Abstract
Low-grade serous ovarian cancer (LGSOC) poses a specific clinical challenge due to advanced presentation at diagnosis and the lack of effective systemic treatments. The aim of this study was to use a precision medicine approach to identify clinically actionable mutations in a patient with recurrent LGSOC. Primary, metastatic and recurrence tissue, and blood samples were collected from a stage IV LGSOC patient. Single-gene testing for clinically actionable mutations (BRAF V600, KRAS and NRAS) and subsequent whole-exome sequencing (WES) were performed. Droplet digital PCR was used to evaluate the presence of an identified BRAF D594G mutation in the matched plasma cell-free DNA (cfDNA). No clinically actionable mutations were identified using single-gene testing. WES identified a BRAF D594G mutation in six of seven tumor samples. The patient was commenced on a MEK inhibitor, trametinib, but with minimal clinical response. A newly designed ddPCR assay detected the BRAF alteration in the matched tissues and liquid biopsy cfDNA. The identification and sensitive plasma detection of a common “druggable” target emphasises the impact of precision medicine on the management of rare tumors and its potential contribution to novel monitoring regimens in this field.
Keywords: Low-grade serous ovarian cancer, Cell-free circulating DNA, Targeted therapy, Next-generation sequencing, Digital droplet PCR
Highlights
-
•
First report of BRAF D594G mutation in multiple samples of a recurrent LGSOC patient.
-
•
Discovery of a BRAF actionable mutation expands the range of LGSOC therapeutic options.
-
•
ddPCR assay allows sensitive detection of the mutation in tissue and plasma samples.
1. Introduction
Ovarian cancer (OC) is the most lethal gynaecological malignancy, accounting for over 180,000 yearly cancer deaths worldwide [1]. Serous OC is the most common histological subtype of OC, and while high-grade serous ovarian cancer (HGSOC) accounts for most serous epithelial OCs (~95%), a small number of women present with low-grade serous ovarian cancer (LGSOC) [2]. Most women present with advanced disease at diagnosis, that is generally resistant to systemic chemotherapy and anti-endocrine therapy, and eventually succumb to their disease [3]. There is thus a significant unmet need for more effective therapeutic approaches for LGSOC.
To improve the treatment modalities currently used in LGSOC, research has focused on understanding the molecular landscape of this rare cancer type. LGSOC is typically characterised by a high frequency of activating mutations in upstream regulators of the mitogen-activated protein (MAPK) pathway, such as KRAS, NRAS and BRAF [4,5]. This suggests that the use of BRAF or MEK inhibitors may have some efficacy in LGSOC [6].
In addition to the observed chemoresistance and dearth of effective therapeutic options, LGSOCs also lack an appropriate follow-up regimen, as the use of standard clinical prognostic and predictive tools used in the context of OC have shown limited utility in LGSOC [7]. Liquid biopsies offer a minimally invasive and sensitive technique to complement existing monitoring tools, via cell-free circulating tumor DNA (ctDNA) or circulating tumor cells (CTCs) [8].
In this case report, we demonstrate how the identification of a clinically actionable genomic alteration led to the development of a novel liquid biopsy assay that could potentially enable surveillance of LGSOC patients on targeted therapy.
