Summary
Background
The FilmArray Meningitis/Encephalitis(FA/ME) panel brings benefits in clinical practice, but its diagnostic test accuracy (DTA) remains unclear. We aimed to determine the DTA of FA/ME for the aetiological diagnostic in patients with suspected central nervous system(CNS) infection.
Methods
We performed a systematic review with DTA meta-analysis (PROSPERO: CRD42020139285). We searched Embase, Medline (Ovid), and Web of Science from inception until September 1st, 2021. We assessed the study-level risk of bias with the QUADAS-2 tool and applied the GRADE approach to assess the certainty of the synthesised evidence. We included studies that simultaneously measured the reference test (CSF/blood culture for bacteria, and specific polymerase chain reaction for viruses) and the FA/ME in patients with suspected CNS infection. We performed random-effects bivariate meta-analysis models of combined sensitivity and specificity using CSF/blood cultures(reference test 1) and a final diagnosis adjudication based on clinical/laboratory criteria (reference test 2).
Findings
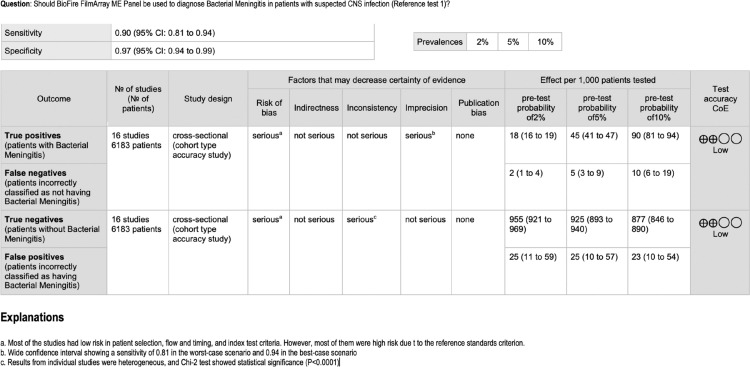
We included 19 studies (11,351 participants). For all bacteria with reference test 1 (16 studies/6183 patients) sensitivity was estimated at 89·5% (95%CI 81·1–94·4), and specificity at 97·4% (95%CI 94–98·9). With reference test 2 (15 studies/5,524 patients), sensitivity was estimated at 92·1%(95%CI 86·8–95·3) and specificity at 99.2(95%CI 98·3–99·6) For herpes simplex virus-2(HSV-2), enteroviruses, and Varicella-Zoster virus (VZV), we obtained sensitivities between 75·5 and 93·8%, and specificities above 99% (reference test 1). Certainty of the evidence was low.
Interpretation
FA/ME may have acceptable-to-high sensitivities and high specificities for identifying bacteria, especially for S.pneumoniae, and viruses, especially for HSV-2, and enteroviruses. Sensitivities for L.monocytogenes, H.influenzae, E.coli, and HSV-1 were suboptimal.
Funding
None.
Keywords: CNS infection, Meningitis, Encephalitis, Film array, Multiplex PCR, Meta-analysis, Diagnostic accuracy
Research in context.
Evidence before this study
The FilmArray Meningitis/Encephalitis panel (BioFire Diagnostics®) (FA/ME) simultaneously detects 14 pathogens in cerebrospinal fluid (CSF). A previous systematic review identified in MEDLINE searches concluded that the entire FA/ME panel has a sensitivity of 90% and a specificity of 97% for identifying any microorganism in the CSF. However, the review did not assess the certainty of the evidence, only included 13 studies, considered evidence from studies in which the reference and the index tests were not performed simultaneously and independently, did not perform meta-analyses according to specific microorganisms (viruses and bacteria) or based on specific clinical subgroups.
Added value of this study
This review is the most updated and rigorous systematic review on the diagnostic test accuracy of the FA/ME. We used state-of-art methods for conducting diagnostic test accuracy meta-analysis (including sensitivity and subgroup analyses), and we provide the certainty of the evidence for both reference tests. We provide the DTA measures of the FA/ME panel discriminated by the most important bacteria and viruses causing central nervous system (CNS) infections in immunocompetent patients, and we identified those microorganisms in which the panel may have better diagnostic accuracy.
Implications of all the available evidence
We found moderate sensitivities and high specificities for the diagnosis of bacterial CNS infection (mainly for identifying any bacteria and S. pneumoniae) and of enterovirus, HSV-1, and HSV-2, in immunocompetent patients. The validity for identifying L.monocytogenes, H.influenzae and E.coli was found to be suboptimal. In general, the FA/ME test seems to be an excellent tool for ruling in but very limited for ruling out CNS infections.
Alt-text: Unlabelled box
Introduction
Central Nervous system (CNS) infections are a major cause of morbidity and mortality.1,2 A timely and accurate aetiological diagnosis in CNS infections is the cornerstone for targeted, appropriate treatments that will allow better clinical outcomes such as a lower hospital stay and mortality and lower health care expenses.3, 4, 5 However, appropriate aetiological diagnoses of meningitis and encephalitis with traditional microbiological methods, i.e., cultures, is challenging due to low diagnostic performance, long time to complete, and the previous use of antibimicrobials.6 Consequently, different molecular biology techniques have recently been developed.7 The use of molecular panels for the simultaneous diagnosis of several microorganisms employing multiplex PCR has opened up new possibilities and new challenges.8
The FilmArray Meningitis/Encephalitis panel (BioFire Diagnostics®) (FA/ME), approved by the Food and Drug Administration in 2015, is a panel that simultaneously detects 14 pathogens in cerebrospinal fluid (CSF). A recent meta-analysis reported that FA/ME has a sensitivity of 90% and a specificity of 97% for detecting microorganisms.9 However, this review has some limitations. Authors included some studies in which the reference and the index tests were not performed simultaneously and independently, they did not perform combined analyses according to specific microorganisms, and they did not present sensitivity and specificity values for some microorganisms such as N.meningitidis and L.monocytogenes for which the largest studies did not have enough information to establish the accuracy for these pathogens,10 and they did not assess the certainty of the summarized body of the evidence. Due to the gaps that still exist in the literature and practice regarding the use of this panel, we decided to conduct this systematic review to determine the diagnostic validity of FA/ME in immunocompetent patients for identifying any microorganism, and for identifying specific bacteria and viruses in suspected acute meningitis and encephalitis.
Methods
We performed a systematic review and meta-analysis of diagnostic test accuracy (DTA) studies. We registered the protocol in the PROSPERO database (CRD42020139285), and a copy of this register is provided as supplemental material. This manuscript follows the reporting guidelines of the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) extension for DTA studies.11
Literature search
Two authors performed searches (JT,IF) with liaison with an experienced librarian in Embase, Medline, and Web of Science. In addition, we conducted manual searches using references of the included studies, and gray literature through WorldWideScience, National Technical Information Service (NTIS), and OpenGrey databases. The search was carried out until September 1st, 2021, and we present it in the Supplemental Material Appendix 1
Eligibility criteria
Prospective or retrospective studies with a diagnostic test or cross-sectional design were included. Studies had to simultaneously apply the reference test (CSF/blood culture for bacteria, specific PCR, or Laboratory Developed Test for viruses) and the index test (i.e. FA/ME), in patients with suspected CNS infection. Studies also had to include reports of the detected microorganisms and sufficient data to calculate the DTA measures, i.e., true positives (TP), true negatives (TN), false positives (FP) and false negatives (FN). We excluded studies with no clear information and whose authors did not respond to e-mail contact; included only immunocompromised patients; or patients with ventriculoperitoneal shunts (or other intracranial devices).
Study selection
Two researchers (JT,MJ) independently and in duplicate reviewed the titles and abstracts. References considered eligible by at least one reviewer were selected for full-text review. The full-texts obtained were independently reviewed in duplicate by two reviewers (JT,MJ). Reviewers resolved discrepancies through discussion.
Data extraction
Two researchers (JT,IF) performed the data extraction independently and in duplicate, using a piloted extraction form in a Microsoft Excel sheet. We extracted the following information: publication year, study design, inclusion criteria, mean participants age, number of participants/sample size, funding source, reference tests per microorganism, diagnosis adjudication methods (additional laboratory test, CSF cytochemical, or clinical analysis result), and the required data for estimating TP, TN, FP, and FN.
