Abstract
Highly pathogenic avian influenza viruses (HPAIV) can be carried long distances by migratory wild birds and by poultry trade. Highly pathogenic avian influenza (HPAI) is often lethal in domestic poultry and can sporadically infect and cause severe respiratory or systemic disease in other species including humans. Since 2003, the H5 subtype of HPAIV have spread from epicenters in China to neighboring regions in East and Southeast Asia, and across Central Asia to the Indian subcontinent, Europe, Africa, and North America. Outbreaks of H5N1 HPAIV struck poultry in Ukraine in 2005. In 2016, A H5N8 clade 2.3.4.4b HPAIV outbreaks occurred in wild and domestic birds in Ukraine concurrently with outbreaks in Central Europe, Russia, and the Middle East. We report outbreaks of HPAI in domestic backyard poultry in (2016–2017) in the southern region of Ukraine, in proximity to mass gathering sites for migratory waterfowl including mute swans (Cygnus olor). All eight genome segments of three novel H5N8 HPAIV isolated in November 2016 from two domestic backyard chickens (Gallus gallus) and one backyard mallard duck (Anas platyrhynchos) found dead of HPAI in Azov-Black Sea region of Ukraine were cladistically related to H5N8 2.3.4.4b HPAI viruses isolated from wild shelduck (Tadorna tadorna) and white-fronted goose (Anser albifrons) in Askania Nova Biopreserve (Kherson district, Ukraine) in 2016–2017 and to other contemporary H5N8 HPAIV strains sequenced from wild birds and poultry in Eurasia. Amino acid variations in hemagglutinin were outside of the polybasic cleavage site (PLREKRRKR/GLF), and D224G suggested avian-like receptor binding specificity; neuraminidase did not have mutations characteristic of oseltamivir drug resistance. Outbreaks of HPAI in Ukraine highlight the continual need for biosurveillance and genomic sequencing of avian influenza viruses along wild bird flyways and interfaces with domestic poultry in Eurasia.
Keywords: highly pathogenic avian influenza virus, H5N8, genetic analysis, phylogenetics, Ukraine
Introduction
The H5N8 subtype of highly pathogenic avian influenza viruses (HPAIV) have been detected in wild birds and poultry in East Asia since 2010 (Verhagen et al. 2015). This subtype spread widely along wild bird migratory flyways across Eurasia to Europe, the Middle East, and Africa, and to North America (Lee et al. 2015, Verhagen et al. 2015). Of note, H5N8 (clade 2.3.4.4) strains infected both wild birds and domestic poultry and, in some cases, reassorted with local low pathogenic avian influenza viruses (LPAIV). The clade 2.3.4, which is one of the major genotypes in Asia, continues to evolve as subclades, resulting in generation of 2.3.4.1, 2.3.4.2, 2.3.4.3, and 2.3.4.4.
In addition, clade 2.3.4.4 appears in various neuraminidase (NA) subtypes such as H5N2, H5N5, and H5N8 (Jackson et al. 2008, Gao et al. 2011, Xu et al. 2012, Lee et al. 2015, Li et al. 2017, Selim et al. 2017). In North America, a reassortant H5N2 HPAIV had led to a widespread outbreak in chickens in the United States, which resulted in the culling of over 44 million birds (Lee et al. 2015). In May 2016, novel reassortant H5N8 viruses of clade 2.3.4.4b were detected in wild birds at Qinghai Lake in China and Lake Uvs-Nuur at the Russia–Mongolia border, and once again spread to many countries in Europe, Asia, and the Middle East (Verhagen et al. 2015, Lee et al. 2017, Li et al. 2017, Selim et al. 2017).
With extensive wetlands in the south (Azov-Black Sea and Danube Delta regions) that serve as spring and autumn stopover sites for mass migration of wild birds, and large populations of resident or semi-resident waterfowl, Ukraine lies at an apparent ecological “hotspot” for emergence of avian pathogens including avian influenza viruses (AIV) and avian paramyxoviruses carried by wild birds (Muzyka et al. 2014, 2018). Moreover, Ukraine has widespread commercial and backyard poultry farms, and outbreaks of highly pathogenic avian influenza (HPAI) in poultry have been reported including H5N1 in 2005 and H5N8 in 2014 and 2016–2017 (Muzyka et al. 2018, OIE 2021).
In this study we report the genomic analysis of three H5N8 HPAIV isolates from domestic birds found dead of HPAI in a rural village in Ukraine in 2016–2017 and compare the viruses with other HPAIV from similar studies in Ukraine and other European countries to understand the origin and genetic relationships among strains.
Materials and Methods
Outbreak sample sources and PCR diagnostics
On November 14, 2016, mass mortality of chickens and ducks was observed in private backyard farms in Novooleksandrivka village, Kherson oblast (a province-level administrative region) in the south of Ukraine. Entire carcasses of dead birds in good condition were submitted for necropsy to the regional state laboratory at the oblast level. Using standard biosafety procedures established for HPAI outbreak response in Ukraine, the tissue samples (internal organs), from dead birds were transported to the State Research Institute of Laboratory Diagnostics and Veterinary and Sanitary Expertise (SSRILDVSE, Kyiv, Ukraine), the Ukrainian national reference laboratory.
All procedures were followed in accordance with the state Instruction for the Prevention and Elimination of Avian Influenza in Birds (Verkhovna Rada of Ukraine 2011), which establishes the procedure for conducting laboratory diagnosis and veterinary and sanitary measures in cases of suspected avian influenza in the country.
Diagnosis of HPAIV was performed by quantitative real-time RT-PCR (qRT-PCR). First, viral RNA was extracted from subsamples of internal organs (lung) using QIAamp cador Pathogen Mini Kit (Qiagen). Positive AIV diagnosis was based on qRT-PCR using universal primers targeting the Matrix Protein (M) gene (Spackman et al. 2003). Samples positive for AIV were typed for H5 and H7 HPAIV subtypes by qRT-PCR using commercial test kits: Avian Influenza Virus RNA Test Kit (VetMax Gold AIV Detection Kit), SureFast Influenza A H5/H7 3plex; LSI RT-PCR TaqMan Avian Influenza H5 Typing Kit; and/or LSI RT-PCR TaqMan Avian Influenza H7 Typing Kit.
