Abstract
Objective
To create and validate a simple and transferable machine learning model from electronic health record data to accurately predict clinical deterioration in patients with covid-19 across institutions, through use of a novel paradigm for model development and code sharing.
Design
Retrospective cohort study.
Setting
One US hospital during 2015-21 was used for model training and internal validation. External validation was conducted on patients admitted to hospital with covid-19 at 12 other US medical centers during 2020-21.
Participants
33 119 adults (≥18 years) admitted to hospital with respiratory distress or covid-19.
Main outcome measures
An ensemble of linear models was trained on the development cohort to predict a composite outcome of clinical deterioration within the first five days of hospital admission, defined as in-hospital mortality or any of three treatments indicating severe illness: mechanical ventilation, heated high flow nasal cannula, or intravenous vasopressors. The model was based on nine clinical and personal characteristic variables selected from 2686 variables available in the electronic health record. Internal and external validation performance was measured using the area under the receiver operating characteristic curve (AUROC) and the expected calibration error—the difference between predicted risk and actual risk. Potential bed day savings were estimated by calculating how many bed days hospitals could save per patient if low risk patients identified by the model were discharged early.
Results
9291 covid-19 related hospital admissions at 13 medical centers were used for model validation, of which 1510 (16.3%) were related to the primary outcome. When the model was applied to the internal validation cohort, it achieved an AUROC of 0.80 (95% confidence interval 0.77 to 0.84) and an expected calibration error of 0.01 (95% confidence interval 0.00 to 0.02). Performance was consistent when validated in the 12 external medical centers (AUROC range 0.77-0.84), across subgroups of sex, age, race, and ethnicity (AUROC range 0.78-0.84), and across quarters (AUROC range 0.73-0.83). Using the model to triage low risk patients could potentially save up to 7.8 bed days per patient resulting from early discharge.
Conclusion
A model to predict clinical deterioration was developed rapidly in response to the covid-19 pandemic at a single hospital, was applied externally without the sharing of data, and performed well across multiple medical centers, patient subgroups, and time periods, showing its potential as a tool for use in optimizing healthcare resources.
Introduction
Risk stratification models that provide advance warning of patients at high risk of clinical deterioration during hospital admission could help care teams manage resources, including interventions, hospital beds, and staffing.1 2 For example, knowing how many and which patients will require ventilators could prompt hospitals to increase ventilator supply while care teams start to allocate ventilators to patients most in need.3 Beyond identifying high risk patients, such models could also help to identify low risk patients (eg, those who are unlikely to deteriorate) as candidates for early discharge (<48 hours from admission), potentially freeing up hospital resources.4 5 6 7
Despite the potential use of risk stratification models in resource allocation, few successful examples exist. Most notably, strong generalization performance (that is, how well a model will perform across different patient populations) is fundamental to realizing the potential benefits of risk models in clinical care. Yet generalization performance is often entirely overlooked when predictive models are developed and validated in healthcare.8 9 10 11 12 13 14 For example, recent work found that only 5% of articles on predictive modeling in PubMed mention external validation in either the title or the abstract.9 This is partly because most approaches to external validation require data sharing agreements.15 16 17 18 In the small numbers of cases in which data sharing agreements have been successfully established, validation was either limited in scope19 20 21 22 (eg, focused on a single geographical region) or the model performed poorly once applied to a population that differed from the development cohort.23 24 Thus, a critical need exists for an accurate, simple, and open source method for patient risk stratification that can generalize across hospitals and patient populations.
In this study, we developed and validated an open source model, the Michigan Critical Care Utilization and Risk Evaluation System (M-CURES), to predict clinical deterioration in patients using routinely available data extracted from electronic health records. The model is designed to be embedded into an electronic health record system, automatically producing updated risk scores over the course of a patient’s hospital admission in set intervals based on available data. We externally validated this risk model across multiple dimensions while preserving data privacy and forgoing the need for data sharing across healthcare institutions. To evaluate the effectiveness of the model in settings where risk stratification could be highly beneficial, we focused on patients admitted to hospital with covid-19 in 13 US medical centers. This disease represents an important case study, given that the increases in hospital admissions during the pandemic have strained hospital resources on a global scale25 26 27; some hospitals have been forced to cancel as much as 85% of elective surgical procedures to free up resources.28 29 Owing to the limited number of people with covid-19 at the beginning of the pandemic, we trained our model on a different (but related) cohort of patients—those with respiratory distress. We hypothesized that a simple model based on a handful of variables would generalize across diverse patient cohorts.
Methods
Model development and reporting followed the Transparent Reporting of a multivariable prediction model for Individual Prognosis Or Diagnosis (TRIPOD) guidelines.30 31 The eMethods 1 section in the supplemental file provides additional details on the methodology.
Outcome
The model was trained to predict a composite outcome of clinical deterioration, defined as in-hospital mortality or any of three treatments indicating severe illness: invasive mechanical ventilation, heated high flow nasal cannula, or intravenous vasopressors. The outcome time was defined as the earliest (if any) of these events within the first five days of hospital admission. Supplemental eMethods 2 describes additional implementation details. As critical care treatments can often be administered throughout a hospital, we focused on a definition centered around what care indicates potential critical illness and deterioration rather than intensive care unit (ICU) transfers. In a sensitivity analysis, we also considered a stricter definition of deterioration where heated high flow nasal cannulation was not included among the outcomes (see supplemental eFigure 4).
Study cohorts
Development cohort—The model was trained on adults (≥18 years) admitted to hospital at Michigan Medicine, the academic medical center of the University of Michigan, during the five years from 1 January 2015 to 31 December 2019. Specifically, the model was trained on unique hospital admissions rather than unique patients, as a particular patient might have multiple admissions. We included all admissions pertaining to patients with respiratory distress—that is, those admitted through the emergency department who received supplemental oxygen support. We excluded hospital admissions in which the patient met the outcome before or at the time of receiving supplemental oxygen, as no prediction of clinical decompensation was needed.
Internal validation cohort—The model was internally validated on adults (≥18 years) admitted to hospital at Michigan Medicine from 1 March 2020 to 28 February 2021 who required supplemental oxygen and had a diagnosis of covid-19. To identify hospital admissions pertaining to patients with covid-19 from retrospective data, we included those with either a positive laboratory test result for SARS-CoV-2 or a recorded ICD-10 code (international classification of diseases, 10th revision) for covid-19 without a negative laboratory test result to identify transfer patients who received a diagnosis of covid-19 at another healthcare facility. A randomly selected subset of 100 hospital admissions was used for variable selection and excluded from evaluation.
External validation cohorts—The external validation cohorts included adults (≥18 years) admitted to hospital at 12 external medical centers from 1 March 2020 to 28 February 2021 who required supplemental oxygen and had a diagnosis of covid-19. These medical centers represent both large academic medical centers and small to mid-size community hospitals in regions geographically distinct from the development institution (Midwest), including the northeast, west, and south regions of the US. Inclusion criteria were similar to those used for the internal validation cohort. Six sites with fewer than 100 patient admissions that met the primary outcome were combined into a single cohort when performing evaluation, resulting in a total of seven external validation cohorts (see supplemental eMethods 2). Institution specific results were anonymized.
Cohort comparison—We compared the internal validation cohort with the development cohort and with each of the external validation cohorts across personal characteristics and outcomes, using χ2 tests for homogeneity with a Bonferroni correction for multiple comparisons, at a significance level of α=0.001.
