring of CRC patients.
However, the sensitivity of cfDNA based markers for early-stage disease is lower than for advanced stages30,31. To our knowledge, this is problem that has not been overcome yet. Although in our intended studies we would like to address this problem and attempt to detect identified pathogenic mutations in cfDNA isolated from both plasma and stool of patients with adenoma and early cancer stages.
Conclusion
Our data confirms the Vogelstein's theory of gradual accumulation of mutations, especially in the driver genes, such as APC, TP53 and KRAS. The set of adenomas and in situ carcinomas only very rarely exhibited MSI-H phenotype and the role of mutations in POLE and PI3KCA genes in adenoma to carcinoma transition warrants further investigations.
Material and methods
Tissue samples
Fresh frozen tissue samples from adenomas and in situ carcinomas were collected consecutively at three different institutes during the planned colonoscopy (Thomayer University Hospital, University Hospital Kralovske Vinohrady, and Mediconas, all in Prague, Czech Republic). The study included individuals with adenomas of tubular, villous or tubulo-villous histology and individuals with in situ carcinomas who underwent colonoscopy examination as a part of CRC screening or for intestinal symptoms. There were no age, gender, and ethnicity restrictions. The exclusion criteria were proven hereditary CRC syndromes, inflammatory bowel disease (IBD), histology of hyperplastic polyps, and size smaller than 5 mm. Patients with any personal history of previous malignancy, or with colorectal cancer-associated well-defined inherited syndromes (including Lynch syndrome, familial adenomatous, and MUTYH-associated polyposis) were also excluded from the study.
The study was approved by Ethical committees of all institutions (Institute of Experimental Medicine, Prague, Czech Republic, IKEM and Thomayer hospital) and all individuals agreed with participation in the study and signed an informed consent in accordance with the World Medical Association Declaration of Helsinki.
Colorectal adenomas were histologically classified according to the revised Vienna classification32. Low-grade dysplasia was marked Category 3, while category 4.1 was assigned to high-grade dysplasia. Accordingly, category 4.2 was as assigned to carcinomas in situ. Adenomas with dysplasia (categories 3 and 4.1) or with carcinoma (other categories, such as 4.2) were analyzed separately.
All patients were monitored with a regular follow-up until December 31, 2019. Follow-up data were recorded by clinicians to determine if patients had normal findings on follow-up colonoscopy, recurrence of adenoma, or occurrence of CRC.
DNA and RNA isolation and quality control
Total DNA was isolated using AllPrep DNA/RNA Isolation kit according to the manufacturer’s protocol (Qiagen, Germany). Quantity and purity of DNA was measured using Nanodrop. OD260/280 ratios of all samples ranged between 1.8 and 2.0. After the isolation, DNA was stored at -80 °C.
Bisulfite modification
200 ng of DNA from each sample were treated with sodium bisulfite using the ‘‘EpiTect Bisulfite Kit’’ (Qiagen, Germany) according to the manufacturer’s protocol.
MS-HRM
For the MS-HRM of the APC gene, methylation independent primers, based on Migheli et al.33, were employed. Primer sequences for MLH1 gene were described earlier34. All analyses were run according to the following conditions: 1 cycle of 95 °C for 12 min, 60 cycles of 95 °C for 30 s, Ta for 30 s and 72 °C for 15 s; followed by an HRM step of 95 °C for 10 s and 50 °C for 1 min, 65 °C for 15 s, and continuous acquisition to 95 °C at one acquisition per 0.2 °C. PCR was performed in a final volume of 25 µl, containing 12.5 µl of master mix (Qiagen), 10 pmol of each primer and 1 µl (almost 10 ng) of bisulfite-modified DNA template. Each reaction was performed in triplicate. We analyzed 10% of the samples independently on separate occasions to verify the inter-assay variability and observed a good reproducibility.
Fully methylated and unmethylated DNA (EpiTectH methylated and unmethylated human control DNA, bisulfite converted, Qiagen, Germany) were mixed to obtain the following ratios of methylation: 0%, 12.5%, 25%, 50%, 75%, 100%. Standard curves with known methylation ratios were included in each assay and were used to deduce the methylation ratio of each tumor and reference sample.