Sample selection and sequencing
As subtyping RT-PCR assays do not provide additional genetic information to differentiate HPAI from LPAI, full genome sequence analysis was performed. In this study, three HPAIV H5N8-positive samples from domestic birds found dead (two chickens and one duck) were selected for sequencing; the major selection criteria identified samples that were of sufficient RNA quality and titer (estimated by RT-PCR Ct value) such that the samples were likely to yield high-quality short-read data to sequence the whole virus genome. The samples were collected in November 2016 at the start of the outbreak in Novooleksandrivka village, Kherson oblast. Sample origin details are provided in Table 1.
Table 1.
Origin of Ukrainian Highly Pathogenic Avian Influenza Viruses Isolates Sequenced in This Study
| No. | Strain name | Subtype | GISAID accession numbers | Location | Collection date | Sample material | Type of farm |
|---|---|---|---|---|---|---|---|
| 1 | A/chicken/Ukraine/1/2016 | H5N8 | EPI_ISL_1121143 | Kherson Oblast, Kalanchatskiy District, Novooleksandrivka village | November 14, 2016 | Lungs | Backyard poultry |
| 2 | A/chicken/Ukraine/3/2016 | H5N8 | EPI_ISL_1121144 | ||||
| 3 | A/duck/Ukraine/4/2016 | H5N8 | EPI_ISL_1121145 |
An oblast is a province-level administrative region in Ukraine.
GISAID, Global Influenza Sharing Database.
Virus genome sequencing
Next-generation sequencing of full genomes (eight segments) of the three putative HPAIV isolates was performed at the Animal and Plant Health Agency (APHA, Weybridge, United Kingdom). In brief, cDNA libraries were amplified from viral RNA with end repair, A-tailing, adaptor ligation, and barcoded (NEBNext; New England Biolabs, and NexteraXT; Illumina, Cambridge, United Kingdom) according to the manufacturer's instructions, and paired-end sequenced on an Illumina MiSeq (v2; Illumina, Inc., San Diego, CA) platform as described (Venkatesh et al. 2018). Complete AIV genomes were constructed by reference-based assembly following previous protocols, using contemporary H5 clade 2.3.4.4 reference genomes (Lee et al. 2017, Li et al. 2017).
Sequence alignment and phylogenetics
Consensus sequences were analyzed using Geneious R11 software and submitted to the Global Influenza Sharing Database (GISAID) EpiFlu™ nucleotide sequence database (accession numbers: EPI_ISL_1121143; EPI_ISL_1121144; and EPI_ISL_1121145). The consensus sequences obtained for the three Ukrainian isolates were aligned using the MAFFT v7 online service (Kuraku et al. 2013, Katoh et al. 2019) with a broad set of H5N8 HPAIV genomic sequences identified in the GISAID EpiFlu and NCBI GenBank databases. Contemporary H5N8 HPAIV genomes used in the tree construction set included three H5N8 HPAIV strains from Askania Nova (AN) Biopreserve (Kherson oblast, Ukraine), which were isolated from wild birds and sequenced previously (Muzyka et al. 2018) by the Freidrich Loeffleur Institute, Germany: A/white-fronted goose/AN/1-15-12/2016(H5N8), A/ruddy shelduck/AN/2-14-12/2016(H5N8), and A/environmental (EM)/AN/2/17(H5N8); sequences for these three strains are available in GISAID EpiFlu database under accession numbers EPI_ISL_300547, EPI_ISL_300548, and EPI_ISL_300562, respectively. Phylogenetic trees for each gene were generated by the maximum likelihood (ML) method using IQ-TREE (Nguyen et al. 2015), phylogenomic web-server by ML with the ultrafast bootstrap (1000) branch supports. For each gene segment the substitution model was determined using ModelFinder through the IQ-TREE (Kalyaanamoorthy et al. 2017) and the best-fit models according to Bayesian information criteria were used. The best-fit substitution models and input sequence data varied for each segment (Table 2). Phylogenetic trees were summarized, visualized, and annotated in FigTree v1.4.4.
Table 2.
Substitution Models Which Were Used in the Molecular Phylogenetic Analyses
| No. | Segment | Best-fit substitution model according to BIC | Input sequence data |
|---|---|---|---|
| 1 | PB2 | GTR+F+G4 | 123 sequences with 2280 nt sites |
| 2 | PB1 | GTR+F+G4 | 127 sequences with 2274 nt sites |
| 3 | PA | GTR+F+I+G4 | 124 sequences with 2151 nt sites |
| 4 | HA | GTR+F+G4 | 103 sequences with 1602 nt sites |
| 5 | NP | TIM2+F+I+G4 | 106 sequences with 1497 nt sites |
| 6 | NA | TPM2+F+G4 | 82 sequences with 1413 nt sites |
| 7 | MP | K3P+G4 | 112 sequences with 982 nt sites |
| 8 | NS | K3Pu+F+G4 | 98 sequences with 838 nt sites |
BIC, Bayesian information criteria; HA, hemagglutinin; MP, matrix protein; NA, neuraminidase; NP, nucleoprotein; NS, nonstructural; PA, polymerase acidic; PB1, polymerase basic 1; PB2, polymerase basic 2.
Phylogenetic assignment of H5 HPAIV clades followed established H5 HPAIV nomenclature standards (Lee et al. 2017, Li et al. 2017, Muzyka et al. 2018, Venkatesh et al. 2018).
