Highlights
-
•
SELENBP1 localizes to vessels and is suppressed in tumor vessels.
-
•
SELENBP1 inhibits in vivo and in vitro angiogenesis.
-
•
SELENBP1 antagonizes tumor angiogenesis by blocking the DLL4/Notch1 signaling pathway.
-
•
SELENBP1 is a candidate target to treat bevacizumab-resistance in colorectal cancer.
Keywords: Selenium binding protein, Colorectal cancer, Angiogenesis, Delta-like ligand 4, Notch signaling pathway
Abbreviations: ANOVA, one-way analysis of variance; CCK8, cell counting kit 8; CM, conditional medium; CoIP, coimmunoprecipitation; CRC, colorectal cancer; DLL4, Delta-like ligand 4; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GEO, gene expression omnibus; GSEA, gene set enrichment analysis; HUVEC, human umbilical vascular endothelial cell; IF, immunofluorescence; IHC, immunohistochemical; IP, immunoprecipitation; SELENBP1, selenium binding protein 1; TCGA COAD and READ, the cancer genome atlas colon adenocarcinoma and rectal adenocarcinoma program; TMA, tissue microarray; VEGF, vascular endothelial growth factor
Abstract
Background
Selenium binding protein 1 (SELENBP1) is frequently downregulated in malignancies such as colorectal cancer (CRC), however, whether it is involved in tumor angiogenesis is still unknown.
Methods
We analyzed the expression and localization of SELENBP1 in vessels from CRC and neighboring tissues. We investigated the in vitro and in vivo activity of SELENBP1 in angiogenesis and explored the underlying mechanism.
Results
SELENBP1 was localized to endothelial cells in addition to glandular cells, while its vascular expression was decreased in tumor vessels compared to that in vessels from neighboring non-tumor tissues. Gain-of-function and loss-of-function experiments demonstrated that SELENBP1 inhibited angiogenesis in vitro, and blocked communications between HUVECs and CRC cells. Overexpression of SELENBP1 in CRC cells inhibited tumor growth and angiogenesis, and enhanced bevacizumab-sensitivity in a mouse subcutaneous xenograft model. Mechanic analyses revealed that SELENBP1 may suppress tumor angiogenesis by binding with Delta-like ligand 4 (DLL4) and antagonizing the DLL4/Notch1 signaling pathway. The inhibitory effects of SELENBP1 on in vitro angiogenesis could largely be rescued by DLL4.
Conclusion
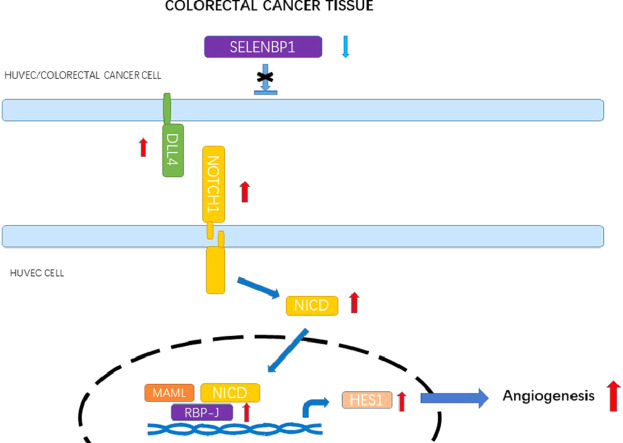
These results revealed a novel role of SELENBP1 as a potential tumor suppressor that antagonizes tumor angiogenesis in CRC by intervening the DLL4/Notch1 signaling pathway.
Graphical abstract
.
Introduction
Colorectal cancer (CRC) constitutes one of the most predominant and deadly malignancies in the world [1]. Although the diagnosis and treatment have been greatly improved during the past decades, many patients still suffer from disease recurrence and metastasis [2]. Some contributing factors have been identified for carcinogenesis and progression of CRC [3], [4], [5], nevertheless our understanding of these processes is still far from enough. Characterizing the underlying mechanisms of carcinogenesis and tumor progression, and identifying potential therapeutic targets for CRC, are therefore of great importance.
Inducing angiogenesis is one of the most well-known hallmarks of cancer. By generating tumor-associated neovasculature, angiogenesis provides sustained supply of nutrients and oxygen to tumor cells [6]. Among modulators of angiogenesis, vascular endothelial growth factor-A (VEGF-A) and its receptors are of particular importance, which regulate endothelial cell proliferation, migration, survival and vascular permeability during vasculogenesis and angiogenesis [7]. By targeting VEGF and blocking its binding with the receptors, bevacizumab (Avastin®) effectively inhibits tumor angiogenesis and thus has been authorized to treat a variety of malignancies including metastatic CRC [7], [8], [9]. However, subsequent studies revealed unresponsiveness in certain patients and acquired drug resistance also appeared inevitable [8,10]. As one of the major molecular executioners of VEGF-mediated angiogenesis, the Delta-like ligand 4 (DLL4)/Notch signaling pathway also exert negative feedback effects on angiogenesis [7]. The possibility of combined therapies against both VEGF and DLL4 has been explored and preliminary clinical trials are now undergoing [11]. Although the regulatory roles of these signaling pathways in mediating tumor angiogenesis have been well-established, the unmet needs for more efficacious antiangiogenic targets necessitate further investigation.
Selenium binding protein 1 (SELENBP1), one of the proteins that directly bind to selenium, is encoded by a gene located at chromosome 1q21.3 near the epidermal differentiation complex, which is closely related to terminal differentiation of the human epidermis [12]. Previous evidences showed that SELENBP1 participated in a variety of physiological processes such as cell differentiation and maturation [13,14], protein transport and degradation [15,16], as well as H2S biosynthesis and adipogenesis [17], while mutations in SELENBP1 resulted in dysregulated methanethiol oxidation and extraoral halitosis [18]. As a binding partner of selenium, SELENBP1 may contribute to carcinogenesis associated with selenium deficiency [19]. Actually, suppression of SELENBP1 has been associated with carcinogenesis and disease progression in CRC [13,20] and many other malignancies [21], [22], [23], [24], [25], [26], [27], [28]; however, whether SELENBP1 regulates tumor angiogenesis remains to be elucidated.
In the current study, we characterized the in vitro and in vivo regulation of tumor angiogenesis by SELENBP1 in CRC. We then explored the underlying mechanism and uncovered the regulatory effects of SELENBP on the DLL4/Notch signaling pathway. The aim of our study was to elucidate the regulatory roles of SELENBP1 in CRC angiogenesis and explore its underlying mechanism.
Materials and methods
Access to public datasets
We retrieved two CRC datasets from the gene expression omnibus (GEO) database [29], including GSE21510 [30] and GSE87211 [31]. We also downloaded the TCGA COAD and READ datasets from UCSC Xena (https://xenabrowser.net/heatmap/) and combined them into one CRC dataset2. These three datasets were utilized to conduct a Gene Set Enrichment Analysis (GSEA) to explore the potential involvement of SELENBP1 in tumor angiogenesis [32,33]. Gene sets with a false discovery rate q-value < 0.25 and nominal p value < 0.05 were regarded as significantly enriched. We retrieved another GEO dataset (GSE104645) which included CRC patients who received oxaliplatin based therapy. Some patients in this dataset also took bevacizumab concurrently and their transcriptomic data was utilized to screen differentially expressed genes that may relate to bevacizumab resistance.
CRC tissue microarray (TMA) and immunofluorescence (IF) staining
This study was approved by the Institutional Ethics Committee at Shanghai Fifth People's Hospital and adhered to the principles listed in the Declaration of Helsinki. Informed consent was obtained from all patients. Collection of clinical samples and preparation of TMA were performed as described previously [34]. The TMA was stained with antibodies against SELENBP1 and CD31 (see Table S1 for detailed antibody information) by Wuhan Servicebio Technology CO., LTD (Wuhan, China) according to their standard protocols as previously described [35]. The microvessel density (MVD) and percent of SELENBP1-positive vessels were quantified using ImageJ 1.44p (NIH, USA).
