Summary
ETV6 transcriptional activity is critical for proper blood cell development in the bone marrow. Despite the accumulating body of evidence linking ETV6 malfunction to hematological malignancies, its regulatory network remains unclear. To uncover genes that modulate ETV6 repressive transcriptional activity, we performed a specifically designed, unbiased genome-wide shRNA screen in pre-B acute lymphoblastic leukemia cells. Following an extensive validation process, we identified 13 shRNAs inducing overexpression of ETV6 transcriptional target genes. We showed that the silencing of AKIRIN1, COMMD9, DYRK4, JUNB, and SRP72 led to an abrogation of ETV6 repressive activity. We identified critical modulators of the ETV6 function which could participate in cellular transformation through the ETV6 transcriptional network.
Subject areas: Molecular biology, Cancer systems biology, Omics
Graphical abstract

Highlights
-
•
We develop a genome-wide shRNAs screen for ETV6 modulators
-
•
The screen uncovered 13 novel putative ETV6 modulator genes
-
•
The modulators demonstrated a broad impact on the ETV6 transcriptional network
-
•
T-ALL cells results suggest modulators are conserved in other cellular contexts
Molecular biology; Cancer systems biology; Omics
Introduction
ETV6 is a member of the ETS (E26 transformation-specific) superfamily of transcription factors (Mavrothalassitis and Ghysdael, 2000) located on the short arm of chromosome 12. ETS transcription factors have been associated with multiple biological processes and pathways with, among others, critical implications in hematopoiesis (Ciau-Uitz et al., 2013). Their dysregulation correlates with hematological diseases such as lymphomas, leukemia, and myelodysplastic syndrome. Chromosomal abnormalities of the 12p13 band are frequently found in these malignancies. So far, more than 30 fusion partners have been identified in ETV6-derived translocations (De Braekeleer et al., 2012). The mechanisms through which these fusions induce transformation are highly variable and depend on the function of the partner as well as the retained domains of ETV6. In some cases, the expected impact of the translocation is to inhibit ETV6 normal function as proposed for the translocations t(1; 12)(q21; p13) (Otsubo et al., 2010) and t(12; 21)(p13; q22) (Romana et al., 1995a; Golub et al., 1995; Gunji et al., 2004).
The t(12; 21)(p13; q22) translocation is one of the most frequent chromosomal abnormalities found in childhood B-cell acute lymphoblastic leukemia (ALL) and is characteristic of the ETV6-RUNX1 ALL subtype (Romana et al., 1995b; Shurtleff et al., 1995; Bhojwani et al., 2015). It fuses all functional domains encoded by the AML1 (RUNX1) gene to the first five exons of ETV6 encoding its Pointed (PD) and repression domains. Despite the high recurrence of t(12; 21) in B-cell ALL and the resulting expression of the ETV6-AML1 fusion protein in patients with t(12; 21) positive leukemic (Agape et al., 1997; Poirel et al., 1998), the t(12; 21) was shown to be insufficient to induce leukemic transformation (Andreasson et al., 2001; van der Weyden et al., 2011; Mori et al., 2002). Interestingly, the non-rearranged residual allele of ETV6 is targeted by deleterious events in most cases of t(12; 21) positive ALL (Poirel et al., 1998; Patel et al., 2003; Lilljebjorn et al., 2010; Montpetit et al., 2004), suggesting that ETV6 inactivation could play a critical role in leukemogenesis. In addition, several groups recently reported germline mutations in ETV6 that were associated with familial hematological disorders (Moriyama et al., 2015; Topka et al., 2015; Noetzli et al., 2015; Zhang et al., 2015). Several of these mutations were in the ETS DNA-binding domain of ETV6 and impaired its nuclear localization and/or repressor activity. These findings strongly support a role for ETV6-mediated transcription in these malignancies.
Despite the consequences of ETV6 dysfunction, very little is known about its regulatory network. To negatively regulate transcription, ETV6 cooperates with co-repressors such as the N-CoR and mSin3A complexes and the histone deacetylase HDAC3 (Lee et al., 2004; Wang and Hiebert, 2001; Guidez et al., 2000). ETV6 interaction with HDAC3 was shown to be modulated through its binding with the IRF-8 transcription factor (Kuwata et al., 2002). This interaction was further associated with a change in ETV6 binding and transcriptional activity. Although indicating that ETV6-mediated transcription can be regulated through complex interactions, these studies were conducted by candidate gene or interaction-based approaches and did not investigate the full intricacy of ETV6 function.
Small hairpin RNAs (shRNAs) are widely used to silence gene expression (Azlan et al., 2016). In this study, we designed a cellular screening strategy to functionally interrogate the impact of over 140,000 shRNAs on ETV6-mediated gene repression. A panel of high priority genes selected from the screen results were further examined for their capacity to efficiently regulate ETV6 activity by monitoring its transcriptional network in pre-B ALL cells (Neveu et al., 2016, 2018). Validation experiments unveiled key modulators of ETV6 repressive function in leukemic cells.
Results
Generating ETV6-dependent Blasticidin sensitive cells
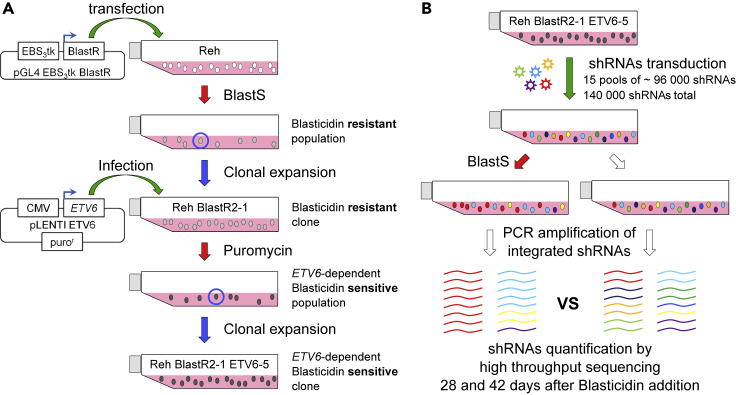
To gain insights into the network of ETV6 modulators, we modified the Reh pre-B ALL cell line (Figure 1A) to develop a genome-wide shRNAs screen for ETV6 modulators (Figure 1B). A Blasticidin-resistance gene (BlastR) was stably integrated and expressed through an artificial ETV6-responsive promoter (EBS3tk) composed of three tandem ETV6 binding sites (EBS) (Szymczyna and Arrowsmith, 2000) followed by the HSV-TK promoter. Reh cells were selected because they lack endogenous ETV6 expression as a result of a t(12; 21)(p13; q22) translocation and the 12p13 deletion of the non-translocated allele (Uphoff et al., 1997). In absence of functional endogenous ETV6, EBS3tk is active and promotes Blasticidin resistance in this modified cell line. Wild-type ETV6 was then re-introduced through lentiviral transduction in isolated blasticidin resistant clones. Ectopically expressed ETV6 is expected to bind and repress the EBS3tk promoter leading to reduce BlastR expression. As shown in Figure 2A, BlastR expression in the Reh EBS3tk BlastR clone2-1 is significantly reduced upon ETV6 expression compared to the mock and pLENTI control clones (2.7-fold compared to pLENTI; p value ≤ 0.001). We derived a final clone from the Reh EBS3tk BlastR clone2-1 expressing ETV6 to obtain a robust and homogeneous cellular system which we refer to as clone 2-1 ETV6-5.
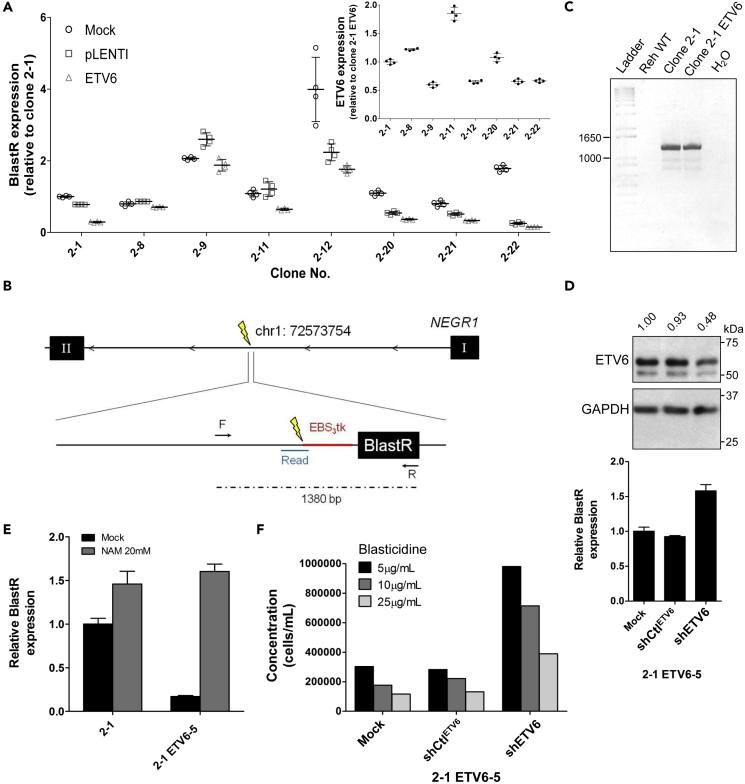
Figure 1.
Overview of the genome-wide shRNA screening design
(A) To identify putative modulators of ETV6-mediated transcriptional repression, a pre-B acute lymphoblastic leukemia cell line (Reh) was modified sequentially to obtain an ETV6-dependent Blasticidin sensitive clone. These cells are expressing the Blasticidin-resistance gene (BlastR) under the control of the ETV6-responsive artificial promoter (EBS3tk).
(B) These cells were then transduced with shRNAs (n ≈ 140,000) and selected with Blasticidin. Over-represented shRNAs in the Blasticidin condition compared to the mock condition are expected to target genes required for ETV6-mediated repression of BlastR.
Figure 2.
Establishment of an ETV6-dependent Blasticidin-sensitive leukemia cell system
(A) BlastR expression was evaluated by qRT-PCR in each Reh EBS3tk BlastR clone (clones 2-1 to 2-22) following ETV6 expression (inset). Data points for technical replicates are shown with lines indicating mean ± SD (n = 4).
(B) ETV6 ChIP-seq data obtained from clone 2-1 located the insertion site of EBS3tk BlastR (yellow mark). One sequencing read (indicated in blue) contained the partial sequence of EBS3tk promoter together with 95 nucleotides of the first intron of the NEGR1 gene. The F/R arrows depict the primers used in the PCR analysis. The dash line represent the amplified fragment (1380 bp) generated by PCR.
(C) PCR analysis of the clone 2-1 confirmed the insertion site of EBS3tk BlastR in the first intron of the NEGR1 gene.
(D) BlastR expression was evaluated by qRT-PCR in 2-1 ETV6-5 cells after shRNA-mediated silencing of ETV6 (quantified Western blot) or (E) after nicotinamide (NAM) treatment. Data are shown as mean ± SD, n = 4.
(F) 2-1 ETV6-5, shCtlETV6−GFP, and shETV6-GFP sorted cells were maintained in culture for 12 days with various Blasticidin concentrations and counted to evaluate proliferation.