2. Case Presentation
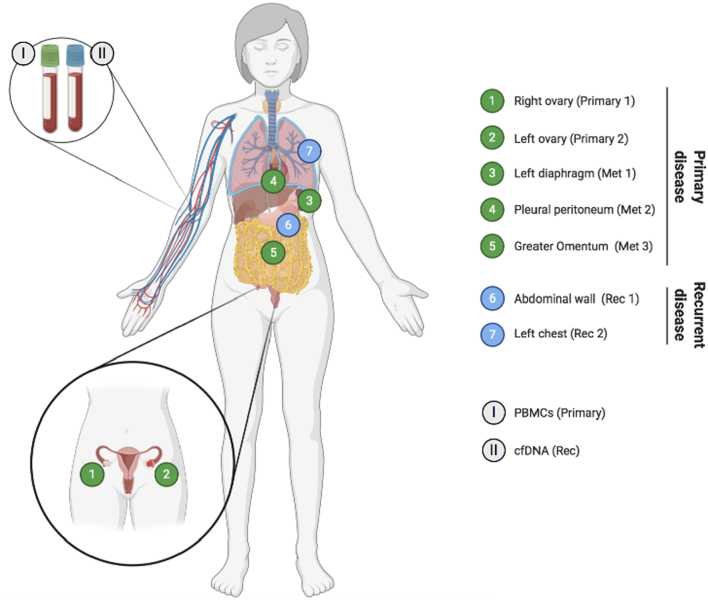
A 43-year-old woman presented with large-volume ascites, bilateral pleural effusions and pelvic masses, and a large-volume omental disease. A laparoscopy showed widespread peritoneal carcinomatosis, and a biopsy confirmed the diagnosis of LGSOC. Immunohistochemistry showed a p53 wild type pattern, with low proliferation index, expression of oestrogen and progesterone receptors, microsatellite stable tumor (Fig. 1A-E). She proceeded to have primary cytoreductive surgery, which included a complete abdomeno-pelvic peritonectomy, greater and lesser omentectomy, splenectomy, cholecystectomy, total colectomy and abdominal hysterectomy, and bilateral salpingo-oophorectomy. Residual disease was 0.5 cm, located on the small bowel mesentery. The completeness of cytoreduction score (CC score) was 2. Tissues from the right and left ovaries (primary tumor) and three other metastatic sites (greater omentum, left diaphragm and pleural peritoneum) were acquired (Fig. 2) and post-operative histology confirmed LGSOC in all specimens. The patient subsequently completed six cycles of adjuvant carboplatinum and paclitaxel and was started on maintenance tamoxifen.
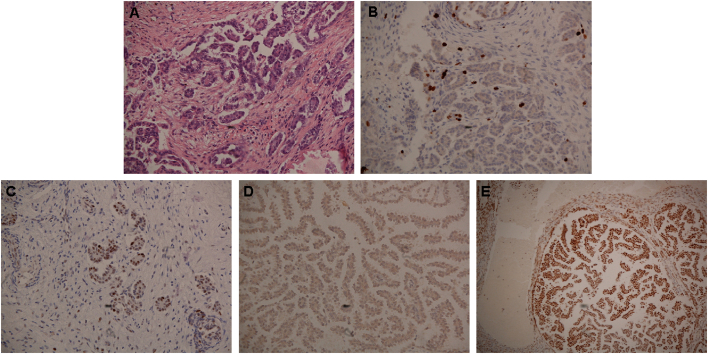
Fig. 1.
Immunohistochemical (IHC) staining of tumor sections.
A) Hematoxylin and eosin staining of patient's tumor section showing histological findings that are compatible with low-grade serous ovarian cancer. B) Low immunoexpression of Ki-67 protein, which is associated with low proliferative tumors. Wild type like and negative expression patterns were observed for C) p53 and D) BRAF V600E proteins, respectively. E) Microsatellite instability testing demonstrated high expression of mismatch repair proteins, indicative of a MSI stable tumor. IHC of MLH1 protein is shown as an example.
Fig. 2.
Overview of analysed specimens.
Anatomical location of samples collected during either primary cytoreductive surgery (green) or second surgery (upon recurrence; blue). Whole blood was also collected during both surgeries. Abbreviations: cfDNA – cell-free DNA; Met – metastasis; PBMC – peripheral blood mononuclear cell; Rec – recurrence. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
Eleven months later, she presented with increasing shortness of breath. A CT scan of the thorax/abdomen/pelvis demonstrated a new left-sided pleural effusion and an abdominal wall mass, which biopsy confirmed as recurrent LGSOC. She underwent a left-sided video-assisted thoracoscopic surgery (VATS) procedure and talc pleurodesis, and tissues from two other sites (abdominal wall and left chest) were collected (Fig. 2). A decision was made to change her anti-endocrine therapy from tamoxifen to letrozole.