Cell culture
Two human CRC cell lines HCT116 and HCT-15 were obtained from the Cell Bank of Chinese Academy of Sciences (Shanghai, China), and were cultured in DMEM medium supplemented with 10% FBS, 100 µg/ml of penicillin, and 100 mg/ml of streptomycin. The Human Umbilical Vascular Endothelial Cells (HUVECs) were purchased from Kelei Corporation (Shanghai China) and cultured on plates coated with 30 μg/ml vitrogen (Collagen Biomaterials, Palo Alto, CA) in ECM medium supplemented with 10% fetal bovine serum, endothelial cell growth supplements and antibiotic solution (Sciencell Research Laboratories, USA). All cells had been validated by short tandem repeat (STR) profiling. Cells were cultured at 37℃ with 5% CO2 in a humidified incubator (Thermo, Waltham, MA) [34].
Ectopic expression or silencing of SELENBP1 and DLL4
Lentiviral plasmids expressing SELENBP1 (using GV367 vector), DLL4 (using GV358 vector), short hairpin RNA (shRNA) oligos of SELENBP1 or DLL4 (using GV248 vector), or respective controls were constructed by Shanghai Genechem Co., LTD (Shanghai, China). The target sequences were CACTTATATGTATGGGACT (shSELENBP1); ACCAGAAGAAGGAGCTGGAAGTGGACTGT (shDLL4) and TTCTCCGAACGTGTCACGT (scramble control). Transfection and construction of stable transfectants were performed as previously reported [34].
Cell proliferation and colony formation assays
These assays were conducted as described in our previously study [34].
Transwell® migration and invasion assays
These assays were performed as previously described by Pijuan et al [36]. Briefly, cells (4 × 105 cells/ml) were seeded in serum-free DMEM or ECM medium in the top chamber of a Transwell® insert coated without (migration assay) or with (invasion assay) Matrigel. The medium containing 20% FBS in the lower chamber served as a chemoattractant. After incubation for 24 h at 37 °C, the cells on the top side of the membrane were removed with a cotton swab, and those on the bottom side were fixed with methanol for 20 min and then stained with crystal violet (0.1% in PBS) for 15 min. Five randomly selected fields per well were photographed, and the numbers of migrated cells were counted.
Tuber formation assay
The tuber formation assay was performed as previously described by Qin et al [37]. Briefly, serum-starved HUVECs transduced with indicated lentiviruses were seeded at a density of 1 × 105 cells in 12-well plates coated with 0.5 mL of growth-factor–reduced Matrigel and incubated for 6 h, then washed with PBS and photographed. Total tube lengths and Nb-junctions were calculated using ImageJ 1.44p (NIH, USA).
Preparation of conditioned medium (CM)
CMs from HUVECs or HCT-15 cells transduced with indicated lentiviruses were collected as previously described [38]. Briefly, stably transfected cells (1 × 106) were seeded into 100 mm dishes containing 10 mL of respective complete culture medium for 24 h and washed twice with serum-free medium. The cells were then cultured in serum-free DMEM or ECM for another 24 h, and the supernatants were collected, centrifuged, filtered, and stored at -20 °C until use.
Protein extraction and western blotting
Proteins were extracted and plotted as previously described [39]. Antibodies used are listed in Table S1. GAPDH (1:2000 dilutions) served as a loading control.
The subcutaneous xenograft model
Female athymic BALB/c nude mice of 6–8 weeks old were purchased from Charles River Laboratories (Beijing, China) and maintained in the Animal Experimental Facility of Normal University of Eastern China in a pathogen-free environment. HCT-116 cells (2 × 106/mouse) stably expressing SELENBP1 or the vector were seeded subcutaneously into flanks of mice (n = 5 per group) and tumor growth was closely monitored twice a week (tumor volume = length × width2× 3.14/6). One month after inoculation, tumors were isolated and growth curves were drawn. Tumor samples were prepared for further use. After the inhibitory effects of SELENBP1 on tumor angiogenesis had been confirmed, we inoculated HCT-116 cells (2 × 106/mouse) stably expressing SELENBP1 or the vector to another two groups of mice (n = 5 per group). Once tumors became observable, bevacizumab was administered to the mice (4 mg/kg, i.p.), twice a week for five weeks. Tumor growth was monitored and compared between the two groups. All experiment procedures were conducted according to the Animal Care and Use guideline and were approved by the Animal Care Committee at Normal University of Eastern China.
Immunohistochemical (IHC) staining
Tumor samples from the mice were formalin-fixed and paraffin-embedded, then cut into 5 μm slides and underwent IHC staining by Wuhan Servicebio Technology CO., LTD (Wuhan, China), using an antibody against CD31. Numbers of CD31-labelled microvessels were counted using ImageJ 1.44p (NIH, USA) to generate the MVD.
Coimmunoprecipitation (CoIP)
For the CoIP assay, we used both tag-labeled antibodies and IP-specific antibodies according to the immunoprecipitation (IP) technical guide and protocols of Thermo Scientific (Tech tip #64, Rockford, IL, USA). Briefly, HCT-15 cells and HUVECs were transduced with HA-SELENBP1 and/or Flag-DLL4 for 48 h, and then cell lysates were collected by IP lysis buffer (SB-BR040, Sharebio, Shanghai, China) and subjected to overnight incubation with tag antibodies at 4 °C under constant vibration. The beads (YJ003, EPIZYME, Shanghai, China) were added, and precipitated proteins were collected for subsequent western blot analysis. Alternatively, untreated HCT-15 cells and HUVECs were lysated and incubated with IP-specific antibodies against SELENBP1 and DLL4 to determine their endogenous binding.
Statistical analyses
Analyses were performed using GraphPad Prism7 (GraphPad, San Diego, CA, USA) and Microsoft Excel 2010 (Microsoft, Redmond, WA, USA). Independent sample t-test was performed for comparison of continuous variables between two groups. One-way analysis of variance (ANOVA) or two-way ANOVA was performed for comparisons of continuous variables among three or more groups, with Dunnett's post-hoc multiple comparison. Nonparametric tests were performed if data did not follow a normal distribution. Statistical significance was defined as a value of p <0.05. All statistical tests were two-sided.
Results
SELENBP1 is a potential modulator of angiogenesis in CRC
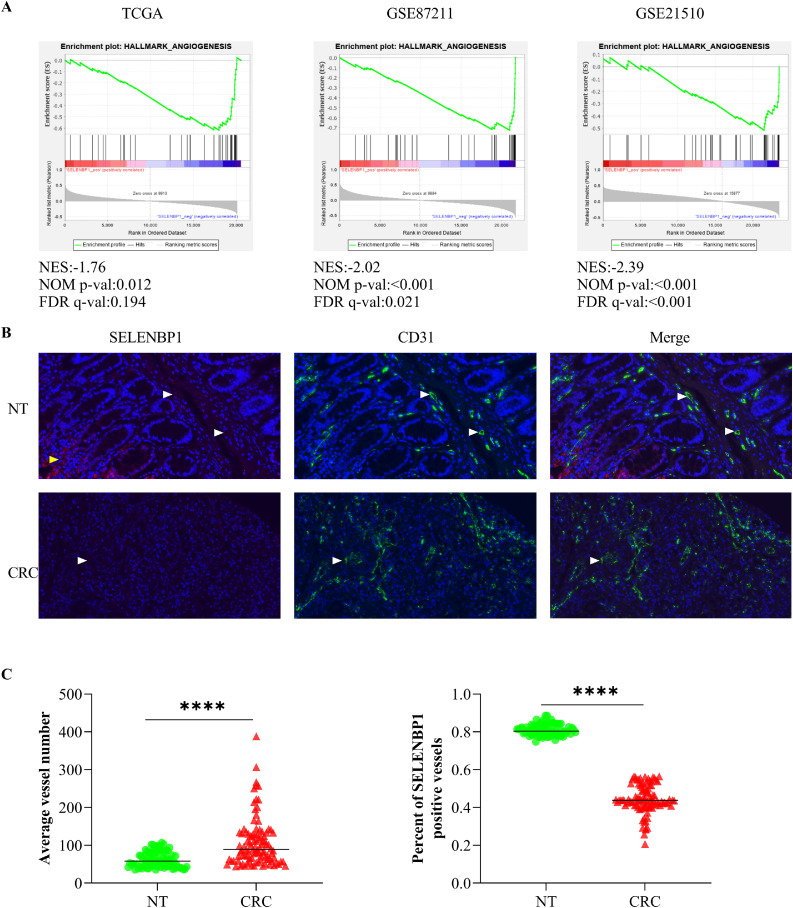
The GSEA program is developed by Broad Institute of Massachusetts Institute of Technology and Harvard, which greatly facilitates gene function annotations by helping evaluate microarray data at the level of gene sets [32]. To uncover the modulating role of SELENBP1 in tumor angiogenesis, we first conducted GSEA analyses using three public transcriptomic datasets. We divided CRC samples in each dataset into high and low expression groups according to expression of SELENBP1 in these samples, and then explored correlation between SELENBP1 expression and predefined gene sets in the program. The results demonstrated that higher expression of SELENBP1 in CRCs was negatively correlated with the hallmark gene set Angiogenesis in all three datasets (Fig. 1A). Beside, higher expression of SELENBP1 was also consistently and negatively correlated with hallmark gene set Epithelial-Mesenchymal Transition (data not shown), indicating that SELENBP1 may suppress CRC progression mainly by inhibiting angiogenesis and EMT. Next, to confirm the localization and expression of SELENBP1 in blood vessels, we stained a TMA with IF using antibodies against SELENBP1 and CD31, respectively. As shown in Fig. 1B, SELENBP1 was localized to endothelial cells in addition to glandular cells, but its expression was much weaker in tumor vessels than that in vessels from nontumor tissues. Beside, the number of microvessels was higher, while the percent of SELENBP1 positive microvessels was lower in CRCs than that in nontumor tissues (Fig. 1C). Taken together, these observations suggest that SELENBP1 may be involved in regulation of tumor angiogenesis.