Results and Discussion
HPAI outbreaks in southern Ukraine
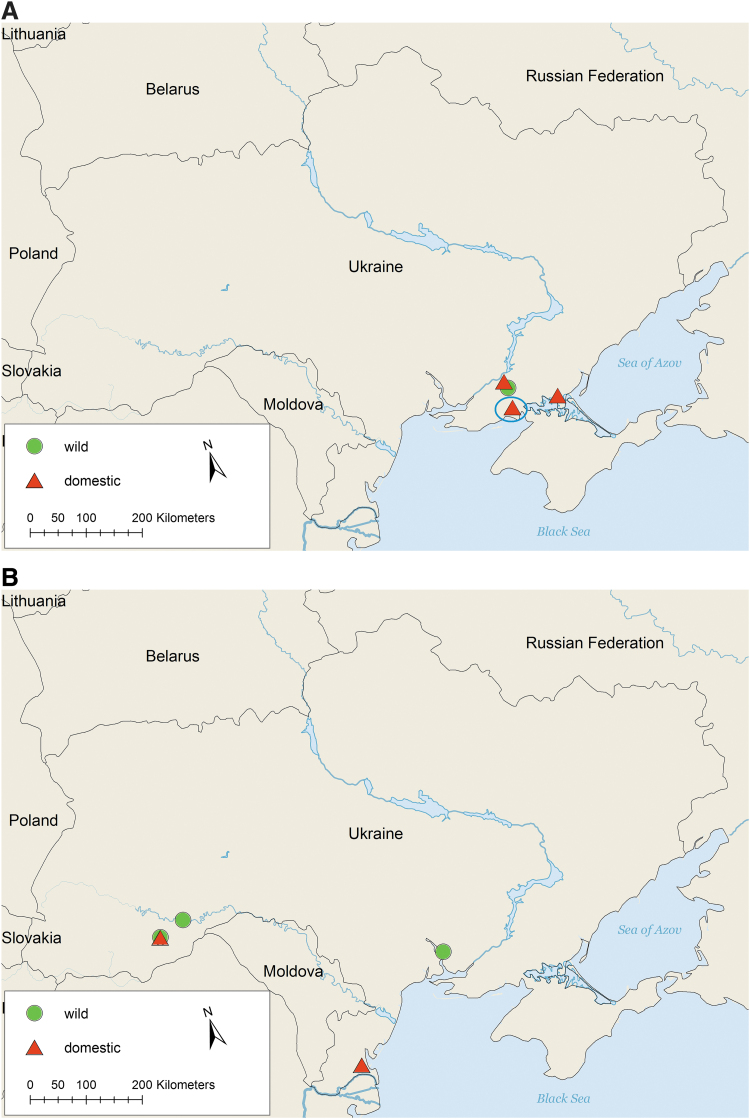
The threat of the emerging H5N8 subtype of HPAI to wild birds and poultry in Eurasia was well known in Ukraine and nearby countries in Eurasia (Verhagen et al. 2015). Typically, HPAI outbreaks had occurred in late autumn and were often detected in poultry. In Ukraine, in November 2016, HPAI was first detected in domestic birds (backyard poultry) found dead in Kherson oblast, Ukraine (Fig. 1A and Table 3). After 2 weeks the Ukrainian veterinary service was informed of mass mortality of birds in the village, and an outbreak investigation was undertaken, including inquiry with local population, and biosecurity measures for transport of carcasses and specimens to a veterinary laboratory. The first clinical signs of disease appeared about 2 weeks before collection in a backyard farm with mixed poultry (chickens, Muscovy ducks, geese, and turkeys) in the village Novooleksandrivka, and were consistent with HPAI. Signs and symptoms included body dampness, elevated temperature (>42°C), severe gastrointestinal distress (green and white diarrhea), and death. Autopsy revealed single pinprick hemorrhage in the gizzard, stomach, intestine, and heart; red swollen lungs; and enlarged kidneys and spleen. Samples were taken from ∼10 dead birds and analyzed by qRT-PCR diagnostic assays for AIV and HPAIV, which confirmed AIV diagnosis and HPAIV of H5 pathotype (OIE 2021) for all 10 dead birds. There were ∼2500 captive birds in the Novooleksandrivka outbreak zone (outbreak no. 1, Table 3).
FIG. 1.
(A) HPAI H5N8 cases in wild and domestic birds in Ukraine, November–December 2016. Locations of wild bird (green circle) and domestic poultry (red triangle) HPAI outbreaks. The first reported outbreak in domestic poultry in Novooleksandrivka village, Kalanchatskiy district (Kherson oblast), is marked (blue circle). Three HPAIV from this outbreak were sequenced. (B) Continuation of HPAI H5N8 outbreaks in wild and domestic birds in Ukraine, January–February 2017. Locations of wild bird (green circle) and domestic poultry (red triangle) HPAI outbreaks. HPAI, highly pathogenic avian influenza; HPAIV, highly pathogenic avian influenza viruses. Color images are available online.
Table 3.
Highly Pathogenic Avian Influenza Disease Outbreak Summary, Ukraine, 2016–2017
| No. | Outbreak location | Start date | Affected species | Susceptible | HPAI cases | Deaths | Culled |
|---|---|---|---|---|---|---|---|
| 1 | Kherson Oblast, Kalanchatskiy District, Novooleksandrivka village | November 14, 2016 | Domestic poultry (backyard) | 2500 | 410 | 410 | 0 |
| 2 | Kherson Oblast, Genicheskiy District, Novodmytrivka village | December 16, 2016 | Domestic poultry (backyard) | 98 | 6 | 6 | 92 |
| 3 | Kherson Oblast, Kakhovskiy District, Tsukury village | December 16, 2016 | Domestic poultry (backyard) | 30 | 2 | 2 | 28 |
| 4 | Kherson Oblast, Kakhovskiy District, Novokamyanka village | December 16, 2016 | Wild birds; mute swan (Cygnus olor) | Not available | 10 | 10 | 0 |
| 5 | Chernovtsy Oblast, Kitsmanskiy District, Chortoryia village | January 3, 2017 | Domestic poultry (backyard) | 37 | 14 | 14 | 23 |
| 6 | Chernovtsy Oblast, Kitsmanskiy District, Chortoryia village | January 1, 2017 | Wild birds; mute swan (C. olor) | Not available | 23 | 23 | 0 |
| 7 | Odesa Oblast, Kiliyskiy District, Myrne village | January 3, 2017 | Domestic poultry (commercial farm) | 10,251 | 1099 | 1099 | 9152 |
| 8 | Ternopil Oblast, Borschivskiy District, Vilkhovets village | January 15, 2017 | Wild birds; mute swan (C. olor) | Not available | 21 | 21 | 0 |
| 9 | Mykolayiv Oblast, Mykolayiv city | February 14, 2017 | Captive zoo birds; Indian Peafowl (Pavo cristatus) | 931 | 10 | 10 | 0 |
Domestic poultry were predominantly chickens, with a lesser number of ducks and geese; zoo birds included 931 birds 104 species, only Indian peafowl was affected by HPAI; an oblast is a province-level administrative region in Ukraine. Data source: OIE (2021).