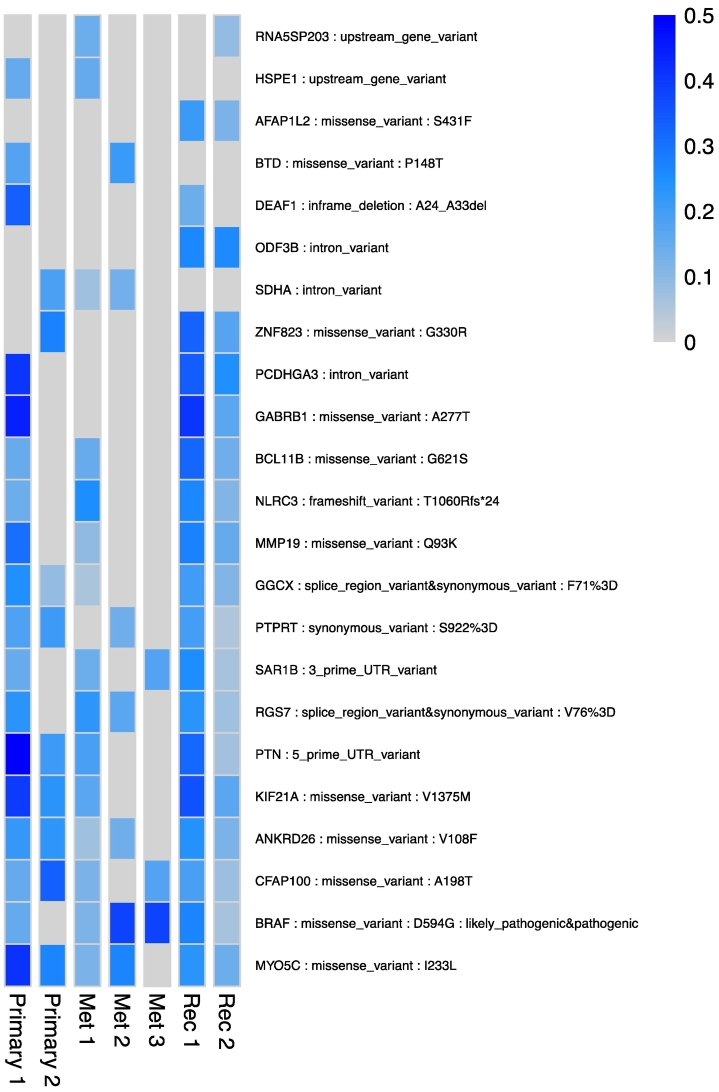
To investigate possible causes for the observed therapy resistance and identify new therapeutic options, we employed a multistep genomic approach to determine the molecular characteristics of this patient's tumor (Supplementary Material). While single-gene testing revealed no alterations for EGFR, NRAS, KRAS and BRAF in all analysed tissue samples (n = 7; 2 primary, 3 metastatic and 2 recurrent sites), whole-exome sequencing (WES) identified 23 protein-coding somatic SNVs across all studied samples (Fig. 3). The majority (90%) were classified as missense variants, including a clinically relevant BRAF mutation (D594G), which has previously been linked to several cancers (i.e., melanoma, colorectal and lung) [9,10], but has been previously described only once in a patient-derived LGSOC cell line [11,12]. Unfortunately, the disease continued to progress while the patient was on letrozole and, given the minimal therapeutic options available and the known presence of a BRAF mutation, a decision was made that this patient should commence the MEK1/2 inhibitor trametinib. However, she showed minimal clinical response and succumbed to her disease two months later.
Fig. 3.
SNVs detected in primary and recurrent LGSOC samples.