Fig. 1.
SELENBP1 is closely related to angiogenesis in CRC. The GSEA analyses revealed that higher expression of SELENBP1 was negatively correlated with angiogenesis in CRCs from three transcriptomic datasets (A). IF staining of a TMA consisting 80 NTs and 100 CRCs demonstrated that SELENBP1 was most abundant in glandular cells (yellow arrow head) and endothelial cells (white arrow head), and its abundance was more remarkable in microvessels from NTs than that from CRCs (B). Quantification of MVD using CD31 as the microvascular indicator showed that the average vessel number increased while the percent of SELENBP1 positive vessels decreased in CRCs, compared to that in NTs (C). Abbreviations: CRC, colorectal cancer; FDR, false discover rate; IF, immunofluorescence; NES, normalized enrichment score; NT, normal tissue; SELENBP1, selenium binding protein 1; TCGA, The Cancer Genome Atlas. ⁎⁎⁎⁎p < 0.0001 vs. the control.
SELENBP1 inhibits angiogenesis in vitro
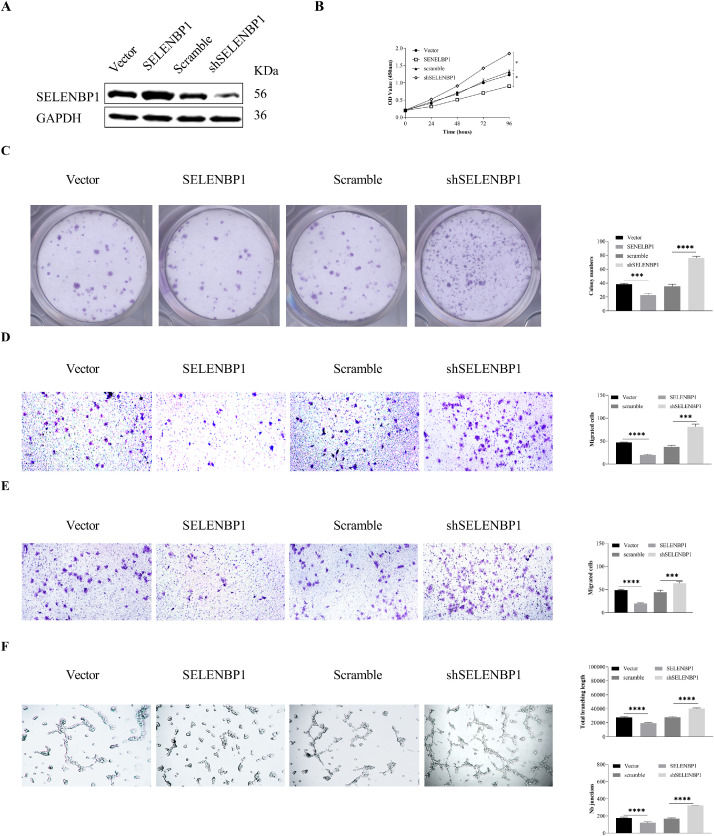
As SELENBP1 was found located in microvessels from both tumor and adjacent nontumor tissues, we next sought to determine its biological activity in angiogenesis in vitro. We ectopically overexpressed or silenced SELENBP1 in HUVECs (Fig. 2A) to perform CCK8, colony formation, Transwell® migration and invasion, and tuber formation assays. As shown in Fig. 2B–F, overexpression of SELENBP1 impeded, whereas silencing of SELENBP1 promoted proliferation, colony formation, migration, invasion, and tuber formation of HUVECs. Taken together, these results indicate that SELENBP1 has in vitro antiangiogenic capacities.
Fig. 2.
SELENBP1 inhibits in vitro angiogenesis. HUVECs were transduced with lentiviruses carrying SELENBP1, shSELENBP1, or respective controls (A), and underwent CCK8 (B), colony formation (C), Transwell migration (D) and invasion (E), and tube formation (F) assays. Abbreviations: GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HUVECs, human umbilical vascular endothelial cells; SELENBP1, selenium binding protein 1. *p < 0.05; ⁎⁎p < 0.01; ⁎⁎⁎p < 0.001; ⁎⁎⁎⁎p < 0.0001 vs. the control group.
SELENBP1 blocks the crosstalk between HUVECs and CRC cells
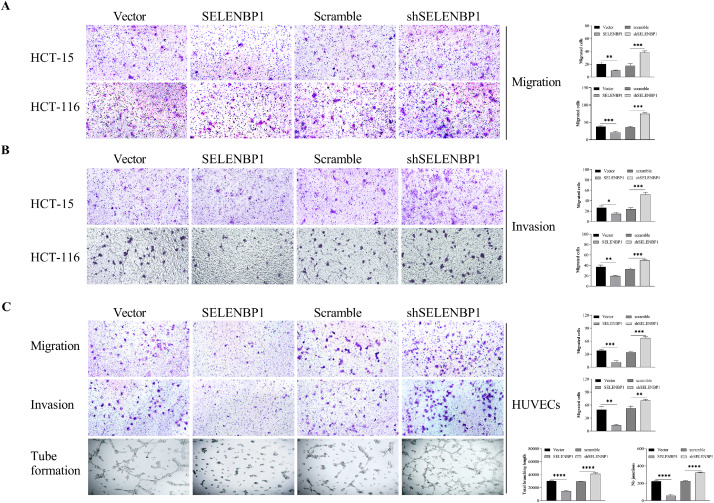
As endothelial cells belong to the microenvironment of tumor cells, we would anticipate that SELENBP1 breaks the endothelial cell-tumor cell interactions. We collected CMs from both HUVECs and HCT-15 cells transduced with SELENBP1, shSELENBP1, or respective controls, and utilized these CMs to cultivate CRC cells or HUVECs, respectively (Fig. S1A, B). CM from HUVECs transduced with SELENBP1 prohibited proliferation (Fig. S1C), colony formation (Fig. S1D), migration (Fig. 3A) and invasion (Fig. 3B) of HCT-15 and HCT-116 cells, while CM from HUVECs transduced with shSELENBP1 yielded opposite results (Figs. S1C, D, and 3A, B), compared to respective controls. In a similar way, CM from HCT-15 cells transduced with SELENBP1 suppressed proliferation (Fig. S1C), colony formation (Fig. S1D), migration, invasion, and tube formation (Fig. 3C) of HUVECs, whereas CM from HCT-15 cells transduced with shSELENBP1 promoted these processes (Figs. S1C, D, and 3C). Taken together, these experiments demonstrated that SELENBP1 could block the communications between endothelial cells and CRC cells.
Fig. 3.
SELENBP1 inhibits aggressiveness of CRC cells and HUVECs by blocking intercellular communications. HUVECs and HCT-15 cells were transduced with lentiviruses carrying SELENBP1, shSELENBP1, or respective controls to prepare conditional medium. HCT-15 and HCT-116 cells were cultured in CMs from HUVECs and underwent migration and invasion assays (A and B). HUVECs were cultured in CMs from HCT-15 cells and underwent migration, invasion, and tube formation assays (C). Abbreviations: CM, conditional medium; HUVEC, human umbilical vascular endothelial cell; SELENBP1, selenium binding protein 1. *p < 0.05; ⁎⁎p < 0.01; ⁎⁎⁎p < 0.001; ⁎⁎⁎⁎p < 0.0001 vs. the control.