Model development and evaluation
Variable selection and feature engineering—Based on data extracted from the electronic health record, we developed a model to predict the primary outcome every four hours (at set time points; see supplemental eFigure 1). All variables in the electronic health record were automatically extracted without conditioning on the outcome of the patient encounter. The model was intentionally designed to be easily integrated into the electronic health record and perform automated risk calculation at intervals of four hours using clinical data as the information becomes available. We used clinical knowledge and data driven feature selection to reduce the input space in the electronic health record from 2686 variables (including personal characteristics, laboratory test results, and data recorded in nursing flowsheets) to nine variables. First, we excluded variables with a high level of missingness (see supplemental eMethods 1). Next, based on clinical expertise, we removed variables with the potential to be spuriously correlated with the outcome.32 In addition, variables that relied on existing deterioration indices or composite scores (eg, the SOFA (sequential organ failure assessment) score33) were removed, owing to the potential for inconsistencies or lack of availability across healthcare systems. Then, using 100 randomly selected patient admissions from the internal validation cohort, we used permutation importance34 35 and forward selection36 to further reduce the variable set (see supplemental eMethods 1). The final nine variables included age, respiratory rate, oxygen saturation, oxygen flow rate, pulse oximetry type (eg, continuous, intermittent), head-of-bed position (eg, at 30°), position of patient during blood pressure measurement (standing, sitting, lying), venous blood gas pH, and partial pressure of carbon dioxide in arterial blood. We used FIDDLE (Flexible Data Driven Pipeline),37 an open source preprocessing pipeline for structured electronic health record data, to map the nine data elements to 88 binary features (each with a value of 0 or 1) describing every four hour window. The features were used as input to the machine learning model and included summary information about each variable (eg, the minimum, maximum, and mean respiratory rate within a window) and indicators for missingness (eg, whether respiratory rate was measured within a window). This form of preprocessing allowed for a variable’s missingness to be explicitly encoded in the model prediction, without the need for imputing missing values using data from previous windows or from other patients (see supplemental eMethods 1).
Model training—An ensemble of regularized logistic regression models was trained to map patient features from each four hour window to an estimate of clinical deterioration risk. From the development cohort, a single four hour window was randomly sampled for each hospital admission to train a logistic regression model. For patient hospital admissions in which the outcome occurred, only windows prior to the one before the outcome were used for training, ensuring the outcome (or any proxies) had not been observed in the training data. We repeated the process 500 times, leading to 500 models, the outputs of which were averaged to create a final prediction. Models were trained to predict whether a patient admitted to hospital would experience the primary outcome within five days of admission (see supplemental eMethods 1 for further details).
Internal validation—We measured the discriminative performance of the model using the area under the receiver operating characteristics curve (AUROC) and the area under the precision-recall curve. Models were evaluated from the first full window of data, with model predictions beginning in the window with the first vital signs recorded for a patient admitted to hospital. The model aims to support clinical decision making prospectively, during which a risk score is recomputed every four hours, and the care team decides whether to intervene once the admitted patient reaches a certain score. For this reason, we performed all evaluations at the hospital admission level, rather than at the level of four hour windows (see supplemental eMethods 1). We assessed model calibration using reliability curves and expected calibration error based on quintiles of predicted risk—that is, the average absolute difference between predicted risk and observed risk.38 39 Calibration was evaluated at the level of four hour windows to measure how well each prediction aligned with absolute risk. As a baseline, in the internal validation cohort we compared the model with a common proprietary model, the Epic Deterioration Index. This index is currently implemented in hundreds of hospitals across the US40 and is also designed to be automatically calculated in the background of an electronic health record system. Though the index was developed before the pandemic, its availability has resulted in widespread use and validation efforts for patients with covid-19.41 42 43 44
External validation —Research teams at each collaborating institution applied the inclusion and exclusion criteria locally to identify an external validation cohort at their institution, and they applied the outcome definition to determine which of the patients admitted to hospital experienced clinical deterioration (see supplemental eMethods 2). They were then given the names and descriptions of the nine clinical and personal characteristic variables, as well as the expected values and categories of these variables (see supplemental eMethods 3). These teams then independently extracted and mapped these variables to match the expected values and categories, so that the data might be saved in a format to enable identical preprocessing. In most cases these mappings were straightforward—for example, vital signs such as respiratory rate were recorded in a consistent manner across institutions. In cases when variables could not be mapped exactly, however, we worked together toward reasonable mappings. For example, head-of-bed positions of less than 20° at certain institutions were mapped to a head-of-bed position of 15° to be compatible with the preprocessing and model code. After preprocessing had taken place, each team independently applied the same model and evaluation code and reported results as summary statistics. As with the internal validation, the model was evaluated for both discriminative and calibration performance in each external cohort. Internal performance was compared with external performance using a bootstrap resampling test by computing 95% confidence intervals of the difference in performance, adjusted by Bonferroni correction. For all cohorts, we also conducted an analysis of lead time—that is, how long in advance our model could identify a patient before he or she experienced the outcome (see supplemental eFigure 5).
Assessing model generalizability across time and subgroups —To further evaluate model performance across time, we measured the AUROC and area under the precision-recall curve scores for every quarter (three month periods) between March 2020 and February 2021 within each validation cohort. Performance was also evaluated across different subgroups as the mean (and standard deviation) of AUROC scores across cohorts for subgroups of sex, age, race, and ethnicity (see supplemental eMethods 1 for categorizations). Within each cohort, we used the bootstrap resampling test to compare subgroup performance with overall performance.
Identifying low risk patients—To further examine how the model might be applied in hospitals for resource allocation, we evaluated the model for its ability to identify hospital admissions in which patients did not develop the outcome (throughout the remainder of the hospital stay) after 48 hours of observation. For these patients, we considered the average of their first 11 risk scores (representing 48 hours, excluding the first incomplete four hour window) since admission. This average risk score was then used to identify patients who were low risk throughout the remainder of their hospital stay and could be considered good candidates for early discharge to facilities providing lower acuity care, such as a temporary (field) hospital, which can be especially helpful in surge settings.45 For each validation cohort, the percentage of patient hospital admissions correctly identified as low risk was calculated subject to a negative predictive value ≥95% (ie, of the patient hospital admissions identified as low risk, ≤5% met the outcome). From this estimate, the number of bed days that potentially could be saved if these patients had been discharged at 48 hours was reported (see supplemental eMethods 1).
Implementation details and code sharing statement
All analyses were performed in Python 3.5.246 using the numpy,47 pandas,48 49 and sklearn50 packages. Code for data preprocessing and model evaluation was packaged, and each institution ran the same pipeline locally and independently. So that other institutions can validate and use the model, all code and documentation are available online at https://github.com/MLD3/M-CURES.
Patient and public involvement
This study was conducted in rapid response to the covid-19 pandemic, a public health emergency of international concern. Neither patients nor members of the public were directly involved in the design, conduct, or reporting of this research.
Results
The development cohort (n=24 419 patients) included 35 040 hospital admissions pertaining to patients admitted with respiratory distress during 2015-19 at a single institution, 3757 (10.7%) of whom experienced the primary outcome, a composite of in-hospital mortality or any of three treatments indicating severe illness: mechanical ventilation, heated high flow nasal cannula, and intravenous vasopressors (see supplemental eTable 2). The internal validation cohort (n=887 patients) included 956 hospital admissions for covid-19, 206 (21.6%) of which concerned the primary outcome (table 1). Patients admitted to hospital in the internal validation cohort were similar in age and sex to those of the development cohort but were more likely to self-report their race as Black (19.6% v 11.3%) (see supplemental eTable 2). Combined, the external validation cohorts consisted of 8335 hospital admissions, 1304 (15.6%) of which concerned the primary outcome. The external validation cohorts differed from the internal validation cohort in at least one personal characteristic dimension (sex, age, race, or ethnicity) (table 1; supplemental eTable 4). For example, the proportions of Hispanic or Latino patients were significantly higher, ranging from 13.5% to 29.0%, compared with 3.6% in the internal validation cohort; in four external cohorts a significantly larger proportion were very elderly patients (>85 years), with one cohort skewed towards being much older (22.3% v 7.3%). Externally, primary outcome rates varied from 13.4% to 19.5%. In addition, the reason for meeting the primary outcome varied significantly across hospitals (see supplemental eTable 5).
Table 1.