Although each Reh EBS3tk BlastR clone significantly expressed ETV6 after transduction (Figure 2A, inset), the inconsistent effect on BlastR repression suggests that the chromatin context where EBS3tk BlastR inserted is rather influential. To identify the integration site of EBS3tk BlastR in the ETV6-responsive clone 2-1, we used data from a previous ETV6 chromatin immunoprecipitation sequencing experiment (ChIP-seq) performed with clone 2-1 (Neveu et al., 2018). Several reads mapped on the EBS3tk promoter, suggesting that ETV6 is effectively recruited to the artificial promoter. One read contained the partial sequence of the EBS3tk promoter merged with a sequence from the first intron of the NEGR1 gene, indicating the integration site (Figure 2B). PCR analysis of the Reh EBS3tk BlastR clone 2-1 confirmed the integration site of EBS3tk BlastR in the intron one of NEGR1 (Figure 2C). Based on previous transcriptome data (Neveu et al., 2016), this integration did not impact NEGR1 expression compared to non-modified Reh cells (53.1 and 51.3 FPKM, respectively). Furthermore, NEGR1 is not significantly modulated by ETV6 re-expression (logFoldChange = −0.46, FDR = 0.50). Together, these results indicate that EBS3tk BlastR insertion at an active locus enables stable BlastR expression, accessibility to ETV6, and concomitant ETV6-mediated gene repression through the EBS3tk promoter.
To validate that ETV6-mediated repression of BlastR is reversible, we introduced in the 2-1 ETV6-5 clone an shRNA against ETV6 (shETV6) or negative control (shCtlETV6) with GFP coexpression. The GFP positive fraction of cells was retrieved and ETV6 knockdown was confirmed at the protein level (Figure 2D). As expected, BlastR expression was positively modulated by ETV6 knockdown (Figure 2D). Treatment with, nicotinamide (NAM), which is used as an inhibitor of epigenetic repression (Schmidt et al., 2004; Bitterman et al., 2002; Sanders et al., 2010), increased BlastR expression in the 2-1 ETV6-5 clone (Figure 2E), further indicating that the repression is reversible. Finally, as expected the BlastR expression observed in shETV6 cells (Figure 2D) considerably increased their proliferation in the presence of Blasticidin (Figure 2F). Altogether, these results demonstrate that the modified Reh cell lines EBS3tk BlastR 2-1 ETV6-5 clone is a suitable model to screen for modulators of ETV6 repressive activity.
Identification of putative ETV6 modulators through a genome-wide shRNA screen
We designed a genome-wide loss-of-function screen to identify ETV6 modulators (Figure 1B). Cells expressing shRNAs impairing ETV6 repressive activity are expected to increase BlastR expression and be selectively enriched following a blasticidin treatment. Fourteen days following transduction with the complete Mission lentiviral shRNA library (∼140,000 unique shRNAs split into 15 pools), deep sequencing genomic DNA allowed the identification of 126,042 unique integrated shRNAs, which were evenly distributed among the 15 pools. By applying a series of thresholds outlined in Figure 3A and the STAR Methods section, we selected 2,858 positive shRNAs that were over-represented in presence of Blasticidine (BlastS condition) at both the 28 and 42-day time points compared to their respective control population without selection. From those, 1,241 shRNAs were retained (Figure 3B and Table S1) after discarding those with a non-significant over-representation (i.e. ∑Counts(BlastS vs ØBlastS) less than 2 SD of the SHC202 control; see STAR Methods section) as well as those targeting unexpressed genes (FPKM ≤0.21 (Hart et al., 2013)) in Reh cells (Neveu et al., 2016). As expected, among these selected shRNAs, one shRNA against ETV6 was enriched in the BlastS conditions. 1,075 genes were targeted by a single shRNA (Figure 3A) and, with few exceptions (see below), were not further considered to avoid potential off-target effects. Nonetheless, 77 and four genes (total of 81 targets) were targeted by two and three distinct shRNAs, respectively, and were thus prioritized (Figure 3A, and Table S2). Interestingly, 33 of these 81 genes were targeted by shRNAs from different pools (for example TACC1 and CAPN7, Figures 3C and 3D, respectively; see also Table S2), suggesting that their selection was not dependent on the other shRNAs present in their respective pool.
Figure 3.
Genome-wide shRNA screening to identify ETV6 modulators
(A) Flowchart illustrating the stepwise procedure for the analysis of the genome-wide shRNA screen sequencing.
(B) Normalized read counts distribution of the 1241 selected shRNAs shown for the five time points. Whiskers represent minimum and maximum.
(C–D). Examples of selected shRNAs normalized counts for the five time points.
(E) Normalized count distributions are shown separately for the best and second-best shRNAs of a pair or trio. Whiskers represent minimum and maximum; statistical significance is calculated with a paired Student t test (n = 81) (∗∗∗p < 0.001, ∗∗p < 0.01).
(F) Relation between the best (y axis) and second-best (x axis) shRNAs of a pair or trio according to their respective Blasticidin count difference.
(G) The same relation between the best (y axis) and second-best (x axis) shRNAs of a pair or trio when their Blasticidin count difference is divided by their initial count at T14. See also Tables S1 and S2.
The shRNA with the greater difference in normalized counts between Blasticidin and control conditions, for a given pair or trio, was selected for further validation (Figures 3E and 3F). Interestingly, although the differences between the two best shRNAs are mostly attributed to their representations in Blasticidin conditions (T28 BlastS and T42 BlastS), the best shRNAs are better represented in control conditions (T28 and T42) as well as in their initial populations prior to Blasticidin selection (T14) (Figure 3E). To attenuate biases, the difference in normalized counts between Blasticidin and control conditions for each shRNA were divided by their representation at T14 (Figure 3G). The relationship between the best and second-best shRNAs is, indeed, less divergent after implementing this correction (compare Figures 3F and 3G), indicating that two shRNAs targeting the same gene are likely to promote a similar impact on Blasticidin resistance. Accordingly, only the shRNAs with the strongest impact were selected (Table S2).
To identify additional candidates among the 1,075 single shRNAs, we used a complementary approach based on structure predictions (Singh et al., 2010). We identified 136 proteins predicted to interact with at least three of the 81 selected ETV6 modulators identified above. Nine of these were part of the group of 1,075 genes targeted by a single shRNAs and were likely valid candidates. They were thus added to the 81 candidates for a total of 90 genes (Figure 3A and Table S2).
Validation of putative ETV6 modulators on known endogenous transcriptional targets
To validate the impact of the putative modulators on ETV6-mediated regulation, we constructed distinct Reh ETV6 cell lines expressing an shRNA against each of these candidates and evaluated the ETV6 transcriptional network (Neveu et al., 2016) by targeted RNA sequencing (refer to STAR Methods) using a custom panel of genes. Of the 84 putative ETV6 target genes included in the panel, 40 were confirmed as their expression was significantly reduced in Reh in an ETV6 dependent manner (Figure 4A and Table S3). Of those, 22 are associated with ETV6 ChIP-seq signal (Neveu et al., 2018) and are considered direct ETV6 targets. Inversely, the expression of 64 genes previously shown to be independent of ETV6 (Neveu et al., 2016) behaves similarly in the current targeted RNA-seq experiment (Figure 4B and Table S3). As expected most (62/64) were not associated with ETV6 ChIP-seq signals (Neveu et al., 2018) and were used as negative controls (Table S3).
Figure 4.
Impact of putative ETV6 modulators on its transcriptional regulatory network
(A) Graphical representation of the relative expression of ETV6 target genes or (B) ETV6-independent control genes in the pLENTI control sample compared to the ETV6-expressing controls.
(C) The mean relative expression of ETV6 targets (x axis) is compared to the relative expression of control genes obtained with the same shRNA; the Student’s t test p values are shown on the y axis. Colored dots (green: overexpressed; red: underexpressed) are significant shRNAs. The blue dot represents the Reh pLENTI sample which lacks ETV6 expression and serves as a positive control.
(D) Relative expression of the shRNA-targeted genes according to the category of knockdown. Best knockdown (Best KD), n = 31; Knockdown (KD), n = 32; No knockdown (No KD), n = 22). Whiskers represent minimum and maximum; statistical significance is calculated with an unpaired Student’s t test (∗∗∗p < 0.001).
(E) Relative COMMD9 expression is shown for all shRNA samples. shCOMMD9 sample is shown in green and displays the lowest expression. Lines represent mean ± SD.
(F) Relative KLF5 expression is diminished in the shKLF5 sample (green) but several other shRNAs also reduce its expression. Lines represent mean ± SD.
(G) Distribution of the relative expressions of ETV6 targets (n = 22) and control genes (n = 62) in the 13 significant shRNA samples. Reh pLENTI is also shown as a control. Whiskers represent minimum and maximum.
(H) Heatmap representation of the normalized read counts of ETV6 targets (lanes) in the 13 significant shRNA samples (and control samples; columns). See also Figure S1 and Table S3.
We calculated the number of direct ETV6 targets (n = 22) affected by the shRNA tested, as compared to the negative shRNA controls (SHC002 and SHC202). 24 shRNAs led to a specific overexpression of ETV6 targets (n = 22) compared to ETV6-unrelated genes (n = 62) (Figure 4C), indicating that their effects on gene expression are likely mediated by impairing ETV6 repressive function.
Analyses of the expression level of shRNAs-targeted genes by RNA sequencing revealed that from the 90 candidate genes, 85 have a detectable expression (informative amplicons). The shRNAs were categorized into three groups based on their silencing patterns (Figure 4D). The ''Best knockdown'' group (n = 31) led to the lowest expression of their target gene (mean silencing of 71%) among all samples (e.g. shCOMMD9, Figure 4E). The shRNAs in the ''Knockdown'' group (n = 32) demonstrate a silencing effect on their target gene but are not the shRNA with the strongest effect on their target (e.g. shKLF5, Figure 4F). Consequently, their effects could not be clearly associated with their target gene. Finally, the ''No knockdown'' group (n = 22) failed to repress its target gene.
From the 31 shRNA in the ''best knockdown'' group, 12 led to significantly overexpressed ETV6 targets and one was validated by qPCR (DYRK4, 73% knockdown) because its RNA targeted sequencing was too low for interpretation (Figure S1). Altogether, we identified 13 shRNAs that efficiently knocked down their target genes and impaired ETV6 transcriptional activity (Figure 4G). However, the overexpression levels and the number of ETV6 targets affected by the silencing of these putative modulators are variable, suggesting that they could act in a gene-specific manner (Figures 4G and 4H). To determine the most influential ETV6 modulators, we compared the expression of ETV6 targets in each shRNA sample to SHC controls. Among the 13 selected shRNAs, 5 (shAKIRIN1, shCOMMD9, shDYRK4, shJUNB, and shSRP72) significantly impacted the expression of ETV6 target genes (Table 1). Moreover, an unsupervised clustering analysis based on the expression of the 22 ETV6 targets revealed the critical influence of AKIRIN1, DYRK4, and SRP72 depletion on ETV6 function as these shRNA samples clustered in a distinct group including Reh pLENTI sample lacking ETV6 expression (Figure 4H).
Table 1.