SELENBP1 inhibits the VEGF/VEGFR2 and DLL4/Notch1 signaling pathways
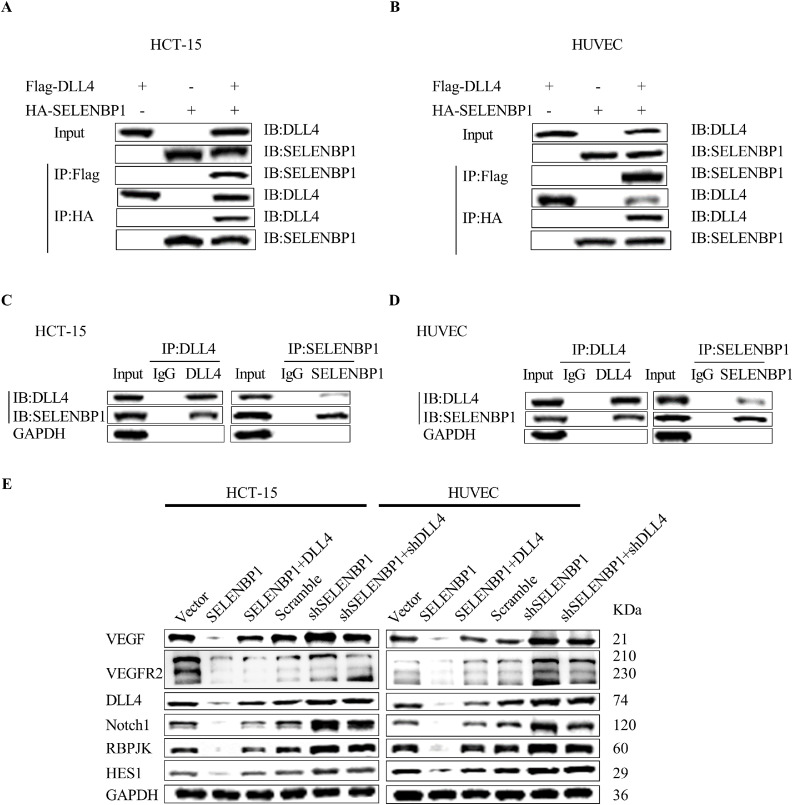
To characterize the potential mechanism of SELENBP1 in inhibiting tumor progression, we previously transduced HCT-15 cells with SELENBP1 or vector control and conducted the immunoprecipitation/mass spectrometry (IP/MS) assays to explore potential interaction proteins of SELENBP1 (Table S2). Among candidate interacting proteins, we selected DLL4 for further validation, as DLL4 and the Notch signaling pathway are well known for their roles in mediating angiogenesis and other tumor–stroma interactions [40]. We first transduced both HCT-15 cells and HUVECs with HA-SELENBP1 and/or Flag-DLL4, and then used tag antibodies to conduct the CoIP assay. The results demonstrated that SELENBP1 bound to DLL4 under exogenous stimulation in both cells (Fig. 4A, B). We then confirmed their endogenous binding in HCT-15 and HUVECs using IP antibodies against SELENBP1 and DLL4 (Fig. 4C, D). We next sought to determine whether SELENBP1 could inhibit the signaling pathways that mediate angiogenesis. HCT-15 cells and HUVECs were transduced with indicated lentiviruses, and then cellular proteins were extracted and processed to undergo western blot analyses. As shown in Fig. 4E, overexpression of SELENBP1 inhibited the VEGF/VEGFR2 and DLL4/Notch1 signaling pathways in both HCT-15 cells and HUVECs, which was recovered by simultaneous transduction of DLL4; while silencing of SELENBP1 activated these two signaling pathways, and this effect was neutralized by co-silencing of DLL4. Taken together, these observations revealed a novel mechanism that SELENBP1 binds to DLL4 and inhibits the DLL4/Notch1 signaling pathway, while it may also block the VEGF/VEGFR2 signaling indirectly.
Fig. 4.
SELENBP1 inhibits the DLL4/Notch signaling pathway. HCT-15 cells and HUVECs were transduced with HA-SELENBP1 and Flag-DLL4, and then underwent the CoIP assay using respective tag antibodies (A and B). HCT-15 cells and HUVECs underwent the CoIP assay using IP antibodies against SELENBP1 and DLL4, respectively (C and D). HCT-15 cells and HUVECs were transduced with indicated lentiviruses, and then cellular proteins were extracted and processed to undergo west blotting analyses using antibodies against VEGF, VEGFR2, DLL4, Notch1, RBP-JK, HES1, and GAPDH, respectively (E). Abbreviations: DLL4, Delta-like ligand 4; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; HES1, Hes Family BHLH Transcription Factor 1; HUVEC, human umbilical vascular endothelial cell; IB, immunoblotting; IP, immunoprecipitation; RBP-JK, immunoglobulin kappa J region; SELENBP1, selenium binding protein 1; VEGF, vascular endothelial growth factor; VEGFR2, VEGF receptor 2.
SELENBP1 inhibits in vivo tumor angiogenesis
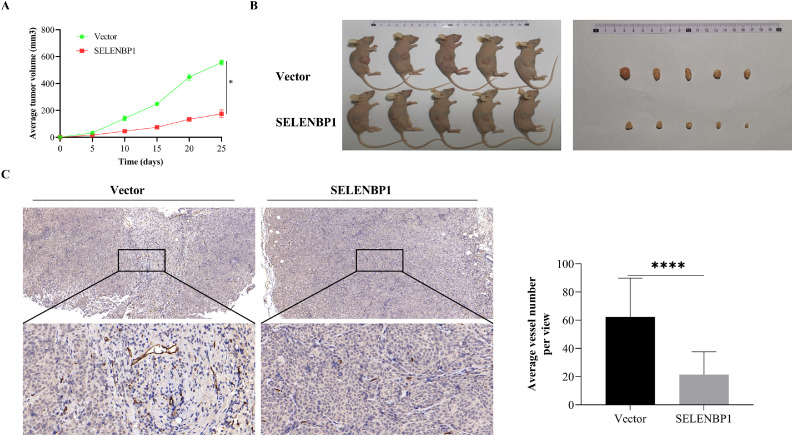
To confirm whether SELENBP1 also suppress CRC angiogenesis in vivo, we inoculated HCT-116 cells that stably overexpressed SELENBP1 or the control subcutaneously into the flanks of nude mice (n= 5/group). As shown in Fig. 5, SELENBP1 significantly inhibited tumor growth (A, B) and angiogenesis (C) in tumor tissues. In addition, SELENBP1 was co-localized with DLL4 in certain microvessels and suppressed the vascular expression of DLL4 (Fig. S2A). By contrast, the cytoplasmic co-localization of SELENBP1 and DLL4 in tumor tissues had no remarkable influence on local DLL4 expression (Fig. S2B). Taken together, these results demonstrated that SELENBP1 has in vivo antiangiogenic capacities, which may be mediated by suppression of DLL4.
Fig. 5.
SELENBP1 inhibits tumor angiogenesis in mice. HCT-116 cells stably overexpressing SELENBP1 or vector were inoculated subcutaneously into the right flank of nude mice (n = 5 per group). Tumor growth was monitored twice a week (A). On Day 25 after inoculation, mice were euthanized and tumors were photographed (B). Formalin-fixed and paraffin-embedded tumor blocks were cut into 5 μm sections and stained with an anti-CD31 antibody for MVD quantification (C). Abbreviations: MVD, microvascular density; SELENBP1, selenium binding protein 1. ⁎⁎⁎⁎p < 0.0001 vs. the control group.