Characteristics of internal and external validation cohorts of adults admitted to hospital with covid-19 (see supplemental eTable 1 for characteristics of the development cohort). Values are numbers (percentages) unless stated otherwise
| Cohort | Internal validation cohort* (n=887) | External validation cohorts† | ||||||
|---|---|---|---|---|---|---|---|---|
| A (n=2161) | B (n=1252) | C (n=1180) | D (n=1009) | E (n=909) | F (n=747) | G (n=555) | ||
| No of hospital admissions | 956 | 2320 | 1320 | 1256 | 1073 | 965 | 794 | 607 |
| Median (IQR) age (years) | 64 (52-75) | 63 (50-76) | 62 (50-73) | 68 (56-79) | 65 (53-76) | 69 (58-80) | 73 (59-84) | 62 (48-75) |
| Age group (years): | ||||||||
| 18-25 | <25 | 52 (2.2) | <25 | <25 | <25 | <25 | <25 | <25 |
| 26-45 | 129 (13.5) | 398 (17.2) | 225 (17.1) | 159 (12.7) | 159 (14.8) | 77 (8.0) | 74 (9.3) | 114 (18.8) |
| 46-65 | 374 (39.1) | 800 (34.5) | 518 (39.2) | 380 (30.3) | 358 (33.4) | 327 (33.9) | 204 (25.7) | 215 (35.4) |
| 66-85 | 365 (38.2) | 873 (37.6) | 497 (37.7) | 539 (42.9) | 435 (40.5) | 412 (42.7) | 331 (41.7) | 184 (30.3) |
| >85 | 70 (7.3) | 197 (8.5) | 57 (4.3) | 159 (12.7) | 97 (9.0) | 145 (15.0) | 177 (22.3) | 74 (12.2) |
| Sex: | ||||||||
| Women | 420 (43.9) | 993 (42.8) | 612 (46.3) | 564 (44.9) | 533 (49.7) | 445 (46.1) | 363 (45.7) | 313 (51.6) |
| Men | 536 (56.1) | 1327 (57.2) | 709 (53.7) | 692 (55.1) | 540 (50.3) | 520 (53.9) | 431 (54.3) | 294 (48.4) |
| Race‡: | ||||||||
| White | 649 (67.9) | 1364 (58.8) | 733 (55.6) | 935 (74.4) | 589 (54.9) | 636 (65.9) | 584 (73.6) | 214 (35.3) |
| Black | 187 (19.6) | 190 (8.2) | 332 (25.2) | 123 (9.8) | 234 (21.8) | 135 (14.0) | 49 (6.2) | 62 (10.2) |
| Asian§ | 30 (3.1) | 80 (3.4) | 29 (2.2) | 51 (4.1) | 39 (3.6) | <25 | 39 (4.9) | 135 (22.2) |
| Other or unknown¶ | 90 (9.4) | 686 (29.6) | 226 (17.1) | 147 (11.7) | 211 (19.7) | 168 (17.4) | 122 (15.4) | 196 (32.3) |
| Ethnicity: | ||||||||
| Hispanic or Latino | 34 (3.6) | 587 (25.3) | 379 (28.7) | 350 (27.9) | 210 (19.6) | 138 (14.3) | 107 (13.5) | 176 (29.0) |
| Non-Hispanic or non-Latino | 883 (92.4) | 1569 (67.6) | 915 (69.3) | 875 (69.7) | 841 (78.4) | 783 (81.1) | 637 (80.2) | 414 (68.2) |
| Other or unknown | 39 (4.1) | 164 (7.1) | 26 (1.8) | 31 (2.5) | <25 | 44 (4.6) | 50 (6.3) | <25 |
| Median (IQR) length of stay (hours) | 138 (83-261) | 160 (95-284) | 141 (96-257) | 136 (93-235) | 167 (100-287) | 143 (92-234) | 154 (95-256) | 183 (113-324) |
| Outcome ever: | ||||||||
| Death | 60 (6.3) | 197 (8.5) | 108 (8.2) | 125 (10.0) | 96 (8.9) | 93 (9.6) | 123 (15.5) | 42 (6.9) |
| Mechanical ventilation | 98 (10.3) | 259 (11.2) | 142 (10.7) | 135 (10.7) | 116 (10.8) | 69 (7.2) | 69 (8.7) | 52 (8.6) |
| Intravenous vasopressors | 87 (9.1) | 299 (12.9) | 152 (11.5) | 139 (11.1) | 125 (11.6) | 65 (6.7) | 74 (9.3) | 70 (11.5) |
| Heated high flow nasal cannula | 218 (22.4) | 132 (5.7) | 263 (19.9) | 121 (9.6) | 95 (8.9) | 99 (10.3) | 106 (13.4) | 101 (16.6) |
| Primary outcome ≤5 days | 206 (21.6) | 311 (13.4) | 249 (18.8) | 206 (16.4) | 155 (14.4) | 136 (14.1) | 155 (19.5) | 92 (15.2) |
| Reason for primary outcome (% of outcomes): | ||||||||
| Death | 5 (2.4) | 34 (10.9) | 4 (1.6) | 21 (10.2) | 16 (10.3) | 25 (18.4) | 37 (23.9) | 2 (2.2) |
| Mechanical ventilation | 20 (9.7) | 89 (28.6) | 25 (10.0) | 52 (25.2) | 52 (33.5) | 22 (16.2) | 18 (11.6) | 8 (8.7) |
| Intravenous vasopressors | 9 (4.4) | 95 (30.5) | 18 (7.2) | 33 (16.0) | 26 (16.8) | 10 (7.4) | 21 (13.5) | 16 (17.4) |
| Heated high flow nasal cannula | 172 (83.5) | 93 (29.9) | 202 (81.1) | 100 (48.5) | 61 (39.4) | 79 (58.1) | 79 (51.0) | 66 (71.7) |
IQR=interquartile range.
Patients with covid-19 admitted to one institution during 2020-21.
Patients admitted with covid-19 during 2020-21 at 12 external medical centers. Six sites with fewer than 100 patients that met the primary outcome were combined into a single cohort when performing evaluation, resulting in seven external validation cohorts.
Race was self-identified by patients or their guardian, with options: American Indian or Alaska Native, Asian, Black, native Hawaiian or other Pacific Islander, White, other, patient refused, or unknown.
As defined by the US Census Bureau,51 the Asian race refers to people having origins in any of the original peoples of the Far East, Southeast Asia, or the Indian subcontinent including, for example, Cambodia, China, India, Japan, Korea, Malaysia, Pakistan, the Philippine Islands, Thailand, and Vietnam.
Includes American Indian or Alaskan, native Hawaiian or other Pacific Islander, other, unknown, or patient refused.
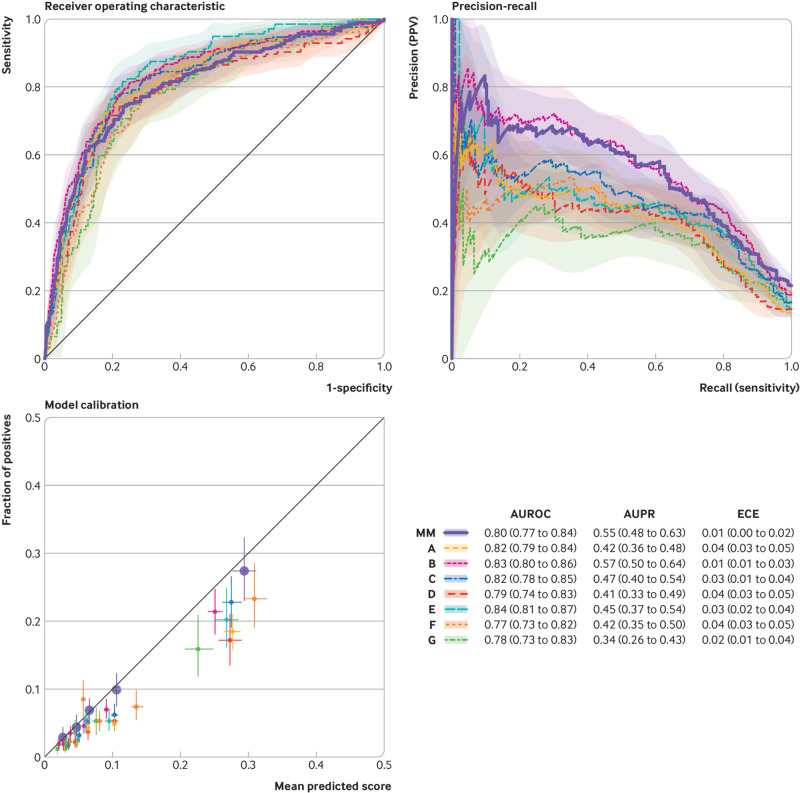
Supplemental eFigure 2 presents the parameters of the final learnt model, and eTable 1 shows all model coefficients as a comma separated values file. This file can be loaded into a computer program and used to automate model prediction and is not intended to be readable by humans (hence the number of digits after the decimal place). The model showed good overall performance in both internal and external validation. When the model was applied to the internal validation cohort, it substantially outperformed the Epic Deterioration Index, achieving an AUROC of 0.80 (95% confidence interval 0.77 to 0.84) v 0.66 (0.62 to 0.70), area under the precision-recall curve of 0.55 (95% confidence interval 0.48 to 0.63) v 0.31 (0.26 to 0.36), and expected calibration error of 0.01 (95% confidence interval 0.00 to 0.02) v 0.31 (0.30 to 0.32) (see supplemental eFigure 3). External validation resulted in similar performance, with AUROCs ranging from 0.77 to 0.84, area under the precision-recall curve ranging from 0.34 to 0.57, and expected calibration errors ranging from 0.02 to 0.04 (fig 1). The AUROC across external institutions did not differ significantly from the internal validation AUROC (supplemental eTable 6) and had an average of 0.81.