Tests’ definition and results
The index test was the FA/ME. We focused on the most frequent microorganisms involved in acute CNS infections (Table 1). For bacteria, since CSF/blood cultures results can be affected by the quality of the samples or by previous antimicrobials use,6,12 they can be of limited value in some cases. Thus, some authors have considered a combination of additional factors to adjudicate the presence of infection. For example, in cases with positive FA/ME and negative culture (disagreement), an additional test or a final diagnosis adjudication following a clinical analysis by the researchers was applied. We, therefore, performed two analyses, using a reference test 1 (positive CSF/blood culture and viral PCR for bacteria and viruses, respectively) and a reference test 2 (final diagnosis was adjudicated following an additional test or a clinical analyses of the cases). Table 1 provides details of both reference tests and the definitions used for TP, TN, FP, and FN.
Table 1.
Definition of tests and results used.
| Index test | Reference test 1/RT1(Cultures or viral PCR) | Reference test 2/ RT2(Adjudicated diagnosis) † | Definitions of results according to reference test. | |
|---|---|---|---|---|
| Bacteria | FA/ME positive for: - Streptococcus pneumoniae - Escherichia coli K1 - Haemophilus influenzae - Listeria monocytogenes - Neisseria meningitidis - Streptococcus agalactiae |
Aerobic CSF cultures/Blood culture for included microorganisms. In case of polymicrobial scenarios each microorganism was analyzed separately. For “all bacteria”, we considered as positive the isolation of one (or more) microorganism). |
Applied only in cases where there was disagreement between FA/ME and reference test 1 (blood/CSF cultures). Final diagnosis adjudication was done through one of the following methods:
|
Reference test 1:
|
| Viruses | FA/ME positive for: - Enterovirus (EV) - Herpes simplex virus 1 (HSV-1) - Herpes simplex virus 2 (HSV-2) - Varicella-zoster virus (VZV) |
- HSV-1 y HSV-2: PCR Simplex a HSV 1&2 Direct (Focus Diagnostics) or MultiCode RTx HSV 1&2 kit ((Luminex Corporation, or PCR LDT or PCR in house with previously validated primers. - Enterovirus: Cepheid Xpert EV or with PCR LDT tests or PCR in house with previously validated primers - VZV: PCR LDT tests or PCR in house with previously validated primers. |
Applied only in cases where there was disagreement between FA/ME and reference test 1 (viral PCR). Final diagnosis adjudication was done through one of the following methods:
|
CSF: Cerebrospinal fluid; FA/ME: FilmArray Meningitis/Encephalitis panel; PCR: polymerase chain reaction; RT: Reference Test; TP: True positive; FP: False positive; TN: True Negative; FN: False Negative.
It was applied only in case of disagreement of RT1; in case there is no disagreement between FA/ME y RT1, the same result for RT2 was considered.
In general, when the FA/ME and reference test 1 matched, this was the same result in reference test 2. When the FA/ME result and reference test 1 did not match, reference test 2 was the final adjudication of the diagnosis through a retrospective analysis of each case by the authors based on additional molecular testing, the findings and clinical evolution, and/or the results of the CSF study.
Risk of bias assessment
We assessed the risk of bias (RoB) of the included studies with the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2)13 by two researchers (JT,IF). In Appendix 2 (Supplemental Material), we detail the methods used the RoB assessment with the QUADAS-2 tool
Diagnostic test accuracy measures
For both reference tests, we created 2 × 2 contingency tables of the number of TP, TN, FP, and FN for “all bacteria” and for each individual microorganism. In the studies that only applied CSF culture (and not alternative viral tests to all samples regardless of the FA/ME result), only the accuracy for bacteria was analysed. Additionally, in those studies in which the data was very clear for only a subgroup of participants, and not for others, we only analysed the information for the former, and discarded the rest of the data if we could not guarantee the other subgroups met our eligibility criteria. We performed analyses for both reference tests 1 and 2 for “all bacteria” (any of our eligible bacteria), for each individual bacterium and virus.
Statistical analyses
We used the bivariate random-effects model to estimate a summary sensitivity and specificity with their corresponding 95% confidence intervals (95%CI).14 We present study-specific sensitivities and specificities in forest plots and crosshair plots, study-specific positive (LR+), and negative likelihood ratios (LR-), with their 95%CI for all the defined groups in scatterplots, and summary sensitivity and specificity estimate in summary receiver operating characteristic (SROC) plots.14 Likelihood ratios were calculated from the combined sensitivity and specificities. LR+ was calculated as the combined sensitivity divided by 1- specificity. LR- was calculated as 1- the combined sensitivity divided by the combined specificity.
We assessed between-study heterogeneity through visual inspection of forest plots for sensitivity and specificity separately. We also visually inspected the study-specific effects in a ROC plot (1-specificity against sensitivity), in which the higher the scatter of the study-specific effects, the larger the prediction ellipse, and hence the higher the heterogeneity. We attempted to explore whether visual variability was based on study characteristics, including sample size and test variations. Analyses for both, reference tests 1 and 2, were performed for “all bacteria” (any bacteria), for each of the six bacteria, and for four viruses. Lastly, we performed the chi2-test and corresponding p-values to assess the presence of statistical heterogeneity and consider p- values < 0.05 as significant.
We aimed to assess the influence of some covariates on the DTA measures. We, therefore, conducted subgroup analyses in children/infants, abnormal CSF (according to authors’ definitions), and in patients with previous antimicrobial use (defined as ≥ 70% of patients with previous antimicrobial use). We also planned to explore heterogeneity if we obtained more than 10 studies, by adding covariate terms in a meta-regression to assess effects by mean age, but very few studies reported this information. Furthermore, we performed sensitivity analyses to evaluate the impact of high RoB studies in the estimates. We conducted two sensitivity analyses by restricting only to studies that had three or more QUADAS criteria judged as low, and with those studies that were judged as low RoB in all the QUADAS criteria.
All analyses were conducted using the ‘mada’ package in R software (version 3.3.3).15 We developed a figure summarizing the calculation of post-test probabilities of having correct or incorrect diagnosis (FP, FN, TP, and TN) according to three different prevalence rates of CNS infection for both reference tests using the interactive version of the summary of findings (GRADEpro GDT: GRADEpro Guideline Development Tool [Software]. McMaster University, 2015 (developed by Evidence Prime, Inc.).
Certainty of the evidence
We summarised key findings in a Grading of Recommendations Assessment, Development and Evaluation (GRADE) ‘Summary of findings’ table indicating the certainty of the evidence for the index test according to both reference tests. The GRADE approach encompasses the assessment of the following criteria: RoB(judged by an overall assessment of QUADAS-2), imprecision, inconsistency (also known as heterogeneity), indirectness and publication bias,16 and summarises the certainty on the evidence for the pooled sensitivity and specificity. The certainty of the evidence can be one of four levels: high, moderate, low, or very low.
Role of the funding source
There was no funding source for this study.
Results
Study selection
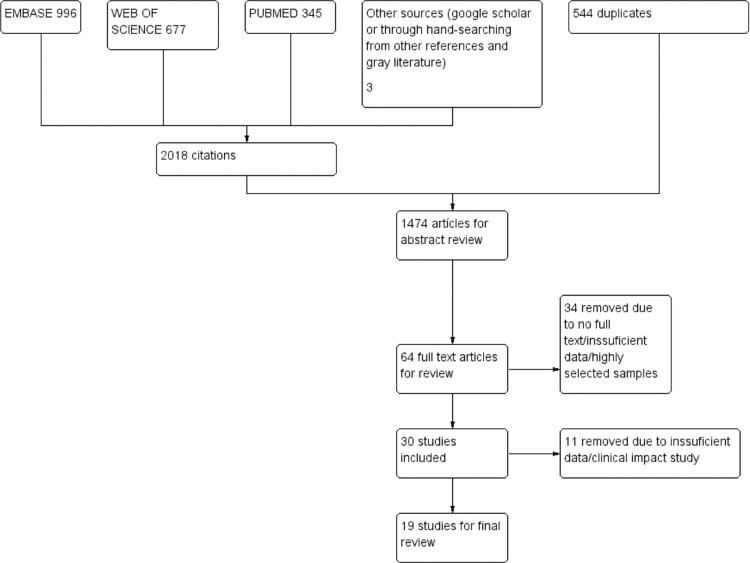
We retrieved 2018 references, and after removing duplicates, 1474 titles and abstracts were screened, of which 64 were selected for full-text review. We excluded 45 studies for multiple reasons (Figure 1), and we included 19 studies. Appendix 3(e-component 3) contains the excluded studies along with reasons for exclusions.
Figure 1.
Flow diagram for study selection.
Characteristics of the included studies
The Appendix 4 in Supplemental Material details the characteristics of the included studies. The 19 studies included 11,351 patients.10,17, 18, 19, 20, 21, 22, 23, 24, 25, 26, 27, 28, 29, 30, 31, 32, 33, 34 Four studies enrolled only children,18,19,27,28 two included only adults, and the rest included both. Six studies10,17,18,26,29,32 used a laboratory method to resolve the disagreements between FA/ME and reference test 1, and the rest used clinical information or CSF cytochemical parameters to resolve disagreements. In total, there were 219 bacterial isolates (105 S.pneumoniae, 27 E.coli, 31 S.agalactiae, 24 H.influenzae, 21 N.meningitidis, and 11 L.monocytogenes).