SELENBP1 enhances bevacizumab sensitivity in CRC xenografts
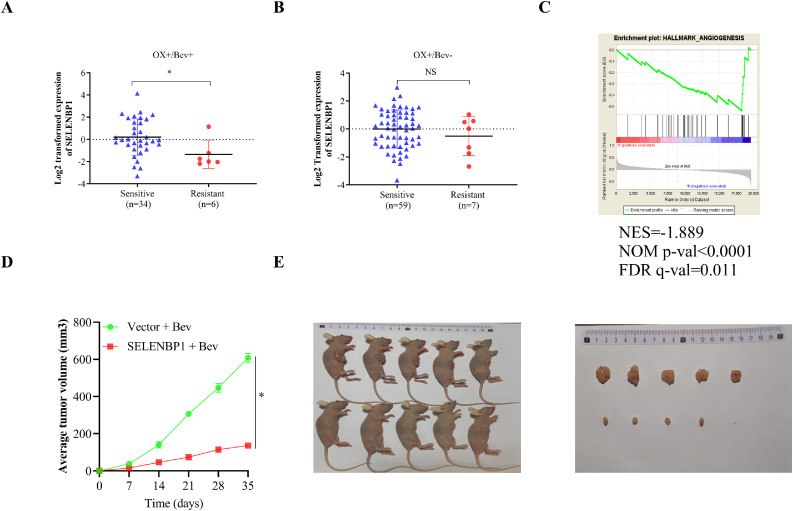
In a previous study, Okita et al investigated the predictive role of the consensus molecular subtypes (CMS) classification for efficacy of standard chemotherapies in patients with metastatic CRC using comprehensive gene expression profiles [41]. A subgroup of their patients also received concurrent therapy of bevacizumab and presented with distinct responses. By analyzing data from this subgroup, we found that the expression of SELENBP1 was significantly dropped in patients who were resistant to oxaliplatin plus bevacizumab compared to that in those who were sensitive (Fig. 6A). By contrast, no significant difference was observed between patients who were resistant to oxaliplatin-based therapy without bevacizumab and those who were sensitive to that therapy (Fig. 6B). A GSEA using gene expression data from those patients who received concurrent bevacizumab revealed that bevacizumab-efficacy was negatively correlated with the hallmark gene set Angiogenesis (Fig. 6C). These results suggest that SELENBP1 might be suppressed during acquisition of bevacizumab-resistance in later stage CRCs. To further elucidate the role of SELENBP1 in bevacizumab-resistance, we inoculated HCT-116 cells that stably overexpressed SELENBP1 or the control subcutaneously into the flanks of nude mice (n = 5/group). After tumors became observable, we injected bevacizumab to the mice (4 mg/kg, i.p.), twice a week for five weeks. As shown in Fig. 6D, E, tumor volumes increased in the vector group despite the administration of bevacizumab, by contrast, tumor growth was significantly inhibited in the SELENBP1 group. Taken together, these results demonstrated that SELENBP1 could be a potential therapeutic target for bevacizumab-resistance in CRC.
Fig. 6.
SELENBP1 enhances bevacizumab-sensitivity in mice. A GEO dataset was downloaded (GSE104645) and the expression of SELENBP1 was compared between CRC patients who were sensitive to OX/Bev combination therapy and those who were not (A), as well as between CRC patients who were sensitive to Oxaliplatin-based therapy without bevacizumab and those who were not (B). A GSEA was conducted using patients who received bevacizumab-containing therapy from the same dataset and patients were divided into bevacizumab-sensitive and -resistant in the analysis (C). HCT-116 cells that stably overexpressed SELENBP1 or the control were inoculated subcutaneously into the flanks of nude mice (n = 5/group). After tumors became observable, bevacizumab was administered to the mice (4 mg/kg, i.p.), twice a week for five weeks. Tumor growth was monitored and compared between the two groups (D, E). Abbreviations: Bev, bevacizumab; CRC, colorectal cancer; FDR, false discover rate; NES, normalized enrichment score; NS, nonsignificant; OX, oxaliplatin; SELENBP1, selenium binding protein 1. *p < 0.05 vs. the control group.
The antiangiogenic effects of SELENBP1 are dependent on its inhibition of DLL4
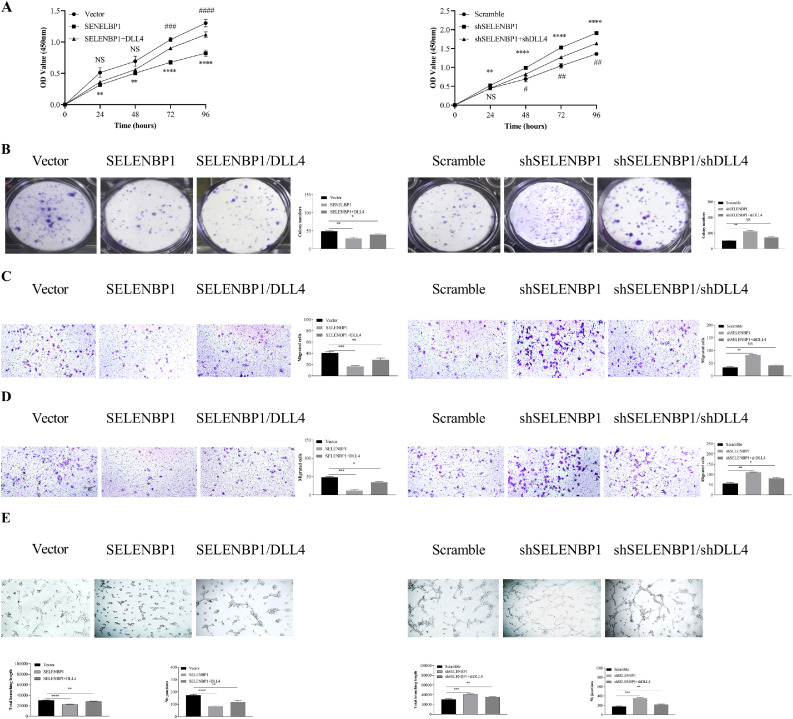
To further determine the relationship between SELENBP1 and DLL4 in mediating tumor angiogenesis, we transduced HUVECs with indicated lentiviruses to perform CCK8, colony formation, Transwell® migration and invasion, and tuber formation assays. As shown in Fig. 7A–E, overexpression of SELENBP1 impeded, whereas silencing of SELENBP1 promoted proliferation, colony formation, migration, invasion, and tuber formation of HUVECs, which could be reversed by transducing DLL4 or shDLL4, respectively. Taken together, these results demonstrated that SELENBP1 suppresses angiogenesis by inhibiting DLL4.
Fig. 7.
SELENBP1 may inhibits angiogenesis in CRC by suppressing DLL4. HUVECs were transduced with indicated lentiviruses and underwent CCK8 (A), colony formation (B), migration (C), invasion (D), and tube formation (E) assays. Abbreviations: CRC, colorectal cancer; DLL4, Delta-like ligand 4; NS, nonsignificant; SELENBP1, selenium binding protein 1. *p < 0.05; ⁎⁎p < 0.01; ⁎⁎⁎p < 0.001; ⁎⁎⁎⁎p < 0.0001 vs. the control. #p < 0.05; ##p < 0.01; ###p < 0.001; ####p < 0.0001 vs. the control for rescue experiments in the CCK8 assay.
Discussion
In the current study, we uncovered a novel function of SELENBP1 that suppressed tumor angiogenesis by binding to DLL4 and inhibiting the DLL4/Notch1 signaling pathway. To our knowledge, this is the first to build a connection between the potential tumor suppressor SELENBP1 and the proangiogenic Notch signaling pathway.
SELENBP1 is most abundant in the colon and rectum under physiological conditions, as suggested by data from the Human Protein Atlas (www.proteinatlas.org) [42], although the significance of its tissue distribution is not fully understood. By contrast, the expression of SELENBP1 is dramatically suppressed in CRCs compared to that in adjacent nontumor tissues, and its suppression has been correlated with increased tumor malignancy and unfavorable patient prognosis [13,20,43,44]. These observations, along with those from other malignancies [[21], [22], [23],28,45], suggest that suppression of SELENBP1 might be a common event during carcinogenesis and tumor progression across different malignancies. Thus, further investigation of the biological and pathological roles of SELENBP1 is warranted.