Data summary of the five top ETV6 modulators
| Gene symbol |
AKIRIN1 |
COMMD9 |
DYRK4 |
JUNB |
SRP72 |
||
|---|---|---|---|---|---|---|---|
| shRNA | TRCN0000134954 | TRCN0000167758 | TRCN0000197116 | TRCN0000014943 | TRCN0000151445 | ||
| Genome-wide shRNA screen | Pool | 12 | 11 | 13 | 2 | 15 | |
| Normalized counts | T14 | 2822 | 1196 | 5864 | 178 | 5250 | |
| T28 | 1563 | 1538 | 15,253 | 1131 | 14,583 | ||
| T28 BlastS | 41,303 | 16,880 | 56,508 | 9936 | 163,516 | ||
| T42 | 1306 | 1835 | 14,864 | 656 | 17,490 | ||
| T42 BlastS | 24,629 | 27,894 | 37,829 | 12,352 | 199,618 | ||
| ∑BlastS-ØBlastS counts | 63,063 | 41,401 | 64,221 | 20,500 | 331,062 | ||
| T14 corrected ratio | 22.35 | 34.61 | 10.95 | 115.39 | 63.06 | ||
| Targeted sequencing validation | KD efficiency (remaining expression) | 38.73% | 18.78% | 26.84% | 19.44% | 48.82% | |
| Number of overexpressed targets (≥1 SD) | 15 | 10 | 16 | 11 | 10 | ||
| Mean targets relative expression | SD corrected | 4.35 | 2.73 | 6.03 | 1.67 | 2.85 | |
| Ratio | 2.10 | 1.93 | 3.21 | 1.99 | 2.55 | ||
| Mean controls relative expression | SD corrected | −1.07 | −1.13 | −0.76 | −1.04 | −0.15 | |
| Ratio | 0.78 | 0.71 | 0.77 | 0.76 | 0.93 | ||
| Student’s t test p value | Targets vs no targets | 0.00133 | 0.00127 | 0.00106 | 0.00129 | 0.00434 | |
| Targets SHC vs shRNA (paired) | 0.00669 | 0.01284 | 0.00281 | 0.02227 | 0.00564 | ||
T14, T28, and T42 represent the timepoints (days after transduction), KD, knockdown; SD, standard deviation.
ETV6-modulated transcription of t(12; 21)-associated genes
Leukemic cells from patients with t(12; 21) positive have distinct and clear expression profiles (Yeoh et al., 2002). This molecular signature can be, at least partly, explained by the absence of ETV6 expression as some of its target genes are specifically overexpressed in these cells (Neveu et al., 2016). Interestingly, nine of the 22 ETV6 target genes included in the validation process were identified as being specifically overexpressed in patients t(12; 21) positive (Neveu et al., 2016).
Unsupervised clustering analysis with the 9 t(12; 21) associated target genes showed that four of the 13 selected modulators, AKIRIN1, DYRK4, SRP72, and JUNB clustered with the pLENTI sample (Figure 5A). Of the five top modulators (Table 1), only COMMD9 showed a moderate impact on t(12; 21)-associated ETV6 targets (Figure 5A).
Figure 5.
Impact of the top ETV6 modulators on the transcription of t(12; 21)-associated overexpressed genes
(A) Heatmap representation of the normalized read counts of the 9 t(12; 21)-associated ETV6 targets (lanes) in the 13 significant shRNA samples (and control samples; columns).
(B) Relative expression of the five top ETV6 modulators after shRNAs transduction. Data are shown as mean ± SD, n = 6.
(C) Relative expression of the t(12; 21)-associated ETV6 target genes in both Reh WT and Reh ETV6 cells upon silencing of ETV6 modulators. Data are shown as mean ± SD, n = 6.
(D) SRP72 (left) and OSTalpha (right) expression in leukemic patient samples. The green dot represents the same patient. Lines represent mean ± SD.
(E) Impact of SRP72 on the ETV6 protein. ETV6 expression and localization (C: cytoplasm; N: nucleus) was evaluated by Western blotting in Reh ETV6 shSRP72 and SHC202 control cells. See also Figures S2 and S3, and Table S4.
We confirmed that the 9 t(12; 21)-associated ETV6 targets were downregulated in Reh ETV6 compared to Reh wild-type (WT) cells (Figure S2). We then silenced each of the top five ETV6 modulators in both cellular backgrounds (Figure 5B) and further evaluated their impact on gene expression (Figure 5C). The silencing of AKIRIN1, DYRK4, JUNB, and SRP72 upregulated most of the ETV6 targets, whereas COMMD9 knockdown showed less impact (Figure 5C). Of note, OSTalpha is the only ETV6 target to be significatively overexpressed by the silencing of all five modulators. Overexpression of the t(12; 21)-associated ETV6 target genes was mostly restricted to Reh ETV6 cells (Figure 5C, Reh ETV6 vs Reh WT), indicating that the observed upregulations are, indeed, owing to the impairment of ETV6-mediated repression.
We next investigated the expression status of the ETV6 modulators in childhood ALL patients. No clear association could be made between the expression of AKIRIN1, COMMD9, DYRK4, and JUNB and t(12; 21)-associated ETV6 targets (data not shown). However, the expression of SRP72 was significantly lower (Figure 5D, left) and, the expression of OSTalpha strikingly upregulated (Figure 5D, right) in one of the t(12; 21) negative samples. This result strengthens the role of SRP72 in ETV6 repressive function in patients with pre-B ALL. However, ETV6 protein level and localization remain unchanged after SRP72 silencing (Figure 5E), suggesting that SRP72 plays a role in ETV6-mediated repression through an alternate pathway.
Recently the ALL subtype ETV6-RUNX1-like has been identified (Lilljebjorn et al., 2016). It is characterized by expression profiles similar to the ETV6-RUNX1 subtype in absence of the defining ETV6-RUNX1 t(12; 21) rearrangement. Using a balanced set of patients’ RNA-seq data (n = 16) from the Mullighan study (Gu et al., 2019) we investigated the difference in expression between ETV6 modulators expression in both subtypes (Figure S3). Only DNAJC10 is differentially expressed in the two subtypes (Wilcoxon p value = 0.0011). However, the difference in expression between the two subtypes remains small.
Implications of ETV6 and its modulators in pre-T acute lymphoblastic leukemia
ETV6 transcriptional function is associated with childhood pre-B ALL (Bhojwani et al., 2015) but remains elusive in other cancers. We thus investigated ETV6-mediated transcription in pre-T ALL with particular attention to its newly identified modulators.
We measured the expression of known ETV6 targets in patients with pre-T ALL (Lajoie et al., 2017). Interestingly, one of them showed significant overexpression of CLIC5 (Figure 6A, arrow), a known ETV6 target gene strongly upregulated in patients with t(12; 21) positive pre-B ALL (Figure 6A) (Neveu et al., 2016). Strikingly, an ETV6 deletion was found in this patient (Spinella et al., 2016), suggesting that CLIC5 overexpression is attributed to ETV6 depletion. To test this, we knocked down ETV6 in the pre-T leukemic cell line MOLT4 and found that CLIC5 expression, but not that of other t(12; 21)-associated ETV6 targets, is significantly upregulated (Figure 6B, inset), demonstrating that CLIC5 gene is an ETV6 target in pre-T cells. We then evaluated the implication of the five top ETV6 modulators on CLIC5 expression. shRNA-mediated silencing was validated for each modulator (Figure S4) and knockdowns were equivalent to those obtained in Reh cells (Figure 5B). Remarkably, repression of all five ETV6 modulators induced a significant upregulation of CLIC5 in MOLT4 cells (Figure 6C) without any marked downregulation of ETV6 expression (Figure S4). These results strongly support the role of these genes as modulators of ETV6-mediated repression and suggest that ETV6 modulators are conserved in various cellular contexts.
Figure 6.
Top ETV6 modulators in childhood pre-T ALL
(A) Expression of CLIC5 in leukemic patient samples according to their molecular subtypes. The arrow indicates a pre-T ALL patient with upregulated CLIC5 expression. Expression in Reh cells is shown for reference (purple).
(B) qRT-PCR analysis of 9 t(12; 21)-associated ETV6 target genes after shRNA-mediated silencing of ETV6 in MOLT4 cells (inset). Two of these genes were not expressed (C6orf222 and DSC3; not shown). Data are shown as mean ± SD, n = 6.
(C) Relative expression of CLIC5 after the knockdown of ETV6 modulators in MOLT4 cells. Data are shown as mean ± SD, n = 6. See also Figure S4.
Discussion
ETV6 loss of function is a key event in the development of hematological malignancies (Moriyama et al., 2015; Noetzli et al., 2015; Topka et al., 2015; Zhang et al., 2015; Bohlander, 2005). To better understand the ETV6-mediated repression process we conducted a functional genome-wide shRNA screen. We uncovered 13 novel putative ETV6 modulator genes Including AKIRIN1, COMMD9, DYRK4, JUNB, and SRP72 that demonstrated a broad impact on the ETV6 transcriptional network in leukemic cells. Alterations in such modulators could disrupt ETV6 function and participate in hematological diseases.
While the mechanisms of action of these regulators in ETV6-mediated transcription remain to be established, several lines of evidence regarding their role in transcriptional regulation are worth mentioning. AKIRIN1 impact on gene expression was highly restricted to ETV6 targets, either suggesting an alternative function in human cells or an ETV6-specific chromatin remodeling activity. AKIRIN1 function in human cells remains largely unknown, but it had been found to participate in transcription-associated chromatin remodeling in Drosophila melanogaster (Nowak et al., 2012; Bonnay et al., 2014). COMMD9 was recently associated with TFDP1/E2F1 transcriptional activity in lung cancer (Zhan et al., 2017), suggesting a role in transcription regulation. Moreover, two other COMMD genes, COMMD8 and COMMD10, were also retrieved from the genome-wide screen with a single shRNA, suggesting that this gene family could be implicated in ETV6 function. This gene family is known to be involved in NF-κB-mediated regulation of transcription (Bartuzi, et al., 2013). The most significant overexpression of ETV6 target genes was observed when DYRK4 was knocked down. DYRK4 is the least studied member of the serine/threonine dual-specificity kinase family (Papadopoulos et al., 2011; Becker et al., 1998) and no role in transcriptional regulation had been associated with this gene so far. Notably, another member of this family, DYRK3, was also identified in the primary shRNA screen. DYRK3 phosphorylates histone H2B (Becker et al., 1998) and SIRT1, a protein deacetylase that targets histones and other transcription factors such as E2F1 and NF-κB (Guo et al., 2010). This suggests that SIRT-mediated histone deacetylation could contribute to ETV6 gene repression. This is also supported by the capture of shRNAs against SIRT3 and SIRT4 in the primary screen. ETV6 phosphorylation is known to modulate its transcriptional function (Lopez et al., 2003; Poirel et al., 1997; Arai et al., 2002). Thus, DYRK3 and DYRK4 could be involved in this process. Determining whether these kinases directly phosphorylate ETV6, its modulators or participate in ETV6-mediated repression through histones modifications would be of great interest because of their substantial impact on ETV6 function. JUNB, a subunit of the AP-1 transcription factor, is an ETV6 modulator that was studied in the context of multiple lymphoid malignancies such as lymphomas and T-cell leukemias (Papoudou-Bai et al., 2016). FOSL2, another subunit of AP-1, was also captured on the screen with a single shRNA, suggesting that AP-1 participates in the ETV6 function. Although ETV6 binding to AP-1 was never tested, physical interactions between ETS and AP-1 factors have been described (Bassuk and Leiden, 1995; Verger and Duterque-Coquillaud, 2002). These interactions were dependent on the conserved ETS domain, suggesting that similar interactions could occur with ETV6 and modulate its transcriptional activity.