HPAI, highly pathogenic avian influenza.
Poultry in Novooleksandrivka village had been vaccinated against Newcastle disease virus (NDV) 5 months before the outbreak using dry NDV La-Sota SPF strain (series 10, control 10) vaccine. On December 16, 2016, 10 dead wild mute swans (Cygnus olor) were found in the same region (Fig. 1A).
The outbreaks among domestic poultry and wild birds continued and spread toward the southwestern and western parts of the country (Fig. 1B) and were reported by veterinary authorities in Ukraine to alert the international community (OIE 2021). As of January 2017, a large commercial poultry farm was also affected (outbreak no. 7, Table 3). In February 2017, H5N8 virus was detected in Indian peafowl (Pavo cristatus) from a zoo in Mykolaiv oblast (outbreak no. 9, Table 3). In the zoo there were 931 birds of 104 different species; 10 Indian peafowl died.
In total, during 2016–2017, there were nine outbreaks of HPAI in wild and domestic birds (Table 3) in Kherson, Odessa, Mykolaiv, Chernivtsi, and Ternopil oblasts in the southern region of Ukraine (Fig. 1A, B). Among them, 10 positive samples were collected from wild mute swan in Chernivtsi and Ternopil oblasts, 12 positive samples from poultry in Chernivtsi and Odessa oblasts, and 5 positive samples from Indian peafowl in the zoo in Mykolayiv city. In domestic poultry and zoo HPAI outbreaks, the rapid onset of disease and spread of HPAI, and standard biosecurity measures implemented to restrict international trade in live birds in Ukraine, suggests that contact with wild migratory birds were the most likely route of virus introduction.
Genetic analysis of HPAIV
All eight gene segments were sequenced for three HPAIV study strains from the first reported outbreak in domestic poultry (Table 1) in Novooleksandrivka village, Kalanchatskiy district Kherson oblast (Fig. 1A). These three viruses are named: A/chicken/Ukraine/1/2016 (H5N8); A/chicken/Ukraine/3/2016 (H5N8); and A/duck/Ukraine/4/2016 (H5N8). Genetic analysis showed that these isolates from Ukraine were HPAIV H5N8 clade 2.3.4.4b strains, and closely related to each other. The three study isolates shared 99–100% nucleotide identity across all eight genes: hemagglutinin (HA), NA, polymerase basic 2 (PB2), polymerase basic 1 (PB1), polymerase acidic (PA), nucleoprotein (NP), matrix (M), and nonstructural (NS), suggesting a common origin for the outbreak.
BLAST analysis showed that all gene segments of H5N8 HPAIV study isolates were also closely related to other contemporary (2016–2017) clade H5N8 2.3.4.4b strains from Europe and Asia, suggesting that the HPAI outbreak in poultry in Ukraine was caused by HPAIV introduced through wild birds and occurred contemporaneously with outbreaks in Russia, Europe, and the Middle East in wild birds and poultry (El-Shesheny et al. 2017, Lee et al. 2017, Li et al. 2017, Selim et al. 2017).
Analysis of HA
The receptor-binding specificity of the HA protein is a major determinant of influenza A virus host range. The amino acid sequence of the protease cleavage site of the HA gene revealed multiple basic amino acids, PLREKRRKR/GLF, in all three study strains, supporting observations of a HPAI phenotype in poultry, where proteolytic activation of the HA protein by ubiquitous proteases enables systemic spread of virus (Lee et al. 2017). The receptor binding site (RBS) of the HA protein contained the Gln 222 and Gly 224 residues (H5 numbering; Q226 and G228 in H3 numbering), indicating an avian-like receptor binding specificity (α-2,3-linked sialic acids) (de Vries et al. 2017).
Other mutations in the HA protein such as D183N, K189N, Q192K, and S223R were also found in all H5N8 Ukrainian study isolates. Of note, Koel et al. (2014) identified that HPAI H5N1 viruses of clade 2.1 in Indonesia with amino acid changes at positions immediately adjacent to the RBS (e.g., D183N and R189M) are in part responsible for antigenic changes, and escape from neutralizing antibody response (Koel et al. 2014). An S133A antigenic site mutation in the HA in the three H5N8 study strains might also suggest immune evasion (Matrosovich et al. 2008). However, how the amino acid variations in HA might associate with disease phenotypes is unknown.
Analysis of NA
The enzymatic activity of the catalytic site of influenza A virus NA proteins cleaves α2-3- and α2-6-linked sialic acid and other glycans on the host cell membrane, releasing viral particles containing the glycan-binding HA virion surface glycoprotein from host cells (Colman 1994). Thus, NA has been an attractive target for antiviral drug development. Particularly in human N1 and N2 viruses, increased use of NA inhibitors (NAI) and the error-prone nature of viral RNA polymerase can result in the development of NAI-resistant influenza viruses (Spackman et al. 2003, Venkatesh et al. 2018). In the Ukrainian H5N8 viruses from this study, absence of substitution H274Y, the presence of R152 in the catalytic site, I222, and R292 (N2 numbering) suggest sensitivity to oseltamivir.