Being a selenium-binding protein, SELENBP1 may duplicate some of the roles of selenium (Se), which is an essential trace mineral indispensable to human health [46]. In the form of selenocysteine, selenium constitutes the catalytic center of selenoproteins such as glutathione peroxidases, iodothyronine deiodinases, and thioredoxin reductases. Many of these selenoproteins function as oxidoreductases that help maintain homeostasis of the internal environment by curbing the propagation of oxidative damages [47]. As such, selenium is regarded as an antioxidant, while inadequate selenium intake has been associated with increased cancer incidence and mortality [48]. Although initial clinical trials supported the use of dietary selenium replenishment in reducing the incidence and mortality of cancers [49,50], later studies revealed that high selenium intake did not bring benefit, or even brought harmful effects [51], [52], [53]. The inconsistent efficacy of selenium as a candidate anticancer agent may in part be ascribed to its complex interactions with selenoproteins and selenium-binding proteins [15,19,23,43]. With this regard, the current study provides valuable evidence that SELENBP1 might prohibit tumor progression by inhibiting angiogenesis, which could shed light on future selenium-oriented studies.
One intriguing observation in the present study was that SELENBP1 bound to DLL4 and inhibited the DLL4/Notch1 signaling pathway, the latter of which is crucial to VEGF-mediated angiogenesis [7]. The human Notch signaling pathway has four transmembrane receptors that recognize ligands, such as DLL4, from neighboring cells. The varied distribution of Notch ligands and receptors across cell types dictates communications between contiguous cells, and the DLL4/Notch1 coupling passes signals from endothelial cells to tumor cells in CRC. Binding with the ligands causes mature Notch receptor heterodimers to separate subunits, followed by cleavage of the transmembrane subunit and release of a Notch intracellular domain (NICD). The NICD then enters the nucleus to regulate downstream gene transcription [54,55]. Accumulating evidence has demonstrated the regulatory roles of the Notch pathway in a variety of tumors that boosted the development of Notch receptor-targeted or Notch ligand-targeted therapies. However, most of these agents failed to translate into promising products. Therefore, more specific and less toxic pharmacological modulations of the Notch signaling are warranted and mechanism-oriented rational combinations are preferable than monotherapy with this regard [40]. Being downstream of VEGF, the DLL4/Notch signaling is required for proper angiogenesis by guiding the sprouting of new vessels both physically and pathologically. VEGF-targeted therapy such as bevacizumab may trigger dysregulated DLL4 expression and Notch signaling, while antagonizing DLL4 could stifle tumors refractory to anti-VEGF therapy. These underpin the development of combination therapies against both VEGF and DLL4 [11,56,57]. In the present study, we also found that the expression of SELENBP1 was significantly decreased in CRC patients unresponsive to oxaliplatin/bevacizumab therapy, while overexpression of SELENBP1 inhibited tumor angiogenesis and enhanced bevacizumab sensitivity in a subcutaneous xenograft model. Beside, based on the limited evidence in our study, SELENBP1 may also negatively regulate the VEGF/VEGFR2 signaling pathway in an indirect way. Collectively, these observations suggest that SELENBP1 could be a promising target for combined antiangiogenic therapy.
Although the current study presents some novel findings that are clinically and scientifically meaningful, there are some inherent limitations. First, we did not investigate the more specific influence of SELENBP1 on the VEGF signaling pathway. Second, we did not further explore the regulatory role of SELENBP1 in bevacizumab resistance in a clinical scenario. Third, we did not take into consideration other factors, such as exosomes, noncoding RNAs, and secretory proteins, that may be critically involved in the crosstalk between CRC cells and tumor vessel endothelial cells. In addition, although we identified DLL4 as one of the binding proteins of SELENBP1 in mediating tumor angiogenesis, we did not elucidate the molecular mechanism that leads to inhibition of DLL4 by SELENBP1. Beside, we did not identify the potential factors that contributed to suppression of SELENBP1 in tumor angiogenesis in the current study. Finally, although we uncovered the inhibitory impact of SELENBP1 on tumor angiogenesis using both in vitro and in vivo models, we did not reproduce the in vivo results using transgenic mice that specifically express SELENBP1 in endothelial cells. These limitations should be addressed in future studies.
Conclusion
We uncovered a novel function of SELENBP1 which suppressed tumor angiogenesis by binding to DLL4 and inhibiting the DLL4/Notch1 signaling pathway (refer to the schematic diagram). SELENBP1 is therefore a candidate antiangiogenic target that deserves further investigation.
Declarations of Competing Interest
None.
Acknowledgments
Data availability
The data used and/or analyzed during the current study are available from the corresponding authors upon reasonable request.
Ethical guidelines
The study protocol was approved by the Institutional Ethics Committee at the Fifth People's Hospital of Shanghai, Fudan University (Ethical Approval Form no. 2017–097) and adhered to the principles of the Declaration of Helsinki. Written informed consent was obtained from each patient prior to tissue collection for experimentation.
Acknowledgments
We greatly appreciate the technological help from the Department of Pathology at our hospital for the IHC staining and data analysis. We also appreciate the valuable work done by Dr. Jun Hou at Zhongshan Hospital (Shanghai, China) for her interpretation of the IHC staining.
Funding
This work was supported by the Medical System of Shanghai Minhang District (Grant Nos. 2017MWDXK01 and 2020MWDXK02); the Fuxing Nursing Research Grant of Fudan University (Grant No. FNF202161), and the Shanghai Minhang District Science and Technology Commission (Grant Nos. 2017MHZ02, 2019MHZ054, 2020MHZ080, and 2021MHZ038). The funding sources were not involved in the study design; in the collection, analysis, or interpretation of data; in the writing of the report; or in the decision to submit the manuscript for publication.
Footnotes
The results <published or shown> here are in whole or part based upon data generated by the TCGA Research Network: http://cancergenome.nih.gov/.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2022.101365.
Contributor Information
Gengming Niu, Email: gengming_niu@126.com.
Ying Yue, Email: yy225977@163.com.
Chongwei Ke, Email: dr_kecw@163.com.
Appendix. Supplementary materials
References
- 1.Sung H., Ferlay J., Siegel R.L., Laversanne M., Soerjomataram I., Jemal A., et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2021;71(3):209–249. doi: 10.3322/caac.21660. Epub 2021/02/05PubMed PMID: 33538338. [DOI] [PubMed] [Google Scholar]
- 2.Tauriello D.V., Calon A., Lonardo E., Batlle E. Determinants of metastatic competency in colorectal cancer. Mol. Oncol. 2017;11(1):97–119. doi: 10.1002/1878-0261.12018. Epub 2017/01/14PubMed PMID: 28085225; PubMed Central PMCID: PMCPMC5423222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schmitt M., Greten F.R. The inflammatory pathogenesis of colorectal cancer. Nat. Rev. Immunol. 2021 doi: 10.1038/s41577-021-00534-x. PubMed PMID: 33911231. [DOI] [PubMed] [Google Scholar]