In regard to hematological diseases, SRP72 is an interesting ETV6 modulator. One of our t(12; 21) negative patient samples showed a 50% reduction in SRP72 expression compared to other patients, suggesting a monoallelic expression. Expression of the ETV6 target OSTalpha was also significantly higher in this leukemia patient, further supporting that SRP72 downregulation impacts ETV6 repressive function. Mutations in SRP72 have been linked to familial Aplastic Anemia (AA) and Myelodysplasia (MDS) (Kirwan et al., 2012), both of which are also associated with ETV6 mutations or deletions (Topka et al., 2015). The mechanism(s) through which SRP72 mutations induce these familial forms of aplasia remains unknown but, considering our findings, could involve the deregulation of ETV6 target genes. In addition to SRP72, SRP14 was also found with a single shRNA in the genome-wide screen. Furthermore, SRP9 was identified as an ETV6-bound protein by a proteomic approach (Table S4). Altogether, these results suggest that a complex containing SRP proteins could bind ETV6 to regulate its transcriptional output. SRP proteins’ canonical function is to form a complex that binds nascent peptide signals, induce translational arrest, and transfer the ribosomes to the translocon onto the ER where the translation would resume (Akopian et al., 2013). However, we show that SRP72 silencing does not affect ETV6 protein level and localization, suggesting an alternate pathway in ETV6-mediated repression.
We showed that ETV6 targets were not equally affected by these modulators. For instance, DSC3, a gene significantly repressed by ETV6 and overexpressed in patients with t(12; 21) positive leukemic, was not significantly influenced by ETV6 modulators while OSTalpha, another ETV6 target was regulated by nine of the 13 modulators. ETV6 association with certain modulators could be important to regulate specific transcriptional responses following a given signal. For instance, SEMA3A and SEMA3D, two members of the Semaphorins Class III gene family, are both regulated by ETV6 and the modulators. The specificity given to ETV6 by its modulators could ensure that genes belonging to a particular pathway are adequately co-expressed. This suggests that ETV6 target genes associated with one or more modulators contribute to a common pathway.
Aside from ETV6 modulators and the chromatin context, other factors might contribute to ETV6 specificity. For instance, the ETV6-AML1 fusion protein is known to interact with residual ETV6, modulate its binding, and dictate specificity (Gunji et al., 2004). In this regard, the expression of NTRK1, one of the ETV6 targets was increased rather than downregulated following the silencing of ETV6 modulators. Interestingly, NTRK1 does not contain the ETV6 consensus binding site(Neveu et al., 2018), suggesting that ETV6 acts as a co-repressor at this locus. Thus, ETV6-AML1 or other repressive ETS factors (Mavrothalassitis and Ghysdael, 2000) might endogenously repress NTRK1. Because ETV6-AML1 retains both the PD and repression domains of ETV6 and the repressive ETS factors share a similar DNA-binding domain (Kar and Gutierrez-Hartmann, 2013), the identified modulators could affect their repressive function as well, explaining NTRK1 upregulation following the silencing of ETV6 modulators. This is of great interest as ETV6 is involved in multiple translocations with various fusion partners (De Braekeleer et al., 2012) and that ETS factors are widely involved in human malignancies (Ciau-Uitz et al., 2013; Kar and Gutierrez-Hartmann, 2013; Feng et al., 2014; Sizemore et al., 2017). Additional experiments are needed to determine the implication of the novel ETV6 modulators on other related proteins. In this study, ETV6 repressive function has been interrogated through a specifically designed functional genome-wide shRNA screen. The individual assessment of a large number of putative modulators and their impact on the ETV6 transcriptional network unveiled the complexity of ETV6-mediated repression. Thirteen novel putative modulators of ETV6 activity were identified and associated with the regulation of ETV6 target genes in various contexts. Most of the modulators were involved in a target-specific manner, but some had a broader impact on the ETV6 transcriptional network. The critical role of AKIRIN1, COMMD9, DYRK4, JUNB, and SRP72 on ETV6-mediated regulation makes them interesting genes regarding the etiology of hematological diseases. Overall, this genome-wide functional screen identified important ETV6 modulators and linked them to the ETV6 transcriptional network.
Limitation of the study
This study focuses on the development and analyses of a genome-wide shRNA screen to identify ETV6 modulators. The screen is a fitness assay taking advantage of a specifically developed ETV6-dependent Blasticidin sensitive cell line. The validation of the modulator was achieved in cellular models but the specific roles of the modulators in ALL as well as in other cancers would need further investigation using patients' samples. The ETV6-RUNX1-like subtype has a similar expression profile to the ETV6-RUNX1 subtype, without the characteristic t(12; 21) translocation, it would thus be interesting to investigate modulators in this subtype. Although our analysis did not reveal a drastic change in the expression levels of the modulators between the two subtypes (Figure S3), a more in-depth analysis accounting for residual ETV6 expression might reveal further insight on their functions.
STAR★Methods
Key resources table
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-ETV6 / Tel antibody | Abcam | Cat# ab54705; RRID: AB_941491 |
| GAPDH Antibody (L-20) | Santa Cruz Biotechnology | Cat# sc-31915; RRID: AB_641105 |
| Anti-β-Actin Antibody (ACTBD11B7) | Santa Cruz Biotechnology | Cat# sc-81178; RRID: AB_2223230 |
| Recombinant Anti-PCNA antibody [EPR3821] | Abcam | Cat# ab92552; RRID: AB_10561973 |
| Bacterial and virus strains | ||
| Mission TRC shRNA whole library | Sigma-Aldrich | CP0001 55K Human Pool + CP0003 90K Human Pool |
| Critical commercial assays | ||
| Pierce™ Anti-HA Magnetic Beads | Thermo Fisher | Cat# 88836; RRID: AB_2861399 |
| Deposited data | ||
| RNA-seq data | (Neveu et al., 2016) | GEO: GSE79373 |
| ChIP-seq data | (Neveu et al., 2018) | GEO: GSE102785 |
| LC-MS/MS data | This study | ProteomeXchange: PXD031001 |
| Experimental models: Cell lines | ||
| REH | ATCC | Cat# CRL-8286; RRID: CVCL_1650 |
| HEK293T | ATCC | Cat# CRL-11268; RRID: CVCL_0063 |
| Reh BlastR 2-1 | This study | N/A |
| 2-1 ETV6-5 | This study | N/A |
| MOLT4 | ATCC | Cat# CRL-1582; RRID: CVCL_001 |
| Oligonucleotides | ||
| All primer sequences are listed in Table S5 | ||
| Recombinant DNA | ||
| pLENTI CMV TetR Blast | Addgene | Cat# 17492; RRID: Addgene_17492 |
| pENTR3C | Invitrogen | 11817-012 |
| pGL4 EBS3tk BlastR | This study | N/A |
| pGL3 EBS3tk | This study | N/A |
| pLENTI CMV puro DEST (w118-1) | Dr. Christian Beauséjour, CHU Sainte-Justine, Montreal, QC, Canada | Addgene #17452; RRID: Addgene_17452 |
| pLENTI ETV6 | This study | N/A |
| pcDNA3.1 ETV6 | (Neveu et al., 2018) | N/A |
| pCCL ETV6-HA | (Neveu et al., 2018) | N/A |
| pCCL ETV6 | (Neveu et al., 2018) | N/A |
| Software and algorithms | ||
| GraphPad Prism 5 | GraphPad Software | https://www.graphpad.com/ RRID:SCR_002798 |
| Cufflinks tool version 2.2.1 | (Trapnell et al., 2010) |
https://github.com/cole-trapnell-lab/cufflinks RRID:SCR_014597 |
| Bowtie2 | (Langmead and Salzberg, 2012) |
https://github.com/BenLangmead/bowtie2 RRID:SCR_016368 |
| BEDTools version 2.22.1. | (Quinlan and Hall, 2010) |
https://github.com/arq5x/bedtools2 RRID:SCR_006646 |
| RTCGAToolbox v.2.0.0 | (Samur, 2014) | https://bioconductor.org/packages/release/bioc/html/RTCGAToolbox.html |
| Mascot version 2.5.1 | Matrix Science | https://www.matrixscience.com/ RRID:SCR_014322 |
| Scaffold version 5.0.1 | Proteome Software Inc. |
https://www.proteomesoftware.com/products/scaffold-5 RRID:SCR_014345 |
Resource availability
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Daniel Sinnett (daniel.sinnett@umontreal.ca).
Materials availability
All requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Daniel Sinnett (daniel.sinnett@umontreal.ca). All reagents will be made available on request after completion of a Materials Transfer Agreement.
Experimental model and subject details
Cell lines
The Reh (ATCC, Manassas, VA, USA, CRL-8286) cell line and derived clones were maintained in RPMI 1640 (Wisent, Saint-Bruno, QC, Canada) 10% Fetal Bovine Serum (FBS; Wisent, Saint-Bruno, QC, Canada) in a 5% CO2 incubator at 37°C. The MOLT4 (ATCC, CRL-1582) cell line and derived clones were maintained in RPMI 1640 (Wisent, Saint-Bruno, QC, Canada) 10% Fetal Bovine Serum (FBS; Wisent, Saint-Bruno, QC, Canada) in a 5% CO2 incubator at 37°C.
Method details
Establishment and control of the screening cell line
The Reh (ATCC, Manassas, VA, USA, CRL-8286) cell line and derived clones were maintained in RPMI 1640 (Wisent, Saint-Bruno, QC, Canada) 10% Fetal Bovine Serum (FBS; Wisent, Saint-Bruno, QC, Canada) in a 5% CO2 incubator at 37°C. Reh cells were transfected with pGL4 EBS3tk BlastR using the Lipofectamine 2000 reagent (Thermo Fisher Scientific, Waltham, MA, USA). Media was changed the next day and 2 μg/mL Blasticidin was added 48h post-transfection for selection. Cells were maintained for a month under Blasticidin selective pressure to retrieve cells with a stable integration. Using 6-wells plates, 500 cells from this Blasticidin resistant population were seeded in 1 mL of MethoCult H4230 media (STEMCELL Technologies, Vancouver, BC, Canada) containing 2 μg/mL Blasticidin and grown for an additional month to obtain distinct colonies (clones). Colonies were transferred and amplified in 96-wells liquid cultures with 2 μg/mL Blasticidin. Blasticidin resistant clones were then transduced with pLENTI ETV6 or pLENTI empty vector lentiviral particles. Cells were selected with 1 μg/mL puromycin before assessing ETV6 and BlastR expression by quantitative real-time PCR (qRT-PCR; see related section). The selected clone (Reh EBS3tk BlastR 2-1) was submitted to an additional round of clonal selection in methylcellulose media (1 μg/mL puromycin) to ensure a strict and homogenous cellular system with single EBS3tk BlastR and ETV6 integrations (i.e. Reh EBS3tk BlastR 2-1 ETV6-5).