The NA glycoprotein possesses pockets that consist of several residues with enzymatic activity (Colman 1994). All three study strains had the common mutation G147D (N2 numbering), critical for the conformation of a cavity containing residues 147–152 in the NA enzyme active site that binds to NAI with similar affinities in N1 and N2 subtypes (Wang et al. 2011, Wu et al. 2013). The majority of AIV possess G at residue 147 of NA protein, whereas AIV of the N5 subtype and human viruses of the N2 subtype mainly have N at this position, resulting in an extended form of the cavity (Wang et al. 2011) that confers NAI resistance, but is not found in clinical samples after NAI treatment (Orozovic et al. 2011, Wu et al. 2013). Although the G147R substitution found in the N9 subtype conferred NAI susceptibility similar to that of the parental virus (Song et al. 2015), the novel G147V in the N5 subtype and N147I in the N8 subtype reduced inhibition by zanamivir. Thus, the sensitivity of N8 NA possessing G147D to NAI requires empirical data. Moreover, the genetic stability of variant amino acids and NAI phenotypes at residue 147 is poorly understood (Choi et al. 2017).
Other NAI resistance mutations, for example, E119A, H274Y, and N294S (N2 numbering), were not found.
Analysis of polymerase genes
Polymerase proteins play important roles in restricting viral transmission from avian species to humans (Gabriel et al. 2007, Manz et al. 2013). PB1 amino acid residues N375 and S678, and PA T97 and T552, were characteristic of strains adapted to avian cells (Gabriel et al. 2007).
The H5N8 HPAIV study strains have not acquired adaptive changes (phenotypic markers) in polymerase complex proteins. The PB2 protein of the Ukrainian strains did not have mutations that have been experimentally associated with adaptation to mammals (Gabriel et al. 2007, Bortz et al. 2011). Nor did they carry other changes, such as PB1 (Y436H), and PA (T515A, T97I), that are associated with increased potential for virulence and viral replication in mammals (Cheng et al. 2014). However, the study strains PB1 carried P13 and V473 that have been reported to increase in viral polymerase activity in mammalian cells (Gabriel et al. 2007, Xu et al. 2012).
Analysis of M, NP, and NS genes
Markers of amantadine resistance (V27A and S31N) in M2 protein were not found in the H5N8 HPAIV study strains from Ukraine. The NP is highly conserved among influenza A viruses. NP amino acids were avian type in the Ukrainian study strains, including N319, associated with enhanced viral RNA synthesis in mammalian cells (Gabriel et al. 2007); and G16, L283, F313, and Q357, residues that can modulate NP evasion of MxA in mammalian cells (Gao et al. 2011).
Influenza NS1 is a multifactorial protein that inhibits host immune responses and regulates viral replication. NS1 of the H5N8 HPAIV study strains was 217 a.a. long, possessing a common 13 a.a. truncation [length variation type LVT(−13)]. This NS1 with LVT(−13) has no mammal-adaptive “avian-like” ESEV motif at the C-terminus (Jackson et al. 2008), lacking nuclear location signals and the poly(A)-binding protein II (PABII) binding site (Melén et al. 2007, Jackson et al. 2008). Some H6N1 AIV with LVT(−13) can infect minor poultry species more easily than chickens (Tai et al. 2007). NS1 positions D92, F103, M106, and D125 were avian adaptive (Schrauwen and Fouchier 2014).
Phylogenetic analysis of the Ukrainian H5N8 HPAIV study strains
We conducted phylogenetics analyses by generating trees using ML method for each gene segment, in comparison with a subset of other H5N8 HPAIV and AIV. For all eight gene segments, the Ukrainian study strains had significant homology to other H5N8 HPAIV strains isolated in Europe and Asia during 2016–2017, clustering with strains from Ukraine, Europe (Belgium, Denmark, Switzerland, etc.) and the Russian Federation, as separate groups on the phylogenetic trees.
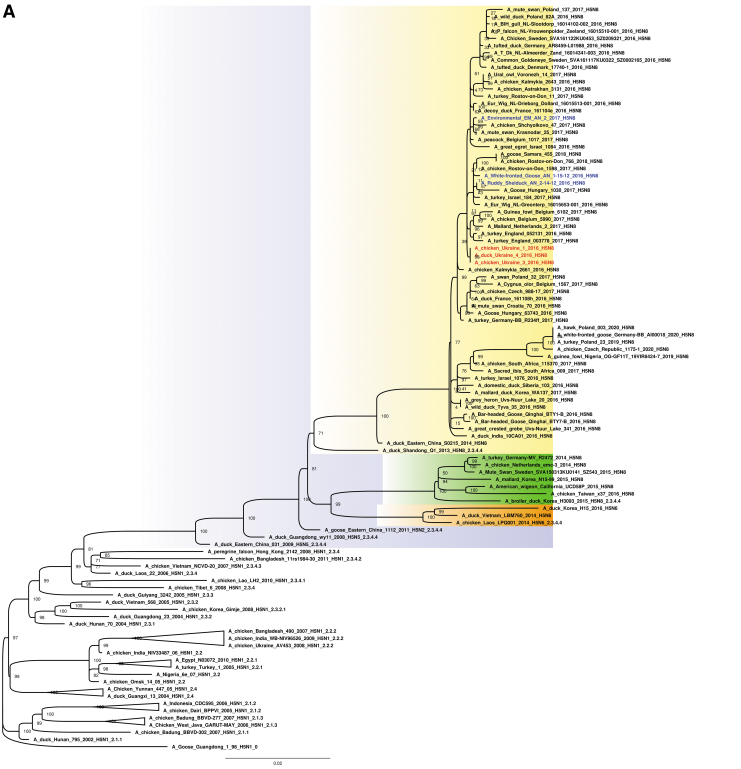
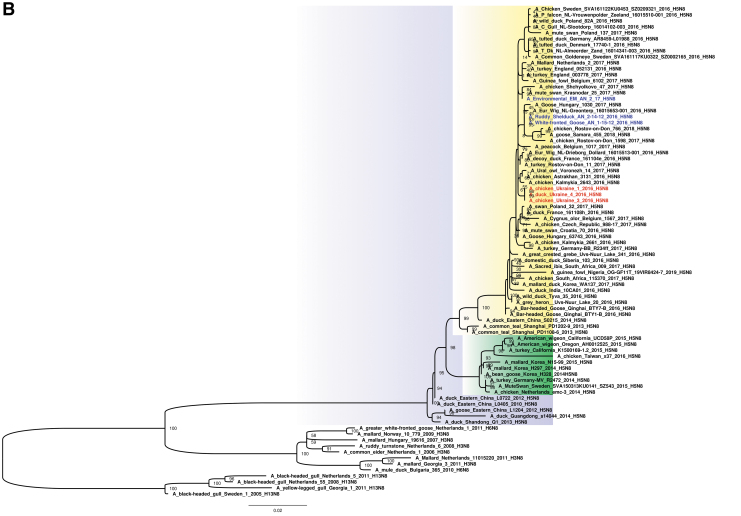