Representation of the 23 somatic single nucleotide variants (SNVs) detected. Differences in mutant allele fraction (MAF) are represented by the blue colour gradient, blue: present, grey: absent. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
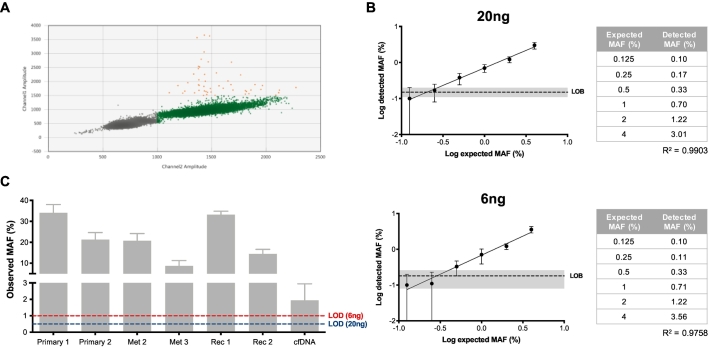
Having identified a novel BRAF mutation in tumor samples, we sought to develop a sensitive droplet digital PCR (ddPCR) assay to detect the BRAF D5945G mutation in plasma cfDNA (Supplementary Material). Evaluation of the sensitivity of the assay was performed with 20 ng (tissue sample input) and 6 ng (cfDNA sample input) and a limit of detection was established (Fig. 4A-B). We were able to detect the BRAF mutation in all tested tissue samples (n = 6; left diaphragm site – Met 1 – was not available) (Fig. 4C). Interestingly, one of the primary tumor samples (left ovary – primary 2), previously revealed to be negative by WES analysis, was positive for the presence of the mutation when assayed with ddPCR. Additionally, we also detected the mutation in a cfDNA sample collected before the patient commenced trametinib (Fig. 4C).
Fig. 4.
Detection of BRAF D594G mutation by ddPCR.
A) Limit of blank (LOB) determination for 20 ng DNA samples. 16-replicate wells of wild type-only template were run. Droplets are classified as wild type-only (green), mutant-only droplets (blue) double wild type-mutant droplets (orange) and double negative (grey). The LOB was set at 0.15% and 0.18% for 20 and 6 ng samples, respectively. B) Limit of detection (LOD) determination for DNA amounts of 20 (upper) and 6 ng (lower). Log of MAF values was used to calculate correlation between expected and detected MAFs (R2). Confidence interval for LOB is shown in grey. Based on obtained values (table), LOD was determined as 0.5% and 1% for 20 and 6 ng, respectively. C) MAF observed for tested tissue and cfDNA samples. Abbreviations: cfDNA – cell-free DNA; MAF – mutant allele fraction. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
3. Discussion
Using a stepwise genomic approach, we identified a BRAF D594G mutation in multiple tissue samples from the same patient, suggesting that altered MAPK signalling was an important aspect of this particular tumor. BRAF mutations are present in up to 14% of LGSOC, with the majority of those corresponding to BRAF V600E alterations [13]. The D594G mutation, designated as a class 3 BRAF mutant, causes inactivation of this protein and subsequent impaired kinase activity [9]. Due to this inactivation, tumors harbouring this mutation are unlikely to respond to established BRAF inhibitors, which generally selectively target a conformational change only present in BRAF V600-mutant activated proteins. Although unable to directly phosphorylate MEK, these “kinase-dead” mutants still exhibit some signalling activity in the MAPK pathway. Heidorn et al. demonstrated that class 3 BRAF mutants have the ability to bind and activate CRAF, in a RAS-dependent manner, leading to CRAF hyperactivation and subsequent elevated MEK and ERK signalling [14]. Prior to this discovery, this mutation had been reported by only one other group, in a patient-derived LGSOC cell line [11,12], which highlights its rarity in this disease. In fact, the authors further report that of all the mutations observed in their established LGSOC cell lines, only D594G and another NRAS mutation were not found in the GENIE cohort, which includes over 97 LGSOC tumor specimens [12,15]. Although MEK inhibitor sensitivity in vitro was also tested in this study, this report gives an insight into the use of MEK inhibitors in vivo, and provides useful information that can be translated to the clinic, and could otherwise be biased by the use of a cell line (i.e. lack of tumor microenvironment or drug-cell interactions). Additionally, this is the first study that identified this particular BRAF variant in plasma cell-free DNA, highlighting the potential of using liquid biopsies as a minimally invasive alternative for identification of new actionable targets, and subsequent therapeutic monitoring in LGSOC, as previously observed for other malignancies [16,17].