A GFP selectable pLKO plasmid containing a shRNA against ETV6 (shETV6) was used for lentiviral production and transduction of the ETV6-dependent Blasticidin sensitive clone 2-1 ETV6-5. A scrambled shRNA sequence (shCtlETV6) was also included as a negative control. GFP positive cells were retrieved by flow cytometry using the FACSAria IIu cell sorter (Becton Dickinson, Franklin Lakes, NJ, USA) and stabilized in culture for several days before validating the ETV6 knockdown by Western Blotting and qRT-PCR (see related sections). GFP positive cells were seeded at an initial concentration of 1 × 105 cells/mL and treated with a range of Blasticidin doses (5,10 and 25 μg/mL). Cells were then cultured for 12 days in these conditions and proliferation was evaluated with the Z1 Particle Counter (Beckman Coulter, Brea, CA, USA). Alternatively, both the original Reh EBS3tk BlastR 2-1 clone and the ETV6 expressing 2-1 ETV6-5 clone were treated with Nicotinamide (NAM; Sigma-Aldrich, Saint Louis, MO, USA) at a final concentration of 20 mM for 72h. BlastR expression was then evaluated by qRT-PCR (see related section).
Recombinant DNA
pGL4 EBS3tk BlastR plasmid was constructed by cloning PCR products of the Blasticidin résistance gene (BlastR) from pLENTI CMV TetR Blast (716-1) plasmid (Addgene, Watertown, MA, USA) between the NcoI and Xba restriction sites and the EBS3tk artificial promoter from the pGL3 EBS3tk vector between the KpnI and NheI restriction sites.
The complete ETV6 complementary DNA (cDNA) sequence (NM_001987) obtained from pcDNA3.1 ETV6 (Neveu et al., 2018) was subcloned into the Gateway compatible vector pENTR 3C and transferred into the pLENTI CMV puro DEST (w118-1) lentiviral vector (kindly provided by Dr. Christian Beauséjour) using LR clonase II reactions according to the manufacturer protocol (Thermo Fisher Scientific, Waltham, MA, USA).
Lentiviral production and transduction
Production of lentiviral particles and cell transduction were performed as previously described (Neveu et al., 2016). Briefly, HEK293T cells (ATCC, CRL-11268) were transfected with the lentiviral expression plasmid (pLENTI backbone) together with the third generation encapsidation plasmids, pRSV-Rev, pMD2.VSVG and pMDL (kindly provided by Dr. Christian Beauséjour) using Polyethylenimine (Polysciences). Media was renewed 16h post-transfection and cells were cultured for 30h. Viral particles from culture media were concentrated by ultracentrifugation (50 000 g) and estimated using the p24 antigen ELISA (Advanced Bioscience Laboratories). The transduction was performed by adding these particles (10-100 ng per 1 × 105 cells) and 8 μg/mL polybrene (Sigma-Aldrich, Saint Louis, MO, USA) to cells for 24h. Media was changed and the selection agent (i.e. 1 μg/mL puromycin) was added 48h post-transduction. Cells were cultured for at least 2 weeks prior to any experiments. (refer to the ''shRNAs preparation'' section for the pLKO shRNAs particles.)
Establishment and control of the screening cell line
The Reh (ATCC, Manassas, VA, USA, CRL-8286) cell line and derived clones were maintained in RPMI 1640 (Wisent, Saint-Bruno, QC, Canada) 10% Fetal Bovine Serum (FBS; Wisent, Saint-Bruno, QC, Canada) in a 5% CO2 incubator at 37°C. Reh cells were transfected with pGL4 EBS3tk BlastR using the Lipofectamine 2000 reagent (Thermo Fisher Scientific, Waltham, MA, USA). Media was changed the next day and 2 μg/mL Blasticidin was added 48h post-transfection for selection. Cells were maintained for a month under Blasticidin selective pressure to retrieve cells with a stable integration. Using 6-wells plates, 500 cells from this Blasticidin resistant population were seeded in 1 mL of MethoCult H4230 media (STEMCELL Technologies, Vancouver, BC, Canada) containing 2 μg/mL Blasticidin and grown for an additional month to obtain distinct colonies (clones). Colonies were transferred and amplified in 96-wells liquid cultures with 2 μg/mL Blasticidin. Blasticidin resistant clones were then transduced with pLENTI ETV6 or pLENTI empty vector lentiviral particles. Cells were selected with 1 μg/mL puromycin before assessing ETV6 and BlastR expression by quantitative real-time PCR (qRT-PCR; see related section). The selected clone (Reh EBS3tk BlastR 2-1) was submitted to an additional round of clonal selection in methylcellulose media (1 μg/mL puromycin) to ensure a strict and homogenous cellular system with single EBS3tk BlastR and ETV6 integrations (i.e. Reh EBS3tk BlastR 2-1 ETV6-5).
A GFP selectable pLKO plasmid containing a shRNA against ETV6 (shETV6) was used for lentiviral production and transduction of the ETV6-dependent Blasticidin sensitive clone 2-1 ETV6-5. A scrambled shRNA sequence (shCtlETV6) was also included as a negative control. GFP positive cells were retrieved by flow cytometry using the FACSAria IIu cell sorter (Becton Dickinson, Franklin Lakes, NJ, USA) and stabilized in culture for several days before validating the ETV6 knockdown by Western Blotting and qRT-PCR (see related sections). GFP positive cells were seeded at an initial concentration of 1 × 105 cells/mL and treated with a range of Blasticidin doses (5,10 and 25 μg/mL). Cells were then cultured for 12 days in these conditions and proliferation was evaluated with the Z1 Particle Counter (Beckman Coulter, Brea, CA, USA). Alternatively, both the original Reh EBS3tk BlastR 2-1 clone and the ETV6 expressing 2-1 ETV6-5 clone were treated with Nicotinamide (NAM; Sigma-Aldrich, Saint Louis, MO, USA) at a final concentration of 20 mM for 72h. BlastR expression was then evaluated by qRT-PCR (see related section).
shRNAs library
Lentiviral particles were prepared from the Mission TRC shRNA whole library (Sigma-Aldrich, Saint Louis, MO, USA) according to the manufacturer’s protocol. Briefly, the library was divided in 15 pools each containing approximately 9600 different shRNAs (10 96-well plates/pool). For lentiviral production, HEK293T cells were transfected (FuGENE 6, Promega, Madison, WI, USA) with the shRNA plasmid pools, psPAX2 (Addgene, Watertown, MA, USA) and pMD2.G (Addgene, Watertown, MA, USA) at a ratio of 1/0.75/0.25. Cell culture media were harvested and filtered (0.45 μm filter) 48 h post-transfection. The lentiviral titer of each pool was evaluated by limiting dilution using NIH3T3 cells.
The multiplicity of infection (MOI) of each shRNAs pool was evaluated by transducing Reh cells with various dilutions of viral particles using 5 μg/mL polybrene. The next day, media was replaced, and cells were selected with 1 μg/mL puromycin for 48h. Cell viability was assessed by propidium iodide (PI) staining (final concentration of 0.2 μg/mL in PBS). 1 × 104 stained cells were analyzed by flow cytometry on a LSRFortessa FACS (Becton Dickinson, Franklin Lakes, NJ, USA). Data was acquired with the FACSDiva software (Becton Dickinson, Franklin Lakes, NJ, USA).
For individual shRNAs, HEK293T cells were transfected with the pLKO shRNA construct together with the third generation encapsidation plasmids, pRSV-Rev, pMD2.VSVG and pMDL using Lipofectamine 2000 (Thermo Fisher Scientific, Waltham, MA, USA). Media was renewed 16h post-transfection and harvested after an additional 30h of culture. Viral particles were used directly for transduction.
Genome wide shRNA screening
1 × 107 ETV6-dependent Blasticidin sensitive 2-1 ETV6-5 cells were transduced with each of the 15 shRNAs pools at a MOI of 0.3-0.4 (determined for each pool; see ''shRNAs preparation'' section) using 5 μg/mL polybrene. In these conditions, each unique shRNA is expected to be integrated by approximately 200 cells. Media was changed the next day and replaced with fresh media containing 1 μg/mL puromycin. Cells were maintained in culture for 14 days to allow knockdowns to modulate ETV6 function and BlastR expression. Each pool of cells was then split into 3 parts: 1 × 107 cells were pelleted and used as the initial time point of the experiment (T14), 2 × 107 cells were maintained as is (no Blasticidin), and 5 × 107 cells were treated with 50 μg/mL Blasticidin. After 14 days, 1 × 107 cells of each shRNAs pool (with and without Blasticidin selection) were pelleted and used as the first time point of selection (T28). Cells were maintained for an additional 14 days in culture in these conditions and pellets were taken for each shRNA pool and used as the final time point of selection (T42).
Genomic DNA (gDNA) was extracted from each sample using the QIAamp DNA Mini Kit according to the manufacturer's protocol (Qiagen, Hilden, Germany). Quantification was performed on Qubit using the Quant-it PicoGreen dsDNA Kit (Broad range; Thermo Fisher Scientific, Waltham, MA, USA). 6 μg of gDNA was used as template for the first PCR with primers flanking the integrated shRNA sequences and containing Illumina-compatible overhang extremities: F- TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCGTAACTTGAAAGTATTTCGATTTCTTGGCTTTATATATCTTGTGG; R- GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGTTGTGGATGAATACTGCCATTTGTCTC. To evaluate the sensitivity of the methodology, duplicates of T14 samples were supplemented with respectively 20 and 200 copies of SHC002 and SHC202 lentiviral vectors prior to the first PCR. Technical duplicates for each T14 samples also allowed the assessment of the reproducibility of the PCR and sequencing methods. For each sample, 12 amplification reactions of 50 μL were performed as follow: gDNA: 10 ng/uL, primers: 0.25 μM each, dNTPs: 0.3 mM and 1U KAPA HiFi DNA polymerase (KAPA Biosystems, Wilmington, MA, USA) in 1X KAPA reaction buffer. Thermocycling method: 95°C, 4 min; [98°C, 20 sec; 70°C, 15 sec; 72°C, 1 min] X 15 cycles; 72°C, 5 min. PCR reactions from each gDNA sample were pooled and 90 μL were purified with 1.8X volume AMPure XP magnetic beads (Beckman Coulter, Brea, CA, USA) and eluted in 30 μL water. The second amplification reaction used Nextera XT index Kit v2 primers (Illumina, San Diego, CA, USA) and was performed in 50 μL as follow: purified PCR#1: 30 μL, primers: 5 μL each, dNTPs: 0.3 mM and 1U KAPA HiFi DNA polymerase in 1X KAPA reaction buffer. Thermocycling method: 95°C, 4 min; [98°C, 20 sec; 68°C, 30 sec; 72°C, 30 sec] X 12 cycles; 72°C, 5 min. Reactions were once again purified with AMPure XP magnetic beads (as above). Amplicons size was confirmed by Bioanalyser on a DNA 1000 chip (Agilent Technologies, Santa Clara, CA, USA) and libraries were sequenced using the HiSeq 2500 system (paired-end 100 b. p.; Illumina, San Diego, CA, USA) with 9 × 106 reads per library.