Risk of bias
Table 2 depicts the RoB assessment. Four studies10,17,31,33 were judged as unclear RoB in the patient selection domain due to uncertainty in the definition of suspected CNS infection. Five studies20,21,23,33,34 were judged as high RoB in the index test criterion due to unclear handling of the sample, storage (recently collected, stored or frozen) or processing of the FA/ME. Six studies were classified as low RoB in the reference test domain,10,17,18,22,26,29 and the remaining were judged as high RoB since it was not clear whether lumbar puncture and CSF sample collection occurred before antimicrobial therapy, and they did not use alternative molecular tests for bacteria. Seventeen and two studies were judged as of low and unclear RoB, respectively, in the flow and timing domain20,21
Table 2.
Risk of bias assessments†.
| Study | Patient selection | Index Test | Reference standard |
Flow and Timing | |||
|---|---|---|---|---|---|---|---|
| Bacteria reference test 1‡ | Bacteria reference test 2§ | Viruses reference test 1 | Viruses reference test 2§ | ||||
| Arora 2016 | Low risk | Low risk | High risk | High risk | Not applicable¶ | Not applicable | Low risk |
| Bailu 2019 | Low risk | Low risk | Low risk | High risk | Not applicable | Not applicable | Low risk |
| Barnes 2018 | Low risk | Low risk | High risk | High risk | Not applicable | Not applicable | Low risk |
| Boudet 2019 | Low risk | High risk | High risk | High risk | Not applicable | Not applicable | Unclear |
| Chong 2021 | Low Risk | Low Risk | Low Risk | High risk | Not applicable | Not applicable | Low Risk |
| Domingues 2019 | Low risk | High risk | High risk | High risk | Not applicable | Not applicable | Unclear |
| Eichinger 2019 | Low risk | Low risk | High risk | High risk | Not applicable | Not applicable | Low risk |
| Ena 2021 | Low Risk | Unclear | High Risk | High risk | Not applicable | Not applicable | Low Risk |
| Hanson 2016 | Unclear | Unclear | Low risk | High risk | Not applicable | Not applicable | Low risk |
| Lindstrom 2021 | Low Risk | High Risk | Not applicable | Not applicable | Low risk | High risk | Low Risk |
| Leber 2016 | Unclear | Low risk | Low risk | High risk | Low Risk | High Risk | Low risk |
| Leli 2019 | Unclear | Low risk | High risk | High risk | Not applicable | Not applicable | Low risk |
| Lopez-Amor 2019 | Low risk | Unclear | High risk | High risk | Not applicable | Not applicable | Low risk |
| Peñata 2020 | Low Risk | Low Risk | High risk | Not applicable | Not applicable | Not applicable | Low Risk |
| Piccirilli 2018 | Low risk | Low risk | Low risk | High risk | Low Risk | High Risk | Low risk |
| Radmard 2019 | Unclear | High risk | High risk | High risk | Not applicable | Not applicable | Low risk |
| Tarai 2019 | Low risk | High risk | High risk | High risk | Not applicable | Not applicable | Low risk |
| Vincent 2020 | Low risk | Low risk | Low risk | High risk | Low risk | High risk | Low risk |
Risk of bias assessment performed with the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2) tool. Reference tests 1 & 2 definitions for bacteria and viruses are detailed in Table 1.
Reference standard item for reference test 1 for bacterial detection was considered as low risk only when it was clear that the cultures samples were taken before the antimicrobial treatment.
All reference test 2 for both bacteria and viruses were judged as high risk because none of these diagnosis adjudications were conducted blinded to the index test results.
Items were not applicable when the reference test was not used for viruses.
Main results
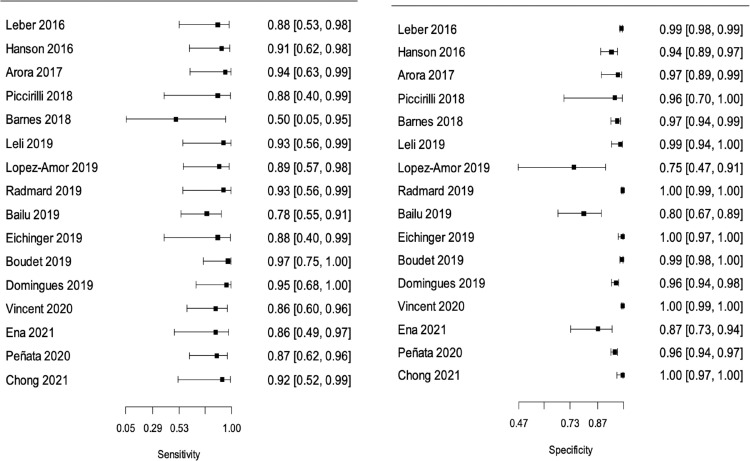
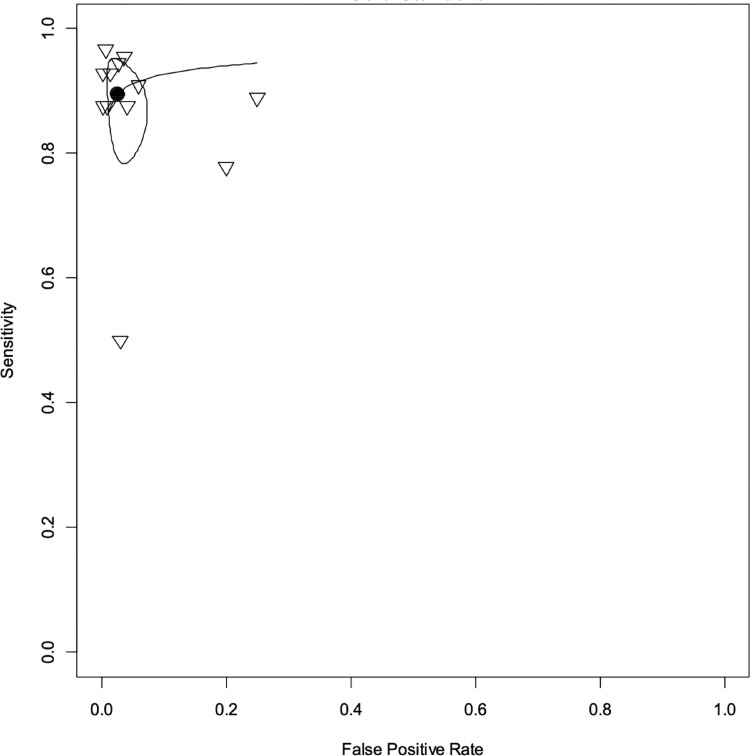
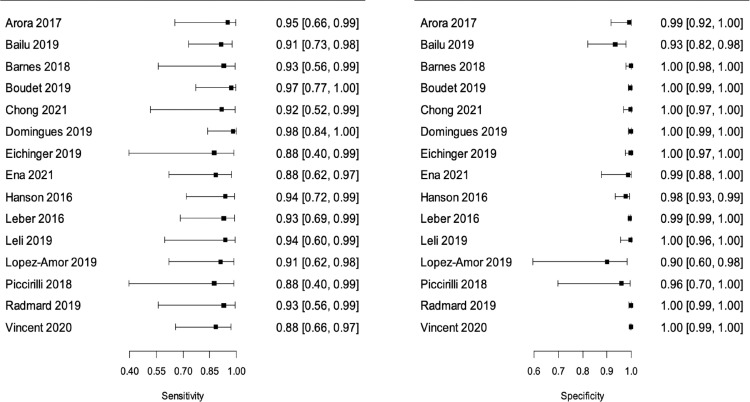
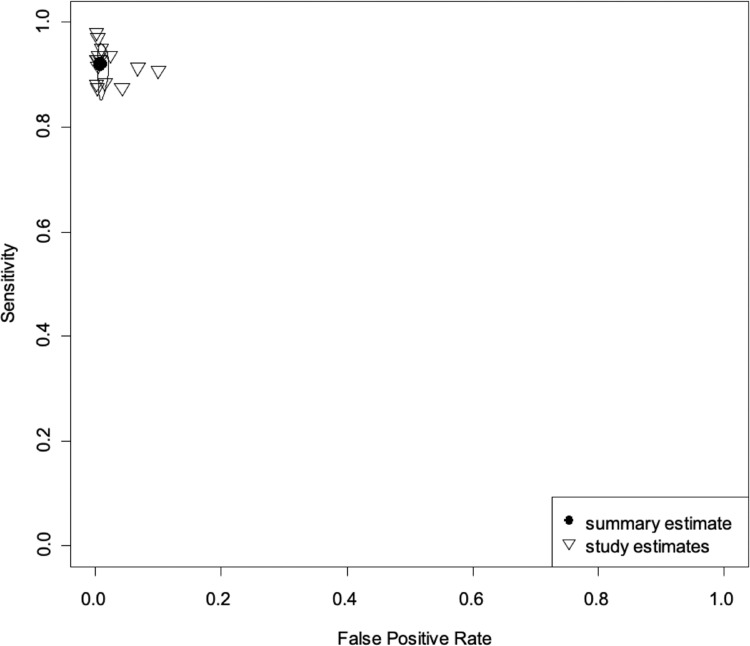
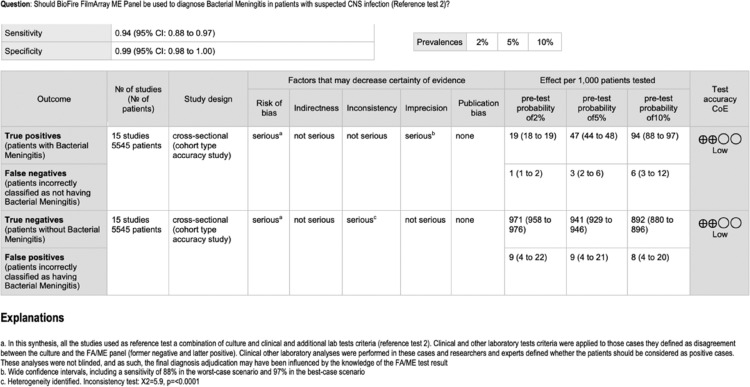