The study suggests that the three H5N8 HPAIV study strains from Ukraine were likely derived from the H5N8 clade 2.3.4.4b group HPAIV circulating among poultry in eastern China in 2015, and shared a common ancestor with A/duck/eastern China/S0215/2014 (H5N8) and descendent strains (Fig. 2A, B and Supplementary Fig. S1). Ukrainian study isolates shared genetically similar HA, NA, PB1, M, and NS segments to those of H5N8 viruses detected in wild birds in Qinghai Lake in China (Li et al. 2017), and prototypical clade 2.3.4.4b strains from Lake Uvs-Nuur at Russia–Mongolia in spring in 2016 (Lee et al. 2017). The PB2, PA, and NP segments of the three study viruses also grouped most closely with these HPAIV; however, related gene segments were found in both H5N5 and H5N6 HPAIV, and LPAIV, in Europe and Asia (Mongolia, Bangladesh, China, and Singapore).
FIG. 2.
(A) Maximum likelihood phylogenetic tree of the HA gene of the H5N8 subtype. H5N8 HPAIV collected in Ukraine from domestic (red) and wild (blue) (Muzyka et al. 2018) birds. Bootstrap supports are indicated next to the nodes, whereas branch lengths are scaled according to the number of nucleotide substitutions per site. H5N8 clade 2.3.4.4b is highlighted in yellow; H5N8 clade 2.3.4.4a in green and H5N6 subtype in orange. (B) Maximum likelihood phylogenetic tree of the NA gene of the H5N8 subtype. H5N8 HPAIV collected in Ukraine from domestic (red) and wild (blue) (Muzyka et al. 2018) birds. Bootstrap supports are indicated next to the nodes, whereas branch lengths are scaled according to the number of nucleotide substitutions per site. H5N8 clade 2.3.4.4b is highlighted in yellow; H5N8 clade 2.3.4.4a in green. HA, hemagglutinin; NA, neuraminidase. Color images are available online.
When the H5N8 clade 2.3.4.4b HPAIV strain reached Ukraine in November 2016, mass mortality in domestic and wild birds was reported (Table 3). For all eight gene segments, other three H5N8 HPAIV contemporary strains (Muzyka et al. 2018) that were identified in wild birds in Askania Nova Biopreserve appear in the same clade as the three Ukrainian poultry strains that we sequenced (Fig. 2A, B and Supplementary Fig. S1). These wild bird strains, A/white-fronted goose/AN/1-15-12/2016 (H5N8), A/ruddy shelduck/AN/2-14-12/2016 (H5N8), and A/environmental (EM)/AN/2/17 (H5N8), were extracted from the virus-containing allantoic fluid after two egg passages, perhaps explaining minor nucleotide changes among the six Ukrainian viruses.
We conducted a comparative analysis of three novel H5N8 clade 2.3.4.4b HPAIV strains in Ukraine from late 2016. Direct or indirect contact with wild birds was the most likely route of virus introduction into backyard poultry farms and a zoo. In 2016–2017, H5N8 HPAIV infections were also reported in Poland, Romania, Austria, Hungary, Germany, Russia, Egypt, Iran, and other countries, suggesting that migration of wild birds carrying H5 clade 2.3.4.4b viruses were the cause of contemporaneous outbreaks along Eurasian flyways (Fig. 2A) (Lee et al. 2015, 2017, Verhagen et al. 2015, Li et al. 2017, Selim et al. 2017, Muzyka et al. 2018). HPAIV outbreaks usually occurred in the late autumn and winter periods (November–February) that coincide with the southern migrations of wild birds.
Although a similar outbreak pattern has again been observed in recent H5 clade 2.3.4.4b HPAI outbreaks in Europe and Asia in 2019–2021 (Lewis et al. 2021), including Ukraine (OIE 2021), resident or semi-resident waterfowl also have been suggested to be reservoirs involved in local amplification of outbreaks in 2016–2017 (Poen et al. 2018). H5 clade 2.3.4.4b viruses have also reassorted with LPAIV strains, with reassortants identified in both wild birds and poultry (Lycett et al. 2020, Lewis et al. 2021); this phenomenon was also observed in this outbreak in Ukraine (E.B., pers. comm.).
Conclusion
In our study we analyzed the phylogenetic proximity of domestic poultry H5N8 HPAIV to wild bird viruses in all eight gene segments, and suggest that wild migratory birds have played a key role in the introduction of H5N8 HPAIV in Ukraine. This is not unprecedented, with wild birds as initial drivers of numerous HPAI outbreaks that subsequently spread and amplify in poultry (Verhagen et al. 2015, Poen et al. 2016, Muzyka et al. 2018, Lewis et al. 2021). Tens of thousands of migrating waterfowl crossing Europe, Asia, and Africa use the wetlands of southern Ukraine as a resting stop, a “hotspot” for avian viruses (Muzyka et al. 2014).