Fig 1.
Model performance across internal and external validation cohorts. Discriminative performance was measured using receiver operating characteristic curves and precision-recall curves. Model calibration is shown in reliability plots based on quintiles of predicted scores. The table summarizes results with 95% confidence intervals. The thick line shows the internal validation cohort at Michigan Medicine (MM) and the different colors represent the external validation cohorts (A-G). PPV=positive predictive value; AUROC=area under the receiver operating characteristics curve; AUPR=area under the precision-recall curve; ECE=expected calibration error
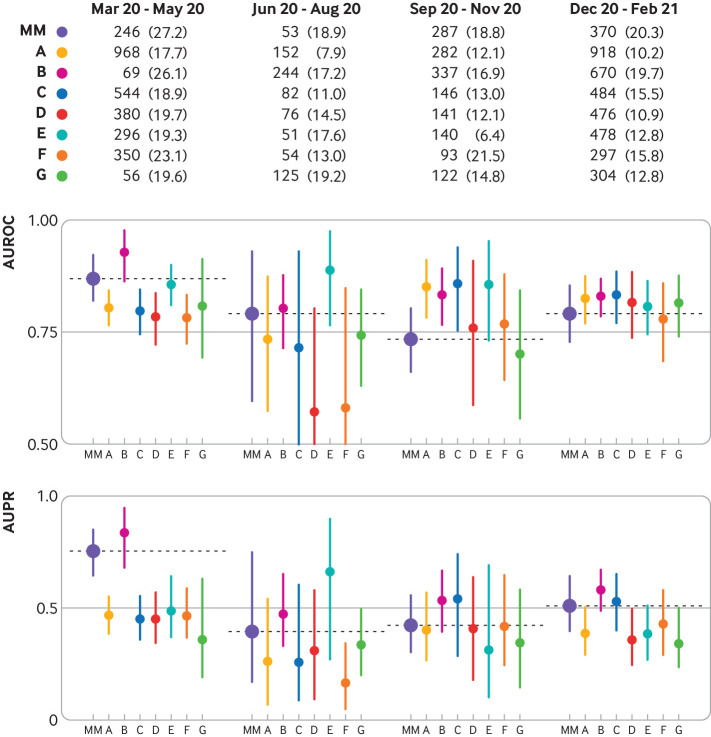
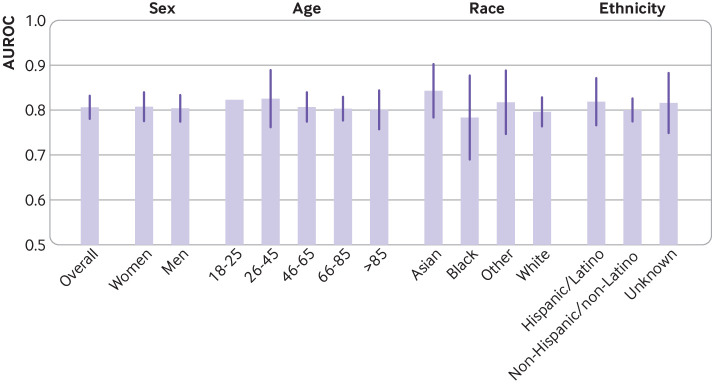
Across time (fig 2; supplemental eTable 7) the model performed consistently in all validation cohorts throughout the four quarters, with AUROCs >0.7 and area under the precision-recall curves >0.2 in most cases. The exception was during June to August 2020, where compared with the overall performance of each cohort, two cohorts showed a decrease in AUROC (from 0.79 to 0.57 and from 0.77 to 0.58) and one cohort showed a decrease in area under the precision-recall curve (from 0.42 to 0.17), but the differences were not statistically significant (see supplemental eTable 8). Across subgroups based on personal characteristics, the model displayed consistent discriminative performance in terms of AUROC (fig 3; supplemental eTable 9); subgroup performance did not vary significantly from the overall performance when evaluated within specific sex, age, and race or ethnicity subpopulations (see supplemental eTable 10). In one external cohort, the model performed significantly better on patients who self-reported their race as Asian (as defined by the US Census Bureau51) compared with patients who self-reported their race as White (see supplementary eTable 11).
Fig 2.
Model discriminative performance (area under the receiver operating characteristics curve (AUROC) and area under the precision-recall curve (AUPR) scores) over the year (March 2020 to February 2021) by quarter. The table shows the number (percentage) of patient hospital admissions in each cohort in each quarter and met the primary outcome of a composite of clinical deterioration within the first five days of hospital admission, defined as in-hospital mortality or any of three treatments indicating severe illness: mechanical ventilation, heated high flow nasal cannula, and intravenous vasopressors. MM=Michigan Medicine; A-G represent the external validation cohorts
Fig 3.
Model discriminative performance (area under the receiver operating characteristics curve (AUROC) scores) evaluated across subgroups. Values are macro-average performance across institutions (error bars are ±1 standard deviation). No error bar shown for age subgroup 18-25 years because only a single institution had enough positive cases to calculate the AUROC score
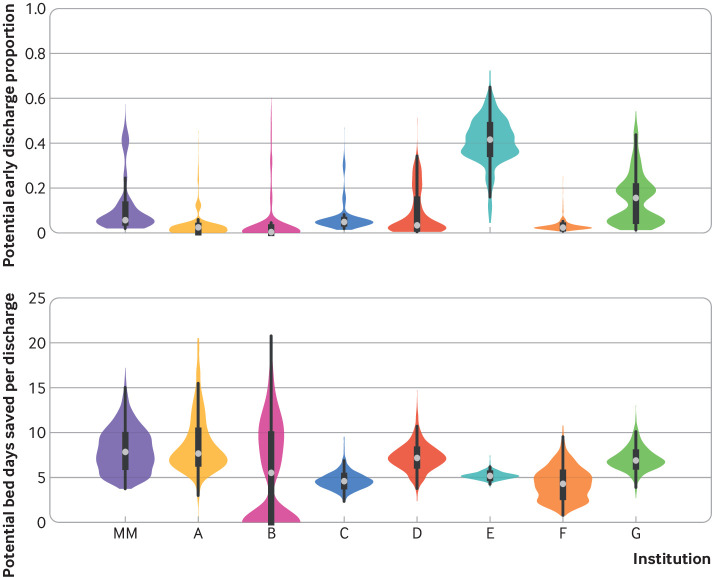
In terms of resource allocation and planning, the model was able to accurately identify low risk patients after 48 hours of observation in both the internal and the external cohorts. At best, the model could correctly triage up to 41.6% of low risk patients admitted to hospital with covid-19 to lower acuity care, with a potential saving of 5.2 bed days for each early discharge. At other institutions, the model could potentially save 7.8 bed days, while correctly triaging fewer patients admitted to hospital as low risk (fig 4). The model achieved this performance level while maintaining a negative predictive value of at least 95%—that is, of those admitted to hospital who were identified as low risk patients, 5% or fewer met the primary outcome.
Fig 4.