In the meta-analysis of “all bacteria” with reference test 1 (16 studies/6183 patients),10,17, 18, 19, 20, 21, 22,24, 25, 26, 27,29, 30, 31, 32, 33 we obtained combined sensitivity and specificity of 89·5% (95%CI 81·1–94·4), and 97·4% (95%CI 94–98·9), respectively. With reference test 2 (15 studies/5524 patients),10,17,29, 30, 31, 32, 33,18, 19, 20, 21, 22,24,26,27 we obtained combined sensitivity and specificity of 92.1% (95%CI 86.8–95.3), and 99.2(95%CI 98.3–99.6), respectively. Table 3 and Figure 2, Figure 3, Figure 4, Figure 5 present all the combined DTA measures and the forest plots and SROC, respectively, for both reference tests. Figure 6, Figure 7 show the GRADE summary of findings table for sensitivity and specificity for reference tests 1 and 2, respectively, which were in all the cases rated as low.
Table 3.
Meta-analyses of all bacteria and per bacteria and viruses, with reference test 1 and reference test 2.
| Ref. Test 1† |
Ref. Test 2‡ |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. Studies /No. Patients (Ref. studies) | Sensitivity (95%CI) X2; p value§ | Specificity(95%CI).X2; p value§ | LR+ (95%CI) |
LR- (95%CI) |
No. Studies/No. Patients(Ref. studies) | Sensitivity(95%CI).X2; p value¥ | Specificity(95%CI)X2; p value¥ | LR+ (95%CI) |
LR- (95%CI) |
|
| All bacteria | 16/618310,17,22,24, 25, 26, 27,29, 30, 31, 32, 33 | 89.5 (81.1–94.4) 6.00; 0.98 | 97.4 (94–98.9) 251.9; <0.0001 |
34 (2.68–137) |
0.11 (0.07–3.64) |
15/5545 (10,17,29–33,18–22,24,26,27) |
93.5 (87.8–96.6) 2.17; 0.99 |
99.1 (97.8–99.6) 60.2; <0.0001 |
104 (7.60–3.98) |
0.07 (0.07–3.58) |
| S. pneumoniae | 16/709010,17, 18, 19, 20, 21, 22,24, 25, 26,30,34 | 87.5 (77–94) 3.71; 0.999 | 98.5 (97–99.3) 144.7; <0.0001 |
58 (4.63–238) |
0.13 (0.07–3.60) |
10/5287 (10,17,30,32,33,34, 18,21,22,24) |
93 (83.3–97.2) 2.64; 0.91 |
99.4 (98.2–99.8) 51.2; <0.0001 |
155 (11–600) |
0.07 (0.07–3.58) |
| H. influenzae | 10/495910,17,18,20, 21, 22,24,25,30,32 | 64.9 (39.5–84) 4.91; 0.842 | 99.4 (98.9–99.6) 22.4; 0.07 |
108 (11–601) |
0.35 (0.07–3.57) |
7/3176 (10,17,18,20,21,24,30) |
81.1 (55.6–93.6) 4.97; 0.42 |
99.8 (99.5–99.9) 5.53; 0.354 |
405 (28–1889) |
0.19 (0.07–3.56) |
| S. agalactiae | 10/526610,17,18,20,22,25, 26, 27,31,33 | 71.5 (49.6–86.5) 7.67; 0.56 | 99.5 (98.5–99.9) 7.67; 0.56 |
143 (13–722) |
0.29 (0.07–3.57) |
5/2543 10,17,18,20,27 |
81.4 (52.3–94.6) 6.71; 0.15 |
99.4 (97.7–99.9) 23.42; <0.0001 |
136 (11–609) |
0.19 (0.07–3.57) |
| E. coli | 11/474310,17, 18, 19, 20, 21,25,27,30,32,33 | 70.9 (50.2–85.5) 4.93; 0.896 | 99.6 (99.1–99.8) 25.5; 0.0043 |
177 (16–909) |
0.29 (0.07–3.56) |
5/2570 10,18,20,30,32 |
76.3 (47.6 – 91.9) 3.56; 0.46 |
99.6 (98.7–99.9) 12.62; 0.01 |
191 (15–925) |
0.24 (0.07–3.56) |
| N. meningitidis | 10/350117,18,20, 21, 22,24,25,29, 30, 31 | 74.5 (52.9–88.4) 2.26; 0.986 | 99.1 (98.6–99.5) 20.9; 0.013 |
83 (7.48–400) |
0.26 (0.07–3.58) |
5/1950 18,21,22,30,31 |
84.4 (53.9–96.2) 0.84; 0.838 |
99.1 (98.8–99.9) 1.17; 0.759 |
281 (19–1265) |
0.16 (0.07–3.56) |
| L. monocytogenes | 7/133218,21,24,25,29,31,32 | 70.4 (40–89.5) 0.504; 0.008 | 98.9 (96.9–99.6) 5.62; 0.22 |
54 (4.99–280) |
0.30 (0.07–3.60) |
3/550 18,21,24) |
80.4 (40.4–96.1) 0.205; 0.903 |
99.5 (97.8–99.9) 2.13; 0.344 |
161 (9.03–796) |
0.20 (0.07–3.57) |
| Enterovirus | 3/688310,22,23 | 93.8 (87–97.2) 2.91; 0.23 | 99.3 (98.7–99.7) 28.53; <0.001 |
313 (22–1209) |
0.06 (0.07–3.56) |
3/6883 10, 22, 23 |
99.8 (86.1–97.4) 4.18; 0.123 |
99.9 (99.7–100) 3.43; 0.179 |
998 (58–3763) |
0.04 (0.07–3.55) |
| HSV-1 | 3/688310,22,23 | 75.5 (51.2–90.1) 1.18;0.554 | 99.9 (94.7–100) 2.55;0.28 |
755 (58–3763) |
0.25 (0.07–3.55) |
3/6883 10, 22, 23 |
78.2 (58.1–90.3) 0.69;0.706 |
99.9 (99.8–100) 1.8;0.405 |
782 (58–3763) |
0.22 (0.07–3.55) |
| HSV-2 | 3/688310,22,23 | 94.4 (83.9–98.2) 0.435;0.804 | 99.9 (99.7–100) 1.36;0.507 |
944 (58–3763) |
0.06 (0.07–3.55) |
3/6883 10, 22, 23 |
94.5 (84.2–98.2) 0.46;0.79 |
99.9 (99.8–100) 1.36;0.507 |
945 (58–3763) |
0.06 (0.07–3.55) |
| VZV | 4/689710,21,23,29 | 91.4 (78.9–96.9) 0.82;0.84 | 99.8 (98.7–100) 23.55;<0.001 |
457 (32–1832) |
0.09 (0.07–3.56) |
4/6897 10,21,23,29 |
93.3 (83.6–97.4) 0.16;0.91 |
99.9 (99.6–100) 6.23;0.004 |
933 (58–3763) |
0.07 (0.07–3.55) |
CSF: Cerebro-Spinal fluid; LR+: Positive Likelihood ratio; LR-: Negative Likelihood ratio; X2: Chi-2 test.
