Abstract
A PCR specific for spacer regions 33 and 34 of the direct repeat region of the Mycobacterium tuberculosis complex was developed to complement the biochemical differentiation of M. tuberculosis, Mycobacterium bovis, M. bovis BCG, and Mycobacterium africanum subtypes I and II. In addition, this approach was incorporated into a multiplex PCR that included primers specific for IS6110 and the 65-kDa antigen gene in order to differentiate members of the M. tuberculosis complex from atypical mycobacteria.
There is value in the rapid differentiation of cultured Mycobacterium tuberculosis complex (MTC) from atypical mycobacteria. This confirms the initial diagnosis and treatment regimen being used. In addition, there is some concern that members of the MTC may not always be accurately distinguished from one another, which confounds accurate epidemiology and could prevent important outbreaks of infection from being observed or sources of infection from being identified. Traditionally, identification relies on a battery of biochemical tests (4), which are slow and time-consuming to set up. Commercial molecular tests are available for testing of cultured isolates, but they have some disadvantages. The tests are usually specific for a given species such that a negative result could be the result of inadequate biomass, as can occur with liquid cultures, or the inhibition of the test by that sample. In addition, these tests do not differentiate between members of the MTC (1, 2, 10). There is, therefore, a need to improve the existing methods.
A number of PCR assays based on observed genetic differences between mycobacterial species have been developed. One PCR assay for distinguishing between M. tuberculosis and Mycobacterium bovis was dependent on the fact that the former contains more IS6110 copies than do M. bovis strains (14). However, further studies revealed that some strains of M. bovis have a high IS6110 copy number, and the reverse is true for some M. tuberculosis strains (17). The gene mtp40 was reported previously to be present only in M. tuberculosis and not in M. bovis (12), which seemed to offer an alternative approach for distinguishing these species, but further studies revealed that this gene is found in most, though not all, M. tuberculosis strains and is also found in some M. bovis strains (19).
In 1997, Kamerbeek et al. (9) reported a PCR-based method, spoligotyping, for the fingerprinting of MTC strains. This assay was based on earlier work that revealed polymorphism in the direct repeat (DR) region of the MTCs that depends on the presence or absence of specific spacer region sequences between two DR sequences (7). It has the added advantage of differentiating between M. bovis and M. tuberculosis (9), as M. bovis contains spacer regions 33 and 34, which are absent in M. tuberculosis. In addition, we have observed from published data (3, 16) that M. bovis BCG has two copies of spacer region 33 but only one of spacer region 34. We have used the observation to design a PCR that could differentiate among M. bovis, M. bovis BCG, and M. tuberculosis. We also incorporated this PCR into a multiplex PCR for use on cultured isolates, which could identify the genus Mycobacterium; identify members of the MTC; and distinguish M. tuberculosis, M. bovis, and M. bovis BCG.
All the mycobacterial strains used in this study were obtained from the Mycobacterium Reference Unit of the Public Health Laboratory Service at Dulwich Hospital. The numbers of strains of each species tested are shown in Table 1. Cultures had already been identified to species level by conventional, culturing, and biochemical procedures.
TABLE 1.
Results of multiplex PCR
| Species | No. of strains tested | Presence of band generated following multiplex PCR
|
|||
|---|---|---|---|---|---|
| 99 bp | 172 bp | 439 bp | 550 bp | ||
| M. tuberculosis | 30 | − | − | + | + |
| M. bovis | 18 | + | − | + | + |
| M. bovis BCG | 18 | + | + | + | + |
| M. africanum I | 5 | + | − | + | + |
| M. africanum II | 5 | − | − | + | + |
| NTMa | 54 | − | − | + | − |
NTM, nontuberculous mycobacteria.
The mycobacteria were inactivated by being heated at 80°C for 20 min prior to DNA extraction. DNA was extracted by modification of a simple, rapid method (20) using chloroform to assist in disrupting cells and to precipitate proteins. With a clean loop (1 μl), a small quantity of mycobacteria grown on solid agar (Lowenstein-Jensen) was harvested and placed into a microcentrifuge tube containing 100 μl of sterile distilled water. One hundred microliters of chloroform was then added and vortexed for 10 s. The mixture was then heated at 80°C for 20 min, followed by brief freezing at −20°C. The tubes were then removed from the freezer and allowed to thaw. Centrifugation at 13,000 × g for 3 min pelleted the cell debris to the chloroform aqueous interface. The clear lysate above the chloroform was used directly in PCR.