Data analysis
Reads from each sample were aligned on a custom reference containing all the target sequences of the MISSION Human shRNA library using the Bowtie2 alignment tool version 2.2.3 (Langmead and Salzberg, 2012). For a given shRNAs pool and for each condition (T14, T28 with and without Blasticidin, T42 with and without Blasticidin), read counts per shRNA were obtained using BEDTools version 2.22.1 (Quinlan and Hall, 2010). Note that for T14 samples, the technical duplicates were merged at this point. Every shRNA counts in a sample were normalized per the total number of reads in that sample to reflect their relative abundance. shRNAs with no coverage (0) in at least one of the time points were removed from further analyses. To be considered a positive hit, a shRNA had to have a greater normalized count in the Blasticidin condition compared to the untreated control condition at both time points (T28 and T42). The normalized count also had to be greater at the end of the experiment (T42 with Blasticidin) compared to the initial count at T14. These positive shRNA hits were further refined using additional thresholds. We filtered out weakly over-represented shRNAs (∑Counts(BlastS - ØBlastS) is less than 2 standard deviation of SHC202 control vector; ≤ 9,770;) as well as shRNAs which target non-expressed genes (Hart et al., 2013) in Reh cells (Neveu et al., 2016).
Validation of candidate shRNAs by targeted RNA sequencing
The shRNAs selected for validation (Table S2) and two negative controls from the MISSION library (SHC002 and SHC202) were used individually to produce lentiviral particles and transduce Reh ETV6 cells (as described in the '' shRNAs preparation'' section). All shRNA particles were used at a 1/10 to 1/20 dilution to reach a MOI >1. All shRNA cell lines were maintained with 1 μg/mL puromycin for 2 weeks.
Total RNA was extracted from these shRNA populations using the RNeasy Mini kit (Qiagen, Hilden, Germany). Two biological replicates of Reh ETV6 cells and one of Reh pLENTI were also included as additional controls. 50 ng of RNA was then processed through the TruSeq Targeted RNA expression kit according to the manufacturer's protocol (Illumina, San Diego, CA, USA). Amplicons included ETV6 target genes (Neveu et al., 2016), non-ETV6 target genes (apoptosis panel; Illumina), genes targeted by shRNAs (Table S2) and housekeeping genes (Eisenberg and Levanon, 2013). Amplicons size was confirmed by Bioanalyser on a DNA 1000 chip (Agilent Technologies, Santa Clara, CA, USA). Amplicons were sequenced on a HiSeq 2500 system (paired-end 100 b.p.; Illumina, San Diego, CA, USA) with 1.8 × 106 reads per sample. Mapping was performed using the Illumina BaseSpace SequenceHub Informatics suite and raw counts were obtained and further processed.
Targeted RNA sequencing analysis
Raw counts for each gene in a sample were first normalized per the number of total reads in that sample. Genes considered very weakly expressed (mean normalised counts < 10 and/or normalised pLENTI counts < 10) were discarded from the analyses. Expression in each sample was further normalised using 12 housekeeping genes (Eisenberg and Levanon, 2013). For each gene, the relative expression in each shRNA sample was calculated compared to the SHC002 and SHC202 controls by including the variability of the 4 ETV6 replicates: ((shRNA sample count - SHC002 and SHC202 mean count) / standard deviation of the 4 ETV6 controls). The relative expression of a gene was also calculated as a simple ratio against the SHC002 and SHC202 controls: (shRNA sample count / SHC002 and SHC202 mean count). Using standard deviation-corrected relative expressions, shRNAs inducing a specific re-expression of validated ETV6 targets, but not ETV6 independent genes, according to a Welch's corrected Student t test, were considered positives. Knockdown efficiency for each shRNA sample was calculated similarly. Heatmaps were generated with the pheatmap R package.
Protein extraction
1 × 107 cells were washed in PBS and disrupted for 15 min on ice in a hypotonic cytoplasm extraction buffer (CEB; 10 mM HEPES pH 7.5, 10mM KCl, 1X Protease Inhibitors Set III Animal-free (Calbiochem, San Diego, CA, USA); 100 μL/1 × 107 cells). NP-40 was added to a final concentration of 0.05% prior to centrifugation. Supernatant containing the cytoplasmic proteins was collected and the nuclei pellet was washed in CEB buffer. Nuclei were then disrupted in a hypertonic nuclear extraction buffer (NEB; 20 mM HEPES pH 7.5, 50 mM KCl, 300 mM NaCl, 5% Glycerol, 0.05% NP-40, 1X Protease Inhibitors Set III (with EDTA); 100 μL/1 × 107 cells) for 1 hour with agitation at 4°C. The supernatant obtained after centrifugation contained soluble nuclear proteins.
Western blotting
Protein immunodetection was performed as previously described (Neveu et al., 2016). Briefly, 20 μg of protein samples were diluted in Laemmli buffer and migrated on SDS-denaturating 10% polyacrylamide gels. Transfer was performed overnight at 4°C on polyvinylidene difluoride membranes. Membranes were then blocked in a milk solution prior to immunoblotting using the following antibodies anti-ETV6 (ab54705, Abcam, Cambridge, UK), anti- GAPDH (sc-31915, Santa Cruz Biotechnology, Dallas, TX, USA), Anti-β-Actin (sc-81178, Santa Cruz Biotechnology, Dallas, TX, USA), Anti-PCNA (ab92552, Abcam, Cambridge, UK). Immunodetection was performed by enhanced chemiluminescence with Western Lightning Plus-ECL (PerkinElmer, Waltham,MA, USA) according to the manufacturer's protocol.
Mass spectrometry
Pre-washed anti-HA magnetic beads (Thermo Fisher Scientific, Waltham, MA, USA) were added directly to nuclear and cytoplasm protein extracts from 5× 107 Reh pCCL ETV6 and ETV6-HA cells. After 4 hours of incubation with agitation at 4°C, beads were washed on a magnetic stand using 50mM ammonium bicarbonate. The on-beads digest and mass spectrometry experiments were performed by the Proteomics platform of the CHU de Quebec Research Center, Quebec, Canada.
Briefly, proteins on beads were washed 3 times with 50mM ammonium bicarbonate buffer and digested with Trypsin (1μg) 5 hours at 37°C then desalted on stage tip (C18) and vacuum dried before MS injection.
Peptide samples were separated by LC-MS/MS on an Ekspert NanoLC425 (Eksigent) coupled to a 5600+ mass spectrometer (AB Sciex, Framingham, MA,USA) equipped with a nanoelectrospray ion source. Peptides were separated with a linear gradient from 5-35% solvent B (acetonitrile, 0.1% formic acid) in 35 minutes, at 300 nL/min on a picofrit columns (Reprosil 3u, 120A C18, 15 cm × 0.075 mm internal diameter). Mass spectra were acquired using a data dependent acquisition mode using Analyst software version 1.7. Each full scan mass spectrum (400 to 1250 m/z) was followed by collision-induced dissociation of the twenty most intense ions. Dynamic exclusion was set for a period of 3 sec and a tolerance of 100 ppm
Database searching
MGF peak list files were created using Protein Pilot version 5.0 software (Sciex) then searched was performed using Mascot (Matrix Science, London, UK; version 2.5.1) on the CP_HomoSapiens_9606 database (92237 entries) assuming the digestion enzyme trypsin. Mascot was searched with a fragment ion mass tolerance of 0,100 Da and a parent ion tolerance of 0,100 Da. Deamidation of asparagine and glutamine and oxidation of methionine were specified in Mascot as variable modifications.
Criteria for protein identification
Scaffold (version Scaffold_5.0.1, Proteome Software Inc., Portland, OR) was used to validate MS/MS based peptide and protein identifications. Peptide and protein identifications were filtered with a FDR (false discovery rate) less than 1,0 % by the Scaffold Local FDR algorithm. Proteins that contained similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony.
qRT-PCR
Total RNA was extracted from cells using the RNeasy Mini kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol and quantified by Nanodrop (Thermo Fisher Scientific, Waltham,MA, USA). 750 ng of RNA were used for reverse transcription using the M-MLV reverse transcriptase (Life Technologies, Carlsbad, CA, USA). These cDNAs were then subjected to quantitative real-time PCR. Reactions were performed in triplicates using the SYBR Green PCR Master Mix (Life technologies, Carlsbad, CA, USA) and the primers listed in Table S5 on the ABI PRISM 7000 sequence detection system (Life Technologies, Carlsbad, CA, USA). Relative expression was calculated by the 2-(ΔΔCt) comparative method (Livak and Schmittgen, 2001) using GAPDH and β2M as reference genes.
Transcriptome analysis of cancer patients
Transcriptome data from childhood ALL patients and controls were obtained from a previous study (Lajoie et al., 2017). Additional childhood B-ALL patients (11 t(12;21) positive and 14 t(12;21) negative) and controls (3 CD19+CD10+ pre-B cells isolated from healthy cord blood samples) were also included. Briefly, white cells from bone marrow samples were enriched with Ficoll-Paque PLUS (GE Healthcare Life Sciences, Piscataway, NJ, USA). DNA and RNA from these cells were then extracted using the Allprep DNA/RNA mini kit (Qiagen, Hilden, Germany). 1 μg of RNA with RIN ranging from 5 to 10 were treated with DNAse (Ambion, Austin, TX, USA). Libraries were generated using the Illumina TruSeq Stranded Total RNA rRNA removal GOLD kit according to the manufacturer protocol. Sequencing was performed on the HiSeq 2500 system (paired-end 100 b.p.; Illumina, San Diego, CA, USA) with 8 × 107 expected reads per sample. The pipeline to produce the RNAseq binary alignment files (BAM) is based on the GATK Best Practice for germline SNPs and Indels in RNAseq. The gene expression values (FPKM) were calculated using the Cufflinks tool version 2.2.1 (Trapnell et al., 2010) on hard-clipped BAM files.
TCGA normalized expression values (rundate '20160128') were obtained using the R package RTCGAToolbox v.2.0.0 (Samur, 2014). For each cancer, a log fold change was calculated for each gene using their median expression in the top and bottom 10% of ETV6 expressing samples. 20 genes with the most negative fold changes were retained for further analysis. Additionally, 20 random genes with log fold changes near 0 (over -0.001 and under 0.001) were used as control genes uncorrelated to ETV6 expression. Log fold changes for those cancer-specific ETV6-correlated and non-correlated genes as well as for known ETV6 target genes were calculated similarly with the top and bottom 10% of samples according to the expression of each modulator.
Quantification and statistical
Data management and statistical analysis were performed using Excel (Microsoft, Redmond, WA, USA) and GraphPad Prism 5 software (GraphPad Software, San Diego, CA, USA). Data are reported as mean ± standard deviation (SD). Wilcoxon and Student t-test and were used to statistically compare two groups (as noted in the figure legend) and significance was set to p < 0.05 (∗∗∗p < 0.001, ∗∗p < 0.01, ∗p < 0.05).