Of importance, the identification of closely related HPAIV in both domestic poultry and wild birds in the same ecogeographic region highlights the risk of wild bird:poultry spillover. The veterinary sector in Ukraine has an integrated network of regional (oblast or provincial level) laboratories, and central laboratories in the capital (Kyiv). Ukraine, like other countries in the region, is part of the World Organization for Animal Health (OIE), and maintains close relationships with reference laboratories in the EU and the United States. Recent Ukrainian, U.S. and E.U.-funded programs have supported systematic surveillance in wild and domestic birds, and built genomic sequencing capacity for AIV genotyping, to inform outbreak control measures and potentially provide early warning of the introduction of HPAI.
Data Availability Statement
Sequence accession numbers in GISAID (“global initiative on sharing avian influenza data”) are as follows: EPI_ISL_1121143 for A/chicken/Ukraine/1/2016; EPI_ISL_1121144 for A/chicken/Ukraine/3/2016; and EPI_ISL_1121145 for A/duck/Ukraine/4/2016.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge the authors, originating and submitting laboratories of the sequences from GISAIDs EpiFlu™ and NCBI GenBank databases, for making tremendous data publicly available and largely benefiting the science community. Special thanks are conferred to Dr. Martin Beer (Freidrich Loeffler Institute, Germany) for sequences of contemporary H5N8 sequences from Ukraine available in GISAID for phylogenetics analyses. Additional thanks are conferred to Lyudmila Maruschak (Ukraine) for assistance with sample isolation; Xiao Bai (University of Alaska) for insight into methodology for AIV genomics analyses; and to Dr. Greg Glass and the scientific staff at BV/Metabiota (Kyiv, Ukraine) for critical reading and assistance with preparation of the article.
Authors' Contributions
M.S., G.K., M.S., M.B., and E.B.—study design, data collection, data interpretation, article preparation, literature search; S.E., N.S.L.—sequencing, analysis; D.M., N.U., A.M., and A.A.—data collection and literature search; G.K., M.S., M.S., N.S.L., and E.B.—writing—original draft preparation, review and editing. All authors read and approved the final version of the article.
Author Disclosure Statement
No conflicting financial interests exist.
Funding Information
This work was supported under a program for state routine monitoring of avian diseases by the Ukraine State Service of Ukraine for Food Safety and Consumer Protection, and State Scientific and Research Institute of Laboratory Diagnostics and Sanitary and Veterinary Expertise; sequence data analysis by U.S. National Institute of Allergy and Infectious Disease (NIAID) Centers of Excellence for Influenza Research and Surveillance (CEIRS) contract HHSN272201400008C (to Nicola S. Lewis and Eric Bortz); a bioinformatics pilot award (to Eric Bortz) from the National Institute of General Medical Sciences (NIGMS) Institutional Development Award (IDeA) program (Alaska INBRE P20GM103395); and funding support by the U.K. Department for the Environment, Food and Rural Affairs (Defra) and the devolved Scottish and Welsh governments (under grant SV3006).
The authors would also like to acknowledge the U.S. Department of Defense, Defense Threat Reduction Agency (DTRA) Cooperative Biological Engagement Program (CBEP) Scientific Writing Mentorship Program (SWMP) for their support in providing resources for writing this manuscript. DTRA/CBEP did not directly support the research described herein. The contents of this publication are the responsibility of the authors and do not necessarily reflect the views of the U.S. National Institutes of Health, Department of Defense, or the U.S. Government.
Supplementary Material
References
- Bortz E, Westera L, Maamary J, Steel J, et al. Host- and strain-specific regulation of influenza virus polymerase activity by interacting cellular proteins. mBio 2011; 2:e00151-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng K, Yu Z, Chai H, Sun W, et al. PB2-E627K and PA-T97I substitutions enhance polymerase activity and confer a virulent phenotype to an H6N1 avian influenza virus in mice. Virology 2014; 468:207–213. [DOI] [PubMed] [Google Scholar]
- Choi WS, Jeong JH, Kwon JJ, Ahn SJ, et al. Screening for neuraminidase inhibitor resistance markers among Avian influenza viruses of the N4, N5, N6, and N8 neuraminidase subtypes. J Virol 2017; 92:e01580-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colman PM. Influenza virus neuraminidase: Structure, antibodies, and inhibitors. Protein Sci 1994; 3:1687–1696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Vries RP, Peng W, Grant OC, Thompson AJ, et al. Three mutations switch H7N9 influenza to human-type receptor specificity. PLoS Pathog 2017; 13:e1006390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Shesheny R, Barman S, Feeroz M, Hasan M, et al. Genesis of influenza A (H5N8) viruses. Emerg Infect Dis 2017; 23:1368–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriel G, Abram M, Keiner B, Wagner R, et al. Differential polymerase activity in avian and mammalian cells determines host range of influenza virus. J Virol 2007; 81:9601–9604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao S, von der Malsburg A, Dick A, Faelber K, et al. Structure of myxovirus resistance protein a reveals intra-and intermolecular domain interactions required for the antiviral function. Immunity 2011; 35:514–525. [DOI] [PubMed] [Google Scholar]