- 4.Saus E., Iraola-Guzmán S., Willis J.R., Brunet-Vega A., Gabaldón T. Microbiome and colorectal cancer: roles in carcinogenesis and clinical potential. Mol. Asp. Med. 2019:69. doi: 10.1016/j.mam.2019.05.001. PubMed PMID: 31082399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jung G., Hernández-Illán E., Moreira L., Balaguer F., Goel A. Epigenetics of colorectal cancer: biomarker and therapeutic potential. Nat. Rev. Gastroenterol. Hepatol. 2020;17(2):111–130. doi: 10.1038/s41575-019-0230-y. PubMed PMID: 31900466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hanahan D., Weinberg R.A. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013. PubMed PMID: 21376230. [DOI] [PubMed] [Google Scholar]
- 7.Apte R.S., Chen D.S., Ferrara N. VEGF in signaling and disease: beyond discovery and development. Cell. 2019;176(6):1248–1264. doi: 10.1016/j.cell.2019.01.021. PubMed PMID: 30849371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia J., Hurwitz H.I., Sandler A.B., Miles D., Coleman R.L., Deurloo R., et al. Bevacizumab (Avastin®) in cancer treatment: a review of 15 years of clinical experience and future outlook. Cancer Treat. Rev. 2020;86 doi: 10.1016/j.ctrv.2020.102017. PubMed PMID: 32335505. [DOI] [PubMed] [Google Scholar]
- 9.Hurwitz H., Fehrenbacher L., Novotny W., Cartwright T., Hainsworth J., Heim W., et al. Bevacizumab plus irinotecan, fluorouracil, and leucovorin for metastatic colorectal cancer. N. Engl. J. Med. 2004;350(23):2335–2342. doi: 10.1056/NEJMoa032691. Epub 2004/06/04PubMed PMID: 15175435. [DOI] [PubMed] [Google Scholar]
- 10.Lambrechts D., Lenz H.J., de Haas S., Carmeliet P., Scherer S.J. Markers of response for the antiangiogenic agent bevacizumab. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013;31(9):1219–1230. doi: 10.1200/jco.2012.46.2762. Epub 2013/02/13PubMed PMID: 23401453. [DOI] [PubMed] [Google Scholar]
- 11.Comunanza V., Bussolino F. Therapy for cancer: strategy of combining anti-angiogenic and target therapies. Front. Cell Dev. Biol. 2017;5:101. doi: 10.3389/fcell.2017.00101. Epub 2017/12/23PubMed PMID: 29270405; PubMed Central PMCID: PMCPMC5725406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lioumi M., Olavesen M.G., Nizetic D., Ragoussis J. High-resolution YAC fragmentation map of 1q21. Genomics. 1998;49(2):200–208. doi: 10.1006/geno.1998.5234. PubMed PMID: 9598307. [DOI] [PubMed] [Google Scholar]
- 13.Li T., Yang W., Li M., Byun D.S., Tong C., Nasser S., et al. Expression of selenium-binding protein 1 characterizes intestinal cell maturation and predicts survival for patients with colorectal cancer. Mol. Nutr. Food Res. 2008;52(11):1289–1299. doi: 10.1002/mnfr.200700331. Epub 2008/04/26PubMed PMID: 18435490. [DOI] [PubMed] [Google Scholar]
- 14.Steinbrenner H., Micoogullari M., Hoang N.A., Bergheim I., Klotz L.O., Sies H. Selenium-binding protein 1 (SELENBP1) is a marker of mature adipocytes. Redox. Biol. 2019;20:489–495. doi: 10.1016/j.redox.2018.11.004. PubMed PMID: 30469030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeong J.Y., Wang Y., Sytkowski A.J. Human selenium binding protein-1 (hSP56) interacts with VDU1 in a selenium-dependent manner. Biochem. Biophys. Res. Commun. 2009;379(2):583–588. doi: 10.1016/j.bbrc.2008.12.110. PubMed PMID: 19118533. [DOI] [PubMed] [Google Scholar]
- 16.Porat A., Sagiv Y., Elazar Z. A 56-kDa selenium-binding protein participates in intra-Golgi protein transport. J. Biol. Chem. 2000;275(19):14457–14465. doi: 10.1074/jbc.275.19.14457. PubMed PMID: 10799528. [DOI] [PubMed] [Google Scholar]
- 17.Randi E.B., Casili G., Jacquemai S., Szabo C. Selenium-binding protein 1 (SELENBP1) supports hydrogen sulfide biosynthesis and adipogenesis. Antioxidants. 2021;10(3) doi: 10.3390/antiox10030361. PubMed PMID: 33673622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pol A., Renkema G.H., Tangerman A., Winkel E.G., Engelke U.F., de Brouwer A.P.M., et al. Mutations in SELENBP1, encoding a novel human methanethiol oxidase, cause extraoral halitosis. Nat. Genet. 2018;50(1):120–129. doi: 10.1038/s41588-017-0006-7. PubMed PMID: 29255262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang M., Sytkowski A.J. Differential expression and androgen regulation of the human selenium-binding protein gene hSP56 in prostate cancer cells. Cancer Res. 1998;58(14):3150–3153. [PubMed] [Google Scholar]
- 20.Kim H., Kang H.J., You K.T., Kim S.H., Lee K.Y., Kim T.I., et al. Suppression of human selenium-binding protein 1 is a late event in colorectal carcinogenesis and is associated with poor survival. Proteomics. 2006;6(11):3466–3476. doi: 10.1002/pmic.200500629. Epub 2006/04/29PubMed PMID: 16645984. [DOI] [PubMed] [Google Scholar]
- 21.Huang K.C., Park D.C., Ng S.K., Lee J.Y., Ni X., Ng W.C., et al. Selenium binding protein 1 in ovarian cancer. Int. J. Cancer. 2006;118(10):2433–2440. doi: 10.1002/ijc.21671. Epub 2005/12/29PubMed PMID: 16380993. [DOI] [PubMed] [Google Scholar]
- 22.Ha Y.S., Lee G.T., Kim Y.H., Kwon S.Y., Choi S.H., Kim T.H., et al. Decreased selenium-binding protein 1 mRNA expression is associated with poor prognosis in renal cell carcinoma. World J. Surg. Oncol. 2014;12:288. doi: 10.1186/1477-7819-12-288. Epub 2014/09/18PubMed PMID: 25227434; PubMed Central PMCID: PMCPMC4176564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhang S., Li F., Younes M., Liu H., Chen C., Yao Q. Reduced selenium-binding protein 1 in breast cancer correlates with poor survival and resistance to the anti-proliferative effects of selenium. PLoS One. 2013;8(5):e63702. doi: 10.1371/journal.pone.0063702. Epub 2013/05/25PubMed PMID: 23704933; PubMed Central PMCID: PMCPMC3660592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang J., Dong W.G., Lin J. Reduced selenium-binding protein 1 is associated with poor survival rate in gastric carcinoma. Med. Oncol. 2011;28(2):481–487. doi: 10.1007/s12032-010-9482-7. PubMed PMID: 20354826. [DOI] [PubMed] [Google Scholar]
- 25.Chen G., Wang H., Miller C.T., Thomas D.G., Gharib T.G., Misek D.E., et al. Reduced selenium-binding protein 1 expression is associated with poor outcome in lung adenocarcinomas. J. Pathol. 2004;202(3):321–329. doi: 10.1002/path.1524. PubMed PMID: 14991897. [DOI] [PubMed] [Google Scholar]
- 26.Silvers A.L., Lin L., Bass A.J., Chen G., Wang Z., Thomas D.G., et al. Decreased selenium-binding protein 1 in esophageal adenocarcinoma results from posttranscriptional and epigenetic regulation and affects chemosensitivity. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2010;16(7):2009–2021. doi: 10.1158/1078-0432.CCR-09-2801. PubMed PMID: 20332323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang C., Ding G., Gu C., Zhou J., Kuang M., Ji Y., et al. Decreased selenium-binding protein 1 enhances glutathione peroxidase 1 activity and downregulates HIF-1α to promote hepatocellular carcinoma invasiveness. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2012;18(11):3042–3053. doi: 10.1158/1078-0432.CCR-12-0183. PubMed PMID: 22512980. [DOI] [PubMed] [Google Scholar]
- 28.Zhang X.Y., Gao P.T., Yang X., Cai J.B., Ding G.Y., Zhu X.D., et al. Reduced selenium-binding protein 1 correlates with a poor prognosis in intrahepatic cholangiocarcinoma and promotes the cell epithelial-mesenchymal transition. Am. J. Transl. Res. 2018;10(11):3567–3578. Epub 2019/01/22. PubMed PMID: 30662608; PubMed Central PMCID: PMCPMC6291736. [PMC free article] [PubMed] [Google Scholar]
- 29.Barrett T., Wilhite S.E., Ledoux P., Evangelista C., Kim I.F., Tomashevsky M., et al. NCBI GEO: archive for functional genomics data sets—update. Nucleic Acids Res. 2013;41(D1):D991–D9D5. doi: 10.1093/nar/gks1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tsukamoto S., Ishikawa T., Iida S., Ishiguro M., Mogushi K., Mizushima H., et al. Clinical significance of osteoprotegerin expression in human colorectal cancer. Clin. Cancer Res. 2011;17(8):2444–2450. doi: 10.1158/1078-0432.Ccr-10-2884. Epub 2011/01/29PubMed PMID: 21270110. [DOI] [PubMed] [Google Scholar]