Acknowledgments
Next-generation sequencing was performed at the Integrated Clinical Genomic Center in Pediatrics at the CHU Sainte-Justine Research Center. LC-MS/MS samples were run and analyzed at the Proteomics Platform of the CHU de Quebec Research Center, Quebec, Canada. Computations were performed on the Briarée supercomputer at the Université de Montréal, managed by Calcul Québec and Compute Canada. The operation of this supercomputer is funded by the Canada Foundation for Innovation (CFI), NanoQuébec, RMGA, and the Fonds de recherche du Québec - Nature et technologies (FRQNT). This study was supported by research funds provided by the Terry Fox Research Institute and the Canadian Institutes of Health Research. BN is the recipient of a Cole Foundation scholarship. DS holds the François-Karl Viau Research Chair in Pediatric Oncogenomics.
Author contributions
DS and SG are co-principal investigators and share primary responsibility for the article. BN, CR, SG, and DS contributed to the conception and design of the study. BN, CR, and PC performed most of the experiments. MC analyzed the patients and TCGA expression data. CJC generated the data from the MOLT4 cell line. PSO was involved in the processing of the sequencing data. CF performed some of the Western blots. NG contributed to setting up the genome-wide shRNA screening approach. BN performed data integration and analyses. BN, CR, SG, and DS contributed to data interpretation. BN drafted the article and SG and DS were involved in the critical revision of the article. All authors approved the final version.
Declaration of interests
The authors declare no competing interests.
B.N. current affiliation: Molecular Diagnostic Laboratory, Montreal Heart Institute, Montreal, Quebec, Canada.
P.C. current affiliations: Program in Infectious Diseases and Global Health, The Research Institute of the McGill University Health Center, Montréal, Quebec, Canada, and McGill International TB Center, Department of Medicine, Faculty of Medicine, McGill University, Montreal, Quebec, Canada.
P.S.O. current affiliation: Center for Integration and Analysis of Medical Data (CITADEL), Department of Medicine, University of Montreal, Center de Recherche du Center Hospitalier de l'Université de Montréal, Montreal, Canada.
N.G current affiliation: Patient Advocacy Lead, Global Product Development, Pfizer Rare Disease, Philadelphia, PA, USA.
D.S is also Scientific Director of CIUSSS du Nord-de-l’Île-de-Montréal (integrated university health and social services centers), Montreal, Canada.
Published: March 18, 2022
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.isci.2022.103858.
Contributor Information
Stéphane Gobeil, Email: stephane.gobeil@crchudequebec.ulaval.ca.
Daniel Sinnett, Email: daniel.sinnett@umontreal.ca.
Supplemental information
Data and code availability
-
•
The RNA-seq and ChIP-seq data used in this study have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE79373 and GSE102785 respectively. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD031001.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
References
- Agape P., Gerard B., Cave H., Devaux I., Vilmer E., Lecomte M.C., Grandchamp B. Analysis of ETV6 and ETV6-AML1 proteins in acute lymphoblastic leukaemia. Br. J. Haematol. 1997;98:234–239. doi: 10.1046/j.1365-2141.1997.1973014.x. [DOI] [PubMed] [Google Scholar]
- Akopian D., Shen K., Zhang X., Shan S.O. Signal recognition particle: an essential protein-targeting machine. Annu. Rev. Biochem. 2013;82:693–721. doi: 10.1146/annurev-biochem-072711-164732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreasson P., Schwaller J., Anastasiadou E., Aster J., Gilliland D.G. The expression of ETV6/CBFA2 (TEL/AML1) is not sufficient for the transformation of hematopoietic cell lines in vitro or the induction of hematologic disease in vivo. Cancer Genet. Cytogenet. 2001;130:93–104. doi: 10.1016/s0165-4608(01)00518-0. [DOI] [PubMed] [Google Scholar]
- Arai H., Maki K., Waga K., Sasaki K., Nakamura Y., Imai Y., Kurokawa M., Hirai H., Mitani K. Functional regulation of TEL by p38-induced phosphorylation. Biochem. Biophys. Res. Commun. 2002;299:116–125. doi: 10.1016/s0006-291x(02)02588-3. [DOI] [PubMed] [Google Scholar]
- Azlan A., Dzaki N., Azzam G. Argonaute: the executor of small RNA function. J. Genet. Genomics. 2016;43:481–494. doi: 10.1016/j.jgg.2016.06.002. [DOI] [PubMed] [Google Scholar]
- Bartuzi P., Hofker M.H., van de Sluis B. Tuning NF-kappaB activity: a touch of COMMD proteins. Biochim. Biophys. Acta. 2013;1832:2315–2321. doi: 10.1016/j.bbadis.2013.09.014. [DOI] [PubMed] [Google Scholar]
- Bassuk A.G., Leiden J.M. A direct physical association between ETS and AP-1 transcription factors in normal human T cells. Immunity. 1995;3:223–237. doi: 10.1016/1074-7613(95)90092-6. [DOI] [PubMed] [Google Scholar]
- Becker W., Weber Y., Wetzel K., Eirmbter K., Tejedor F.J., Joost H.G. Sequence characteristics, subcellular localization, and substrate specificity of DYRK-related kinases, a novel family of dual specificity protein kinases. J. Biol. Chem. 1998;273:25893–25902. doi: 10.1074/jbc.273.40.25893. [DOI] [PubMed] [Google Scholar]
- Bhojwani D., Yang J.J., Pui C.H. Biology of childhood acute lymphoblastic leukemia. Pediatr. Clin. North Am. 2015;62:47–60. doi: 10.1016/j.pcl.2014.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bitterman K.J., Anderson R.M., Cohen H.Y., Latorre-Esteves M., Sinclair D.A. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- Bohlander S.K. ETV6: a versatile player in leukemogenesis. Semin. Cancer Biol. 2005;15:162–174. doi: 10.1016/j.semcancer.2005.01.008. [DOI] [PubMed] [Google Scholar]
- Bonnay F., Nguyen X.H., Cohen-Berros E., Troxler L., Batsche E., Camonis J., Takeuchi O., Reichhart J.M., Matt N. Akirin specifies NF-kappaB selectivity of Drosophila innate immune response via chromatin remodeling. EMBO J. 2014;33:2349–2362. doi: 10.15252/embj.201488456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciau-Uitz A., Wang L., Patient R., Liu F. ETS transcription factors in hematopoietic stem cell development. Blood Cells Mol. Dis. 2013;51:248–255. doi: 10.1016/j.bcmd.2013.07.010. [DOI] [PubMed] [Google Scholar]
- De Braekeleer E., Douet-Guilbert N., Morel F., Le Bris M.J., Basinko A., De Braekeleer M. ETV6 fusion genes in hematological malignancies: a review. Leuk. Res. 2012;36:945–961. doi: 10.1016/j.leukres.2012.04.010. [DOI] [PubMed] [Google Scholar]
- Eisenberg E., Levanon E.Y. Human housekeeping genes, revisited. Trends Genet. 2013;29:569–574. doi: 10.1016/j.tig.2013.05.010. [DOI] [PubMed] [Google Scholar]
- Feng F.Y., Brenner J.C., Hussain M., Chinnaiyan A.M. Molecular pathways: targeting ETS gene fusions in cancer. Clin. Cancer Res. 2014;20:4442–4448. doi: 10.1158/1078-0432.CCR-13-0275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golub T.R., Barker G.F., Bohlander S.K., Hiebert S.W., Ward D.C., Bray-Ward P., Morgan E., Raimondi S.C., Rowley J.D., Gilliland D.G. Fusion of the TEL gene on 12p13 to the AML1 gene on 21q22 in acute lymphoblastic leukemia. Proc. Natl. Acad. Sci. U S A. 1995;92:4917–4921. doi: 10.1073/pnas.92.11.4917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z., Churchman M.L., Roberts K.G., Moore I., Zhou X., Nakitandwe J., Hagiwara K., Pelletier S., Gingras S., Berns H., et al. PAX5-driven subtypes of B-progenitor acute lymphoblastic leukemia. Nat. Genet. 2019;51:296–307. doi: 10.1038/s41588-018-0315-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidez F., Petrie K., Ford A.M., Lu H., Bennett C.A., MacGregor A., Hannemann J., Ito Y., Ghysdael J., Greaves M., et al. Recruitment of the nuclear receptor corepressor N-CoR by the TEL moiety of the childhood leukemia-associated TEL-AML1 oncoprotein. Blood. 2000;96:2557–2561. [PubMed] [Google Scholar]
- Gunji H., Waga K., Nakamura F., Maki K., Sasaki K., Nakamura Y., Mitani K. TEL/AML1 shows dominant-negative effects over TEL as well as AML1. Biochem. Biophys. Res. Commun. 2004;322:623–630. doi: 10.1016/j.bbrc.2004.07.169. [DOI] [PubMed] [Google Scholar]
- Guo X., Williams J.G., Schug T.T., Li X. DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J. Biol. Chem. 2010;285:13223–13232. doi: 10.1074/jbc.M110.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hart T., Komori H.K., LaMere S., Podshivalova K., Salomon D.R. Finding the active genes in deep RNA-seq gene expression studies. BMC Genomics. 2013;14:778. doi: 10.1186/1471-2164-14-778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar A., Gutierrez-Hartmann A. Molecular mechanisms of ETS transcription factor-mediated tumorigenesis. Crit. Rev. Biochem. Mol. Biol. 2013;48:522–543. doi: 10.3109/10409238.2013.838202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirwan M., Walne A.J., Plagnol V., Velangi M., Ho A., Hossain U., Vulliamy T., Dokal I. Exome sequencing identifies autosomal-dominant SRP72 mutations associated with familial aplasia and myelodysplasia. Am. J. Hum. Genet. 2012;90:888–892. doi: 10.1016/j.ajhg.2012.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuwata T., Gongora C., Kanno Y., Sakaguchi K., Tamura T., Kanno T., Basrur V., Martinez R., Appella E., Golub T., Ozato K. Gamma interferon triggers interaction between ICSBP (IRF-8) and TEL, recruiting the histone deacetylase HDAC3 to the interferon-responsive element. Mol. Cell Biol. 2002;22:7439–7448. doi: 10.1128/MCB.22.21.7439-7448.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lajoie M., Drouin S., Caron M., St-Onge P., Ouimet M., Gioia R., Lafond M.H., Vidal R., Richer C., Oualkacha K., et al. Specific expression of novel long non-coding RNAs in high-hyperdiploid childhood acute lymphoblastic leukemia. PLoS One. 2017;12:e0174124. doi: 10.1371/journal.pone.0174124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B., Salzberg S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee Y.J., Kim J.H., Bae S., Rho S.K., Choe S.Y. Mechanism of transcriptional repression by TEL/RUNX1 fusion protein. Mol. Cells. 2004;17:217–222. [PubMed] [Google Scholar]