- Jackson D, Hossain MDJ, Hickman D, Perez D, et al. A new influenza virus virulence determinant: The NS1 protein four C-terminal residues modulate pathogenicity. Proc Natl Acad Sci USA 2008; 105:4381–4386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, et al. ModelFinder: Fast model selection for accurate phylogenetic estimates. Nat Methods 2017; 14:587–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katoh K, Rozewicki J, Yamada KD. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 2019; 20:1160–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koel BF, van der Vliet S, Burke DF, Bestebroer TM, et al. Antigenic variation of clade 2.1 H5N1 virus is determined by a few amino acid substitutions immediately adjacent to the receptor binding site. mBio 2014; 5:e01070-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuraku S, Zmasek CM, Nishimura O, Katoh K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res 2013; 41:W22–W28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Sharshov K, Swayne DE, Kurskaya O, et al. Novel reassortant clade 2.3.4.4 avian influenza A(H5N8) virus in wild aquatic birds, Russia, 2016. Emerg Infect Dis 2017; 23:359–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee DH, Torchetti MK, Winker K, Ip HS, et al. Intercontinental spread of Asian-origin H5N8 to North America through Beringia by migratory birds. J Virol 2015; 89:6521–6524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis NS, Banyard AC, Whittard E, Karibayev T, et al. Emergence and spread of novel H5N8, H5N5 and H5N1 clade 2.3.4.4 highly pathogenic avian influenza in 2020. Emerg Microbes Infect 2021; 10:148–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Liu H, Bi Y, Sun J, et al. Highly Pathogenic Avian Influenza A(H5N8) virus in wild migratory birds, Qinghai Lake, China. Emerg Infect Dis 2017; 23:637–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycett SJ, Pohlmann A, Staubach C, Caliendo V, et al. Genesis and spread of multiple reassortants during the 2016/2017 H5 avian influenza epidemic in Eurasia. Proc Natl Acad Sci USA 2020; 117:20814–20825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manz B, Schwemmle M, Brunotte L. Adaptation of avian influenza A virus polymerase in mammals to overcome the host species barrier. J Virol 2013; 87:7200–7209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matrosovich MN, Gambaryan AS, Klenk H-D. Receptor specificity of influenza viruses and its alteration during interspecies transmission. In: Klenk H-D, Matrosovich MN, Stech J, eds. Avian Influenza. Monogr Virol. Basel: Karger, 2008:134–155. [Google Scholar]
- Melén K, Kinnunen L, Fagerlund R, Ikonen N, et al. Nuclear and nucleolar targeting of influenza A virus NS1 protein: Striking differences between different virus subtypes. J Virol 2007; 81:5995–6006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzyka D, Pantin-Jackwood M, Stegniy B, Rula O, et al. Wild bird surveillance for avian paramyxoviruses in the Azov-black sea region of Ukraine (2006 to 2011) reveals epidemiological connections with Europe and Africa. Appl Environ Microbiol 2014; 80:5427–5438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzyka D, Rula O, Tkachenko S, Muzyka N, et al. Highly pathogenic and low pathogenic Avian influenza H5 subtype viruses in wild birds in Ukraine. Avian Dis 2018; 63(Suppl 1):219–229. [DOI] [PubMed] [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum likelihood phylogenies. Mol Biol Evol 2015; 32:268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orozovic G, Orozovic K, Lennerstrand J, Olsen B. Detection of resistance mutations to antivirals oseltamivir and zanamivir in avian influenza A viruses isolated from wild birds. PLoS One 2011; 6:e16028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poen MJ, Bestebroer TM, Vuong O, Scheuer RD, et al. Local amplification of highly pathogenic avian influenza H5N8 viruses in wild birds in the Netherlands, 2016 to 2017. Euro Surveill 2018; 23:17-00449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrauwen EJ, Fouchier RA. Host adaptation and transmission of influenza A viruses in mammals. Emerg Microbes Infect 2014; 3:e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selim AA, Erfan AM, Hagag N, Zanaty A, et al. Highly pathogenic Avian influenza virus (H5N8) Clade 2.3.4.4 infection in migratory birds, Egypt. Emerg Infect Dis 2017; 23:1048–1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song M-S, Marathe BM, Kumar G, Wong S-S, et al. Unique determinants of neuraminidase inhibitor resistance among N3, N7, and N9 avian influenza viruses. J Virol 2015; 89:10891–10900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spackman E, Senne DA, Bulaga LL, Myers TJ, et al. Development of real-time RT-PCR for the detection of avian influenza virus [published correction appears in Avian Dis 2005;49(2):313]. Avian Dis 2003; 47(3 Suppl):1079–1082. [DOI] [PubMed] [Google Scholar]
- Tai H, Wang J, Poon LL, Peiris JS, et al. Establishment of influenza A virus (H6N1) in minor poultry species in southern China. J Virol 2007; 81:10402–10412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venkatesh D, Poen MJ, Bestebroer TM, Scheuer RD, et al. Avian influenza viruses in wild birds: Virus evolution in a multihost ecosystem. J Virol 2018; 92:e00433-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verhagen JH, van der Jeugd HP, Nolet BA, Slaterus R, et al. Wild bird surveillance around outbreaks of highly pathogenic avian influenza A(H5N8) virus in the Netherlands, 2014, within the context of global flyways. Euro Surveill 2015; 20:21069. [DOI] [PubMed] [Google Scholar]
- Verkhovna Rada of Ukraine. Legislation of Ukraine. Instruction for the Prevention and Elimination of Avian Influenza in Birds. Document z1277-11, valid, current version—Adoption on October 17, 2011. Available at https://zakon.rada.gov.ua/laws/show/z1277-11?lang=en#Text
- Wang M, Qi J, Liu Y, Vavricka CJ, et al. Influenza A virus N5 neuraminidase has an extended 150-cavity. J Virol 2011; 85:8431–8435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- World Organisation for Animal Health (OIE). Avian Influenza Portal. REF OIE 21731 (30/11/2016); REF OIE 22326 (12/01/2017); REF OIE 22345 (12/01/2017); REF OIE 21731 (30/11/2016); REF OIE 32998 (20/01/2020); REF OIE (Vol. 34-No. 04, January 28, 2021); Outbreak Reports; Ukraine. Available at https://www.oie.int/en/animal-health-in-the-world/update-on-avian-influenza/
- Wu Y, Qin G, Gao F, Liu Y, et al. Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding. Sci Rep 2013; 3:1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Hu WB, Xu K, He YX, et al. Amino acids 473V and 598P of PB1 from an avian-origin influenza A virus contribute to polymerase activity, especially in mammalian cells. J Gen Virol 2012; 93:531–540. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence accession numbers in GISAID (“global initiative on sharing avian influenza data”) are as follows: EPI_ISL_1121143 for A/chicken/Ukraine/1/2016; EPI_ISL_1121144 for A/chicken/Ukraine/3/2016; and EPI_ISL_1121145 for A/duck/Ukraine/4/2016.