This data, combined with increasingly large cohorts demonstrating the prevalence of genomic alterations in BRAF, NRAS and KRAS in LGSOC [18,19], suggests that a variety of tyrosine kinase inhibitors warrant further investigation in LGSOC. A number of clinical trials have sought to exploit this therapeutic approach in recurrent LGSOC, with a particular focus in MEK inhibitors. An open-label, single-arm phase II trial of the MEK inhibitor selumetinib showed a 15% response rate and a median progression-free survival time of 11 months. Despite mutational data being available for the majority of the cohort, no correlation was found between any BRAF (codon 599 only) or KRAS (codons 12/13) mutation and therapeutic response [20]. Two other phase II/III trials compared two MEK inhibitors against physician's choice of chemotherapy/hormonal therapy in women with persistent or recurrent LGSOC. The MILO/ENGOT-ov11 study failed to demonstrate an improvement in progression-free survival for binimetinib; however, a post-hoc analysis suggested that responses rates were higher in KRAS mutated compared with KRAS wild type LGSOC (objective response rate 44% vs 19%) [21]. A second randomised phase II/III trial compared trametinib to physician's choice, and demonstrated a significant improvement in objective response rates (26% v 6%) and improved progression-free survival (13 vs 7.2 months) [22]. Based on these findings, we decided to administer trametinib to the patient highlighted in this report.
The advent of liquid biopsies has contributed towards the potential integration of personalised cancer medicine in the clinical management of OC patients. In fact, the use of liquid biopsy approaches has been associated with superior levels of prediction of therapeutic outcome in the OC setting. For example, Parkinson et al. reported that a reduction of ≤60% in TP53 MAF in plasma after one cycle of cytotoxic platinum-based chemotherapy was associated with a shorter time to progression [23]. Although the vast majority of liquid biopsy studies have been conducted in HGSOC patients, these reports highlight the reliability of using ctDNA for prediction of recurrence and treatment response. We were able to use a novel ddPCR assay to detect the presence of the BRAF D594G mutation in both tissue and plasma samples from a patient with LGSOC. Notably, one of the tissue samples that was determined as negative by WES (primary 2) was positive when evaluated by ddPCR. Compared with ddPCR, WES has lower detection sensitivity (~0.001% for ddPCR and ~ 5–10% for WES). Although the ddPCR-determined MAF for this sample was above the usual WES LOD, we believe that this was a false-negative result which stemmed from low sequencing coverage. Compared with the MAF obtained with the tissue samples, detection of the mutation in plasma cfDNA was lower, especially for a recurrent patient. This observation might be a result of cfDNA containing DNA from multiple cell sources, of which tumor is only a portion, as opposed to samples derived from the tumor itself. Nevertheless, detection was still above the determined limit for this assay and these results are on par with other reports evaluating the presence of alterations in cfDNA [24]. Thus, these results provide support for further evaluation of liquid biopsy-based therapeutic response monitoring in OC.
Technological advances have improved our understanding of the underlying molecular features of rare tumors such as LGSOC and, as a result, the clinical management of LGSOC has been slowly evolving from a “one-size-fits-all” regimen to a more personalised approach. Here, the clinical application of WES and ddPCR as useful tools for managing a recurrent LGSOC case are demonstrated. Further analysis, in a series of patients, is required to understand how to fully implement these personalised approaches into the clinical management of OC.