MSI Status
MSI status was determined by molecular testing of five mononucleotide repeat markers (Bethesda consensus panel, BAT-25, BAT-26, NR-21, NR-24, and NR-27) that were run as a pentaplex, using fluorescently labeled primers and standard PCR as described in Kroupa et al.35. Fragment analysis was performed on ABI 3130 (Applied Biosystems). A comparison between the adenoma or in situ carcinoma and adjacent mucosa DNA short tandem repetition profiles were analyzed with GeneMapper v4.1 software (Applied Biosystems). When one or more markers were instable, the sample was interpreted as MSI, all other samples were classified as microsatellite stable (MSS). Further, when one instable marker was presented, the sample was indicated as MSI-Low (MSI-L), in the case that 2 or more markers were instable, the sample was marked as MSI-High (MSI-H).
Mutation analysis
DNA concentrations were measured prior to amplification, using the Qubit® dsDNA HS assay (Life Technologies) and diluted to a concentration of 5 ng/μl.
Mutations were determined using a custom multiplex PCR sequencing panel consisted of M13-tailed primer pairs, as described previously36. The custom primers cover mutational hotspots in the genes BRAF, EGFR, KRAS, NRAS, PIK3CA and SMAD4, the exonuclease domain of POLE and POLD1 and the entire coding sequence of APC, PTEN and TP53 (primer sequences available upon request).
PCR was performed with FastStart Hifi Enzyme Blend (Sigma-Aldrich, St. Louis, MO, USA) in two PCR pools with non-overlapping M13-tailed primers. PCR products per sample were combined and purified with Agencourt AMPure XP beads (Beckman Coulter Life Sciences, Brea, CA, USA). In a second PCR barcodes and sequencing primers A and P1 were added. The reads generated by the Ion Torrent PGM sequencer (Thermo Fisher) were mapped against the human reference genome (GRCh37/hg19) using the TMAP 5.0.7 software with default parameters (https://github.com/iontorrent/TS). Variants were called with VarScan with more conservative coverage and minimum variant allele frequency cut-off values for indels (min-coverage = 20, min-var-freq = 0.2) than for single nucleotide variants (min-coverage = 8, min-var-freq = 0.1).
Statistical analysis
The occurrence of individual mutations (independently) between groups was assessed by the Fisher's exact test. Similarly, their possible associations with the localization, gender, or histology type and mutually with each other were determined by the Fisher's exact test. Associations of mutations with age, Vienna classification and APC methylation were analyzed using the Mann–Whitney U test. The statistical analysis was performed using STATISTICA (version 11Cz; TIBCO Software Inc., Palo Alto, CA, USA), Matlab (version 2019b; The MathWorks, Inc., Natick, MA, USA), SISA (https://www.quantitativeskills.com/sisa/statistics/fiveby2.htm) and JVenn (http://jvenn.toulouse.inra.fr/app/index.html,37). Confidence intervals of mutation frequencies were calculated according to Agresti and Coull38. All reported p-values are two-tailed and the level of statistical significance was set at α = 0.05.
Supplementary Information
Author contributions:
Conceptualization, T.W, L.V, P.V, and V.V.; methodology, M.U., A.B., P.B., A.S., J.S., T.W, V.V ; formal analysis, M.U., P.H., A.B.; sample resources, J.J., M.K., D.T., S.S., Z.B., T.B., P.K., T.H., R.M.; data curation, T.W, P.V, V.V.,; writing—original draft preparation, V.V., J.J.; writing—review and editing, J.J.,M.U., A.B., P.H., P.B., A.S., J.S., M.K., D.T., S.S., Z.B., T.B., P.K., T.H., M.R., L.V., T.W., P.V., V.V.; supervision, T.W., P.V.; project administration, L.V.; funding acquisition, T.W, P.V., V.V., P.K., T.H.. All authors have read and agreed to the published version of the manuscript. We thank Ludovit Bielik for his excellent technical support.
Funding
This project was supported by the Czech Health Research council of the Ministry of Health of the Czech Republic (AZV NV18-03-00199), the Grant Agency of the Czech Republic (GACR 18-09709S). We are thankful to Charles University Research Centre program UNCE/MED/006 “University Centre of Clinical and Experimental Liver Surgery” and by project No. CZ.02.1.01/0.0/0.0/16_019/0000787 „Fighting INfectious Diseases “, awarded by the MEYS CR “, financed from EFRR. This publication is part of a project that has received funding from the European Union's Horizon 2020 research and innovation program under grant agreement Nº856620. This article is based upon work from COST Action CA17118 (www.transcoloncan.eu), supported by COST (European Cooperation in Science and Technology). www.cost.eu.