The following primers were used: spacer region-specific primers, spacer region 33 specific (5′ACACCGACATGACGGCGG3′) and spacer region 34 specific (5′CGACGGTGTGGGCGAGG3′); IS6110, MTC-specific primers (20), TB284 (sequence 5′GGACAACGCCGAATTGCG3′) and TB850 (sequence 5′TAGGCGTCGGTGACAAAGGCCAC3′); and Mycobacterium genus-specific (65-kDa antigen gene) primers (15), TB11 (sequence 5′ACCAACGATGGTGTGTCCAT3′) and TB12 (sequence 5′CTTGTCGAACCGCATACCCT3′).
PCR mixtures contained 20 μl of 2× PCR mix (20), 10 μl of primer mix with each primer at 0.66 pmol/μl, 0.2 μl of Taq polymerase enzyme (Roche Diagnostics Ltd.), and 10 μl of extracted DNA. The PCR conditions were 95°C for 3 min; 30 cycles of 95°C for 20 s, 65°C for 30 s, and 72°C for 30 s; and 72°C for 7 min. After PCR, the products were analyzed by electrophoresis on a high-resolution 2% (wt/vol) Metaphor agarose matrix (Flowgen).
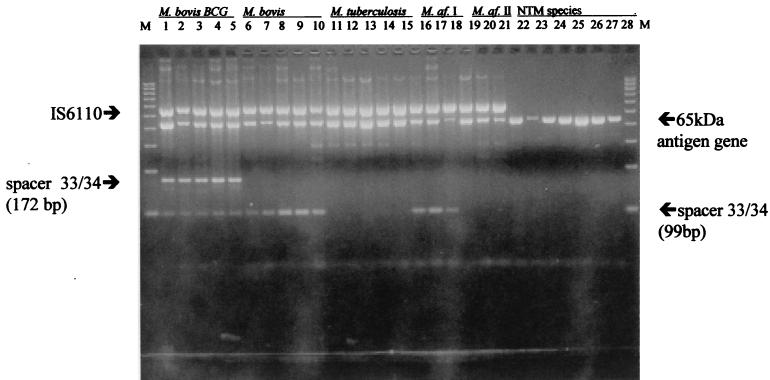
As expected, all 18 strains of M. bovis BCG produced two bands of 172 and 99 bp corresponding to amplification products from both of the spacer regions 33 in conjunction with spacer region 34 (Fig. 1 and Table 1). All 18 strains of M. bovis, which has only one spacer region 33, produced only the 99-bp band, whereas these bands were absent in all 30 strains of M. tuberculosis, as M. tuberculosis does not contain either spacer region. Interestingly, when Mycobacterium africanum strains were tested, all five strains of M. africanum I produced the 99-bp M. bovis-specific band, but all five strains of M. africanum II, similarly to M. tuberculosis, did not produce any band in this region. All the MTC strains produced a band of 550 bp corresponding to amplification of IS6110, and the mycobacterium-specific 65-kDa antigen gene resulted in a band of 439 bp (Fig. 1 and Table 1).
FIG. 1.
Products of multiplex PCR assay using the IS6110, 65-kDa antigen gene, and spacer 33 and spacer 34 specific primers. M. af., M. africanum; NTM, nontuberculous mycobacteria. The 100-bp size markers are in the lanes labeled M (from left to right, the lane labels shown at the top of the figure have a one-to-one correspondence with the lanes shown).
A number of strains of six different atypical mycobacterial species including Mycobacterium chelonae (a total of 10 strains tested), Mycobacterium malmoense (a total of 10 strains tested), the Mycobacterium avium complex (a total of 12 strains tested), Mycobacterium fortuitum (a total of 10 strains tested), Mycobacterium marinum (a total of 2 strains tested), and Mycobacterium kansasii (a total of 10 strains tested) were also analyzed. None of these species produced either of the M. bovis-specific bands of 99 and 172 bp or the IS6110-specific band (Fig. 1 and Table 1). However, all the atypical mycobacterial species produced the mycobacterium-specific 65-kDa antigen gene band.