Acknowledgments
Contributors
R. Silva contributed to formal analysis, investigation, writing the original draft, and review and editing.
B. Moran contributed to data curation, formal analysis, investigation, and review and editing.
S. Das contributed to investigation and review and editing.
N. Mulligan contributed to investigation and review and editing.
M. Doughty contributed to investigation and review and editing.
A. Treacy contributed to investigation and review and editing.
K. Sheahan contributed to investigation and review and editing.
C. M. Kelly contributed to review and editing.
A. G. Duffy contributed to review and editing.
A. S. Perry contributed to writing the original draft, and review and editing.
D. J. Brennan contributed to conceptualization, patient care, project administration, writing the original draft, and review and editing.
Funding
Funding for this work is acknowledged from the Ireland East Hospital Group; National Maternity Hospital Foundation; Science Foundation Ireland Strategic Partnership Programme Precision Oncology Ireland [18/SPP/3522]; Irish Cancer Society [CRS17SIL]; and Irish Association of Cancer Research [AOIFA award 2019].
Patient consent
Informed consent was obtained, from both the patient and her husband, for the use of collected clinical specimens and publication of this case report.
Provenance and peer review
Peer review was directed by Professor Margaret Rees, Editor-in-Chief, independently of D. J. Brennan, one of the authors and a member of the editorial board of Case Reports in Women's Health, who was blinded to the process.
Acknowledgments
Acknowledgements
The authors wish to acknowledge the technical support of Accuscience and BioRad, in particular Mr Jake Gill and Dr Petar Podlesniy. We especially wish to thank the patient involved in this study, and her family, without whom this research would not have been possible.
Conflict of interest statement
The authors declare that they have no conflict of interest regarding the publication of this case report.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.crwh.2022.e00395.
Appendix A. Supplementary data
Supplementary material
References
- 1.Bray F., Ferlay J., Soerjomataram I., et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Slomovitz B., Gourley C., Carey M.S., et al. Low-grade serous ovarian cancer: state of the science. Gynecol. Oncol. 2020;156:715–725. doi: 10.1016/j.ygyno.2019.12.033. [DOI] [PubMed] [Google Scholar]
- 3.Grabowski J.P., Harter P., Heitz F., et al. Operability and chemotherapy responsiveness in advanced low-grade serous ovarian cancer. An analysis of the AGO study group metadatabase. Gynecol. Oncol. 2016;140:457–462. doi: 10.1016/j.ygyno.2016.01.022. [DOI] [PubMed] [Google Scholar]
- 4.Jones S., Wang T.L., Kurman R.J., et al. Low-grade serous carcinomas of the ovary contain very few point mutations. J. Pathol. 2012;226:413–420. doi: 10.1002/path.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunter S.M., Anglesio M.S., Ryland G.L., et al. Molecular profiling of low grade serous ovarian tumours identifies novel candidate driver genes. Oncotarget. 2015;6:37663–37677. doi: 10.18632/oncotarget.5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sanchez J.N., Wang T., Cohen M.S. BRAF and MEK inhibitors: use and resistance in BRAF-mutated cancers. Drugs. 2018;78:549–566. doi: 10.1007/s40265-018-0884-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fader A.N., Java J., Krivak T.C., et al. The prognostic significance of pre- and post-treatment CA-125 in grade 1 serous ovarian carcinoma: a gynecologic oncology group study. Gynecol. Oncol. 2014;132:560–565. doi: 10.1016/j.ygyno.2013.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Alix-Panabieres C., Schwarzenbach H., Pantel K. Circulating tumor cells and circulating tumor DNA. Annu. Rev. Med. 2012;63:199–215. doi: 10.1146/annurev-med-062310-094219. [DOI] [PubMed] [Google Scholar]