Competing interests
The authors declare no competing interests.
Footnotes
The original online version of this Article was revised: In the original version of this Article, the Supplementary Table 1 was omitted from the Supplementary Information section.
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
These authors contributed equally: Jungwirth Jiri and Urbanova Marketa.
Change history
10/21/2022
A Correction to this paper has been published: 10.1038/s41598-022-21956-0
Change history
4/4/2022
A Correction to this paper has been published: 10.1038/s41598-022-09561-7
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-022-06498-9.
References
- 1.Siegel RL, et al. Cancer statistics, 2021. CA Cancer J. Clin. 2021;71(1):7–33. doi: 10.3322/caac.21654. [DOI] [PubMed] [Google Scholar]
- 2.Grady WM, Markowitz SD. The molecular pathogenesis of colorectal cancer and its potential application to colorectal cancer screening. Dig. Dis. Sci. 2015;60(3):762–772. doi: 10.1007/s10620-014-3444-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carethers JM, Jung BH. Genetics and genetic biomarkers in sporadic colorectal cancer. Gastroenterology. 2015;149(5):1177–1190 e3. doi: 10.1053/j.gastro.2015.06.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell. 1990;61(5):759–767. doi: 10.1016/0092-8674(90)90186-I. [DOI] [PubMed] [Google Scholar]
- 5.Siskova A, et al. Colorectal adenomas-genetics and searching for new molecular screening biomarkers. Int. J. Mol. Sci. 2020;21(9):3260. doi: 10.3390/ijms21093260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Beggs AD, et al. A study of genomic instability in early preneoplastic colonic lesions. Oncogene. 2013;32(46):5333–5337. doi: 10.1038/onc.2012.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Voorham QJ, et al. Comprehensive mutation analysis in colorectal flat adenomas. PLoS One. 2012;7(7):e41963. doi: 10.1371/journal.pone.0041963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wolff RK, et al. Mutation analysis of adenomas and carcinomas of the colon: Early and late drivers. Genes Chromosom. Cancer. 2018;57(7):366–376. doi: 10.1002/gcc.22539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liang TJ, et al. APC hypermethylation for early diagnosis of colorectal cancer: A meta-analysis and literature review. Oncotarget. 2017;8(28):46468–46479. doi: 10.18632/oncotarget.17576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tate JG, et al. COSMIC: The catalogue of somatic mutations in cancer. Nucleic Acids Res. 2019;47(D1):D941–D947. doi: 10.1093/nar/gky1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fokkema IF, et al. LOVD v.2.0: The next generation in gene variant databases. Hum. Mutat. 2011;32(5):557–63. doi: 10.1002/humu.21438. [DOI] [PubMed] [Google Scholar]
- 12.Stenson PD, et al. Human gene mutation database (HGMD): 2003 update. Hum. Mutat. 2003;21(6):577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 13.Loeve F, et al. National polyp study data: Evidence for regression of adenomas. Int. J. Cancer. 2004;111(4):633–639. doi: 10.1002/ijc.20277. [DOI] [PubMed] [Google Scholar]
- 14.Cross W, et al. The evolutionary landscape of colorectal tumorigenesis. Nat. Ecol. Evol. 2018;2(10):1661–1672. doi: 10.1038/s41559-018-0642-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Muller MF, Ibrahim AE, Arends MJ. Molecular pathological classification of colorectal cancer. Virchows Arch. 2016;469(2):125–134. doi: 10.1007/s00428-016-1956-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee-Six H, et al. The landscape of somatic mutation in normal colorectal epithelial cells. Nature. 2019;574(7779):532–537. doi: 10.1038/s41586-019-1672-7. [DOI] [PubMed] [Google Scholar]
- 17.Lewandowska MA, Jozwicki W, Zurawski B. KRAS and BRAF mutation analysis in colorectal adenocarcinoma specimens with a low percentage of tumor cells. Mol. Diagn. Ther. 2013;17(3):193–203. doi: 10.1007/s40291-013-0025-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wang JY, et al. Molecular mechanisms underlying the tumorigenesis of colorectal adenomas: Correlation to activated K-ras oncogene. Oncol. Rep. 2006;16(6):1245–1252. [PubMed] [Google Scholar]
- 19.Zauber P, Marotta S, Sabbath-Solitare M. KRAS gene mutations are more common in colorectal villous adenomas and in situ carcinomas than in carcinomas. Int. J. Mol. Epidemiol. Genet. 2013;4(1):1–10. [PMC free article] [PubMed] [Google Scholar]