Many reference laboratories perform a molecular test on cultured isolates so that identification of MTC is not delayed by subculturing for biochemical testing. These tests do not have a mycobacterium-specific internal control and do not differentiate between the members of the MTC (2, 10). Our multiplex PCR approach complements the biochemical testing in rapidly differentiating MTC from other mycobacteria and in confirming subsequent species-level identification of MTC. The procedure is simple, using a simple chemical extraction for preparation of the sample. In addition, the method is also rapid, taking a total of 5 h when using a conventional thermal cycler, which could be reduced to 3 h if a rapid capillary cycler were used. The sample throughput is limited only by the capacity of the cycler used, which could range from 24 to 96 samples per run. Unlike other PCR-based protocols, this method does not require hybridization or restriction enzyme analysis (PCR-restriction fragment length polymorphism) (5, 8, 13, 15).
This study confirms and complements the observations of Niemann et al. (11), who used PCR-restriction fragment length polymorphism of the gyrB gene to investigate members of the MTC. Using this method, in contrast to our study, M. africanum I could be differentiated from other MTC members but M. bovis could not be differentiated from M. bovis BCG. As in our study, M. tuberculosis could not be differentiated from M. africanum II.
The fact that in this study M. africanum I was similar to M. bovis whereas M. africanum II, in both this and the study by Niemann et al. (11), was similar to M. tuberculosis is an interesting observation that perhaps strengthens the view that these species are intermediate between M. bovis and M. tuberculosis. A recent publication also concluded that the spoligotyping pattern produced by the analysis of the DR region of M. africanum isolates was intermediate between that of M. bovis and M. tuberculosis (18). Our assay could not differentiate between M. africanum I and M. bovis or M. africanum II and M. tuberculosis, but M. africanum is rare in western Europe (6), and our test was designed to complement rather than replace biochemical testing and to aid in workload management. It is envisaged that our test would be used after confirmation of mycobacterial cultures by Ziehl-Neelsen staining. The results of the multiplex PCR enable a rapid identification of MTC, indicate whether the result is likely to be M. bovis or M. bovis BCG, and facilitate the setting up of the appropriate biochemical test.
REFERENCES
- 1.Abe C, Hirano K, Wada M, Kazumi Y, Takahashi M, Fukasawa Y, Yoshimura T, Miyagi C, Goto S. Detection of Mycobacterium tuberculosis in clinical specimens by polymerase chain reaction and Gen-Probe Amplified Mycobacterium Tuberculosis Direct Test. J Clin Microbiol. 1993;31:3270–3274. doi: 10.1128/jcm.31.12.3270-3274.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alcaide F, Benítez M A, Escribà J M, Martin R. Evaluation of the BACTEC MGIT 969 and the MB/BacT systems for recovery of mycobacteria from clinical specimens and for species identification by DNA AccuProbe. J Clin Microbiol. 2000;38:398–401. doi: 10.1128/jcm.38.1.398-401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Beggs M L, Cave M D, Marlowe C, Cloney L, Duck P, Eisenach K D. Characterization of Mycobacterium tuberculosis complex direct repeat sequence for use in cycling probe reaction. J Clin Microbiol. 1996;34:2985–2989. doi: 10.1128/jcm.34.12.2985-2989.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Collins C H, Grange J M, Yates M D. Tuberculosis bacteriology: organisation and practise. 2nd ed. 1997. p. 5. and 69–97. Butterworth Heinemann, Oxford, United Kingdom. [Google Scholar]