Model used to identify potential patients with covid-19 for early discharge after 48 hours of observation. A decision threshold was chosen that achieves a negative predictive value of ≥95%. Figure depicts both the proportion of patients who could be discharged early and the number of bed days saved, normalized by the number of correctly discharged patients in each validation cohort. Results are computed over 1000 bootstrap replications. MM=Michigan Medicine; A-G represent the external validation cohorts
Discussion
Accurately predicting the deterioration of patients can assist clinicians in risk assessment during a patient’s hospital admission by identifying those who might need ICU level care in advance of deterioration.52 53 54 In scenarios with a surge in admissions, hospitals might use predictions to manage limited resources, such as beds, by triaging low risk patients to lower acuity care. This has spurred considerable efforts in developing prediction models for the prognosis of covid-19, as shown in a living systematic review.12 Despite these efforts, however, generalization performance, or the performance of the model on new patient populations, is often overlooked when such models are developed and evaluated. To this end, we developed an open source patient risk stratification model that uses nine routinely collected personal characteristic and clinical variables from a patient’s electronic health record for prediction of clinical deterioration. Compared with previous deterioration indices that have failed to generalize across multiple patient cohorts,23 55 the model achieved excellent discriminative performance in five validation cohorts, and acceptable discriminative performance in the remaining three, all while achieving strong calibration performance.56
External validation can highlight blind spots when the validation cohort differs substantially from the development cohort, including clinical conditions (eg, covid-19 is a new disease); personal characteristics, such as race and ethnicity; clinical workflows; and number of beds in the hospital. Ensuring consistency of features across both patient populations and different institutions remains challenging, even in the most basic settings. For example, differences in clinical workflows across hospitals could result in different documentation practices or different monitoring strategies (eg, intermittent versus continuous pulse oximetry measurement), which could in turn affect the usefulness of these variables. Despite the likely differences in clinical practice across hospitals, our proposed model performed well across institutions, suggesting that these variables capture certain aspects of illness severity that are generalizable. The model’s strong generalizability might be attributed to several design choices. First, we utilized a separate but related development cohort for training. This idea, known as transfer learning, allowed us to utilize a large cohort of patients for training.57 58 Moreover, the clinician-informed data driven approach to feature selection and a rigorous approach to internal validation contributed to the strong generalization performance of the model.
We also evaluated performance on specific subgroups (based on age, sex, race, and ethnicity) and across time.59 60 Ensuring consistent performance across such subgroups can help mitigate biases against certain vulnerable populations.61 62 63 Despite an underrepresentation of Hispanic and Latino patients in the development cohort compared with the external validation cohorts, model performance in this subgroup was consistent with performance in people of non-Hispanic and Latino ethnicity. At several points during the pandemic, changes in the patient population presenting with severe disease and changes to clinical workflows could have impacted model performance. For example, timings of surges in admissions and outcome rates differed throughout regions of the US owing to factors such as local policies and lockdown timings.64 65 66 67 These changes could have resulted in a modest decline in model performance at two sites in the summer of 2020. Beyond surge settings, the treatments, availability of vaccines, and outcome rates likely have an impact on how risk models might perform.68 69 70 71 72 73 In particular, model performance stabilized in the autumn and winter surges, which could indicate a convergence in treatment of covid-19.
Our evaluation of the model’s performance focused on two relevant clinical use cases: identifying high risk patients who might need critical care interventions and identifying low risk patients who might be candidates for transfer to lower acuity settings. As a clinical risk indicator, the model could be displayed within the electronic health record near vital signs to provide clinicians with summary information about a patient’s status without prespecifying a threshold recommending action. Alternatively, an institution might decide to use the model to support a rapid response team that evaluates patients at high risk for clinical decompensation. In such a scenario, the threshold chosen to trigger an evaluation would depend, in part, on the number of evaluations the team could perform during a shift. Ultimately, decisions on how the model will inform patient care should be largely driven by local needs, resource constraints, and available interventions, as well as by an institution’s tolerance of false positives and false negatives.
Strengths and limitations of this study
Unlike previous work on the external validation of patient risk stratification models,22 our approach did not rely on sharing data across multiple sources. Instead, we developed the model using data from a single institution and then shared the code with collaborators in external institutions who then applied the model to their data using their own computing platforms. This approach has many benefits. The sharing and aggregation of data that contain protected health information (eg, dates) from 12 healthcare systems into a single repository would have required extensive data use agreements and additional computational infrastructure and added substantial delays to model evaluation. Maintaining patient data internally further mitigates the potential risk of data access breaches. In addition to distributing the workload and evaluation process, this approach reduced the chance of errors because each team was most familiar with its own data and thus less likely to make incorrect assumptions when identifying the cohort, model variables, and outcomes.
The success of this paradigm relied on several design decisions early in the process as well as continued collaboration throughout. First, the number of variables used by the model was limited, ensuring that all variables could be reliably identified and validated at each institution. Beyond model inputs, it was equally crucial to validate inclusion and exclusion criteria and outcome definitions. To this end, we worked closely with both clinicians and informaticists from each institution to establish accurate definitions. Finally, we developed a code workflow with common input and output formats and shared detailed documentation. This in turn allowed for quick iteration among institutions, facilitating debugging.
The data driven approach for feature selection resulted in features that might not immediately align with clinical intuition, though still represent important aspects of a patient’s illness, and can help in predicting the outcome. For example, both head-of-bed position and the patient’s position during blood pressure measurement might indicate aspects of patient illness severity that are not captured by other data. A blood pressure reading taken in a standing position might indicate a healthy patient who can tolerate such a maneuver. Strong external validation performance ensured that these variables captured aspects of illness that generalized across multiple institutions.
The current analysis should be interpreted in the context of its study design. Importantly, a single electronic health record software provider (Epic Systems; Verona, WI) was used across all medical centers. This commonality between institutions facilitated model validation. Despite a common electronic health record vendor being used, however, local implementation of each electronic health record system requires local knowledge of institutions, which was a feat of our multisite team approach. To further ensure the model can generalize to more institutions, researchers should focus on validating the model in healthcare systems utilizing different electronic health record systems. Moreover, the model was developed and validated on adults with respiratory distress and a diagnosis of covid-19 in distinct geographical regions across the US. We focused on covid-19 owing to the ongoing strain on hospital resources created by the pandemic.25 26 27 28 29 The model may or may not apply to patients with respiratory distress without a covid-19 diagnosis, in other regions of the US (eg, mountain west and northwest) or other countries. Furthermore, when we estimated potential bed days saved resulting from the triage of low risk patients, we assumed that those patients could be safely discharged at 48 hours. Other reasons might, however, exist as to why a patient needs to remain in hospital, preventing early discharge. The model may be particularly effective in identifying those patients who can be discharged especially when lower acuity care centers are available for transfer of patients. Finally, the composite outcome we considered was developed early in the pandemic based on clinical workflows and treatments at the time. As treatments evolve, outcome definitions might change that could affect model performance. Without implementation into clinical practice, it remains unknown whether the use of such a model has an impact on clinical or operational outcomes, such as early discharge planning.
Comparison with other studies
As a baseline, we compared our model with the Epic Deterioration Index in the internal validation cohort and found favorable performance. Although additional baselines (such as the 4C mortality and deterioration models21 22) exist, they are not directly comparable with our proposed model. Most importantly, the intended use of the 4C models differs from that of our model. The 4C models were designed as a bedside calculator for estimating a patient’s risk at one point in time and inputs must be provided by the clinician (allowing for potential subjectivity for some features) and are not automatically extracted from the electronic health records. In contrast, our model automatically estimates risk at regular intervals throughout a patient’s hospital admission without any extra effort from a clinician. Despite the perceived simplicity of the 4C models, it is challenging to collect some of the necessary variables in an automated fashion. For example, extracting comorbidities from electronic health record data through ICD codes can be error prone and inconsistent across institutions.74 75 Therefore, we focused on the comparison with the Epic Deterioration Index, which operates in a similar manner to our model and was already implemented at the development institution.
Conclusions and policy implications
This study represents an important step toward building and externally validating models for identifying patients at both high and low risk of clinical deterioration during their hospital stay. The model generalized across a variety of institutions, subgroups, and time periods. Our method for external validation alleviates potential concerns surrounding patient privacy by forgoing the need for data sharing while still allowing for realistic and accurate evaluations of a model within different patient settings. Thus, the implications are twofold; the work here can help develop models to predict patient deterioration within a single institution, and the work can promote external validation and multicenter collaborations without the need for data sharing agreements.
What is already known on this topic
Risk stratification models can augment clinical care and help hospitals better plan and allocate resources in healthcare settings
A useful risk stratification model should generalize across different patient populations, though generalization is often overlooked when models are developed because of the difficulty in sharing patient data for external validation
Models that have been externally validated have failed to generalize to populations that differed from the cohort on which the models were built
What this study adds
This study presents a paradigm for model development and external validation without the need for data sharing, while still allowing for quick and thorough evaluations of a model within different patient populations
The findings suggest that the use of data driven feature selection combined with clinical judgment can help identify meaningful features that allow the model to generalize across a variety of patient settings
Acknowledgments
We thank the staff of the Data Office for Clinical and Translational Research at the University of Michigan for their help in data extraction and curation, and Melissa Wei, Ian Fox, Jeeheh Oh, Harry Rubin-Falcone, Donna Tjandra, Sarah Jabbour, Jiaxuan Wang, and Meera Krishnamoorthy for helpful discussions during early iterations of this work.