- Lilljebjorn H., Henningsson R., Hyrenius-Wittsten A., Olsson L., Orsmark-Pietras C., von Palffy S., Askmyr M., Rissler M., Schrappe M., Cario G., et al. Identification of ETV6-RUNX1-like and DUX4-rearranged subtypes in paediatric B-cell precursor acute lymphoblastic leukaemia. Nat. Commun. 2016;7:11790. doi: 10.1038/ncomms11790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lilljebjorn H., Soneson C., Andersson A., Heldrup J., Behrendtz M., Kawamata N., Ogawa S., Koeffler H.P., Mitelman F., Johansson B., et al. The correlation pattern of acquired copy number changes in 164 ETV6/RUNX1-positive childhood acute lymphoblastic leukemias. Hum. Mol. Genet. 2010;19:3150–3158. doi: 10.1093/hmg/ddq224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lopez R.G., Carron C., Ghysdael J. v-SRC specifically regulates the nucleo-cytoplasmic delocalization of the major isoform of TEL (ETV6) J. Biol. Chem. 2003;278:41316–41325. doi: 10.1074/jbc.M306435200. [DOI] [PubMed] [Google Scholar]
- Mavrothalassitis G., Ghysdael J. Proteins of the ETS family with transcriptional repressor activity. Oncogene. 2000;19:6524–6532. doi: 10.1038/sj.onc.1204045. [DOI] [PubMed] [Google Scholar]
- Montpetit A., Larose J., Boily G., Langlois S., Trudel N., Sinnett D. Mutational and expression analysis of the chromosome 12p candidate tumor suppressor genes in pre-B acute lymphoblastic leukemia. Leukemia. 2004;18:1499–1504. doi: 10.1038/sj.leu.2403441. [DOI] [PubMed] [Google Scholar]
- Mori H., Colman S.M., Xiao Z., Ford A.M., Healy L.E., Donaldson C., Hows J.M., Navarrete C., Greaves M. Chromosome translocations and covert leukemic clones are generated during normal fetal development. Proc. Natl. Acad. Sci. U S A. 2002;99:8242–8247. doi: 10.1073/pnas.112218799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moriyama T., Metzger M.L., Wu G., Nishii R., Qian M., Devidas M., Yang W., Cheng C., Cao X., Quinn E., et al. Germline genetic variation in ETV6 and risk of childhood acute lymphoblastic leukaemia: a systematic genetic study. Lancet Oncol. 2015;16:1659–1666. doi: 10.1016/S1470-2045(15)00369-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveu B., Caron M., Lagace K., Richer C., Sinnett D. Genome wide mapping of ETV6 binding sites in pre-B leukemic cells. Sci. Rep. 2018;8:15526. doi: 10.1038/s41598-018-33947-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveu B., Spinella J.F., Richer C., Lagace K., Cassart P., Lajoie M., Jananji S., Drouin S., Healy J., Hickson G.R., Sinnett D. CLIC5: a novel ETV6 target gene in childhood acute lymphoblastic leukemia. Haematologica. 2016;101:1534–1543. doi: 10.3324/haematol.2016.149740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noetzli L., Lo R.W., Lee-Sherick A.B., Callaghan M., Noris P., Savoia A., Rajpurkar M., Jones K., Gowan K., Balduini C.L., et al. Germline mutations in ETV6 are associated with thrombocytopenia, red cell macrocytosis and predisposition to lymphoblastic leukemia. Nat. Genet. 2015;47:535–538. doi: 10.1038/ng.3253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak S.J., Aihara H., Gonzalez K., Nibu Y., Baylies M.K. Akirin links twist-regulated transcription with the Brahma chromatin remodeling complex during embryogenesis. PLoS Genet. 2012;8:e1002547. doi: 10.1371/journal.pgen.1002547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otsubo K., Kanegane H., Eguchi M., Eguchi-Ishimae M., Tamura K., Nomura K., Abe A., Ishii E., Miyawaki T. ETV6-ARNT fusion in a patient with childhood T lymphoblastic leukemia. Cancer Genet. Cytogenet. 2010;202:22–26. doi: 10.1016/j.cancergencyto.2010.07.121. [DOI] [PubMed] [Google Scholar]
- Papadopoulos C., Arato K., Lilienthal E., Zerweck J., Schutkowski M., Chatain N., Muller-Newen G., Becker W., de la Luna S. Splice variants of the dual specificity tyrosine phosphorylation-regulated kinase 4 (DYRK4) differ in their subcellular localization and catalytic activity. J. Biol. Chem. 2011;286:5494–5505. doi: 10.1074/jbc.M110.157909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoudou-Bai A., Hatzimichael E., Barbouti A., Kanavaros P. Expression patterns of the activator protein-1 (AP-1) family members in lymphoid neoplasms. Clin. Exp. Med. 2016;17:291–304. doi: 10.1007/s10238-016-0436-z. [DOI] [PubMed] [Google Scholar]
- Patel N., Goff L.K., Clark T., Ford A.M., Foot N., Lillington D., Hing S., Pritchard-Jones K., Jones L.K., Saha V. Expression profile of wild-type ETV6 in childhood acute leukaemia. Br. J. Haematol. 2003;122:94–98. doi: 10.1046/j.1365-2141.2003.04399.x. [DOI] [PubMed] [Google Scholar]
- Poirel H., Lacronique V., Mauchauffe M., Le Coniat M., Raffoux E., Daniel M.T., Erickson P., Drabkin H., MacLeod R.A., Drexler H.G., et al. Analysis of TEL proteins in human leukemias. Oncogene. 1998;16:2895–2903. doi: 10.1038/sj.onc.1201817. [DOI] [PubMed] [Google Scholar]
- Poirel H., Oury C., Carron C., Duprez E., Laabi Y., Tsapis A., Romana S.P., Mauchauffe M., Le Coniat M., Berger R., et al. The TEL gene products: nuclear phosphoproteins with DNA binding properties. Oncogene. 1997;14:349–357. doi: 10.1038/sj.onc.1200829. [DOI] [PubMed] [Google Scholar]
- Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Romana S.P., Mauchauffe M., Le Coniat M., Chumakov I., Le Paslier D., Berger R., Bernard O.A. The t(12;21) of acute lymphoblastic leukemia results in a tel-AML1 gene fusion. Blood. 1995;85:3662–3670. [PubMed] [Google Scholar]
- Romana S.P., Poirel H., Leconiat M., Flexor M.A., Mauchauffe M., Jonveaux P., Macintyre E.A., Berger R., Bernard O.A. High frequency of t(12;21) in childhood B-lineage acute lymphoblastic leukemia. Blood. 1995;86:4263–4269. [PubMed] [Google Scholar]
- Samur M.K. RTCGAToolbox: a new tool for exporting TCGA Firehose data. PLoS One. 2014;9:e106397. doi: 10.1371/journal.pone.0106397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanders B.D., Jackson B., Marmorstein R. Structural basis for sirtuin function: what we know and what we don't. Biochim. Biophys. Acta. 2010;1804:1604–1616. doi: 10.1016/j.bbapap.2009.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt M.T., Smith B.C., Jackson M.D., Denu J.M. Coenzyme specificity of Sir2 protein deacetylases: implications for physiological regulation. J. Biol. Chem. 2004;279:40122–40129. doi: 10.1074/jbc.M407484200. [DOI] [PubMed] [Google Scholar]
- Shurtleff S.A., Buijs A., Behm F.G., Rubnitz J.E., Raimondi S.C., Hancock M.L., Chan G.C., Pui C.H., Grosveld G., Downing J.R. TEL/AML1 fusion resulting from a cryptic t(12;21) is the most common genetic lesion in pediatric ALL and defines a subgroup of patients with an excellent prognosis. Leukemia. 1995;9:1985–1989. [PubMed] [Google Scholar]
- Singh R., Park D., Xu J., Hosur R., Berger B. Struct2Net: a web service to predict protein-protein interactions using a structure-based approach. Nucleic Acids Res. 2010;38:W508–W515. doi: 10.1093/nar/gkq481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sizemore G.M., Pitarresi J.R., Balakrishnan S., Ostrowski M.C. The ETS family of oncogenic transcription factors in solid tumours. Nat. Rev. Cancer. 2017;17:337–351. doi: 10.1038/nrc.2017.20. [DOI] [PubMed] [Google Scholar]
- Spinella J.F., Cassart P., Richer C., Saillour V., Ouimet M., Langlois S., St-Onge P., Sontag T., Healy J., Minden M.D., Sinnett D. Genomic characterization of pediatric T-cell acute lymphoblastic leukemia reveals novel recurrent driver mutations. Oncotarget. 2016;7:65485–65503. doi: 10.18632/oncotarget.11796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szymczyna B.R., Arrowsmith C.H. DNA binding specificity studies of four ETS proteins support an indirect read-out mechanism of protein-DNA recognition. J. Biol. Chem. 2000;275:28363–28370. doi: 10.1074/jbc.M004294200. [DOI] [PubMed] [Google Scholar]
- Topka S., Vijai J., Walsh M.F., Jacobs L., Maria A., Villano D., Gaddam P., Wu G., McGee R.B., Quinn E., et al. Germline ETV6 mutations confer susceptibility to acute lymphoblastic leukemia and thrombocytopenia. Plos Genet. 2015;11:e1005262. doi: 10.1371/journal.pgen.1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uphoff C.C., MacLeod R.A., Denkmann S.A., Golub T.R., Borkhardt A., Janssen J.W., Drexler H.G. Occurrence of TEL-AML1 fusion resulting from (12;21) translocation in human early B-lineage leukemia cell lines. Leukemia. 1997;11:441–447. doi: 10.1038/sj.leu.2400571. [DOI] [PubMed] [Google Scholar]
- van der Weyden L., Giotopoulos G., Rust A.G., Matheson L.S., van Delft F.W., Kong J., Corcoran A.E., Greaves M.F., Mullighan C.G., Huntly B.J., Adams D.J. Modeling the evolution of ETV6-RUNX1-induced B-cell precursor acute lymphoblastic leukemia in mice. Blood. 2011;118:1041–1051. doi: 10.1182/blood-2011-02-338848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verger A., Duterque-Coquillaud M. When Ets transcription factors meet their partners. Bioessays. 2002;24:362–370. doi: 10.1002/bies.10068. [DOI] [PubMed] [Google Scholar]
- Wang L., Hiebert S.W. TEL contacts multiple co-repressors and specifically associates with histone deacetylase-3. Oncogene. 2001;20:3716–3725. doi: 10.1038/sj.onc.1204479. [DOI] [PubMed] [Google Scholar]
- Yeoh E.J., Ross M.E., Shurtleff S.A., Williams W.K., Patel D., Mahfouz R., Behm F.G., Raimondi S.C., Relling M.V., Patel A., et al. Classification, subtype discovery, and prediction of outcome in pediatric acute lymphoblastic leukemia by gene expression profiling. Cancer Cell. 2002;1:133–143. doi: 10.1016/s1535-6108(02)00032-6. [DOI] [PubMed] [Google Scholar]
- Zhan W., Wang W., Han T., Xie C., Zhang T., Gan M., Wang J.B. COMMD9 promotes TFDP1/E2F1 transcriptional activity via interaction with TFDP1 in non-small cell lung cancer. Cell Signal. 2017;30:59–66. doi: 10.1016/j.cellsig.2016.11.016. [DOI] [PubMed] [Google Scholar]
- Zhang M.Y., Churpek J.E., Keel S.B., Walsh T., Lee M.K., Loeb K.R., Gulsuner S., Pritchard C.C., Sanchez-Bonilla M., Delrow J.J., et al. Germline ETV6 mutations in familial thrombocytopenia and hematologic malignancy. Nat. Genet. 2015;47:180–185. doi: 10.1038/ng.3177. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
-
•
The RNA-seq and ChIP-seq data used in this study have been deposited in NCBI’s Gene Expression Omnibus and are accessible through GEO Series accession number GSE79373 and GSE102785 respectively. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD031001.
-
•
This paper does not report original code.
-
•
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.