Reference Test 1: Aerobic CSF cultures/Blood culture or viral PCR.
Reference Test 2: It was applied only in case of disagreement of RT1. Final adjudication of the diagnosis through a retrospective analysis of each case by the authors based on additional molecular testing, findings, and clinical evolution, and/or the results of the cerebrospinal fluid study.
Chi2-test and corresponding p-value to assess presence of statistical heterogeneity.
Figure 2.
Forest Plot for “all bacteria” with reference test 1. Sensitivities (left) and specificities (right) of FA/ME per study for the detection of any bacteria in Cerebrospinal (CSF) fluid when the reference standard was a positive CSF or a blood culture (reference test 1).
Figure 3.
Summary receiver operating characteristic (SROC) curve for "all bacteria" with reference test 1. Each study is identified with a small reverse triangle. Back dot denotes the combined sensitivity and specificity. The figure also shows 95% confidence contour and 95% prediction contour. Reference test 1 means the standard was a positive cerebrospinal or a blood culture.
Figure 4.
Forest Plot for “all bacteria” with reference test 2. Sensitivities (left) and specificities (right) of FA/ME for the detection of any bacteria with reference test 2 per study. Reference test 2 means the standard (final diagnosis of the infection in cases where cerebrospinal fluids SF/blood cultures or viral tests were negative) was defined by the researchers through a final diagnosis adjudication using molecular tests, an analysis of the clinical manifestations or based on the cerebrospinal fluid findings.
Figure 5.
Summary receiver operating characteristic (SROC) curve for "all bacteria" with reference test 2. Each study is identified with a small reverse triangle. Back dot denotes the combined sensitivity and specificity. The figure also shows 95% confidence contour and 95% prediction contour. Reference test 2 means the standard (final diagnosis of the infection in cases where cerebrospinal fluids SF/blood cultures or viral tests were negative) was defined by the researchers through a final diagnosis adjudication using molecular tests, an analysis of the clinical manifestations or based on the CSF findings.
Figure 6.
GRADE Summary of findings table for reference test 1. The table summarises the certainty of the evidence according to the Grading of Recommendations Assessment, Development and Evaluation (GRADE) methodology, also called “quality of evidence”. The certainty can be one of four levels: High, moderate, low, or very low. The interpretation of these levels should be performed s follows; High: we are very confident that the true effect lies close to that of the estimate of the effect we found; Moderate: we are moderately confident in the effect estimate: the true effect is likely to be close to the estimate of the effect that we found, but there is a possibility that it is substantially different. Low: our confidence in the effect estimate we found is limited: the true effect may be substantially different from the estimate of the effect; Very low: we have very little confidence in the effect estimate: the true effect is likely to be substantially different from the estimate of effect we found. The results presented in this table should not be interpreted in isolation from the results of individual included studies contributing to each summary test accuracy measure. Reference test 1 means the standard was a positive CSF or a blood culture.
Figure 7.
GRADE Summary of findings table for reference test 2. The table summarises the certainty of the evidence according to the Grading of Recommendations Assessment, Development and Evaluation (GRADE) methodology, also called “quality of evidence”. The certainty can be one of four levels: High, moderate, low or very low. The interpretation of these levels should be performed s follows; High: we are very confident that the true effect lies close to that of the estimate of the effect we found; Moderate: we are moderately confident in the effect estimate: the true effect is likely to be close to the estimate of the effect that we found, but there is a possibility that it is substantially different. Low: our confidence in the effect estimate we found is limited: the true effect may be substantially different from the estimate of the effect; Very low: we have very little confidence in the effect estimate: the true effect is likely to be substantially different from the estimate of effect we found. The results presented in this table should not be interpreted in isolation from the results of individual included studies contributing to each summary test accuracy measure. Reference test 2 means the standard (final diagnosis of the infection in cases where cerebrospinal fluids SF/blood cultures or viral tests were negative) was defined by the researchers through a final diagnosis adjudication using molecular tests, an analysis of the clinical manifestations or based on the CSF findings.
For S. pneumoniae (16 studies/7090 participants)10,17,29, 30, 31, 32, 33, 34,18, 19, 20, 21, 22,24, 25, 26 we obtained combined sensitivity and specificity of 87·5% (95%CI 77–94), and 98·5% (95%CI 97–99·3), respectively, for reference test 1, and 93.4% (95%CI 85.4–97.1) and 99·5% (95%CI 98.6–99.8), respectively, for reference test 2. The main DTA measures for the rest of the microorganisms are detailed in Table 3. In general, LR+ were optimal for all the bacteria, being L.monocytogenes, E.coli, S.agalactiae and H.influenzae, the ones with higher values (LR+ >100) for reference test 1. The LR- values, on the other hand, were acceptable, and none of the bacteria had very low values (i.e., LR- all > 0·1). The Forest plots and SROC curves for all these microorganisms are presented in the Appendix 5 of the Supplemental Material).
In the Appendix 6 of the Supplemental Material, we discriminate the total positive results for FA/ME by each bacterium and study, along with TP and FP rates for both reference tests. For a total of 211 bacteria (16 studies, 6514 patients) detected by FA/ME or reference test, 113 (53·5%) were considered true positives and 98 (46·4%) false positives based on reference test 1. Based on reference test 2, 191 (90·5%) were considered true positives and 20 (9·4%) false positives. Since predictive values of tests depend on the prevalence of the disease, we have summarised and presented three different clinical scenarios according to different prevalence values (2, 5 and 10%), to display the expected TP, FP, TN, and FN probabilities for both reference tests for bacteria detection, in Figure 8. As expected, the higher the prevalence, the lower the FP rate for both reference tests.
Figure 8.
Post-test probabilities of correct or incorrect diagnoses of meningitis according to three prevalence scenarios for both reference tests. The figure display three potential scenarios based on three different meningitis prevalence rates, for each reference test. Readers could choose a potential prevalence (low 2%, medium 5%, or high 10%) of meningitis in a patient with suspected meningitis and based on an example of 1000 patients in which we would apply the FA/ME test, the figure shows the expected positive and negative results, and the correspondent true and false positives and negatives. The larger the prevalence, the fewer the expected false positives.
As for the viruses, we performed meta-analyses for the detection of enterovirus, Herpes simplex virus 1 and 2 (HSV-1, HSV-2) and Varicella-Zoster virus (VZV). For enterovirus, HSV-1 and HSV-2, three studies were analysed (6883 patients).10,22,23 For enterovirus, we obtained a combined sensitivity and specificity of 93·8% (95%CI 87–97·2) and 99·7% (95%CI 98·1%−100%), respectively, for reference test 1, and 99·8 (95%CI 86·1–97·4) and 99·9% (95%CI 99·7–100), for reference test 2. The DTA measures of all the viruses analysed, for both reference tests, are presented in Table 3. Forest plots and the SROC curves for the rest of the viruses are presented in the Appendix 7 of the Supplemental Material.
Additional analyses
In the RoB sensitivity analysis 1, for all bacteria, we combined results from 7 studies, and we obtained a lower combined sensitivity and very similar specificity: 84·4% (95%CI 72–92%), and 98% (95%CI 93·5 to 99·4), respectively. In the RoB sensitivity analysis 2, for all bacteria, we combined results from 3 studies, and we also obtained a lower combined sensitivity and a lower specificity: 82·5% (95%CI 65·3–92·3), and 98·7% (95%CI 67·8–99·8), respectively (Appendix 8, Supplemental Material).
Subgroup analyses are presented in Table 4. In the analysis of studies including only infants and children, we obtained a combined sensitivity and specificity of 83·6% (95%CI 65·6–93·2) and 97·4% (95%CI 84·8–99·6), respectively. In patients with abnormal CSF (defined by the authors as >10 CSF cells in one study and with no clear definition in another study), with two studies, the combined sensitivity and specificity were 94·4% (95%CI 65·6–99·3) and 99·6% (95% CI 93·7–100), respectively. Forest plots for all subgroups analyses are presented in the Appendix 9 of the Supplemental Material. Although LR+ are high for all the subgroups (except for previous antimicrobials’ use), their 95%CI were wide related with high uncertainty. LR- values were acceptable, except in the patients with abnormal CSF which showed to be very low, but with wide 95%CI.
Table 4.