Web extra.
Extra material supplied by authors
Supplementary information: eMethods 1-3, eTables 1-11, and eFigures 1-5
Supplementary table: spreadsheet showing eTable 1 data
Contributors: FK and ST are co-first authors of equal contribution. MWS and JW are co-senior authors of equal contribution. JZA, BKN, MWS, and JW conceptualized the study. FK, ST, EO, DSM, SNS, JG, BYL, SD, XL, RJM, TSV, LRW, KS, SB, JPD, ESS, MWS, and JW acquired, analyzed, or interpreted the data. FK, ST, DSM, SNS, JG, XL, MWS, and JW had access to study data pertaining to their respective institutions and took responsibility for the integrity of the data and the accuracy of the data analysis. FK, ST, and EO drafted the manuscript. FK, ST, EO, DSM, SNS, JG, BYL, SD, XL, RJM, TSV, LRW, KS, SB, JPD, ESS, JZA, BKN, MWS, and JW critically revised the manuscript for important intellectual content. BKN, MWS, and JW supervised the conduct of this study. FK and ST are guarantors. The corresponding author attests that all listed authors meet authorship criteria and that no others meeting the criteria have been omitted.
Funding: This work was supported by the National Science Foundation (NSF; award IIS-1553146 to JW), by the National Institutes of Health (NIH) -National Library of Medicine (NLM; grant R01LM013325 to JW and MWS), -National Heart, Lung, and Blood Institute (NHLBI; grant K23HL140165 to TSV; grant K12HL138039 to JPD), by the Agency for Healthcare Research and Quality (AHRQ; grant R01HS028038 TSV), by the Centers for Disease Control and Prevention (CDC) -National Center for Emerging and Zoonotic Infectious Diseases (NCEZID; grant U01CK000590 to SB and RJM), by Precision Health at the University of Michigan (U-M), and by the Institute for Healthcare Policy and Innovation at U-M. The funding sources had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication. The views and conclusions in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of NSF, NIH, AHRQ, CDC, or the US government.
Competing interests: All authors have completed the ICMJE uniform disclosure form at www.icmje.org/coi_disclosure.pdf and declare: support from National Science Foundation (NSF), National Institutes of Health (NIH) -National Library of Medicine (NLM) and -National Heart, Lung, and Blood Institute (NHLBI), Agency for Healthcare Research and Quality (AHRQ), Centers for Disease Control and Prevention (CDC) -National Center for Emerging and Zoonotic Infectious Diseases (NCEZID), Precision Health at the University of Michigan, and the Institute for Healthcare Policy and Innovation at the University of Michigan. JZA received grant funding from National Institute on Aging, Michigan Department of Health and Human Services, and Merck Foundation, outside of the submitted work; JZA also received personal fees for consulting at JAMA Network and New England Journal of Medicine, honorariums from Harvard University, University of Chicago, and University of California San Diego, and monetary support for travel reimbursements from NIH, National Academy of Medicine, and AcademyHealth, during the conduct of the study; JZA also served as a board member of AcademyHealth, Physicians Health Plan, and Center for Health Research and Transformation, with no compensation, during the conduct of the study. SB reports receiving grant funding from NIH, outside of the submitted work. JPD reports receiving personal fees from the Annals of Emergency Medicine, during the conduct of the study. RJM reports receiving grant funding from Verily Life Sciences, Sergey Brin Family Foundation, and Texas Health Resources Clinical Scholar, outside of the submitted work; RJM also served on the advisory committee of Infectious Diseases Society of America - Digital Strategy Advisory Group, during the conduct of the study. BKN reports receiving grant funding from NIH, Veterans Affairs -Health Services Research and Development Service, the American Heart Association (AHA), Janssen, and Apple, outside of the submitted work; BKN also received compensation as editor in chief of Circulation: Cardiovascular Quality and Outcomes, a journal of AHA, during the conduct of the study; BKN is also a co-inventor on US Utility Patent No US15/356 012 (US20170148158A1) entitled “Automated Analysis of Vasculature in Coronary Angiograms,” that uses software technology with signal processing and machine learning to automate the reading of coronary angiograms, held by the University of Michigan; the patent is licensed to AngioInsight, in which BKN holds ownership shares and receives consultancy fees. EÖ reports having a patent pending for the University of Michigan for an artificial intelligence based approach for the dynamic prediction of health states for patients with occupational injuries. SNS reports serving on the editorial board for the Journal of the American Medical Informatics Association, and on the student editorial board for Applied Informatics Journal, during the conduct of the study. KS reports receiving grant funding from Blue Cross Blue Shield of Michigan, and Teva Pharmaceuticals, outside of the submitted work; KS also serves on a scientific advisory board for Flatiron Health, where he receives consulting fees and honorariums for invited lectures, during the conduct of the study. MWS reports serving on the planning committee for the Machine Learning for Healthcare Conference (MLHC), a non-profit organization that hosts a yearly academic meeting. JW reports receiving grant funding from Cisco Systems, D Dan and Betty Kahn Foundation, and Alfred P Sloan Foundation, during the conduct of the study outside of the submitted work; JW also served on the international advisory board for Lancet Digital Health, and on the advisory board for MLHC, during the conduct of the study. No other disclosures were reported that could appear to have influenced the submitted work. SD, JG, FK, BYL, XL, DSM, ESS, ST, TSV, and LRW all declare: no additional support from any organization for the submitted work; no additional financial relationships with any organizations that might have an interest in the submitted work in the previous three years; and no other relationships or activities that could appear to have influenced the submitted work.
FK, ST, MWS, and JW affirm that the manuscript is an honest, accurate, and transparent account of the study being reported; that no important aspects of the study have been omitted; and that any discrepancies from the study as originally planned (and, if relevant, registered) have been explained.
Dissemination to participants and related patient and public communities: The results of this study will be disseminated to the general public, primarily engaging with print and internet press, blog posts, and twitter. As this study is related to inpatient admissions for covid-19, it is important that the model can be used by clinicians to guide decision making in a reliable way. The model coefficients and validation code are publicly available online at https://github.com/MLD3/M-CURES.
Provenance and peer review: Not commissioned; externally peer reviewed.
Ethics statements
Ethical approval
This study was approved by the institutional review boards of all participating sites (University of Michigan, Michigan Medicine HUM00179831, Mass General Brigham 2012P002359, University of Texas Southwestern Medical Center STU-2020-0922, University of California San Francisco 20-31825), with a waiver of informed consent.
Data availability statement
To guarantee the confidentiality of personal and health information, only the authors have had access to the data during the study in accordance with the relevant license agreements. The full model (including model coefficients and supporting code) are available online at https://github.com/MLD3/M-CURES.