- 5.Evans K D, Nakasone A S, Sutherland P A, de la Maza L M, Peterson E M. Identification of Mycobacterium tuberculosis and Mycobacterium avium-intracellulare directly from primary BACTEC cultures by using acridinium-ester-labeled DNA probes. J Clin Microbiol. 1992;31:2427–2431. doi: 10.1128/jcm.30.9.2427-2431.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Grange J M, Yates M D. Incidence and nature of human tuberculosis due to Mycobacterium africanum in South-East England: 1977–1987. Epidemiol Infect. 1989;103:127–132. doi: 10.1017/s0950268800030429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hermans P W, van Soolingen D, Bik E M, de Haas P E, Dale J W, van Embden J D. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot spot integration region for insertion elements in M. tuberculosis complex strains. Infect Immun. 1991;59:2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ichiyama S, Iinuma Y, Tawada Y, Yamori S, Hasegawa Y, Shimokata K, Nakashima N. Evaluation of Gen-Probe Amplified Mycobacterium Tuberculosis Direct Test and Roche PCR-microwell plate hybridization method (AMPLICOR MYCOBACTERIUM) for direct detection of mycobacteria. J Clin Microbiol. 1996;34:130–133. doi: 10.1128/jcm.34.1.130-133.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J D. Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol. 1997;35:907–914. doi: 10.1128/jcm.35.4.907-914.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Katila L, Katila P, Erkinjuntti-Perkkanen R. Accelerated detection and identification of mycobacteria with MGIT 960 and COBAS AMPLICOR systems. J Clin Microbiol. 2000;38:960–964. doi: 10.1128/jcm.38.3.960-964.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Niemann S, Harmsen D, Rüsch-Gerdes S, Richter E. Differentiation of clinical Mycobacterium tuberculosis complex isolates by gyrB DNA sequence polymorphism analysis. J Clin Microbiol. 2000;38:3231–3234. doi: 10.1128/jcm.38.9.3231-3234.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parra C A, Londono L P, Del Portillo P, Patarroyo M E. Isolation, characterization, and molecular cloning of a specific Mycobacterium tuberculosis antigen gene: identification of a species-specific sequence. Infect Immun. 1991;59:3411–3417. doi: 10.1128/iai.59.10.3411-3417.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patel J B, Leonard D G, Pan X, Musser J M, Berman R E, Nachamkin I. Sequence-based identification of Mycobacterium species using the MicroSeq 500 16S rDNA bacterial identification system. J Clin Microbiol. 2000;38:246–251. doi: 10.1128/jcm.38.1.246-251.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Plikaytis B B, Eisenach K D, Crawford J T, Shinnick T M. Differentiation of Mycobacterium tuberculosis and Mycobacterium bovis BCG by a polymerase chain reaction assay. Mol Cell Probes. 1991;5:215–219. doi: 10.1016/0890-8508(91)90043-j. [DOI] [PubMed] [Google Scholar]
- 15.Telenti A, Marchesi F, Balz M, Bally F, Böttger E C, Bodmer T. Rapid identification of mycobacteria to species level by polymerase chain reaction and restriction enzyme analysis. J Clin Microbiol. 1993;31:175–178. doi: 10.1128/jcm.31.2.175-178.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van Embden J D, van Gorkom T, Kremer K, Jansen R, van Der Zeijst B A, Schouls L M. Genetic variation and evolutionary origin of the direct repeat locus of Mycobacterium tuberculosis complex bacteria. J Bacteriol. 2000;182:2393–2401. doi: 10.1128/jb.182.9.2393-2401.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Soolingen D, de Haas P E, Hermans P W, Groenen P M, van Embden J D. Comparison of various repetitive DNA elements as genetic markers for strain differentiation and epidemiology of Mycobacterium tuberculosis. J Clin Microbiol. 1993;31:1987–1995. doi: 10.1128/jcm.31.8.1987-1995.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Viana-Niero C, Gutierrez C, Sola C C, Filliol I, Boulahbal F, Vincent V, Rastogi N. Genetic diversity of Mycobacterium africanum clinical isolates based on IS6110-restriction fragment length polymorphism analysis, spoligotyping, and variable number of tandem DNA repeats. J Clin Microbiol. 2001;39:57–65. doi: 10.1128/JCM.39.1.57-65.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Weil A, Plikaytis B B, Butler W R, Woodley C L, Shinnick T M. The mtp40 gene is not present in all strains of Mycobacterium tuberculosis. J Clin Microbiol. 1996;34:2309–2311. doi: 10.1128/jcm.34.9.2309-2311.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wilson S M, McNerney R, Nye P M, Godfrey-Faussett P D, Stoker N G, Voller A. Progress toward a simplified polymerase chain reaction and its application to diagnosis of tuberculosis. J Clin Microbiol. 1993;31:776–782. doi: 10.1128/jcm.31.4.776-782.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]