Subgroup analyses for bacterial microorganisms.
| Ref. Test 1 |
Ref. Test 2 |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. studies/ No. patients (Ref studies) | Sensitivity (95%CI) X2; p value§ | Specificity (95%CI). X2; p value§ | LR+ (95%CI) | LR- (95%CI) | No. studies/ No. patients (Ref studies) | Sensitivity (95%CI) X2; p value§ | Specificity (95%CI). X2; p value§ | LR+ (95%CI) | LR- (95%CI) | |
| Infants and children (All bacteria) | 4/46218,19,27,28 | 83.6 (65.6–93.2) 1.99; 0.57 |
97.4 (84.8–99.6) 54.64; <0.0001 |
28 (2.13–122) |
0.18 (0.07–3.66) |
4/462 (18,19,27,28) |
90.7 (69.4–97.7) 3.39; 0.33 |
98.4 (93.6–99.6) 11.6; 0.008 |
45 (3.04–186) |
0.1 (0.07–3.63) |
| Infants and children (S. pneumoniae) | 3/40018,19,28 | 76.6 (39.2–94.3) 0.51; 0.77 |
98.3 (90.7–99.7) 12.05; 0.00247 |
38 (2.98–187) |
0.24 (0.07–3.63) |
3/400 (18,19,28) |
82 (41.7–96.6) 0; 1 |
98.8 (95.8–99.6) 0; 1 |
41 (2.61–193) |
0.18 (0.07–3.63) |
| 0–3 months (All bacteria) | 2/20727,28 | 86.4 (38.8–98.5) 0; 1.0 |
98.3 (94.9–99.5) 0.0546; 0.815 |
43 (2.59–194) |
0.14 (0.07–3.63) |
NA | NA | NA | NA | NA |
| Patients with abnormal CSF (All bacteria) † | 2/48221,24 | 94.4 (65.6–99.3) 0.26; 0.6 |
99.6 (93.7–100) 0; 1.0 |
94 (5.38–383) |
0.06 (0.07–3.7) |
2/482 (21,24) |
94.4 (65.6–99.3) 0.26; 0.6 |
99.6 (93.7–100) 0; 1.0 |
94 (5.38–383) |
0.06 (0.07–3.7) |
| Patients with previous use of antibiotics‡ (All bacteria) | 2/13018,27 | 87.6 (53.7–97.8) 0.3; 0.584 |
91.5 (55.6–98.9) 6.33; 0.0118 |
10 (0.82–42) |
0.14 (0.08–3.88) |
2/130 (18,27) |
92.2 (76.5–97.7) 0; 1 |
95 (86.6–98.2) 0; 0.32 |
18 (1.37–72) |
0.08 (0.07–3.74) |
CSF: Cerebro-spinal fluid; LR+: Positive Likelihood ratio; LR-: Negative Likelihood ratio NA: Not applicable; X2: Chi-2 test.
Chi2-test and corresponding p-value to assess presence of statistical heterogeneity.
Defined by the authors as >10 CSF cells in one study (21) and with no definition in the other study (24).
We defined it as studies with more than 70% of patients with previous antimicrobial therapy.
Discussion
In this systematic review (19 studies; 11,351), we found that the FA/ME panel has moderate sensitivities and very high specificities for identifying the selected bacteria and viruses. However, sensitivity values for bacteria (“all bacteria” and individual bacteria) were lower than 90% in all cases of reference test 1 (CSF/blood culture). These sensitivities increase when clinical and other test analyses complement cultures results (reference test 2). Namely, for the detection of any bacteria, the sensitivity of FA/ME seems to range between 89·5% (reference test 1) and 93·5% (reference test 2). Nonetheless, sensitivity values are lower when we consider only low RoB studies (between 82·5%−84·4%), and the certainty of the evidence was low. The only analyses in which we obtained sensitivity values higher than 90% and LR- lower than 0.1, for bacteria, were “all bacteria” and S.pneumoniae with reference test 2. The worst sensitivity values for bacteria were found for L.monocytogenes, H.influenzae and E.coli. Sensitivities and LR- were also suboptimal for HSV-1 but were high for VZV, HSV-2 and specially, for enteroviruses. Nonetheless, specificity and LR+ values were optimal for bacteria and viruses. Thus, in summary, FA/ME seems to be excellent for ruling in all the analysed bacteria and viruses, very limited for ruling out bacteria (acceptable for ruling out S. pneumoniae) HSV-1 and VZV, and excellent for ruling out enterovirus, VZV, and HSV-2.
However, most studies were judged as high RoB due to issues related to the reference test domain. In reference test 1, this is explained by the limitations of the CSF/blood cultures, which are easily affected by previous antimicrobial use and due to limitations in the samples’ management protocols. Reference test 2 would be an ideal approach as it can incorporate clinical and other tests analyses for diagnosis adjudication. Nonetheless, in all the studies, this adjudication was unblinded to the index test results. Moreover, the criteria used by researchers for the adjudication varied among the studies.
Our planned a priori subgroup analyses showed some interesting findings. Sensitivities for bacteria were lower in children than the obtained in the complete analyses, while specificities remained high. Moreover, the DTA measures in patients with abnormal CSF were remarkably high, which may suggest that using the FA/ME in this population might be an alternative to testing all the patients. While it is true that there are cases of viral CNS infection with normal CSF, such as early stages of herpetic encephalitis,35, 36, 37 and enterovirus meningitis in neonates,38,39 these are rare and occur in precise clinical settings.40 Considering the limitations of FA/ME for some microorganisms and its relatively high cost, defining diagnostic algorithms approaches to define the group of patients who can benefit the most from this test is an urgent need.
Very high specificity values were found for all the microorganisms, which shows that the FP of FA/ME may be irrelevant. However, when analysing in detail the proportion of FP of the total of positive results, the former values are not negligible; that is, the positive predictive values (PPV) are not high. For instance, for reference test 1, we found FP of 46·4% (Appendix 29). FP are higher for S.pneumoniae and S.agalactiae. PPV and negative predictive values (NPV) depend on the disease prevalence in each context, and the positive result should always be analysed in combination with clinical manifestation and additional tests to make a final diagnosis. The prevalence of CNS infections in the complete sample of the meta-analysis for all bacteria was 2%. The low pre-test probability of CNS infections as a result of a significant decrease in recent years,6,41 explains the low prevalence and a high FP rate. Clinical scenarios where prevalence rates are higher such as 5 or 10% will yield PPV of 33% and NPV of 18%. Causes of FP are related to sample contamination and cross-reactivity with other bacteria.8,10 Limiting the use of FA/ME to more selected cases with higher pre-test probabilities will yield lower FP.
Furthermore, we found that the FP were much lower (9·4%) with reference test 2 (Appendix 29), which means that the clinical analysis to complement the FA/ME resulted in some cases adjudicated as a CNS infection. However, we should add a caveat here, as the reference standard 2 assessment was judged as of high risk in all the studies because it was unblinded. Therefore, we cannot be certain about how many of those adjudicated cases were due to a real specific infection or could have been biased due to the knowledge of the FA/ME results. For bacteria, we think the true FP rate may be between 9·4 and 46·4%. Further studies would need to apply a reference test 2 in a blinded fashion, so we can obtain more certain DTA measures. Regardless of these results, in the clinical context, the best approach to dealing with cases in which a FA/ME FP is suspected is to assess the clinical scenario (i.e., considering clinical manifestations and history of antimicrobial use) and laboratory findings (CSF results).
To date, there is only one systematic review on the diagnostic validity of FA/ME9. In this review, the authors found higher sensitivity and specificity for the entire FA/ME panel. Their results differ from ours in several aspects. We provide the DTA measures discriminated by the most important bacteria and viruses, and therefore, we could highlight those microorganisms in which the FA/ME panel may have higher DTA measures, such as enteroviruses, or those with suboptimal performance of the FA/ME, such as L.monocytogenes, H.influenzae and E.coli. Also, in the mentioned review authors included some studies we excluded because they failed to demonstrate they performed index and reference tests independently, an essential element for a high-quality DTA analysis. Moreover, our RoB assessment was performed according to the characteristics of both reference tests, and as a result, the findings from the adjudication diagnosis (reference test 2), were judged to have high RoB. Lastly, we have doubled the number of included studies providing more updated evidence and we have applied cutting-edge methods for DTA synthesis, including conducting sensitivity and subgroup analyses, following the PRISMA-DTA guidance, and applying the GRADE approach to assess the certainty of the evidence.
Our results have important implications for clinical practice. The FA/ME has the advantages of a higher speed of the results, the ability to test for multiple organisms simultaneously, and it is not affected by previous antimicrobials use. These benefits may translate into targeted and timely treatments to initiate, change, or dismantle an antimicrobial treatment, which in turn can be reflected in fewer adverse effects and shorter hospital stays.42, 43, 44, 45, 46, 47 However, the performance of the test varies among microorganisms and depends on the intended purpose. FA/ME seems better for ruling in, than for ruling out, the disease. Therefore, clinicians should be very cautious about their results given the relatively high LR- values and high FP in scenarios with a low prevalence of CNS infections.