References
- 1. Shah NH, Milstein A, Bagley PhD SC. Making Machine Learning Models Clinically Useful. JAMA 2019;322:1351-2. 10.1001/jama.2019.10306. [DOI] [PubMed] [Google Scholar]
- 2. Peterson ED. Machine Learning, Predictive Analytics, and Clinical Practice: Can the Past Inform the Present? JAMA 2019;322:2283-4. 10.1001/jama.2019.17831. [DOI] [PubMed] [Google Scholar]
- 3. White DB, Lo B. A Framework for Rationing Ventilators and Critical Care Beds During the COVID-19 Pandemic. JAMA 2020;323:1773-4. 10.1001/jama.2020.5046. [DOI] [PubMed] [Google Scholar]
- 4. Coley CM, Li Y-H, Medsger AR, et al. Preferences for home vs hospital care among low-risk patients with community-acquired pneumonia. Arch Intern Med 1996;156:1565-71. 10.1001/archinte.1996.00440130115012. [DOI] [PubMed] [Google Scholar]
- 5. Page K, Barnett AG, Graves N. What is a hospital bed day worth? A contingent valuation study of hospital Chief Executive Officers. BMC Health Serv Res 2017;17:137. 10.1186/s12913-017-2079-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Razavian N, Major VJ, Sudarshan M, et al. A validated, real-time prediction model for favorable outcomes in hospitalized COVID-19 patients. NPJ Digit Med 2020;3:130. 10.1038/s41746-020-00343-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Pericàs JM, Cucchiari D, Torrallardona-Murphy O, et al. Hospital Clínic 4H Team (Hospital at Home-Health Hotel) . Hospital at home for the management of COVID-19: preliminary experience with 63 patients. Infection 2021;49:327-32. 10.1007/s15010-020-01527-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Habib AR, Lin AL, Grant RW. The epic sepsis model falls short-the importance of external validation. JAMA Intern Med 2021;181:1040-1. 10.1001/jamainternmed.2021.3333. [DOI] [PubMed] [Google Scholar]
- 9. Ramspek CL, Jager KJ, Dekker FW, Zoccali C, van Diepen M. External validation of prognostic models: what, why, how, when and where? Clin Kidney J 2020;14:49-58. 10.1093/ckj/sfaa188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Siontis GCM, Tzoulaki I, Castaldi PJ, Ioannidis JP. External validation of new risk prediction models is infrequent and reveals worse prognostic discrimination. J Clin Epidemiol 2015;68:25-34. 10.1016/j.jclinepi.2014.09.007. [DOI] [PubMed] [Google Scholar]
- 11. Steyerberg EW, Harrell FE, Jr. Prediction models need appropriate internal, internal-external, and external validation. J Clin Epidemiol 2016;69:245-7. 10.1016/j.jclinepi.2015.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Wynants L, Van Calster B, Collins GS, et al. Prediction models for diagnosis and prognosis of covid-19: systematic review and critical appraisal. BMJ 2020;369:m1328. 10.1136/bmj.m1328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cummings BC, Ansari S, Motyka JR, et al. Predicting Intensive Care Transfers and Other Unforeseen Events: Analytic Model Validation Study and Comparison to Existing Methods. JMIR Med Inform 2021;9:e25066. 10.2196/25066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Shamout FE, Shen Y, Wu N, et al. An artificial intelligence system for predicting the deterioration of COVID-19 patients in the emergency department. NPJ Digit Med 2021;4:80. 10.1038/s41746-021-00453-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Price WN, 2nd, Cohen IG. Privacy in the age of medical big data. Nat Med 2019;25:37-43. 10.1038/s41591-018-0272-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Panch T, Mattie H, Celi LA. The “inconvenient truth” about AI in healthcare. NPJ Digit Med 2019;2:77. 10.1038/s41746-019-0155-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Luengo-Oroz M, Hoffmann Pham K, Bullock J, et al. Artificial intelligence cooperation to support the global response to COVID-19. Nat Mach Intell 2020;2:295-7. 10.1038/s42256-020-0184-3. [DOI] [Google Scholar]
- 18. Peiffer-Smadja N, Maatoug R, Lescure F-X, et al. Machine Learning for COVID-19 needs global collaboration and data-sharing. Nat Mach Intell 2020;2:293-4. 10.1038/s42256-020-0181-6. [DOI] [Google Scholar]
- 19. Xie J, Hungerford D, Chen H, et al. Development and External Validation of a Prognostic Multivariable Model on Admission for Hospitalized Patients with COVID-19. 2020. 10.2139/ssrn.3562456 [DOI]
- 20. Chow DS, Glavis-Bloom J, Soun JE, et al. Development and external validation of a prognostic tool for COVID-19 critical disease. PLoS One 2020;15:e0242953. 10.1371/journal.pone.0242953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Knight SR, Ho A, Pius R, et al. ISARIC4C investigators . Risk stratification of patients admitted to hospital with covid-19 using the ISARIC WHO Clinical Characterisation Protocol: development and validation of the 4C Mortality Score. BMJ 2020;370:m3339. 10.1136/bmj.m3339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gupta RK, Harrison EM, Ho A, et al. ISARIC4C Investigators . Development and validation of the ISARIC 4C Deterioration model for adults hospitalised with COVID-19: a prospective cohort study. Lancet Respir Med 2021;9:349-59. 10.1016/S2213-2600(20)30559-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Carmichael H, Coquet J, Sun R, et al. Learning from past respiratory failure patients to triage COVID-19 patient ventilator needs: A multi-institutional study. J Biomed Inform 2021;119:103802. 10.1016/j.jbi.2021.103802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barish M, Bolourani S, Lau LF, et al. External validation demonstrates limited clinical utility of the interpretable mortality prediction model for patients with COVID-19. Nat Mach Intell 2021;3:25-7. 10.1038/s42256-020-00254-2. [DOI] [Google Scholar]
- 25. Eriksson CO, Stoner RC, Eden KB, Newgard CD, Guise JM. The Association Between Hospital Capacity Strain and Inpatient Outcomes in Highly Developed Countries: A Systematic Review. J Gen Intern Med 2017;32:686-96. 10.1007/s11606-016-3936-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Emanuel EJ, Persad G, Upshur R, et al. Fair Allocation of Scarce Medical Resources in the Time of Covid-19. N Engl J Med 2020;382:2049-55. 10.1056/NEJMsb2005114. [DOI] [PubMed] [Google Scholar]
- 27. Vergano M, Bertolini G, Giannini A, et al. Clinical ethics recommendations for the allocation of intensive care treatments in exceptional, resource-limited circumstances: the Italian perspective during the COVID-19 epidemic. Crit Care 2020;24:165. 10.1186/s13054-020-02891-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Carenzo L, Costantini E, Greco M, et al. Hospital surge capacity in a tertiary emergency referral centre during the COVID-19 outbreak in Italy. Anaesthesia 2020;75:928-34. 10.1111/anae.15072. [DOI] [PubMed] [Google Scholar]
- 29. COVIDSurg Collaborative . Elective surgery cancellations due to the COVID-19 pandemic: global predictive modelling to inform surgical recovery plans. Br J Surg 2020;107:1440-9. 10.1002/bjs.11746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Moons KGM, Altman DG, Reitsma JB, et al. Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD): explanation and elaboration. Ann Intern Med 2015;162:W1-W73. 10.7326/M14-0698. [DOI] [PubMed] [Google Scholar]
- 31. Riley RD, Ensor J, Snell KIE, et al. Calculating the sample size required for developing a clinical prediction model. BMJ 2020;368:m441. 10.1136/bmj.m441. [DOI] [PubMed] [Google Scholar]
- 32. Kaufman S, Rosset S, Perlich C, et al. Leakage in data mining: Formulation, detection, and avoidance. ACM Trans Knowl Discov Data 2012;6:1-21. 10.1145/2382577.2382579. [DOI] [Google Scholar]
- 33. de Mendonça A, Vincent J-L, Suter PM, et al. Acute renal failure in the ICU: risk factors and outcome evaluated by the SOFA score. Intensive Care Med 2000;26:915-21. 10.1007/s001340051281. [DOI] [PubMed] [Google Scholar]
- 34. Breiman L. Random Forests. Mach Learn 2001;45:5-32. 10.1023/A:1010933404324. [DOI] [Google Scholar]
- 35. Hooker G, Mentch L. Please Stop Permuting Features: An Explanation and Alternatives. arXiv. 2019.https://arxiv.org/abs/1905.03151
- 36. Ferri FJ, Pudil P, Hatef M, et al. Comparative study of techniques for large-scale feature selection. In: Gelsema ES, Kanal LS, eds. Machine Intelligence and Pattern Recognition 1994.;16:403-13. 10.1016/B978-0-444-81892-8.50040-7 [DOI] [Google Scholar]
- 37. Tang S, Davarmanesh P, Song Y, Koutra D, Sjoding MW, Wiens J. Democratizing EHR analyses with FIDDLE: a flexible data-driven preprocessing pipeline for structured clinical data. J Am Med Inform Assoc 2020;27:1921-34. 10.1093/jamia/ocaa139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naeini MP, Cooper GF, Hauskrecht M. Obtaining Well Calibrated Probabilities Using Bayesian Binning. In: Proceedings of the Twenty-Ninth AAAI Conference on Artificial Intelligence. Austin, Texas: AAAI Press 2015. 2901-2907. [PMC free article] [PubMed] [Google Scholar]
- 39. Huang Y, Li W, Macheret F, Gabriel RA, Ohno-Machado L. A tutorial on calibration measurements and calibration models for clinical prediction models. J Am Med Inform Assoc 2020;27:621-33. 10.1093/jamia/ocz228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Systems E. Artificial Intelligence Triggers Fast, Lifesaving Care for COVID-19 Patients. Epic Systems. 2020. https://www.epic.com/epic/post/artificial-intelligence-epic-triggers-fast-lifesaving-care-covid-19-patients (accessed 1 Jan 2022).