- 31.Hu Y., Gaedcke J., Emons G., Beissbarth T., Grade M., Jo P., et al. Colorectal cancer susceptibility loci as predictive markers of rectal cancer prognosis after surgery. Genes Chromosomes Cancer. 2018;57(3):140–149. doi: 10.1002/gcc.22512. Epub 2017/11/10PubMed PMID: 29119627; PubMed Central PMCID: PMCPMC5778444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Subramanian A., Tamayo P., Mootha V.K., Mukherjee S., Ebert B.L., Gillette M.A., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mootha V.K., Lindgren C.M., Eriksson K.F., Subramanian A., Sihag S., Lehar J., et al. PGC-1α-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 2003;34:267. doi: 10.1038/ng1180. https://www.nature.com/articles/ng1180#supplementary-information [DOI] [PubMed] [Google Scholar]
- 34.Niu G., Deng L., Zhang X., Hu Z., Han S., Xu K., et al. GABRD promotes progression and predicts poor prognosis in colorectal cancer. Open Med. Wars. 2020;15(1):1172–1183. doi: 10.1515/med-2020-0128. PubMed PMID: 33336074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen G., Wang Y., Wu P., Zhou Y., Yu F., Zhu C., et al. Reversibly stabilized polycation nanoparticles for combination treatment of early- and late-stage metastatic breast cancer. ACS Nano. 2018;12(7):6620–6636. doi: 10.1021/acsnano.8b01482. [DOI] [PubMed] [Google Scholar]
- 36.Pijuan J., Barceló C., Moreno D.F., Maiques O., Sisó P., Marti R.M., et al. In vitro cell migration, invasion, and adhesion assays: from cell imaging to data analysis. Front. Cell Dev. Biol. 2019;7 doi: 10.3389/fcell.2019.00107. 107-PubMed PMID: 31259172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Qin L., Zhao D., Xu J., Ren X., Terwilliger E.F., Parangi S., et al. The vascular permeabilizing factors histamine and serotonin induce angiogenesis through TR3/Nur77 and subsequently truncate it through thrombospondin-1. Blood. 2013;121(11):2154–2164. doi: 10.1182/blood-2012-07-443903. PubMed PMID: 23315169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Liao R., Wu H., Yi Y., Wang J.X., Cai X.Y., He H.W., et al. Clinical significance and gene expression study of human hepatic stellate cells in HBV related-hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2013;32:22. doi: 10.1186/1756-9966-32-22. Epub 2013/04/23PubMed PMID: 23601182; PubMed Central PMCID: PMCPMC3654985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hong R., Gu J., Niu G., Hu Z., Zhang X., Song T., et al. PRELP has prognostic value and regulates cell proliferation and migration in hepatocellular carcinoma. J. Cancer. 2020;11(21):6376–6389. doi: 10.7150/jca.46309. PubMed PMID: 33033521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Majumder S., Crabtree J.S., Golde T.E., Minter L.M., Osborne B.A., Miele L. Targeting Notch in oncology: the path forward. Nat. Rev. Drug Discovery. 2021;20(2):125–144. doi: 10.1038/s41573-020-00091-3. PubMed PMID: 33293690. [DOI] [PubMed] [Google Scholar]
- 41.Okita A., Takahashi S., Ouchi K., Inoue M., Watanabe M., Endo M., et al. Consensus molecular subtypes classification of colorectal cancer as a predictive factor for chemotherapeutic efficacy against metastatic colorectal cancer. Oncotarget. 2018;9(27):18698–18711. doi: 10.18632/oncotarget.24617. Epub 2018/05/04PubMed PMID: 29721154; PubMed Central PMCID: PMCPMC5922348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Uhlen M., Fagerberg L., Hallstrom B.M., Lindskog C., Oksvold P., Mardinoglu A., et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347(6220) doi: 10.1126/science.1260419. Epub 2015/01/24PubMed PMID: 25613900. [DOI] [PubMed] [Google Scholar]
- 43.Hughes D.J., Kunicka T., Schomburg L., Liska V., Swan N., Soucek P. Expression of selenoprotein genes and association with selenium status in colorectal adenoma and colorectal cancer. Nutrients. 2018;10(11) doi: 10.3390/nu10111812. Epub 2018/11/25PubMed PMID: 30469315; PubMed Central PMCID: PMCPMC6266908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pohl N.M., Tong C., Fang W., Bi X., Li T., Yang W. Transcriptional regulation and biological functions of selenium-binding protein 1 in colorectal cancer in vitro and in nude mouse xenografts. PLoS One. 2009;4(11):e7774. doi: 10.1371/journal.pone.0007774. Epub 2009/11/20PubMed PMID: 19924303; PubMed Central PMCID: PMCPMC2774949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xia Y.J., Ma Y.Y., He X.J., Wang H.J., Ye Z.Y., Tao HQ. Suppression of selenium-binding protein 1 in gastric cancer is associated with poor survival. Hum. Pathol. 2011;42(11):1620–1628. doi: 10.1016/j.humpath.2011.01.008. Epub 2011/04/19PubMed PMID: 21497372. [DOI] [PubMed] [Google Scholar]
- 46.Rayman M.P. The importance of selenium to human health. Lancet North Am. Ed. 2000;356(9225):233–241. doi: 10.1016/S0140-6736(00)02490-9. [DOI] [PubMed] [Google Scholar]
- 47.Hatfield D.L., Tsuji P.A., Carlson B.A., Gladyshev V.N. Selenium and selenocysteine: roles in cancer, health, and development. Trends Biochem. Sci. 2014;39(3):112–120. doi: 10.1016/j.tibs.2013.12.007. Epub 2014/02/04PubMed PMID: 24485058; PubMed Central PMCID: PMCPMC3943681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rayman M.P. The importance of selenium to human health. Lancet. 2000;356(9225):233–241. doi: 10.1016/s0140-6736(00)02490-9. Epub 2000/08/30PubMed PMID: 10963212. [DOI] [PubMed] [Google Scholar]
- 49.Clark L.C., Combs G.F., Turnbull B.W., Slate E.H., Chalker D.K., Chow J., et al. Effects of selenium supplementation for cancer prevention in patients with carcinoma of the skin. A randomized controlled trial. Nutritional prevention of cancer study group. JAMA. 1996;276(24):1957–1963. Epub 1996/12/25. PubMed PMID: 8971064. [PubMed] [Google Scholar]
- 50.Yu S.Y., Zhu Y.J., Li W.G. Protective role of selenium against hepatitis B virus and primary liver cancer in Qidong. Biol. Trace Elem. Res. 1997;56(1):117–124. doi: 10.1007/BF02778987. Epub 1997/01/01. PubMed PMID: 9152515. [DOI] [PubMed] [Google Scholar]
- 51.Vinceti M., Filippini T., Del Giovane C., Dennert G., Zwahlen M., Brinkman M., et al. Selenium for preventing cancer. Cochrane Database Syst. Rev. 2018;1 doi: 10.1002/14651858.CD005195.pub4. Epub 2018/01/30PubMed PMID: 29376219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lippman S.M., Klein E.A., Goodman P.J., Lucia M.S., Thompson I.M., Ford L.G., et al. Effect of selenium and vitamin E on risk of prostate cancer and other cancers: the selenium and vitamin E cancer prevention trial (SELECT) JAMA. 2009;301(1):39–51. doi: 10.1001/jama.2008.864. Epub 12/09PubMed PMID: 19066370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vinceti M., Vicentini M., Wise L.A., Sacchettini C., Malagoli C., Ballotari P., et al. Cancer incidence following long-term consumption of drinking water with high inorganic selenium content. Sci. Total Environ. 2018;635:390–396. doi: 10.1016/j.scitotenv.2018.04.097. Epub 2018/04/21PubMed PMID: 29674262. [DOI] [PubMed] [Google Scholar]
- 54.Bray S.J. Notch signalling in context. Nat. Rev. Mol. Cell Biol. 2016;17(11):722–735. doi: 10.1038/nrm.2016.94. PubMed PMID: 27507209. [DOI] [PubMed] [Google Scholar]
- 55.Meurette O., Mehlen P. Notch signaling in the tumor microenvironment. Cancer Cell. 2018;34(4):536–548. doi: 10.1016/j.ccell.2018.07.009. PubMed PMID: 30146333. [DOI] [PubMed] [Google Scholar]
- 56.Pitulescu M.E., Schmidt I., Giaimo B.D., Antoine T., Berkenfeld F., Ferrante F., et al. Dll4 and Notch signalling couples sprouting angiogenesis and artery formation. Nat. Cell Biol. 2017;19(8):915–927. doi: 10.1038/ncb3555. PubMed PMID: 28714968. [DOI] [PubMed] [Google Scholar]
- 57.Ridgway J., Zhang G., Wu Y., Stawicki S., Liang W.C., Chanthery Y., et al. Inhibition of Dll4 signalling inhibits tumour growth by deregulating angiogenesis. Nature. 2006;444(7122):1083–1087. doi: 10.1038/nature05313. PubMed PMID: 17183323. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data used and/or analyzed during the current study are available from the corresponding authors upon reasonable request.