Future DTA studies interested in filling some of the identified gaps need to perform a blind adjudication of the diagnosis to reduce biases associated with the reference test and need to apply the reference test (specific PCR) for each of the viruses, independent of the result of the panel in patients with suspected CNS infection. Likewise, studies focused on specific subpopulations such as neonates, patients with previous antimicrobial use, or with abnormal CSF are required.
Our review has several strengths. This is the first review that presents DTA measures discriminated by microorganisms (six bacteria and four viruses). As mentioned above, we followed the highest methodological standards for a systematic review and DTA meta-analysis, and followed the PRISMA-DTA guidelines.11 We only included studies that performed both index and reference tests simultaneously, i.e., we did not include studies in which FA/ME was performed after results from the cultures or other molecular tests were known. We conducted analyses by type of reference test, type of microorganism, sensitivity analyses, and by some subgroups, we assessed the certainty of the evidence with the GRADE approach, and we present figures that facilitate results interpretation and contextualization for users according to different pre-test probabilities.
Our study is not free of limitations. The criteria used for resolving disagreements between the FA/ME and the cultures varied among the studies, which may have introduced some heterogeneity. We did not consider other microorganisms included in the FA/ME panel, such as cytomegalovirus, HSV-6, human parechovirus and Cryptococcus neoformans/gattii because their role in CNS infections in immunocompetent patients is not clear. We are, therefore, unable to provide conclusions of the performance of the test in immunocompromised patients.
In conclusion, the FA/ME panel may be a valid diagnostic tool to identify different microorganisms in CNS infections. FA/ME may have acceptable to high sensitivity, and high specificity for identifying bacteria and viruses in CNS infections, in immunocompetent patients. However, the certainty of the results was low mostly due to the high RoB of the studies. Moreover, in the context of low and high prevalence rates of bacterial infections, the FP and the FN, respectively, can be relevant. As a result, the FA/ME validity may be reduced in some scenarios (e.g., low prevalence rates) as the only diagnostic tool, and clinicians should apply an integral analysis, including the CSF findings and the clinical manifestations.
FA/ME diagnostic validity was high for most of the studied microorganisms. Nonetheless, attention should be put, mainly, into those microorganisms with less available evidence, that had the less suboptimal DTA measures. The diagnostic validity of FA/ME to detect L.monocytogenes, H.influenzae, E.coli, and HSV-1, may be suboptimal. More targeted DTA research on these microorganisms, on other viruses, and in specific subgroups (children and previous antimicrobial use) and with blinded appropriate diagnosis adjudication are encouraged. Furthermore, there is an urgent need for developing protocols and diagnosis algorithms to determine the patients who can benefit the most from this FA/ME, and to conduct economic evaluations to optimize its usefulness and relevance in different contexts.
Funding
None.
Contributors
JT and IDF conceptualised and designed the study; IDF supervised the project; JTG and MJJ screened literature, JT and IDF extracted data and performed the RoB and the certainty of the evidence assessment. JT and IDF had access to and verified the data. ST and AAV did the statistical analysis. JT, IF and ST and AAV interpreted the data. JT and IDF wrote the first draft of the manuscript, and all authors provided critical review and revision of the text and approved the final version. JT and IDF had final responsibility for the decision to submit for publication.
Declaration of interests
Authors declare no competing interests.
Acknowledgments
Data sharing statement
All data relevant to the study come from published studies and are included in the article or uploaded as supplementary information.
Acknowledgments
We would like to thank Dr Javier Sierra for his invaluable help in the analysis of the results.
Footnotes
Supplementary material associated with this article can be found in the online version at doi:10.1016/j.eclinm.2022.101275.
Appendix. Supplementary materials
References
- 1.Charles R., Diane E. In: Central Nervous System Infections in Childhood. Singhi P., Griffin D.E., Newton C.R., editors. Mac Keith; Londons: 2014. Burden of cerebral nervous system infections; pp. 1–7. [Google Scholar]
- 2.Singhi S., Angurana S.K. Principles of management of central nervous system infections. Indian J Pediatr. 2018;86:52–59. doi: 10.1007/s12098-017-2583-y. [DOI] [PubMed] [Google Scholar]
- 3.Cunha B.A. The clinical and laboratory diagnosis of acute meningitis and acute encephalitis. Expert Opin Med Diagn. 2013;7(4):1–21. doi: 10.1517/17530059.2013.804508. [DOI] [PubMed] [Google Scholar]
- 4.Binnicker M.J., Espy M.J., Irish C.L. Rapid and direct detection of herpes simplex virus in cerebrospinal fluid by use of a commercial real-time PCR assay. J Clin Microbiol. 2014;52(12):4361–4362. doi: 10.1128/JCM.02623-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Erdem H., Cag Y., Ozturk-engin D., et al. Results of a multinational study suggest the need for rapid diagnosis and early antiviral treatment at the onset of herpetic meningoencephalitis. Antimicrob Agents Chemother. 2015;59(6):3084–3089. doi: 10.1128/AAC.05016-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brouwer M.C., Tunkel A.R., Van De Beek D. Epidemiology, diagnosis, and antimicrobial treatment of acute bacterial meningitis. Clin Microbiol Rev. 2010;23(3):467–492. doi: 10.1128/CMR.00070-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Houlihan C.F., Bharucha T., Breuer J. Advances in molecular diagnostic testing for central nervous system infections. Curr Opin Infect Dis. 2019;32(3):244–250. doi: 10.1097/QCO.0000000000000548. [DOI] [PubMed] [Google Scholar]
- 8.Ramanan P., Bryson A.L., Binnicker M.J., Pritt B.S. Syndromic Panel-Based Testing in Clinical Microbiology. Clin Microbiol Rev. 2018;31(1):1–28. doi: 10.1128/CMR.00024-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tansarli G.S., Chapin K.C. Diagnostic test accuracy of the BioFire® FilmArray® Meningitis/Encephalitis panel: a systematic review and meta-analysis. Clin Microbiol Infect. 2020;26(3):281–290. doi: 10.1016/j.cmi.2019.11.016. [DOI] [PubMed] [Google Scholar]
- 10.Leber A.L., Everhart K., Balada-llasat J., et al. Multicenter evaluation of BioFire FilmArray meningitis /encephalitis panel for detection of bacteria, viruses, and yeast in cerebrospinal fluid specimens. J Clin Microbiol. 2016;54(9):2251–2261. doi: 10.1128/JCM.00730-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McInnes M.D.F., Moher D., Thombs B.D., et al. Preferred reporting items for a systematic review and meta-analysis of diagnostic test accuracy studies the PRISMA-DTA statement. JAMA J Am Med Assoc. 2018;319(4):388–396. doi: 10.1001/jama.2017.19163. [DOI] [PubMed] [Google Scholar]
- 12.Nigrovic L.E., Malley R., Macias C.G., et al. Effect of antibiotic pretreatment on cerebrospinal fluid profiles of children with bacterial meningitis. Pediatrics. 2008;122(4):726–730. doi: 10.1542/peds.2007-3275. [DOI] [PubMed] [Google Scholar]
- 13.Whitng P., Rutjes A., Marie W., et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–536. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 14.Reitsma J., Glas A., Rutjes A., Scholten R., Bossuyt P., Zwinderman A. Bivariate analysis of sensitivity and specificity produces informative summary measures in diagnostic reviews. J Clin Epidemiol. 2005;58:982–990. doi: 10.1016/j.jclinepi.2005.02.022. [DOI] [PubMed] [Google Scholar]
- 15.Doebler P. Mada: meta-analysis of diagnostic accuracy. R package version 0.5. 2015;7:1419.