- 41. Singh K, Valley TS, Tang S, et al. Evaluating a Widely Implemented Proprietary Deterioration Index Model among Hospitalized Patients with COVID-19. Ann Am Thorac Soc 2021;18:1129-37. 10.1513/AnnalsATS.202006-698OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ross C. Hospitals are using AI to predict the decline of Covid-19 patients — before knowing it works. STAT. 2020. https://www.statnews.com/2020/04/24/coronavirus-hospitals-use-ai-to-predict-patient-decline-before-knowing-it-works/ (accessed 1 Jan 2022).
- 43.Robbins R. ‘Human experts will make the call’: Stanford launches an accelerated test of AI to help care for Covid-19 patients. STAT. 2020. https://www.statnews.com/2020/04/01/stanford-artificial-intelligence-coronavirus/ (accessed 14 Aug 2021).
- 44.Strickland E. AI May Help Hospitals Decide Which COVID-19 Patients Live or Die. IEEE Spectrum. 2020. https://spectrum.ieee.org/ai-can-help-hospitals-triage-covid19-patients (accessed 1 Jan 2022).
- 45.Carmody S. U of M health system planning to open COVID-19 field hospital. Michigan Radio. 2020. https://www.michiganradio.org/health/2020-03-31/u-of-m-health-system-planning-to-open-covid-19-field-hospital (accessed 1 Jan 2022).
- 46. Python Software Foundation . Python. https://www.python.org/
- 47. Harris CR, Millman KJ, van der Walt SJ, et al. Array programming with NumPy. Nature 2020;585:357-62. 10.1038/s41586-020-2649-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Reback J, McKinney W, et al. pandas-dev/pandas: Pandas 1.3.2. Zenodo 2021. 10.5281/ZENODO.3509134 [DOI]
- 49.McKinney W. Data Structures for Statistical Computing in Python. In: Proceedings of the 9th Python in Science Conference. SciPy 2010. 10.25080/Majora-92bf1922-00a [DOI] [Google Scholar]
- 50. Pedregosa F, Varoquaux G, Gramfort A, et al. Scikit-learn: Machine Learning in Python. J Mach Learn Res 2011;12:2825-30. [Google Scholar]
- 51.US Census Bureau. About the Topic of Race. www.census.gov/topics/population/race/about.html
- 52. Rajkomar A, Dean J, Kohane I. Machine Learning in Medicine. N Engl J Med 2019;380:1347-58. 10.1056/NEJMra1814259. [DOI] [PubMed] [Google Scholar]
- 53. Angus DC. Randomized Clinical Trials of Artificial Intelligence. JAMA 2020;323:1043-5. 10.1001/jama.2020.1039. [DOI] [PubMed] [Google Scholar]
- 54. Escobar GJ, Liu VX, Schuler A, Lawson B, Greene JD, Kipnis P. Automated Identification of Adults at Risk for In-Hospital Clinical Deterioration. N Engl J Med 2020;383:1951-60. 10.1056/NEJMsa2001090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wong A, Otles E, Donnelly JP, et al. External Validation of a Widely Implemented Proprietary Sepsis Prediction Model in Hospitalized Patients. JAMA Intern Med 2021;181:1065-70. 10.1001/jamainternmed.2021.2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Hosmer DW, Jr, Lemeshow S, Sturdivant RX. Applied logistic regression. John Wiley & Sons, 2013. 10.1002/9781118548387. [DOI] [Google Scholar]
- 57. Pan SJ, Yang Q. A Survey on Transfer Learning. IEEE Trans Knowl Data Eng 2010;22:1345-59. 10.1109/TKDE.2009.191. [DOI] [Google Scholar]
- 58. Wiens J, Guttag J, Horvitz E. A study in transfer learning: leveraging data from multiple hospitals to enhance hospital-specific predictions. J Am Med Inform Assoc 2014;21:699-706. 10.1136/amiajnl-2013-002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Davis SE, Lasko TA, Chen G, et al. Calibration drift among regression and machine learning models for hospital mortality. In: AMIA Annual Symposium Proceedings. American Medical Informatics Association 2017. 625. [PMC free article] [PubMed] [Google Scholar]
- 60. Van Calster B, Wynants L, Timmerman D, Steyerberg EW, Collins GS. Predictive analytics in health care: how can we know it works? J Am Med Inform Assoc 2019;26:1651-4. 10.1093/jamia/ocz130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Obermeyer Z, Powers B, Vogeli C, Mullainathan S. Dissecting racial bias in an algorithm used to manage the health of populations. Science 2019;366:447-53. 10.1126/science.aax2342. [DOI] [PubMed] [Google Scholar]
- 62.Buolamwini J, Gebru T. Gender Shades: Intersectional Accuracy Disparities in Commercial Gender Classification. In: Friedler SA, Wilson C, eds. Proceedings of the 1st Conference on Fairness, Accountability and Transparency. New York, NY, USA: PMLR 2018. 77-91. https://proceedings.mlr.press/v81/buolamwini18a.html
- 63. Ashana DC, Anesi GL, Liu VX, et al. Equitably Allocating Resources during Crises: Racial Differences in Mortality Prediction Models. Am J Respir Crit Care Med 2021;204:178-86. 10.1164/rccm.202012-4383OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. James N, Menzies M. COVID-19 in the United States: Trajectories and second surge behavior. Chaos 2020;30:091102. 10.1063/5.0024204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Huang X, Shao X, Xing L, Hu Y, Sin DD, Zhang X. The impact of lockdown timing on COVID-19 transmission across US counties. EClinicalMedicine 2021;38:101035. 10.1016/j.eclinm.2021.101035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hale T, Atav T, Hallas L, et al. Variation in US states responses to COVID-19. Blavatnik School of Government, 2020. [Google Scholar]
- 67. Dave D, Friedson AI, Matsuzawa K, Sabia JJ. When Do Shelter-in-Place Orders Fight COVID-19 Best? Policy Heterogeneity Across States and Adoption Time. Econ Inq 2020;59:29-52. 10.1111/ecin.12944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nguyen NT, Chinn J, Nahmias J, et al. Outcomes and Mortality Among Adults Hospitalized With COVID-19 at US Medical Centers. JAMA Netw Open 2021;4:e210417. 10.1001/jamanetworkopen.2021.0417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rosenberg ES, Dufort EM, Udo T, et al. Association of Treatment With Hydroxychloroquine or Azithromycin With In-Hospital Mortality in Patients With COVID-19 in New York State. JAMA 2020;323:2493-502. 10.1001/jama.2020.8630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Wang M, Zhang J, Ye D, et al. Time-dependent changes in the clinical characteristics and prognosis of hospitalized COVID-19 patients in Wuhan, China: A retrospective study. Clin Chim Acta 2020;510:220-7. 10.1016/j.cca.2020.06.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Angeli F, Bachetti T, Maugeri Study Group . Temporal changes in co-morbidities and mortality in patients hospitalized for COVID-19 in Italy. Eur J Intern Med 2020;82:123-5. 10.1016/j.ejim.2020.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Kip KE, Snyder G, Yealy DM, et al. Temporal changes in clinical practice with COVID-19 hospitalized patients: Potential explanations for better in-hospital outcomes. bioRxiv. 2020. 10.1101/2020.09.29.20203802 [DOI]
- 73. Sands KE, Wenzel RP, McLean LE, et al. Changes in hospitalized coronavirus disease 2019 (COVID-19) patient characteristics and resource use in a system of community hospitals in the United States. Infect Control Hosp Epidemiol 2021;42:228-9. 10.1017/ice.2020.1264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wiens J, Saria S, Sendak M, et al. Do no harm: a roadmap for responsible machine learning for health care. Nat Med 2019;25:1337-40. 10.1038/s41591-019-0548-6. [DOI] [PubMed] [Google Scholar]
- 75. O’Malley KJ, Cook KF, Price MD, Wildes KR, Hurdle JF, Ashton CM. Measuring diagnoses: ICD code accuracy. Health Serv Res 2005;40:1620-39. 10.1111/j.1475-6773.2005.00444.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary information: eMethods 1-3, eTables 1-11, and eFigures 1-5
Supplementary table: spreadsheet showing eTable 1 data
Data Availability Statement
To guarantee the confidentiality of personal and health information, only the authors have had access to the data during the study in accordance with the relevant license agreements. The full model (including model coefficients and supporting code) are available online at https://github.com/MLD3/M-CURES.