- 20.Rayner E, et al. A panoply of errors: Polymerase proofreading domain mutations in cancer. Nat. Rev. Cancer. 2016;16(2):71–81. doi: 10.1038/nrc.2015.12. [DOI] [PubMed] [Google Scholar]
- 21.Palles C, et al. Germline mutations affecting the proofreading domains of POLE and POLD1 predispose to colorectal adenomas and carcinomas. Nat. Genet. 2013;45(2):136–144. doi: 10.1038/ng.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boot A, et al. Characterization of colibactin-associated mutational signature in an Asian oral squamous cell carcinoma and in other mucosal tumor types. Genome Res. 2020;30(6):803–813. doi: 10.1101/gr.255620.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Augert A, et al. Small cell lung cancer exhibits frequent inactivating mutations in the histone methyltransferase KMT2D/MLL2: CALGB 151111 (Alliance) J. Thorac. Oncol. 2017;12(4):704–713. doi: 10.1016/j.jtho.2016.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poulos RC, Olivier J, Wong JWH. The interaction between cytosine methylation and processes of DNA replication and repair shape the mutational landscape of cancer genomes. Nucleic Acids Res. 2017;45(13):7786–7795. doi: 10.1093/nar/gkx463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chan TL, et al. BRAF and KRAS mutations in colorectal hyperplastic polyps and serrated adenomas. Cancer Res. 2003;63(16):4878–4881. [PubMed] [Google Scholar]
- 26.Yuen ST, et al. Similarity of the phenotypic patterns associated with BRAF and KRAS mutations in colorectal neoplasia. Cancer Res. 2002;62(22):6451–6455. [PubMed] [Google Scholar]
- 27.Velho S, et al. The prevalence of PIK3CA mutations in gastric and colon cancer. Eur. J. Cancer. 2005;41(11):1649–1654. doi: 10.1016/j.ejca.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 28.Marcuello M, et al. Circulating biomarkers for early detection and clinical management of colorectal cancer. Mol. Aspects Med. 2019;69:107–122. doi: 10.1016/j.mam.2019.06.002. [DOI] [PubMed] [Google Scholar]
- 29.Cervena K, et al. Mutational landscape of plasma cell-free DNA identifies molecular features associated with therapeutic response in patients with colon cancer. A pilot study. Mutagenesis. 2021;36(5):358–368. doi: 10.1093/mutage/geab024. [DOI] [PubMed] [Google Scholar]
- 30.Cohen JD, et al. Detection and localization of surgically resectable cancers with a multi-analyte blood test. Science. 2018;359(6378):926–930. doi: 10.1126/science.aar3247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mazouji O, et al. Updates on clinical use of liquid biopsy in colorectal cancer screening, diagnosis, follow-up, and treatment guidance. Front Cell Dev Biol. 2021;9:660924. doi: 10.3389/fcell.2021.660924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schlemper RJ, et al. The Vienna classification of gastrointestinal epithelial neoplasia. Gut. 2000;47(2):251–255. doi: 10.1136/gut.47.2.251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Migheli F, et al. Comparison study of MS-HRM and pyrosequencing techniques for quantification of APC and CDKN2A gene methylation. PLoS One. 2013;8(1):e52501. doi: 10.1371/journal.pone.0052501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vymetalkova VP, et al. Molecular characteristics of mismatch repair genes in sporadic colorectal tumors in Czech patients. BMC Med. Genet. 2014;15:17. doi: 10.1186/1471-2350-15-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kroupa M, et al. Relationship of telomere length in colorectal cancer patients with cancer phenotype and patient prognosis. Br. J. Cancer. 2019;121(4):344–350. doi: 10.1038/s41416-019-0525-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schubert SA, et al. Evidence for genetic association between chromosome 1q loci and predisposition to colorectal neoplasia. Br. J. Cancer. 2017;117(6):1215–1223. doi: 10.1038/bjc.2017.240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bardou P, et al. JVENN: An interactive Venn diagram viewer. BMC Bioinform. 2014;15:293. doi: 10.1186/1471-2105-15-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Agresti A, Coull BA. Approximate is Better than "exact" for interval estimation of binomial proportions. Am. Stat. 1998;52:119–126. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.