- 16.Brozek J.L., Akl E.A., Jaeschke R., et al. Grading quality of evidence and strength of recommendations in clinical practice guidelines: part 2 of 3. the GRADE approach to grading quality of evidence about diagnostic tests and strategies. Allergy Eur J Allergy Clin Immunol. 2009;64(8):1109–1116. doi: 10.1111/j.1398-9995.2009.02083.x. [DOI] [PubMed] [Google Scholar]
- 17.Hanson K.E., Slechta E.S., Killpack J.A., et al. Preclinical assessment of a fully automated multiplex PCR panel for detection of central nervous system pathogens. J Clin Microbiol. 2016;54(3):785–787. doi: 10.1128/JCM.02850-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bailu D., Hua C., Xia Y., et al. Evaluation of the BioFire FilmArray Meningitis/Encephalitis panel for the detection of bacteria and yeast in Chinese children. Ann Transl Med. 2019;7(18):437. doi: 10.21037/atm.2019.08.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eichinger A., Hagen A., Meyer-Bühn M., Huebner J. Clinical benefits of introducing real-time multiplex PCR for cerebrospinal fluid as routine diagnostic at a tertiary care pediatric center. Infection. 2019;47(1):51–58. doi: 10.1007/s15010-018-1212-7. [DOI] [PubMed] [Google Scholar]
- 20.Boudet A., Pantel A., Carles M.J., et al. A review of a 13-month period of FilmArray Meningitis/Encephalitis panel implementation as a first-line diagnosis tool at a university hospital. PLoS One. 2019;14(10):1–14. doi: 10.1371/journal.pone.0223887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Domingues R.B., dos Santos M.V., de Leite F.B.V.M., Senne C. FilmArray Meningitis/Encephalitis (ME) panel in the diagnosis of bacterial meningitis. Braz J Infect Dis. 2019;23(6):468–470. doi: 10.1016/j.bjid.2019.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vincent J.J., Zandotti C., Baron S., et al. Point-of-care multiplexed diagnosis of meningitis using the FilmArray® ME panel technology. Eur J Clin Microbiol Infect Dis. 2020;39(8):1573–1580. doi: 10.1007/s10096-020-03859-y. [DOI] [PubMed] [Google Scholar]
- 23.Lindström J., Elfving K., Lindh M., Westin J., Studahl M. Assessment of the FilmArray ME panel in 4199 consecutively tested cerebrospinal fluid samples. Clin Microbiol Infect. 2021 doi: 10.1016/j.cmi.2021.05.017. [DOI] [PubMed] [Google Scholar]
- 24.Ena J., Afonso R., Bou M., et al. Evaluation of FilmArray ME panel for the rapid diagnosis of meningitis-encephalitis in emergency. Intern Emerg Med. 2021;16:1289–1295. doi: 10.1007/s11739-020-02593-9. [DOI] [PubMed] [Google Scholar]
- 25.Peñata A., Mesa S., Leal A., Castaño T., Bustamante J., Ospina S. Molecular diagnosis of meningitis and meningoencephalitis with an automated real-time multiplex polymerase chain reaction in a tertiary reference complex in Medellín, Colombia. Rev Inst Med Trop São Paulo. 2020;62(e77):1–9. doi: 10.1590/S1678-9946202062077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chong B.S.W., Kennedy K.J. Comparison of a commercial real-time PCR panel to routine laboratory methods for the diagnosis of meningitis-encephalitis. Pathology. 2021;53(5):635–638. doi: 10.1016/j.pathol.2020.09.029. [DOI] [PubMed] [Google Scholar]
- 27.Arora H.S., Asmar B.I., Salimnia H., Agarwal P., Chawla S., Abdel-Haq N. Enhanced identification of group B streptococcus and escherichia coli in young infants with meningitis using the BioFire FilmArray Meningitis/Encephalitis panel. Pediatr Infect Dis J. 2017;36(7):685–687. doi: 10.1097/INF.0000000000001551. [DOI] [PubMed] [Google Scholar]
- 28.Blaschke A.J., Holmberg K.M., Daly J.A., et al. Retrospective evaluation of infants aged 1 to 60 days with residual cerebrospinal fluid (CSF) tested using the FilmArray Meningitis/Encephalitis (ME) panel. J Clin Microbiol. 2018;56(7):1–9. doi: 10.1128/JCM.00277-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piccirilli G., Chiereghin A., Gabrielli L., et al. Infectious Meningitis/Encephalitis: evaluation of a rapid and fully automated multiplex PCR in the microbiological diagnostic workup. New Microbiol. 2018;41(2):118–125. [PubMed] [Google Scholar]
- 30.Bårnes G.K., Gudina E.K., Berhane M., et al. New molecular tools for meningitis diagnostics in ethiopia - a necessary step towards improving antimicrobial prescription. BMC Infect Dis. 2018;18(1):684. doi: 10.1186/s12879-018-3589-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leli C., Gotta F., Vay D., et al. Diagnostic accuracy of a commercial multiplex PCR for the diagnosis of meningitis and encephalitis in an italian general hospital. Infez Med. 2019;27(2):141–148. [PubMed] [Google Scholar]
- 32.López-Amor L., Escudero D., Fernández J., et al. Diagnóstico de meningitis/encefalitis en UCI con sistema de PCR múltiple. ¿Es tiempo de cambio? Rev Esp Quimioter. 2019;32(3):246–253. [PMC free article] [PubMed] [Google Scholar]
- 33.Radmard S., Reid S., Ciryam P., et al. Clinical utilization of the FilmArray Meningitis/Encephalitis (ME) multiplex polymerase chain reaction (PCR) assay. Front Neurol. 2019;10:281. doi: 10.3389/fneur.2019.00281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tarai B., Das P. FilmArray® Meningitis/Encephalitis (ME) panel, a rapid molecular platform for diagnosis of CNS infections in a tertiary care hospital in North India: one-and-half-year review. Neurol Sci. 2019;40(1):81–88. doi: 10.1007/s10072-018-3584-y. [DOI] [PubMed] [Google Scholar]
- 35.Polage C.R., Cohen H. State-of-the-art microbiologic testing for community-acquired mningitis and encephalitis. J Clin Microbiol. 2016;54(5):1197–1202. doi: 10.1128/JCM.00289-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chaudhuri A., Kennedy P.G.E. Diagnosis and treatment of viral encephalitis. Postgrad Med J. 2002;78:575–583. doi: 10.1136/pmj.78.924.575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kneen R., Michael B.D., Menson E., Mehta B. National guideline for the management of suspected viral encephalitis in children. J Infect. 2011:1–31. [Google Scholar]
- 38.de Crom SC, van Furth MA, Peeters MF, Rossen JW, Obihara CC. Characteristics of pediatric patients with enterovirus meningitis and no cerebral fluid pleocytosis. European journal of pediatrics. 2012 May 1;171(5):795-80 [DOI] [PubMed]
- 39.Tan NW, Lee EY, Khoo GM, Tee NW, Krishnamoorthy S, Choong CT. Cerebrospinal fluid white cell count: discriminatory or otherwise for enteroviral meningitis in infants and young children?. Journal of neurovirology. 2016 Apr 1;22(2):213-7. [DOI] [PubMed]
- 40.Precit M.R., Yee R., Pandey U., et al. Cerebrospinal fluid findings are poor predictors of appropriate FilmArray Meningitis/Encephalitis panel utilization in pediatric patients. J Clin Microbiol. 2020;58(3):1–13. doi: 10.1128/JCM.01592-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agrawal S., Nadel S. Acute bacterial meningitis in infants and children: epidemiology and management. Paediatr Drugs. 2011;13(6):385–400. doi: 10.2165/11593340-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 42.Vikram A., HOlmen J., Neely M., Pia S.P., Dien Bardb J. Neonatal meningitis caused by listeria monocytogenes diagnosed by multiplex molecular panel. J Clin Microbiol. 2016;54(12):2846–2849. doi: 10.1128/JCM.01159-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mina Y., Schechner V., Savion M., et al. Clinical benefits of FilmArray meningitis-encephalitis PCR assay in partially-treated bacterial meningitis in Israel. BMC Infect Dis. 2019;19(1):1–8. doi: 10.1186/s12879-019-4348-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wootton S.H., Aguilera E., Salazar L., Hemmert A.C., Hasbun R. Enhancing pathogen identification in patients with meningitis and a negative gram stain using the BioFire FilmArray ®meningitis /encephalitis panel. Ann Clin Microbiol Antimicrob. 2016;15(1):26–29. doi: 10.1186/s12941-016-0137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.O'Brien M.P., Francis J.R., Marr I.M., Baird R.W. Impact of cerebrospinal fluid multiplex assay on diagnosis and outcomes of central nervous system infections in children: a before and after cohort study. Pediatr Infect Dis J. 2018;37(9):868–871. doi: 10.1097/INF.0000000000001936. [DOI] [PubMed] [Google Scholar]
- 46.Messacar K., Breazeale G., Robinson C.C., Dominguez S.R. Potential clinical impact of the FilmArray meningitis encephalitis panel in children with suspected central nervous system infections. Diagn Microbiol Infect Dis. 2017;86(1):118–120. doi: 10.1016/j.diagmicrobio.2016.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fleischer E., Aronson P.L. Rapid diagnostic tests for meningitis and encephalitis - Biofire. Pediatr Emerg Care. 2020;36(8):397–403. doi: 10.1097/PEC.0000000000002180. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.