- 9.Yao Z., Yaeger R., Rodrik-Outmezguine V.S., et al. Tumours with class 3 BRAF mutants are sensitive to the inhibition of activated RAS. Nature. 2017;548:234–238. doi: 10.1038/nature23291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nieto P., Ambrogio C., Esteban-Burgos L., et al. A Braf kinase-inactive mutant induces lung adenocarcinoma. Nature. 2017;548:239–243. doi: 10.1038/nature23297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fernandez M.L., Dawson A., Hoenisch J., et al. Markers of MEK inhibitor resistance in low-grade serous ovarian cancer: EGFR is a potential therapeutic target. Cancer Cell Int. 2019;19:10. doi: 10.1186/s12935-019-0725-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shrestha R., Llaurado Fernandez M., Dawson A., et al. Multiomics characterization of low-grade serous ovarian carcinoma identifies potential biomarkers of MEK inhibitor sensitivity and therapeutic vulnerability. Cancer Res. 2021;81:1681–1694. doi: 10.1158/0008-5472.CAN-20-2222. [DOI] [PubMed] [Google Scholar]
- 13.Moujaber T., Etemadmoghadam D., Kennedy C.J., et al. BRAF mutations in low-grade serous ovarian Cancer and response to BRAF inhibition. JCO Precision Oncol. 2018:1–14. doi: 10.1200/PO.17.00221. [DOI] [PubMed] [Google Scholar]
- 14.Heidorn S.J., Milagre C., Whittaker S., et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140:209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Consortium APG AACR project GENIE: powering precision medicine through an international consortium. Cancer Discov. 2017;7:818–831. doi: 10.1158/2159-8290.CD-17-0151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yaeger R., Kotani D., Mondaca S., et al. Response to anti-EGFR therapy in patients with BRAF non-V600-mutant metastatic colorectal cancer. Clin. Cancer Res. 2019;25:7089–7097. doi: 10.1158/1078-0432.CCR-19-2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang Y., Shen X., Li R., et al. The detection and significance of EGFR and BRAF in cell-free DNA of peripheral blood in NSCLC. Oncotarget. 2017;8:49773–49782. doi: 10.18632/oncotarget.17937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grisham R.N., Garg K., Zhou Q., et al. A comprehensive analysis of BRAF and KRAS mutation status in low-grade serous (LGS) and serous borderline (SB) ovarian cancer (OC) J. Clin. Oncol. 2012;30:5030. [Google Scholar]
- 19.Gershenson D.M., Sun C.C., Wong K.K. Impact of mutational status on survival in low-grade serous carcinoma of the ovary or peritoneum. Br. J. Cancer. 2015;113:1254–1258. doi: 10.1038/bjc.2015.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Farley J., Brady W.E., Vathipadiekal V., et al. Selumetinib in women with recurrent low-grade serous carcinoma of the ovary or peritoneum: an open-label, single-arm, phase 2 study. Lancet Oncol. 2013;14:134–140. doi: 10.1016/S1470-2045(12)70572-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monk B.J., Grisham R.N., Banerjee S., et al. MILO/ENGOT-ov11: binimetinib versus physician’s choice chemotherapy in recurrent or persistent low-grade serous carcinomas of the ovary, fallopian tube, or primary peritoneum. J. Clin. Oncol. 2020;0 doi: 10.1200/JCO.20.01164. JCO.20.01164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gershenson D.M., Miller A., Brady W. 2020. A randomized phase II/III study to assess the efficacy of trametinib in patients with recurrent or progressive low-grade serous ovarian or peritoneal cancer. For presentation at: 2020 Society of Gynecologic Oncology Annual Meeting; March 28-31, 2020; Toronto, Canada. Abstract 42. [Google Scholar]
- 23.Parkinson C.A., Gale D., Piskorz A.M., et al. Exploratory analysis of TP53 mutations in circulating tumour DNA as biomarkers of treatment response for patients with relapsed high-grade serous ovarian carcinoma: a retrospective study. PLoS Med. 2016;13 doi: 10.1371/journal.pmed.1002198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bettegowda C., Sausen M., Leary R.J., et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014;6:224ra224. doi: 10.1126/scitranslmed.3007094. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material